- 1Qinghai-Tibetan Plateau Grass-Feeding Livestock Engineering Technology Research Center of Sichuan Province, Southwest Minzu University, Chengdu, China
- 2Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization Key Laboratory of Sichuan Province, Southwest Minzu University, Chengdu, China
- 3Sichuan Academy of Grassland Sciences, Chengdu, China
- 4Institute of Qinghai-Tibetan Plateau, Southwest Minzu University, Chengdu, China
Background: The long interspersed nuclear element 1 (LINE1) retrotransposon has been identified as a specific substrate for fat mass and obesity-related gene (FTO), which facilitates the removal of N6-methyladenosine modifications from its targeted RNAs.
Methods: This study examined the dynamic interaction between FTO and LINE1 in yak tissues and muscle satellite cells, utilizing RT-qPCR, RNA immunoprecipitation (RIP), immunofluorescence staining, and techniques involving overexpression and interference of FTO and LINE1 to elucidate the relationship between FTO and LINE1 in yak tissues and muscle satellite cells.
Results: Cloning and analysis of the FTO coding sequence in Jiulong yak revealed a conserved protein structure across various Bos breeds, with notable homology observed with domestic yak, domestic cattle, and Java bison. Comprehensive examination of FTO and LINE1 gene expression patterns in Jiulong yaks revealed consistent trends across tissues in both sexes. FTO mRNA levels were markedly elevated in the heart and kidney, while LINE1 RNA was predominantly expressed in the heart. Immunoprecipitation confirmed the direct interaction between the FTO protein and LINE1 RNA in yak tissues and muscle satellite cells. The FTO–LINE1 axis was confirmed by a significant decrease in LINE1 RNA enrichment following its expression interference in yak muscle satellite cells. Overexpression of FTO substantially reduced the expression of recombinant myogenic factor 5 (MYF5). However, FTO interference had no discernible effect on MYF5 and myoblast determination protein 1 (MYOD1) mRNA levels. Immunofluorescence analysis revealed no alterations in Ki-67 protein expression following FTO interference or overexpression. However, phalloidin staining demonstrated enhancement in the myotube fusion rate of yak muscle satellite cells upon LINE1 interference.
Conclusion: This comprehensive mapping of the FTO and LINE1 mRNA expression patterns establishes a direct interaction between the FTO protein and LINE1 RNA in yak. The findings suggest that FTO overexpression promotes muscle satellite cells differentiation, whereas LINE1 negatively regulates myotube fusion. The study provides fundamental insights into the role of the FTO–LINE1 axis in determining the fate of muscle satellite cells in yak, laying a solid theoretical foundation for future investigations.
1 Introduction
The yak (Bos grunniens) is endemic to the Qinghai–Tibet Plateau and adjacent regions, serving as a crucial herbivorous livestock species. Its remarkable resilience has made the yak central to the livelihood of local farmers and herdsmen (1). The growth and development of skeletal muscle are pivotal determinants of meat yield and quality in livestock (2). Recent studies have implicated N6-methyladenosine (m6A) methylation in mammalian skeletal muscle growth (3). m6A is the most prevalent form of mRNA methylation in eukaryotes, playing a key role in regulating gene expression (4, 5). This modification process involves several proteins, specifically writer/methyltransferase (6), erasers/demethylase (7), and readers (8), collectively forming a dynamic and reversible regulatory network (9). Among these, the fat mass and obesity-related gene (FTO) is a vital m6A demethylase that profoundly impacts mammalian development (10). Studies involving chicken myoblasts have demonstrated that FTO regulates myogenic differentiation, influencing the expression of myogenic proteins and myosin (11). Despite increasing evidence highlighting the role of FTO in regulating skeletal muscle development in other mammals (12), its direct effect on yak muscle satellite cell proliferation and differentiation remains largely unexplored. Consequently, the specific mechanism through which FTO regulates the proliferation and differentiation of yak muscle satellite cells remains elusive.
Transposable elements, commonly known as mobile or jumping genes, are DNA segments with the unique ability to insert and excise themselves within the genome, altering their genomic location (13, 14). Among these elements, long interspersed nuclear element 1 (LINE1) is a retrotransposon that constitutes a considerable fraction of mammalian DNA, accounting for approximately 18% of mouse DNA, 23% of rat DNA, and 17% of human DNA (15, 16). LINE1 profoundly impacts mammalian embryonic development, as indicated by the severe disruption of mouse embryonic growth following the deletion of LINE1 RNA (17). The transient high expression of LINE1 during zygotic genome activation in early embryonic development of humans and mice suggests that LINE1 expression is under stringent regulation, playing a crucial role in important physiological processes (18). Recent research indicates that LINE1 RNA serves as a specific substrate for FTO protein to remove m6A modification. In mouse embryos and mouse embryonic stem cells, FTO mediates the demethylation of LINE1 RNA, thereby modulating downstream gene expression by altering LINE1 RNA levels and local chromatin status (19). However, whether FTO protein exerts similar effects on LINE1 in other tissues and cell types remains unknown.
This study aimed to investigate the interaction between FTO and LINE1 in yak tissues and muscle stem cells and explore whether FTO regulates muscle-derived gene expression and myogenic differentiation of yak muscle satellite cells through LINE1. The FTO coding sequence (CDS) region from the Jiulong yak was cloned, sequenced, and analyzed for bioinformatics analysis. Various tissues, including the heart, liver, spleen, lung, kidney, longissimus dorsi muscle, and pectoral muscle from the Jiulong yak, were used examine the expression of FTO and LINE1. RIP, immunofluorescence staining, and techniques involving overexpression and interference of FTO and LINE1 were employed to elucidate the relationship between FTO and LINE1 in yak tissues and muscle satellite cells and explore their roles in regulating muscle-derived gene expression and satellite cell differentiation.
2 Materials and methods
2.1 Animal materials
Jiulong yaks were sourced from Jiulong County in the Ganzi Tibetan Autonomous Prefecture of Sichuan Province. Tissue samples, including the heart, lung, liver, kidney, spleen, longissimus dorsi muscle, and pectoral muscle, were collected from three adult males and three adult females. Immediately after collection, the samples were labeled, submerged in liquid nitrogen, transported to the laboratory, and stored at −80°C. All animal experimental procedures strictly adhered to the guidelines established by the Regional Ethics Committee for Animal Experimentation and complied with the care regulations approved by the Animal Protection and Utilization Committee of Southwest Minzu University.
2.2 Isolation and culture of yak muscle satellite cells
The extensor carpi radialis muscles from 5-month-old yaks were surgically isolated under sterile environment. Following disinfection with 75% alcohol, the samples were placed in phosphate-buffered saline (PBS) containing penicillin/streptomycin antibiotics and transported to the laboratory. Blood vessels, fat, and connective tissue were carefully removed, and the muscle tissues were minced finely. The minced samples were resuspended in Dulbecco’s modified Eagle’s medium (DMEM, 11965092; Gibco) mixed with pronase XIV (P5147; Sigma-Aldrich) at a concentration of 1 mg/mL, using a 2:1 v/v ratio of DMEM to pronase XIV solution. This mixture was digested at 37°C for 2 h in a water bath shaker. After digestion, the tissue suspension was allowed to settle for 5 min to collect the supernatant. The supernatant was then filtered through a cell sieve with pore sizes ranging from 70 to 40 μm and centrifuged at 500 × g for 10 min to harvest the satellite cells. The isolated cells were resuspended in DMEM supplemented with 20% fetal bovine serum and cultured in a standard culture incubator with humidified air containing 5% CO2 at 37°C. The differential adherent method was employed to eliminate myofibroblasts, allowing for the collection and transfer of pure muscle satellite cells to a new Petri dish for further cultivation. Cells reaching 70–80% confluence were either passaged or cryopreserved using trypsin.
2.3 FTO gene cloning and bioinformatics analysis
Primers were designed using Primer5.0 based on the FTO gene sequence of the wild yak (ID: XM_005890937.1), with the following sequences: F: TTAGTAGTGGCGAAGGC, R: ATACCGCCCTT GCCTAA. Yak longissimus dorsi muscle cDNA served as the template for polymerase chain reaction (PCR) amplification. The PCR conditions included an initial denaturation at 95°C for 30 s, followed by cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 2 min. The amplified PCR product was purified and transformed into Escherichia coli DH5-α (TSC-C14; Tsingke). Positive clones were screened by plating the bacterial solution on Luria–Bertani solid medium containing ampicillin and subsequently identified by Tsingke.
Bioinformatics analysis of the cloned CDS sequence of the FTO gene from Jiulong yak was conducted using various online tools. ORF finder (NIH) was used to predict open reading frames. ProtParam and ProtScale (Expasy) were utilized to analyze the physicochemical properties of protein. SignalP-4.1 and TMHMM (QIAGEN) were employed to predicting the signal peptide and transmembrane domain, respectively. SOPMA was used to provide insights into the secondary structure, while Swiss-Model was used for tertiary structure prediction. Protein–protein interactions were explored using STRING. Additionally, a phylogenetic tree was constructed using MEGA 11 software.
2.4 Reverse transcription quantitative PCR (RT-qPCR) and tissue expression analysis
Total RNA was extracted from yak heart, liver, spleen, lung, kidney, longissimus dorsi, and pectoral muscle tissues using TRIzol reagent (15596018CN; Invitrogen). RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Beijing), and RNA integrity was assessed via 1.5% agarose gel electrophoresis. The extracted RNA from each tissue served as a template for reverse transcription to cDNA using a reverse transcription kit (639,506; TaKaRa Bio). The cDNA was stored at −20°C until further use.
RT-qPCR primers were designed based on the FTO gene sequence of wild yak (ID: XM_005890937.1) and the LINE1 gene sequence of cattle (ID: DQ000238.1). The primer sequences for FTO and LINE1 were as follows: qFTO F: CAGGTGCCAGTCTCGAATTG, R: TGGTTTCCAGAAGCAGACCT; LINE1 F: GCTGGGAAATCT GGTCAACC, R: TCTGGGAGGTGGGTCATAGA. β-Actin was used as an internal reference, with the primer sequence F: GCAGG TCATCACCATCGG, R: CCGTGTTGGCGTAGAGGT. Additionally, primers for MYF5 (ID: XM_014480707.1) and MYOD1 (ID: XM_005896772.1) from wild yak were designed with the following sequences: MYF5 F: ACGATGGACATGATGGACGG, R: AAACT CGTCCCCGAACTCAC; MYOD1 F: TCAGACCCTCAGTGC TTTGC, R: CGACAGCAGCTCCATATCCC. qPCR experiments were conducted using TB Green® Premix EX Taq™ II kit (RR820A, TaKaRa Bio). The reaction conditions included pre-denaturation at 95°C for 30 s, denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, with a total of 40 cycles. Relative expression levels were calculated using the 2-ΔΔCt method.
2.5 RIP
The RIP experiment was performed as described previously (20). Yak heart tissues were lysed using immunoprecipitation (IP) lysate (P0013; Beyotime), and the supernatant was ground on ice. IP experiments were performed using the Dynabeads™ Protein A Immunoprecipitation Kit (10006D; Invitrogen) along with an FTO-specific polyclonal antibody (27226-1-AP; Proteintech) derived from rabbit. Rabbit immunoglobulin G (IgG) loading control antibody (AC005; ABclonal) served as the negative control. After incubating the antibody and magnetic beads at room temperature, the antigen-containing supernatant was introduced and further incubated at room temperature. Following the removal of the supernatant, the bound antibody was eluted using protease K. After a 30 min incubation, the mixture was placed on a magnetic frame, allowing the supernatant to be transferred to a fresh test tube. RNA was extracted using the TRIzol method and subsequently reverse transcribed into cDNA for qPCR analysis.
2.6 Interference with FTO and LINE1 genes
A specific small interfering RNA (siRNA) sequence was designed and synthesized based on the yak FTO gene sequence (ID: XM005890937.1). The interference sequences were F: GUGGCA GUGUACAGUUAUATT, R: UAUAACUGUACACUGCCACTT. Yak muscle satellite cells were cultured in 12-well plates. When cell growth reached 70–80% confluence, the medium was replaced with antibiotic- and serum-free DMEM. The si-FTO and Lipo8000 transfection reagents (C0533; Beyotime) were combined in a sterile Eppendorf tube and subsequently added to the wells. Six hours post-transfection, the medium was replaced with complete medium, and the cells were incubated for an additional 48 h. The cells were harvested and examined via qPCR. Similarly, a specific siRNA sequence targeting the bovine LINE1 gene (ID: DQ000238.1) was designed and synthesized. The si-LINE1 sequences were F: GGACCUAAUUAACCUUAAATT and R: UUUAAGGUUAAUUAGGUCCTT. Yak muscle satellite cells were seeded in 12-well plates. When cell growth was 70–80% confluence, the cells were transfected with si-LINE1.
2.7 FTO gene overexpression
Using the yak FTO gene sequence (ID: XM_005890937.1), we designed an overexpression primer sequence incorporating a homologous arm (homologous arm sequences are shown in bold): F: CTTGGTACCGAGCTCGGATCCATGAAGCGGACCCCGACG, R: TGCTGGATATCTGCAGAATTCCTAGGGCCTGGTTTCCAGAAG. The target fragment was amplified using cDNA from the longissimus dorsi muscle as a template. The OK Clon ligation kit (AG11807; Agbio) was employed to ligate the amplified fragment into a vector. Positive clones were screened and verified through sequencing. Plasmids were extracted using an endotoxin-free plasmid extraction kit (DP120-01; TIANGEN) and stored at −20°C. Yak muscle satellite cells were cultured in 12-well plates. Once cell growth reached 70–80% confluence, the complete medium was replaced with antibiotic- and serum-free DMEM. Plasmid DNA containing the FTO target fragment was combined with Lipo8000 transfection reagent in a sterile Eppendorf tube and introduced into the wells. After 6 h of transfection, the medium was substituted with complete medium, and the cells were incubated for an additional 48 h. The cells were then harvested, and qPCR was performed to assess transfection efficiency and the expression of muscle-derived genes.
2.8 Phalloidin staining
Yak muscle satellite cells were seeded in 24-well plates and cultured until they reached 70–80% confluence. At this stage, the complete medium was substituted with differentiation medium consisting of 2% horse serum (16,050,122; GIBCO) and 1% penicillin/streptomycin. The medium was refreshed every 2 days, and cell differentiation was assessed on day 4. Microfilaments within the cells were visualized using Actin-Tracker Red (C2203S; Beyotime) staining. The cells were observed under fluorescence microscopy and images were collected for further analysis.
2.9 Immunofluorescence of Ki-67
Yak muscle satellite cells were seeded in 24-well plates and cultured until they reached 70–80% confluence. The cells were then transfected with either pcDNA3.1-FTO or si-FTO. Following 48 h of incubation, the cells were fixed with 100 μL of 4% paraformaldehyde (BL539A; Biosharp) per ell for 10–20 min. Following fixation, the cells were incubated in PBS containing 0.5% Triton X-100 for 20 min. Blocking was performed with 3% bovine serum albumin at room temperature for 12 h. The cells were then incubated overnight at 4°C in the dark with a diluted Ki-67 rabbit polyclonal primary antibody (PA5-19462; Invitrogen). The secondary antibody was added dropwise and incubated at room temperature for 2 h in the dark. 4′,6-Diamidino-2-phenylindole staining was performed at room temperature for 3–5 min. After a final wash with PBS containing Tween, an anti-quenching agent was applied, and the samples were observed through fluorescence microscope.
2.10 Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software. One-way analysis of variance and multiple comparison tests were performed to determine statistically significance (p < 0.05) among the groups. Complete data visualization mapping was also performed using the same software. All experiments were conducted in triplicate.
3 Results
3.1 Sequence characteristics of the yak FTO CDS region
The CDS region of the FTO gene was cloned using cDNA from the longissimus dorsi muscle of Jiulong yak. The cloned sequence, 1,518 bp in length, was assigned GenBank accession number of PP764744. Bioinformatics analysis revealed that the FTO protein has a molecular formula of C2606H4047N709O770S27 and a relative molecular mass of 58495.71 kDa. Leucine was the most prevalent amino acid in the FTO protein, comprising 11.3%, while cysteine and histidine were the least abundant at 2.6% each. The FTO protein had an instability index of 50.37 and an estimated half-life of 30 h. Additionally, the protein exhibited a fat coefficient of 80.34. Analysis of the hydrophilicity/hydrophobicity of the protein predicted a maximum value of 2 at position 438 and a minimum of −3.22 at position 12, with an overall average hydrophilicity of −0.528, classifying the FTO protein as hydrophilic (Figure 1A). SignalP (version 4.1) analysis indicated that the FTO protein lacked a signal peptide region, suggesting that it is not a secreted protein (Figure 1B). TMHMM predictions confirmed the absence of a transmembrane structure in the protein (Figure 1C). Secondary structure analysis revealed that α-helix comprised the largest proportion of the FTO protein at 43.17%, followed by random coil (38.61%), extended chain (11.49%), and β-turn (6.73%) (Figure 1D). Tertiary structure prediction revealed that the main components of FTO protein were still α-helix, extended chain, and random coil (Figure 1E).
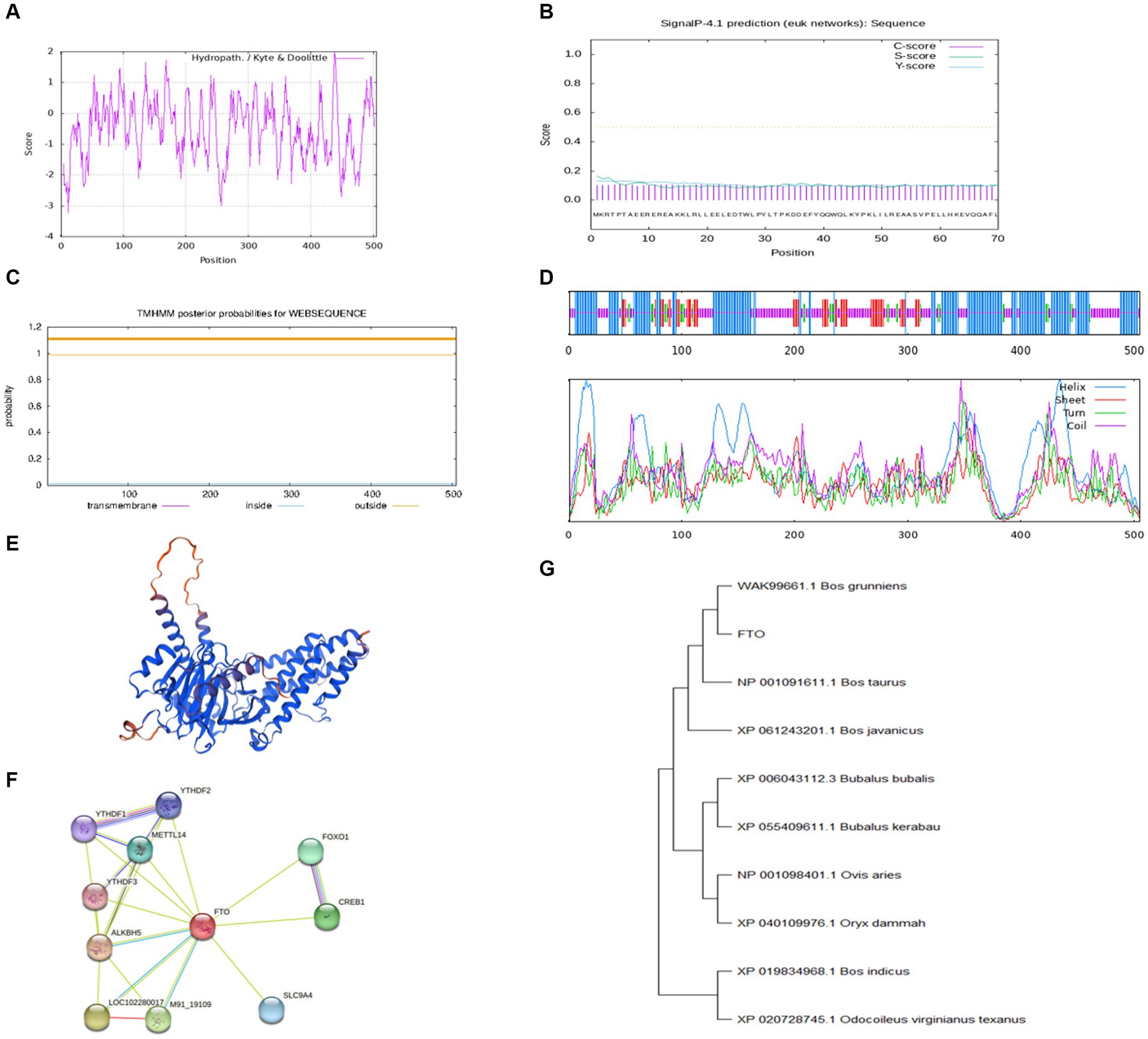
Figure 1. Bioinformatics analysis of FTO CDS region in Jiulong yak. (A) Hydrophilic/hydrophobic prediction analysis of the protein. (B) Signal peptide prediction analysis. (C) Transmembrane domain prediction analysis. (D) Prediction analysis of protein secondary structure. (E) Prediction of protein tertiary structure. (F) Prediction analysis of protein interactions. (G) The phylogenetic tree of FTO.
Analysis conducted using STRING software identified a strong correlation between yak FTO and several other genes, including AlkB homolog 5, cAMP responsive element binding protein 1, methyltransferase-like 14, forkhead box protein O1, and YTH domain family member 1 (Figure 1F). Furthermore, BLAST nucleotide sequence alignment revealed remarkable similarity (up to 99.74%) between the FTO gene of Jiulong yak and its counterparts in wild yak (ID: XM_005890937.1) and domestic yak (ID: OM640141.1). Significant similarities were also noted with hybrids of North American wild yak (ID: XM_010861935.1; 99.41%), domestic cattle (ID: NM_001098142.1; 99.41%), and zebu and yellow cattle (99.34%). The FTO gene sequences of Jiulong yak were compared with those of nine distinct species retrieved from GenBank using a phylogenetic tree constructed by the MEGA (version 11) software. The results indicated the highest homology between Jiulong yak and B. grunniens, followed by Bos taurus and Bos javanicus (Figure 1G).
3.2 Expression analysis of FTO and LINE1 in yak tissues
The expression levels of FTO and LINE1 were evaluated in the heart, liver, spleen, lung, kidney, longissimus dorsi, and pectoral muscle tissues of adult male and female Jiulong yaks. Both FTO and LINE1 exhibited consistent expression patterns across these tissues, with no observable sex-specific differences (Figure 2). In female yak tissues, FTO expression was notably higher in the heart and kidney tissues than in the other tissues, while LINE1 expression was predominantly elevated in the heart. Similarly, in male yak tissues, the heart tissue demonstrated significantly increased expression levels of both FTO and LINE1 compared to the spleen and lung tissues.
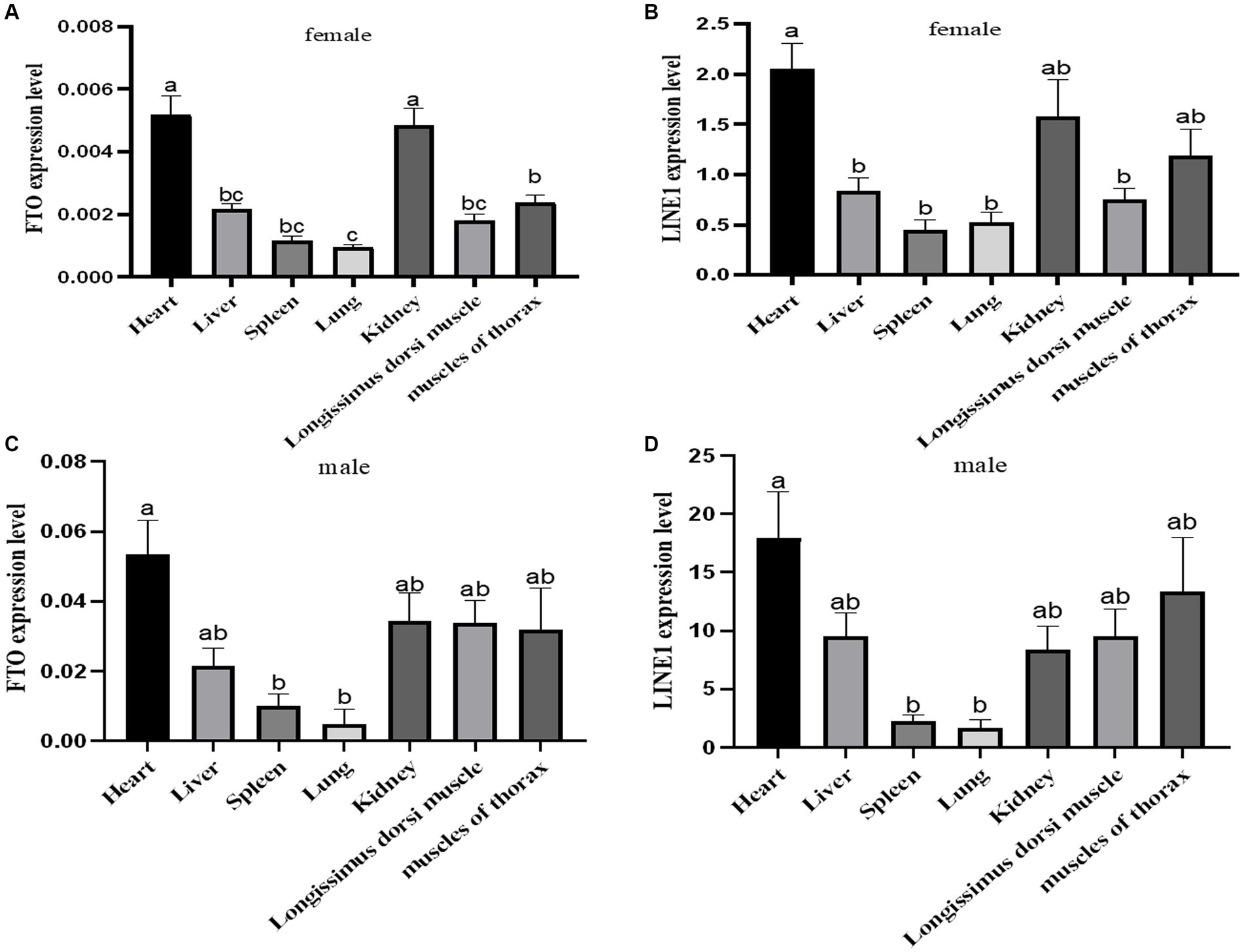
Figure 2. Expression analysis of FTO and LINE1 in yaks. (A) Analysis of FTO expression in female yak tissues. (B) Analysis of LINE1 expression in female yak tissues. (C) Analysis of FTO expression in male yak tissues. (D) Analysis of LINE1 expression in male yak tissues.
3.3 Interaction between FTO and LINE1 in yak tissues
To explore the interaction between FTO protein and LINE1 RNA in yak tissues, RIP using an FTO antibody was performed on yak heart tissues, followed by qPCR analysis of LINE1 expression, with IgG as a negative control. Our results indicated a direct interaction between FTO protein and LINE1 RNA specifically in yak heart tissue (Figure 3A). To further validate this interaction, LINE1 expression was knocked down in yak muscle satellite cells by transfecting them with si-LINE1 and repeating the RIP assay. A significant decrease in LINE1 expression was evident after si-LINE1 transfection, accompanied by a reduction in the binding affinity of FTO to LINE1 (Figure 3B).
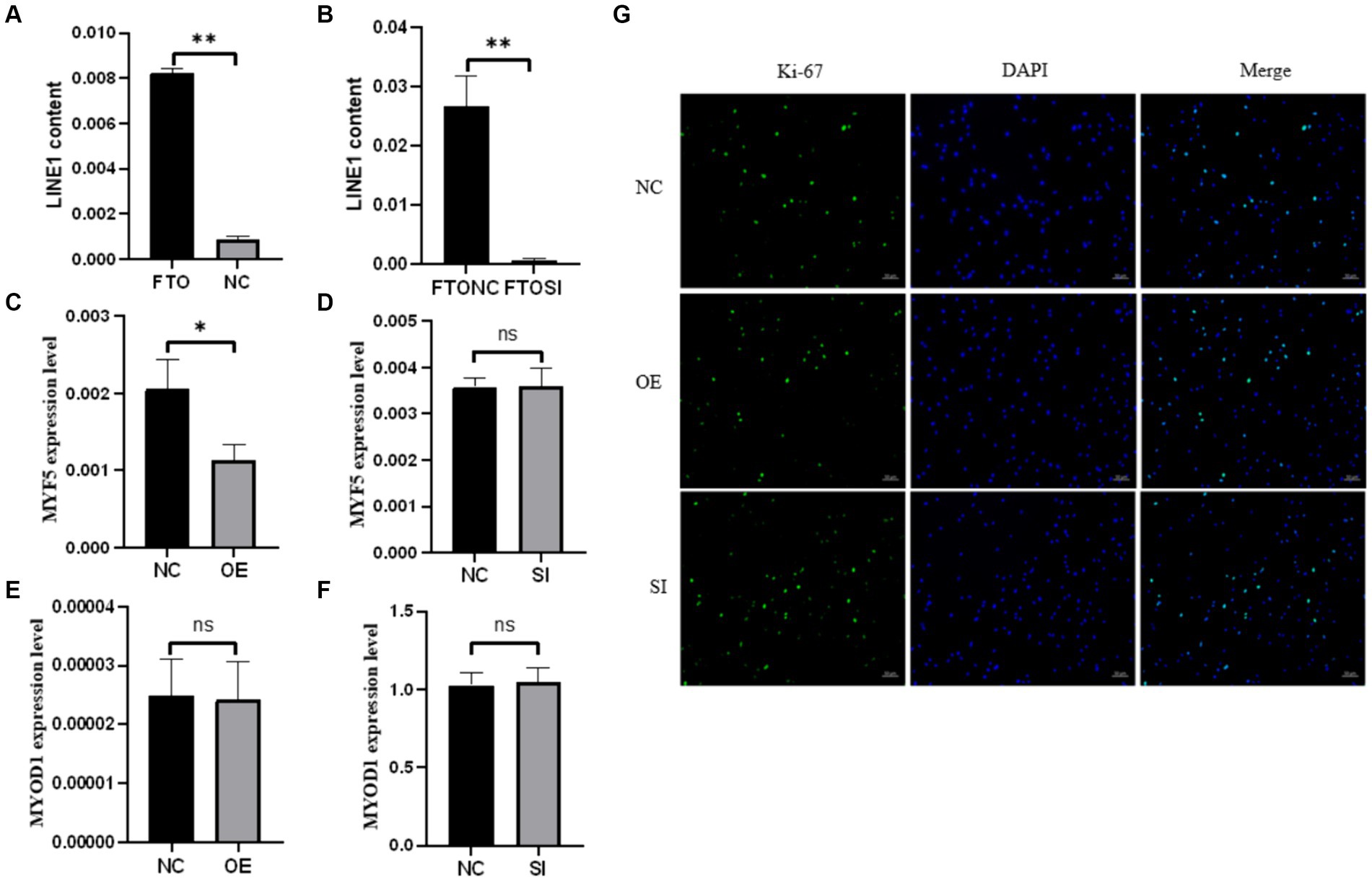
Figure 3. The RIP experiment of FTO and the effects of FTO modulation on the proliferation and differentiation of muscle satellite cells. (A) RIP assay in heart tissue. (B) RNA immunoprecipitation analysis using the FTO antibody after LINE1 interference. Impact of FTO modulation on the expression of LINE1 and muscle-specific genes in muscle satellite cells. (C) Analysis of MFY5 expression upon FTO overexpression. (D) Assessment of MYF5 expression after FTO interference. (E) MYOD1 expression analysis following FTO overexpression. (F) MYOD1 expression profiling after FTO interference. (G) Immunofluorescence-based detection of Ki-67.
Upon modulating FTO levels in muscle satellite cells, MYF5 expression remained unaffected by FTO interference (Figure 3C), whereas FTO overexpression significantly downregulated MYF5 expression (Figure 3D). However, MYOD1 expression was not influenced by FTO knockdown (Figure 3E) or overexpression (Figure 3F). Additionally, cell immunofluorescence results indicated that Ki-67 expression was not significantly affected by changes in FTO levels (Figure 3G). These results suggest that altering FTO expression does not affect the proliferation of yak skeletal muscle satellite cells; however, FTO overexpression tends to promote the myogenic differentiation of these cells.
3.4 Impact of LINE1 downregulation on myotube formation in yak muscle satellite cells
Following siRNA-induced downregulation of LINE1 expression in yak muscle satellite cells, the differentiation process was visualized on day 5 through phalloidin staining. Comparative analysis between the si-LINE1 and si-NC groups revealed a significant enhancement in the myotube fusion rate upon LINE1 downregulation (Figure 4).
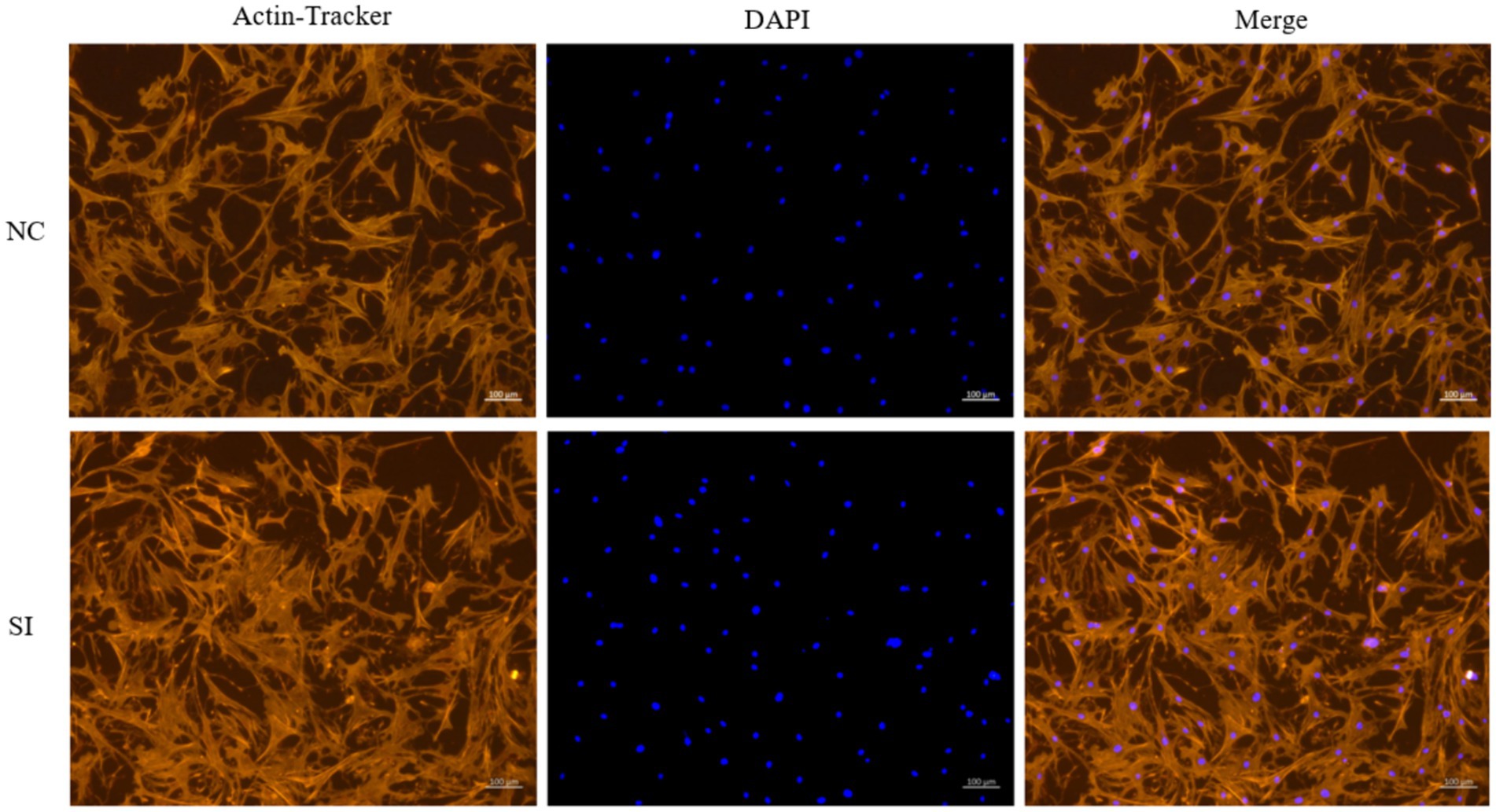
Figure 4. Examination of muscle tube fusion via AcN-tracker Red staining after LINE1 interference. Yak muscle satellite cells were cultured in differentiation medium for four days, and the formation of myotubes was detected by phalloidin staining, and the nucleus were stained with DAPI.
4 Discussion
FTO plays a pivotal role as an m6A demethylase in mammalian growth and development (10), with LINE1 recently identified as a key substrate, enabling FTO to eliminate m6A modifications from the target RNA (19). During mouse embryonic development, FTO regulates the m6A modification of chromatin-related RNA, particularly LINE1 RNA, transcribed from transposon elements in mouse embryonic stem cells (19). This modification of FTO shapes the local chromatin state, thereby affecting the opening and closing of genes containing LINE1 elements. This study aimed to elucidate the interaction between FTO and LINE1 in yak tissues and muscle satellite cells to determine the regulatory mechanism within the FTO–LINE1 axis in muscle development.
In this study, we successfully cloned the 1,518 bp CDS region of the FTO gene from the Jiulong yak, encoding 505 amino acids. Our bioinformatics analysis indicated that the secondary structure of FTO protein predominantly consists of α-helices, with no transmembrane domains or signal peptides, suggesting that FTO is neither a transmembrane nor a secretory protein, aligning with previous observations in goat (21) and pig FTO (22). This cross-species consistency underscores the remarkable conservation of the FTO gene. Tissue-specific expression analysis of FTO and LINE1 in adult yaks revealed distinct patterns. In female yaks, FTO expression was significantly higher in the heart and kidney than in the other tissues. In male yaks, heart tissue exhibited predominantly higher FTO expression followed by the spleen and lung. These findings diverge from those of a previous study on 18-month-old Datong yaks, where heart tissue exhibited peak FTO expression, exceeding the expressions in the liver, spleen, lung, kidney, and longissimus dorsi (23). Comparable patterns of elevated FTO expression were observed in the liver and subcutaneous fat of 3-year-old Guangling donkeys (24). Varying FTO expression patterns, with high expression in the liver, have been reported in Tan sheep (25) and the hypothalamus and liver of Qianbei Brown goat (26). Our study identified consistent trends in FTO expression across tissues in both male and female yaks, although the actual expression levels differed. Our findings contribute to the broader understanding of FTO expression patterns and regulatory mechanisms, offering valuable insights into its potential functions in various biological processes.
Transposable elements constitute a significant portion of many eukaryotic genomes, with nearly half of the human genome consisting of transposons (14). LINE1, as a retrotransposon, plays a pivotal role in facilitating chromatin opening within cells (27). Jachowicz et al. (28) demonstrated that premature silencing of the LINE1 element in mouse embryonic cells reduces chromatin accessibility, while its prolonged activation helps counteract natural chromatin compression during developmental processes. This LINE1 activation influences global chromatin accessibility in early developmental stages, suggesting that retrotransposon activation is essential for developmental progression. We observed, both FTO and LINE1 were widely expressed across various tissues in both male and female yaks. LINE1 expression was significantly higher in the heart, kidney, and chest muscle of female yaks than in the other four tissues examined. Similarly, in male yaks, LINE1 expression was markedly increased in the heart compared to that in the spleen and lung. Consistent expression patterns of FTO and LINE1 across diverse tissues, independent of sex, suggest a possible interaction between these molecules in yak tissues.
Although a direct relationship between FTO and LINE1 remains limited, indirect evidence suggests a significant connection between them. FTO has been associated with skeletal muscle development and aging. In older individuals, FTO mRNA expression in the skeletal muscle is reduced by approximately 28% compared to that in younger individuals (29), a pattern also observed in aging Tibetan sheep (30). Exercise has been shown to attenuate the impact of FTO gene expression in skeletal muscle (31). Similarly, LINE1 expression correlates with skeletal muscle development and aging. For instance, the expression of LINE1 mRNA in the skeletal muscle of 24- and 36-month-old mice exceeds that of 5-month-old mice (32). In mice, rats, and humans, LINE1 expression increases with age, while prolonged exercise decreases its expression in human skeletal muscle (33). These findings suggests a potential correlation between FTO and LINE1 expression changes during skeletal muscle development and aging. Therefore, investigating the interplay between the demethylase FTO and its substrate LINE1 holds significant promise in elucidating the molecular mechanism underlying skeletal muscle development and aging.
To investigate the potential interaction between FTO and LINE1 in yak tissues, we performed RIP analysis on heart tissue, where both FTO and LINE1 expression were highest. Our results demonstrated a direct interaction between FTO protein and LINE1 RNA, which was further validated in yak muscle satellite cells. A significant decrease in the binding of FTO protein to LINE1 RNA was observed by performing RIP experiments following siRNA-mediated interference of LINE1 expression in these cells, further confirming the direct interaction between the two molecules.
During aging, individuals experience muscle mass loss, increased chronic inflammation, and cellular senescence (34), factors that may influence FTO gene expression. In Tibetan sheep, aging is accompanied by alterations in muscle fiber composition from slow to fast fibers, along with changes in energy metabolism (30). The elevated expression of LINE1 in older mice, rats, and humans relative to their younger counterparts is potentially attributable to age-related modifications in chromatin status, augmenting to higher transcriptional activity of LINE1 (33). FTO contributes to the preservation of slow muscle fibers in mice, both in vivo and in vitro, through its demethylation activity (12). These results suggest a correlation between the trends of FTO and LINE1 expression changes during skeletal muscle development and aging.
FTO also plays a crucial role in myogenic differentiation (35). MYF5 and MYOD1 are essential myogenic regulatory factors that regulate muscle development and regeneration by directing the activation and differentiation of muscle precursor cells into mature muscle fibers (36). During muscle stem cell differentiation, MYF5 expression decreases (37), while MYOD1 initiates the transcription of skeletal muscle-specific genes and induces cell cycle arrest, essential for muscle cell differentiation and initiating myogenesis (38). FTO knockdown enhances bovine myoblast proliferation while inhibiting differentiation, whereas its overexpression stimulates myoblast differentiation (39). In our study, we observed a significant decrease in MYF5 expression upon FTO overexpression, while MYOD1 expression remained unchanged, suggesting the involvement of compensatory regulatory mechanisms. Immunofluorescence analysis of Ki-67 revealed no significant changes following FTO expression modification, suggesting that FTO does not significantly influence the proliferation of yak muscle satellite cells. This finding contradicts previous studies where FTO knockdown increased bovine muscle stem cell proliferation (39) and decreased Ki-67 expression in mouse tumor cells (40). Further examination is needed to elucidate the role and mechanism of FTO in regulating cell proliferation.
In this study, we further manipulated LINE1 expression, the primary substrate for FTO-mediated m6A demethylation, using siRNA. Phalloidin staining revealed a significant increase in myotube fusion following LINE1 downregulation. This myotube fusion process is intricately linked to chromatin dynamics, particularly nuclear mechanics regulation and chromatin accessibility (41). Disruption of actomyosin complexes can alter nuclear morphology and decrease chromatin accessibility, which is crucial for myogenic differentiation (42), as it enables regulatory proteins, such as transcription factors, to access genes and activating the expression of muscle-specific genes. Interfering with microtubules in mouse embryonic fibroblasts can partially restore nuclear morphology and chromatin accessibility without compromising cellular force generation (43). However, in certain contexts, low chromatin accessibility is essential to maintain cellular function stability. The observed decrease in LINE1 mRNA expression may positively regulate chromatin openness, promoting myogenic differentiation and myotube fusion in yak muscle satellite cells. This function of LINE1 appears to be dependent on the m6A demethylase activity of FTO, given the direct interaction between FTO protein and LINE1 RNA.
5 Conclusion
The cloning and analysis of the Jiulong yak FTO gene CDS sequence revealed its conservation across species. Tissue analysis showed similar expression patterns of FTO and LINE1 RNA across yak tissues, with a direct FTO–LINE1 RNA interaction confirmed experimentally. The overexpression of FTO downregulated MYF5 mRNA expression in yak muscle cells without affecting MYOD1 expression or cell proliferation, while disrupting LINE1 expression enhanced muscle cell fusion. These findings provide valuable insights into the role of the FTO–LINE1 axis in yak muscle development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PP764744.
Ethics statement
The animal study was approved by the Animal Protection and Utilisation Committee of Southwest Minzu University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZM: Data curation, Writing – original draft, Writing – review & editing. ZC: Data curation, Supervision, Writing – review & editing. HY: Data curation, Validation, Writing – review & editing. XZ: Writing – review & editing. HZ: Writing – review & editing. XL: Supervision, Writing – review & editing. JZ: Funding acquisition, Supervision, Writing – review & editing. ZW: Data curation, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grands from the Fundamental Research Funds for the Central Universities, Southwest Minzu University (grant no.: ZYN2023101), the Sichuan Science and Technology Program (grant no.: 2024NSFSC0391), the Scientific and Technological Innovation Team for Qinghai-Tibetan the Plateau Research in Southwest Minzu University (grant no.: 2024CXTD13), the Basic Scientific Research Project in Sichuan (evaluation on the effect of introduction of Maiwa yak and its offspring performance) and the Program of National Beef Cattle and Yak Industrial Technology System (CARS-37).
Acknowledgments
The authors thank the Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization and the Qinghai-Tibetan Plateau Grass-Feeding Livestock Engineering Technology Research Center of Sichuan Province of Southwest Minzu University for providing the experimental platform and conditions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ayalew, W, Chu, M, Liang, C, Wu, X, and Yang, P. Adaptation mechanisms of yak (Bos grunniens) to high-altitude environmental stress. Animals. (2021) 11:2344. doi: 10.3390/ani11082344
2. Mo, M, Zhang, Z, Wang, X, Sheng, WJ, Zhang, L, and Lin, X. Molecular mechanisms underlying the impact of muscle fiber types on meat quality in livestock and poultry. Front Vet Sci. (2023) 10:1284551. doi: 10.3389/fvets.2023.1284551
3. Yang, X, Wang, J, Ma, X, Du, J, Mei, C, and Zan, L. Transcriptome-wide N (6)-Methyladenosine Methylome profiling reveals m(6)a regulation of skeletal myoblast differentiation in cattle (Bos taurus). Front Cell Dev Biol. (2021) 9:785380. doi: 10.3389/fcell.2021.785380
4. Zhang, T, Zhang, SW, Zhang, SY, Gao, S, Chen, Y, and Huang, Y. m6A-express: uncovering complex and condition-specific m6A regulation of gene expression. Nucleic Acids Res. (2021) 49:e116. doi: 10.1093/nar/gkab714
5. He, PC, and He, C. M(6) a RNA methylation: from mechanisms to therapeutic potential. EMBO J. (2021) 40:e105977. doi: 10.15252/embj.2020105977
6. Nilsen, TW. Molecular biology. Internal mRNA methylation finally finds functions. Science. (2014) 343:1207–8. doi: 10.1126/science.1249340
7. Jiang, X, Liu, B, Nie, Z, Duan, L, Xiong, Q, Jin, Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
8. Du, H, Zhao, Y, He, J, Zhang, Y, Xi, H, and Liu, M. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. (2016) 7:12626. doi: 10.1038/ncomms12626
9. Fang, Z, Mei, W, Qu, C, Lu, J, Shang, L, Cao, F, et al. Role of m6A writers, erasers and readers in cancer. Exp Hematol Oncol. (2022) 11:45. doi: 10.1186/s40164-022-00298-7
10. Azzam, SK, Alsafar, H, and Sajini, AA. FTO m6A demethylase in obesity and Cancer: implications and underlying molecular mechanisms. Int J Mol Sci. (2022) 23:3800. doi: 10.3390/ijms23073800
11. Huang, H, Liu, L, Li, C, Liang, Z, Huang, Z, Wang, Q, et al. Fat mass- and obesity-associated (FTO) gene promoted myoblast differentiation through the focal adhesion pathway in chicken. 3 Biotech. (2020) 10:403. doi: 10.1007/s13205-020-02386-z
12. Wang, W, Du, X, Luo, M, and Yang, N. FTO-dependent m(6)A regulates muscle fiber remodeling in an NFATC1-YTHDF2 dependent manner. Clin Epigenetics. (2023) 15:109. doi: 10.1186/s13148-023-01526-5
13. Zhang, X, Zhao, M, Mccarty, DR, and Lisch, D. Transposable elements employ distinct integration strategies with respect to transcriptional landscapes in eukaryotic genomes. Nucleic Acids Res. (2020) 48:6685–98. doi: 10.1093/nar/gkaa370
14. Feschotte, C. Transposable elements: McClintock's legacy revisited. Nat Rev Genet. (2023) 24:797–800. doi: 10.1038/s41576-023-00652-3
15. Romero, MA, Mumford, PW, Roberson, PA, Osburn, SC, Parry, HA, Kavazis, AN, et al. Five months of voluntary wheel running downregulates skeletal muscle LINE-1 gene expression in rats. Am J Physiol Cell Physiol. (2019) 317:C1313–23. doi: 10.1152/ajpcell.00301.2019
16. Lu, JY, Chang, L, Li, T, Wang, T, Yin, Y, Zhan, G, et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. (2021) 31:613–30. doi: 10.1038/s41422-020-00466-6
17. Percharde, M, Lin, CJ, Yin, Y, Guan, J, Peixoto, GA, Ayda, BK, et al. A LINE1-Nucleolin partnership regulates early development and ESC identity. Cell. (2018) 174:e19:391–405. doi: 10.1016/j.cell.2018.05.043
18. Li, X, Bie, L, Wang, Y, Hong, Y, Zhou, Z, Fan, Y, et al. LINE-1 transcription activates long-range gene expression. Nat Genet. (2024) 56:1494–502. doi: 10.1038/s41588-024-01789-5
19. Wei, J, Yu, X, Yang, L, Liu, X, Gao, B, Huang, B, et al. FTO mediates LINE1 m(6)a demethylation and chromatin regulation in mESCs and mouse development. Science. (2022) 376:968–73. doi: 10.1126/science.abe9582
20. Martindale, JL, Gorospe, M, and Idda, ML. Ribonucleoprotein immunoprecipitation (RIP) analysis. Bio Protoc. (2020) 10:e3488. doi: 10.21769/BioProtoc.3488
21. Lin, Y, Liao, H, He, Q, Li, Q, and Wang, Y. Cloning and expression profiling of FTO gene of goat. Acta Veterinaria et Zootechnica Sinica. (2016) 47:888–98. doi: 10.11843/j.issn.0366-6964.2016.05.005
22. Fu, Y, Li, L, Wang, X, Li, B, Fang, X, Zhao, F, et al. The mRNA expression, cloning and sequencing of FTO gene and bioinformatics analysis in Suzhong pig. Acta Agriculturae Boreali-Sinica. (2013) 28:128–34. (in Chinese). doi: 10.3969/j.issn.1000-7091.2013.01.024
23. Gu, Y, Ma, L, Wang, F, Zhang, Y, Yan, P, and Pan, H. Cloning and expression analysis of FTO gene in yak. J Agric Biotechnol. (2022) 30:667–75. (in Chinese). doi: 10.3969/j.issn.1674-7968.2022.04.005
24. Qiu, L, Guan, J, Li, L, Li, W, and Du, M. Cloning, sequence analysis and tissue differential expression of FTO gene in Guangling donkey. China Anim Husb Vet Med. (2022) 49:12–22. (in Chinese). doi: 10.16431/j.cnki.1671-7236.2022.01.002
25. Ma, Y, Zhang, T, Mei, S, Zhou, X, Zhao, Z, Feng, D, et al. Cloning,molecular characteristics and expression patterns of FTO gene in Tan sheep. J Fujian Agric Forestry Univ. (2023) 52:505–11. (in Chinese). doi: 10.13323/j.cnki.j.fafu(nat.sci.).2023.04.012
26. Weng, J. Gene with meat quality traits in Qianbei Brown goat association study for polymorphism and expression of FTO. Guiyang, Guizhou: Guizhou University (2019).
27. Kohlrausch, FB, Berteli, TS, Wang, F, Navarro, PA, and Keefe, DL. Control of LINE-1 expression maintains genome integrity in germline and early embryo development. Reprod Sci. (2022) 29:328–40. doi: 10.1007/s43032-021-00461-1
28. Jachowicz, JW, Bing, X, Pontabry, J, Bošković, A, Rando, OJ, and Torres-Padilla, ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet. (2017) 49:1502–10. doi: 10.1038/ng.3945
29. Reddon, H, Gerstein, HC, Engert, JC, Mohan, V, Bosch, J, Desai, D, et al. Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study. Sci Rep. (2016) 6:18672. doi: 10.1038/srep18672
30. Bao, G, Liu, X, Wang, J, Hu, J, Shi, B, Li, S, et al. Effects of slaughter age on myosin heavy chain isoforms, muscle fibers, fatty acids, and meat quality in longissimus Thoracis muscle of Tibetan sheep. Front Vet Sci. (2021) 8:689589. doi: 10.3389/fvets.2021.689589
31. Danaher, J, Stathis, GS, Wilson, RA, Moreno-Asso, A, Wellard, RM, and Cooke, MB. High intensity exercise downregulates FTO mRNA expression during the early stages of recovery in young males and females. Nutr Metab (Lond). (2020) 17:68. doi: 10.1186/s12986-020-00489-1, eCollection 2020
32. De, CM, Criscione, SW, Peterson, AL, Neretti, N, Sedivy, JM, and Kreiling, JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. (2013) 5:867–83. doi: 10.18632/aging.100621
33. Lai, Y, Ramírez-Pardo, I, Isern, J, An, J, Perdiguero, E, Serrano, AL, et al. Multimodal cell atlas of the ageing human skeletal muscle. Nature. (2024) 629:154–64. doi: 10.1038/s41586-024-07348-6
34. Li, X, Li, C, Zhang, W, Wang, Y, Qian, P, and Huang, H. Inflammation and aging: signaling pathways and intervention therapies. Signal Transduct Target Ther. (2023) 8:239. doi: 10.1038/s41392-023-01502-8
35. Wang, X, Huang, N, Yang, M, Wei, D, Tai, H, Han, X, et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1alpha pathway-mediated mitochondria biogenesis. Cell Death Dis. (2017) 8:e2702. doi: 10.1038/cddis.2017.122
36. Conerly, ML, Yao, Z, Zhong, JW, Groudine, M, and Tapscott, SJ. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev Cell. (2016) 36:375–85. doi: 10.1016/j.devcel.2016.01.021
37. Zammit, PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. (2017) 72:19–32. doi: 10.1016/j.semcdb.2017.11.011
38. Adhikari, A, Kim, W, and Davie, J. Myogenin is required for assembly of the transcription machinery on muscle genes during skeletal muscle differentiation. PLoS One. (2021) 16:e0245618. doi: 10.1371/journal.pone.0245618
39. Yang, X, Mei, C, Ma, X, Du, J, Wang, J, and Zan, L. M(6)a Methylases regulate myoblast proliferation, apoptosis and differentiation. Anim Open Access J MDPI. (2022) 12:773. doi: 10.3390/ani12060773
40. Tan, Z, Shi, S, Xu, J, Liu, X, Lei, Y, Zhang, B, et al. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m(6)A-YTHDF2-dependent manner. Oncogene. (2022) 41:2860–72. doi: 10.1038/s41388-022-02306-w
41. Geng, J, Kang, Z, Sun, Q, Zhang, M, Wang, P, Li, Y, et al. Microtubule assists Actomyosin to regulate cell nuclear mechanics and chromatin accessibility. Research (Washington, DC). (2023) 6:0054. doi: 10.34133/research.0054
42. Lilja, KC, Zhang, N, Magli, A, Gunduz, V, Bowman, CJ, Arpke, RW, et al. Pax7 remodels the chromatin landscape in skeletal muscle stem cells. PLoS One. (2017) 12:e0176190. doi: 10.1371/journal.pone.0176190
Keywords: FTO , LINE1 , muscle satellite cell, RIP, cell differentiation
Citation: Ma Z, Chai Z, Yang H, Zhang X, Zhao H, Luo X, Zhong J and Wu Z (2024) Comprehensive analysis of the expression patterns and function of the FTO–LINE1 axis in yak tissues and muscle satellite cells. Front. Vet. Sci. 11:1448587. doi: 10.3389/fvets.2024.1448587
Edited by:
Wansheng Liu, The Pennsylvania State University (PSU), United StatesReviewed by:
Yongzhen Huang, Northwest A&F University, ChinaLin Xiong, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Ma, Chai, Yang, Zhang, Zhao, Luo, Zhong and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jincheng Zhong, emhvbmdqaW5jaGVuZzUxOEAxMjYuY29t; Zhijuan Wu, d3pqZHJlYW0yMDA1QDE2My5jb20=
 Zongliang Ma
Zongliang Ma Zhixin Chai
Zhixin Chai Huan Yang1,2
Huan Yang1,2 Xiangfei Zhang
Xiangfei Zhang Jincheng Zhong
Jincheng Zhong Zhijuan Wu
Zhijuan Wu