
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 18 July 2024
Sec. Veterinary Clinical, Anatomical, and Comparative Pathology
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1421153
This article is part of the Research TopicEmerging Diseases and Diagnostics in Poultry ProductionView all 7 articles
 Amany Adel1*
Amany Adel1* Ali Zanaty1
Ali Zanaty1 Zienab Mosaad1
Zienab Mosaad1 Karim Selim1
Karim Selim1 Naglaa M. Hagag1
Naglaa M. Hagag1 Mona Badr2
Mona Badr2 Hany Ellakany2
Hany Ellakany2 Momtaz Shahien1
Momtaz Shahien1 Ahmed Samy1,3*
Ahmed Samy1,3*The very virulent infectious bursal disease virus (vvIBDV) induces an acute, highly contagious and immunosuppressive disease in younger chicken causing massive economic losses globally. A major challenge in the field’s clinical diagnosis is distinguishing gross lesions caused by vvIBDV from those induced by classic IBDV (cIBDV), commonly used as live attenuated vaccines. This study introduces a one-step multiplex real-time PCR assay designed to distinguish between vvIBDV and non-vvIBDV viruses. Via simultaneously targeting the VP2 sequence for vvIBDV detection and the VP1 sequence for non-vvIBDV identification, including classic, American variant and the recently emerged novel variant IBDV (nvarIBDV), the assay’s specificity was validated against common avian viral diseases and nonspecific IBDV strains without any observed cross-reactions. It effectively differentiated between vvIBDV and non-vvIBDV field samples, including nvarIBDV, as confirmed by genotyping based on VP2 sequencing. The assay demonstrated a limit of detection ranging from 1.9×1010 to 103 DNA copies for vvIBDV-VP2, 9.2×1010 to 103 DNA copies for classic strains, and 1.2×1011 to 104 DNA copies for nvarIBDV in VP1 detection of non-vvIBDV. In conclusion, this study presents a specific, sensitive, and straight forward multiplex real-time PCR assay.
Infectious bursal disease (IBD) is an acute, highly contagious, immunosuppressive virus affecting young chickens. This disease is characterized by the extensive destruction of immature B lymphocytes and severe damage to the cloacal bursa. Typically, high mortality rates are observed in chickens at 3 weeks of age and older for certain strains. The immune suppression caused by IBD leads to diminished responsiveness to commonly used vaccines and increases the impact of other diseases (1). It is caused by the IBD virus (IBDV), a non-enveloped, double-stranded RNA (dsRNA) virus that belongs to the genus Avibirnavirus under the family Birnaviridae and is composed of two segments (A and B) (2). Segment A (3.2 kb) is formed from two overlapped open reading frames: the first encodes VP5, while the second encodes the VP2, VP3, and VP4 polyprotein, among which VP2 and VP3 are structural proteins. VP2 has a hypervariable region (HVR) that is responsible for virus antigenicity and virulence. Segment B (2.8 Kb) encodes a viral RNA polymerase (VP1) (3, 4).
IBDV is classified into two serotypes based on antigenicity, with only serotype 1 being pathogenic in chickens (5). Serotype 1 viruses are further categorized based on pathogenicity into classical IBDV (cIBDV), variant IBDV (varIBDV), very virulent IBDV (vvIBDV), attenuated IBDV, and novel variant IBDV (nVarIBDV) (6). The virus’s bi-segmented double-stranded RNA genome and the presence of viral RNA-dependent RNA polymerase (RdRP) contribute to continuous evolution through mutations, reassortment, and recombination (7). This evolution challenges classification based on pathogenicity and hinders accurate and rapid diagnosis. Therefore, improved genotyping schemes, mainly based on VP2-HVR region sequences along with partial sequences of VP1, have been proposed to describe the genetic and pathogenic diversity of IBDV (7–10). While accurate and rapid diagnosis remains challenging, especially in differentiating between vvIBDV and classic virulent strains, including vaccinal strains, due to their similarity under field conditions. Both genotypes are associated with characteristic postmortem lesions, such as muscular hemorrhage, inflammatory exudation, hemorrhage, and yellow staining of the bursae, while mortality rates vary based on the pre-existing immunity and strains (4, 11) making differentiation challenging, especially considering the widespread use of live classical strains-based vaccines for clinical protection against vvIBDV (12).
Rapid and accurate differentiation between vvIBDV and non-vvIBDV strains is crucial to understanding the epidemiology of viral spread and the efficiency of used control strategies. Here, we propose using Real-time RT-PCR, targeting the phylogenetically relevant genes of the IBDV genome, VP1 and VP2 genes, for molecular diagnosis of IBDV infection. These approaches will enable faster, more sensitive, and specific detection and discrimination of IBDV strains compared to the currently used approach relying on RT-PCR and gene sequencing.
The primers and probes were designed in silico using the nucleotide sequences of IBDV’s VP1 and VP2 genes. In brief, several aligned nucleotide sequences for each of the VP1 and VP2 genes from the classic, variation, variant, and vvIBDV strains of Egyptian viruses under research, as well as additional reference strains retrieved from the NCBI database, were employed. The sequences were aligned with Bioedit software (13) and examined to identify the most specific locations for vvIBDV on the VP2 gene and classic strains on the VP1 gene (Figure 1). The specificity, secondary structures, and other physical properties of the proposed primers and probes were analyzed in silico using BLAST on the NCBI and OligoCalc servers.

Figure 1. Alignment of forward, reverse, and probe sequences of the real-time RT-PCR. Various representative vvIBDV, classical (cIBDV), and variant (nVarIBDV) strains’ VP2 sequences (A) and VP1 sequences (B). The primers and probes target sites are indicated using the European vvIBDV strain 89163 for VP2 and the classical strain CT for VP1.
Bursa samples from forty-six vaccinated broiler farms that were submitted to our laboratory to confirm clinical diagnoses have been used to evaluate the efficiency of the assay. Sample preparation, following the method outlined in (14), Briefly, homogenizing the bursae with phosphate-buffered saline supplemented with Kanamycin (700 μg/mL). The homogenate was then subjected to three cycles of freezing and thawing, followed by centrifugation at 8,000 rpm for five minutes to remove cell debris. The resulting supernatant was collected, filtered through a 0.45 μm filter (Sartorius, Germany), and stored in a −80°C freezer for the next step. Also, Leukert, Winterfeild, JOVAC (D78) and Bursin vaccinal strains were used in validation of the assay.
Viral RNA extraction was conducted using the EasyPure® Viral DNA/RNA Kit (TransGen Biotechnology, Inc., Beijing, China), following the kit instructions. Subsequently, cDNA synthesis and qPCR were performed using the TransScript® Probe One-Step qRT-PCR SuperMix (TransGen Biotechnology, Inc., Beijing, China), following the manufacturer’s instructions. The one-step real-time PCR was performed and analyzed in a Stratagene Mx3000P instrument (Agilent Technologies Inc., Santa Clara, CA, USA) using primers and probes targeting VP2-HVR and VP1 (Table 1). The thermal profile was as follows: Reverse transcription at 45°C for 15 min, initial denaturation at 94°C for 5 min, followed by 40 cycles of 94°C for 40 s and 52°C for 60 s.
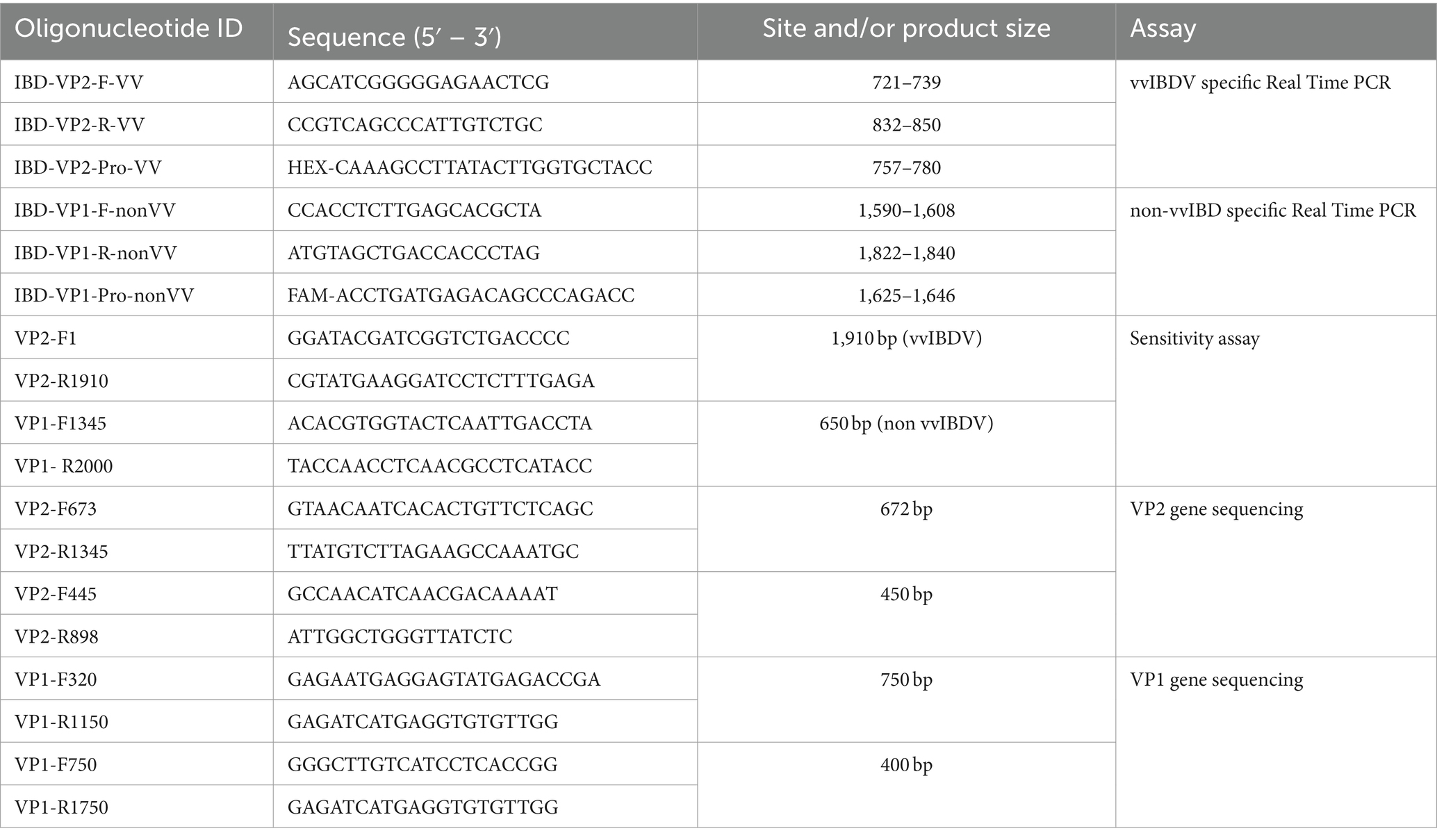
Table 1. primers and probes of the multiplex real time RT-PCR assay and the primers of nucleotide sequencing assay.
The specificity of the designed primers and probes for the multiplex assay was tested using RNA from known IBDV strains representing different genotypes, as well as other common viral pathogens in the poultry population such as LPAI-H9N2, HPAI-H5N8, IBV, and NDV, employing one-step real-time PCR. To assess the sensitivity of the assay, specific purified PCR products were amplified using specific primers (Table 1) and the Easyscript® One-Step RT-PCR kit (TransGen Biotech) following the manufacturer’s recommendations. The copy numbers in the purified products were calculated as described in,1 followed by tenfold serial dilution of one or multiple samples, which were then subjected to single or multiplex real-time PCR assays. The purified amplified PCR products of VP2 from the sample IBD-2668-VP2-2021-vvIBDV and VP1 from IBD-632-VP1-2021-classic and IBD-2530-VP1-2023-new variant were utilized to determine the sensitivity limit of the assay.
The developed multiplex one-step real-time PCR was employed to test 46 clinical samples, and results were recorded. To validate the results, representative samples of each genotype underwent partial gene sequencing of the VP1 and VP2-HVR genes. For the amplification, specific primers (Table 1) and the Easyscript® One-Step RT-PCR kit (TransGen Biotech) were used following the manufacturer’s recommendations. Subsequently, the PCR products at the expected size were purified using the QIAquick Gel Extraction Kit (QIAGEN, USA) in accordance with the manufacturer’s instructions. The BigDye Terminator kit (Applied Biosystems, CA, USA) was utilized for Sanger sequencing of the purified amplified products of both VP1 and VP2 genes. The prepared sequence reactions were then read by ABI (Applied Biosystems 3500xl Genetic Analyzers, USA).
Concordance between the results of both systems and the results of the nucleotide sequences has been determined by Cohen’s Kappa agreement measure at a p-value <0.05. Additionally, the coefficient of determination (R2) in linear regression was calculated for the standard curves of the real-time qRT-PCR systems. Statistical analysis has been done by SPSS software (IBM).
Sequence analysis was conducted on the VP2-HVR sequences of several IBDV strains to identify nucleotide polymorphisms that could distinguish between vvIBDV and non-vvIBDV sequences. Sequences spanning from nucleotide 721 to 850 has some unique nucleotide sequences to vvIBDV strains and can be employed for differentiation (Figure 1A). The selected probe, along with the forward and reverse primer sequences, exhibited 3 to 6, 3 to 5, and 2 to 4 single nucleotide polymorphisms (SNPs), respectively, in selected representative non-vvIBDV strains when compared to vvIBDV, including the Egyptian antigenically atypical vvIBDV (Figure 1A). On the other hand, sequences spanning from nucleotide 1,476 to 1,726 in segment B can be used to distinguish between classic and variant strains compared to vvIBDV, with 2 to 6 single nucleotide polymorphisms (SNPs) in vvIBDV compared to classical and variant strains (Figure 1B). It’s worth mentioning that when aligning the designed primers and probes used to distinguish non-VVIBDV with nVarIBDV sequences, there are 1–2 SNPs in the primer and probe sequences (Figure 1B). Since the system was primarily established to differentiate between vvIBDV and non-vvIBDV, which can manifest with similar clinical symptoms and lesions, nVarIBDV, known to induce subclinical infection, can be easily distinguished from vvIBDV. Technically, the developed multiplex qrt-PCR assay showed no signal with viruses and live attenuated vaccines commonly found on poultry farms, including HPAI-H5, LPAI-H9N2, IBV, and NDV, and the negative or no template controls in the assay were also not amplified. Additionally, the uniplex assay that was specific for vvIBDV showed negative results for caIBDV and nVarIBDV, and vice versa (Figure 2). Since the antigenically atypical vvIBDV has been only recognized by HEX fluorescence channel, while the classic and the nvarIBDV have been recognized only by FAM fluorescence channel, indicating no nonspecific binding with non-matching primer and probe combinations (Figure 2).
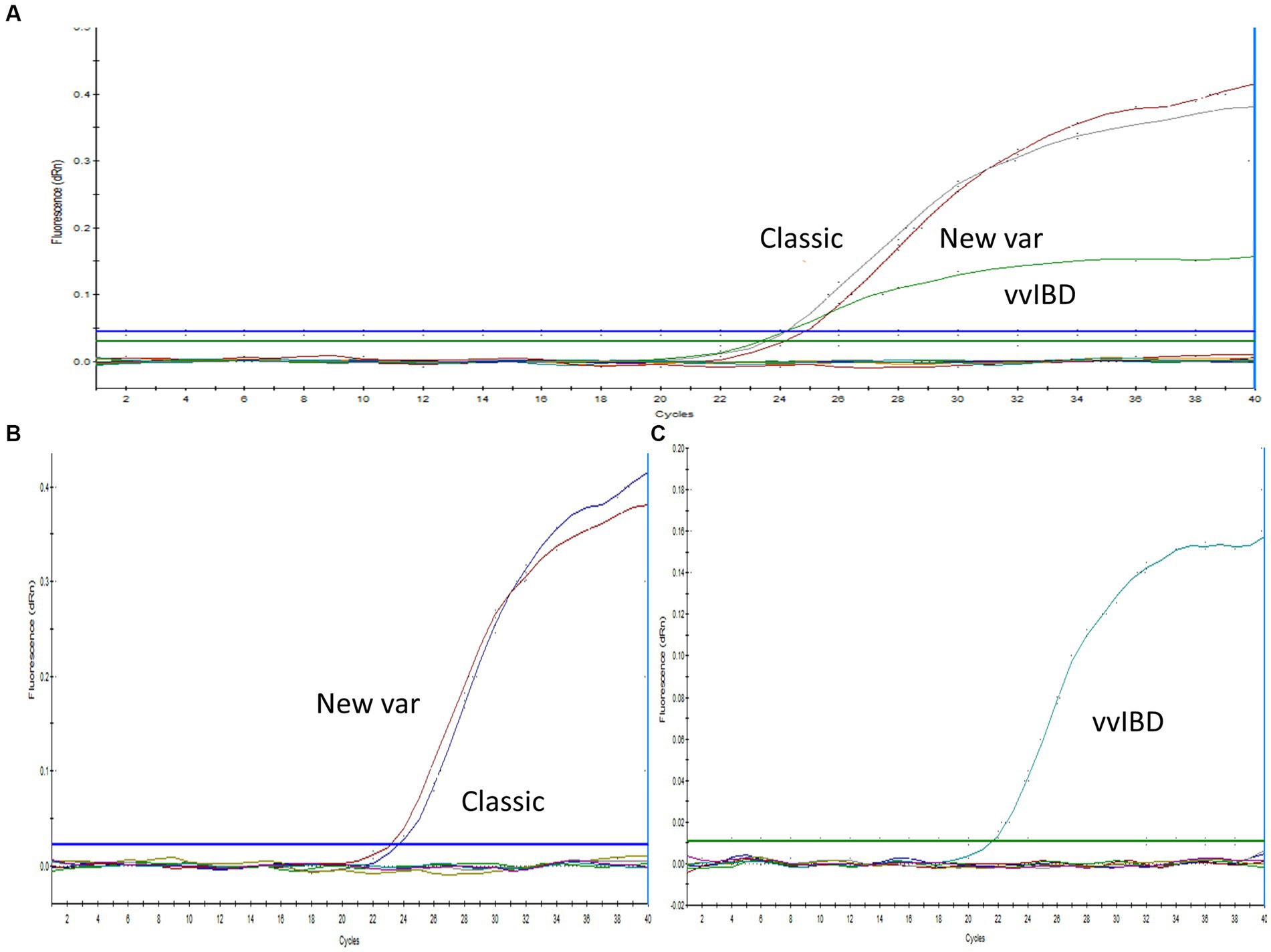
Figure 2. Specificity of the one-step real-time TaqMan RT-PCR assay. (A) Multiplex PCR analysis with positive samples representing vvIBDV, classical, and variant strains (as indicated in the figure), in addition to IBV, NDV, H9N2, and a no-template control. IBDV-negative field samples exhibited no amplification, with vvIBDV specifically recognized by the HEX fluorescence channel, while classical and variant IBDV were identified solely by the FAM fluorescence. (B) Uniplex PCR with positive samples representing classical and variant IBDV strains (as indicated in the figure), along with IBV, NDV, H9N2, and a no-template control. IBDV-negative field samples showed no amplification, with classical and variant IBDV uniquely identified by FAM fluorescence. (C) Uniplex PCR with positive samples representing vvIBDV strains, along with IBV, NDV, H9N2, and a no-template control. IBDV-negative field samples did not exhibit amplification, and vvIBDV was specifically recognized by HEX fluorescence.
Tenfold serial dilutions of known concentrations of genomic material for each genotype were tested using the multiplex one-step real time-PCR to establish a correlation between the cycle threshold (CT) and the quantity of IBDV genetic material. The results revealed linear correlation between CT and IBDV genetic quantity with an average R2 of 0.99 for vvIBDV and caIBDV strains and 0.979 for nVarIBDV (Figure 3). The detection limit for vvIBDV was in the range of 1.9×1010 to 1.9×103 DNA copies/μL. Regarding the non-vvIBDV, the limit of detection ranged from 9.2×1010 to 9.2×103 DNA copies/μL for the classic strain, while for the nVarIBDV, the range was 1.2×1011 to 1.2×104 DNA copies/μL (Figure 3). Each dilution of each strain was tested in triplicate, and then the averages of these reads have been calculated and used as points for evaluation of the limit of detection for each strain, as illustrated in (Figure 3).
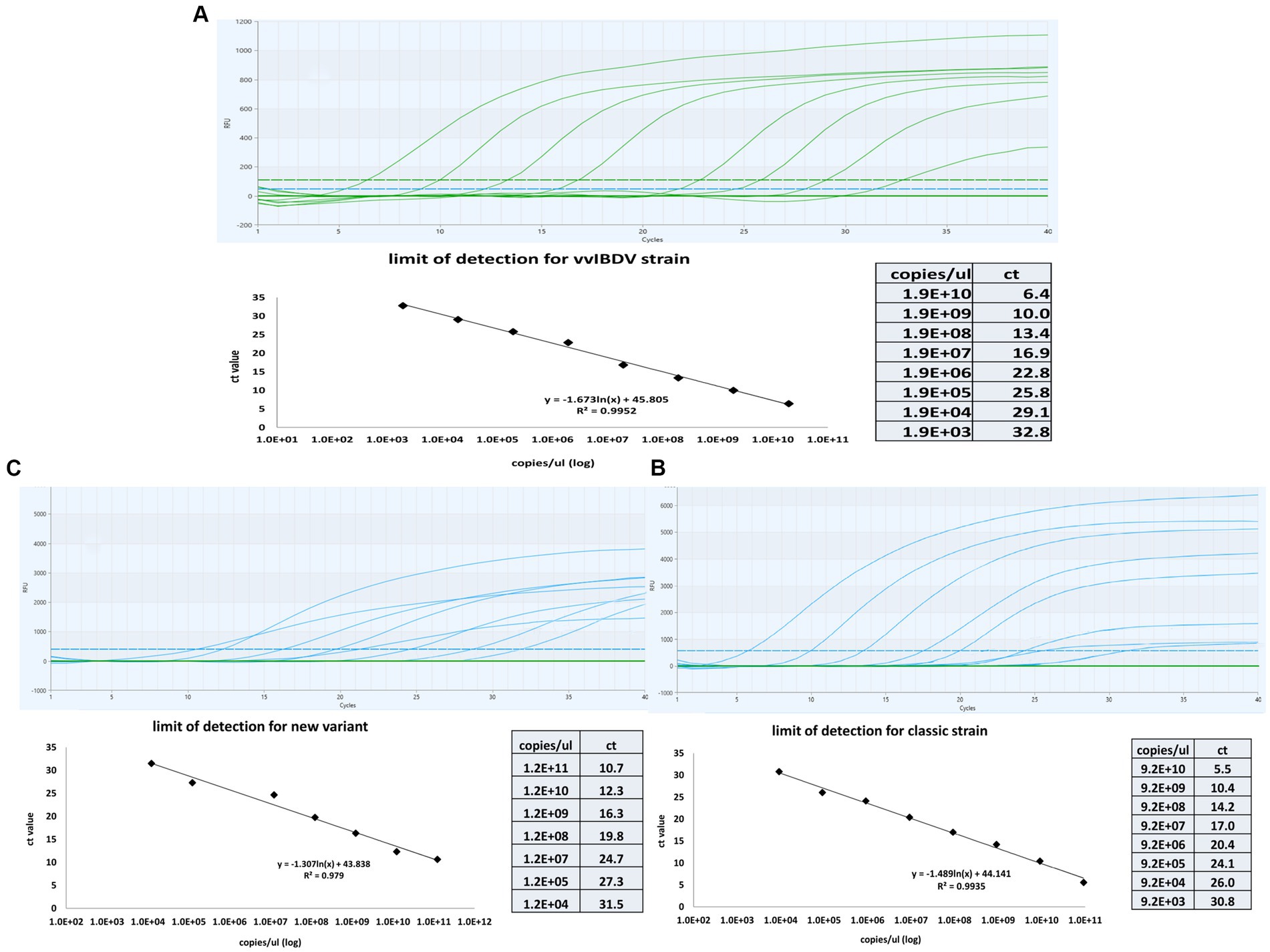
Figure 3. Sensitivity and standard curves for the multiplex one-step real-time-TaqMan RT-PCR assay. The curves were constructed by employinga 10-fold serial dilution of known concentrations of genetic material from samples representing very virulent (A), classical (B), and new variant IBDV (C). The figure demonstrates the linear correlation between Cycle Threshold (CT) and the quantity of genetic material for different IBDV genotypes. The coefficient of determination (R2) and efficiency of each linear regression curve are indicated.
Forty-six bursa samples, along with vaccinal strains commonly used for vaccination in Egyptian poultry farms, including Lukert, Winterfield, Bursin, and D78 strains, have been examined by a multiplex one-step real-time TaqMan RT-PCR assay developed in the present manuscript (Table 2). Samples were collected from farms with ages ranging from 20 to 34 days, exhibiting varied clinical lesions—from severe hemorrhage in the bursa, thigh, and breast muscles accompanied by typical IBDV clinical signs to lesions limited to atrophy in the bursa with no apparent clinical symptoms. Based on the multiplex one-step real-time TaqMan RT-PCR results, 8 samples were positive for vvIBDV, 37 were positive for non-vvIBDV, and 5 samples were positive for both vvIBDV and non-vvIBDV (Table 2).
The results of the phylogenetic analysis based on VP2-HVR revealed that, out of the 17 successfully sequenced samples, 3 clustered within the A3 genogroup representing vvIBDV, 4 clustered within the A1 genogroup representing classic virulent IBDV, and 9 clustered within the A2 genogroup representing variant IBDV and subgroup d, which represents the newly emerged variant strains (nvarIBDV) (Figure 4).
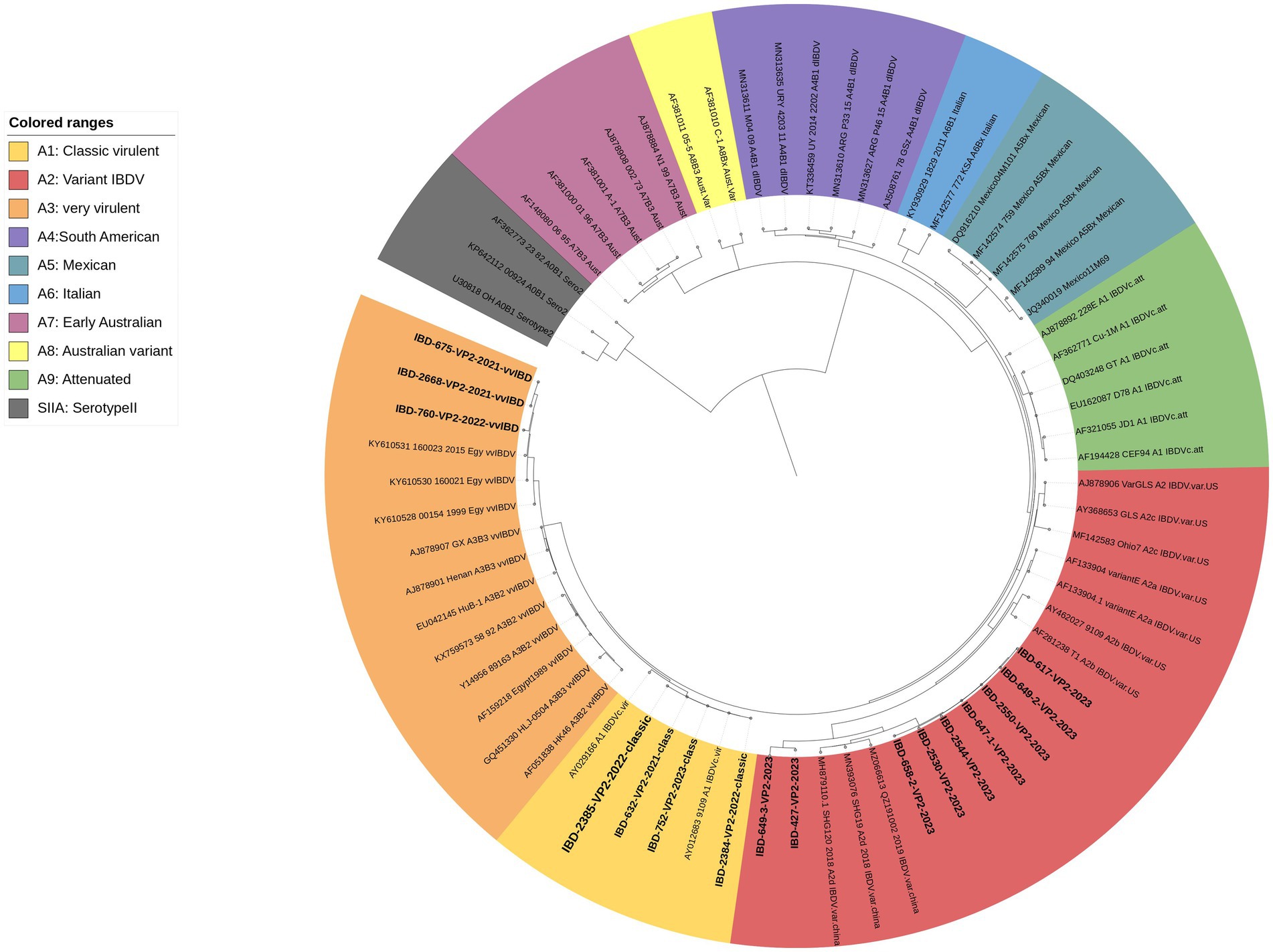
Figure 4. A concise circular phylogenetic tree based on the VP2-HVR sequence. The tree was generated using a maximum likelihood (ML) with 1,000 bootstrap replications, using the IQ-TREE program (17) and the (GTR + F + G4) substitution model embedded within IQ-Tree. The tree was annotated in iTOL (https://itol.embl.de) with genogroup information displayed in colored circular using scheme proposed by Gao et al. (7). Samples tested in the present study are in bold.
The deduced amino acid sequences of VP2 for the studied samples were aligned with reference samples representing each genogroup (Figure 5). All Egyptian variant samples exhibited typical residues found in variant IBDV, including 222T, 249K, 286I, and 318D, along with the three conserved amino acid residues found only in Chinese variant IBDVs: 217K, 252I, and 299S. However, two samples, namely 427 and 649, possessed 318G and 323D, resembling the GLS strain at these residues. In addition, sample 649-3 exhibited 299N, resembling GLS a9ariantntE strains. Apart from samples 427 and 649, all Egyptian variant strains possessed 321V (Figure 5).
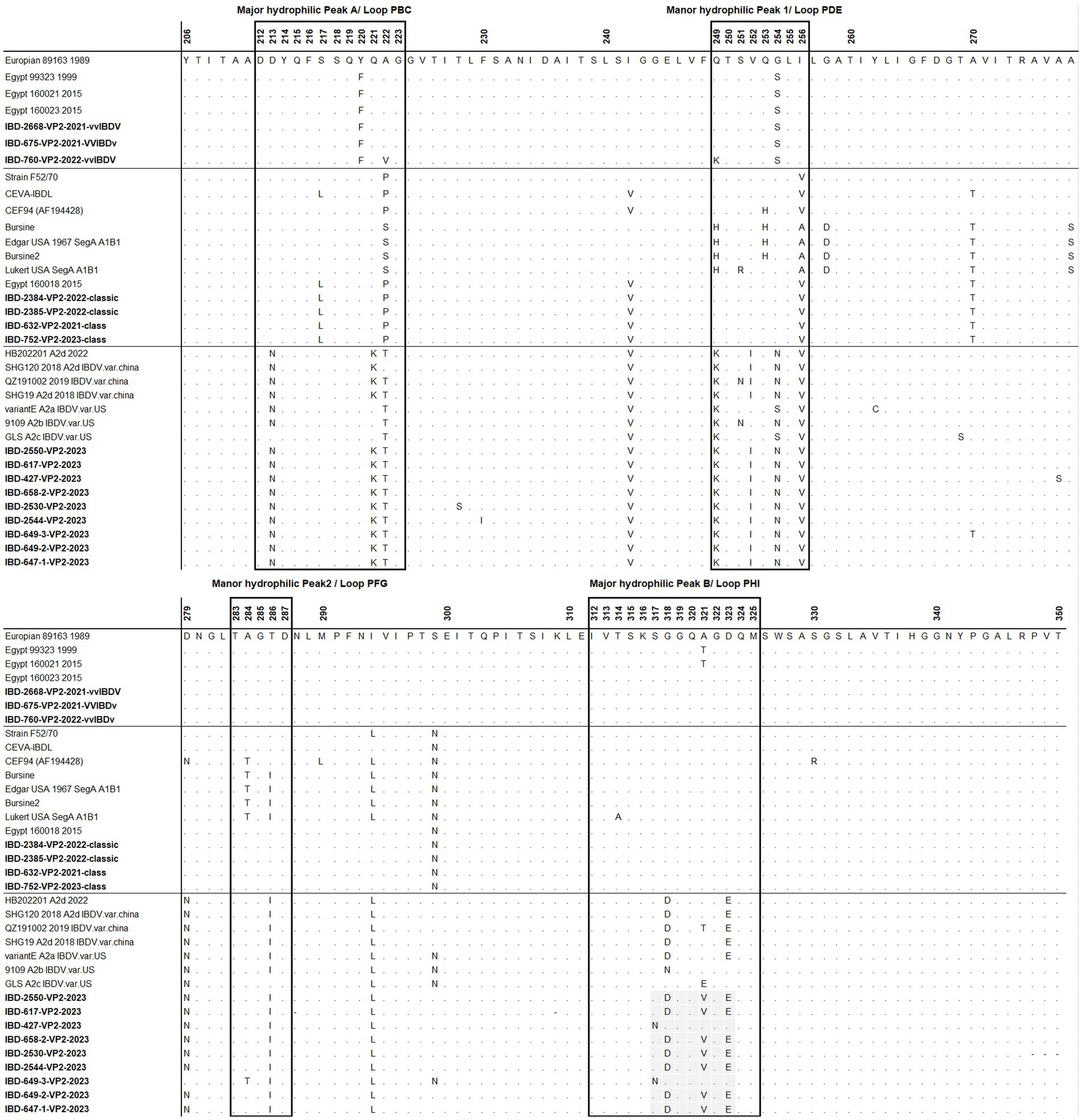
Figure 5. Amino acid alignment of VP2 HVR for the successfully sequenced samples in comparison with reference sequences. Amino acids surrounded by black line boxes indicate the most exposed peaks of the VP2 P domain and amino acids surrounded by red dotted lines red boxes represent unique amino acid mutation for the newly emerged nVarIBDV in Egypt. Samples tested in the present study are in bold.
Regarding classical strains, the amino acid sequences for VP2-HVR of cIBDV showed typical amino acid similarity with IBDL, a commonly used vaccine strain in Egypt (Figure 5). In comparison to the Egyptian antigenic atypical vvIBDV strain isolated in 1999, all the vvIBDV samples detected in the present study possessed Y220F and G254S located in major hydrophilic peak A and manor hydrophilic peak 1, respectively, but not A321T in major hydrophilic peak B (Figure 5).
Only 17 samples were successfully sequenced to cover the entire VP2-HVR, which is required for genotyping (Figure 4; Table 2). Comparing the results of genotyping with the established one-step real-time PCR revealed 100% concordance with genotyping based on gene sequencing, and it also agreed with the clinical findings. It is worth mentioning here that the established one-step real-time PCR was able to detect the nVarIBDV samples newly emerging in Egypt with no marked CT value difference between nVarIBDV and cIBDV.
Infectious bursal disease represents a significant threat to poultry production globally, characterized by mortality rates that can reach up to 100% and immunosuppression in young chicks (15). With the emergence of vvIBDV and classical strain-based live IBDV vaccines, they have become widely utilized in numerous countries, including Egypt (12, 16). Some of these strains target the bursa and cause lesions that add more complications to field diagnosis, especially if we take into account the common lesions induced by classic IBDV and vvIBDV (4, 11). The differentiation between various IBDV field strains and vaccinal strains relies mainly on gene sequencing of the VP2-HVR region. However, gene sequencing is a laborious, time-consuming, and expensive process, further complicated by supply chain issues, especially in low- and middle-income countries. Here, we designed and validated a one-step real-time qRT-PCR using two distinct and specific probes, along with two sets of primers simultaneously, to distinguish between vvIBDV and non-vvIBDV strains. One set of primers and a HEX-labeled TaqMan probe are specific to vvIBDV VP2-HVR, while the non-vvIBDV strains are identified by a FAM-labeled TaqMan probe and two primers specific to the VP1 gene of the classic and variant IBDV strains. The proposed real-time qRT-PCR assay not only saves time and is affordable but also can detect mixed infections with more than one genotype.
Numerous trials and assays have been conducted for the rapid detection and identification of classic and very virulent strains of IBDV. These include nanoparticle-assisted PCR, SYBR green, and TaqMan-based real-time RT-PCRs (RT-qPCR) (18–21). Including a recent study developed a real-time one-step qRT-PCR assay, this assay can distinguish the nVarIBDV from other non-nVarIBDV strains based on nucleotide polymorphisms in the 5’-UTR and the vp5/vp2 overlapping region of the segment A sequences (22). Despite the widespread use of the nVarIBDV in several countries, including China, Japan, South Korea, Malaysia, and Egypt (6, 23–26) the vvIBDV remains a significant threat to poultry production (25, 27) due to its high replication efficiency and rapid spread (4) and its rapid accurate differentiation from classical strains ultimately required to assess the vaccination efficiency. Previous studies, such as those by Gao et al. (7) and Michel and Jackwood (9), have also focused on distinguishing IBDV strains. However, in our assay in addition to that our assay successfully differentiates newly emerged variant strains, providing a rapid and accurate single-assay solution for field diagnostics.
Results showed that the designed assay had no cross-reaction with other circulating viruses in the field, such as HPAI H5N8, LPAI H9N2, IBV, and NDV. Using a tenfold serial dilution of known amounts of VP1 and VP2 sequences, the assay detected up to 1.9×103, 9.2×103, and 1.2×104 DNA copies/μL at CT 32.8, 30.78, and 31.45 for vvIBDV, classic IBDV, and nvarIBDV, respectively. The assay successfully differentiated between vvIBDV and non-vvIBDV field samples with a zero-error rate compared to sanger sequencing results, regardless of the mismatches, especially in the probe in different vvIBDV strains.’ It’s essential to note that, due to the high variability of VP2-HVR, some mismatches occurred with different vvIBDV strains, including some Egyptian antigenically atypical and typical vvIBDV. Yet, there was no marked impact on the assay sensitivity, as indicated by the CT values of the samples tested. Despite potential mismatches in viral field isolates that could impact quantification, leading to false negatives, the impact of mismatches on PCR efficiency varies based on sequence context and reaction conditions. For example, mismatches toward the end of the primers can dramatically impact the PCR efficiency, but in some cases, they can also contribute to the amplification efficiency and may avoid false priming (28).
While our work is ongoing, a new Chinese variant (nVarIBDV) emerged in Egypt in early 2023. This variant, first recorded in China in 2019, belongs to genotype A2dB1 (6, 7). Despite mismatches, especially in forward primers and probes, the FAM-labeled assay successfully detected the Chinese nVarIBDV. In general, the assay was able to detect all field samples with a relatively low CT ranging from 18–28, indicating its suitability for field situations.
VP2-HVR sequencing is considered phylogenetically representative of the full genome length of IBDV and is used for genotypic classification (7–10). Here in the present study we used genotyping scheme proposed by Gao et al. (7) to confirm the results of real time PCR. Phylogenetic analysis revealed three samples clustered with vvIBDV and other Egyptian antigenically atypical vvIBDV that originally emerged in 1999. Four samples clustered with classic virulent strains, showing high similarity with hot strain (IBDL) live vaccines. Nine samples clustered with the nvarIBDV genogroup that emerged in Egypt in 2023, showing high similarity with Chinese variant strains (25). It’s worth mentioning that only 17 samples out of 46 were successfully sequenced from field material, highlighting the challenge of genotyping IBDV based on Sanger sequencing. However, our in-house, one-step real-time qRT-PCR was found to be capable of detecting single and mixed infections of various genotypes directly from field samples.
The persistent threat posed by vvIBDV to poultry production necessitates the development of a rapid and sensitive assay to distinguish between vvIBDV and non-vvIBDV strains that are commonly used as vaccination strains. Here, we propose a real-time qRT-PCR assay using two sets of primers and probes simultaneously. The first set detects vvIBDV based on VP2-HVR, and the second set detects non-vvIBDV genotypes based on VP1 sequencing of classic and American variant IBDV. Despite the high variability of HVR, the designed primers and probes showed higher sensitivity with the circulated vvIBDV in Egypt. We also report the emergence of nvarIBDV that can be recognized by non-vvIBDV primers and probes. It is necessary to highlight here that the assay has been thoroughly tested using vvIBDV samples collected from Egypt, considered an antigenic atypical vvIBDV. More samples representing different vvIBDV strains are required to be tested.
The Nucleotide sequence of the tested field samples presented in this study have been deposited in GenBank. The accession numbers are provided in the article.
The animal study was approved by the Scientific and Ethics Committee of the Animal Health Research Institute (AHRI), Agriculture Research Center (ARC), Egypt. The study was conducted in accordance with the local legislation and institutional requirements.
AA: Conceptualization, Data Curation, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. AZ: Formal analysis, Methodology, Writing – original draft. ZM: Formal analysis, Software, Validation, Writing – original draft, Writing – review & editing. KS: Investigation, Writing – original draft. NH: Investigation, Writing – review & editing. MB: Methodology, Writing – original draft, Writing – review & editing. HE: Writing – review & editing. MS: Writing – review & editing. AS: Formal analysis, Data Curation, Investigation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. All wet lab work was conducted at the NLQP-AHRI, a governmental institute funded by the Agriculture Research Center, Ministry of Agriculture, and Land Reclamation, Egypt. AS was supported by the BBSRC/UKRI fund (project codes: BBS/E/PI/230002C and BBS/E/PI/23NB0003).
We would like to thank Cairo3A company for providing the field samples to accomplish this study.
The authors declare that the research was conducted in the absence of any ‑commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
vvIBDV, Very virulent infectious bursal disease virus; RT-PCR, Reverse transcriptase polymerase chain reaction; nVarIBD, New variant infectious bursal disease virus.
1. Eterradossi, N, and Saif, Y. Infectious bursal disease. 13th ed. Hoboken, NJ: Blackwell Publishing Ltd. (2013).
2. Sajid, S, Ur Rahman, S, Nayab, S, Khan, IU, and Javeed, A. Infectious bursal disease virus cloning and structural protein (VP2) expression in Escherichia coli. Pure Appl Biol. (2020) 9:743–9. doi: 10.19045/bspab.2020.90080
3. Jackwood, DJ, Sommer-Wagner, SE, Crossley, BM, Stoute, ST, Woolcock, PR, and Charlton, BR. Identification and pathogenicity of a natural reassortant between a very virulent serotype 1 infectious bursal disease virus (IBDV) and a serotype 2 IBDV. Virology. (2011) 420:98–105. doi: 10.1016/j.virol.2011.08.023
4. Van den Berg, T, Morales, D, Eterradossi, N, Rivallan, G, Toquin, D, Raue, R, et al. Assessment of genetic, antigenic and pathotypic criteria for the characterization of IBDV strains. Avian Pathol. (2004) 33:470–6. doi: 10.1080/03079450400003650
5. McFerran, J, McNulty, M, McKillop, E, Connor, T, McCracken, R, Collins, D, et al. Isolation and serological studies with infectious bursal disease viruses from fowl, turkeys and ducks: demonstration of a second serotype. Avian Pathol. (1980) 9:395–404. doi: 10.1080/03079458008418423
6. Fan, L, Wu, T, Hussain, A, Gao, Y, Zeng, X, Wang, Y, et al. Novel variant strains of infectious bursal disease virus isolated in China. Vet Microbiol. (2019) 230:212–20. doi: 10.1016/j.vetmic.2019.01.023
7. Gao, H, Wang, Y, Gao, L, and Zheng, SJ. Genetic insight into the interaction of IBDV with host—a clue to the development of novel IBDV vaccines. Int J Mol Sci. (2023) 24:8255. doi: 10.3390/ijms24098255
8. Islam, MR, Nooruzzaman, M, Rahman, T, Mumu, TT, Rahman, MM, Chowdhury, EH, et al. A unified genotypic classification of infectious bursal disease virus based on both genome segments. Avian Pathol. (2021) 50:190–206. doi: 10.1080/03079457.2021.1873245
9. Michel, LO, and Jackwood, DJ. Classification of infectious bursal disease virus into genogroups. Arch Virol. (2017) 162:3661–70. doi: 10.1007/s00705-017-3500-4
10. Wang, YL, Fan, LJ, Jiang, N, Li, GAO, Kai, LI, Gao, YL, et al. An improved scheme for infectious bursal disease virus genotype classification based on both genome-segments a and B. J Integr Agric. (2021) 20:1372–81. doi: 10.1016/S2095-3119(20)63424-4
11. Berg, TP, Gonze, M, and Meulemans, G. Acute infectious bursal disease in poultry: isolation and characterisation of a highly virulent strain. Avian Pathol. (1991) 20:133–43. doi: 10.1080/03079459108418748
12. Samy, A, Courtillon, C, Briand, F-X, Khalifa, M, Selim, A, Hegazy, A, et al. Continuous circulation of an antigenically modified very virulent infectious bursal disease virus for fifteen years in Egypt. Infect Genet Evol. (2020) 78:104099. doi: 10.1016/j.meegid.2019.104099
13. Hall, T, Biosciences, I, and Carlsbad, C. BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
14. Yovel, Y, Franz, MO, Stilz, P, and Schnitzler, HU. Plant classification from bat-like echolocation signals. PLoS Comput Biol. (2008) 4:e1000032. doi: 10.1371/journal.pcbi.1000032
15. Eterradossi, N, and Saif, YM. Infectious bursal disease. Dis Poultry. (2020):257–83. doi: 10.1002/9781119371199.ch7
16. Zanaty, A, Mossad, Z, Said, M, Samy, M, Amer, F, Rabie, N, et al. Genetic characterization of co-circulated classic and very virulent infectious bursal disease viruses in commercial broiler flocks of Egypt. J World’s Poultry Res. (2022) 12:124–32. doi: 10.36380/jwpr.2022.14
17. Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A, and Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molec Biol Evol. (2015) 32:268–274.
18. Kong, LL, Omar, AR, Bejo, MH, Ideris, A, and Tan, SW. Development of SYBR green I based one-step real-time RT-PCR assay for the detection and differentiation of very virulent and classical strains of infectious bursal disease virus. J Virol Methods. (2009) 161:271–9. doi: 10.1016/j.jviromet.2009.06.023
19. Luan, Q, Jiang, Z, Wang, D, Wang, S, Yin, Y, and Wang, J. A sensitive triple nanoparticle-assisted PCR assay for detection of fowl adenovirus, infectious bursal disease virus and chicken anemia virus. J Virol Methods. (2022) 303:114499. doi: 10.1016/j.jviromet.2022.114499
20. Techera, C, Tomás, G, Panzera, Y, Banda, A, Perbolianachis, P, Pérez, R, et al. Development of real-time PCR assays for single and simultaneous detection of infectious bursal disease virus and chicken anemia virus. Mol Cell Probes. (2019) 43:58–63. doi: 10.1016/j.mcp.2018.11.004
21. Tomás, G, Hernández, M, Marandino, A, Panzera, Y, Maya, L, Hernández, D, et al. Development and validation of a TaqMan-MGB real-time RT-PCR assay for simultaneous detection and characterization of infectious bursal disease virus. J Virol Methods. (2012) 185:101–7. doi: 10.1016/j.jviromet.2012.06.012
22. Wang, C, Hou, B, Shao, G, and Wan, C. Development of a one-step real-time TaqMan reverse transcription polymerase chain reaction (RT-PCR) assay for the detection of the novel variant infectious bursal disease virus (nVarIBDV) circulating in China. Viruses. (2023) 15:1453. doi: 10.3390/v15071453
23. Aliyu, HB, Hair-Bejo, M, Omar, AR, and Ideris, A. Genetic diversity of recent infectious bursal disease viruses isolated from vaccinated poultry flocks in Malaysia. Front Vet Sci. (2021) 8:643976. doi: 10.3389/fvets.2021.643976
24. Myint, O, Suwanruengsri, M, Araki, K, Izzati, UZ, Pornthummawat, A, Nueangphuet, P, et al. Bursa atrophy at 28 days old caused by variant infectious bursal disease virus has a negative economic impact on broiler farms in Japan. Avian Pathol. (2021) 50:6–17. doi: 10.1080/03079457.2020.1822989
25. Shahein, MA, Sultan, HA, Zanaty, A, Adel, A, Mosaad, Z, Said, D, et al. Emergence of the novel infectious bursal disease viruse variant in vaccinated poultry flocks in Egypt. bioRxiv. (2023). doi: 10.1101/2023.11.13.566865
26. Thai, TN, Jang, I, Kim, H-A, Kim, H-S, Kwon, Y-K, and Kim, H-R. Characterization of antigenic variant infectious bursal disease virus strains identified in South Korea. Avian Pathol. (2021) 50:174–81. doi: 10.1080/03079457.2020.1869698
27. Zhang, W, Wang, X, Gao, Y, and Qi, X. The Over-40-years-epidemic of infectious bursal disease virus in China. Viruses. (2022) 14:2253. doi: 10.3390/v14102253
Keywords: VP2, VP1, cIBDV, vvIBDV, nVarIBDV, real-time PCR
Citation: Adel A, Zanaty A, Mosaad Z, Selim K, Hagag NM, Badr M, Ellakany H, Shahien M and Samy A (2024) Advancing IBDV diagnostics: a one-step multiplex real-time qRT-PCR for discriminating between vvIBDV and non-vvIBDV viruses, including the newly emerged IBDV variant. Front. Vet. Sci. 11:1421153. doi: 10.3389/fvets.2024.1421153
Received: 21 April 2024; Accepted: 21 June 2024;
Published: 18 July 2024.
Edited by:
Chunhe Wan, Fujian Academy of Agricultural Sciences, ChinaReviewed by:
Xiaole Qi, Harbin Veterinary Research Institute (CAAS), ChinaCopyright © 2024 Adel, Zanaty, Mosaad, Selim, Hagag, Badr, Ellakany, Shahien and Samy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amany Adel, YS5hZGVsMTg3ODRAZ21haWwuY29t; Ahmed Samy, YWhtZWQuaWJyYWhpbUBwaXJicmlnaHQuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.