- 1College of Veterinary Medicine, Yunnan Agricultural University, Kunming, China
- 2Dongguan Center for Animal Disease Prevention and Control, Dongguan, Guangdong, China
- 3Dongguan Key Laboratory of Zoonosis, Dongguan, Guangdong, China
Edwardsiella tarda (E. tarda) can infect humans and a variety of animals, including fish, amphibians, reptiles, birds, and mammals. However, a more highly sensitive, specific, and repeatable test for its detection is lacking. The objective of this study was to develop a highly sensitive, specific, and repeatable droplet digital polymerase chain reaction (ddPCR)-based method for the quantitative detection of E. tarda. The gyrB gene was selected as the target gene, and primers and probe were designed and synthesized. Using E. tarda genomic DNA as templates, the reaction method was optimized to establish a linear relationship with real-time PCR detection methods. The sensitivity, specificity, and repeatability of the method were analyzed, and clinical samples were tested. When the primer and probe concentrations were 900 and 300 nM, respectively, and the annealing temperature was 57°C, the efficiency of the ddPCR amplification reaction was highest and the boundary between positive and negative droplet distribution was clearest. The sensitivity was high, with detection limit being as low as 0.56 copies·μL−1; additionally, and a good linear relationship (R2 = 0.9962) between ddPCR and real-time PCR detection, within the range of 1–25,000 copies·μL−1, was evident. The repeatability was good, with a detection coefficient of variation of 2.74%. There was no cross-reactivity with 15 other common pathogenic microorganisms in aquatic animals (Streptococcus agalactiae, Streptococcus iniae, Streptococcus suis type 2, Nocardia seriolae, Vibrio parahaemolyticus, Aeromonas sobria, red sea bream iridovirus, decapod iridescent virus 1, enterocytozoon hepatopenaei, carp edema virus, Koi herpesvirus, goldfish hematopoietic necrosis virus, tilapia lake virus, viral nervous necrosis virus, or grass carp reovirus) in positive samples. Among the 48 clinical samples, including Bahaba taipingensis and its live food fish, pond water samples, and routine monitoring samples (Koi), 21 were positive for E. tarda, consistent with the bacterial isolation and identification results. The E. tarda ddPCR detection method has high specificity, sensitivity, and repeatability, can more accurately quantify E. tarda, and provides a useful reference for research related to this bacterium.
1 Introduction
Edwardsiella tarda (E. tarda) is a Gram-negative bacillus that was first isolated from the Japanese eel (Anguilla japonica) by Hoshina in 1962 (1). E. tarda has a wide range of hosts, can infect aquatic animals (2–4) and birds (5), and has been isolated from the feces of South China tigers (Panthera tigris ssp. amoyensis) (6). Infection by this bacterium can occur in all seasons; the higher the water temperature, the longer the disease cycle and the greater the damage. Infection presents mainly as skin hemorrhage, ascites, hepatosplenic and renal swelling, congestion, and hemorrhagic sepsis (7, 8). The geographical scope of these infections has been expanding, with reports of severe economic losses to the aquaculture industry in the United States (9), Germany (10), Italy (11), and South Africa (12). E. tarda has been reported to cause disease in various economically important fish species, including Chinese soft-shelled turtles, carp, tilapia, flounder, and turbot (1, 2, 13), have been a matter of concern for the Chinese aquaculture industry since 1989 (2).
Edwardsiella tarda is an important zoonotic bacterium. Exposure to water contaminated with E. tarda or undercooked, infected food causes symptoms such as low-grade fever, gastroenteritis, liver abscess, meningitis, and sepsis and even lead to death (14–16). Therefore, it is important to establish a method for detecting E. tarda with high specificity and sensitivity and good reproducibility to ensure public health safety and prevent and control aquatic animal diseases.
Research on the detection of E. tarda initially focused on traditional bacterial isolation and culture methods (17), followed by enzyme staining (18), enzyme-linked immunosorbent assay (ELISA) (19, 20), PCR (21), real-time PCR (22), loop-mediated isothermal amplification (LAMP) (23, 24), and gene chips (25). However, a more highly-sensitive, specific, and repeatable test is still lacking. The objective of the current study was to develop a droplet digital PCR (ddPCR)-based method for detecting E. tarda to provide technical support for the prevention and treatment of infections caused by E. tarda.
2 Materials and methods
2.1 Standards and training sources
Edwardsiella tarda (DG20230920, Dongguan, China) was isolated and identified at the Laboratory of the Dongguan Animal Disease Prevention and Control Center. E. tarda was aseptically collected from the spleen, liver, and kidney of Bahaba taipingensis, and inoculated onto tryptone soybean Agar (TSA) plate for 24 h at 30°C. Streptococcus iniae (ATCC29178, American, Virginia) was obtained from the American Type Culture Collection (ATCC). Streptococcus agalactiae, Nocardia seriolae, Vibrio parahaemolyticus, Aeromonas sobria, and Streptococcus suis type 2 were preserved in the Dongguan Animal Disease Prevention and Control Center laboratory (Dongguan, China). Nucleic acids positive for viral nervous necrosis virus (VNNV), grass carp reovirus (GCRV), tilapia lake virus (TiLV), red sea bream iridovirus (RSIV), decapod iridescent virus 1(DIV1), and enterocytozoon hepatopenaei (EHP) were obtained from the Guangdong Provincial Center for Animal Disease Prevention and Control (Guangzhou, China). Nucleic acids of carp edema virus (CEV) were obtained from the Beijing Aquatic Technology Extension Station (Beijing, China). Nucleic acids of koi herpesvirus (KHV) and goldfish hematopoietic necrosis virus (GFHNV) were obtained from the Chinese Academy of Inspection and Quarantine (Beijing, China).
2.2 Sample collection
A total of 48 clinical samples were collected from the rescue base of the Dongguan Bahaba Taipingensis Nature Reserve (Bahaba Taipingensis, live food fish, and pond water).
2.3 Design of primers and probe
The whole gene sequence was accessed from the GenBank (Accession number: MG026726.1). Given that gyrB is more suitable for distinguishing and identifying E. tarda, gyrB was selected as the target gene (26). The primer and probe sequences are detailed in Table 1.
2.4 DNA extraction
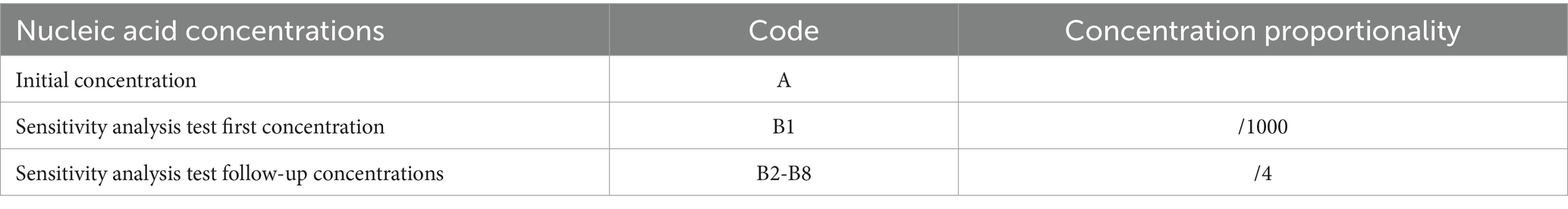
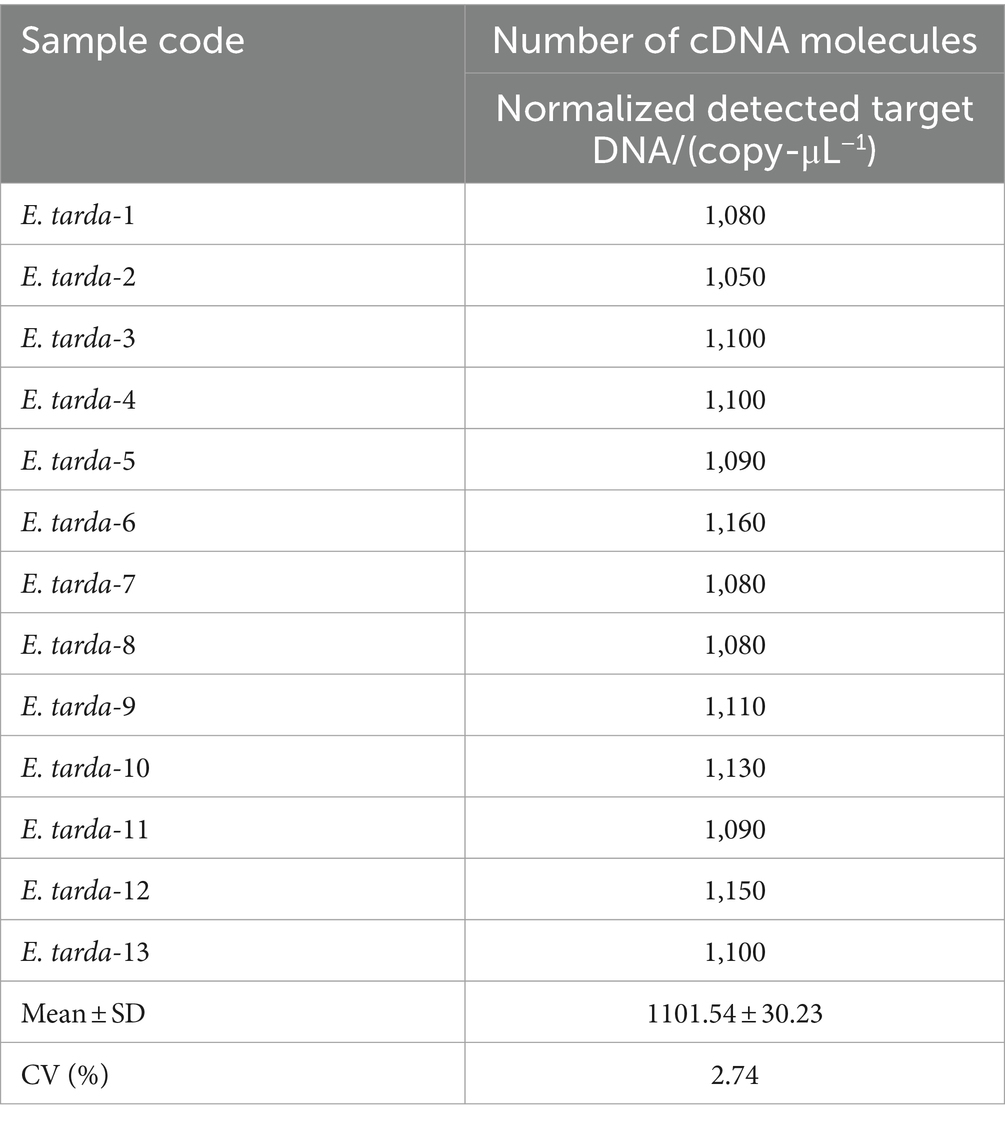
DNA was extracted from the liver of Bahaba Taipingensis (or its forage fish) using a DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions, the initial concentration of DNA obtained was 2.5*106 copies/L. The DNA of E. tarda was diluted 1,000-fold with double-distilled water (dd-H2O). This was followed by four-fold serial dilutions, with eight dilutions for the sensitivity analysis; the third dilution was selected for the repeatability analysis (Table 2).
2.5 Development of ddPCR-based detection method for Edwardsiella tarda
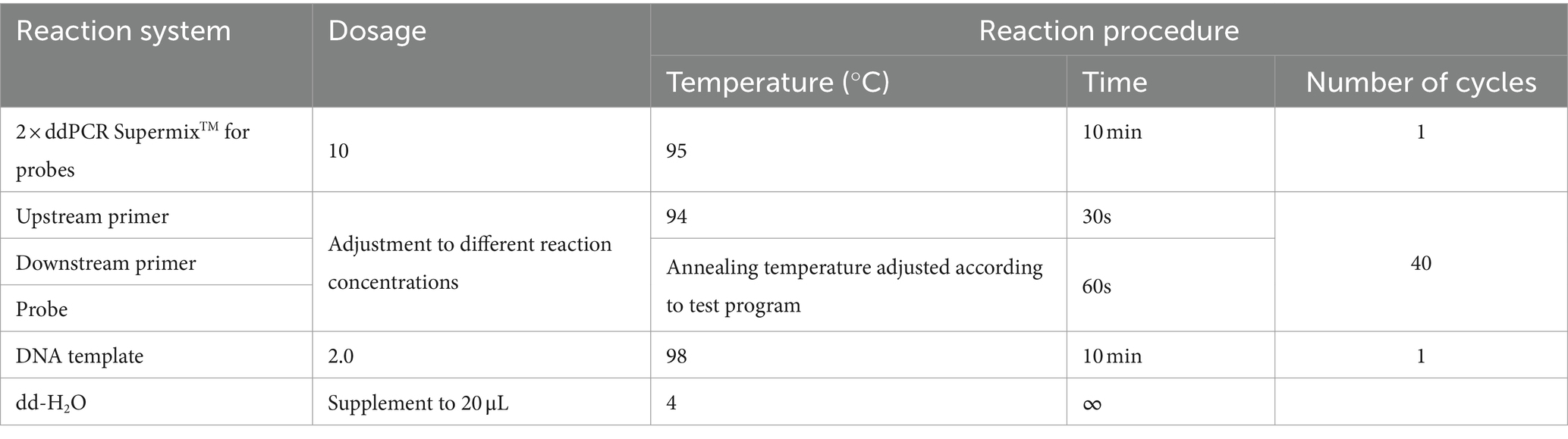
The E. tarda ddPCR reaction system consisted of 2 × ddPCR Supermix for probe, upstream and downstream primers, probe, dd-H2O, and template. The 20 μL ddPCR reaction-generated droplets were transferred to a 96-well plate, placed in a thermal cycler for amplification, and then placed in a droplet reader to analyze the results. The ddPCR reaction system and procedures are listed in Table 3.
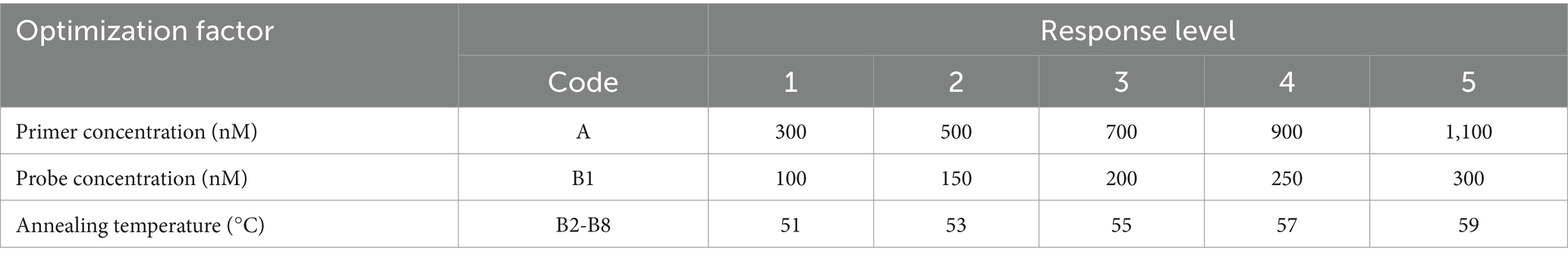
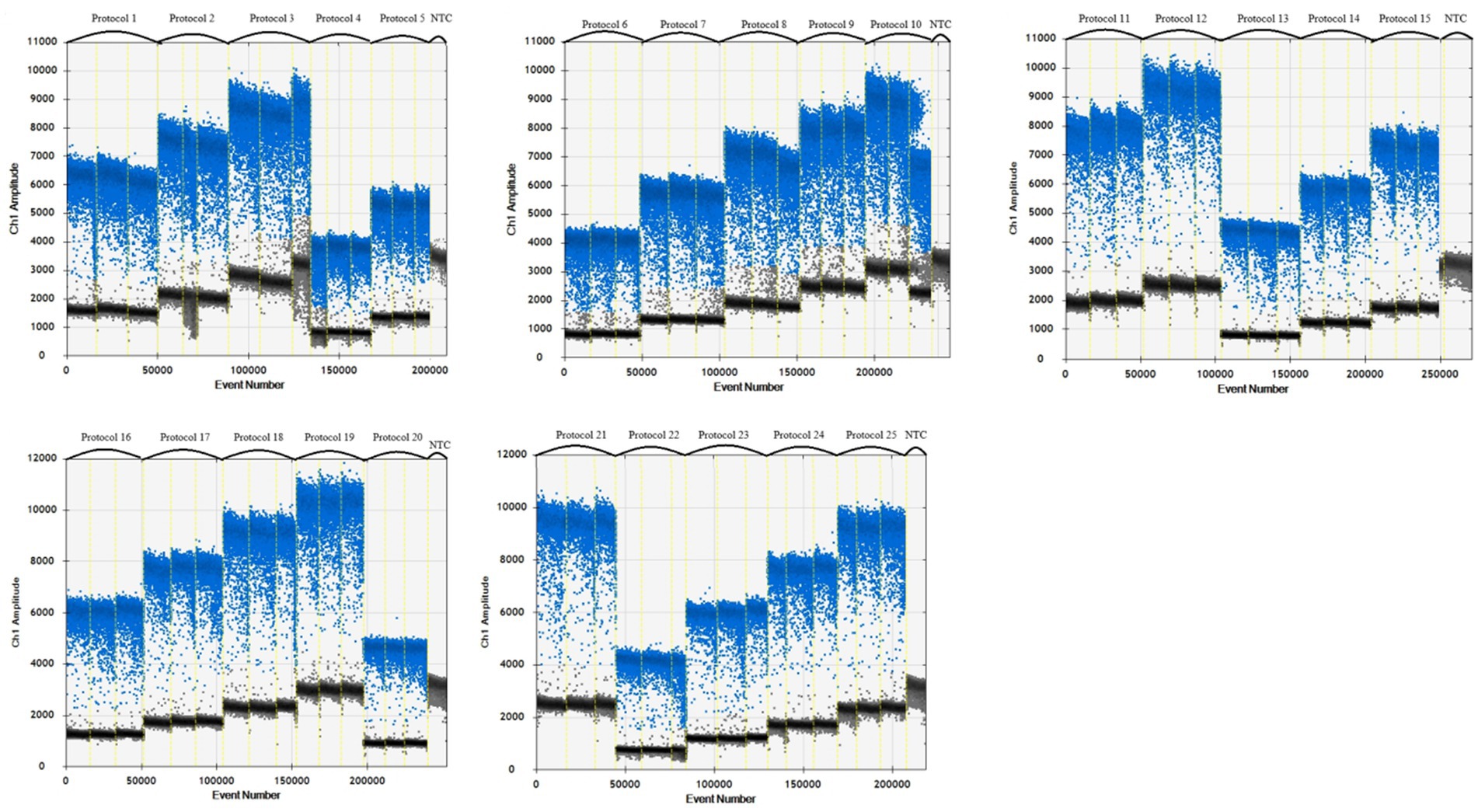
A 1,000-fold dilution gradient of E. tarda nucleic acids was used as a template to optimize the ddPCR conditions, including primers, probe concentration, and annealing temperature; each factor was set at five levels (Table 4). The SPSS software (version 16.0) was used to design an orthogonal array of optimization factors, arranged according to the annealing temperature, from small to large, for a total of 25 test protocols (Table 5), and each test protocol had three replicates. The main strategy for optimization was to maximize the difference in the fluorescence amplitude between the negative and positive droplet partitions and minimize the number of partitions with moderate fluorescence intensity.
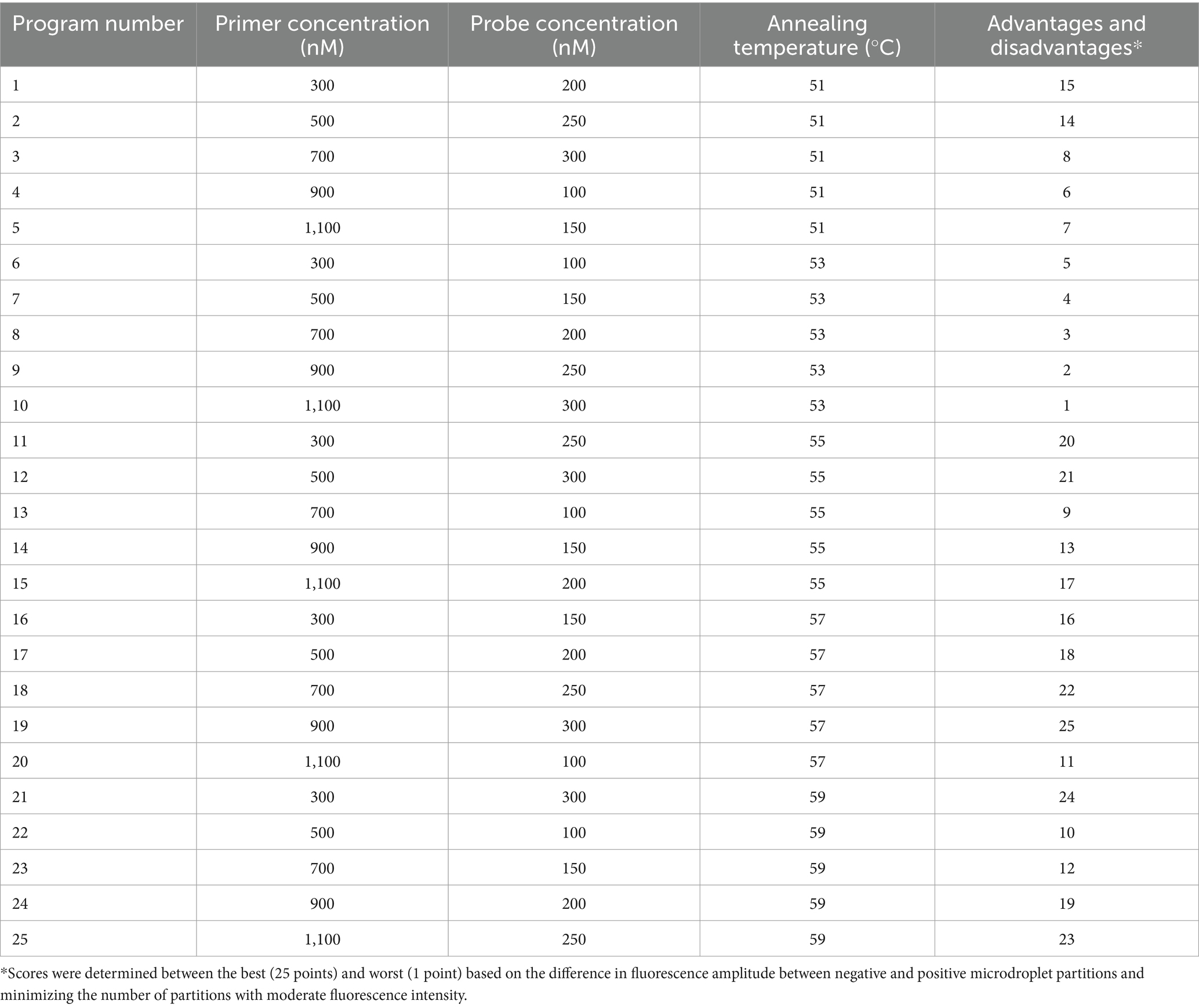
Table 5. Orthogonal array design and advantages and disadvantages of the optimization of reaction conditions for E. tarda ddPCR.
2.6 Establishment of standard curves
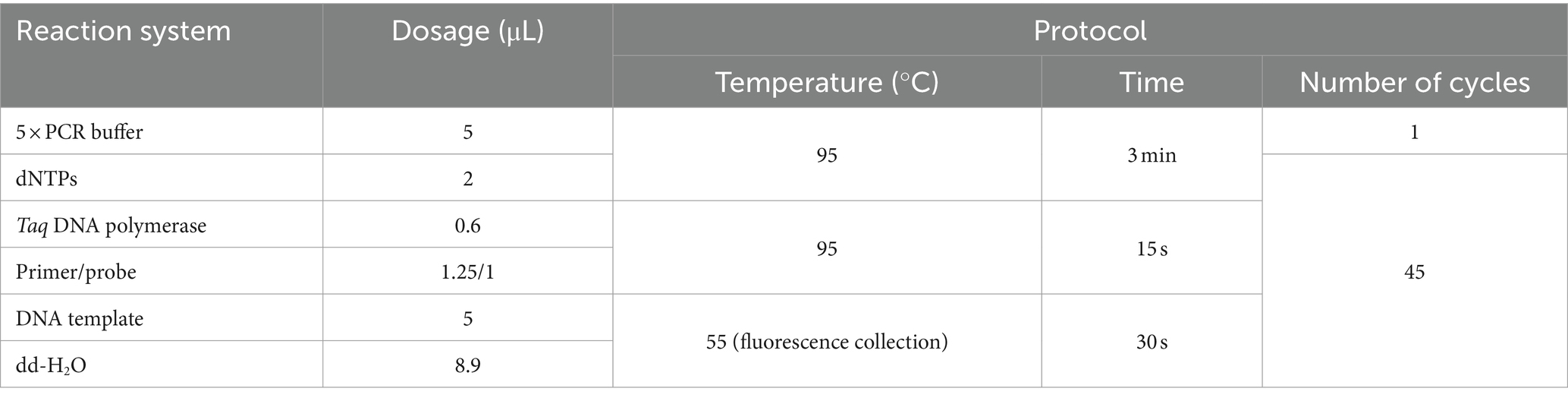
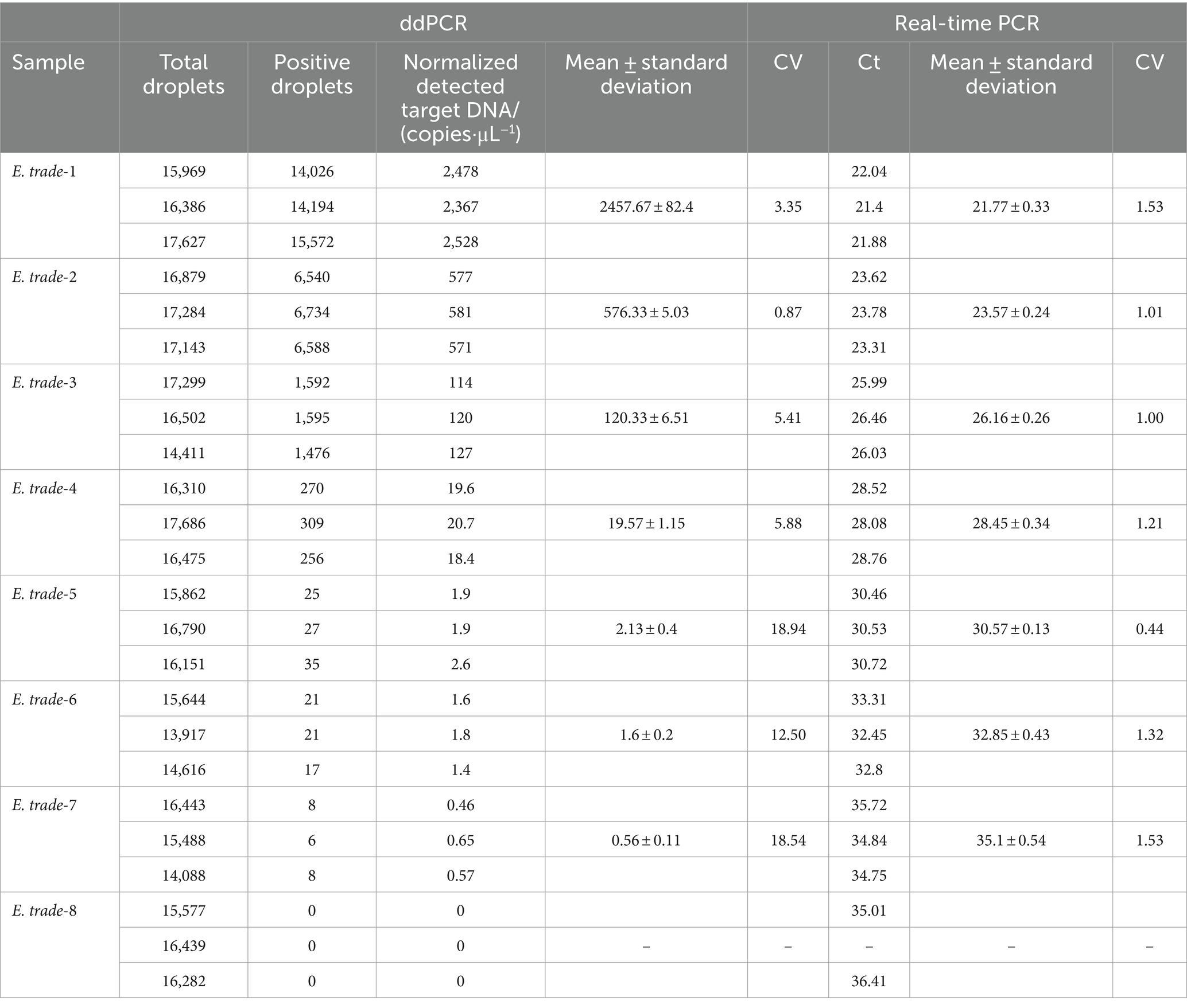
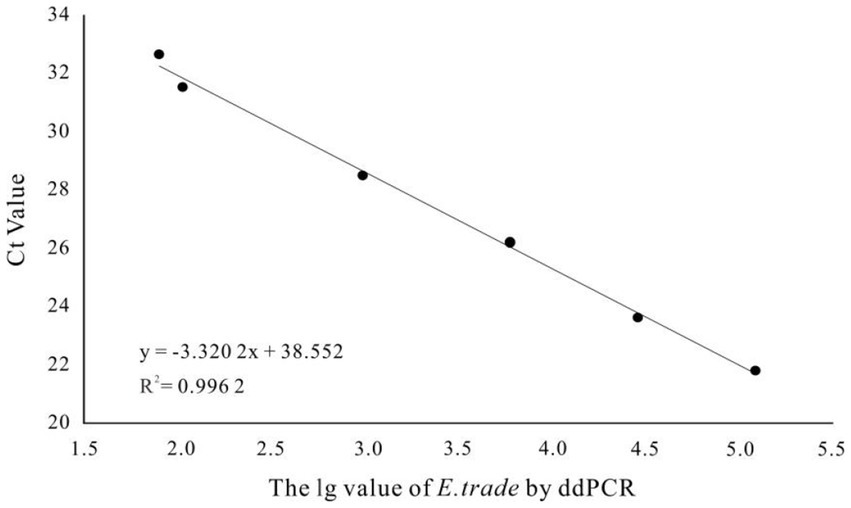
After 1,000-fold dilution of E. tarda strain nucleic acid with dd-H2O, a four-fold serial dilution was performed for eight gradients and three replicates of each gradient were included. The ddPCR and real-time PCR-based detection was performed simultaneously. Negative and blank controls were used. The logarithmic value of the number of E. tarda DNA molecules measured using ddPCR was used as the abscissa, and the cycle threshold (Ct) value of real-time PCR was used as the ordinate for constructing the standard curve. The primers, probe sequences, reaction systems, and real-time PCR reaction conditions are shown in Tables 1, 6.
2.7 Specificity test
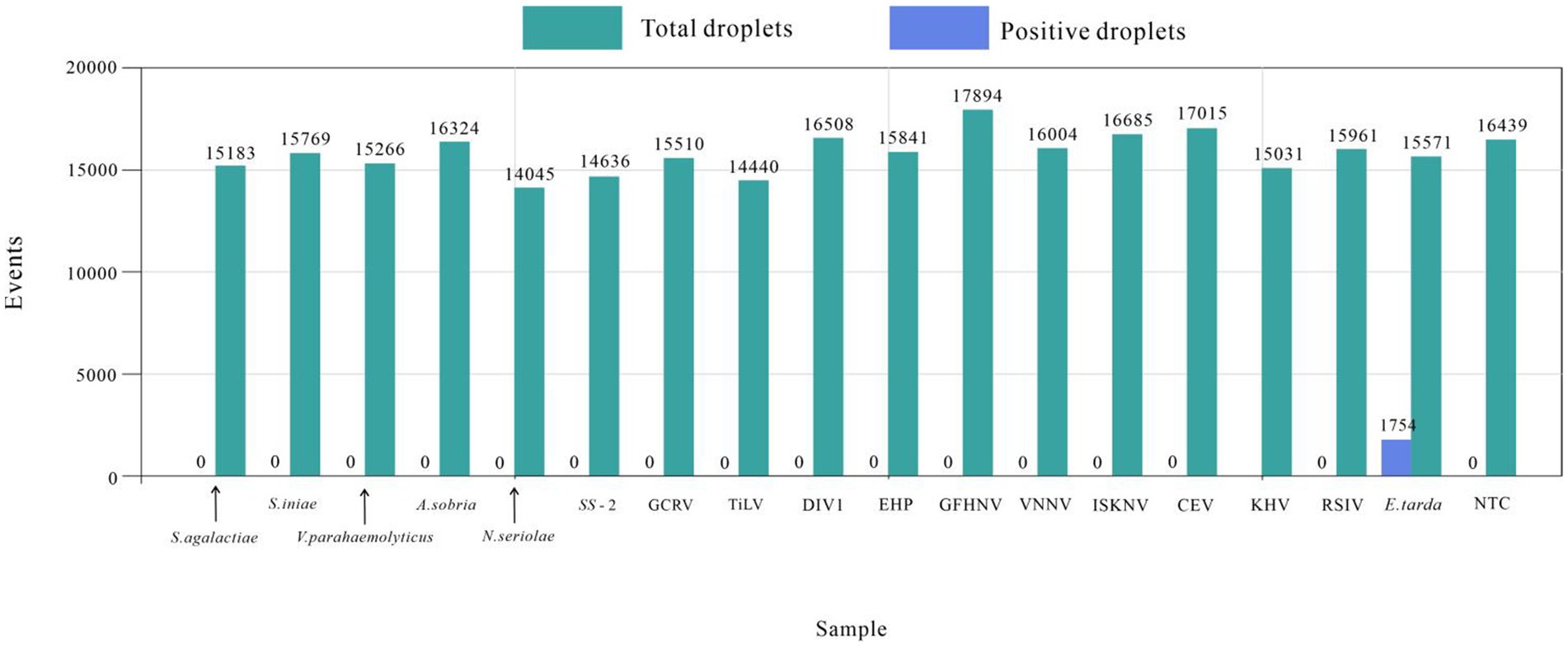
The DNA or cDNA of S. agalactiae, S. iniae, S. suis type 2, N. seriolae, V. parahaemolyticus, A. sobria, RSIV, DIV1, EHP, CEV, KHV, GFHNV, VNNV, ISKNV, TiLV, and GCRV were tested to evaluate the specificity of ddPCR for E. tarda.
2.8 Reproducibility test
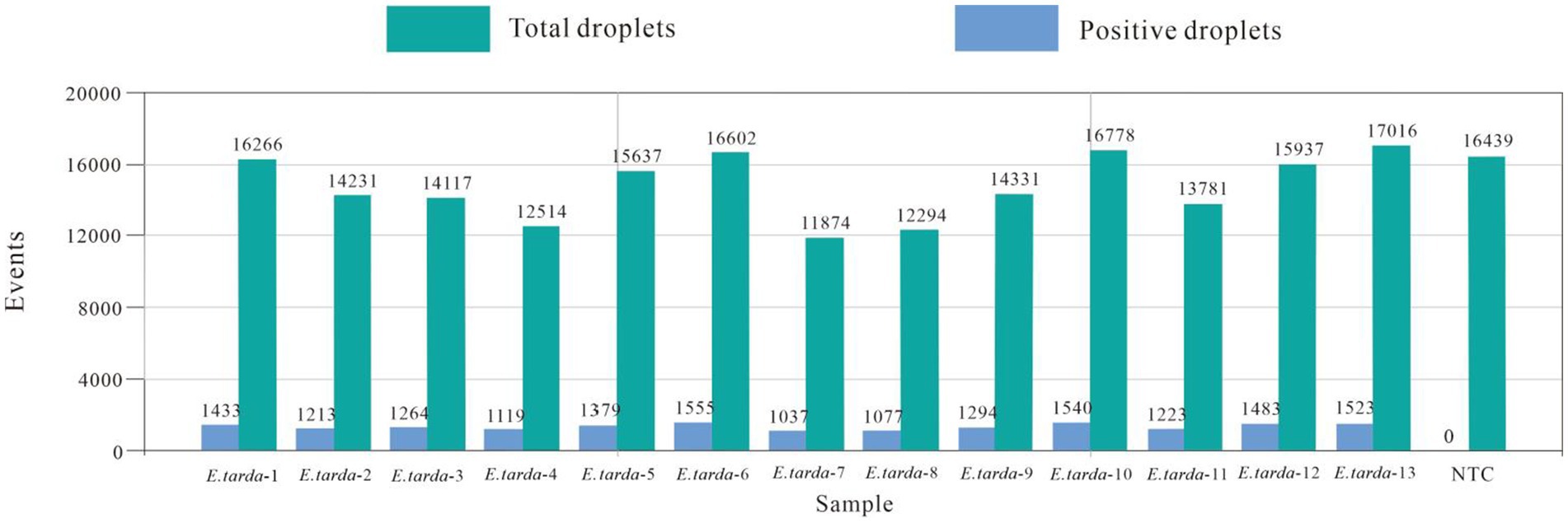
The nucleic acids of the E. tarda strain were diluted 1,000-fold with dd-H2O, followed by a four-fold serial dilution for a total of eight gradients. The third dilution of DNA was detected according to the (optimized item 1.2.4), and 13 replicates were used to calculate the coefficient of variation to evaluate the stability of the E. tarda ddPCR (Table 7).
2.9 Testing of clinical samples
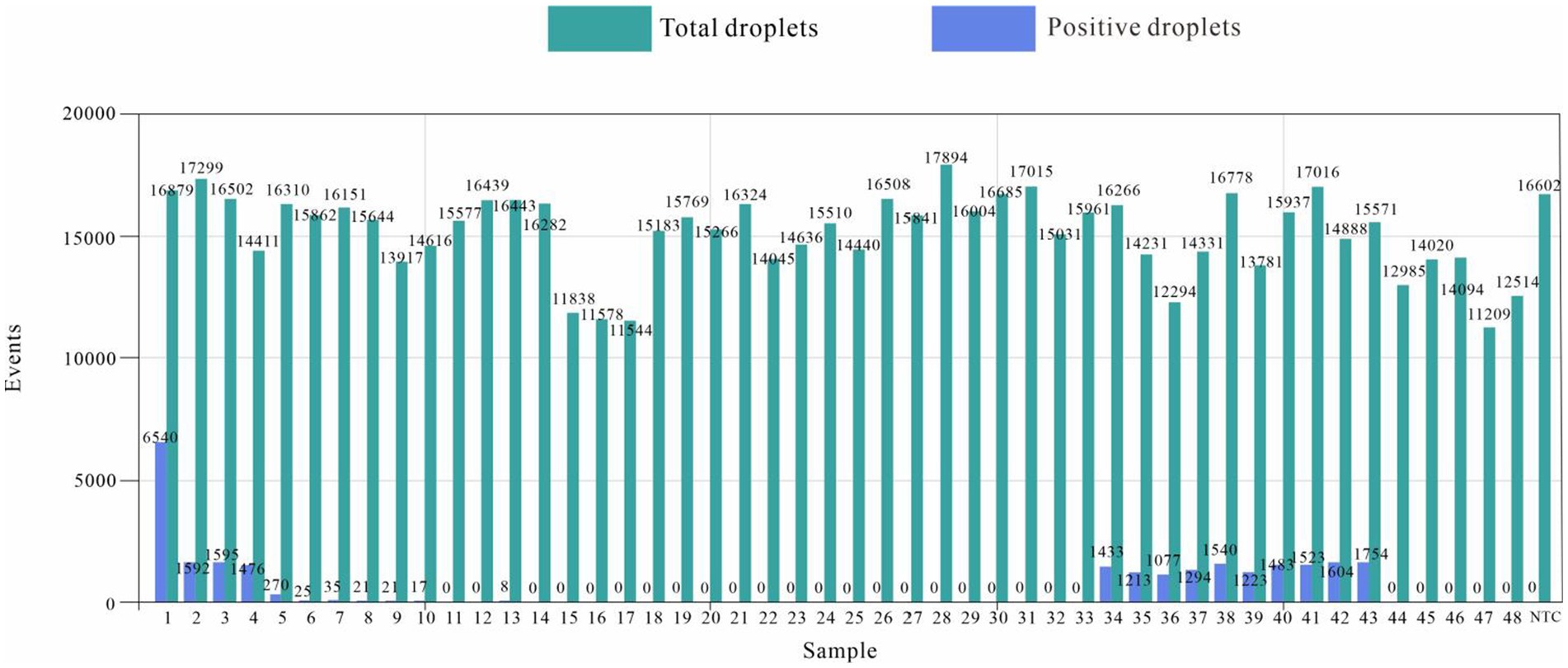
The nucleic acids of 48 samples of Bahaba taipingensis, live food fish, and ponds water were detected using the established E. tarda ddPCR method, and the results were compared with those obtained using real-time PCR and standard bacterial isolation and culture identification methods (Table 8).
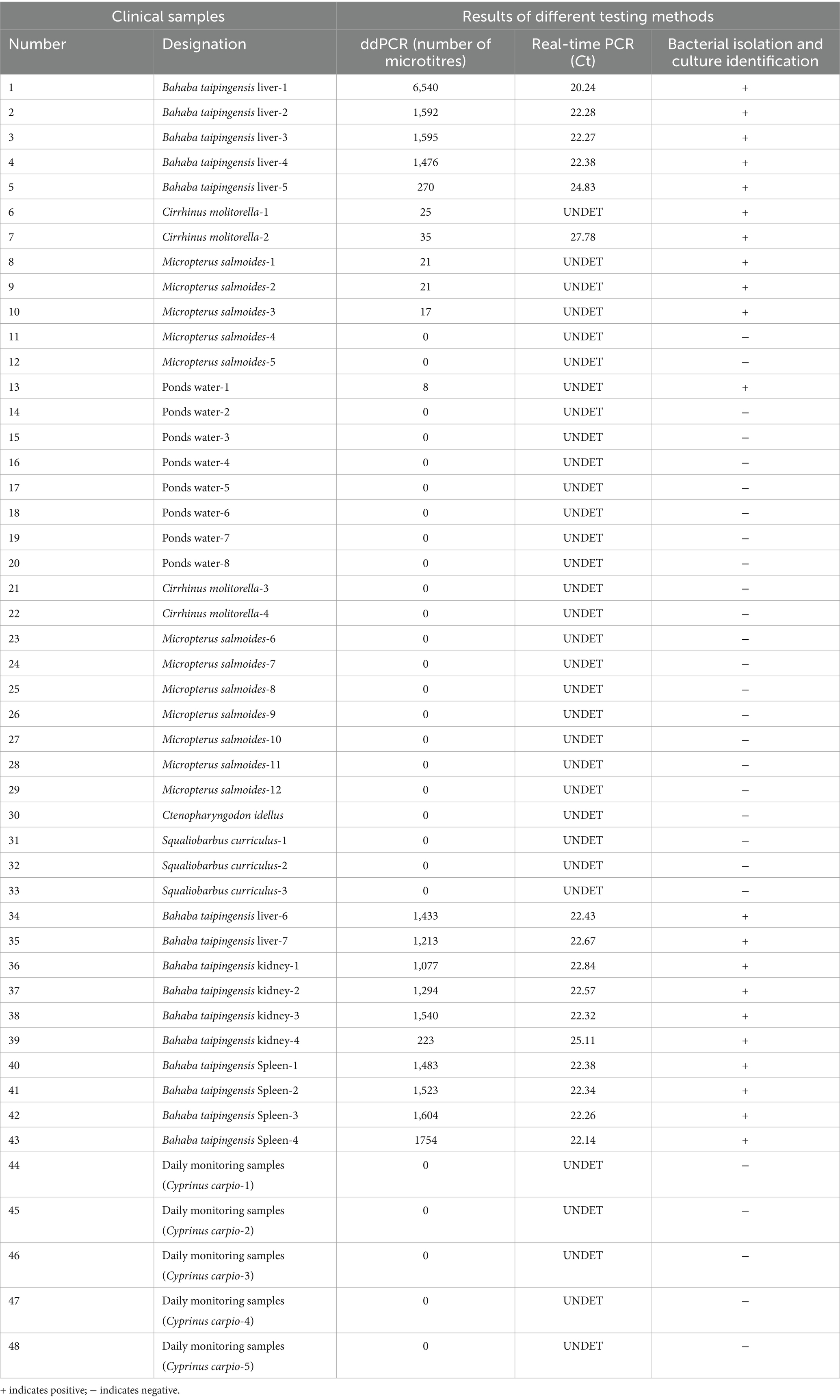
Table 8. Detection of E. tarda ddPCR, real-time PCR, and three methods of bacterial isolation and culture identification of clinical samples.
2.10 Statistical analyses
The ddPCR data for E. tarda were used for image processing and analysis using the Quantasoft software (26). A total number of droplets >10,000 was used as the criterion for ddPCR. No positive droplets were detected in the negative or blank controls, indicating that the system was not contaminated or specifically amplified. The key strategy for optimization was to maximize the difference in the fluorescence amplitude between the negative and positive droplet partitions and minimize the number of partitions with moderate fluorescence intensity. After PCR amplification of all the droplets, the droplets containing the target were amplified, and the droplets with higher fluorescence intensity were judged as positive droplets; the droplets that did not contain the target were not amplified and those with lower fluorescence intensity were considered to be negative droplets.
After the droplet reader interpretation, the droplet population would have a positive rate value, P. Upon combining this result with the Poisson distribution algorithm, the copy number of each positive droplet would be −ln (1 − p), and the concentration (copies μL−1) of the sample could be converted to a fixed and known volume of each droplet. Experimental data are presented as the mean ± standard deviation (X ± SD).
Number of DNA molecules = number of copies per microliter (copies·μL−1) × 20 μL ddPCR reaction system/DNA template dosage.
3 Results
3.1 Establishment of a ddPCR method for the detection of Edwardsiella tarda
When the primer concentration was 900 nM, probe concentration was 300 nM, and annealing temperature was 57°C, the resultant fluorescence intensity was the highest, the amplification reaction had the highest efficiency, and the boundary between the distribution of positive and negative droplets was most obvious. Test scheme 19 (primer concentration, probe concentration, and annealing temperature of 900 nM, 300 nM, 57°C, respectively) was therefore close as the optimal permutation of conditions (Table 5 and Figure 1).
3.2 Sensitivity tests and construction of standard curves
More than 13,000 results of the ddPCR tests on the DNA of the eight strains of E. tarda with different concentration gradients were obtained. The average lower limit of ddPCR detection was 0.56 copies·μL−1 (Figure 2). The log value of the molecular number of E. bradynia DNA measured using ddPCR was used as the abscissa, and the Ct value of real-time PCR was used as the ordinate to construct the standard curve. The gradients of ddPCR showed excellent correlation over the detection range, y = −3.3202x + 38.552, R2 = 0.9962. This standard curve was used to calculate the number of nucleic acid molecules in a clinical sample (Table 9 and Figure 3).
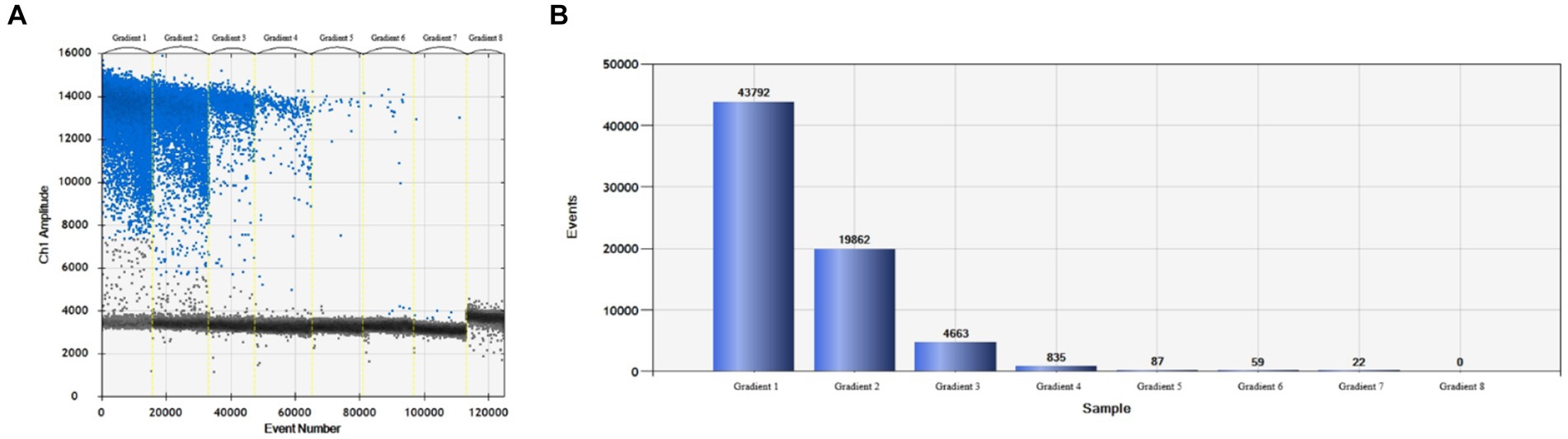
Figure 2. Edwardsiella tarda ddPCR sensitivity test. (A) Microdroplet 1D, (B) microdroplet histogram.
3.3 Specificity test
The established ddPCR method for E. tarda was used to test the DNA or cDNA of S. agalactiae, S. iniae, S. suis type 2, N. seriolae, V. parahaemolyticus, A. sobria, RSIV, DIV1, EHP, CEV, KHV, GFHNV, VNNV, ISKNV, TiLV, and GCRV. The number of droplets amplified by ddPCR was more than 14,000, and the results were valid. Except for the E. tarda-positive samples (1,754), none of the other 16 viral nucleic acid tests showed positive droplets. These findings indicated that the developed ddPCR method had good specificity for E. tarda (Figure 4).
3.4 Reproducibility test
The ddPCR detected nucleic acids at the B3 dilution with 13 replicates. The number of droplets amplified by ddPCR was more than 11,000. The coefficient of variation (CV) of the within-group assay was 2.74%, demonstrating that the established ddPCR for E. tarda detection had favorable reproducibility (Table 7 and Figure 5).
3.5 Test results of clinical samples
The number of droplets amplified via ddPCR was more than 11,000, and the results were positive; 21 samples were positive for E. tarda, and the remaining 27 samples were negative for E. tarda nucleic acid (Figure 6). However, sample 6, 8, 9, 10, and 13 were not confirmed to be positive for E. tarda nucleic acids by 16S rRNA or real-time PCR after 24 h of incubation in Trypticase Soy Broth liquid medium (Table 8). The ddPCR-based method for E. tarda detection was feasible and more sensitive than quantitative PCR and was suitable for detection in clinical samples as well as for calibrating standard E. tarda samples.
4 Discussion
Edwards is a collective term for aquatic animal diseases caused by E. tarda. The genus Edwardsiella includes three species- Edwardsiella ictaluri, Edwardsiella hoshinae, and E. tarda (14). There have been reports of more than 20 species of aquatic animals, reptiles, and other animals infected by E. tarda, which has caused huge economic losses to the aquaculture industry. More importantly, E. tarda is the only species in the genus Edwardsiella that infects humans (27), posing a serious threat to public health and safety. Antibiotics are predominantly used to prevent and control aquatic bacterial diseases caused due to the non-standard and unscientific use of drugs in fisheries. In recent years, aquatic product quality and safety incidents have occasionally occurred, and the resistance of pathogenic bacteria to drugs has gradually increased. This has created challenges for the prevention and control of diseases.
In the early stage of E. tarda infection in aquatic animals, the main manifestations are external, such as individual congestion or hemorrhage, anal redness, swelling, and protrusion (8, 9), which cannot be detected in time because of the particularity of the living environment. The onset of the disease is acute, and morbidity and mortality rates are high (28). Therefore, it is important to develop a highly sensitive and specific detection method to prevent and control the infections caused by E. tarda. Currently, the diagnosis of E. tarda includes characterization and observation of diseased aquatic animals, isolation and culture of bacteria, serological testing, and molecular biological testing. Characterization and observation cannot accurately confirm E. tarda infection in aquatic animals; therefore, further testing is required. Bacterial isolation and culture assays are labor-intensive, time-consuming, and do not allow timely treatment or control of infections. Molecular biological detection methods include PCR based on 16S rRNA, qPCR, and LAMP, based on the hemolysin gene. Ordinary PCR detection is cumbersome, with a risk of EB contamination. The qPCR-based detection cannot achieve absolute quantification.
The ddPCR is a next-generation technology based on real-time PCR that can be used for absolute quantification of nucleic acids of interest. In the droplet generator, the reaction system is separated into 10,000 ~ 20,000 small water-in-oil droplets that act as PCR bioreactors. After conventional PCR amplification, each reaction chamber contains zero to multiple copies of the nucleic acid of interest. The droplets are analyzed separately using a droplet reader similar to the flow cytometry for fluorescence. The Poisson distribution is used to determines the copy number. This technique has been applied to detect aquatic animal diseases (29). In the current study, specific primers and probe were designed, and the ddPCR system and amplification program were optimized, providing a new theoretical and practical means for the rapid, accurate, and sensitive detection of diseased by E. tarda.
In the current study, ddPCR detection technology was used for the first time to detect E. tarda. The gyrB gene, suitable for the differentiation and identification of strains, was selected as the target gene (30) and applied to the detection of clinical samples. The minimum detection limit of the ddPCR method for E. tarda was 0.56 copies·μL−1, which was higher than that of other molecular biological detection methods reported (22, 31). Sun et al. (22) used nested PCR to detect E. tarda at least 10 fg of E. tarda, but this approach was only suitable for quantitative detection using multiple dilutions when the detection object was known. Li et al. (32) combined recombinase polymerase amplification (RPA) technology with a lateral flow strip (LFS) to establish an RPA-LFS method for the detection of E. tarda; 1 × 101 CFU/g was the lowest detection amount, but this concentration of bacteria needed to be enriched and cultured for 4 h before being detected.
Chen and Lai (24) established a method for detecting E. tarda using LAMP; however, false positives were observed. The ddPCR method established in the current study for detecting E. tarda was highly specific. It showed no cross-reactivity with 16 microorganisms, including S. agalactiae, S. iniae, S. suis type 2, N. seriolae, V. parahaemolyticus, A. sobria, RSIV, DIV1, EHP, CEV, KHV, GFHNV, VNNV, ISKNV, TiLV, and GCRV. The DNA from E. tarda was used as the template. When the primer concentration was 900 nmol·L−1, probe concentration was 300 nmol·L−1 and annealing temperature was 57°C, the distribution boundary of positive and negative droplets in the ddPCR amplification reaction was the most obvious. The coefficient of E. tarda established in this study was 2.74%, which demonstrates good stability. The lowest detection limit was 0.56 copies·μL−1 in the range of 1–25,000 copies·μL−1.
5 Conclusion
The E. tarda ddPCR established in this study exhibits high specificity, high sensitivity and good reproducibility and can be used for clinical diagnosis in the early stages of E. tarda infection in aquatic animals. Application to testing in reptiles and humans can be investigated subsequently. This study provides technical support for the early detection of E. tarda infections.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Yunnan Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ML: Writing – original draft. XL: Writing – original draft, Data curation. YY: Writing – original draft, Data curation, Writing – review & editing. JY: Conceptualization, Writing – review & editing. ZM: Investigation, Writing – review & editing. HX: Writing – original draft. YZ: Writing – original draft. LZ: Writing – original draft, Methodology. XZ: Writing – original draft. JB: Writing – review & editing, Data curation, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by Yunnan Province High-level Scientific and Technological Talents and Innovative Teams Selection Special Project-Technological Innovation Talents Cultivation Objects (Project no. 202105AD160036); Dongguan 2021 Provincial Rural Revitalization Strategy Special Funds (“Big Special Project+Task List”) (Project no. 20211800400112); and Yunnan Provincial Local Universities Joint Special Project-Face-up (Project no. 202101BA070001-185).
Acknowledgments
We thank our students and technicians for their contributions to this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Goh, KW, Abdul Kari, Z, Wee, W, Zakaria, NNA, Rahman, MM, Kabir, MA, et al. Exploring the roles of phytobiotics in relieving the impacts of Edwardsiella tarda infection on fish: a mini-review. Front. Vet. Sci. (2023) 10:1149514. doi: 10.3389/fvets.2023.1149514
2. Park, SB, Aoki, T, and Jung, TS. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res. (2012) 43:67. doi: 10.1186/1297-9716-43-67
3. Yu, JE, Yoo, AY, Choi, KH, Cha, J, Kwak, I, and Kang, HY. Identification of antigenic Edwardsiella tarda surface proteins and their role in pathogenesis. Fish Shellfish Immunol. (2013) 34:673–82. doi: 10.1016/j.fsi.2012.11.019
4. Owens, DR, Nelson, SL, and Addison, JB. Isolation of Edwardsiella tarda from swine. Appl Microbiol. (1974) 27:703–5. doi: 10.1128/am.27.4.703-705.1974
5. Xie, H, Li, W, Zhang, R, Li, H, Zhang, Y, Guo, R, et al. Edwardsiella tarda causing septicemia in a wild crested Ibis (Nipponia nippon). J Wildl Dis. (2024) 60:232–5. doi: 10.7589/JWD-D-23-00103
6. Iveson, JB. Strontium chloride B and E.E. Enrichment broth media for the isolation of Edwardsiella, Salmonella and Arizona species from tiger snakes. J Hyg. (1971) 69:323–30. doi: 10.1017/s0022172400021562
7. Su, L, Guo, H, Guo, B, Yi, J, Yang, Z, Zhou, S, et al. Efficacy of bivalent vaccine against Aeromonas salmonicida and Edwardsiella tarda infections in turbot. Fish Shellfish Immunol. (2023) 139:108837. doi: 10.1016/j.fsi.2023.108837
8. Sun, XM, Yoshida, A, Toutani, F, Shimizu, T, Oda, T, and Osatomi, K. Cloning, DNA sequence, and expression of flagellins from high and low virulence strains of Edwardsiella tarda and their macrophage-stimulating activities. Microb Pathog. (2023) 176:105993. doi: 10.1016/j.micpath.2023.105993
9. White, FH, Simpson, CF, and Williams, LE Jr. Isolation of Edwardsiella tarda from aquatic animal species and surface waters in Florida. J Wildl Dis. (1973) 9:204–8. doi: 10.7589/0090-3558-9.3.204
10. Fuhrmann, H, and Savvidis, G. On the occurrence of economically important viral and bacterial diseases in German fish hatcheries and the fight against them. EMOP4. (1984) 14:248–9.
11. Pirollo, T, Perolo, A, Mantegari, S, Barbieri, I, Scali, F, Alborali, GL, et al. Mortality in farmed European eel (Anguilla anguilla) in Italy due to Streptococcus iniae. Acta Vet Scand. (2023) 65:5. doi: 10.1186/s13028-023-00669-y
12. Bragg, RR. First isolation of Edwardsiella tarda from fish in South Africa. Bull Eur Ass Fish Pathol. (1998) 8:87–8.
13. Xu, T, and Zhang, XH. Edwardsiella tarda: an intriguing problem in aquaculture. Aquaculture. (2014) 431:129–35. doi: 10.1016/j.aquaculture.2013.12.001
14. Janda, JM, Abbott, SL, Kroske-Bystrom, S, Cheung, WK, Powers, C, Kokka, RP, et al. Pathogenic properties of Edwardsiella species. J Clin Microbiol. (1991) 29:1997–2001. doi: 10.1128/jcm.29.9.1997-2001.1991
15. Wen, X, Cui, L, Morrisroe, S Jr, Emlet, D, Watkins, S, Hukriede, NA, et al. A zebrafish model of infection-associated acute kidney injury. Am J Physiol Renal Physiol. (2018) 315:F291–9. doi: 10.1152/ajprenal.00328.2017
16. Thune, RL, Stanley, LA, and Cooper, RK. Pathogenesis of gram-negative bacterial infections in warm water fish. Annu Rev Fish Dis. (1993) 3:37–68. doi: 10.1016/0959-8030(93)90028-A
17. Wu, JH, Li, DL, Tan, XH, Chen, XW, Liu, YL, Munang’andu, HM, et al. Functional proteomics analysis of Norfloxacin-resistant Edwardsiella tarda. J Proteome Res. (2023) 22:3489–98. doi: 10.1021/acs.jproteome.3c00365
18. Kumar, G, Rathore, G, Sengupta, U, Kapoor, D, and Lakra, WS. Production of monoclonal antibodies specific to major outer membrane protein of Edwardsiella tarda. Comp Immunol Microbiol Infect Dis. (2010) 33:133–44. doi: 10.1016/j.cimid.2008.08.008
19. Swain, P, Mukherjee, SC, Sahoo, PK, Das, BK, Pattnaik, P, Murjani, G, et al. Dot-enzyme-linked immunosorbent assay (dot-ELISA) for the diagnosis of Edwardsiella tarda infection in fish. Asian Fish Sci. (2001) 14:89–93. doi: 10.33997/j.afs.2001.14.1.010
20. Swain, P, and Nayak, SK. Comparative sensitivity of different serological tests for seromonitoring and surveillance of Edwardsiella tarda infection of Indian major carps. Fish Shellfish Immunol. (2003) 15:333–40. doi: 10.1016/s1050-4648(02)00178-x
21. Horenstein, S, Smolowitz, R, Uhlinger, K, and Roberts, S. Diagnosis of Edwardsiella tarda infection in oyster toadfish (Opsanus tau) held at the marine resources center. Biol Bull. (2004) 207:171. doi: 10.1086/BBLv207n2p171a
22. Sun, Z, Deng, J, Wu, H, Wang, Q, and Zhang, Y. Selection of stable reference genes for real-time quantitative PCR analysis in Edwardsiella tarda. J Microbiol Biotechnol. (2017) 27:112–21. doi: 10.4014/jmb.1605.05023
23. Savan, R, Igarashi, A, Matsuoka, S, and Sakai, M. Sensitive and rapid detection of edwardsiellosis in fish by a loop-mediated isothermal amplification method. Appl Environ Microbiol. (2004) 70:621–4. doi: 10.1128/AEM.70.1.621-624.2004
24. Chen, JD, and Lai, SY. PCR for direct detection of Edwardsiella tarda from infected fish and environmental water by application of the hemolysin gene. Zool Stud. (1998) 37:169–76. doi: 10.5555/19982218634
25. Pires, NMM, Dong, T, Yang, Z, and da Silva, LFBA. Recent methods and biosensors for foodborne pathogen detection in fish: progress and future prospects to sustainable aquaculture systems. Crit Rev Food Sci Nutr. (2021) 61:1852–76. doi: 10.1080/10408398.2020.1767032
26. Tzonev, S. Fundamentals of counting statistics in digital PCR: I just measured two target copies-what does it mean? Methods Mol Biol. (2018) 1768:25–43. doi: 10.1007/978-1-4939-7778-9_3
27. Phillips, AD, Trabulsi, LR, Dougan, G, and Frankel, G. Edwardsiella tarda induces plasma membrane ruffles on infection of HEp-2 cells. FEMS Microbiol Lett. (1998) 161:317–23. doi: 10.1111/j.1574-6968.1998.tb12963.x
28. Leung, KY, Siame, BA, Tenkink, BJ, Noort, RJ, and Mok, YK. Edwardsiella tarda – virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. (2012) 14:26–34. doi: 10.1016/j.micinf.2011.08.005
29. Shibata, D. PCR diagnostics of herpes-virus-group infections. Ann Med. (1992) 24:221–4. doi: 10.3109/07853899209147826
30. Mugimba, KK, Chengula, AA, Wamala, S, Mwega, ED, Kasanga, CJ, Byarugaba, DK, et al. Detection of tilapia lake virus (TiLV) infection by PCR in farmed and wild Nile tilapia (Oreochromis niloticus) from Lake Victoria. J Fish Dis. (2018) 41:1181–9. doi: 10.1111/jfd.12790
31. Xie, GS, Huang, J, Zhang, QL, Han, NN, Shi, CY, Wang, XH, et al. A real-time PCR targeted to the upstream regions of HlyB for specific detection of Edwardsiella tarda. Chin J Ocean Limnol. (2012) 30:731–7. doi: 10.1007/s00343-012-1244-2
Keywords: Edwardsiella tarda, droplet digital PCR, linear relationship, specificity, clinical application
Citation: Li M, Li X, Ye Y, Yin J, Mo Z, Xie H, Zhu Y, Zhong L, Zhang X and Bi J (2024) Establishment and clinical application of a droplet digital PCR method for the detection of Edwardsiella tarda. Front. Vet. Sci. 11:1439743. doi: 10.3389/fvets.2024.1439743
Edited by:
Zhe Zhao, Hohai University, ChinaCopyright © 2024 Li, Li, Ye, Yin, Mo, Xie, Zhu, Zhong, Zhang and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junlong Bi, anVubG9uZ2JpQGZveG1haWwuY29t; Xianpeng Zhang, eGlhbi5wZW5nQDE2My5jb20=
†These authors have contributed equally to this work
 Min Li1,2,3†
Min Li1,2,3† Junlong Bi
Junlong Bi