
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 18 July 2024
Sec. Veterinary Infectious Diseases
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1415685
This article is part of the Research Topic Bacteriophages, a weapon against animal bacterial pathogens and biofilms View all 4 articles
 Yuzhi Wang1†
Yuzhi Wang1† Guixiang Tong2†
Guixiang Tong2† Xinglong Jiang3
Xinglong Jiang3 Chuandeng Tu4
Chuandeng Tu4 Hongjiao Cai1
Hongjiao Cai1 Wenhong Fang5
Wenhong Fang5 Honglian Tan2
Honglian Tan2 Qibiao Weng6
Qibiao Weng6 Xinxian Wei2*‡
Xinxian Wei2*‡ Mao Lin1,3*‡
Mao Lin1,3*‡Introduction: Aeromonas hydrophila is particularly harmful to freshwater aquaculture, and the search for phage is an effective biological control method, but reports of possible temperate phages and their mutants are rare in this field. In this study, a virulent phage highly homologous to prophage in the genomes of A. hydrophila was collected and preliminary biological characterization was carried out to understand its nature.
Materials and methods: Water samples taken from eel ponds in Fujian, China were combined with the strain. Spot test method and double-layer agar plate assay was used for confirmation and purification. Phage virions were observed using transmission electron microscope. A total of 68 strains of Aeromonas spp. were used to determine the host range. MOI groups of 1,000, 100, 10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001 were prepared to detect the optimal MOI. The conditions of thermal stability assay were set as 30, 40, 50, 60, 70 and 80°C for 1 h, respectively, and conditions of acid and alkali stability assay were set as 2.0, 4.0, 6.0, 8.0, 10.0 and 12.0 of pH. MOI of 0.01 and 0.1, respectively, are set to determine the inhibitory capacity of phage.
Results: A novel virulent A. hydrophila phage designated phiA051 has been isolated from aquaculture water. Electron microscopic observation showed that the phage phiA051 was composed of an icosahedral capsid. The phage phiA051 possesses an optimal multiplicity of infection (MOI) of 0.01, and its burst size was 108 PFU/cell. The phage maintained a high viability at temperatures of 30–50°C or pH 6.0–10.0 for 1 h. Phage phiA051 has certain potentials in rapidly inhibiting the spread of pathogen early in the outbreak, and it has a linear dsDNA with GC content of 60.55% and a total length of 32,212 bp, including 46 ORFs.
Discussion: The phage phiA051 behaved as a virulent phage. However, the BLASTN result showed that 23 of the top 25 hits were genomes of Aeromonas strains. It was suggested that phiA051 was probably derived from some prophage in the chromosome of Aeromonas. Further investigation of the mechanism how phage phiA051 transforms from a temperate phage to a virulent phage will provide a unique perspective and idea to explore the potential of prophages.
Diseases caused by bacterial infections are one of the major threats to the health of animals and human beings (1, 2). Antibiotics have been extensively used to control bacterial infections for a long time. However, the emergence of antimicrobial-resistant bacteria has greatly reduced their effectiveness and led to numerous deaths. Therefore, researchers have been committed to finding alternative therapies to antibiotics (3, 4). One potential option is the use of phages, which are viruses that exclusively infect bacteria (5). Phages are categorized into virulent and temperate phages based on the difference in their lysis cycle. Virulent phages do not enter a stage of integration with the host bacterial genome, while temperate phages integrate their genome into the host bacterial chromosome and may re-enter the lytic cycle under certain conditions. The integrated DNA is referred to as a prophage. Temperate phages are difficult to apply clinically as they are unable to kill bacteria stably, and the integration of them can lead to horizontal gene transfer, including transfer of antibiotic resistance genes or virulence factors. Such gene transfer could inadvertently increase pathogenicity or resistance of bacterial population, so only virulent phages are prioritized for screening and consideration for phage therapy (6–12).
Aeromonas hydrophila, a Gram-negative facultative anaerobic bacterium, is a common pathogen of freshwater farmed animals (13–16). It is pathogenic to a wide range of fish, amphibians and reptiles and can cause systemic and ulcerative infections, including septicemia, gill rot and kidney disease (17–19). In addition, it can infect terrestrial animals and even humans (20–22). Given the effectiveness of phage therapy against bacteria (8, 10, 12, 23, 24), some researchers have also carried out studies on Aeromonas phages, mainly focusing on the elaboration of phage screening and bactericidal capabilities, for example the host ranges of several virulent Aeromonas sp. phages were delineated (25–27), and methodologies for the augmentation of aquatic phages have also been investigated by researchers (9, 28–32). Despite a number of studies, the amount of phage species that have been discovered and studied remain relatively small. There are currently 322 genomes of A. hydrophila strains in the National Center for Biotechnology Information (NCBI) database, whereas there are only 300 genome records for phages targeting this bacterium, and even fewer phages with documented taxonomic status. Phages of A. hydrophila are now known to belong mostly to the families Autographiviridae, Chaseviridae, Demerecviridae, Ackermannviridae, Straboviridae, Casjensviridae and Peduoviridae. The continued exploration of phage bioresources is necessary due to the limitations of many known phage species for applications such as rather narrow lysis spectrum, low lysis volumes and short inhibition times (8, 12, 33–35). Since it has been shown in recent years that some virulent phages can be obtained by gene editing of temperate phages (36, 37), it has become very important to study the mechanism of natural and artificial transitions from temperate to virulent phages, which can not only help people to gain a deeper understanding of the survival mode and properties of phages, but also help them to obtain potentially virulent phages as much as possible by means of mutation.
In this study, a virulent phage with high homology to a series of prophages was obtained during the screening of virulent phage targeting A. hydrophila. This phage is not capable of lysogeny switching, and from this it should be considered as a virulent phage. However, during the later genome sequence comparison we found that this phage has a high degree of homology with many Aeromonas prophage genomes, suggesting that this phage may originate from a temperate phage, but the mechanism of this transition is not clear. In the future it can be an important material to study the evolution of temperate phage into virulent phage.
Water samples were taken from eel ponds in Fujian, China, aiming to detect phages against A. hydrophila A051, a strain isolated from diseased eels (38). The host A051 was inoculated into 100 mL of LB broth and incubated at 30°C, 150 rpm for 12 h to logarithmic phase, at which time the concentration of the bacterial suspension was approximately 109 CFU/mL. Water samples (1 L) were combined with 50 mL culture of the strain A051 and 800 mL of Luria–Bertani (LB) broth, then the mixture was cultured for 24 h at 30°C with intermittent stirring to enrich possible phages. Following this, 10 mL of the mixture was centrifuged for 5 min at 4°C, 10,000 rpm. The supernatant was filtered with a 0.22 μm filter membrane, and spot test method was applied to seek out phages according to the method from Zhang et al. (39). Once a phage plaque had formed, a double-layer agar plate assay was used for confirmation and purification of the phage according to the method from Ye et al. (38). Purification procedure was repeated 5 times. Purified phage was stored at 4°C.
The purified phage stock (approximately 1.0 × 109 PFU/mL) was placed on paraffin film with a copper mesh and left for 30 min. The film was negatively stained with 20 μL 1% (w/w) phosphotungstic acid solution for 3 min subsequently. Excess filtrate and staining solution were then removed. Phage virions were observed using transmission electron microscope (JEOL Co., Tokyo, Japan) and the head length, head width and tail length were measured.
A total of 68 strains of Aeromonas spp. respectively designated as A001 ~ A068 were used to determine the host range, which included 24 strains of A. hydrophila, 20 strains of A. veronii and 24 strains of other Aeromonas spp. Spot test was used to verify if the strain could be a host. Both strains source and test method were identical to those used by Ye et al. (38). The formation of phage plaques confirmed the strain as a host.
The bacterial suspension was estimated by OD600 value based on the OD-concentration standard curve in the pretest, and the titer of stock phage was determined using spot test. Host bacterial dilutions and phage dilutions for MOI groups of 1,000, 100, 10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001 were prepared according to Liu et al. (23). Thereafter, 100 μL of both bacterial and phage dilutions of each group were taken, respectively, and mixed with 800 μL of LB broth. The mixture was incubated for 5 h at 30°C, 180 rpm. The double-layer agar assay was employed to calculate the phage titer. The MOI giving the highest final phage titer was deemed the optimal MOI (OMOI). All groups were performed in triplicate.
Following the method from Zhang et al. with some modifications, host A051 was cultured to the logarithmic phase, then 5 mL of phage suspension and 5 mL of A051 was then mixed with an equal volume of it at the optimal MOI of 0.01, incubated at 30°C for 10 min. The bacteria-phage mixture was centrifuged for 2 min at 10,000 rpm, then the supernatant was discarded and the sediment was resuspended with 10 mL LB broth and cultured at 30°C, 150 rpm. The 100 μL of culture was pipetted out at 10, 20, 30, 40, 60, 90, 120 and 150 min, respectively, and diluted in PBS prior to enumeration using the double-layer plate assay. The phage titer of each sample was determined in triplicate.
For thermal stability assay, 1 mL of phage suspension with a titer of 5 × 107 PFU/mL was incubated at 30, 40, 50, 60, 70 or 80°C for 1 h, respectively. For acid and alkali stability assay, the solution with pH value of 2.0, 4.0, 6.0, 8.0, 10.0 or 12.0 were prepared using PBS buffer, 0.2 mol/L Na2HPO4·12H2O and 0.1 mol/L citric acid. 900 μL of each solution was mixed with 100 μL phage suspension with a titer of 5 × 107 PFU/mL, respectively, and incubated for 1 h at 30°C. Phage titers of all samples were determined using double-layer plate assay and performed in triplicate.
Aeromonas hydrophila A051 was inoculated into 100 mL of LB broth and cultured until the OD600 value of the culture is 0.14 (approximately 1.2 × 108 CFU/mL of bacterial density), then was mixed with phage phiA051 at a 1:1 volume ratio at a starting MOI of 0.01 and 0.1, respectively. 100 μL of mixture was transferred to each well of a 96-well plate with 100 μL of LB broth. The culture of bacteria alone with 100 μL of LB broth was used as a control group. The plate was incubated in a multi-plate reader at 30°C for 24 h with shaking, and the OD600 value of each well was measured at the interval of 30 min during incubation. Each treatment was performed in triplicate.
Phage DNA was extracted following the directions provided by the TIANamp Virus DNA/RNA Kit (Tiangen Biotech Co., Ltd., Beijing) and was sequenced by Illumina Hiseq sequencing system at Majorbio (Shanghai, China). The bacterial genome scaffolds were assembled by splicing the optimized sequences after second-generation sequencing with multiple K-mer parameters using the short sequence assembly software SOAPdenovo21 (41) to obtain the optimal contigs results, and then comparing the reads to the contigs, local assembly and optimization of the assembly results were carried out to form the scaffolds according to the paired-end and overlap relationships of the reads. The completed genome map was assembled by using the assembly software unicycler v0.4.8 (42) to assemble the three-generation sequences, and the sequences were corrected with the help of the software pilonjin. Coding sequences (CDS) in the genome were predicted using Glimmer (43).2 Prodigal. tRNAscan-SE v2.03 (44) was used to predict tRNAs contained in the genome. ResFinder (45) was used to predict genes mediating antimicrobial resistance in phage DNA, and VFDB (46) was used to predict the presence of virulence factors.
The phage concentration data used for coordinate mapping were converted to common logarithms and plotted using GraphPad Prism 9.0.0 (121). Each coordinate point represented the mean value of triplicates, and the error bar indicated the standard deviation. GeneMarkS4 and EasyFig (47) were used for gene prediction and Co-linearity analysis of phage genome, respectively. The predicted genes were annotated using the online BLASTP tool (NCBI)5 and GeneMarkS (see text footnote 4, respectively). The whole genome circle map was graphed using Proksee.6 MEGA 11.0 (48) and VIPtree7 were used to construct and visualize the phylogenetic tree. PHASTER and Prophage Hunter were used to indicate prophage (49, 50).
A phage targeting A. hydrophila A051 was isolated and designated as phiA051. Without any inducer, the phage could be cultured continuously for more than 20 passages, with stable formation of phage plaques and no bacterial colonies in the phage plaques (Figure 1A). Diameter of the plaque was 0.32 ± 0.06 cm on the double layer plate (Figure 1B). The phage phiA051 was composed of an icosahedral capsid with an isometric dimension of 71.3 ± 2.0 nm, and a 73.1 ± 2.7 nm tail, imaged by transmission electron microscope (TEM) (Figure 1C).
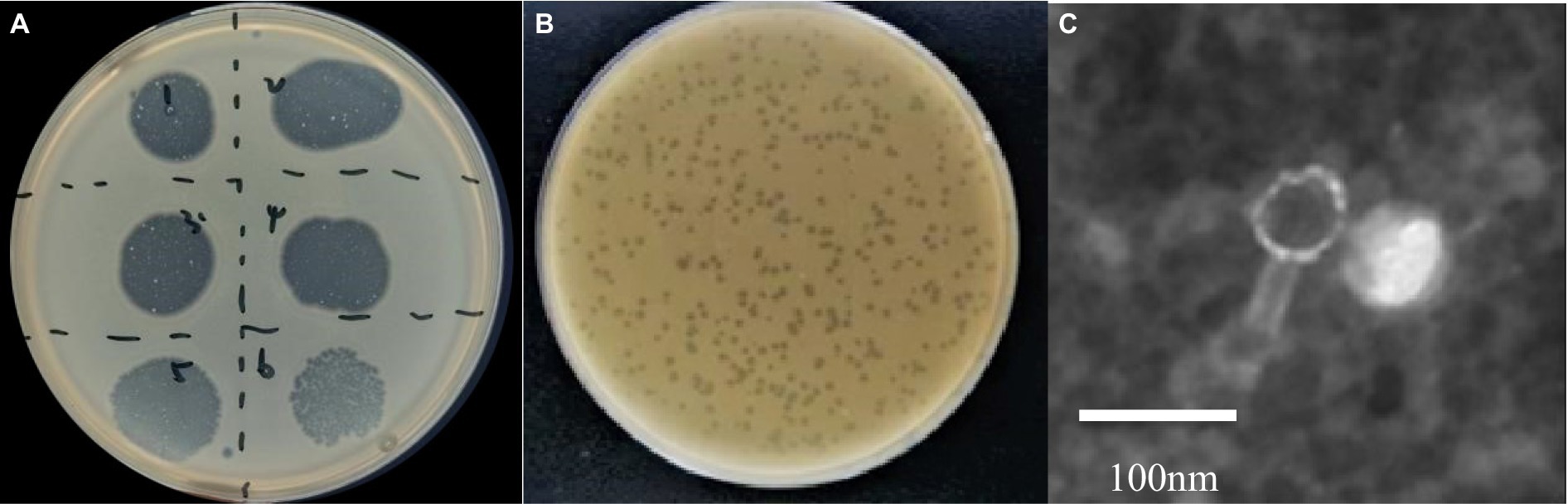
Figure 1. Morphology of phage phiA051. (A) Plaque formation by spot test, dilutions from number 1–6 are 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, respectively. (B) The appearance of plaque on the double-layer agar. (C) The TEM graphs of negatively-stained. Bar, 100 nm.
Phage phiA051 could only lyse A. hydrophila A009, A013, A019 and A051 out of 68 tested Aeromonas strains (including 24 strains of A. hydrophila), implying that it has a narrow host range compare with some of phages that have been reported (38, 51–59).
The optimal MOI for phage phiA051 was 0.01, as indicated by the fact that phage phiA051 achieved the highest final titer (1.21 × 109 PFU/mL) in this infected group (Table 1).
The one-step growth curve of phage phiA051 indicates that the phage first undergoes a latent phase of 20 min after absorption with the host bacterium, followed by a lysis phase of 70 min, and finally enters a plateau phase at 90 min. The burst size of this phage was 108 PFU/cell, indicating that each infected host cell could produce 108 phage progenies on average (Figure 2A).

Figure 2. One-step growth curve (A), thermal (B) and acid-alkali (C) stability of phage phiA051. Error bars represent the standard deviation of the mean.
The phage had a high survival rate in the range of temperature between 30°C and 50°C. The viability decreased with the increase of temperature. When the temperature rose to 70°C, all phages were inactivated (Figure 2B). Phage phiA051 had an acid-alkali tolerance range of pH 6.0–10.0 (Figure 2C).
Under the co-culture conditions of phage and A. hydrophila A051, the MOI = 0.01 group did not have significant inhibition and only showing limited suppression in the pre-logarithmic period of growth of A051, whereas the MOI = 0.1 group suppressed the growth of A051 throughout the whole process after 4 h. After 24 h of incubation, the OD600 of MOI = 0.1 group was only 62.4% of that of the control group. The result indicate that the amount of phage has an effect on the inhibition effect, and a larger amount of phage within a certain limit is more effective (Figure 3).
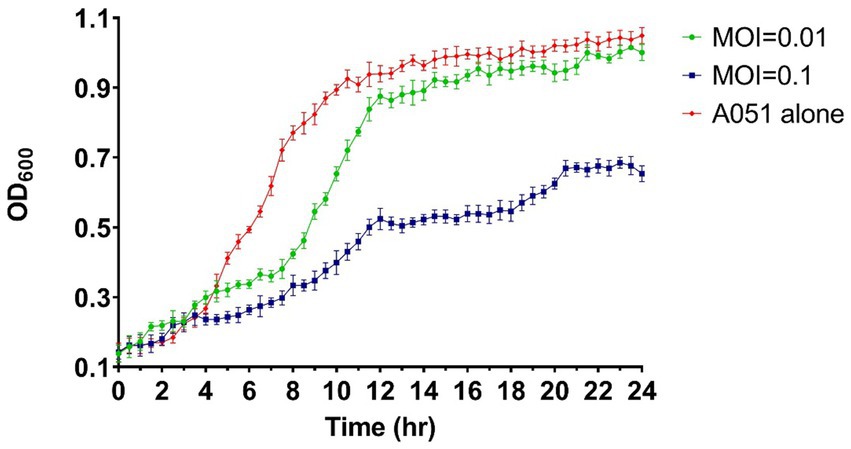
Figure 3. Bacteriostatic effect of phage phiA051 against A. hydrophila A051 at MOI of 0.1 and 0.01 in vitro. The concentration of bacteria was indirectly reflected by the OD600 value. Error bars represent the standard deviation of the mean.
Next,-generation sequencing results show that the genome of phage phiA051 is a linear double-stranded DNA (dsDNA) comprising 32,212 bp and a GC content of 60.55% (Figure 4). There were 46 open reading frames (ORFs), including 26 functional ORFs assigned a function until now and 20 hypothetical proteins (Table 2). Among the functional ORFs, there are many important proteins for phage, such as the integrase (ORF19) which may play an important role in regulating the transition between the lysis and lysogenic cycles, the two terminase subunits (ORF39, 42) involving in DNA translocation and head-filling, Rz1-like lysis system protein LysC (ORF07) and holin (ORF04) associated with lytic infestation, TraR/DksA family protein (ORF03) and helix- turn- helix regulator (ORF20) related to the replication of DNA, tail protein (ORF01,14), portal protein (ORF38), capsid protein (ORF40, 41), head protein (ORF44) and virion morphogenesis protein (ORF46) as structure proteins. No putative toxins and antimicrobial resistance could be found.
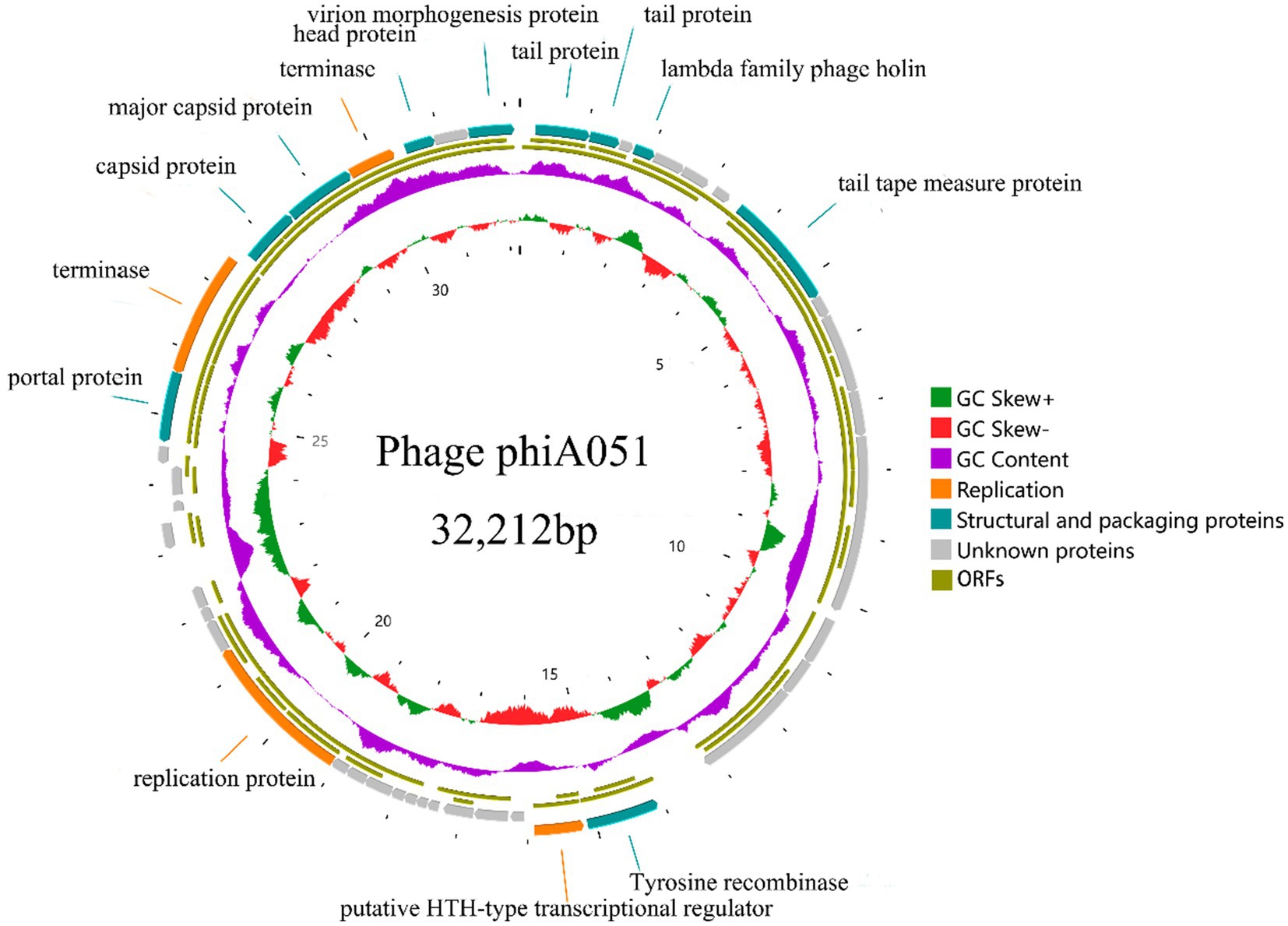
Figure 4. Circularized genomic map of phage phiA051. The number at the innermost circle denotes positions. The inward peaks of GC skew ± (known as G-C/G + C) indicating a greater proportion of G. The clockwise arrow with an inner position indicates the forward reading frame.
The genome of phage phiA051 was analyzed using BLASTN and most of the query hits were prophages, displaying as bacterial genomic sequences of Aeromonas spp. Of the top 25 hits, there were 23 prophages in bacterial chromosomes and only 2 phage isolates (Table 3). The most similar nucleotide sequence was the prophage coming from the 3,003,688 - 3,033,875 bp fragment of the Aeromonas caviae SS332 genome, with an overall similarity of 84.9% (% identity multiplied by % coverage). And the closest phage isolate was Aeromonas phage phiO18P, with an overall similarity of 70.5%. The nucleotide sequences of them were selected for synthetic analysis with phiA051, respectively. Co-linearity analysis of phiA051 and phiO18P genomes showed two homologous fragments for phiA051 and phiO18P. Co-linearity analysis of phiA051 and SS332 showed that phiA051 and SS332 have three homologous fragments (Figure 5).
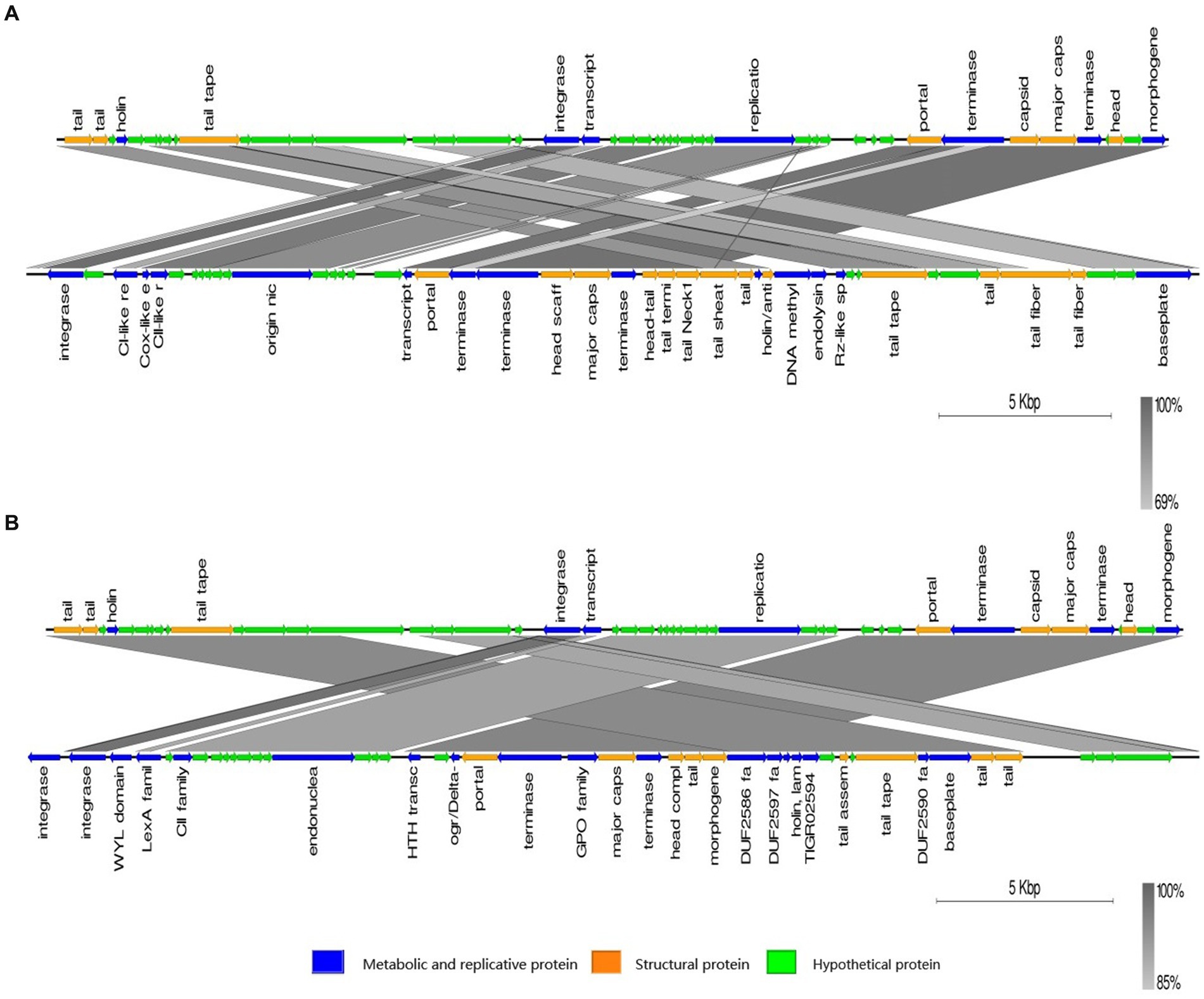
Figure 5. Visualized co-linearity analysis analysis between phage phiA051 (upper) and phage phiO18P (A), phage phiA051 (upper) and prophage in Aeromonas caviae SS332 (B). Right arrow indicates the forward reading frame.
Proteomic trees were constructed by VIPTree (Figure 6). Phage phiA051 was clustered in the family Peduoviridae with only two Aeromonas phages phiO18P and vB_AsaM_LPM4 (Figure 6B). The largest number of Aeromonas phages were clustered in the familys Straboviridae and Autographviridae.
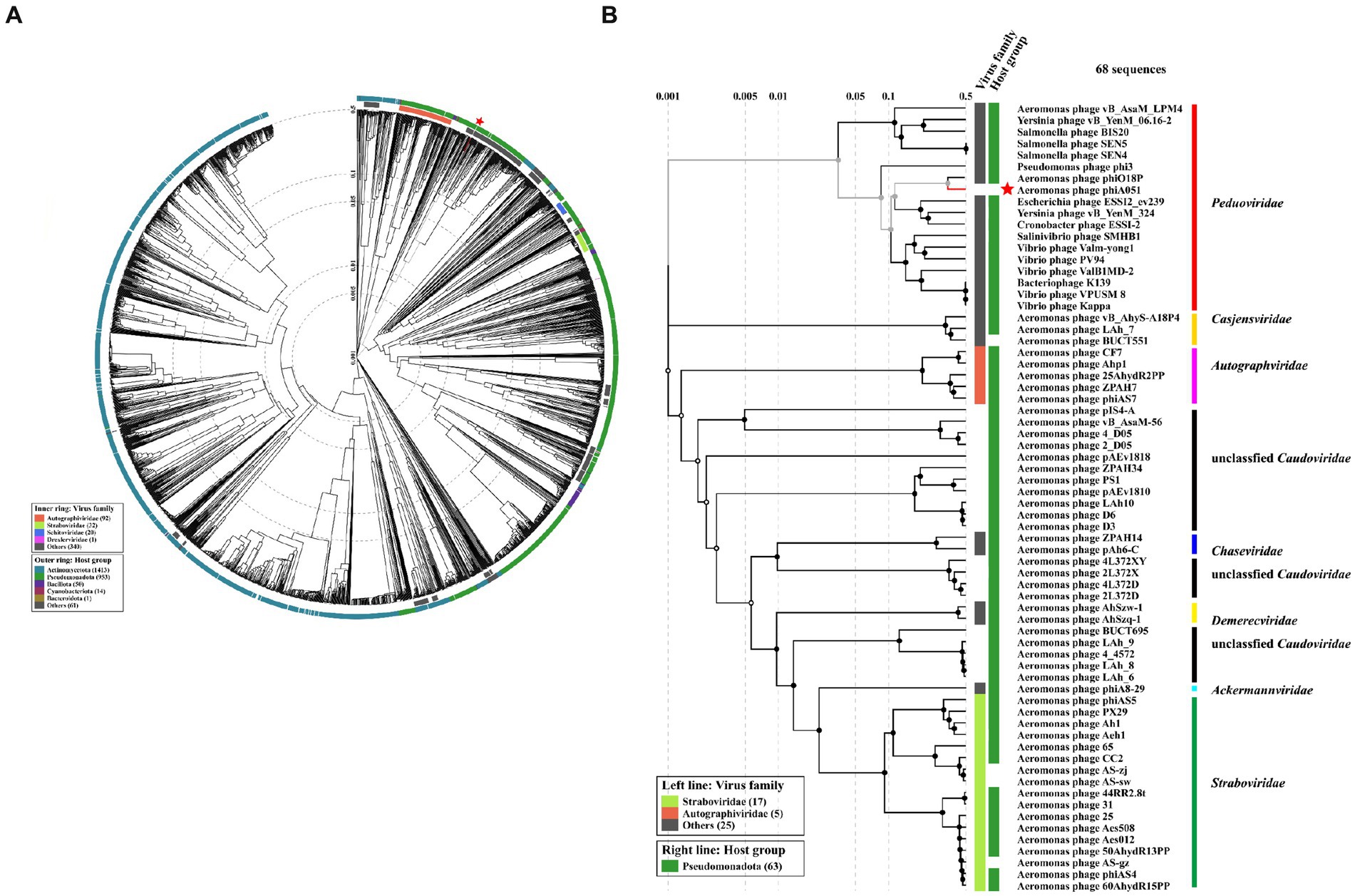
Figure 6. Proteomic trees based on proteomic alignment using VIPTree. (A) The viral proteomic circular tree involving phage phiA051 and 2,564 related phages. (B) The viral proteomic rectangular tree involving all Aeromonas phages and some phages against other hosts. Phage phiA051 is labeled with a red star.
The phylogenetic tree constructed using both the major capsid protein gene and the integrase gene showed that phiA051, phiO18P and SS332 were clustered in the closest branch. However, phylogenetic trees constructed using the terminase large subunit gene show that phiA051 and phiO18P remained on the adjacent branch, while SS332 was on the more distant branch (Figure 7).
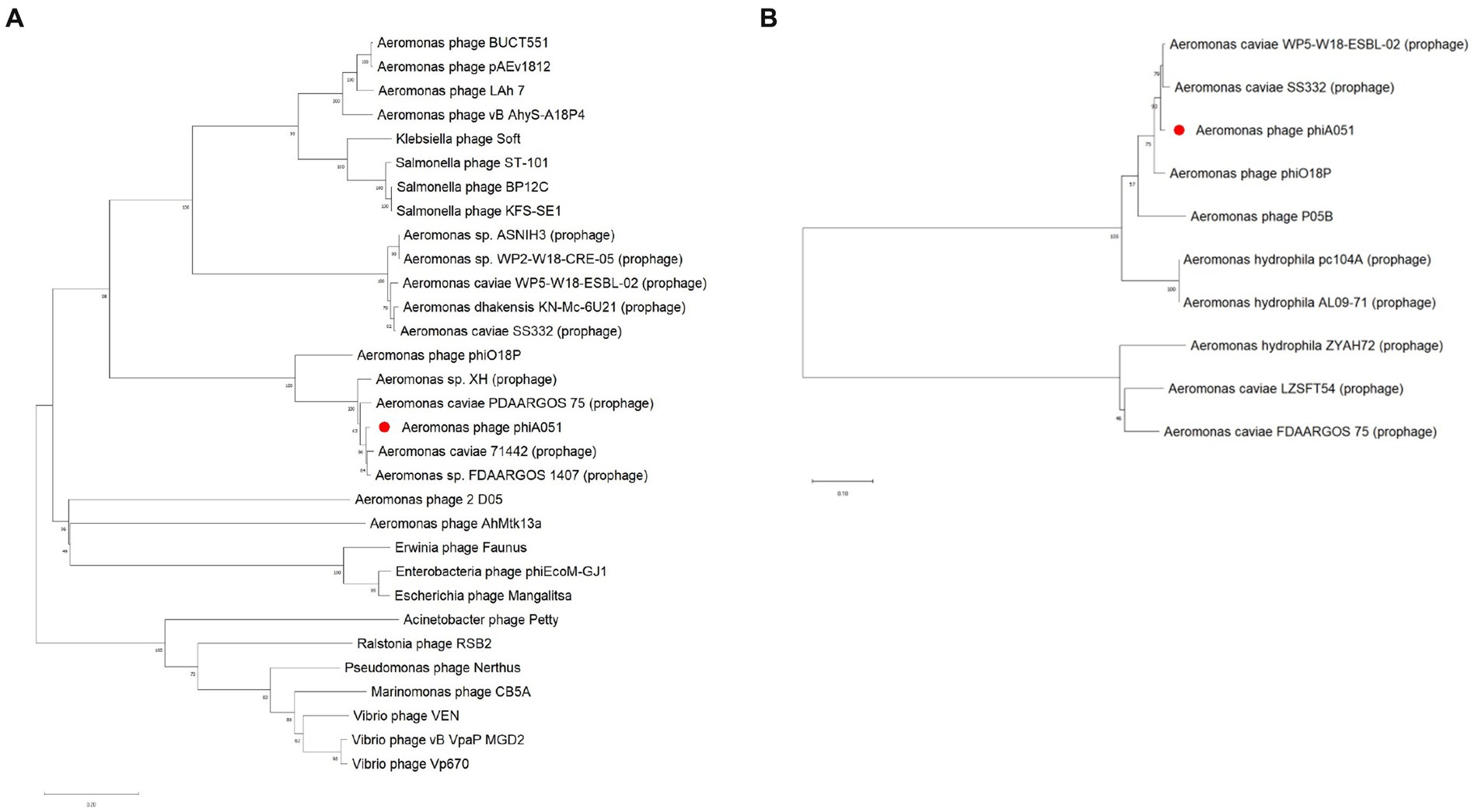
Figure 7. Neighbor-joining phylogenetic tree constructed using the terminase large subunit gene (A) and the integrase gene (B) with 1,000 bootstrap replicates.
According to the latest demarcation criteria of the International Committee on Classification of Viruses (ICTV) (60), 70 and 95% nucleotide identity of the full genome length were established as the cut-off for genera and species, respectively. The query result by BlastN on the NCBI database showed that phiO18P was the closest isolated phage with phiA051, with an overall similarity of 70.5%. Phage phiO18P had been classified as Heunggongvirae, Uroviricota, Caudoviricetes, Peduoviridae, Bielevirus, Bielevirus phiO18P by ICTV, therefore phage phiA051 should be regarded as a novel species in the same genus Bielevirus as phiO18P.
In this study, a novel virulent Aeromonas hydrophila phage phiA051 was discovered and identified. Although it was only able to lyse 4 out of 24 tested strains of A. hydrophila, phage phiA051 could notably inhibit the proliferation of the host bacterium and reduce the final concentration of it, suggesting that this phage has certain potential in rapidly inhibiting the spread of pathogen early in the outbreak. This result likewise suggests that higher phage titers result in better bacterial inhibition, and that the optimal MOI should not be the only reference during practical application. Consistent with previous research findings on bacteriophages, phage phiA051 was found to be not acid tolerant (31, 32, 61–66). The genome of phiA051 comprises 46 open reading frames, of which only 26 are confirmed to be functional. Analyses of the evolutionary position of this phage have shown that it may have originated from a P2-type temperate phage and nonproliferative homologous fragments of this phage exist in several A. hydrophila strains, as phiO18P is also a temperate phage and has a ORF of phage major capsid protein of the P2 family in its genome (67). Integrase drives recombination between the phage and specific attachment sites on the host chromosome known as attP and attB during the integration process (68), while holin can accumulate in the bacterial cytosol and eventually penetrate the peptidoglycan layer, and can achieve control of the lytic cycle through self-regulation of its concentration (69–71). This result suggests that the genome of phiA051 is characterized by both temperate phage and virulent phage.
BLASTN comparisons of phage phiA051 showed that the genome of this phage has high similarity to a variety of prophage, the highest of which is Aeromonas caviae SS332. It is noteworthy that most of the prophage fragments with high total similarity to phiA051 are close in length and structurally very similar to phiA051, so the result suggests that phiA051 most probably originated from some prophages. In contrast to this possible origin of phiA051, there are not many known proteins associated with lysogeny within the genome of this phage, the most notable of which are CII family proteins. CI and CII-like protein are used to determine the timing of lysis cycle onset and to regulate phage polarity. In particularly, CII proteins are thought to play a key initiating role in inhibiting lytic gene expression and the regulation of the lysogenic cycle of temperate phages (72–74). Despite the annotation of CII family proteins within phiA051, the phage was considered to have lost its lysogenicity in this study considering that the phage exhibited stable lytic ability in culture, but the specific molecular mechanism of the lysogenic shift remains to be thoroughly investigated.
The results of the co-linearity analysis show that the genome of phage phiA051 is significantly different in the location of the gene synteny compared to other phages and prophage that are more homologous to it. It has been shown that the lytic cycle can be restarted by DNA recombination and that may be the opportunity to trigger the process of a temperate phage becoming a virulent phage (75). Myron Levine performed lysogenicity assays on a mutant strain of Salmonella temperate phage and demonstrated the possibility of controlling the emergence of lysogenicity by altering the sequence of genes upstream and downstream of the prophage (76), thus regulation of the bacterial genome may impact phage variation. Some studies have since also affirmed the importance of host bacterial genome variation and recombination in the formation of temperate phage mutants (66, 77, 78). Therefore, certain regions in the genome of a strain related to the origin of phiA051 may be similarly involved in the transition between the lytic and lysogenic states. However, in the present study, the upstream and downstream sequences of each prophage with high genomic similarity to phiA051 were examined, and no traces of variation and recombination similar to those reported in the known literature were found, so it was thought that it might be necessary to further explore the origin of phiA051.
Most of the current mainstream research uses induced mutagenesis to isolate possible prophage from known strains to indirectly analyze temperate phages (36, 66, 77–81), and relatively few studies have been conducted on natural mutants, but it was found that phiA051 exhibited the properties of a virulent phage without the need for special treatment, unlike other temperate phages which require stimulation to maintain stable lysis. The discovery of this phage is therefore an important addition to the field. The phage phiA051 was not found potential lysogenic genes such as excisionase and transposase. A number of Aeromonas strains that are highly homologous to phiA051 were explored by PHASTER, and it was found that transposase genes were present in some of the strains with prophage. This fact suggests that this gene in genome of phiA051 might have been lost in some case. A study analyzed the gene map of virulent mutants of the temperate phage of Rhizobium meliloti and concluded that two mutations in loci avirC and avirT are necessary for the phage to acquire virulent feature (80). However, no similar fragment was found within phiA051. Therefore, further analysis of the lysis mechanism of phiA051 is required.
In this study, Aeromonas hydrophila phage phiA051 was identified by biological and genomic analysis. It was proposed a novel species belong to class Caudoviricetes, family Peduoviridae, genus Bielevirus. The phage showed stable virulent characteristics, but its genome has high similarity to many prophages of Aeromonas spp., therefore it is considered to be of probable prophage origin. This discovery has provided a new clue about the prophage in the field of A. hydrophila research on temperate phage, and also provided the basis for further research on the origin of viral gene fragments in A. hydrophila.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material. The datasets sequence of phage phiA051 for this study can be found in the GenBank under GenBank accession No. OP484843.
YW: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. GT: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. XJ: Data curation, Methodology, Writing – review & editing. CT: Data curation, Methodology, Writing – review & editing. HC: Data curation, Methodology, Writing – review & editing. WF: Data curation, Methodology, Writing – review & editing. HT: Data curation, Methodology, Writing – review & editing. QW: Data curation, Methodology, Writing – review & editing. XW: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. ML: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Fund of Xiamen Key Laboratory of Intelligent Fishery (No. XMKLIF-OP-202306), Key Research and Development Program of Guangxi (No. GUIKE-AB23026030), Natural Science Foundation of Fujian (No. 2023 J01143 and 2022 J01802), Science and Technology Project of Quanzhou (No. 2022 N044), Science and Technology Major Project of Xiamen (No. 3502Z20221018), and Fund of Engineering Research Center of the Modern Technology for Eel Industry, Education Ministry of China (No. RE202407).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.The reviewer QY declared a shared affiliation with the authors YW, XJ, HC, ML to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. ^ http://soap.genomics.org.cn/
2. ^ http://ccb.jhu.edu/software/glimmer/index.shtml
3. ^ http://trna.ucsc.edu/software/
4. ^ http://topaz.gatech.edu/GeneMark/
1. Herndon, D, Rodriguez, N, Mitchell, K, and Gallagher, J. Burn wound infections In: D Herndon, editor. Surgical infections : Jaypee Brothers Medical Publishers (P) Ltd. (2013). 133–3.
2. Laupland, KB, and Church, DL. Population-based epidemiology and microbiology of community-onset bloodstream infections. Clin Microbiol Rev. (2014) 27:647–64. doi: 10.1128/CMR.00002-14
3. Freire, MP, De Oliveira, GD, Garcia, CP, Campagnari Bueno, MF, Camargo, CH, Kono Magri, ASG, et al. Bloodstream infection caused by extensively drug-resistant Acinetobacter baumannii in cancer patients: high mortality associated with delayed treatment rather than with the degree of neutropenia. Clin Microbiol Infect. (2016) 22:352–8. doi: 10.1016/j.cmi.2015.12.010
4. Zhang, H, Liu, DW, Wang, XT, Long, Y, and Chen, H. Bloodstream infection with Carbapenem-resistant Klebsiella Pneumoniae and multidrug-resistant Acinetobacter Baumannii: a case report. Chin Med Sci J. (2014) 29:51–4. doi: 10.1016/S1001-9294(14)60025-0
5. Kumar, A, and Yadav, A. Synthetic phage and its application in phage therapy In: A Kumar, editor. Progress in molecular biology and translational science. Amsterdam, Netherlands: Elsevier (2023). 61–89.
6. Feiner, R, Argov, T, Rabinovich, L, Sigal, N, Borovok, I, and Herskovits, AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. (2015) 13:641–50. doi: 10.1038/nrmicro3527
7. Gummalla, VS, Zhang, Y, Liao, YT, and Wu, VCH. The role of temperate phages in bacterial pathogenicity. Microorganisms. (2023) 113:8266–71. doi: 10.3390/microorganisms11030541
8. Ho, K. Bacteriophage therapy for bacterial infections: rekindling a memory from the pre-antibiotics era. Perspect Biol Med. (2001) 44:1–16. doi: 10.1353/pbm.2001.0006
9. Huang, C, Zhang, Y, and Jiao, N. Phage resistance of a marine bacterium, Roseobacter denitrificans OCh114, as revealed by comparative proteomics. Curr Microbiol. (2010) 61:141–7. doi: 10.1007/s00284-010-9588-3
10. Keen, EC. A century of phage research: bacteriophages and the shaping of modern biology. BioEssays. (2015) 37:6–9. doi: 10.1002/bies.201400152
11. Monteiro, R, Pires, DP, Costa, AR, and Azeredo, J. Phage therapy: going temperate? Trends Microbiol. (2019) 27:368–78. doi: 10.1016/j.tim.2018.10.008
12. Ramos-Vivas, J, Superio, J, Galindo-Villegas, J, and Acosta, F. Phage therapy as a focused management strategy in aquaculture. Int J Mol Sci. (2021) 22:436. doi: 10.3390/ijms221910436
13. Daskalov, H. The importance of Aeromonas hydrophila in food safety. Food Control. (2006) 17:474–83. doi: 10.1016/j.foodcont.2005.02.009
14. Grim, CJ, Kozlova, EV, Sha, J, Fitts, EC, Van Lier, CJ, Kirtley, ML, et al. Characterization of Aeromonas hydrophila wound Pathotypes by comparative genomic and functional analyses of virulence genes. Gilligan P, editor. MBio. (2013) 4:e00064. doi: 10.1128/mBio.00064-13
15. Janda, JM, and Abbott, SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. (2010) 23:35–73. doi: 10.1128/CMR.00039-09
16. Xiao, D, Cao, H, Hu, K, and Yang, X. ERIC-PCR genotyping and drug resistant analysis of pathogenic Aeromonas hydrophila from freshwater animals: ERIC-PCR genotyping and drug resistant analysis of pathogenic Aeromonas hydrophila from freshwater animals. J Fish Sci China. (2013) 18:1092–9. doi: 10.3724/SP.J.1118.2011.01092
17. Brouqui, P, and Raoult, D. Endocarditis due to rare and fastidious Bacteria. Clin Microbiol Rev. (2001) 14:177–207. doi: 10.1128/CMR.14.1.177-207.2001
18. Qin, Y, Lin, G, Chen, W, Xu, X, and Yan, Q. Flagellar motility is necessary for Aeromonas hydrophila adhesion. Microb Pathog. (2016) 98:160–6. doi: 10.1016/j.micpath.2016.07.006
19. Qin, Y, Lin, G, Chen, W, Huang, B, Huang, W, and Yan, Q. Flagellar motility contributes to the invasion and survival of Aeromonas hydrophila in Anguilla japonica macrophages. Fish Shellfish Immunol. (2014) 39:273–9. doi: 10.1016/j.fsi.2014.05.016
20. Chang, CY, Thompson, H, Rodman, N, Bylander, J, and Thomas, J. Pathogenic analysis of Aeromonas hydrophila septicemia. Ann Clin Lab Sci. (1997) 27:254–9.
21. Ghatak, S, Blom, J, Das, S, Sanjukta, R, Puro, K, Mawlong, M, et al. Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila. Antonie Van Leeuwenhoek. (2016) 109:945–56. doi: 10.1007/s10482-016-0693-6
22. Guo, W, He, Q, Wang, Z, Wei, M, Yang, Z, Du, Y, et al. Influence of antimicrobial consumption on gram-negative bacteria in inpatients receiving antimicrobial resistance therapy from 2008-2013 at a tertiary hospital in Shanghai. China Am J Infect Control. (2015) 43:358–64. doi: 10.1016/j.ajic.2014.12.010
23. Liu, B, Zheng, T, Quan, R, Jiang, X, Tong, G, Wei, X, et al. Biological characteristics and genomic analysis of a novel Vibrio parahaemolyticus phage phiTY18 isolated from the coastal water of Xiamen China. Front Cell Infect Microbiol. (2022) 12:1035364. doi: 10.3389/fcimb.2022.1035364
24. Schulz, P, Robak, S, Dastych, J, and Siwicki, AK. Influence of bacteriophages cocktail on European eel (Anguilla anguilla) immunity and survival after experimental challenge. Fish Shellfish Immunol. (2019) 84:28–37. doi: 10.1016/j.fsi.2018.09.056
25. Jun, JW, Kim, JH, Shin, SP, Han, JE, Chai, JY, and Park, SC. Protective effects of the Aeromonas phages pAh1-C and pAh6-C against mass mortality of the cyprinid loach (Misgurnus anguillicaudatus) caused by Aeromonas hydrophila. Aquaculture. (2013) 416-417:289–95. doi: 10.1016/j.aquaculture.2013.09.045
26. Kokkari, C, Sarropoulou, E, Bastias, R, Mandalakis, M, and Katharios, P. Isolation and characterization of a novel bacteriophage infecting Vibrio alginolyticus. Arch Microbiol. (2018) 200:707–18. doi: 10.1007/s00203-018-1480-8
27. Paterson, WD, Douglas, RJ, Grinyer, I, and McDermott, LA. Isolation and preliminary characterization of some Aeromonas salmonicida bacteriophages. J Fish Res Bd Can. (1969) 26:629–32. doi: 10.1139/f69-056
28. John, SG, Mendez, CB, Deng, L, Poulos, B, Kauffman, AKM, Kern, S, et al. A simple and efficient method for concentration of ocean viruses by chemical flocculation: corrigendum. Environ Microbiol Rep. (2011) 3:809–9. doi: 10.1111/j.1758-2229.2011.00301.x
29. Matamp, N, and Bhat, S. Phage Endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms. (2019) 7:84. doi: 10.3390/microorganisms7030084
30. Reynes, V. A propos des bacteriemies a bacilles dysenteriques. Med Mal Infect. (1973) 3:166. doi: 10.1016/S0399-077X(73)80074-5
31. Wu, JL, Lin, HM, Jan, L, Hsu, YL, and Chang, LH. Biological control of fish bacterial pathogen, Aeromonas hydrophila, by bacteriophage AH 1. Fish Pathol. (1981) 15:271–6. doi: 10.3147/jsfp.15.271
32. Xiang, Y, Li, W, Song, F, Yang, X, Zhou, J, Yu, H, et al. Biological characteristics and whole-genome analysis of the Enterococcus faecalis phage PEf771. Can J Microbiol. (2020) 66:505–20. doi: 10.1139/cjm-2019-0336
33. Nikolich, MP, and Filippov, AA. Bacteriophage therapy: developments and directions. Antibiotics. (2020) 9:135. doi: 10.3390/antibiotics9030135
34. Burrowes, B, Harper, DR, Anderson, J, McConville, M, and Enright, MC. Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens. Expert Rev Anti-Infect Ther. (2011) 9:775–85. doi: 10.1586/eri.11.90
35. Royer, S, Morais, AP, and Da Fonseca Batistão, DW. Phage therapy as strategy to face post-antibiotic era: a guide to beginners and experts. Arch Microbiol. (2021) 203:1271–9. doi: 10.1007/s00203-020-02167-5
36. Chang, Y, Bai, J, Lee, JH, and Ryu, S. Mutation of a Staphylococcus aureus temperate bacteriophage to a virulent one and evaluation of its application. Food Microbiol. (2019) 82:523–32. doi: 10.1016/j.fm.2019.03.025
37. García, P, Madera, C, Martínez, B, Rodríguez, A, and Evaristo, SJ. Prevalence of bacteriophages infecting Staphylococcus aureus in dairy samples and their potential as biocontrol agents. J Dairy Sci. (2009) 92:3019–26. doi: 10.3168/jds.2008-1744
38. Ye, Y, Tong, G, Chen, G, Huang, L, Huang, L, Jiang, X, et al. The characterization and genome analysis of a novel phage Phia034 targeting multiple species of Aeromonas. Virus Res. (2023) 336:199193. doi: 10.1016/j.virusres.2023.199193
39. Zhang, W, Zhang, R, Hu, Y, Liu, Y, Wang, L, An, X, et al. Biological characteristics and genomic analysis of a Stenotrophomonas maltophilia phage vB_SmaS_BUCT548. Virus Genes. (2021) 57:205–16. doi: 10.1007/s11262-020-01818-5
41. Luo, R, Liu, B, Xie, Y, Li, Z, Huang, W, Yuan, J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. (2012) 1:18. doi: 10.1186/2047-217X-1-18
42. Wick, RR, Judd, LM, Gorrie, CL, and Holt, KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. (2017) 13:e1005595. doi: 10.1371/journal.pcbi.1005595
43. Delcher, AL, Bratke, KA, Powers, EC, and Salzberg, SL. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. (2007) 23:673–9. doi: 10.1093/bioinformatics/btm009
44. Lowe, TM, and Eddy, SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. (1997) 25:955–64. doi: 10.1093/nar/25.5.955
45. Bortolaia, V, Kaas, RS, Ruppe, E, Roberts, MC, Schwarz, S, Cattoir, V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
46. Liu, B, Zheng, D, Zhou, S, Chen, L, and Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. (2022) 50:D912–7. doi: 10.1093/nar/gkab1107
47. Sullivan, MJ, Petty, NK, and Beatson, SA. Easyfig: a genome comparison visualizer. Bioinformatics. (2011) 27:1009–10. doi: 10.1093/bioinformatics/btr039
48. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Battistuzzi FU, editor. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
49. Arndt, D, Grant, JR, Marcu, A, Sajed, T, Pon, A, Liang, Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. (2016) 44:W16–21. doi: 10.1093/nar/gkw387
50. Song, W, Sun, HX, Zhang, C, Cheng, L, Peng, Y, Deng, Z, et al. Prophage hunter: an integrative hunting tool for active prophages. Nucleic Acids Res. (2019) 47:W74–80. doi: 10.1093/nar/gkz380
51. Cai, L, Tian, Y, Li, Z, Yang, Y, Ai, C, and Zhang, R. A broad-host-range lytic phage vB_VhaS-R18L as a candidate against vibriosis. Front Microbiol. (2023) 14:1191157. doi: 10.3389/fmicb.2023.1191157
52. Lu, R, Liu, B, Wu, L, Bao, H, García, P, Wang, Y, et al. A broad-Spectrum phage Endolysin (LysCP28) able to remove biofilms and inactivate Clostridium perfringens strains. Food Secur. (2023) 12:411. doi: 10.3390/foods12020411
53. Piekarowicz, A, Kłyż, A, Adamczyk-Popławska, M, and Stein, DC. Association of host proteins with the broad host range filamentous phage NgoΦ6 of Neisseria gonorrhoeae. Shafer WM, editor. PLoS One. (2020) 15:e0240579. doi: 10.1371/journal.pone.0240579, 15
54. Feng, T, Leptihn, S, Dong, K, Loh, B, Zhang, Y, Li, M, et al. Characterisation of bacteriophage JD419, a staphylococcal phage with an unusual morphology and broad host range. (2020).
55. Loganathan, A, and Nachimuthu, R. Characterization of vB_Sau_S90, and vB_Sau_S165, a broad host range phages, and multiple phage doses as a potential therapeutic strategy to eliminate transient phage resistant mutants in Staphylococcus Aureus. (2021). Available at: https://www.researchsquare.com/article/rs-1070956/v1.
56. Wang, Y, Zhao, G, Wang, J, and Lin, H. Complete genome analysis and biological characterization of a novel Clostridium perfringens phage vB_CP_qdyz_P5. (2023). Available at: https://www.researchsquare.com/article/rs-2532103/v1.
57. Oliveira, A, Melo, LDR, Kropinski, AM, and Azeredo, J. Complete genome sequence of the broad-host-range Paenibacillus larvae phage phiIBB_Pl23. Genome Announc. (2013) 1:e00438–13. doi: 10.1128/genomeA.00438-13
58. Kelly, D, McAuliffe, O, O’Mahony, J, and Coffey, A. Development of a broad-host-range phage cocktail for biocontrol. Bioeng Bugs. (2011) 2:31–7. doi: 10.4161/bbug.2.1.13657
59. Sillankorva, S, Kropinski, AM, and Azeredo, J. Genome sequence of the broad-host-range Pseudomonas phage Φ-S1. J Virol. (2012) 86:10239. doi: 10.1128/JVI.01605-12
60. Zerbini, FM, Siddell, SG, Lefkowitz, EJ, Mushegian, AR, Adriaenssens, EM, Alfenas-Zerbini, P, et al. Correction to: changes to virus taxonomy and the ICTV statutes ratifed by the international committee on taxonomy of viruses (2023). Arch Virol. (2023) 168:269. doi: 10.1007/s00705-023-05880-w
61. Akmal, M, Rahimi-Midani, A, Hafeez-ur-Rehman, M, Hussain, A, and Choi, TJ. Isolation, characterization, and application of a bacteriophage infecting the fish pathogen Aeromonas hydrophila. Pathogens. (2020) 9:215. doi: 10.3390/pathogens9030215
62. Chandrarathna, HPSU, Nikapitiya, C, Dananjaya, SHS, De Silva, BCJ, Heo, GJ, De Zoysa, M, et al. Isolation and characterization of phage AHP-1 and its combined effect with chloramphenicol to control Aeromonas hydrophila. Braz J Microbiol. (2020) 51:409–16. doi: 10.1007/s42770-019-00178-z
63. Gu, J, Xu, W, Lei, L, Huang, J, Feng, X, Sun, C, et al. LysGH15, a novel bacteriophage Lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J Clin Microbiol. (2011) 49:111–7. doi: 10.1128/JCM.01144-10
64. Li, C, Wang, Z, Zhao, J, Wang, L, Xie, G, Huang, J, et al. A novel Vibriophage vB_VcaS_HC containing Lysogeny-related gene has strong lytic ability against pathogenic Bacteria. Virol Sin. (2021) 36:281–90. doi: 10.1007/s12250-020-00271-w
65. Ofir, G, and Sorek, R. Contemporary phage biology: from classic models to new insights. Cell. (2018) 172:1260–70. doi: 10.1016/j.cell.2017.10.045
66. Putzeys, L, Poppeliers, J, Boon, M, Lood, C, Vallino, M, and Lavigne, R. Transcriptomics-driven characterization of LUZ100, a T7-like Pseudomonas phage with temperate features. mSystems. (2023) 8:e01189–22. doi: 10.1128/msystems.01189-22
67. Beilstein, F, and Dreiseikelmann, B. Temperate bacteriophage ΦO18P from an Aeromonas media isolate: characterization and complete genome sequence. Virology. (2008) 373:25–9. doi: 10.1016/j.virol.2007.11.016
68. Ba, F, Zhang, Y, Wang, L, Liu, WQ, and Li, J. Applications of serine integrases in synthetic biology over the past decade. SynBio. (2023) 1:172–89. doi: 10.3390/synbio1020012
69. Gründling, A, Smith, DL, Bläsi, U, and Young, R. Dimerization between the Holin and Holin inhibitor of phage λ. J Bacteriol. (2000) 182:6075–81. doi: 10.1128/jb.182.21.6075-6081.2000
70. Samir, S. Molecular machinery of the triad Holin, Endolysin, and Spanin: key players orchestrating bacteriophage-induced cell lysis and their therapeutic applications. PPL. (2024) 31:85–96. doi: 10.2174/0109298665181166231212051621
71. Young, R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. (1992) 56:430–81. doi: 10.1128/mr.56.3.430-481.1992
72. Hayes, S, and Bull, HJ. Translational frameshift sites within bacteriophage lambda genes rexA and cI. Acta Biochim Pol. (1999) 46:879–84. doi: 10.18388/abp.1999_4109
73. Ho, Y, and Rosenberg, M. Characterization of the phage lambda regulatory protein cII. Ann Microbiol (Paris). (1982) 133:215–8.
74. Kobiler, O, Koby, S, Teff, D, Court, D, and Oppenheim, AB. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci U S A. (2002) 99:14964–9. doi: 10.1073/pnas.222172499
75. Dragoš, A, Priyadarshini, B, Hasan, Z, Strube, ML, Kempen, PJ, Maróti, G, et al. Pervasive prophage recombination occurs during evolution of spore-forming Bacilli. ISME J. (2021) 15:1344–58. doi: 10.1038/s41396-020-00854-1
76. Levine, M. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology. (1957) 3:22–41. doi: 10.1016/0042-6822(57)90021-1
77. Dickinson, L. The behaviour of a temperate phage of Pseudomonas aeruginosa compared with that of a serologically related, virulent mutant. J Gen Microbiol. (1954) 11:105–13. doi: 10.1099/00221287-11-1-105
78. Doskočil, J, Forstová, J, and Štokrová, J. Temperate and virulent forms of phage theta attacking Bacillus licheniformis. Mol Gen Genet. (1978) 160:311–7. doi: 10.1007/BF00332974
79. Chen, Y, Yang, L, Yang, D, Song, J, Wang, C, Sun, E, et al. Specific integration of temperate phage decreases the pathogenicity of host Bacteria. Front Cell Infect Microbiol. (2020) 10:14. doi: 10.3389/fcimb.2020.00014
80. Dallmann, G, Olasz, F, and Orosz, L. Virulent mutants of temperate Rhizobium meliloti phage 16-3: loci avirC and avirT, and increased recombination. Mol Gen Genet. (1980) 178:443–6. doi: 10.1007/BF00270497
Keywords: Aeromonas hydrophila, bacteriophage, biological characteristics, whole genome, prophage
Citation: Wang Y, Tong G, Jiang X, Tu C, Cai H, Fang W, Tan H, Weng Q, Wei X and Lin M (2024) Biologic and genomic characterization of a novel virulent Aeromonas hydrophila phage phiA051, with high homology to prophages. Front. Vet. Sci. 11:1415685. doi: 10.3389/fvets.2024.1415685
Received: 11 April 2024; Accepted: 05 July 2024;
Published: 18 July 2024.
Edited by:
Hang Yang, Chinese Academy of Sciences, ChinaReviewed by:
Robert Atterbury, University of Nottingham, United KingdomCopyright © 2024 Wang, Tong, Jiang, Tu, Cai, Fang, Tan, Weng, Wei and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinxian Wei, YnlhbmcxNUAxMjYuY29t; Mao Lin, bGlubWFvQGptdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.