
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 22 July 2024
Sec. Veterinary Clinical, Anatomical, and Comparative Pathology
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1417244
This article is part of the Research Topic Unraveling inflammaging : A pathway to Prevent Age-related Disease in Animals View all 11 articles
Amidst rising global temperatures, chronic heat stress (CHS) is increasingly problematic for the poultry industry. While mammalian CHS responses are well-studied, avian-specific research is lacking. This study uses in-depth transcriptome sequencing to evaluate the pulmonary response of Cherry Valley ducks to CHS at ambient temperatures of 20°C and a heat-stressed 29°C. We detailed the CHS-induced gene expression changes, encompassing mRNAs, lncRNAs, and miRNAs. Through protein–protein interaction network analysis, we identified central genes involved in the heat stress response—TLR7, IGF1, MAP3K1, CIITA, LCP2, PRKCB, and PLCB2. Subsequent functional enrichment analysis of the differentially expressed genes and RNA targets revealed significant engagement in immune responses and regulatory processes. KEGG pathway analysis underscored crucial immune pathways, specifically those related to intestinal IgA production and Toll-like receptor signaling, as well as Salmonella infection and calcium signaling pathways. Importantly, we determined six miRNAs—miR-146, miR-217, miR-29a-3p, miR-10926, miR-146b-5p, and miR-17-1-3p—as potential key regulators within the ceRNA network. These findings enhance our comprehension of the physiological adaptation of ducks to CHS and may provide a foundation for developing strategies to improve duck production under thermal stress.
The exponential growth of the global population necessitates substantial protein production, placing escalating demands on animal production systems, particularly within tropical and subtropical regions. This factor significantly contributes to ensuring global food security (1, 2). Among these production sectors, the poultry industry emerges as a crucial subsector that substantially fuels economic growth (3, 4). Nevertheless, the implications of global warming have engendered prolonged hyperthermia during the summer months, presenting a formidable challenge to the industry. This challenge has manifested in reduced productivity and substantial economic losses (5). This deleterious phenomenon, recognized as heat stress, has garnered extensive global attention and scrutiny (6).
Heat stress entails an imbalance between heat acquisition and dissipation, resulting in an elevation of poultry body temperature (7–9). It is typically categorized into intense or chronic heat stress (CHS), pertaining to brief or prolonged exposure to elevated ambient temperatures, respectively (10). Unfortunately, both intense and chronic heat stress can exact a toll on avian health (11, 12). This consequence extends to other domestic animals as well. Poultry, in particular, faces heightened vulnerability due to the absence of sweat glands and the majority of their body surface being covered in feathers. Consequently, the role of heat dissipation is largely assumed by the lungs in poultry (13, 14). When the ambient temperature falls within the thermal comfort zone, birds can sustain their body temperature with minimal effort. Typically, the optimal temperature for growing Pekin ducks ranges from 18 to 20°C (15). However, deviations from this range trigger behavioral, physiological, and metabolic adaptations aimed at temperature regulation and mitigating the impact of high temperatures (16–18). These adaptations include panting, reduced food intake, weight loss, and can culminate in undesirable outcomes such as increased feed conversion ratios (FCRs), stunted growth, and compromised meat quality. In cases where efficient heat dissipation is unattainable, multi-organ dysfunction may ensue, potentially leading to fatality (19–21).
Recent strides in high-throughput screening technology have culminated in the routine utilization of transcriptome sequencing for the quantification and identification of RNAs across diverse tissues and cells (22). RNA molecules bear transcribed genetic information that can be translated into proteins or directly/indirectly modulate gene expression levels (23). The variation in the number of different transcripts in response to temperature changes can offer valuable insights into cellular states and stress mechanisms (22, 24). Notably, heat shock proteins (HSPs) and phosphoinositide 3-kinase (PIK3) emerge as key participants in heat stress acclimation, with the genes encoding these proteins showing significant upregulation in heat-treated Fujian shelducks and Shan Ma, Pekin, Muscovy ducks (25–29). Furthermore, non-coding RNAs that play immunoregulatory roles during Salmonella enteritidis infection have been revealed through competing endogenous RNA (ceRNA) regulatory networks in Shaoxing ducks (30). Yet, the landscape of ceRNA networks under heat stress conditions remains largely uncharted.
In this pioneering study, we have created a CHS model in CVds—a breed with significant economic value—to explore the comprehensive transcriptomic alterations (31). Our innovative approach employs environmental control chambers for a precise induction of CHS, facilitating the in-depth examination of gene, miRNA, mRNA, and lncRNA expression variations using advanced sequencing techniques. Furthermore, we have delineated lncRNA-miRNA-mRNA networks to elucidate the intricate molecular dialogs during CHS, marking a novel foray into the full-spectrum transcriptomic impact of heat stress on these ducks.
This study was conducted in strict accordance with the regulations outlined by the Administration of Affairs Concerning Experimental Animals (Decree No. 63 of the Jiangsu Academy of Agricultural Science on 8 July 2014). All experimental procedures involving animals received ethical approval from the Research Committee of the Jiangsu Academy of Agricultural Sciences (Nanjing, China).
Following the approach detailed in (15, 32), a batch of 72 newly-hatched CVds with identical genetic backgrounds were procured from a commercial farm. These ducks were subsequently divided into two groups through random allocation, ensuring that there were no significant disparities in phenotypes or weights among any pair of groups. For brevity, these groups were subsequently referred to as W20 and W29. During the initial phase of rearing, all ducks were individually housed in separate pens equipped with an environmental control chamber, providing access to ad libitum feed and water (Supplementary Figure S1). Figure 1 illustrates that the ambient temperature was sustained at 35°C for the first 4 days and was subsequently decreased to 34°C for the following 3 days. Over days 8–12, the temperature in the W29 enclosure was gradually lowered by 1°C daily to 29°C, which was then sustained for 30 days. In parallel, the ambient temperature for W20 was similarly reduced by 1°C daily throughout this interval, leading to a steady state at 20°C (33). During the entire rearing phase, humidity was consistently maintained at 74% (34), with all ducks having free access to standard commercial pellet feed and water from a drip-nipple system.
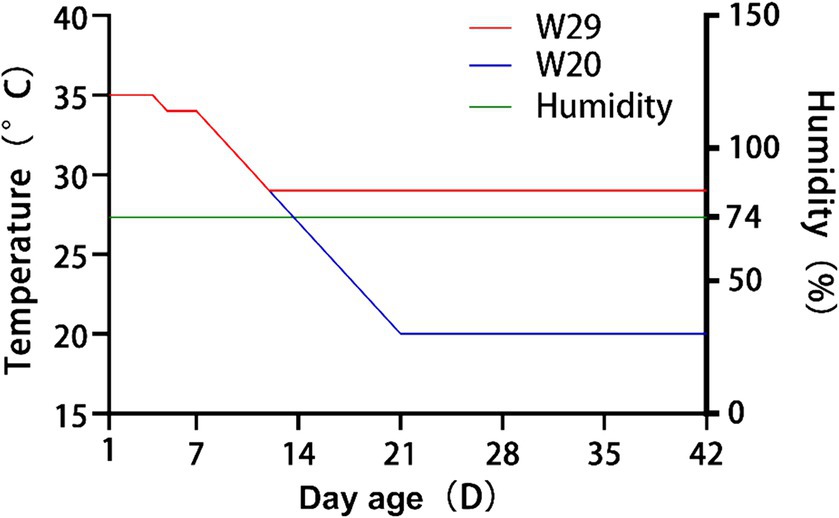
Figure 1. Detailed temperature regimens for W20 and W29 cohorts over the course of the investigation.
Ultimately, at the market-age of day 43, three samples were collected from each group were humanely stunned with a 36 V electrified pool and then euthanized via jugular vein dissection (35). As the second key organ for heat dissipation, their lungs were carefully excised, promptly frozen in liquid nitrogen and then stored at −80°C prior to dispatch to the sequencing company.
The lungs of each selected duck from both groups were examined under a microscope at a magnification of 20×. Lung tissue samples from CVds were carefully extracted and subsequently fixed in 4% paraformaldehyde for a duration of 24 h to preserve the cellular structure for detailed histological analysis. Subsequently, the samples underwent dehydration using a graded alcohol series. These specimens were then infiltrated and embedded in paraffin, before being sectioned into thin serial slices of approximately 5 μm thickness. These sections were mounted onto glass slides, subjected to hematoxylin and eosin staining, and observed using an Olympus microscope (BX53).
For each group, total RNA was isolated from three biological replicates of lung tissue using TRIzol® reagent (Invitrogen, United States), following the manufacturer’s protocol, and genomic DNA was removed with DNase I RNase-free (TaKara, Japan). The RNA concentration and integrity were verified using a NanoDrop 2,100 spectrophotometer (Thermo Fisher Scientific, United States) and an Agilent Bioanalyzer 2,100 (Agilent Technologies, United States), respectively. Only high-quality RNA samples (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, >1 μg) were used for sequencing library construction.
Libraries for mRNA and lncRNA were prepared using a ribosomal RNA depletion strategy with the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (New England Biolabs, United States), starting with 3 μg of RNA per sample. miRNA libraries were generated using the QIAseq miRNA Library Kit (Qiagen, Germany), adhering to the supplier’s instructions. Based on the length distribution characteristic of miRNA, target fragments (16–35 nt) were isolated by gel excision on a 6% Novex TBE PAGE gel (1.0 mm, 10 well) (Invitrogen, United States). Quantification was performed using the Qubit 4.0 fluorometer (Thermo Fisher Scientific, United States), and sequencing was carried out on an Illumina NovaSeq 6,000 system (Illumina, United States) by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China).
After the above deep sequencing, the raw paired-end reads were preprocessed using SeqPrep1 and Sickle2 by removing adaptor sequences and filtering low-quality reads (Supplementary Table S1). Clean reads were aligned using HISAT to the high-resolution reference genome of Anas platyrhynchos (A. platyrhynchos GCF_015476345.1, https://www.ncbi.nlm.nih.gov/genome/2793?genome_assembly_id=1498951) (36, 37). The mapped reads were assembled by StringTie in a reference-based approach (38). Also, the number of reads mapped to each transcript was calculated using RSEM, and the transcripts per million reads (TPM) was estimated to measure the expression level of each gene/transcript (39).
Gene expression level is a broader term that encompasses the levels of all products of gene expression, including mRNA, non-coding RNAs, and proteins. mRNA expression level is a subset of this, focusing only on the messenger RNA produced during transcription. The DESeq2 package within R software was employed to identify genes and mRNAs that were differentially expressed between W20 and W29, with an emphasis on those exhibiting a greater than 1.5-fold change and an adjusted padj below 0.05, marking them as statistically significant (40). These findings were further examined for functional relevance using GO and KEGG pathway enrichment via GOATOOLS and KOBAS (41, 42). Additionally, differentially expressed genes (DEGs) were integrated into STRING (version 12.0) and Cytoscape (version 11.0.13) to delineate protein–protein interaction (PPI) networks, highlighting key modules using the cytoHubba plugin (43–45). Enrichment analysis of interactive gene targets was conducted using the BINGO plugin and the clusterProfiler package (46, 47).
Using the StringTie tool (38) with its default settings, we reassembled transcripts from the aligned clean reads. Subsequently, we cross-referenced these merged transcripts against known reference transcripts annotated in GFF/GTF formats and existing lncRNA databases to identify recognized lncRNAs. In addition, we pinpointed putative novel lncRNA transcripts by their length, requiring over 200 base pairs, and by the presence of two or more exons. To assess the coding potential of these transcripts, we employed a suite of tools: CNCI (48) with a score threshold below zero, CPC (49) with a score under 0.5, CPAT (50) with a score below 0.5, and Pfam with an e-value stricter than 1e-3 but not passing the threshold. Transcripts that were consistently predicted to lack protein-coding potential by these metrics were classified as novel lncRNAs. Potential cis- and trans-acting target mRNAs of lncRNAs were identified through an examination of gene expression patterns and chromosomal positioning, as outlined by (51). For cis-acting targets, genes situated within a 100,000 base-pair range flanking the lncRNA were pinpointed utilizing BEDTOOLS software as described by (52). The analysis of lncRNA-mediated trans-regulation was predicated on the correlation coefficient between the expressions of lncRNA and mRNA, with coefficients exceeding 0.9 signifying a potential trans-regulatory interaction.
Initially, all clean mapped tags were matched to known miRNAs using the miRBase (version 22.1) database3. Subsequently, the remaining tags were cross-referenced with the Rfam and Repbase databases to filter out ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), other non-coding RNAs, and repeats. Finally, unannotated tags were evaluated for potential novel miRNAs with miRdeep2 software (53), based on their genomic location and the formation of hairpin structures. The miRanda algorithm (54) was applied to animal samples, while psRobot (55) was utilized for insect samples to forecast miRNA targets. Predicted miRNA-target RNAs were then determined by identifying the overlap in the outcomes from both tools.
The refined datasets for lncRNAs, miRNAs, and mRNAs were obtained by discarding low-quality reads from the initial raw data, ensuring a Phred quality score of at least 20. RNAs that showed no expression in over three samples were excluded from subsequent analyses to maintain data integrity. Consequently, only high-quality filtered datasets were utilized for further analysis. To quantify RNA expression, the polished reads from the lncRNA and mRNA libraries were mapped to the reference genome via the STAR aligner, while the miRNA sequences were aligned to miRBase using the BOWTIE tool. The R software’s limma package (56) was utilized for identifying differentially expressed mRNAs, lncRNAs, and miRNAs (DEmRNAs, DElncRNAs, and DEmiRNAs). Significant DEmRNAs and DElncRNAs were detected within the comparison groups, applying a threshold of padj <0.05 and an absolute log2FC greater than 1. For DEmiRNAs, the criteria of an absolute Log2FC greater than 0.585 and a p-adjust value below 0.05 were adopted. Functional enrichment analyses for GO terms and KEGG pathways were conducted on the target genes of DEmiRNAs and DElncRNAs, as well as on DEmRNAs, using the GOATOOLS and KOBAS tools (42).
To elucidate the relationships between DEmRNAs, DElncRNAs, and DEmiRNAs, a lncRNA-miRNA-mRNA regulatory network was established rooted in the ceRNA hypothesis. Predictions for miRNA-lncRNA and miRNA-mRNA pairings were conducted utilizing Miranda and Targetscan (57), respectively, while the Spearman correlation coefficient, hinged on expression levels, was employed to assess the interplay among these pairings. Visualization of the intricate network was achieved through Cytoscape software.
The cDNA synthesis for mRNA involved reverse transcription using HiScript III RT SuperMix with gDNA wiper (Vazyme, China) in a thermal cycler, following the protocol provided by the manufacturer. The inverse transcription reaction (ITR) for mRNA was executed in a 20 μL reaction mix, incubated at 37°C for 15 min and 85°C for 5 s, then cooled to 4°C. For lncRNA, cDNA synthesis was performed using the lnRcute lncRNA cDNA First-Strand Synthesis Kit and FastKing One Step First-Strand Synthesis Kit (both from Tiangen, Beijing, China). The cDNA for miRNA was synthesized using the miRNA First Strand cDNA Synthesis (Stem-loop Method) Kit (Sangon Biotech, China), with the ITR for both lncRNA and miRNA conducted as per their respective kits’ guidelines.
Primer sets for mRNA were crafted using Primer Premier 6 software, while primer sets for lncRNA and miRNA were designed via Sangon Biotech’s online tools4. All primers used in this study were synthesized by Genewiz@ (China), based on sequences in GenBank and the reference genome. Then, quantitative real-time PCR (RT-qPCR) was carried out using SYBR Green Master Mix (Yeasen, Shanghai, China) on an ABI 7500 Sequence Detector (Applied Biosystem, United States) according to the manufacturer’s instructions. The reaction mixture included 10 μL of first-strand cDNA, 0.4 μM each of forward and reverse primers, and 10 μL of 2× SYBR Green Master Mix, totalling 20 μL. For miRNA, the MicroRNAs qPCR Kit (SYBR Green Method) from Sangon Biotech was employed. The RT-qPCR protocol consisted of an initial denaturation at 95°C for 2 min, followed by 34 cycles of 95°C for 10 s and 60°C for 30 s. All assays were performed in triplicate, with GAPDH as the internal control gene for mRNA and lncRNA, and U6 snRNA as the internal control for miRNA expression studies (37, 58). Melting curve analysis confirmed the specificity of the amplification, and relative gene expression was quantified using the 2−ΔΔCt method.
Supplementary Figure S2 reveals that at the 43-day collection point, W20 exhibited superior performance over W29 in the majority of carcass characteristics. For a more comprehensive analysis, please consult our previously published work (1). These findings suggest that the growth rate of CVds decelerates when ambient temperatures rise from 20°C to 29°C, aligning with the findings reported in (4).
In the comparative illustration provided by Figure 2, the lungs of W20 exhibited a robust, reddish color and no exudate, with the alveoli retaining their proper form and exhibiting only minor signs of inflammation. The lung interstitium was also free from any signs of excess fluid. In stark contrast, the W29 samples showed significant exudation and altered alveoli that no longer maintained their usual shape. These samples demonstrated considerable changes including thickened alveolar walls, capillary closure, and the presence of inflammatory cells within the alveolar passages, along with interstitial swelling. Yet, these issues were less pronounced than in previous W29 samples.
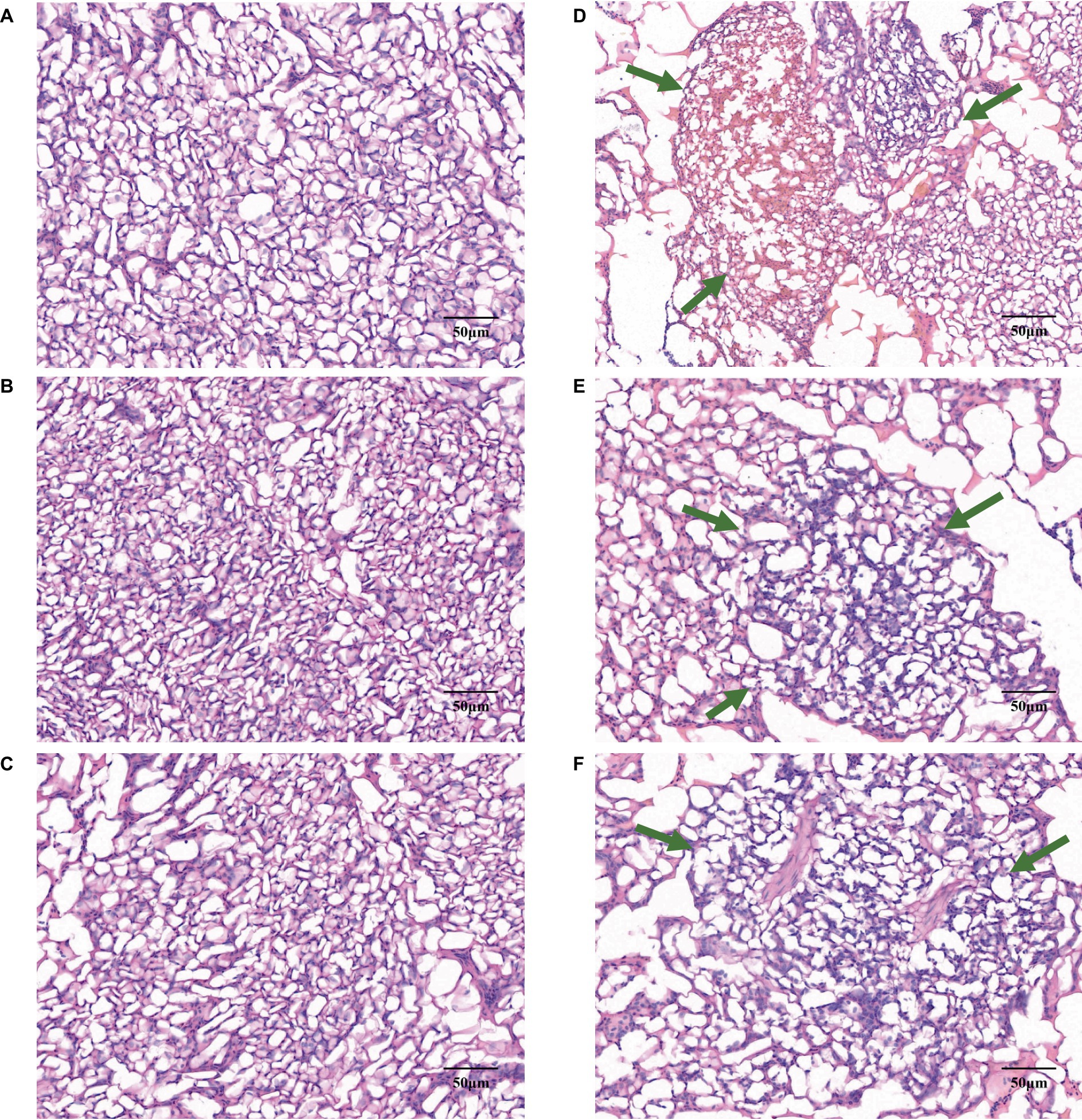
Figure 2. Gene expression profiles in CVds at 43 days of age. The cross-sectional areas of CVds lung tissue reared in an environmental control chamber at (A–C) 20°C and (D–F) 29°C. In panels (D–F), the green arrows point to areas of inflammatory tissue.
To elucidate the genetic underpinnings of heat tolerance in CVds, transcriptomic analyses were conducted on individual lung samples from both W20 and W29 groups using the Illumina NovaSeq 6,000 system. Each of the three cDNA libraries yielded a substantial number of clean reads, amassing billions of nucleotides in total. More than 91% of the clean reads from each library can be mapped to the reference genome of A. platyrhynchos. The expression level of each transcript was measured using TPM method. For the evaluation of gene expression variance, the profiles of the longest transcript isoforms were considered. Followed Principal Component Analysis (PCA) demonstrated a marked distinction between the W20 and W29 sample groups (Figure 3A).
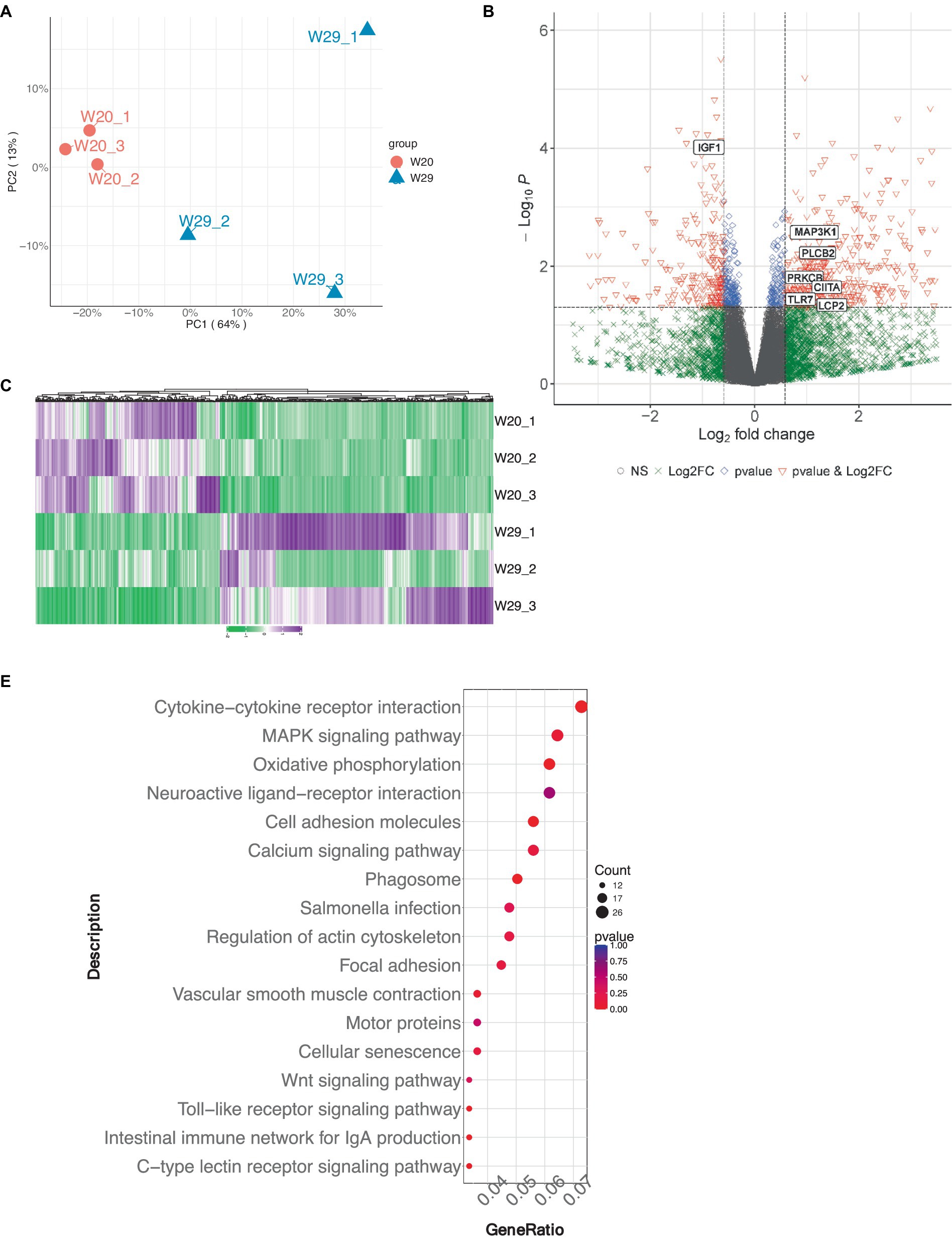
Figure 3. Gene expression profile analysis in CVds at 43 days of age. (A) Principal Component analysis plot for six samples. (B) Volcano plot visualizing the gene expression profile, created using the ComplexHeatmap package in R. (C) Heatmap representation of selected differentially expressed genes. (D) Gene Ontology enrichment analysis of DEGs. (E) Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for DEGs.
Genes exhibiting an absolute fold change of at least 1.5, coupled with a padj under 0.05, were classified as DEGs (Supplementary Table S2). From the W20/W29 comparison, a total of 1,013 DEGs were discovered, among which were 605 up-regulated and 408 down-regulated, as shown in Figures 3B,C. Based on the MCC algorithm and literature research, the top genes were identified as potential hub genes, which were TLR7, IGF1, MAP3K1, CIITA, LCP2, PRKCB, and PLCB2 (Figure 4).
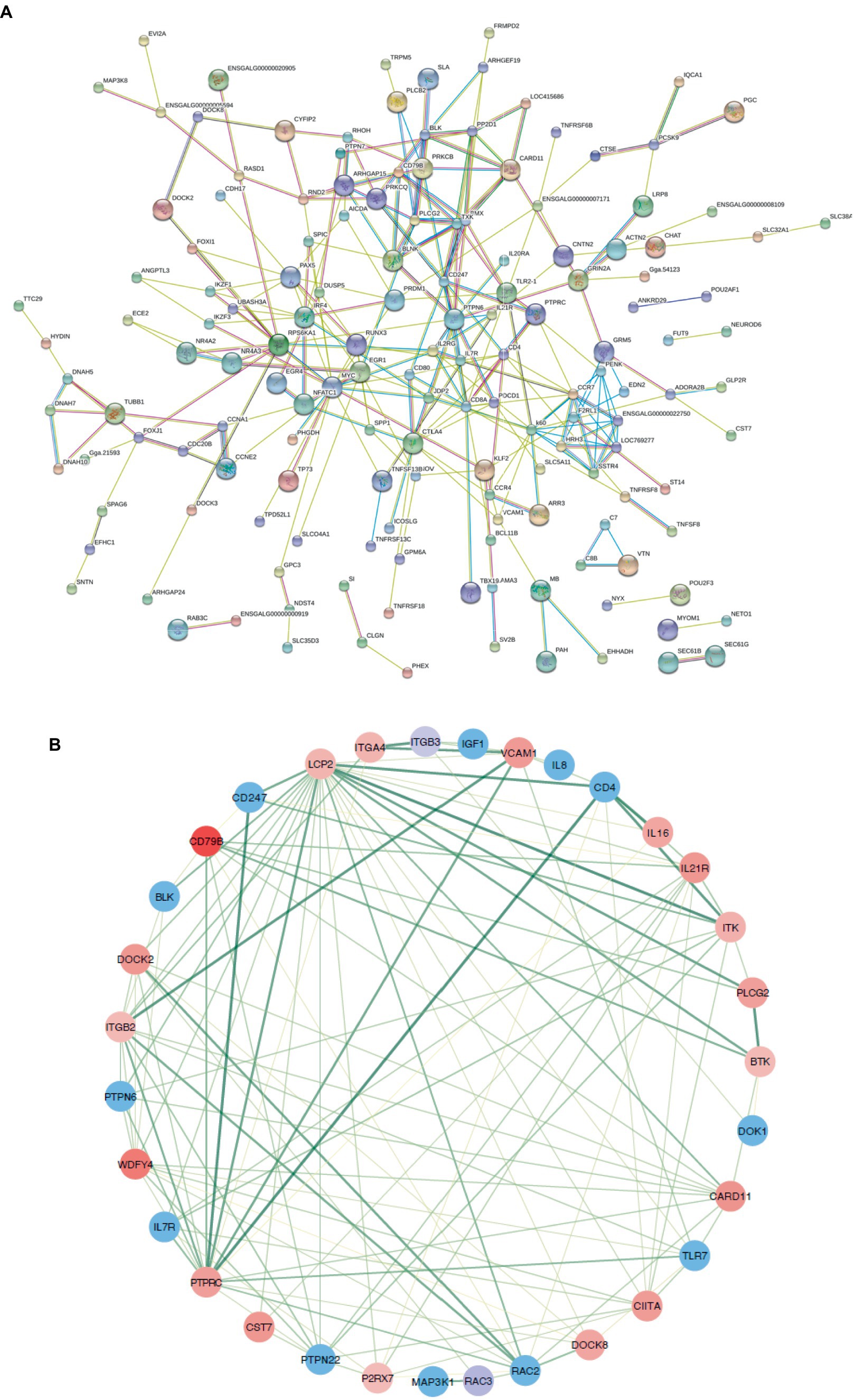
Figure 4. Protein–protein interaction networks. (A) A comprehensive PPI network for all differentially expressed genes, purged of any isolated nodes; (B) a focused subnetwork displayed in a circular layout to emphasize connectivity.
Subsequently, the GOATOOLS software was employed to pinpoint GO terms significantly enriched within these DEGs. A Fisher’s exact test, adjusted for multiple testing with the Benjamin-Hochberg method, flagged GO terms with a false discovery rate less than 0.05 as significantly enriched. Functional annotation of GO terms revealed DEGs predominantly engaged in four biological processes: ‘immune response,’ ‘immune system process,’ ‘regulation of immune system process,’ and ‘response to external stimulus,’ mirroring findings from multiple other studies. In terms of cellular components, the majority of associated GO terms pertained to membrane-related structures, including ‘membrane,’ ‘mitochondrial inner membrane,’ and ‘organelle membrane’ (Figure 3D). These DEGs were predominantly found to be connected to immune system pathways, including ‘intestinal immune network for IgA production,’ ‘Toll-like receptor signaling pathway,’ and ‘C-type lectin receptor signaling pathway.’ Notably, a significant number of these DEGs were also enriched in pathways related to ‘oxidative phosphorylation’ and were implicated in ‘Salmonella infection’ (Figure 3E).
Utilizing quality-assured RNAseq data, we delineated 34,363 protein-encoding mRNAs via custom shell scripts developed in-house. Of which, there were 1,434 mRNA observed to be significantly differentially expressed, with 803 up-regulated and 631 down-regulated (Figures 5A,B).
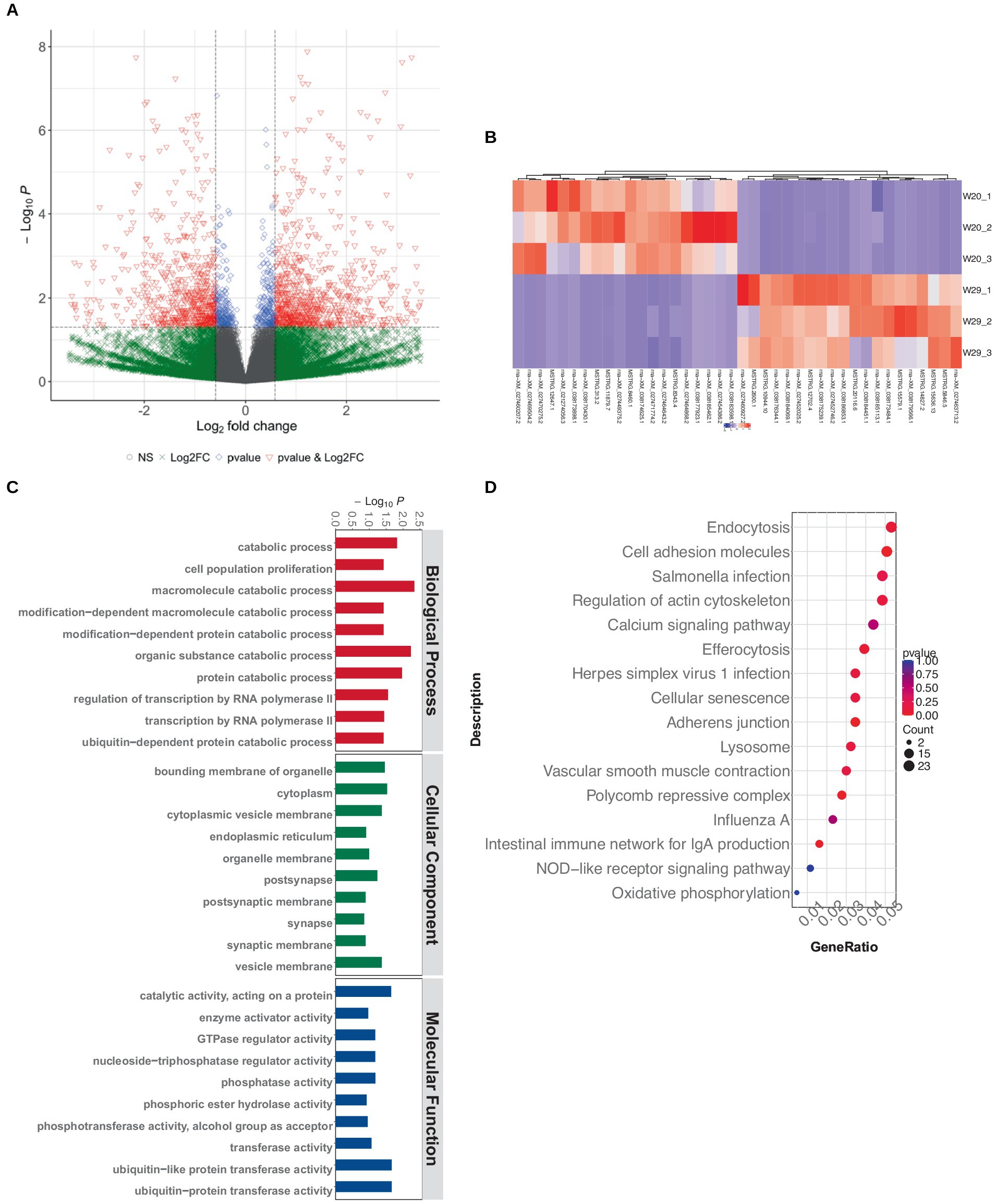
Figure 5. Differentially expressed mRNA profiling in CVds at 43 days. (A) Volcano plot visualization of mRNA expression profiles generated with the ComplexHeatmap package in RStudio. (B) Heatmap depiction of top DEmRNAs. (C) GO enrichment analysis of DE-mRNAs. (D) KEGG pathway enrichment for DEmRNAs.
The GO term functional annotation highlighted that DEmRNAs were actively involved in breakdown processes, including the catabolism of macromolecules and organic substances, as well as protein metabolism. Cellular component analysis showed an abundance of GO terms related to ubiquitin-related enzyme activities, specifically ‘ubiquitin-protein transferase activity’ and ‘ubiquitin-like protein transferase activity’ (Figure 5C). Additionally, these DEmRNAs showed a significant presence in immune system pathways, like ‘Toll-like receptor signaling’ and ‘C-type lectin receptor signaling.’ Notably, a greater number of terms were associated with infectious diseases, such as those related to ‘Herpes simplex virus 1 infection,’ ‘Influenza A’ and ‘Salmonella infection’ (Figure 5D).
In the realm of gene regulation, lncRNAs act as cis-regulators, often influencing proximate protein-coding genes. Utilizing refined RNAseq data subjected to quality control, our tailored shell scripts facilitated the identification of 5,352 (4,387 known and 965 novel) lncRNA transcripts. The genomic analysis revealed that while lncRNAs and mRNAs share similar transcript lengths, lncRNAs are more likely to have longer sequences exceeding 3,000 bp. LncRNAs typically feature a greater proportion with 2–5 exons and possess shorter open reading frames (ORFs) and lower expression levels as quantified by FPKM (Figures 6A–D).
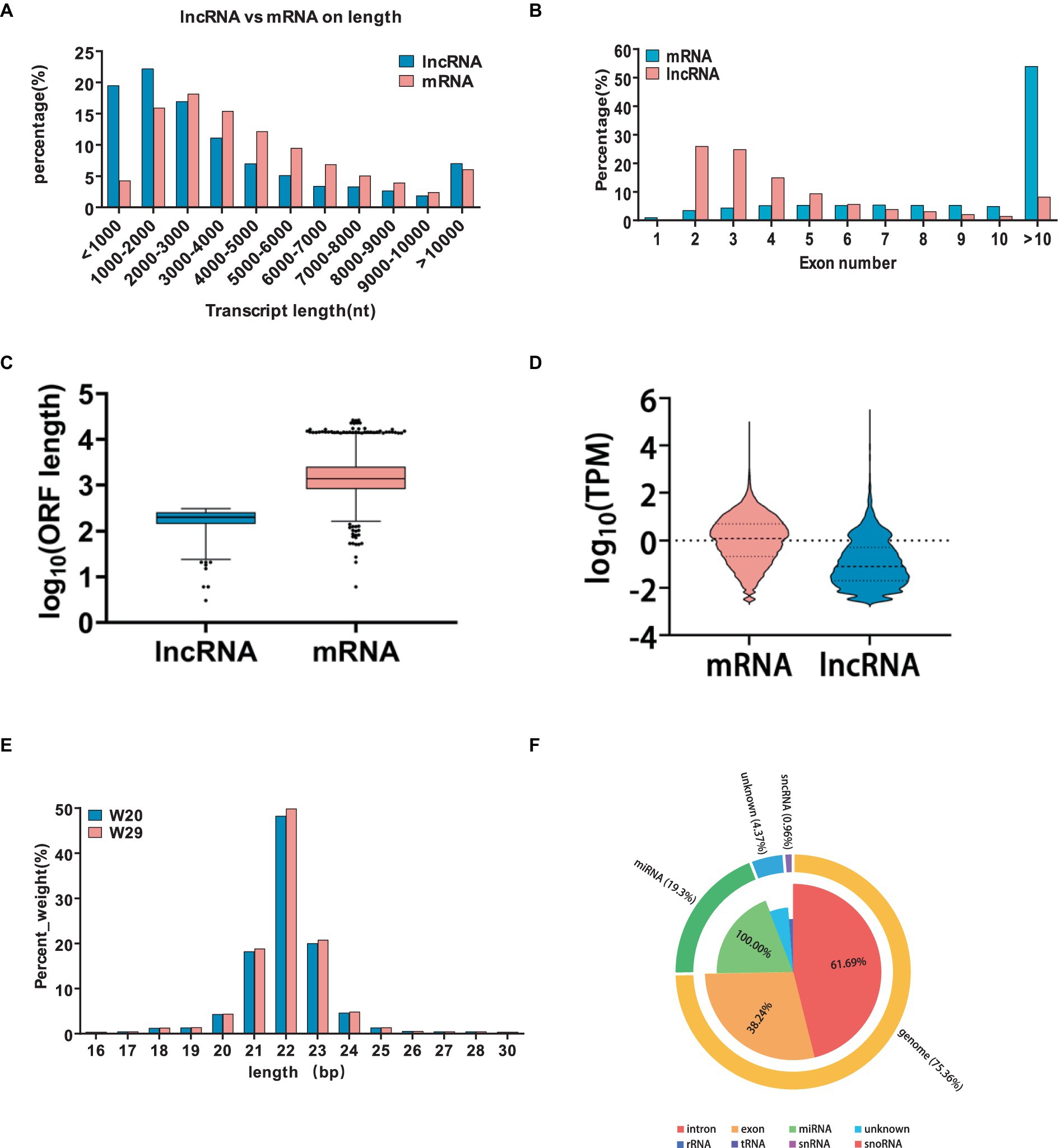
Figure 6. Characterization and comparative analysis of differentially expressed lncRNAs under CHS. (A–D) Comparative metrics between lncRNAs and mRNAs, including transcript length, exon count, open reading frame size, and expression levels measured as transcripts per million. (E) Categorization of the identified small RNAs. (F) Distribution of transcript lengths for all identified miRNAs in the two groups.
Subsequent analysis revealed 217 differentially expressed lncRNAs within the W29 profile, comprising 111 that were up-regulated and the rest displaying down-regulation (Figures 7A,B). Plus, GO and KEGG pathway enrichment analyses were undertaken to decipher the functions and pathways associated with the predicted targets. The enriched GO terms were principally connected to immunoreaction, such as ‘immune response,’ ‘immune system process,’ and ‘regulation of immune system process’ (Figure 7C). The KEGG pathway analysis revealed involvement in immune system (e.g., Intestinal immune network for IgA production) and Infectious disease (e.g., Salmonella infection) (Figure 7D).
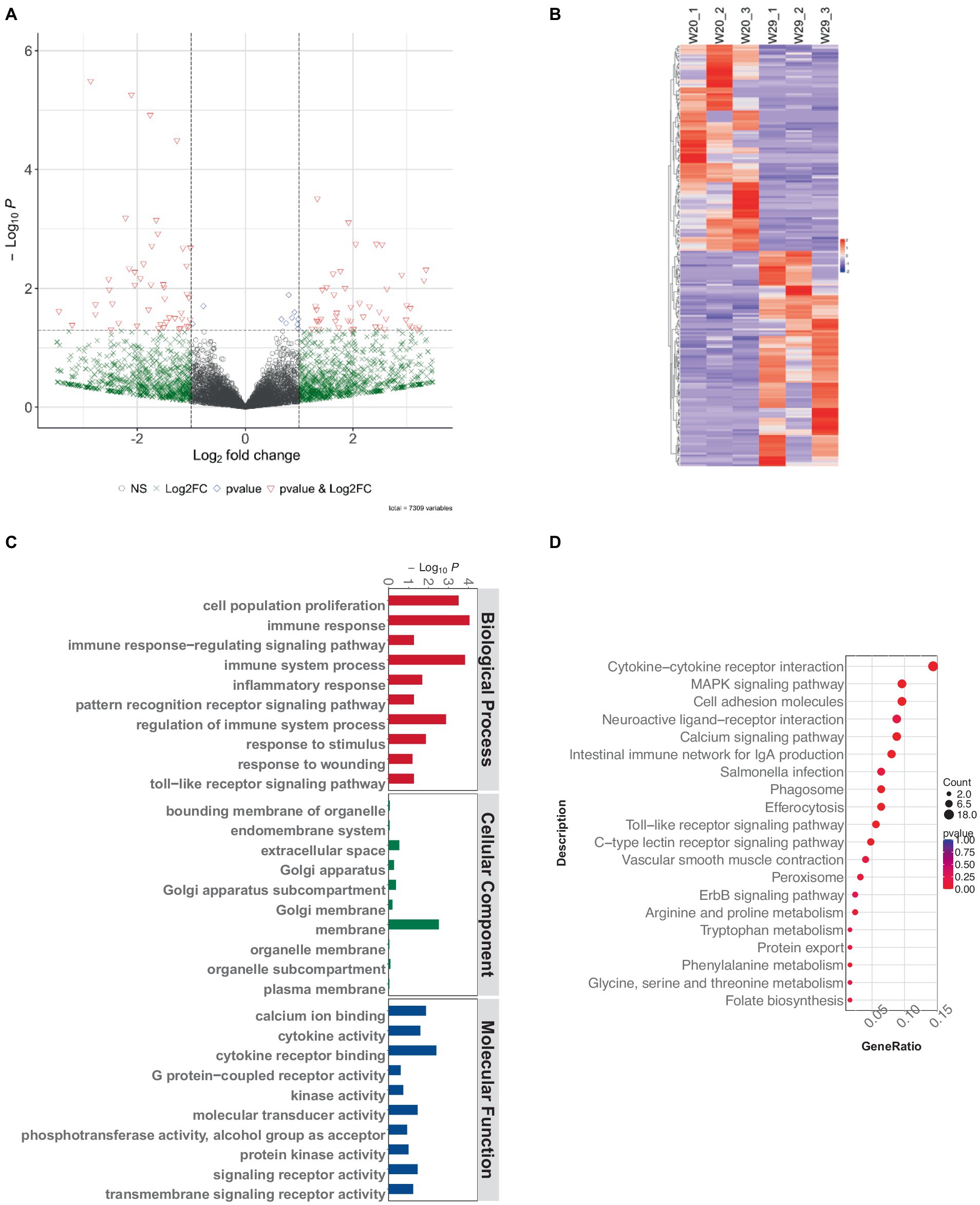
Figure 7. Exploring lncRNA variances in CVds at 43 days. (A) Visualization of lncRNA expression variations using a volcano plot from the ComplexHeatmap package. (B) A heatmap delineating notable DElncRNAs. (C) Analysis of GO term enrichment among DElncRNA targets. (D) Examination of DElncRNA targets within KEGG pathway frameworks.
In this study, we generated a substantial number of raw reads, totaling 12,706,326, 10,121,409, and 9,982,953 for W20, and 12,311,631, 12,412,586, and 10,192,859 for W29, respectively. Following the removal of adaptor sequences, low-quality sequences, and reads outside the length range of 18 to 32 nucleotides, we obtained high-quality clean reads: 12,553,092, 10,032,226, and 9,858,879 for W20, and 11,971,885, 12,250,135, and 9,864,788 for W29. The majority of these clean reads ranged in length from 20 to 24 nucleotides (Figure 6E). Upon classifying the small RNAs, we discovered that 75.3% of the clean reads were attributed to intronic (61.69%) and exonic (38.24%) regions. Additionally, miRNAs accounted for 19.3% of the reads, small non-coding RNAs (sncRNAs) comprised 0.96, and 4.37% were categorized as other types (Figure 6F).
Additionally, for the miRNA expression profiles, 543 DEmiRNAs were identified in W29, including 413 up-regulated and 130 down-regulated miRNAs (Figures 8A,B). In the target study of W29-specific DEmiRNAs, GO annotation revealed a preponderance linked to molecular functions like receptor activities such as ‘neurotransmitter receptor activity,’ ‘signaling receptor activity,’ and ‘transmembrane signaling receptor activity.’ Cellular component annotations centered around structures such as membranes and organelles, while molecular functions primarily involved binding and catalysis. The enriched GO terms for known DEmiRNAs were associated with catabolic processes (Figure 8C). In KEGG pathway analysis, the targets of DEmiRNAs were involved in MAPK signaling pathway, Calcium signaling pathway, and TGF-beta signaling pathway (Figure 8D).
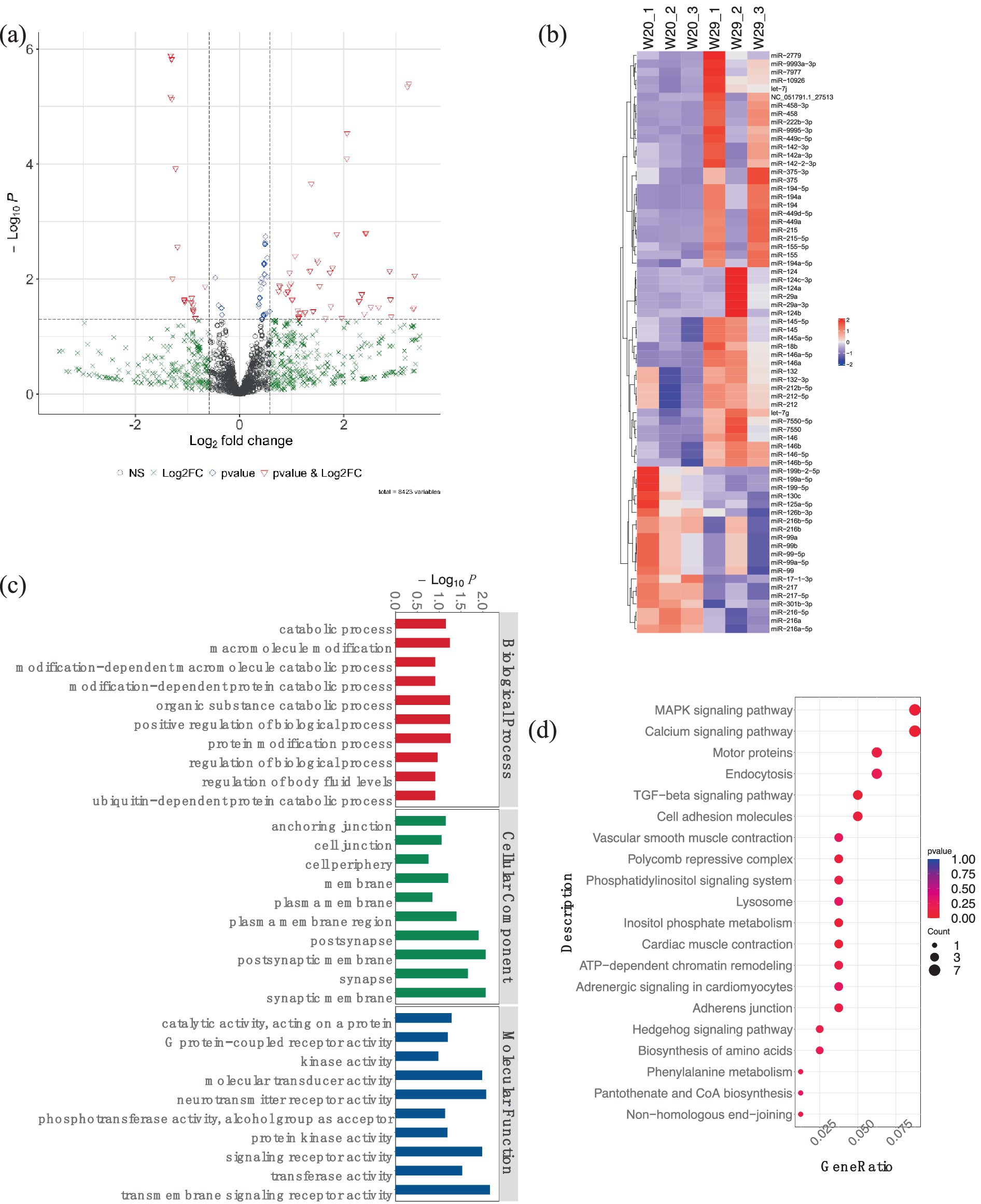
Figure 8. Analysis of differentially expressed miRNAs in CVds at 43 days. (A) A volcano plot generated with the ComplexHeatmap package in RStudio to visualize miRNA expression profiles. (B) Heatmap representation highlighting top DEmiRNAs. (C) GO enrichment analysis for predicted targets of DEmiRNAs. (D) Assessment of DEmiRNAs within the context of KEGG pathways.
To elucidate the comprehensive regulatory matrix involving protein-coding RNAs and non-coding RNAs in response to CHS, a complex ceRNA network was established, integrating differentially expressed miRNAs, mRNAs and lncRNAs. This expansive network predicted thousands of mRNAs and dozens of lncRNAs as miRNA targets in the lungs of W29. Filtering interactions by a strong negative correlation revealed several thousand potential miRNA-mRNA and a handful of miRNA-lncRNA linkages. Certain miRNAs (miR-146, miR-217, miR-29a-3p, miR-10926, miR-146b-5p, and miR-17-1-3p) emerged as central hubs, potentially key to regulatory mechanisms, while multiple lncRNAs (LOC101804558, LOC113841824, LOC101798355, LOC119717605, and LOC110353088) were also pinpointed as significant network participants (Figure 9).
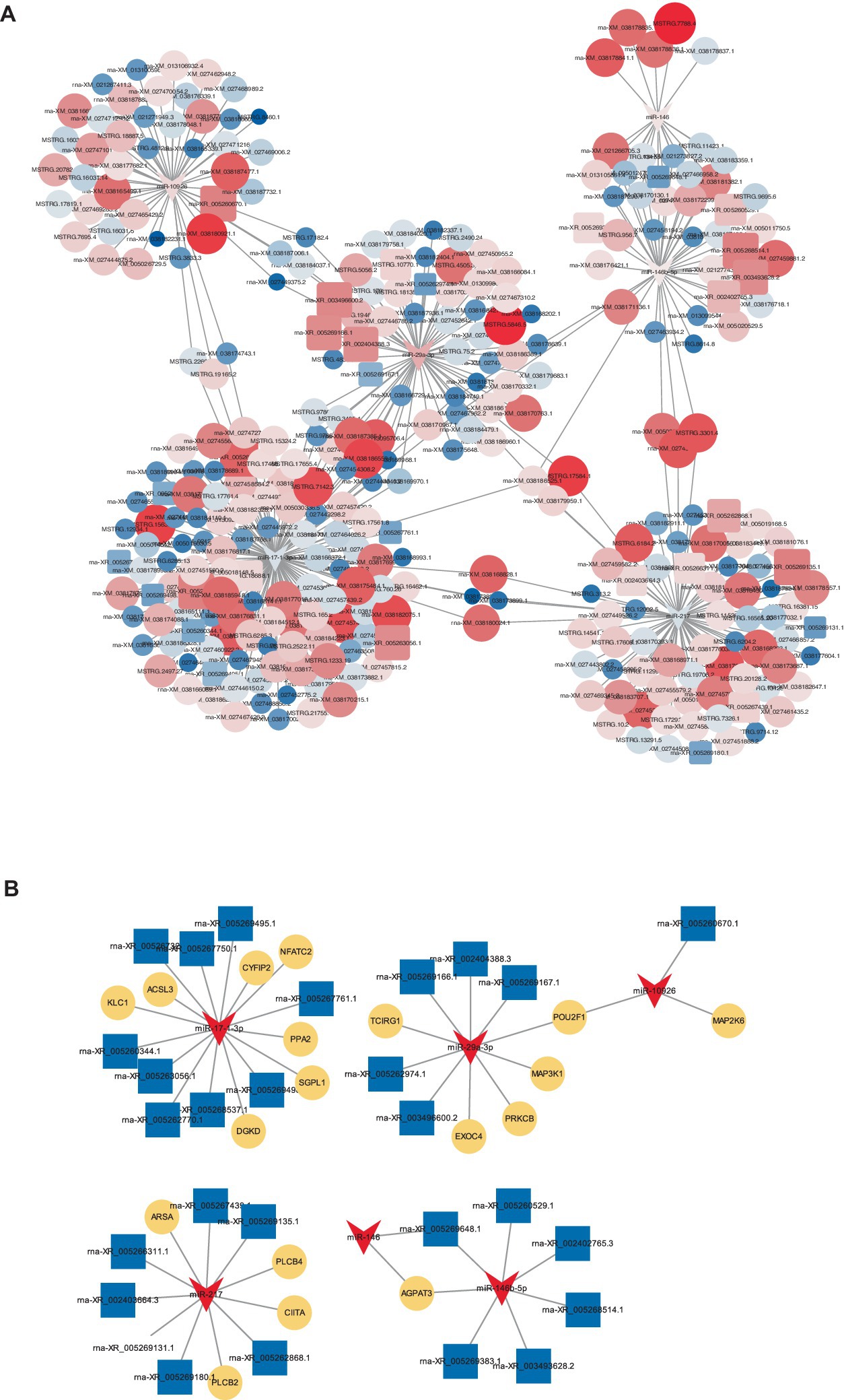
Figure 9. CeRNA network. (A) A constructed ceRNA network centered around miR-217, miR-146b-5p, miR-29a-3p, miR-10926, miR-17-1-3p, and miR-146. In this network, dark blue arrows represent miRNAs, pink triangles indicate mRNAs, light blue circles denote lncRNAs. (B) Depiction of mini-ceRNA network involving six DE-miRNAs and their associated target genes.
To substantiate the RNAseq data and examine the expression correlation between miRNAs and their targets, we selected four key miRNAs—miR-146, miR-217, miR-29a-3p, and miR-10926—and their associated mRNAs and lncRNAs from the ceRNA network for RT-qPCR analysis. The primer sequences for the mRNAs and lncRNAs are itemized in Tables 1, 2, respectively, incorporating GAPDH as the internal reference gene. The primers for the miRNAs are cataloged in Table 3, with U6 employed as the internal normalization gene. The analysis affirmed a predominantly accurate reflection of regulatory dynamics, with miRNAs and their respective targets displaying the expected regulatory trends of either up- or down-regulation (Figure 10). Notably, miR-146 was observed to be up-regulated, with its predicted targets being up-regulated, while miR-217 demonstrated down-regulation, accompanied by up-regulation of all its targets. These results not only validate the RNAseq data’s accuracy but also the postulated inverse relationship between the expression levels of miRNAs and their corresponding ceRNAs.
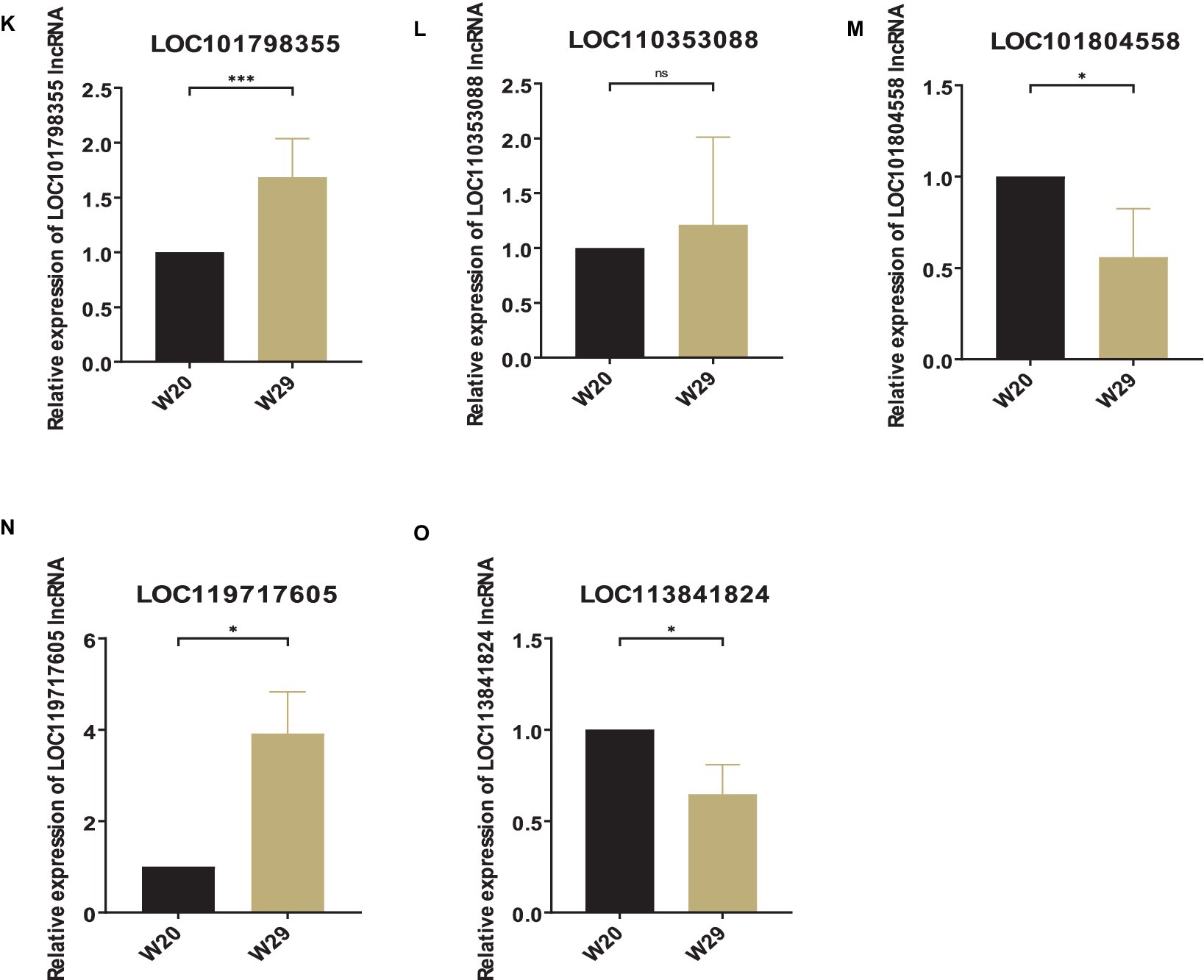
Figure 10. Validation of different expressed RNAs via RT-qPCR. (A–F) mRNA, (G–J) miRNA, (K–O) lncRNA. The relative expressions were calculated in triplicate using the method of 2−△△Ct and presented as mean ± SD. *p-value <0.05, **p-value <0.01, ***p-value <0.001, ****p-value <0.0001.
The detrimental physiological effects of CHS on poultry and livestock industries are well-documented. These effects encompass reductions in feed intake, feed efficiency, growth performance, meat and egg production, meat quality, and survival rates (15, 24, 32, 59–67). Moreover, various studies have explored the impact of CHS on inflammatory responses, dysbiosis, reactive oxidative stress (ROS), signal reactions, and energy metabolism (68–76). Some studies have specifically investigated the health status and well-being of ducks under increasing ambient temperatures, focusing on granulosa cells and the jejunum (25, 77). As the one of the main high-throughput sequencing technologies, transcriptomics has been facilitating the poultry rearing in recent years (78). In the current study, we made full transcriptome profiling of lungs detached from two groups of CVds at 43 days of age reared under 20 and 29°C ambient temperatures. In order to understand the molecular mechanism of response to CHS, comprehensive bioinformatics analysis and intensive RT-qPCR experiments was implemented.
Initial findings indicate that CHS significantly modulates a range of physiological aspects in CVds through pivotal genes including TLR7, IGF1, MAP3K1, CIITA, LCP2, PRKCB, and PLCB2. Notably, TLR7, highly expressed in duck lung tissue, is a key gene in the innate immune defense against viral infections such as influenza (78–80), and also plays a role in egg production (79–83). Immunostimulants are reported to enhance antioxidant and immune efficacy by stimulating TLR7 expression (84, 85). CIITA (Class II major histocompatibility complex trans-activator) is integral to the innate immune response, functioning as a trans-activator that boosts MHC-II expression in both antigen-presenting and virus-infected cells. This activation sparks antiviral responses in the host, serving as a blockade against viral replication and aiding in the clearance of viral infections (86, 87). IGF1 is acknowledged as a key gene influencing growth, body composition, and the development of metabolic and skeletal traits, and plays a significant role in the growth of various tissues, including muscle and bone (88–95). Moreover, its correlation with reproductive efficiency underscores its importance in developmental biology and poultry breeding programs (96–100). Publications concerning MAPK3K1 (mitogen-activated protein kinase kinase kinase 1) (101, 102), LCP2 (103) are limited, yet these genes are known for their roles in lipid metabolism and the development of fatty liver disease. PRKCB (protein kinase C alpha) (104), and PLCB2 (Phospholipase C Beta 2) are less frequently mentioned. The comprehensive analysis incorporating differential expression and RT-qPCR suggests the potential of these genes as biomarkers for heat stress in CVds.
Similarly, it was found that biological profiles, inflammation, and stress protein markers were significantly enriched in miRNA target genes in differential genomes, including L1RAPL2, IL7R, TRAF3, TRAF5, HSPA8, etc. IL1RAPL2 is a molecule in the IL1R family that has different biological effects on immune and inflammatory responses. There is evidence to suggest that IL1RAPL2 is a specific biomarker for kidney injury (105). IL7R is often regulated to varying degrees after virus attacks on poultry. Similarly, there is evidence to suggest that IL7R is associated with cellular responses to heat exposure (106). TRAF3 is a key innate immune regulatory factor that plays a crucial role in defending against viral invasion (107, 108). TRAF5 has been found to regulate inflammation and apoptosis of atherosclerosis, steatosis and melanoma cells, and also plays an important role in regulating myocardial I/R injury (109). HSPA8 can significantly affect the proliferation, apoptosis, and immune function of poultry macrophages, while significantly promoting the proliferation of HD11 cells and inhibiting their apoptosis, with pro-inflammatory effects (110).
Subsequent analysis of gene enrichment pointed to CHS significantly bolstering immunological processes, with a particular focus on catabolic pathways in the case of DEmRNAs and targets of DEmiRNAs. Across the differentially expressed RNAs, pathways in the immune system, especially those involved in IgA production and Toll-like receptor signaling, were highlighted. These enrichments align with the literature on CHS’s impact on animal health (1, 12, 26, 70, 75, 76). The pathway of Salmonella infection consistently appears as one of the most enriched, reflecting the pathogen’s notoriety as a leading poultry-associated foodborne illness. This observation aligns with findings that heat stress may compromise the immune defenses, potentially increasing the risk of Salmonella infection in poultry, as noted in recent studies (31). The calcium signaling pathway, akin to the Salmonella infection pathway, has shown deep involvement in response to heat stress (Figures 3E, 5D, 7D). Elevated temperatures can increase membrane fluidity and permeability, leading to a calcium imbalance and the release of cytochrome c into the cytoplasm, triggering apoptosis through factors like caspases (63). Simultaneously, ROS generated under heat stress can inflict oxidative damage on enzymes responsible for muscle calcium regulation, further disturbing cellular homeostasis. As a result, high temperatures lead to reduced levels of calcium and phosphorus in the plasma of laying hens. These minerals are crucial for egg production and the quality of the eggshell (18, 104, 111).
In addition, this study presents, for the first time, a batch of miRNAs which play a role in CHS response of duck. In the foundational stages of inflammation, miR-146 is upregulated following exposure to lipopolysaccharide LPS, a bacterial element detected by TLR4. This triggers a series of events where NFκB migrates to the nucleus, initiating the expression of genes responsible for inflammatory mediators (112–114). Research has strengthened miR-146’s role in this pathway, underscoring its function in a negative feedback mechanism that tempers the inflammatory response initiated by TLR4 (115). MiR-146b-5p is typically upregulated in response to Salmonella enterica infection, playing a pivotal role in maintaining immune balance by dampening the initiation of the innate immune reaction (116, 117). This miRNA has been shown to facilitate replication of the Duck Tembusu virus through the suppression of the RPS14 gene, demonstrating its negative regulatory capacity in immune processes (118). miR-217 is noted for its regulatory influence on diverse muscle cell types by modulating critical genes, targeting ROCK1 in vascular smooth muscle cells and FGFR2 in skeletal muscle progenitors (119, 120). In the context of chicken liver cancer cells, miR-29a-3p responds to selenium levels and can instigate cell movement and invasion. It does this by focusing on the COL4A2 gene, leading to the suppression of the RhoA/ROCK signaling pathway (121). To date, the roles of miR-10926 and miR-17-1-3p remain unexplored, signaling the need for further investigation into their functions.
However, it’s important to acknowledge the limitations of this study. Firstly, we focused solely on morphological changes in lung tissue, and future research should incorporate biochemical tests of lung tissue and morphological tests of skin tissue, as skin is the largest heat dissipation system. Secondly, while the short-read transcriptome sequencing and data analysis provided valuable insights, they do not provide a complete view. The list of DEGs obtained here includes some unreported genes that likely play crucial roles in the CHS-induced response of ducks. These genes should be characterized in future studies. Furthermore, investigating the mechanisms that regulate the expression of these genes is essential, as is the use of other omics approaches (multi-omics studies). Additionally, the present study could benefit from additional examinations, such as assessing amino acid composition, nutritional value, and protein digestibility (122).
As a brief supplement, several strategies have been proposed to mitigate the adverse effects of CHS, such as improving housing, ventilation, and cooling systems (e.g., using little rearing systems and cage rearing systems) (123, 124), dietary supplementation (e.g., with vitamin A, vitamin C, vitamin E, Glutamine, and Herbs) (125–129), feed additives (e.g., probiotics, prebiotics, polyphenols, and palm oils) (130, 131), and other approaches (e.g., feed restrictions and genetic selection for heat tolerance) (132–136).
In essence, this study aims to delve into the effects of varying ambient temperatures on the growth performance of CVds and shed light on the underlying mechanisms responding to heat stress using cutting-edge high-throughput sequencing technologies. In conclusion, like other poultry, CVds are also sensitive to high ambient temperatures, and environmental control chambers offer advantages in improving their quality of life (20, 137, 138). Considering the predictions of continued climate change in most models (19, 139), further research is urgently needed to uncover the response mechanisms and regulatory networks of ducks to CHS.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was approved by Administration of Affairs Concerning Experimental Animals (Decree No. 63 of the Jiangsu Academy of Agricultural Science on 8 July 2014). The study was conducted in accordance with the local legislation and institutional requirements.
YL: Data curation, Formal analysis, Writing – original draft. DS: Resources, Writing – review & editing. CX: Resources, Visualization, Writing – review & editing. XL: Project administration, Resources, Writing – review & editing. MT: Supervision, Writing – review & editing. SY: Funding acquisition, Project administration, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32002235), Jiangsu Agricultural Science and Technology Innovation Fund (JASTIF) (CX (20) 2010), and China Agriculture Research System (CARS-42-38).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1417244/full#supplementary-material
1. Brugaletta, G, Teyssier, JR, Rochell, SJ, Dridi, S, and Sirri, F. A review of heat stress in chickens. Part I: insights into physiology and gut health. Front Physiol. (2022) 13:934381. doi: 10.3389/fphys.2022.934381
2. Qaid, MM, and Al-Garadi, MA. Protein and amino acid metabolism in poultry during and after heat stress: a review. Animals. (2021) 11:167. doi: 10.3390/ani11041167
3. Perini, F, Cendron, F, Rovelli, G, Castellini, C, Cassandro, M, and Lasagna, E. Emerging genetic tools to investigate molecular pathways related to heat stress in chickens: a review. Animals. (2020) 11:46. doi: 10.3390/ani11010046
4. Wasti, S, Sah, N, and Mishra, B. Impact of heat stress on poultry health and performances, and potential mitigation strategies. Animals. (2020) 10:1266. doi: 10.3390/ani10081266
5. Nawaz, AH, Amoah, K, Leng, QY, Zheng, JH, Zhang, WL, and Zhang, L. Poultry response to heat stress: its physiological, metabolic, and genetic implications on meat production and quality including strategies to improve broiler production in a warming world. Front Vet Sci. (2021) 8:699081. doi: 10.3389/fvets.2021.699081
6. Naranjo-Gomez, JS, Uribe-Garcia, HF, Herrera-Sanchez, MP, Lozano-Villegas, KJ, Rodriguez-Hernandez, R, and Rondon-Barragan, IS. Heat stress on cattle embryo: gene regulation and adaptation. Heliyon. (2021) 7:e06570. doi: 10.1016/j.heliyon.2021.e06570
7. Zheng, Y, Xie, T, Li, S, Wang, W, Wang, Y, Cao, Z, et al. Effects of selenium as a dietary source on performance, inflammation, cell damage, and reproduction of livestock induced by heat stress: a review. Front Immunol. (2021) 12:820853. doi: 10.3389/fimmu.2021.820853
8. Oladimeji, AM, Johnson, TG, Metwally, K, Farghly, M, and Mahrose, KM. Environmental heat stress in rabbits: implications and ameliorations. Int J Biometeorol. (2022) 66:1–11. doi: 10.1007/s00484-021-02191-0
9. Bohler, MW, Chowdhury, VS, Cline, MA, and Gilbert, ER. Heat stress responses in birds: a review of the neural components. Biology. (2021) 10:1095. doi: 10.3390/biology10111095
10. Ahmad, R, Yu, YH, Hsiao, FS, Su, CH, Liu, HC, Tobin, I, et al. Influence of heat stress on poultry growth performance, intestinal inflammation, and immune function and potential mitigation by probiotics. Animals. (2022) 12:2297. doi: 10.3390/ani12172297
11. Salem, HM, Alqhtani, AH, Swelum, AA, Babalghith, AO, Melebary, SJ, Soliman, SM, et al. Heat stress in poultry with particular reference to the role of probiotics in its amelioration: An updated review. J Therm Biol. (2022) 108:103302. doi: 10.1016/j.jtherbio.2022.103302
12. Liu, L, Ren, M, Ren, K, Jin, Y, and Yan, M. Heat stress impacts on broiler performance: a systematic review and meta-analysis. Poult Sci. (2020) 99:6205–11. doi: 10.1016/j.psj.2020.08.019
13. Gonzalez-Rivas, PA, Chauhan, SS, Ha, M, Fegan, N, Dunshea, FR, and Warner, RD. Effects of heat stress on animal physiology, metabolism, and meat quality: a review. Meat Sci. (2020) 162:108025. doi: 10.1016/j.meatsci.2019.108025
14. Uyanga, VA, Oke, EO, Amevor, FK, Zhao, J, Wang, X, Jiao, H, et al. Functional roles of taurine, L-theanine, L-citrulline, and betaine during heat stress in poultry. J Anim Sci Biotechnol. (2022) 13:23. doi: 10.1186/s40104-022-00675-6
15. Sun, PX, Shen, ZJ, Tang, J, Huang, W, Hou, SS, and Xie, M. Effects of ambient temperature on growth performance and carcass traits of male growing white Pekin ducks. Br Poult Sci. (2019) 60:513–6. doi: 10.1080/00071668.2019.1633011
16. Chen, S, Yong, Y, and Ju, X. Effect of heat stress on growth and production performance of livestock and poultry: mechanism to prevention. J Therm Biol. (2021) 99:103019. doi: 10.1016/j.jtherbio.2021.103019
17. Rostagno, MH. Effects of heat stress on the gut health of poultry. J Anim Sci. (2020) 98:98. doi: 10.1093/jas/skaa090
18. Zaboli, G, Huang, X, Feng, X, and Ahn, DU. How can heat stress affect chicken meat quality?- a review. Poult Sci. (2019) 98:1551–6. doi: 10.3382/ps/pey399
19. Kumar, M, Ratwan, P, Dahiya, SP, and Nehra, AK. Climate change and heat stress: impact on production, reproduction and growth performance of poultry and its mitigation using genetic strategies. J Therm Biol. (2021) 97:102867. doi: 10.1016/j.jtherbio.2021.102867
20. Vandana, GD, Sejian, V, Lees, AM, Pragna, P, Silpa, MV, and Maloney, SK. Heat stress and poultry production: impact and amelioration. Int J Biometeorol. (2021) 65:163–79. doi: 10.1007/s00484-020-02023-7
21. Abdel-Moneim, AE, Shehata, AM, Khidr, RE, Paswan, VK, Ibrahim, NS, El-Ghoul, AA, et al. Nutritional manipulation to combat heat stress in poultry – a comprehensive review. J Therm Biol. (2021) 98:102915. doi: 10.1016/j.jtherbio.2021.102915
22. Goel, A, Ncho, CM, and Choi, YH. Regulation of gene expression in chickens by heat stress. J Anim Sci Biotechnol. (2021) 12:11. doi: 10.1186/s40104-020-00523-5
23. Dai, X, Zhang, S, and Zaleta-Rivera, K. RNA: interactions drive functionalities. Mol Biol Rep. (2020) 47:1413–34. doi: 10.1007/s11033-019-05230-7
24. Goel, A. Heat stress management in poultry. J Anim Physiol Anim Nutr. (2021) 105:1136–45. doi: 10.1111/jpn.13496
25. Yang, C, Huang, XB, Chen, SJ, Li, XJ, Fu, XL, Xu, DN, et al. The effect of heat stress on proliferation, synthesis of steroids, and gene expression of duck granulosa cells. Anim Sci J. (2021) 92:e13617. doi: 10.1111/asj.13617
26. Zeng, T, Li, JJ, Wang, DQ, Li, GQ, Wang, GL, and Lu, LZ. Effects of heat stress on antioxidant defense system, inflammatory injury, and heat shock proteins of Muscovy and Pekin ducks: evidence for differential thermal sensitivities. Cell Stress Chaperones. (2014) 19:895–901. doi: 10.1007/s12192-014-0514-7
27. Ma, X, Lin, Y, Zhang, H, Chen, W, Wang, S, Ruan, D, et al. Heat stress impairs the nutritional metabolism and reduces the productivity of egg-laying ducks. Anim Reprod Sci. (2014) 145:182–90. doi: 10.1016/j.anireprosci.2014.01.002
28. Kim, JM, Lim, KS, Byun, M, Lee, KT, Yang, YR, Park, M, et al. Identification of the acclimation genes in transcriptomic responses to heat stress of white Pekin duck. Cell Stress Chaperones. (2017) 22:787–97. doi: 10.1007/s12192-017-0809-6
29. Yu, J, Bao, E, Yan, J, and Lei, L. Expression and localization of Hsps in the heart and blood vessel of heat-stressed broilers. Cell Stress Chaperones. (2008) 13:327–35. doi: 10.1007/s12192-008-0031-7
30. Zhang, Y, Dong, X, Hou, L, Cao, Z, Zhu, G, Vongsangnak, W, et al. Identification of differentially expressed non-coding RNA networks with potential Immunoregulatory roles during Salmonella enteritidis infection in ducks. Front Vet Sci. (2021) 8:692501. doi: 10.3389/fvets.2021.692501
31. Ricke, SC. Strategies to improve poultry food safety, a landscape review. Annual Review of Animal Biosciences. (2021) 9:379–400. doi: 10.1146/annurev-animal-061220-023200
32. Cheng, S, He, Y, Zeng, T, Wang, D, He, J, Xia, Q, et al. Heat stress induces various oxidative damages to myofibrillar proteins in ducks. Food Chem. (2022) 390:133209. doi: 10.1016/j.foodchem.2022.133209
33. Shakeri, M, Oskoueian, E, Le, HH, and Shakeri, M. Strategies to combat heat stress in broiler chickens: unveiling the roles of selenium, vitamin E and vitamin C. Vet Sci. (2020) 7:71. doi: 10.3390/vetsci7020071
34. Sun, D, Xu, C, Liu, Y, Dai, Z, Pan, Z, Chen, R, et al. The impact of different relative humidity levels on the production performance, slaughter performance, and meat quality of white Pekin ducks aged 4 to 42 days. Animals. (2023) 13:3711. doi: 10.3390/ani13233711
35. Cao, Z, Gao, W, Zhang, Y, Huo, W, Weng, K, Zhang, Y, et al. Effect of marketable age on proximate composition and nutritional profile of breast meat from Cherry Valley broiler ducks. Poult Sci. (2021) 100:101425. doi: 10.1016/j.psj.2021.101425
36. Kim, D, Langmead, B, and Salzberg, SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
37. Li, J, Zhang, J, Liu, J, Zhou, Y, Cai, C, Xu, L, et al. A new duck genome reveals conserved and convergently evolved chromosome architectures of birds and mammals. Gigascience. (2021) 10:giaa142. doi: 10.1093/gigascience/giaa142
38. Pertea, M, Pertea, GM, Antonescu, CM, Chang, TC, Mendell, JT, and Salzberg, SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. (2015) 33:290–5. doi: 10.1038/nbt.3122
39. Li, B, and Dewey, CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. (2011) 12:323. doi: 10.1186/1471-2105-12-323
40. Love, MI, Huber, W, and Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
41. Klopfenstein, DV, Zhang, L, Pedersen, BS, Ramirez, F, Warwick Vesztrocy, A, Naldi, A, et al. GOATOOLS: a python library for gene ontology analyses. Sci Rep. (2018) 8:10872. doi: 10.1038/s41598-018-28948-z
42. Xie, C, Mao, X, Huang, J, Ding, Y, Wu, J, Dong, S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. (2011) 39:W316–22. doi: 10.1093/nar/gkr483
43. Szklarczyk, D, Gable, AL, Lyon, D, Junge, A, Wyder, S, Huerta-Cepas, J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47:D607–13. doi: 10.1093/nar/gky1131
44. Chin, CH, Chen, SH, Wu, HH, Ho, CW, Ko, MT, and Lin, CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. (2014) 8:S11. doi: 10.1186/1752-0509-8-s4-s11
45. Shannon, P, Markiel, A, Ozier, O, Baliga, NS, Wang, JT, Ramage, D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
46. Maere, S, Heymans, K, and Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. (2005) 21:3448–9. doi: 10.1093/bioinformatics/bti551
47. Wu, T, Hu, E, Xu, S, Chen, M, Guo, P, Dai, Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. (2021) 2:100141. doi: 10.1016/j.xinn.2021.100141
48. Sun, L, Luo, H, Bu, D, Zhao, G, Yu, K, Zhang, C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. (2013) 41:e166. doi: 10.1093/nar/gkt646
49. Kong, L, Zhang, Y, Ye, ZQ, Liu, XQ, Zhao, SQ, Wei, L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. (2007) 35:W345–9. doi: 10.1093/nar/gkm391
50. Wang, L, Park, HJ, Dasari, S, Wang, S, Kocher, JP, and Li, W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. (2013) 41:e74. doi: 10.1093/nar/gkt006
51. Fu, XZ, Zhang, XY, Qiu, JY, Zhou, X, Yuan, M, He, YZ, et al. Whole-transcriptome RNA sequencing reveals the global molecular responses and ceRNA regulatory network of mRNAs, lncRNAs, miRNAs and circRNAs in response to copper toxicity in Ziyang Xiangcheng (Citrus junos Sieb. Ex Tanaka). BMC Plant Biol. (2019) 19:509. doi: 10.1186/s12870-019-2087-1
52. Quinlan, AR, and Hall, IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. (2010) 26:841–2. doi: 10.1093/bioinformatics/btq033
53. Friedlander, MR, Mackowiak, SD, Li, N, Chen, W, and Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. (2012) 40:37–52. doi: 10.1093/nar/gkr688
54. Enright, AJ, John, B, Gaul, U, Tuschl, T, Sander, C, and Marks, DS. MicroRNA targets in Drosophila. Genome Biol. (2003) 5:R1. doi: 10.1186/gb-2003-5-1-r1
55. Wu, HJ, Ma, YK, Chen, T, Wang, M, and Wang, XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. (2012) 40:W22–8. doi: 10.1093/nar/gks554
56. Ritchie, ME, Phipson, B, Wu, D, Hu, Y, Law, CW, Shi, W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
57. Agarwal, V, Bell, GW, Nam, JW, and Bartel, DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. (2015) 4:4. doi: 10.7554/eLife.05005
58. Mueller, RC, Ellstrom, P, Howe, K, Uliano-Silva, M, Kuo, RI, Miedzinska, K, et al. A high-quality genome and comparison of short- versus long-read transcriptome of the palaearctic duck Aythya fuligula (tufted duck). Gigascience. (2021) 10:giab081. doi: 10.1093/gigascience/giab081
59. Farghly, MFA, Abd El-Hack, ME, Alagawany, M, Saadeldin, IM, and Swelum, AA. Ameliorating deleterious effects of heat stress on growing Muscovy ducklings using feed withdrawal and cold water. Poult Sci. (2019) 98:251–9. doi: 10.3382/ps/pey396
60. Oluwagbenga, EM, Tetel, V, Schober, J, and Fraley, GS. Chronic heat stress part 1: decrease in egg quality, increase in cortisol levels in egg albumen, and reduction in fertility of breeder Pekin ducks. Front Physiol. (2022) 13:1019741. doi: 10.3389/fphys.2022.1019741
61. Luo, X, Zheng, C, Xia, W, Ruan, D, Wang, S, Cui, Y, et al. Effects of constant or intermittent high temperature on egg production, feed intake, and hypothalamic expression of antioxidant and pro-oxidant enzymes genes in laying ducks. J Anim Sci. (2018) 96:5064–74. doi: 10.1093/jas/sky355
62. Chen, XL, Zeng, YB, Liu, LX, Song, QL, Zou, ZH, Wei, QP, et al. Effects of dietary chromium propionate on laying performance, egg quality, serum biochemical parameters and antioxidant status of laying ducks under heat stress. Animal. (2021) 15:100081. doi: 10.1016/j.animal.2020.100081
63. He, J, Xia, C, He, Y, Pan, D, Cao, J, Sun, Y, et al. Proteomic responses to oxidative damage in meat from ducks exposed to heat stress. Food Chem. (2019) 295:129–37. doi: 10.1016/j.foodchem.2019.05.073
64. He, Y, Zhou, M, Xia, C, Xia, Q, He, J, Cao, J, et al. Volatile flavor changes responding to heat stress-induced lipid oxidation in duck meat. Anim Sci J. (2020) 91:e13461. doi: 10.1111/asj.13461
65. Farghly, MFA, Abd El-Hack, ME, Alagawany, M, Saadeldin, IM, and Swelum, AA. Wet feed and cold water as heat stress modulators in growing Muscovy ducklings. Poult Sci. (2018) 97:1588–94. doi: 10.3382/ps/pey006
66. Mashaly, MM, Hendricks, GL 3rd, Kalama, MA, Gehad, AE, Abbas, AO, and Patterson, PH. Effect of heat stress on production parameters and immune responses of commercial laying hens. Poult Sci. (2004) 83:889–94. doi: 10.1093/ps/83.6.889
67. Hashizawa, Y, Kubota, M, Kadowaki, M, and Fujimura, S. Effect of dietary vitamin E on broiler meat qualities, color, water-holding capacity and shear force value, under heat stress conditions. Anim Sci J. (2013) 84:732–6. doi: 10.1111/asj.12079
68. Akbarian, A, Michiels, J, Degroote, J, Majdeddin, M, Golian, A, and De Smet, S. Association between heat stress and oxidative stress in poultry; mitochondrial dysfunction and dietary interventions with phytochemicals. J Anim Sci Biotechnol. (2016) 7:37. doi: 10.1186/s40104-016-0097-5
69. Lian, P, Braber, S, Garssen, J, Wichers, HJ, Folkerts, G, Fink-Gremmels, J, et al. Beyond heat stress: intestinal integrity disruption and mechanism-based intervention strategies. Nutrients. (2020) 12:734. doi: 10.3390/nu12030734
70. Siddiqui, SH, Kang, D, Park, J, Khan, M, and Shim, K. Chronic heat stress regulates the relation between heat shock protein and immunity in broiler small intestine. Sci Rep. (2020) 10:18872. doi: 10.1038/s41598-020-75885-x
71. Huang, C, Jiao, H, Song, Z, Zhao, J, Wang, X, and Lin, H. Heat stress impairs mitochondria functions and induces oxidative injury in broiler chickens. J Anim Sci. (2021) 93:2144–53. doi: 10.2527/jas.2014-8739
72. Quinteiro-Filho, WM, Gomes, AV, Pinheiro, ML, Ribeiro, A, Ferraz-de-Paula, V, Astolfi-Ferreira, CS, et al. Heat stress impairs performance and induces intestinal inflammation in broiler chickens infected with Salmonella enteritidis. Avian Pathol. (2012) 41:421–7. doi: 10.1080/03079457.2012.709315
73. Kim, WS, Ghassemi Nejad, J, Roh, SG, and Lee, HG. Heat-shock proteins gene expression in peripheral blood mononuclear cells as an Indicator of heat stress in beef calves. Animals. (2020) 10:895. doi: 10.3390/ani10050895
74. Hosseindoust, AR, Lee, SH, Kim, JS, Choi, YH, Kwon, IK, and Chae, BJ. Productive performance of weanling piglets was improved by administration of a mixture of bacteriophages, targeted to control coliforms and Clostridium spp. shedding in a challenging environment. J Anim Physiol Anim Nutr. (2017) 101:e98–e107. doi: 10.1111/jpn.12567
75. Cui, Y, and Gu, X. Proteomic changes of the porcine small intestine in response to chronic heat stress. J Mol Endocrinol. (2015) 55:277–93. doi: 10.1530/JME-15-0161
76. Lambert, GP. Stress-induced gastrointestinal barrier dysfunction and its inflammatory effects. J Anim Sci. (2009) 87:E101–8. doi: 10.2527/jas.2008-1339
77. Yang, C, Luo, P, Chen, SJ, Deng, ZC, Fu, XL, Xu, DN, et al. Resveratrol sustains intestinal barrier integrity, improves antioxidant capacity, and alleviates inflammation in the jejunum of ducks exposed to acute heat stress. Poult Sci. (2021) 100:101459. doi: 10.1016/j.psj.2021.101459
78. Li, Z, Sun, Y, He, M, and Liu, J. Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing. Bioengineered. (2021) 12:1026–39. doi: 10.1080/21655979.2021.1899533
79. Wang, B, Zhang, J, Chitrakar, B, Wang, Y, Xu, T, and Zhou, C. Preservation of duck eggs through glycerol monolaurate nanoemulsion coating. Curr Res Food Sci. (2021) 4:752–7. doi: 10.1016/j.crfs.2021.10.008
80. Sun, J, Wang, J, Lin, W, Li, B, Ma, R, Huang, Y, et al. Predict the gelling properties of alkali-induced egg white gel based on the freshness of duck eggs. Food Secur. (2023) 12:4028. doi: 10.3390/foods12214028
81. Cui, H, Yang, H, Abdel-Samie, MA, Siva, S, and Lin, L. Controlled-release casein/cinnamon essential oil nanospheres for the inactivation of Campylobacter jejuni in duck. Int J Food Microbiol. (2021) 341:109074. doi: 10.1016/j.ijfoodmicro.2021.109074
82. Sun, L, Feng, S, Chen, C, Liu, X, and Cai, J. Identification of eggshell crack for hen egg and duck egg using correlation analysis based on acoustic resonance method. J Food Process Eng. (2020) 43:13430. doi: 10.1111/jfpe.13430
83. Xu, F, Huang, X, Tian, X, Yu, S, Zhang, X, and Zareef, M. Application of hyperspectral imaging and colorimetric sensor array coupled with multivariate analysis for quality detection during salted duck eggs processing. J Food Process Eng. (2024) 47:14589. doi: 10.1111/jfpe.14589
84. Gu, T, Lu, L, Xu, W, Zeng, T, Tian, Y, Chen, B, et al. Immunopotentiators improve the antioxidant defense, apoptosis, and immune response in Shaoxing ducklings. Poult Sci. (2022) 101:101641. doi: 10.1016/j.psj.2021.101641
85. An, H, Liu, Y, Fang, L, Shu, M, Zhai, Q, and Chen, J. Placenta-specific 8 facilitates the infection of duck hepatitis a virus type 1 by inhibiting the TLR7 MyD88-dependent signaling pathway. Poult Sci. (2023) 102:102724. doi: 10.1016/j.psj.2023.102724
86. Tosi, G, Forlani, G, Andresen, V, Turci, M, Bertazzoni, U, Franchini, G, et al. Major histocompatibility complex class II transactivator CIITA is a viral restriction factor that targets human T-cell lymphotropic virus type 1 Tax-1 function and inhibits viral replication. J Virol. (2011) 85:10719–29. doi: 10.1128/JVI.00813-11
87. Li, R, Guo, M, Lin, J, Chai, T, and Wei, L. Molecular cloning, characterization, and anti-avian pathogenic Escherichia coli innate immune response of the Cherry Valley duck CIITA gene. Front Microbiol. (2017) 8:1629. doi: 10.3389/fmicb.2017.01629
88. Francoeur, L, Scoville, DM, and Johnson, PA. Effect of IGF1 and FSH on the function of granulosa cells from pre-hierarchal follicles in chickens dagger. Biol Reprod. (2023) 109:498–506. doi: 10.1093/biolre/ioad082
89. Wang, WJ, Guo, YQ, Xie, KJ, Li, YD, Li, ZW, Wang, N, et al. A functional variant in the promoter region of IGF1 gene is associated with chicken abdominal fat deposition. Domest Anim Endocrinol. (2021) 75:106584. doi: 10.1016/j.domaniend.2020.106584
90. Gong, Y, Yang, J, Liu, Q, Cai, J, Zheng, Y, Zhang, Y, et al. IGF1 knockdown hinders myocardial development through energy metabolism dysfunction caused by ROS-dependent FOXO activation in the chicken heart. Oxidative Med Cell Longev. (2019) 2019:1–31. doi: 10.1155/2019/7838754
91. Zhou, H, Mitchell, AD, McMurtry, JP, Ashwell, CM, and Lamont, SJ. Insulin-like growth factor-I gene polymorphism associations with growth, body composition, skeleton integrity, and metabolic traits in chickens. Poult Sci. (2005) 84:212–9. doi: 10.1093/ps/84.2.212
92. Li, X, Qiu, J, Liu, H, Deng, Y, Hu, S, Hu, J, et al. MicroRNA-33a negatively regulates myoblast proliferation by targeting IGF1, follistatin and cyclin D1. Biosci Rep. (2020) 40:BSR20191327. doi: 10.1042/BSR20191327
93. Lin, J, Guan, L, Ge, L, Liu, G, Bai, Y, and Liu, X. Nanopore-based full-length transcriptome sequencing of Muscovy duck (Cairina moschata) ovary. Poult Sci. (2021) 100:101246. doi: 10.1016/j.psj.2021.101246
94. Bhattacharya, TK, Chatterjee, RN, Dushyanth, K, Paswan, C, Shukla, R, and Shanmugam, M. Polymorphism and expression of insulin-like growth factor 1 (IGF1) gene and its association with growth traits in chicken. Br Poult Sci. (2015) 56:398–407. doi: 10.1080/00071668.2015.1041098
95. Hajihoseini, Z, and Eghbalsaied, S. Simultaneous effects of IGF1 and Fadrozole on parthenogenesis and pluripotency markers in chicken embryo. Theriogenology. (2018) 114:317–23. doi: 10.1016/j.theriogenology.2018.04.009
96. Ogunpaimo, OJ, Ojoawo, HT, Wheto, MY, Adebambo, AO, and Adebambo, OA. Association of insulin-like growth factor 1 (IGF1) gene polymorphism with the reproductive performance of three dual-purpose chicken breeds. Transl Anim Sci. (2021) 5:txab215. doi: 10.1093/tas/txab215
97. Hosnedlova, B, Vernerova, K, Kizek, R, Bozzi, R, Kadlec, J, Curn, V, et al. Associations between IGF1, IGFBP2 and TGFss3 genes polymorphisms and growth performance of broiler chicken lines. Animals. (2020) 10:800. doi: 10.3390/ani10050800
98. Ye, Q, Xu, J, Gao, X, Ouyang, H, Luo, W, and Nie, Q. Associations of IGF2 and DRD2 polymorphisms with laying traits in Muscovy duck. PeerJ. (2017) 5:e4083. doi: 10.7717/peerj.4083
99. Zhao, J, Zhao, X, Shen, X, Zhang, Y, Zhang, Y, Ye, L, et al. CircCCDC91 regulates chicken skeletal muscle development by sponging miR-15 family via activating IGF1-PI3K/AKT signaling pathway. Poult Sci. (2022) 101:101803. doi: 10.1016/j.psj.2022.101803
100. Rahaie, A, Toghyani, M, and Eghbalsaied, S. Co-treatment of IGF1 and Fadrozole upregulates the expression of RSPO1, SOX9, and AMH in chicken embryos. Cells Tissues Organs. (2018) 206:218–28. doi: 10.1159/000499079
101. Zhao, Y, Zou, M, Sun, Y, Zhang, K, and Peng, X. Gga-miR-21 modulates Mycoplasma gallisepticum (HS strain)-induced inflammation via targeting MAP3K1 and activating MAPKs and NF-kappaB pathways. Vet Microbiol. (2019) 237:108407. doi: 10.1016/j.vetmic.2019.108407
102. Xie, HL, Zhang, YH, Tan, XD, Zheng, Y, Ni, HY, Dong, LP, et al. miR-375 induced the formation and transgenerational inheritance of fatty liver in poultry by targeting MAP3K1. DNA Cell Biol. (2022) 41:590–9. doi: 10.1089/dna.2022.0078
103. Pesti, D, Hasler-Rapacz, J, Rapacz, J, and McGibbon, WH. Immunogenetic studies on low-density lipoprotein allotypes in chickens (Lcp1 and Lcp2). Poult Sci. (1981) 60:295–301. doi: 10.3382/ps.0600295
104. Tan, GH, Li, JZ, Zhang, YY, You, MF, Liao, CM, and Zhang, YG. Association of PRKCA expression and polymorphisms with layer duck eggshell quality. Br Poult Sci. (2021) 62:8–16. doi: 10.1080/00071668.2020.1817329
105. Lin, L, Ye, K, Chen, F, Xie, J, Chen, Z, and Xu, Y. Identification of new immune subtypes of renal injury associated with anti-neutrophil cytoplasmic antibody-associated vasculitis based on integrated bioinformatics analysis. Front Genet. (2023) 14, 14:1119017. doi: 10.3389/fgene.2023.1119017
106. Habauzit, D, Le Quement, C, Zhadobov, M, Martin, C, Aubry, M, Sauleau, R, et al. Transcriptome analysis reveals the contribution of thermal and the specific effects in cellular response to millimeter wave exposure. PLoS One. (2014) 9:e109435. doi: 10.1371/journal.pone.0109435
107. Zhou, Y, Zhou, Y, Kang, X, Meng, C, Zhang, R, Guo, Y, et al. Molecular cloning and functional characterisation of duck (Anas platyrhynchos) tumour necrosis factor receptor-associated factor 3. Br Poult Sci. (2019) 60:357–65. doi: 10.1080/00071668.2019.1614528
108. Cai, W, Pan, Y, Luo, W, Cheng, A, Wang, M, Chen, S, et al. NS5 hijacks TRAF3 to inhibit type I interferon signaling during duck Tembusu virus infection. Vet Microbiol. (2023) 286:109894. doi: 10.1016/j.vetmic.2023.109894
109. Xu, W, Zhang, L, Ma, S, Zhang, Y, Cai, Z, Zhang, K, et al. TRAF5 protects against myocardial ischemia reperfusion injury via AKT signaling. Eur J Pharmacol. (2020) 878:173092. doi: 10.1016/j.ejphar.2020.173092
110. Tian, H, Ding, M, Guo, Y, Zhu, Z, Yu, Y, Tian, Y, et al. Effect of HSPA8 gene on the proliferation, apoptosis and immune function of HD11 cells. Dev Comp Immunol. (2023) 142:104666. doi: 10.1016/j.dci.2023.104666
111. Scott, TA, and Balnave, D. Comparison between concentrated complete diets and self-selection for feeding sexually-maturing pullets at hot and cold temperatures. Br Poult Sci. (1988) 29:613–26. doi: 10.1080/00071668808417088
112. Testa, U, Pelosi, E, Castelli, G, and Labbaye, C. miR-146 and miR-155: two key modulators of immune response and tumor development. Noncoding RNA. (2017) 3:22. doi: 10.3390/ncrna3030022
113. Schulte, LN, Westermann, AJ, and Vogel, J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. (2013) 41:542–53. doi: 10.1093/nar/gks1030
114. Laanesoo, A, Urgard, E, Periyasamy, K, Laan, M, Bochkov, YA, Aab, A, et al. Dual role of the miR-146 family in rhinovirus-induced airway inflammation and allergic asthma exacerbation. Clin Transl Med. (2021) 11:e427. doi: 10.1002/ctm2.427
115. Lee, HM, Kim, TS, and Jo, EK. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. (2016) 49:311–8. doi: 10.5483/bmbrep.2016.49.6.056
116. Lee, JH, Kim, SW, Han, JS, Shin, SP, Lee, SI, and Park, TS. Functional analyses of miRNA-146b-5p during myogenic proliferation and differentiation in chicken myoblasts. BMC Mol Cell Biol. (2020) 21, 21:40. doi: 10.1186/s12860-020-00284-z
117. Hu, G, Liu, L, Miao, X, Zhao, Y, and Li, X. Research note: IsomiRs of chicken miR-146b-5p are activated upon Salmonella enterica serovar Enteritidis infection. Poult Sci. (2022) 101:101977. doi: 10.1016/j.psj.2022.101977
118. Huang, J, Lei, L, Cui, M, Cheng, A, Wang, M, Liu, M, et al. miR-146b-5p promotes duck Tembusu virus replication by targeting RPS14. Poult Sci. (2023) 102:102890. doi: 10.1016/j.psj.2023.102890
119. Zhou, W, Ye, S, and Wang, W. miR-217 alleviates high-glucose-induced vascular smooth muscle cell dysfunction via regulating ROCK1. J Biochem Mol Toxicol. (2021) 35:e22668. doi: 10.1002/jbt.22668
120. Zhu, M, Chen, G, Yang, Y, Yang, J, Qin, B, and Gu, L. miR-217-5p regulates myogenesis in skeletal muscle stem cells by targeting FGFR2. Mol Med Rep. (2020) 22:850–8. doi: 10.3892/mmr.2020.11133
121. Hu, X, Tan, S, Yin, H, Khoso, PA, Xu, Z, and Li, S. Selenium-mediated gga-miR-29a-3p regulates LMH cell proliferation, invasion, and migration by targeting COL4A2. Metallomics. (2020) 12:449–59. doi: 10.1039/c9mt00266a
122. Chowdhury, VS, Han, G, Eltahan, HM, Haraguchi, S, Gilbert, ER, Cline, MA, et al. Potential role of amino acids in the adaptation of chicks and market-age broilers to heat stress. Front Vet Sci. (2020) 7:610541. doi: 10.3389/fvets.2020.610541
123. Abo Ghanima, MM, Abd El-Hack, ME, Othman, SI, Taha, AE, Allam, AA, and Eid Abdel-Moneim, AM. Impact of different rearing systems on growth, carcass traits, oxidative stress biomarkers, and humoral immunity of broilers exposed to heat stress. Poult Sci. (2020) 99:3070–8. doi: 10.1016/j.psj.2020.03.011
124. Farghly, MFA, Mahrose, KM, Cooper, RG, Ullah, Z, Rehman, Z, and Ding, C. Sustainable floor type for managing Turkey production in a hot climate. Poult Sci. (2018) 97:3884–90. doi: 10.3382/ps/pey280
125. Habibian, M, Ghazi, S, and Moeini, MM. Effects of dietary selenium and vitamin E on growth performance, meat yield, and selenium content and lipid oxidation of breast meat of broilers reared under heat stress. Biol Trace Elem Res. (2016) 169:142–52. doi: 10.1007/s12011-015-0404-6
126. Wu, QJ, Liu, N, Wu, XH, Wang, GY, and Lin, L. Glutamine alleviates heat stress-induced impairment of intestinal morphology, intestinal inflammatory response, and barrier integrity in broilers. Poult Sci. (2018) 97:2675–83. doi: 10.3382/ps/pey123
127. Abd El-Hack, ME, Alagawany, M, and Noreldin, AE. Managerial and nutritional trends to mitigate heat stress risks in poultry farms In: Abdel-Wahab RK, Hikal, WM, editors. Sustainability of Agricultural Environment in Egypt: Part II : The Handbook of Environmental Chemistry. Berlin, Germany: Springer (2018). 325–38.
128. Kucuk, O, Sahin, N, and Sahin, K. Supplemental zinc and vitamin a can alleviate negative effects of heat stress in broiler chickens. Biol Trace Elem Res. (2021) 94:225–36. doi: 10.1385/BTER:94:3:225
129. Sahin, K, Smith, MO, Onderci, M, Sahin, N, Gursu, MF, and Kucuk, O. Supplementation of zinc from organic or inorganic source improves performance and antioxidant status of heat-distressed quail. Poult Sci. (2005) 84:882–7. doi: 10.1093/ps/84.6.882
130. Yadav, S, and Jha, R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J Anim Sci Biotechnol. (2019) 10:2. doi: 10.1186/s40104-018-0310-9
131. Song, J, Xiao, K, Ke, YL, Jiao, LF, Hu, CH, Diao, QY, et al. Effect of a probiotic mixture on intestinal microflora, morphology, and barrier integrity of broilers subjected to heat stress. Poult Sci. (2014) 93:581–8. doi: 10.3382/ps.2013-03455
132. Renaudeau, D, Collin, A, Yahav, S, de Basilio, V, Gourdine, JL, and Collier, RJ. Adaptation to hot climate and strategies to alleviate heat stress in livestock production. Animal. (2012) 6:707–28. doi: 10.1017/S1751731111002448
133. Duah, KK, Essuman, EK, Boadu, VG, Olympio, OS, and Akwetey, W. Comparative study of indigenous chickens on the basis of their health and performance. Poult Sci. (2020) 99:2286–92. doi: 10.1016/j.psj.2019.11.049
134. Ouyang, J-h, Xie, L, Nie, Q-h, Zeng, H, Peng, Z-j, Zhang, D-x, et al. The effects of different sex-linked dwarf variations on Chinese native chickens. J Integr Agric. (2012) 11:1500–8. doi: 10.1016/s2095-3119(12)60150-6
135. Yunis, R, and Cahaner, A. The effects of the naked neck (Na) and frizzle (F) genes on growth and meat yield of broilers and their interactions with ambient temperatures and potential growth rate. Poult Sci. (1999) 78:1347–52. doi: 10.1093/ps/78.10.1347
136. Dong, J, He, C, Wang, Z, Li, Y, Li, S, Tao, L, et al. A novel deletion in KRT75L4 mediates the frizzle trait in a Chinese indigenous chicken. Genet Sel Evol. (2018) 50:68. doi: 10.1186/s12711-018-0441-7
137. Bilal, RM, Hassan, FU, Farag, MR, Nasir, TA, Ragni, M, Mahgoub, HAM, et al. Thermal stress and high stocking densities in poultry farms: potential effects and mitigation strategies. J Therm Biol. (2021) 99:102944. doi: 10.1016/j.jtherbio.2021.102944
138. Fathi, MM, Galal, A, Radwan, LM, Abou-Emera, OK, and Al-Homidan, IH. Using major genes to mitigate the deleterious effects of heat stress in poultry: an updated review. Poult Sci. (2022) 101:102157. doi: 10.1016/j.psj.2022.102157
Keywords: chronic heat stress, duck rearing, environmental temperature, high-throughput sequencing technology, ceRNA
Citation: Liu Y, Sun D, Xu C, Liu X, Tang M and Ying S (2024) In-depth transcriptome profiling of Cherry Valley duck lungs exposed to chronic heat stress. Front. Vet. Sci. 11:1417244. doi: 10.3389/fvets.2024.1417244
Received: 14 April 2024; Accepted: 08 July 2024;
Published: 22 July 2024.
Edited by:
Toshiro Arai, Nippon Veterinary and Life Science University, JapanReviewed by:
Jingjing Ling, Elanco, United StatesCopyright © 2024 Liu, Sun, Xu, Liu, Tang and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijia Ying, MjUxNTEwNjUwQHFxLmNvbQ==; Min Tang, bXQzMTM4QHVqcy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.