- 1Animal Parasitic Diseases Laboratory, NEA, BARC, ARS, USDA, Beltsville, MD, United States
- 2Electron and Confocal Microscopy Unit, SEL, NEA, BARC, ARS, USDA, Beltsville, MD, United States
Eimeria maxima (APU1 and APU2) differ in virulence for chickens, due in part to the greater fecundity of the former. In a previous study, RNA-seq was used to identify a transcripts upregulated in E. maxima APU1 compared to E. maxima APU2. In this study, 2 of these upregulated genes (EMWEY 23530 and EMWEY 48910) were characterized by first confirming upregulation using quantitative RT-PCR. For both EMWEY 23530 and EMWEY 48910, RNA transcription was fairly consistent during sporulation. The extent of differential expression was about 2-fold log2 higher in APU-1 compared to APU-2 (peaking at 18 h for EMWEY 23530 and 0 h for EMWEY 48910). EMWEY 23530 and EMWEY 48910 cDNA were cloned and expressed as polyHis-fusion proteins in Escherichia coli. The observed size of recombinant EMWEY 23530 was 24 kDa; the observed size of recombinant EMWEY 48910 was 35 kDa, which are consistent with the predicted size based on the coding sequences. Immunostaining 2D gel blots of E. maxima APU1 and APU2 oocyst/sporocyst protein with antisera specific for EMWEY 23530 identified a 33.5 kDa protein with a pH 7.4 isoelectric point (Emax p33.5). Similar 2D gel blot analysis with EMWEY 48910 identified a 41 kDa protein with a pH 7.2 isoelectric point (Emax p41). The intensity of Emax p33.5 and Emax p41 was noticeably greater in oocyst/sporocyst proteins from E. maxima APU1 compared to E. maxima APU2. This was corroborated by ELISA wherein equal amounts of total E. maxima APU1 and APU2 protein were probed with serial dilutions of anti-rEmax p33.5 or anti-rEmax p41. Immunofluorescence (IFA) staining of permeabilized unsporulated E. maxima APU1 and APU2 oocysts revealed Emax p33.5 to be localized in one end of oocysts, while Emax p41 appeared on the surface of oocysts. After sporulation, the p33.5 and p41 antigens appeared loosely associated with sporocysts. Taken together, these data confirm excess expression of two proteins in the E. maxima strain characterized by greater fecundity and virulence, and may provide insight into basis for phenotypic differences among different E. maxima.
Introduction
Avian coccidiosis is an intestinal disease that occurs world-wide in poultry causing annual losses in excess of $ 13 billion (1). The losses stem from poor weight gain and feed conversion efficiency in infected chickens and from increased mortalities due to necrotic enteritis (NE) of which coccidiosis is a major predisposing factor. The causative organisms are protozoa in the genus Eimeria that are transmitted through a fecal-oral route of infection arising from the ingestion of an environmentally-resistant oocyst stage that is present in litter. Although coccidiosis in chickens can be caused by any of 7 Eimeria, the most problematic is Eimeria maxima because it infects a region of the gut that is a critical region for nutrient uptake. Moreover, E. maxima disrupts the intestinal epithelium and thereby is predisposing to invasion and subsequent toxin release by Clostridium perfringens leading to NE. In our research, two strains of E. maxima, namely E. maxima APU1 and APU2, were characterized as having different levels of pathogenicity due in part to differences in fecundity, with E. maxima APU1 producing greater numbers of oocysts than E. maxima APU2 (2). The genetic basis for these phenotypic differences has been explored using RNA-seq. This technology has been used to study Eimeria oocyst sporulation (3–5), to compare different Eimeria life cycle stages in vivo and in vitro (6–11), to compare precocious to virulent Eimeria (12–14), and to compare drug-sensitive to drug-resistant isolates (15–23). All of these studies have identified genes that may encode proteins involved in parasite development and drug resistance. In our research, RNA-seq analysis identified several genes whose transcripts were upregulated in the more fecund and virulent E. maxima APU1 (24). The present study describes the molecular characteristics of these two upregulated transcripts.
Materials and methods
Parasites and preparation of native protein
Eimeria maxima APU1 and APU2 were isolated from commercial broiler farms over 10 years ago and have been propagated every 3 mo. since isolation in susceptible chickens. Purity of the isolates was confirmed after each propagation by microscopy and ITS1-PCR (25). Eimeria maxima APU1 and APU2 oocysts were treated for 30 min with 6.5% sodium hypochlorite to remove contaminating bacteria, washed 5 times with diH2O with centrifugation at 1850 g for 10 min and suspended in Saline A (140 mM NaCl, 5 mM KCl, 4.2 mM NaHCO3, 0.1% glucose, pH 7.0). The oocysts were ground in a glass mortar with a Teflon pestle 75 times to release sporocysts. An aliquot of the sporocysts were subjected to in vitro excystation at 41°C by exposure to 0.5% trypsin and 4% sodium taurocholate (Sigma) plus 1 mM dithiothreitol (DTT) for about 45 min. Released sporozoites, intact sporocysts, and oocysts were pelleted by centrifugation for 10 min at 13,800 g, followed by resuspension in PBS [for IFA (see below)] or 8 M urea, 2% CHAPS, and 50 mM DTT for isoelectric focusing (see below).
Quantitative RT-PCR
In the previous study detailing RNA-seq in E. maxima strains APU-1 and APU-2, oocysts during a sporulation time course were collected every 6–12 h up to 48 h (24). RNA and cDNA from oocysts of that study were used in the current study for quantitative reverse transcriptase PCR. Here, we measured transcription of genes EMWEY 23530 (encodes 18 kDa cyclophilin) and EMWEY 48910 (encodes a hypothetical protein) following similar procedures. In brief, RT-qPCR reactions were prepared using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, United States), with 400 nM of each primer (Table 1), and 1 μL of diluted cDNA in a total volume of 10 μL. EMWEY 42350 (Beta tubulin, 200 nM) was used as a reference to normalize the expression levels of target genes. RT-qPCR consisted of an initial denaturing step at 95°C for 30 s, followed by 35 cycles of 95°C for 15 s, 55°C for 20 s. Melt-curve analysis entailed 55–95°C in increments of 0.5°C for 5 s. The abundance of mRNA at time points T0, T18, T36 was compared between strains APU-1 and APU-2 in triplicate reactions. Gene expression was estimated for the reference and target genes after averaging the Cq values for each replicate at each time point. The fold change in expression was calculated using an efficiency-corrected relative expression method (26). Each experiment was performed three times, and the mean of expression change (log2 fold change (FC)) was calculated for each gene and time point. All primers were designed using NCBI Primer-BLAST (27) and synthesized by Integrated DNA Technologies (Coralville, IA, United States).

Table 1. Oligonucleotide primers used in quantitative RT-PCR analysis of RNA from Eimeria maxima APU1 and APU2 during oocyst sporulation.
Expression cloning
EMWEY 23530 and EMWEY 48910 sequences were identified in a previous RNA-seq analysis to be upregulated at all time-points during sporulation of E. maxima oocysts (24). Further comparative transcriptome analysis between two strains of E. maxima (APU1 and APU2) revealed about a two-fold greater number of EMWEY 23530 and EMWEY 48910 transcripts in E. maxima APU1 than in E. maxima APU2. The open reading frame of EMWEY 23530 and EMWEY 48910 were synthesized by a commercial company (GenScript, Piscataway NJ) containing 5’ XhoI and 3’ EcoRI sites for cloning into pBluescript II SK. The cDNA inserts were excised with XhoI and EcoRI, cloned into pTrcHisA (Invitrogen) with DNA ligase, and recombinant plasmid used to transform Escherichia coli BL21. Recombinant clones were expanded in LB-Amp at 37°C and induced at log phase growth (O.D.600 = 0.5) for 4 h with 1 mM IPTG. The induced cells were harvested by centrifugation at 1850 RPM for 10 min. The cell pellets were extracted first for 30 min with NBB containing 0.1 mg/mL lysozyme and protease inhibitors, followed by DNase and RNase treatment for an additional 30 min on a rocker. The soluble fraction after NBB treatment was collected by centrifugation, and the resulting pellet was further extracted with DBB for 30 min on a rocker, followed by centrifugation to retrieve denaturing soluble (DS) supernatant. Recombinant EMWEY 23530 and EMWEY 48910 proteins were purified by affinity chromatography using NiNTA agarose (Invitrogen) and NiNTA eluates were analyzed by SDS-PAGE and immunoblotting (see below).
Preparation of polyclonal anti-EMWEY 23530 and anti-EMWEY 48910 sera
Eluates from NiNTA-column purification of recombinant EMWEY 23530 (rEMWEY 23530) and EMWEY 48910 (rEMWEY48910) were used to immunize New Zealand White rabbits (2/recombinant antigen) (Pacific Immunology, Ramona CA). Immunizations utilized Freund’s Complete Adjuvant in the primary immunization and Freund’s Incomplete Adjuvant in booster immunizations at 3, 6, and 10 weeks post-primary immunization. Blood was collected prior to primary immunization, at various time-points during and after the final booster immunization, and processed for serum using standard procedures (28).
Isoelectric focusing
Eimeria maxima APU1 and APU2 oocyst, sporocyst, and sporozoite protein from 107 oocysts were extracted with 8M urea, 2% CHAPS, and 50 mM DTT at RT for 1 h on a rocker, followed by centrifugation for 10 min at 13,800 g. The supernatant protein concentration was estimated by BCA assay. Eimeria maxima APU1 or APU2 protein (100 μg) was mixed with Rehydration/Sample buffer (Bio-Rad, Hercules CA) and adsorbed to ReadyStrip IPG strips pH 7–10 for 10 h at RT following manufacturer’s instructions (Bio-Rad). The IPG strips were then subjected to isoelectric focusing in the following steps: Step 1–250 v, 20 min, linear ramp speed; Step 2–4,000 v, 2 h, linear ramp speed; Step 3–4,000 v to reach 10,000 v-hr, rapid ramp speed. After IEF, the IPG strips were rinsed with Equilibration Buffers I and II for 10 min each, then inserted into a preparative well of SDS-PAGE and overlain with low melting point agarose containing trace amounts of Bromophenol Blue.
SDS-PAGE/immunoblotting
Unpurified and NiNTA-purified recombinant EMWEY 23530 (rEMWEY 23530) and EMWEY 48910 (rEMWEY 48910) proteins were analyzed by SDS-PAGE followed by transblotting to Immobilon membrane (Millipore-Sigma Burlington MA), followed by immunostaining with mouse anti-His antibodies (Invitrogen-Thermo Scientific, Waltham MA) using standard procedures (29). For 2D gel analysis, IPG strips were subjected to SDS-PAGE for fractionation of IEF-resolved native E. maxima APU1 and APU 2 proteins followed by transblotting to Immobilon membrane. The membranes were immunostained with rabbit anti-rEMWEY 23530 or anti-rEMWEY 48910 sera using standard procedures (29).
Enzyme-linked immunosorbent assay
Eimeria maxima APU1 and APU2 native protein extracted for 2D gel immunoblotting was diluted in carbonate buffer (pH 9.5) and adsorbed for 6 h at 37°C onto wells of Immulon II microtiter plates (Thermo Scientific) at 1.0 μg/well. The wells were washed with PBS-containing 0.05% Tween 20 (BSA-Tw), blocked for 30 min with 1% BSA in PBS-Tw, incubated for 1 h at RT with serial dilutions of rabbit anti-EMWEY 23530 or anti-EMWEY 48910 sera or pre-immunization sera, followed by alkaline-phosphatase labeled anti-rabbit IgG (Sigma, 1:1000 dilution) for an additional 1 h at RT. Binding was visualized by addition of alkaline-phosphatase substrate (1 mg/mL p-nitrophenyl phosphate, disodium) and measured on a microtiter plate reader (SpectraMax 190, Molecular Devices, San Jose, CA) at 405 nm. Antibodies were removed after each step by 3 washes with PBS-Tw.
Immunofluorescence assay
Eimeria maxima APU1 and APU2 oocysts, sporocysts, and sporozoites (see above) were adhered to the surface of 8-well microscope slides (MP Biomedicals, San Diego CA), air-dried, and then treated with cold methanol for 5 min, followed by a brief wash with PBS-Tw. The wells were first treated with PBS-Tw + 1% BSA, followed by a 1 h incubation with a 1:100 or 1:500 dilution of rabbit anti-EMWEY 23530 or anti-EMWEY 48910 sera or preimmunization sera, followed by a 1 h incubation with a 1:100 dilution of FITC-labeled goat anti-rabbit IgG (Sigma). The slides were washed between each incubation step with PBS-Tw, allowed to air dry after the last wash, overlaid with VectaShield (Vector Laboratories, Newark NJ) anti-bleaching medium and a coverslip. The slides were examined on a Zeiss microscope and images captured using software.
Results and discussion
Quantitative RT-PCR
qRT-PCR using primers directed to either EMWEY 23530 or EMWEY 48910 corroborated our previous RNA-seq findings in that both EMWEY 23530 and EMWEY 48910 were transcribed 1.5–2.0 log2 higher at all timepoints in E. maxima APU1 compared to E. maxima APU2 (Figures 1A,B). In the current study, the greatest difference in EMWEY 23530 expression between E. maxima APU1 and APU2 was at 18 h sporulation (Figure 1A); for EMWEY 48910 was at 0 h sporulation (Figure 1B). These data along with the IFA staining pattern suggest that EMWEY 23530 and EMWEY 48910 code for proteins that play some role in oocyst wall formation.
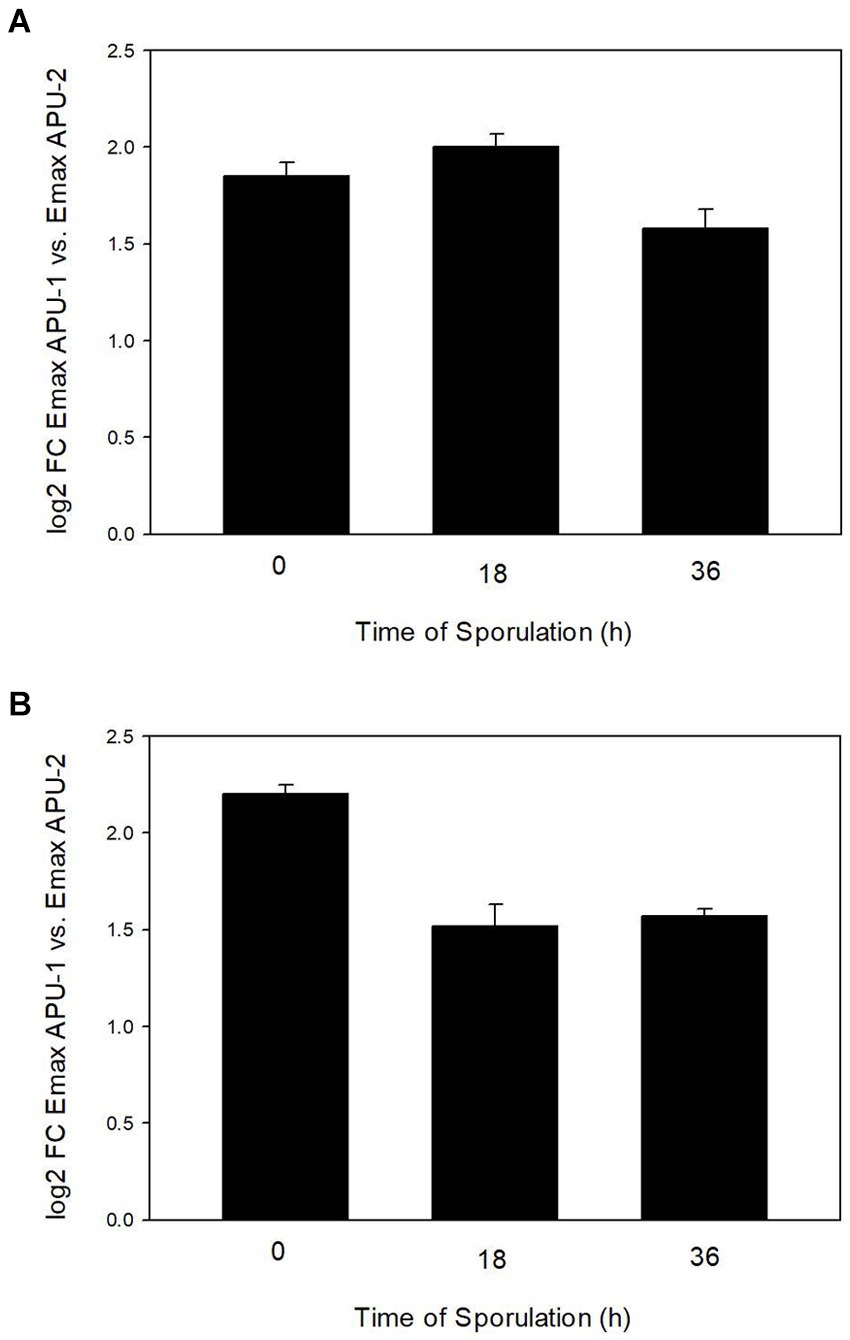
Figure 1. Comparison of increased EMWEY 23530 (A) and EMWEY 48910 (B) transcript levels in Eimeria maxima APU1 vs. E. maxima APU2 at different times of sporulation as measured by quantitative RT-PCR analysis.
Expression of recombinant protein and identification of relevant native protein
Expression cloning of EMWEY 23530 and EMWEY 48910 as polyHis fusion proteins in Es. coli revealed by SDS-PAGE/immunoblotting a 24–26 kDa protein for the former and a 35 kDa protein for the latter (Figure 2). The predicted size of recombinant EMWEY 23530 based on the DNA sequence is 20.5 kDa which with the addition of the upstream polyHis tail (~5 kDa) would give rise to the observed 24–26 kDa protein. The observed (35 kDa) and predicted (29.5 kDa) sizes of recombinant EMWEY 48910 is also in agreement with the DNA sequence. Employing 2D gel blots of native E. maxima protein, polyclonal antisera specific for EMWEY23530 identified a native 33.5 kDa protein with a pI = 7.4 (Figure 3). A similar method using antisera specific for EMWEY 48910 identified a 41 kDa protein with a pI = 7.2 (Figure 3). The difference between observed (33.5) and predicted (20.4) sizes of EMWEY 23530 may be due secondary processing such as glycosylation and to 3D folding of the protein. A similar size discrepancy between observed (41 kDa) and predicted (29.5) EMWEY 48910. Software for predicting N-glycosylation sites1 and O-glycosylation sites2 found 2 potential N-glycosylation sites and 8 potential O-glycosylation sites in EMWEY 23530. Similar analyses for EMWEY 48910 found 2 potential N-glycosylation sites and 5 potential O-glycosylation sites.
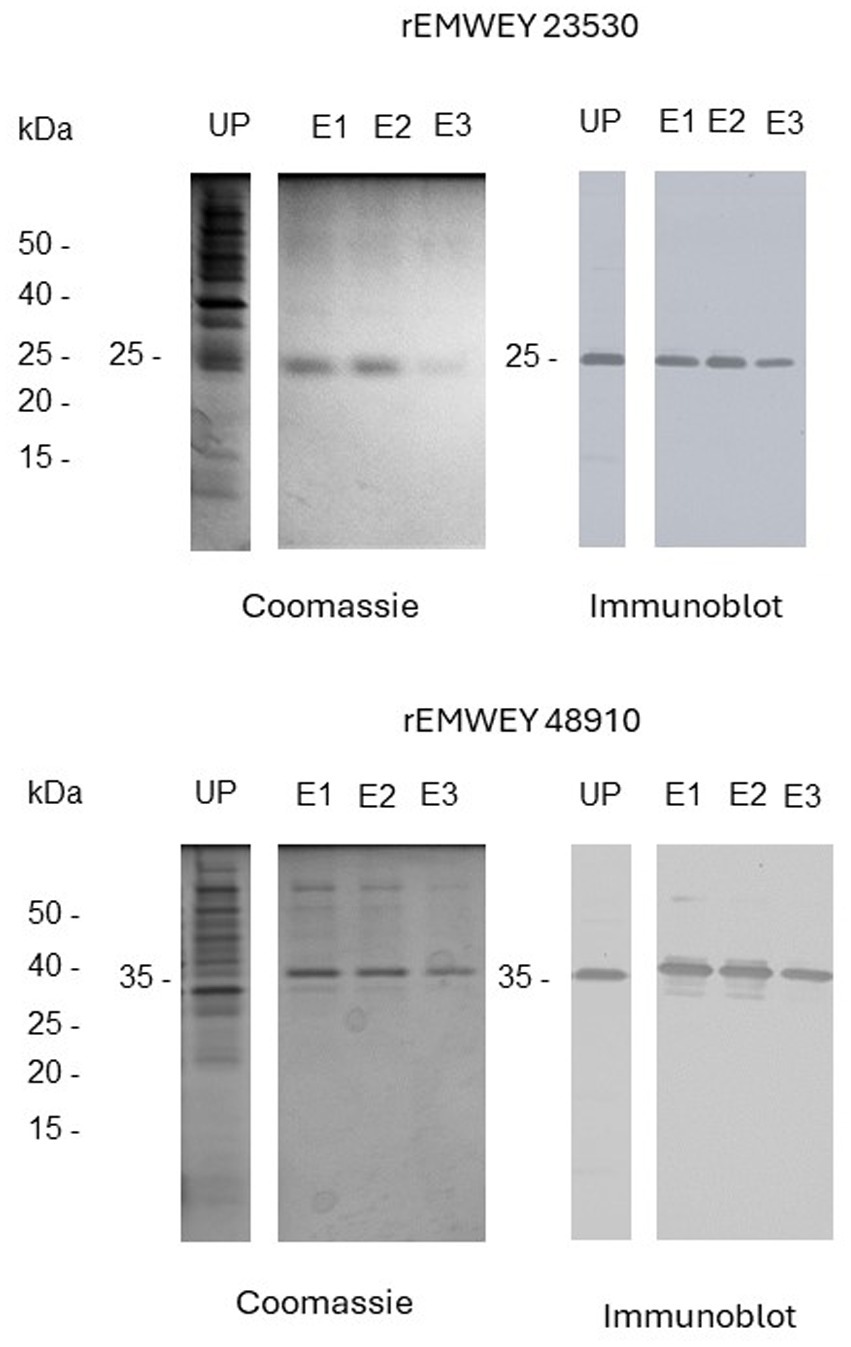
Figure 2. SDS-PAGE analysis of unpurified (UP) and NiNTA affinity chromatography-purified recombinant EMWEY 23530 (upper panel) and recombinant rEMWEY 48910 protein (lower panel) visualized by Coomassie Blue staining (left panels) or immunoblotting with mouse anti-His tag sera (right panels). UP-unpurified, E1-, E2-, E3-NiNTA eluate fractions used in generation of rabbit antisera, kDa-relative molecular size markers.
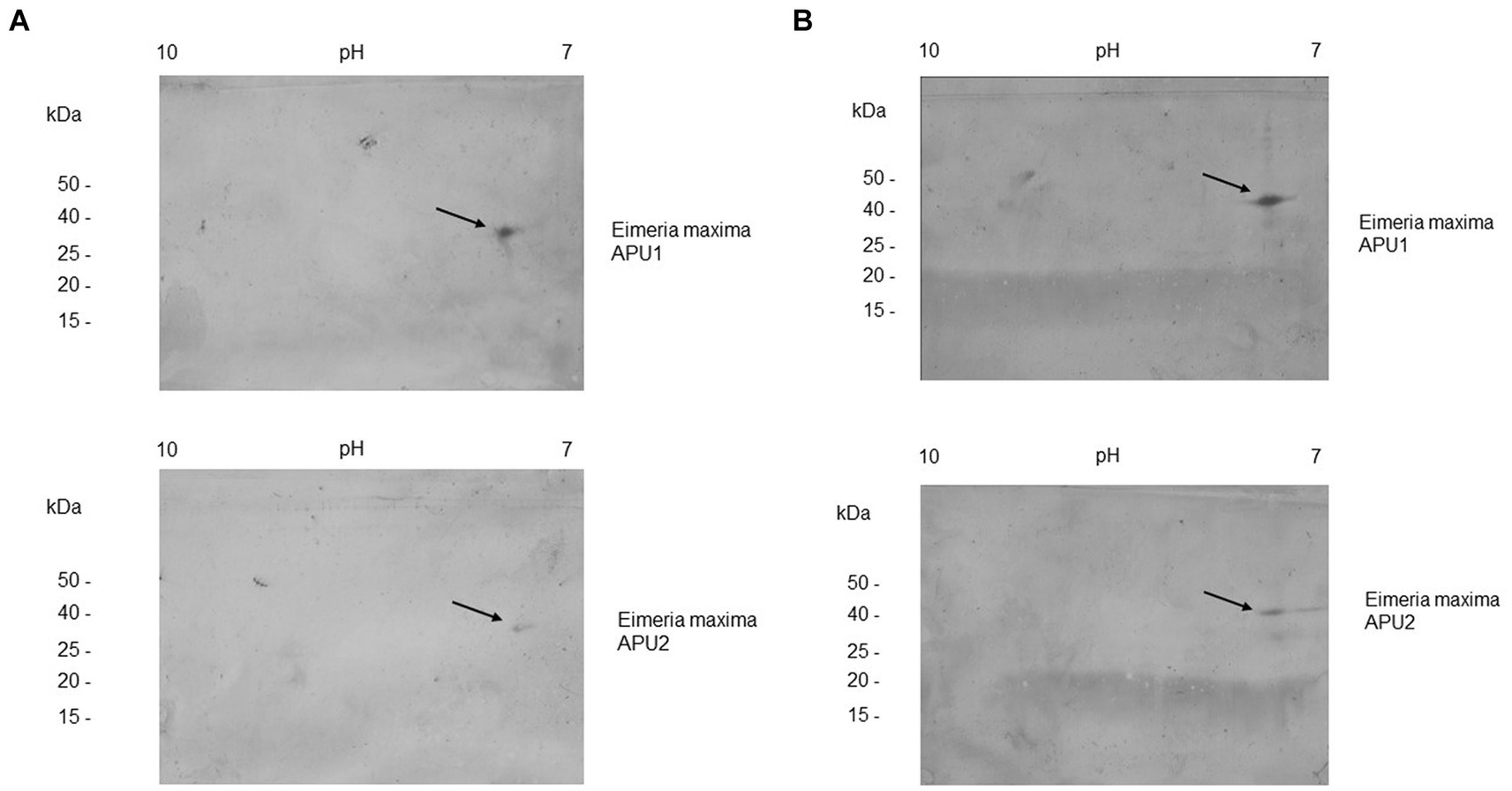
Figure 3. 2D gel immunoblot analysis of Eimeria maxima APU1 or E. maxima APU2 total oocyst, sporocysts, sporozoite protein resolved by isoelectric focusing (IEF) in the first dimension and SDS-PAGE in the second dimension. Equal amounts of total E. maxima APU1 and APU2 protein were subjected to 2D gel electrophoresis and immunostained with antisera specific for recombinant EMWEY 23530 (A), or EMWEY 48910 (B).
The higher transcript levels of EMWEY 23530 and EMWEY 48910 in E. maxima APU1 compared to E. maxima APU2 was also observed at the protein level as observed in 2D immunoblots of E. maxima APU1 or E. maxima APU2 native oocyst/sporocyst protein. Eimeria maxima APU1 displayed greater amounts of the native 33.5 (Figure 3A) and 41 kDa proteins (Figure 3B) than E. maxima APU2. The greater expression of native EMWEY 23530 and EMWEY 48910 was corroborated by ELISA using total native E. maxima APU1 and APU2 protein probed with anti-rEMWEY 23530 or anti-rEMWEY 48910 sera (Figure 4). These data suggest that higher EMWEY 23530 and EMWEY 48910 transcript levels leads to greater amounts of Emax p33.5 and Emax p41 protein in E. maxima APU1 compared to E. maxima APU2.
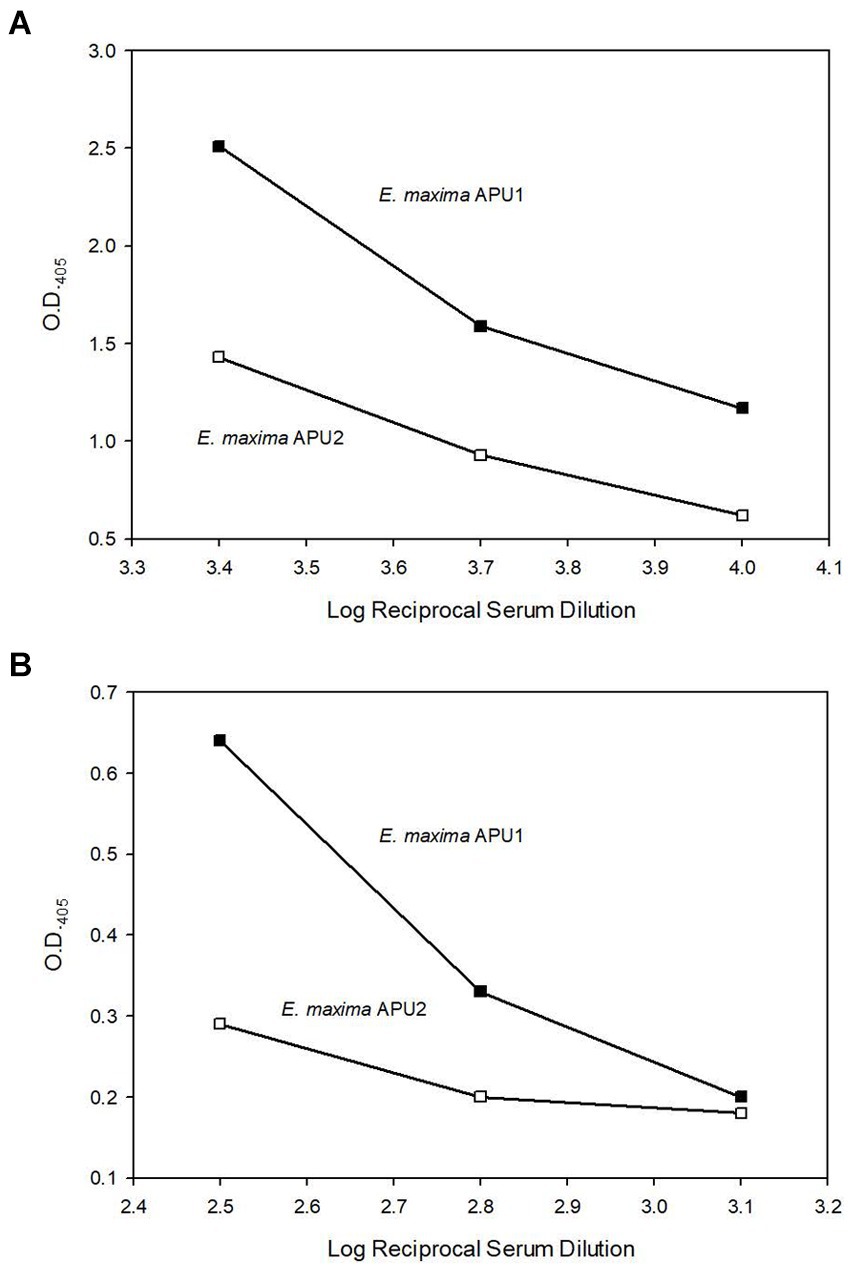
Figure 4. Enzyme-linked immunosorbent assay (ELISA) analysis of equal amounts (1 μg) of total Eimeria maxima APU1 (closed symbols) or E. maxima APU2 (open symbols) protein adhered to Immulon 2 HB plates and probed with serial dilutions of anti-EMWEY 23530 sera (A) (log reciprocal dilutions 3.4, 3.7, and 4.0) or anti-EMWEY 48910 sera (B) (log reciprocal dilutions 2.5, 2.8, and 3.1).
Antigen localization by IFA
EMWEY 23530 appeared to localize to antigens found internal and external to E. maxima APU1 and APU2 oocysts whereas EMWEY 48910 was only found inside oocysts and appeared to be associated with sporocysts (Figure 5). The reactivity of antisera, particularly against rEMWEY48910, to sporocysts inside oocysts may be explained by the variability in morphology of oocysts after grinding and excystation. Many oocysts and sporocysts did not react with the antisera (data not shown) suggesting that the antigen is present inside oocysts and is released when the oocyst wall is completely broken open. No internal labeling of sporozoites was observed with anti-EMWEY 48910 sera and no difference in the staining pattern between E. maxima APU1 and APU2 oocysts, sporocysts, and sporozoites was found (data not shown). The similarity in IFA staining of E. maxima APU1 and APU2 which is different than the increased binding as observed by 2D gel immunoblotting and ELISA probably reflects the more random nature of microscopy staining. 2D immunoblots and ELISA utilize total protein which would be less susceptible to effects on morphology.
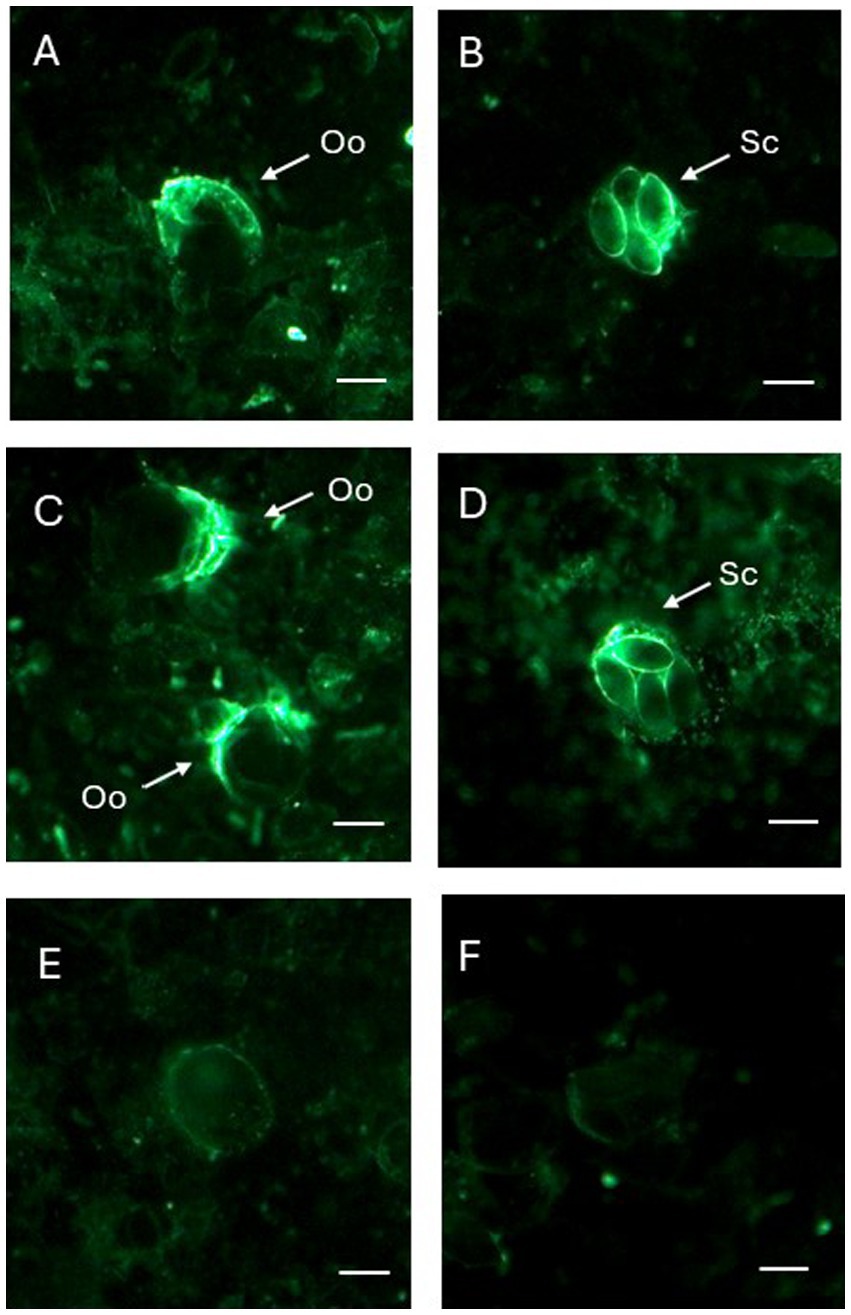
Figure 5. Immunofluorescence (IFA) staining of Eimeria maxima APU1 (A,B,E) or E. maxima APU2 (C,D,F) with anti-recombinant EMWEY 23530 (A,C) or anti-recombinant EMWEY 48910 (B,E) sera. Oo, oocysts; Sc, sporocysts. Preimmunization control IFA staining (E,F). Bar = 10 μm.
Conclusion
Previous studies using RNAseq to compare RNA transcripts between two different strains of E. maxima during sporulation identified a number of upregulated genes in E. maxima APU1 compared to E. maxima APU2. This study employed qRT-PCR to confirm the relatively higher levels of EMWEY 23530 and EMWEY 48910 transcripts in E. maxima APU1. This greater expression was reflected in greater intensity of native Emax p33.5 and Emax 41 protein as observed in 2D gel immunoblots and in ELISA using antisera to recombinant EMWEY 23530 and EMWEY 48910. The former appears to be associated with an antigen inside and outside sporulated oocysts, whereas the latter is only found inside oocysts and loosely associated with sporocysts. Whether these proteins have any role in the differences in fecundity between E. maxima APU1 and APU2 remains unknown.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving animals were reviewed and approved by Beltsville Agricultural Research Center Institutional Animal Care and Use Committee under Protocol No. 2206.
Author contributions
MJ: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. CP: Investigation, Methodology, Writing – review & editing. AJ: Investigation, Methodology, Writing – review & editing. MP: Investigation, Methodology, Writing – review & editing. MT: Formal analysis, Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the USDA-ARS CRIS Project No. 8042–32000-114-00D.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://services.healthtech.dtu.dk/services/NetNGlyc-1.0/
2. ^https://services.healthtech.dtu.dk/services/NetOGlyc-4.0/
References
1. Blake, DP, Knox, J, Dehaeck, B, Huntington, B, Rathinam, T, Ravipati, V, et al. Re-calculating the cost of coccidiosis in chickens. Vet Res. (2020) 51:115. doi: 10.1186/s13567-020-00837-2
2. Jenkins, MC, Dubey, JP, Miska, K, and Fetterer, R. Differences in fecundity of Eimeria maxima strains exhibiting different levels of pathogenicity in its avian host. Vet Parasitol. (2017) 236:1–6. doi: 10.1016/j.vetpar.2017.01.009
3. Matsubayashi, M, Hatta, T, Anisuzzaman, SK, Shimura, K, Isobe, T, Kita, K, et al. High-throughput RNA sequencing profiles and transcriptional evidence of aerobic respiratory enzymes in sporulating oocysts and sporozoites of Eimeria tenella. Infect Genet Evol. (2013) 18:269–76. doi: 10.1016/j.meegid.2013.06.002
4. Tucker, MS, O'Brien, CN, Jenkins, MC, and Rosenthal, BM. Dynamically expressed genes provide candidate viability biomarkers in a model coccidian. PLoS One. (2021) 16:e0258157. doi: 10.1371/journal.pone.0258157
5. Gao, Y, Suding, Z, Wang, L, Liu, D, Su, S, Xu, J, et al. Full-length transcriptome sequence analysis of Eimeria necatrix unsporulated oocysts and sporozoites identifies genes involved in cellular invasion. Vet Parasitol. (2021) 296:109480. doi: 10.1016/j.vetpar.2021.109480
6. Walker, RA, Slapetova, I, Slapeta, J, Miller, CM, and Smith, NC. The glycosylation pathway of Eimeria tenella is upregulated during gametocyte development and may play a role in oocyst wall formation. Eukaryot Cell. (2010) 9:127–35. doi: 10.1128/EC.00255-09
7. Walker, RA, Sharman, PA, Miller, CM, Lippuner, C, Okoniewski, M, Eichenberger, RM, et al. RNA Seq analysis of the Eimeria tenella gametocyte transcriptome reveals clues about the molecular basis for sexual reproduction and oocyst biogenesis. BMC Genomics. (2015) 16:94. doi: 10.1186/s12864-015-1298-6
8. Su, S, Hou, Z, Liu, D, Jia, C, Wang, L, Xu, J, et al. Comparative transcriptome analysis of second-and third-generation merozoites of Eimeria necatrix comparative study. Parasit Vectors. (2017) 10:388. doi: 10.1186/s13071-017-2325-z
9. Su, S, Hou, Z, Liu, D, Jia, C, Wang, L, Xu, J, et al. Comparative transcriptome analysis of Eimeria necatrix third-generation merozoites and gametocytes reveals genes involved in sexual differentiation and gametocyte development comparative study. Vet Parasitol. (2018) 252:35–46. doi: 10.1016/j.vetpar.2018.01.019
10. Sandholt, AKS, Wattrang, E, Lilja, T, Ahola, T, Lundén, A, Troell, K, et al. Dual RNA-seq transcriptome analysis of caecal tissue during primary Eimeria tenella infection in chickens. BMC Genomics. (2021) 22:660. doi: 10.1186/s12864-021-07959-7
11. Sandholt, AKS, Xu, F, Söderlund, R, Lundén, A, Troell, K, Svärd, SG, et al. Dual RNA-Seq transcriptome analysis of chicken macrophage-like cells (HD11) infected in vitro with Eimeria tenella. Parasitology. (2021) 148:712–25. doi: 10.1017/S0031182021000111
12. Matsubayashi, M, Kawahara, F, Hatta, T, Yamagishi, J, Miyoshi, T, Sasai, K, et al. Transcriptional profiles of virulent and precocious strains of Eimeria tenella at sporozoite stage; novel biological insight into attenuated asexual development. Infect Genet Evol. (2016) 40:54–62. doi: 10.1016/j.meegid.2016.02.021
13. Hu, D, Wang, C, Wang, S, Tang, W, Duan, C, Zhang, S, et al. Comparative transcriptome analysis of Eimeria maxima (Apicomplexa: Eimeriidae) suggests DNA replication activities correlating with its fecundity comparative study. BMC Genomics. (2018) 19:699. doi: 10.1186/s12864-018-5090-2
14. Matsubayashi, M, Inaoka, DK, Komatsuya, K, Hatta, T, Kawahara, F, Sakamoto, F, et al. Novel characteristics of mitochondrial Electron transport chain from Eimeria tenella. Genes. (2019) 10:29. doi: 10.3390/genes10010029
15. Chen, T, Huang, B, Zhao, Q, Dong, H, Zhu, S, Zhao, Z, et al. Molecular characterization and functional analysis of Eimeria tenella malate dehydrogenase. Parasitol Res. (2018) 117:2053–63. doi: 10.1007/s00436-018-5875-x
16. Xie, Y, Huang, B, Xu, L, Zhao, Q, Zhu, S, Zhao, H, et al. Comparative transcriptome analyses of drug-sensitive and drug-resistant strains of Eimeria tenella by RNA-sequencing comparative study. J Eukaryot Microbiol. (2020) 67:406–16. doi: 10.1111/jeu.12790
17. Wang, H, Zhao, Q, Zhu, S, Dong, H, Yu, S, Wang, Q, et al. Molecular characterization and functional analysis of Eimeria tenella citrate synthase. Parasitol Res. (2021) 120:1025–35. doi: 10.1007/s00436-020-07014-6
18. Yu, Y, Dong, H, Zhao, Q, Zhu, S, Liang, S, Wang, Q, et al. Molecular characterization and analysis of the ATPase ASNA1 homolog gene of Eimeria tenella in a drug sensitive strain and drug resistant strains. Int J Parasitol Drugs Drug Resist. (2021) 15:115–25. doi: 10.1016/j.ijpddr.2021.02.005
19. Huang, J, Huang, J, Husien, HM, Peng, W, Liu, W, Bo, W, et al. Comparison of endogenous development, invasion ability and apoptotic features between diclazuril resistant and sensitive strains of Eimeria tenella. Vet Parasitol. (2022) 305:109719. doi: 10.1016/j.vetpar.2022.109719
20. Huang, Y, Zhu, S, Chen, T, Zhao, Q, Dong, H, Huang, B, et al. Molecular characterization of glyceraldehyde-3-phosphate dehydrogenase from Eimeria tenella. Parasitol Res. (2022) 121:1749–60. doi: 10.1007/s00436-022-07508-5
21. Zhang, H, Zhang, L, Si, H, Liu, X, Suo, X, and Hu, D. Early transcriptional response to Monensin in sensitive and resistant strains of Eimeria tenella. Front Microbiol. (2022) 13:934153. doi: 10.3389/fmicb.2022.934153
22. Sun, P, Wang, C, Zhang, Y, Tang, X, Hu, D, Xie, F, et al. Transcriptome profile of halofuginone resistant and sensitive strains of Eimeria tenella. Front Microbiol. (2023) 14:1141952. doi: 10.3389/fmicb.2023.1141952
23. Yu, Y, Huang, W, Wang, Q, Dong, H, Zhao, Q, Zhu, S, et al. Molecular characterization and analysis of drug resistance-associated protein enolase 2 of Eimeria tenella. Int J Parasitol Drugs Drug Resist. (2023) 21:81–90. doi: 10.1016/j.ijpddr.2023.01.004
24. Tucker, MS, O'Brien, CN, Johnson, AN, Dubey, JP, Rosenthal, BM, and Jenkins, MC. RNA-Seq of phenotypically distinct Eimeria maxima strains reveals coordinated and contrasting maturation and shared sporogonic biomarkers with Eimeria acervulina. Pathog. (2023) 13:2. doi: 10.3390/pathogens13010002
25. Jenkins, MC, Miska, K, and Klopp, S. Improved polymerase chain reaction technique for determining the species composition of Eimeria in poultry litter. Avian Dis. (2006) 50:632–5. doi: 10.1637/7615-042106R.1
26. Pfaffl, MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. (2001) 29:e45. doi: 10.1093/nar/29.9.e45
27. Ye, J, Coulouris, G, Zaretskaya, I, Cutcutache, I, Rozen, S, and Madden, TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. (2012) 13:134. doi: 10.1186/1471-2105-13-134
28. Harlow, E, and Lane, D. Immunizations In: Antibodies. A laboratory manual : Cold Spring Harbor Laboratory. 53–138.
Keywords: Eimeria maxima, recombinant protein, 2D gel, immunofluorescence, Eimeria, RT-PCR, EMWEY 23530, EMWEY 48910
Citation: Jenkins MC, Parker C, Jansen A, Papadopoulos MD and Tucker MS (2024) Molecular characterization of cDNA coding for 33.5 and 41 kDa oocyst and sporocyst proteins that are differentially regulated in different strains of Eimeria maxima. Front. Vet. Sci. 11:1445646. doi: 10.3389/fvets.2024.1445646
Edited by:
Ruediger Hauck, Auburn University, United StatesReviewed by:
Zhaofeng Hou, Yangzhou University, ChinaQing Liu, Shanxi Agricultural University, China
Copyright © 2024 Jenkins, Parker, Jansen, Papadopoulos and Tucker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark C. Jenkins, bWFyay5qZW5raW5zQHVzZGEuZ292
 Mark C. Jenkins
Mark C. Jenkins Carolyn Parker1
Carolyn Parker1 Andrew Jansen
Andrew Jansen Matthew S. Tucker
Matthew S. Tucker