- 1Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 2The International Joint Laboratory for Cooperation in Agriculture and Agricultural Product Safety, Ministry of Education, Yangzhou, China
The pathogenic nature of bacteria can be increased by cleaving antimicrobial peptides using omptins, to avoid or counter the host’s natural immune defenses. Plasmid-encoded OmpT (pOmpT or ArlC) in avian pathogenic Escherichia coli (APEC), like the chromosome-encoded OmpT (cOmpT), belongs to the omptin family and both exhibit highly similar sequences and structures. Through sequence alignment and physiological examinations, pOmpT has been identified as a virulence factor, distinct from cOmpT in terms of substrate specificity. When pOmpT is compared with cOmpT regarding their proteolytic activities and target substrates, Asp267 and Ser276 on loop 5 of cOmpT are found to be binding sites that facilitate substrate anchoring and enhance substrate cleavage (protamine or synthetic peptide) by the catalytic center. Conversely, the characteristics of residues at positions 267 and 276 on loop 5 of pOmpT inhibit protamine cleavage, yet allow the specific cleavage of the human antimicrobial peptide RNase 7, which plays a role in host defense. This finding suggests a relationship between these two binding sites and substrate specificity. Furthermore, the substrate-binding sites (residues 267 and 276, particularly residue 267) of cOmpT and pOmpT are determined to be critical in the virulence of APEC. In summary, residues 267 and 276 of pOmpT are crucial for the pathogenicity of APEC and offer new insights into the determinants of APEC virulence and the development of antimicrobial drugs.
1 Introduction
Escherichia coli (E. coli) is a common bacterium found in the gastrointestinal tract of mammals and birds, but it can also cause a range of severe diseases in humans and animals due to its multifunctional pathogenic nature. In poultry, avian pathogenic E. coli (APEC), an extraintestinal pathogenic E. coli (ExPEC), acting either as a primary or secondary agent can infect all types of birds at all ages, causing localized and systemic infections, often referred to as avian colibacillosis. At present, avian colibacillosis is one of the leading causes of mortality and morbidity associated with economic losses in the poultry industry throughout the world. The development of bacterial antibiotic resistance and the ineffectiveness of vaccines pose significant risks to public health and the poultry industry. Consequently, there is an urgent need for more effective prevention and treatment strategies.
Antimicrobial peptides (AMPs) represent one of the primary challenges that bacterial pathogens encounter when infecting a host, playing a crucial role in the innate immune system (1). These peptides, small in size (20–50 amino acids), cationic, and amphiphilic, are primarily secreted by the host’s epithelial cells and neutrophils (2–4). Their ability to attach to anionic bacterial membranes, create pores, and consequently cause bacterial death through cell lysis is well-documented (5–7). Beyond their bactericidal capabilities, AMPs also have the capacity to attract immune cells of the host to infection sites, thereby inducing a wide spectrum of immunomodulatory activities to manage bacterial infections (8).
Despite AMPs’ critical role in the host’s defense mechanism, bacteria have developed numerous strategies to counteract the effects of AMPs, such as utilizing LPS modifications, efflux pumps, capsules, and proteases (1). Omptins, a distinct group of outer membrane (OM) proteases with proteolytic capabilities, have garnered significant attention. Found across various Gram-negative bacteria within the Enterobacteriaceae family, these proteases influence bacterial virulence through the modification or degradation of numerous host and bacterial proteins (9–19). Currently identified omptins include OmpT, OmpP, and ArlC in E. coli, Pla in Yersinia pestis, PgtE in Salmonella enteritidis, IcsP in Shigella flexneri, and CroP in Citrobacter rodentium. They share a high degree of amino acid sequence similarity (45–80%) with highly conserved active sites (11, 19–24). OmpT, the first to be characterized and encoded by the E. coli chromosome, is a 37 kDa protein that forms a hollow β-barrel structure with its active sites exposed to the external environment (24, 25). Inhibitory studies on omptin have shown that elements such as Zn2+, Cu2+, and benzamidine can hinder OmpT activity (26–28), while the serine protease inhibitors aprotinin and ulinastatin can also disrupt OmpT’s functionality (29, 30). Additionally, the highly conserved structure of OmpT and the nature of its active site residues (residues Asp83, Asp85, Asp210, and His212) are crucial for its substrate cleavage specificity, often preferring dibasic motifs (24, 31–33). It has been observed that OmpT in urinary pathogenic E. coli (UPEC) aids in bacterial survival within the host by cleaving secreted antimicrobial peptides like protamine P1 and cathelicidin LL-37 from human urethral epithelial cells (13, 34–36). Furthermore, OmpT present in enterohemorrhagic E. coli (EHEC) and enteropathogenic E. coli (EPEC) can cleave and inactivate LL-37 to varying degrees (12, 37). The plasmid-encoded OmpT-like protease OmpP, found in isolates from urinary tract infections (UTIs), has been shown to cleave the AMP protamine, and ArlC is linked to AMP resistance (e.g., human AMP RNase 7); thus, aiding bacterial survival (23, 38, 39).
Omptins, as key virulence factors that play a core role in the host-interface, are potential targets for antimicrobial and vaccine development, so they have research potential. In fact, for most E. coli, OmpT is only encoded by the ompT gene on the chromosome. Previous studies have focused mainly on the proteolytic activity and pathogenicity of OmpT located on the chromosome (24, 25, 32, 40). We found that the avian pathogenic E. coli (APEC) E058 strain has both the OmpT (cOmpT) gene (compT) on its chromosome and the OmpT (pOmpT) gene encoded by the ColV plasmid (pompT), both of which exhibit highly homologous amino acid sequences up to about 76 percent and are involved in the pathogenicity of APEC in the host (40, 41). Although cOmpT has been extensively studied, research on pOmpT is relatively scarce. Here, we aimed to explore the special role of pOmpT in physiology compared with that of cOmpT and to clarify the effect of pOmpT on APEC pathogenicity both in vitro and in vivo. The in-depth study of proteases such as OmpT is conducive to designing new enzyme-resistant antimicrobial peptides and provides a possibility for the exploration of new anti-infection approaches and the screening of new drug targets.
2 Materials and methods
2.1 Bacterial strains, plasmids, antibodies, and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The oligonucleotide primers used are listed in Supplementary Table S2. The APEC strain E058 was isolated from a chicken with the typical clinical symptoms of colibacillosis in China (42). OmpT mouse-origin monoclonal antibody was developed by our laboratory (43). The other details related to the strains are as follows. The strains were grown in Luria–Bertani (LB) broth, on LB agar plates or in N-minimal medium (44) adjusted to pH 7.5 and supplemented with 0.2% glucose and 1 mM MgCl2. Antibiotics such as 50 μg/mL kanamycin, 50 μg/mL spectinomycin, 30 μg/mL chloramphenicol or 50 μg/mL tetracycline were used for selection wherever needed. All the cultures were grown at 37°C or 30°C under aerobic conditions.
2.2 Generation of mutants, revertants, and recombinant expression bacteria
The bacterial strains used in this study are listed in Supplementary Table S1. Single/double gene deletions of the compT and pompT genes of the APEC E058 strain were performed using the lambda red recombinase system (41). Simultaneously, the native compT and pompT genes, alongside their putative promoters, were amplified, cloned, and inserted into the plasmid pACYC184. Following PCR and DNA sequencing, the plasmids p184-ccompT and p184-ppompT were transformed into the compT/pompT single gene deletion strains to create complementation strains through electroporation.
To minimize interference from outer membrane proteins other than the cOmpT/pOmpT protein in the E058 strain, the compT and pompT genes’ entire open reading frame (ORF) was cloned and inserted into the expression plasmid pET-28a. Subsequently, the correct plasmids were transformed into E. coli BL21(DE3), which naturally lacks the ompT gene, after PCR and DNA sequencing. Furthermore, the amino acid sequences of the cOmpT and pOmpT proteins were divided into five loops based on the topological structure of the cOmpT protein. Employing fusion PCR technology, we utilized the same gene cloning method as mentioned earlier to construct a series of recombinant bacteria with corresponding loop interchanges and site-directed mutations in loop 5 (L5). To further determine the importance of L5 and the amino acid at positions 83/85/210/212/267/276 of cOmpT/pOmpT in the wild-type strain E058, mutual interference between cOmpT and pOmpT must be excluded. Therefore, we first inserted chimeric or site-directed mutant DNA fragments via gene deletion and then deleted or inserted another gene on the basis of the compT/pompT single-gene deletion strain using a CRISPR-Cas9 system-based continual genome editing strategy (45). The guide sequence (N20 sequence), which targets the FRT sequence of the compT/pompT single-gene mutant strain and the compT and pompT gene sequences, was used to construct pTarget series plasmids. The donor DNA was amplified correspondingly using the genomic DNA of the compT/pompT single-gene deletion strain and the above correct pET series plasmids with 5 loops interchange and sites mutation/interchange of the compT/pompT gene as templates.
The DNA template of RNase 7 was derived from the experimenter’s nasal swab sample. The rnase 7 gene without a signal peptide and with a His tag and site-directed mutations was amplified by overlap PCR and subsequently cloned and inserted into the expression plasmid pDEST17 after digestion. Then, the correct plasmid pDEST17-RNase 7 was transformed into competent BL21(AI) cells (46, 47).
2.3 Bacterial RNA isolation, RT–PCR/qPCR, sequencing, and alignment analysis
Total RNA was extracted from the APEC strain E058 and reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa, China) according to the manufacturer’s protocol. Primer sets for the PCR amplification of the target genes compT and pompT in cDNA samples are detailed in Supplementary Table S2. Concurrently, PCRs were conducted using strain E058 DNA as positive controls and cDNA samples without reverse transcription (RT) activation as negative controls. The PCR products were separated on 0.8% agarose gels. Then, the fragments corresponding to the PCR-amplified genes compT and pompT were extracted from the agarose gels using an Axygen DNA gel extraction kit (Corning, China) and were subjected to sequencing verification. The qPCR system was followed with ChamQ SYBR qPCR Master Mix (Vazyme, China). The thermal conditions were as follows: 95°C for 30 s, 40 cycles of 95°C for 10 s, and 60°C for 30 s. The gapA gene was used as the internal control for normalization. The primers used for qRT–PCR of the compT and pompT genes are listed in Supplementary Table S2. Sequence alignment was performed among the amino acid sequences of the compT and pompT genes of APEC E058, the ompT gene amino acid sequence located on the chromosome of the E. coli strain K12 substrain MG1655 (NC_000913.3), UPEC CFT073 (CP051263.1), AIEC NRG857C (CP001855.1) and UPEC isolate cystitis 6 (CP041302.1) and the amino acid sequence of the arlC gene located on the AIEC NRG857C (CP001856.1) and UPEC isolate cystitis 6 (CP041301.1) plasmids published in the NCBI.
2.4 Outer membrane protein extraction
Bacteria were cultured overnight in 200 mL N-minimal medium. The isolation of outer membrane fractions was performed as follows (48): bacterial cells were centrifuged at 6,000 rpm for 10 min at 4°C, and the pellets were resuspended in 7 mL HEPES buffer (10 mM HEPES, pH 7.4) and sonicated. A 1 mL aliquot of the lysate was reserved as whole bacterial protein for subsequent use. The remaining samples were then centrifuged at 6,000 rpm for 10 min at 4°C. The supernatants were collected and supplemented with 48 mL of sarcosyl buffer (2% sarcosyl), followed by a 30-min incubation at 4°C. After centrifugation for 1 h at 35,000 rpm, the pellet containing the outer membrane protein was resuspended in 1 mL of buffer A (20 mM Tris–HCl pH 7.5, 10% glycerol) for later analysis.
2.5 Western blotting
A BCA protein concentration determination kit (Beyotime, China) was used to determine the concentration of the total bacterial protein and total outer membrane protein extracted, which were subsequently normalized. Samples were resolved on a 12% SDS–PAGE gel and transferred to a polyvinylidene fluoride membrane. The membranes were blocked overnight in PBST buffer [10 mM phosphate-buffered saline (PBS, pH 7.4), 0.05% Tween-20] supplemented with 5% nonfat milk at 4°C. The membranes were then incubated with OmpT mouse-origin monoclonal antibody for detection and then with secondary antibodies [goat anti-mouse IgG conjugated with horseradish peroxidase (Beyotime, China)]. Finally, the membranes were exposed to a chemiluminescent HRP substrate (Sharebio, China).
2.6 Protein expression and purification of RNase 7
RNase 7 was expressed in E. coli BL21(AI) cells harboring the expression plasmid pDEST17-RNase 7. Cultures were induced with L-arabinose (final concentration 2%) in LB broth medium for 3 h. The cell pellet was collected by centrifugation and resuspended in 20 mL buffer D (20 mM Tris-Cl, pH 7.5). After cell lysis by sonication, the supernatant was collected by centrifugation and purified through a Ni-NTA column (eluent: 50 mM Na2HPO4, 0.3 M NaCl, and 250 mM imidazole, pH 8.0) and a Superdex-200 column (buffer E: 20 mM Tris–HCl, 150 mM NaCl, and 10% glycerol, pH 8.0). RNase 7 was stored in buffer E (20 mM Tris–HCl, 150 mM NaCl, 10% glycerol) at 4°C. The protein concentration, determined by the BCA method, was approximately 0.15 mg/mL.
2.7 Growth kinetics of bacteria co-incubated with protamine
To study the growth kinetics of bacteria incubated with protamine, we cultivated bacteria in N-minimal media to avoid potential interference from some components of complex media on the function of OmpT. Overnight bacterial cultures in N-minimal medium were prepared and centrifuged. The cultures were resuspended in 10 mL of fresh N-minimal medium and normalized to an OD600 of 0.35 (a final concentration of 3.5 × 108 colony forming units [CFU]/mL), and protamine was then added to the bacterial suspension at a final concentration of 100 μg/mL. The mixture was shaken at 37°C, and the OD600 was measured every 2 h. The results were drawn into the growth curve of bacteria, and GraphPad Prim 7 software was used for differential analysis.
2.8 Proteolytic cleavage of AMPs
Bacterial cells grown in N-minimal medium to an OD600 nm of 0.6–0.8 were washed, pelleted by centrifugation, resuspended in PBS (pH 7.4), and normalized to a bacterial density of 3 × 1010 CFU/mL. Bacteria were combined at a 1:4 (v/v) ratio with 2.5 μg/μL protamine or at a 1:12 (v/v) ratio with 0.15 μg/μL RNase 7 to facilitate visualization of degradation products and incubated at 37°C for various time points. Bacteria were separated from peptide cleavage products by centrifugation, and supernatants were mixed with 2× Tricine sample buffer (Beyotime, China) or 5× SDS-PAGE protein loading buffer (Yeasen Biotechnology, China), then boiled and stored at −20°C. Peptide cleavage products were heated at 96°C for 10 min and separated by 16.5% Tris-Tricine SDS-PAGE (Beyotime, China) or 13% SDS-PAGE. After fixation for 30 min in 5% glutaraldehyde and subsequent washing for 30 min with deionized water, the peptides were stained for 1 h with Coomassie blue G-250.
2.9 Fluorescence resonance energy transfer (FRET) activity assay
The synthetic FRET substrate containing ortho-aminobenzoic acid (Abz) as the fluorophore and group 2, 4-nitrophenyl (Dnp) as the quencher and a dibasic motif (RK) in its center (2Abz-SLGRKIQI-K(Dnp)-NH2) was purchased from GL Biochem Ltd. (China)1 (25, 49). To perform the assay, bacteria were grown in N-minimal medium to the mid-exponential phase and normalized to an OD600 of 0.6–0.8. The bacterial cells were centrifuged, resuspended in PBS (pH 7.4), and normalized to 3 × 108 CFU/mL. Bacteria (~2.25× 107 CFU in 75 μL) were mixed in a 96-well plate with 75 μL of FRET substrate (final concentration 40 μM). The fluorescence emission was monitored for 360 min at 25°C using a BioTek Synergy 2 plate reader with an excitation wavelength of 325 nm and an emission wavelength of 430 nm. The initial background measurements were subtracted from the final reaction sample values. The kinetic parameters (Km, Kcat, and Kcat/Km) were calculated by measuring OmpT activity at 0–240 μM substrate and fitting the resulting Michaelis–Menten equation.
2.10 Colorimetric assay
Samples of membrane fractions were diluted in buffer A to appropriate concentrations prior to measurements of OmpT activity. OmpT activity was assessed in a coupled spectrophotometric assay using the chromogenic substrate IAA-Arg-Arg-pNA purchased from GL Biochem Ltd. (China) (49). In a 200 μL reaction system, the assay included 100 μg total outer membrane protein, 0.5 mM IAA-Arg-Arg-pNA, 1 mM Tween 20, 20 mM Mes (pH 7.0), and 0.5 U·mL−1 aminopeptidase M. OmpT specific cleavage between the two arginines results in the release of Arg-pNA, which is subsequently cleaved by aminopeptidase M (Sigma, USA), present in excess to ensure its activity is not rate-limiting. This process releases pNA, detected spectrophotometrically at 405 nm over 12 h at 37°C using a BioTek Synergy 2 plate reader. Initial background measurements were subtracted from the final reaction sample values. The data from these experiments were plotted on a kinetic curve and analyzed using GraphPad Prism 7 software.
2.11 Inhibition of proteolytic activity
To investigate the inhibition of proteolytic activity, three serine protease inhibitors [PMSF (Beyotime, China), leupeptin (Beyotime, China), and aprotinin (all from Beyotime, China)] were used to study the roles of Asp267 (aspartic acid, D) and Ser276 (serine, S) in cOmpT through a FRET activity assay. Bacteria grown in N-minimal medium to mid-exponential phase were normalized to an OD600 nm of 0.6–0.8. Bacterial cells were pelleted by centrifugation, resuspended in PBS (pH 7.4), and adjusted to a bacterial density of 3 × 108 CFU/mL. In a 175 μL reaction system within a 96-well plate, the assay included bacteria (~2.25 × 107 CFU), 40 μM FRET substrate, and either 0.8 mM PMSF, 0.8 mM leupeptin, or 1 mM aprotinin. Fluorescence (with an excitation of 325 nm and an emission of 430 nm) was monitored over 360 min at 25°C using a BioTek Synergy 2 plate reader. Initial background measurements were subtracted from the final reaction sample values. The data were used to construct kinetic curves, and GraphPad Prism 7 software was used for differential expression analysis.
2.12 Molecular docking
The molecular docking method was used to simulate the interactions between protamine and cOmpT/pOmpT and their mutants by using the software Autodock (50). The crystal structure of cOmpT (PDB ID: 1I78) was downloaded from the PDB.2 The structures of the pOmpT, cOmpTD267S/S276T, and pOmpTS267D/T276S mutants were all obtained by online modeling using Swiss-Model based on the crystal structure of cOmpT.3 Protamine (P69015) is based on the predicted structure from the Alpha Fold protein structure database.4 Autodock Tools software was used to delete water, add hydrogens, compute Gasteiger charges, and assign AD4-type atoms to them. The grid box was 126 × 126 × 126 with a grid point spacing of 0.7 Å and centered at 25.946, 58.190, and 13.075 (x, y, z). AutoDock was used to dock protamine with cOmpT, pOmpT, and their double mutants, and the potential binding conformations were identified using a genetic algorithm. The genetic algorithm parameters included a population size of 150, a maximum number of media of 2,500,000, a maximum number of generations of 27,000, a GA crossover mode of twopt, and a number of GA runs of 10. A scoring function was used to evaluate the calculated binding free energy, and clustering was carried out using the root mean square error (RMSD) between the binding modes. The least energetic conformation was chosen as the optimal binding mode for OmpT and protamine.
2.13 Animal experiment
This study received approval from the Institutional Animal Care and Use Committee (IACUC) of Yangzhou University (Chicken: SCXK(Su)2021–0027) and was conducted in accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
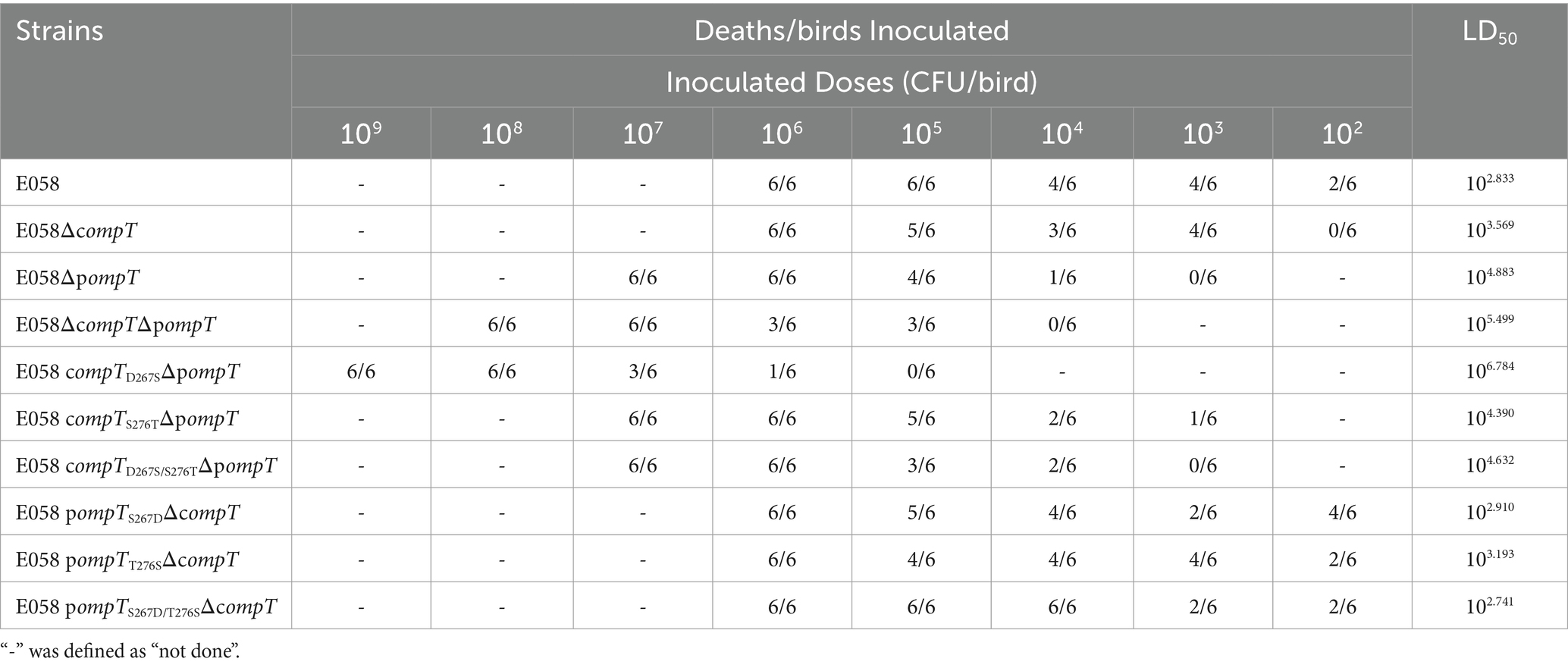
The LD50 assay was conducted on 3-day-old specific pathogen-free (SPF) chickens (White Leghorn; Jinan SPAFAS Poultry Co., Ltd., Jinan, China) to assess the pathogenicity of the wild-type strain E058 and its mutant strains in the compT and pompT genes. Cultures of the wild-type strain and its mutant derivatives were grown to the logarithmic phase at 37°C. The bacteria from each strain were collected, washed twice, and suspended in sterile PBS (containing 10% glycerol) before being diluted to appropriate concentrations (1010 or 109 CFU/mL) and then further diluted to 109, 108, 107, 106, 105, 104, and 103 CFU/mL, respectively. Six birds in each group were challenged via the air sac with 0.1 mL of each culture suspension. The mock group was injected with sterile PBS. The chickens were monitored for 7 days until survival rates stabilized. LD50 results were estimated using the Reed-Muench method and IBM SPSS statistics software. Statistical significance was assessed with the t-test, and differences with p-values <0.05 were considered statistically significant.
3 Results
3.1 pOmpT is a homologous derivative of cOmpT
Sequence alignment revealed that APEC E058 pOmpT shared a highly homologous amino acid sequence with those of cOmpT from UPEC CFT073 and cystitis 6, adherent-invasive E. coli (AIEC) NRG857C, and APEC E058 (≈ 76%) (Figure 1). Notably, pOmpT of E058 exhibited 226 out of 297 amino acid sequence identities with those of cOmpT, while it shared an identical sequence identity (297/297) with those of the ArlC encoded by plasmids in both UPEC cystitis 6 and AIEC NRG857C (Figure 1), indicating a common ancestral origin for these plasmid-encoded OmpTs. The alignment suggested that E058 cOmpT is a typical outer membrane protease, similar to OmpT of E. coli K12 MG1655, UPEC CFT073, AIEC NRG857C, and UPEC cystitis 6 strains. Although pOmpT shared only about 76% sequence identity with cOmpT, the critical residues of OmpT protease between E058 pOmpT and E058 cOmpT, including the catalytic and active sites, are identical. Nonetheless, two residues in the LPS-binding sites of E058 pOmpT differed from those in cOmpT strains. In addition, AlphaFold 3.0 was used to predict the structure of pOmpT based on the amino acid sequence of E058 pOmpT, indicating both cOmpT and E058 pOmpT exhibited a β-barrel structure.
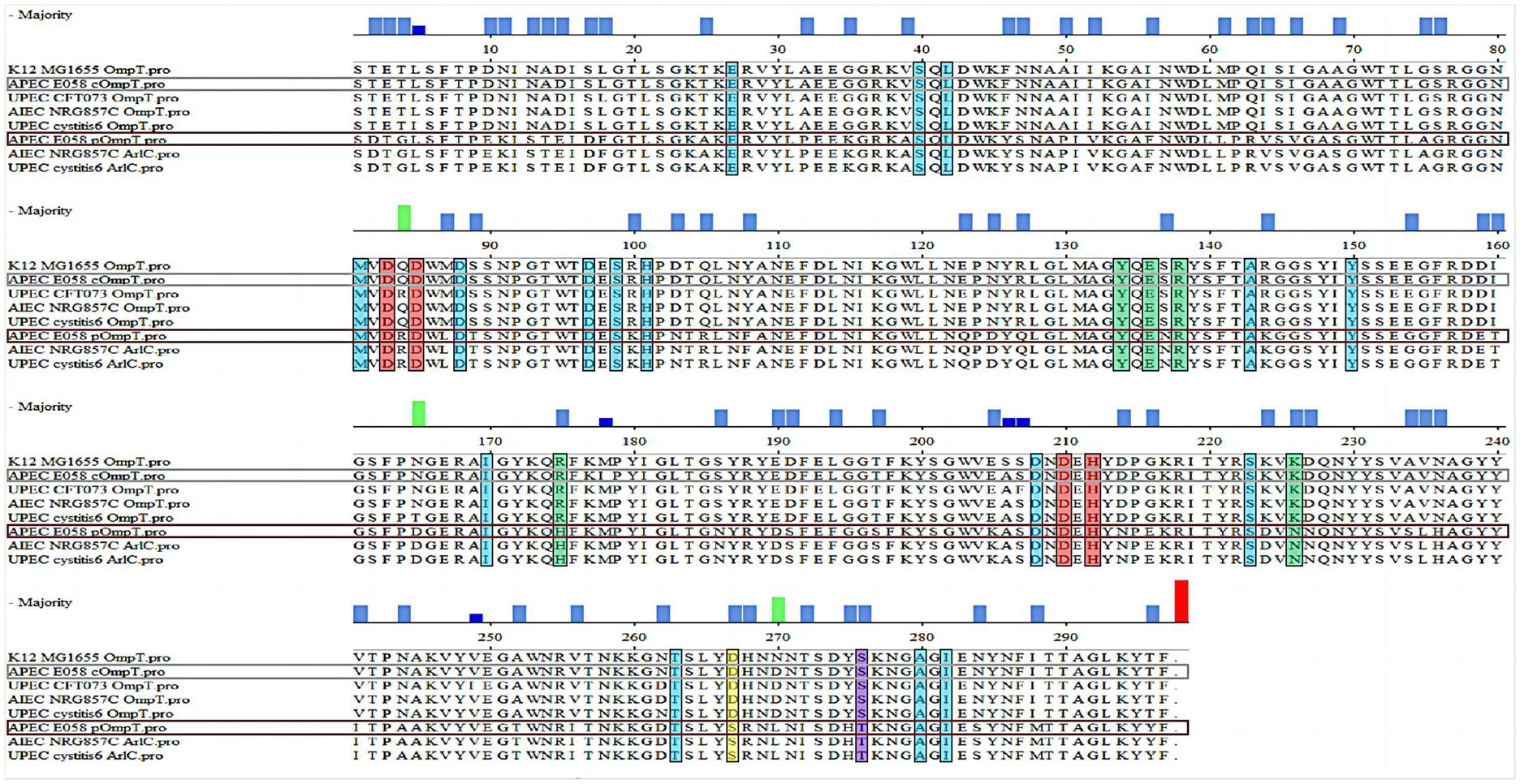
Figure 1. pOmpT is a homologous derivative of cOmpT. Alignment of the amino acid sequences between OmpT-like protein from different strains. The sequences of both the chromosome-encoded OmpT (E058 cOmpT) and the plasmid-encoded OmpT (E058 pOmpT) in the APEC E058 strain were determined by our laboratory; In the figure, the residues marked in the red boxes are the catalytic residues of OmpT outer membrane protease, the residues marked in the blue boxes are the enzyme active sites of OmpT and the green boxes indicated the amino acid residues as the binding site of OmpT and LPS.
Reverse transcription PCR (RT–PCR) revealed that both the compT and pompT genes can be transcribed constitutively and are 954 bp in length in the APEC E058 strain (Figure 2A). The results of quantitative PCR showed that the transcription level of the pompT gene was significantly greater than that of the compT gene (Figure 2B). It is unclear whether pOmpT is the same as cOmpT and is located in the outer membrane, although it shares the same number of nucleotides and highly similar amino acid sequence as cOmpTs up to about 76 percent and is also transcribed in E058. To determine the location of pOmpT, the outer membrane proteins of the wild-type strain E058 and the compT/pompT single/double-gene deletion mutants were extracted and detected by western blotting using an OmpT specific monoclonal antibody. The results showed that a single band could be detected in the E058 compT/pompT single-gene deletion mutants, and the sizes of the bands differed between the two strains, suggesting that two different proteins were expressed in these two deletion mutants (Figure 2C). Moreover, two bands in the wild-type strain E058 were detected, and the positions of the bands were consistent with those observed in the E058 compT/pompT single-gene deletion mutants (Figure 2C). The above results indicated that both pOmpT and cOmpT are expressed on the outer membrane of E058. In addition, the estimated protein molecular weight of pOmpT (≈36 kDa) was less than that of cOmpT (≈37 kDa) based on their amino acid sequences. Moreover, the expression level of pOmpT on the membrane was found to be higher than that of cOmpT, consistent with qPCR results.
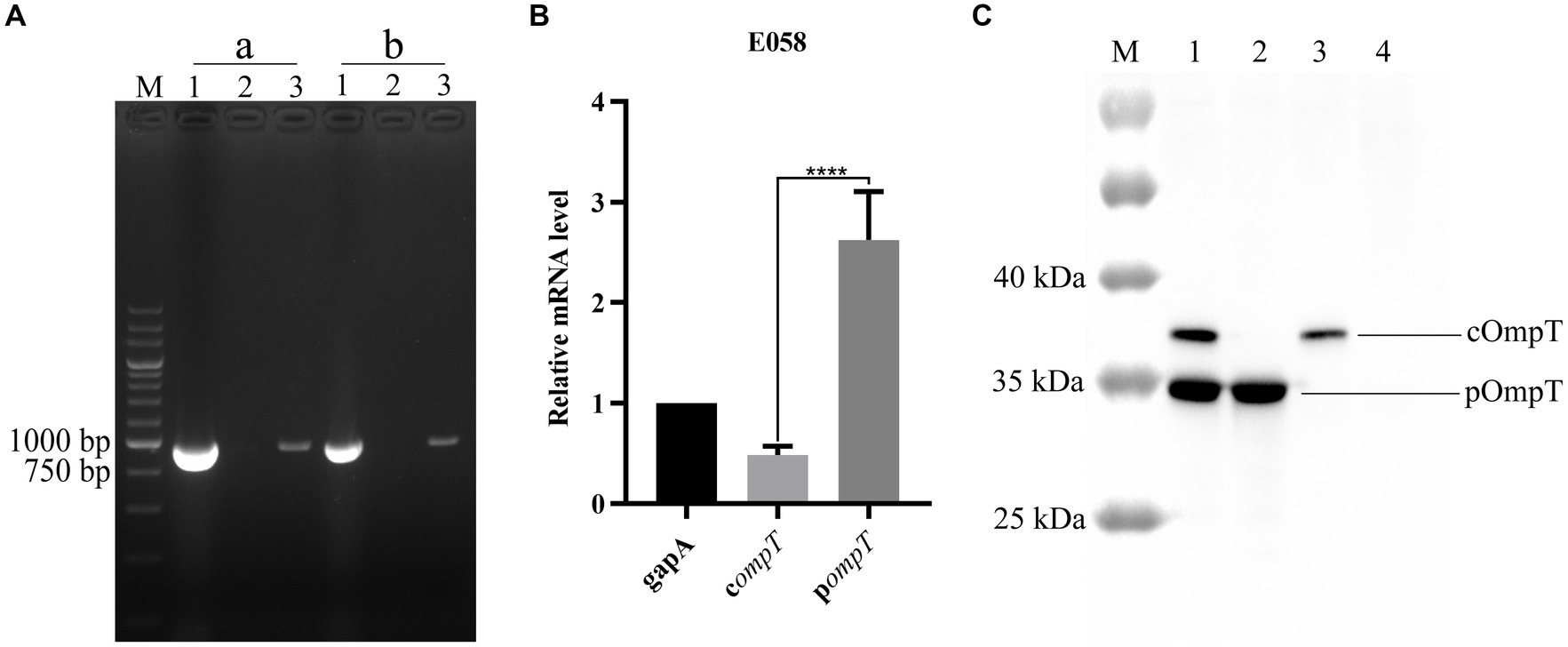
Figure 2. pOmpT is an outer membrane protein. (A) RT-PCR analysis of compT and pompT genes in APEC E058 strain. a: compT gene; b: pompT gene; Lane 1: Genomic DNA from E058; Lane 2: Total RNA from E058; Lane 3: cDNA derived from the total RNA of E058. 200 bp DNA marker (TaKaRa) was used as the molecular size standard (lane M). (B) qPCR analysis of the transcription level of compT and pompT genes in APEC E058 strain. (C) The extracted total outer membrane proteins were also analyzed with OmpT specific monoclonal antibody. Lane 1: Total outer membrane protein extracted from strain E058; Lane 2: Total outer membrane protein extracted from strain E058ΔcompT; Lane 3: Total outer membrane protein extracted from strain E058ΔpompT; Lane 4: Total outer membrane protein extracted from strain E058ΔcompTΔpompT; PageRuler™ prestained protein ladder (Thermo Fisher scientific, USA) was used as the molecular size standard (lane M). All experiments were repeated three times. Statistical significance was determined using the t-test. Differences with p-values <0.05 were considered as statistically significant. ****p < 0.0001.
3.2 pOmpT cannot resist protamine
Protamine cleavage is one of the important roles for cOmpT in host resistance (13, 34–36). To understand the role of pOmpT in resisting the host, growth kinetics were evaluated for the wild-type strain E058, compT/pompT single/double-gene deletion strains and complementation strains incubated with protamine. Compared with that of the wild-type strain, the growth of the strains E058ΔpompT and ReE058ΔcompT-ccompT did not significantly differ when cOmpT was present in the strains (p > 0.05), while the growth ability of the strain was significantly reduced when only pOmpT was present in the deletion strain E058ΔcompT and complementation strain ReE058ΔcompT-ppompT (p < 0.01) (Figure 3A). The same phenomenon was verified in recombinant compT/pompT gene-expressing bacteria (Figure 3B), which indicates that cOmpT can resist protamine but that pOmpT cannot.
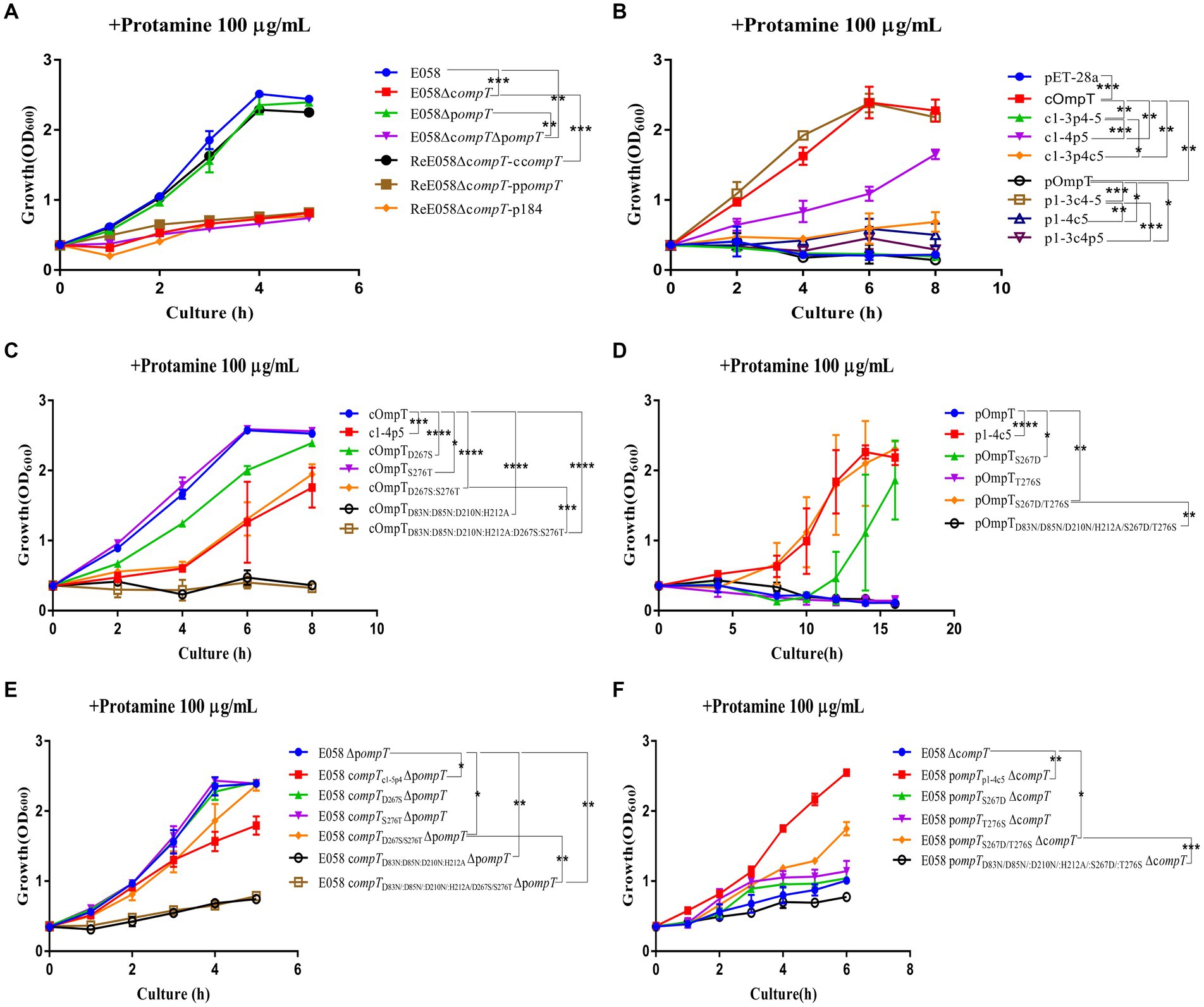
Figure 3. The kinetics of bacterial growth under protamine treatment. (A) Growth profiles of APEC wild-type strain E058, compT/pompT single/double-gene deletion strains and their complementation strains under protamine treatment. (B) Growth profiles of strains E. coli BL21(DE3) expressing cOmpT, pOmpT and the chimeric protein with the interchanged different loops between cOmpT and pOmpT under protamine treatment. (C) Growth profiles of strains E. coli BL21(DE3) expressing site-directed mutagenesis of cOmpT under protamine treatment. (D) Growth profiles of strains E. coli BL21(DE3) expressing site-directed mutagenesis of pOmpT under protamine treatment. (E) Growth profiles of pompT gene deletion strains expressing site-directed mutagenesis of cOmpT of APEC E058 under protamine treatment. (F) Growth profiles of compT gene deletion strains expressing site-directed mutagenesis of pOmpT of APEC E058 under protamine treatment. The bacterial strains used here are listed in Supplementary Table S1. All experiments were repeated three times. Statistical significance was determined using the two-way ANOVA. Differences with p-values <0.05 were considered as statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
3.3 The difference in the kind of residues at positions 267 and 276 discriminates protamine cleavage specificity by cOmpT and pOmpT
To explore which one of the five loops in pOmpT plays a critical role in protamine deactivation, mutants of pOmpT and cOmpT were engineered by swapping the loops between the two. The sensitivity of these modified constructs to protamine was assessed. The findings revealed a significant decrease in protamine resistance for the cOmpT variant containing the pOmpT L5 segment (Figure 3B). Conversely, replacing the L5 region of cOmpT with that of pOmpT in the pOmpT construct substantially increased its protamine resistance (Figure 3B). Further experimentation with mutants involving other loop exchanges between cOmpT and pOmpT indicated no significant impact on protamine resistance, underscoring the pivotal role of the L5 region in mediating the differences in protamine resistance between cOmpT and pOmpT. In pursuit of identifying crucial residues in cOmpT’s L5 contributing to protamine resistance, exchanges of single or multiple L5 residues between cOmpT and pOmpT were conducted. Protamine resistance assays demonstrated that replacing Ser276 in cOmpT with Thr276 from pOmpT significantly reduced protamine resistance (p < 0.05) (Figure 3C). Substituting Asp267 in cOmpT with Ser267 from pOmpT, or concurrently swapping both Asp267 and Ser276 in cOmpT with Ser267 and Thr276 from pOmpT, resulted in a marked decrease in protamine resistance (p < 0.001) (Figure 3C). Compared to pOmpT, a mutant form pOmpTS267D exhibited notably enhanced growth in the presence of protamine resistance (p < 0.05), and the mutant pOmpTS267D/T276S showed significantly increased protamine resistance (p < 0.01) (Figure 3D). The growth kinetics of bacterial resistance to protamine further affirmed the contribution of residues 267 and 276 in cOmpT toward protamine resistance in APEC E058 (Figures 3E,F).
We also analyzed protamine cleavage efficiency in E. coli expressing cOmpT, pOmpT and their various mutants. The cleavage efficiency of protamine by single-mutant D267S and double-mutant D267S/S276T of cOmpT was significantly lower than that of cOmpT (p < 0.001) (Supplementary Figures S1A,C). In contrast, the cleavage efficiency of protamine by the single mutant T276S and double mutant S267D/T276S of pOmpT increased significantly (p < 0.01) (Supplementary Figures S1B,D). Notably, the cleavage efficiency of the single mutant S267D of pOmpT was even greater than that of the other mutants (p < 0.001) (Supplementary Figures S1B,D). The above results indicated that L5 of cOmpT is the key loop that contributes to differences in resistance to protamine between cOmpT and pOmpT, and the key residues are residues 267 and 276.
To investigate the potential interaction between OmpT and protamine, protamine was docked to cOmpT, pOmpT, and their mutants using AutoDock software (AutoDock 4.0, Scripps Research Institute, USA)5 (50). The configuration with the lowest binding energy underwent visual analysis (51). Given that cOmpT and pOmpT are outer membrane proteins, simulation models representing the transmembrane region were excluded (50). Docking simulation results indicated that protamine could not dock into the binding pocket of pOmpT, while it readily docked into that of cOmpT (Supplementary Figures S2A,B). In the cOmpT model, six residues—D159, I160, K216, D267, S273, and D274—were found to form hydrogen bonds with corresponding residues of protamine, highlighting their importance for protamine’s binding to cOmpT (Supplementary Figure S2B). However, the cOmpTD267S/S276T-protamine complex revealed only three anchor residues (E34, D270, and N269) for protamine binding (Supplementary Figures S2C,D), aligning with the diminished cleavage ability of the cOmpTD267S/S276T mutant toward protamine. Furthermore, replacing residues 267 and 276 in pOmpT with those from cOmpT enabled protamine docking into its binding site, forming hydrogen bonds at three residues, including D267 (Supplementary Figures S2E,F). This suggests that D267 in cOmpT is a key residue for anchoring protamine.
3.4 Residues 267 and 276 of cOmpT and pOmpT are involved in substrate affinity
OmpT specifically cleaves substrates at dibasic motifs (RR, RK, and KK) using synthetic peptides as substrates (31, 32). Protamine, due to its abundance of dibasic motifs in the primary structure, is cleaved by cOmpT, explaining why the protamine cleavage products in gels are not a single band. Synthetic peptides containing dibasic motifs, such as the synthetic FRET substrate [2Abz-SLGRKIQI-K(Dnp)-NH2] and chromogenic substrate [IAA-Arg-Arg-pNA], are commonly used to detect OmpT enzyme activity (25, 49). We examined whether cOmpT and pOmpT share similar characteristics and if differences at residues 267 and 276 affect their activity toward synthetic substrate. Digestion results showed that both pOmpT and cOmpT in equal amounts of E. coli BL21(DE3) could cleave the synthetic substrate (Figures 4A–C). Although the cleavage efficiency of cOmpT remains higher than that of pOmpT, this differs from the cleavage activity toward the substrate protamine, which is exclusively digested by cOmpT. Single and double replacement mutations of cOmpT (S276T and D267S/S276T) significantly decreased cOmpT activity in equal amounts of E. coli BL21(DE3) (p < 0.01). In contrast, replacement mutation of pOmpT significantly increased cleavage efficiency compared to pOmpT (p < 0.001) in equal amounts of E. coli BL21(DE3) (Figure 4A). The notable efficiency in cleaving substrates by the pOmpTS276T variant underscores the critical role of residue 276. This observation further supports the contribution of residues 267 and 276 to the cleavage differences observed between cOmpT and pOmpT.
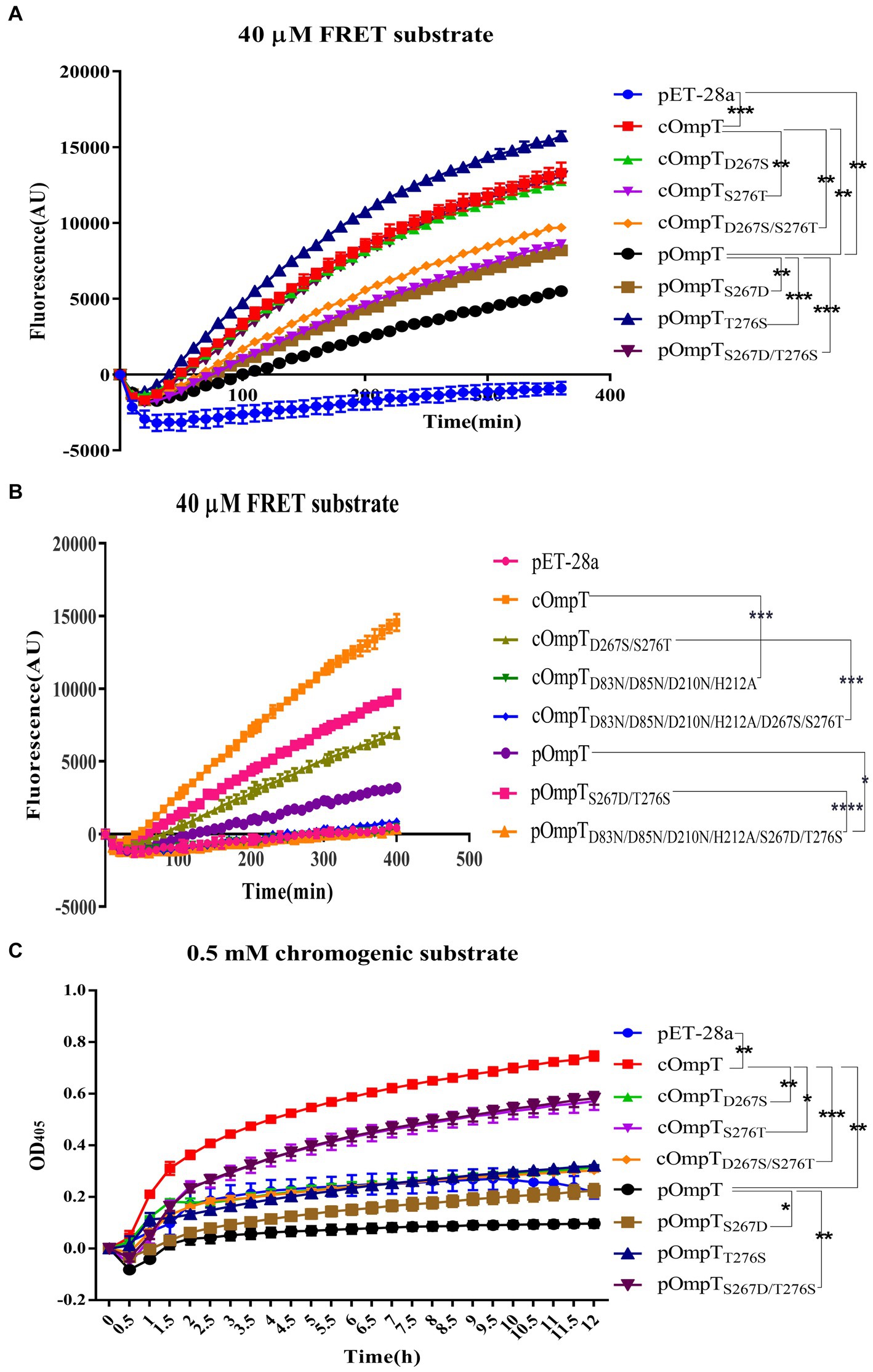
Figure 4. Determination of enzyme activity of cOmpT, pOmpT and its site-directed mutants. (A,B) Enzyme activity of cOmpT, pOmpT and their variants expressed in E. coli BL21(DE3) by utilizing a FRET substrate (2Abz-SLGRKIQI-K(Dnp)-NH2). (C) Enzyme activity of cOmpT, pOmpT and their variants expressed in E. coli BL21(DE3) by utilizing a chromogenic substrate (IAA-Arg-Arg-pNA). Statistical significance was determined using the two-way ANOVA. Differences with p-values <0.05 were considered as statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
It has been documented that cOmpT can be inhibited by serine protease inhibitors (27, 52). Experiments were conducted to ascertain whether pOmpT is similarly affected by serine protease inhibitors as cOmpT. The findings demonstrated that pOmpT in E. coli BL21(DE3) was inhibited by aprotinin (Figure 5) and leupeptin (Supplementary Figure S3), along with mutants of residues 267 and 276 in both cOmpT and pOmpT variants (Figure 5; Supplementary Figure S3). This indicates that the difference between pOmpT and cOmpT does not influence their inhibition by serine protease inhibitors. Residues Asp83, Asp85, Asp210, and His212 have been proposed as the catalytic sites for cOmpT (24, 25). Mutations at these sites, along with mutations at residues 267 and 276 in both cOmpT and pOmpT, resulted in a loss of activity toward synthetic substrates, both with and without the presence of inhibitors. This suggests these residues are essential for the activity of pOmpT as well (Figures 4B; 5C,D; Supplementary Figures S3C,D; S4C,D). Furthermore, it implies that residues 267 and 276 may not act as catalytic sites in either protein variant. To elucidate the roles of residues 267 and 276 further, the Km values of cOmpT, pOmpT, and their respective mutants were determined. The Km values for the mutants of cOmpT were found to be higher than those of the wild type (1.56–4.44 times) (Table 1), indicating a decrease in substrate binding affinity. Conversely, the Km value of pOmpT was higher than that of its mutants (2.37–4.24 times) (Table 1), suggesting that residues 267 and 276 are involved in substrate binding rather than in catalysis. The Kcat/Km values for cOmpT and pOmpT and their mutants further revealed that the catalytic efficiency of cOmpT is significantly higher than that of pOmpT. Mutations at residues 267 and 276 improved the catalytic efficiency of pOmpT, whereas they dramatically decreased that of cOmpT (Table 1), indicating that changes in binding affinity due to these residues also affect catalytic efficiency.
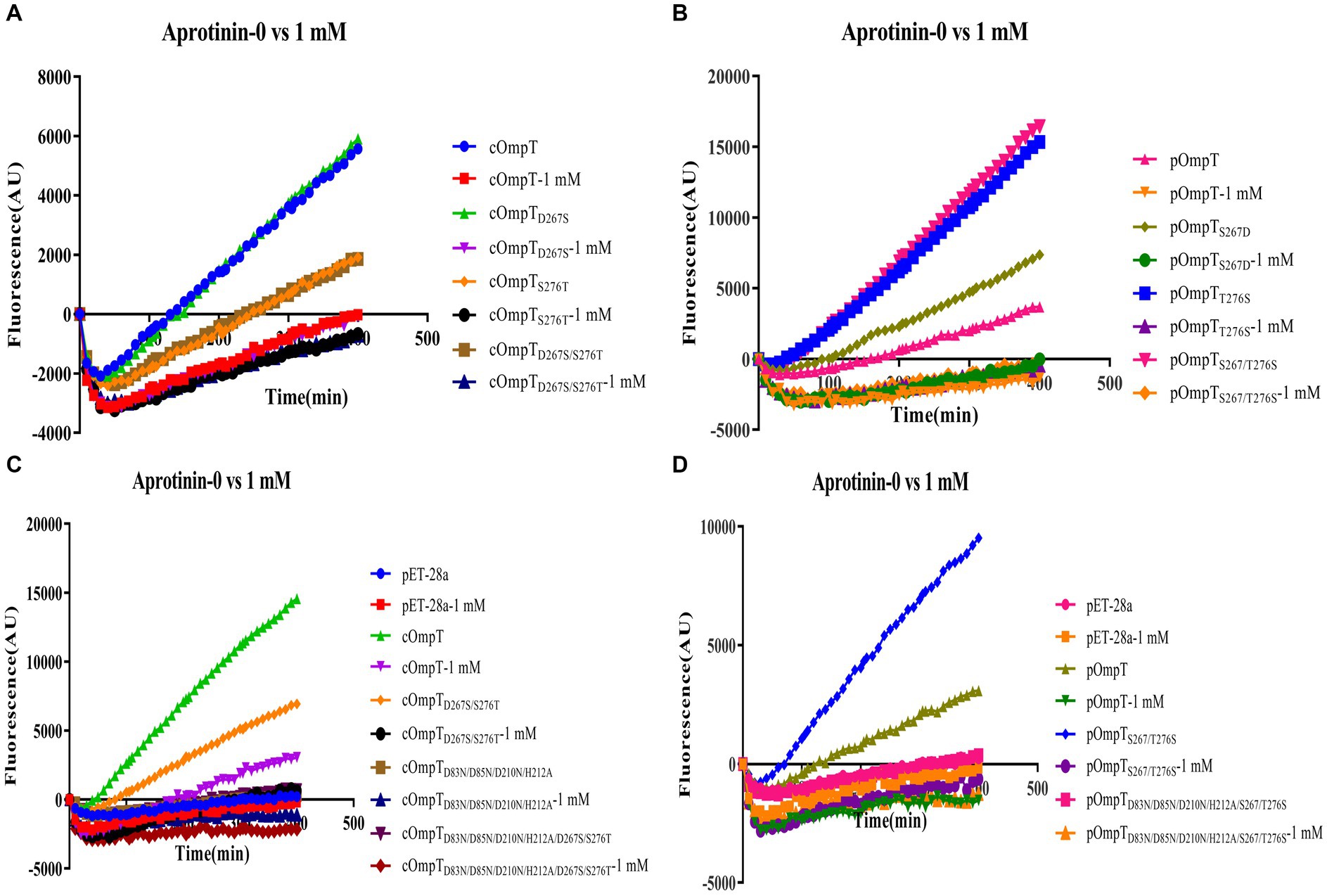
Figure 5. Inhibition of enzyme activity of cOmpT, pOmpT and its site-directed mutants expressed in E. coli BL21 by aprotinin. (A–D) FRET assays were performed with cOmpT, pOmpT and their site-directed mutants in PBS or in the presence of aprotinin (1 mM).
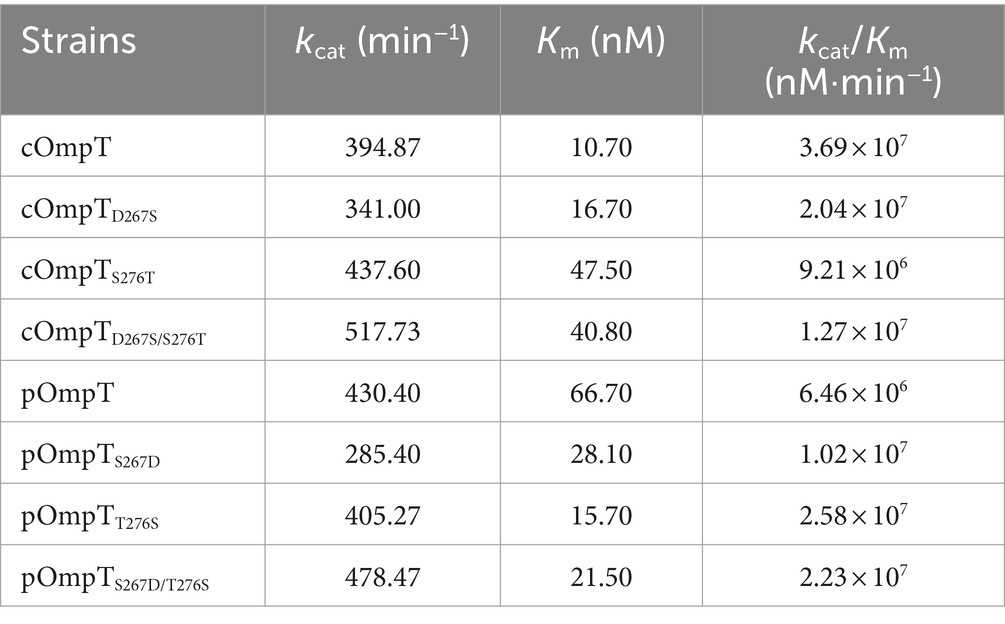
Table 1. Kinetic parameters of (c/p)OmpT and their site-directed mutants expressed in E. coli BL21 using synthetic fluorimetric peptide as substrate.
3.5 Residues 267 and 276 of pOmpT contribute to the cleavage of human RNase 7
ArlC in UPEC, which shares 100% identity with pOmpT, can promote bacterial resistance to the host by cleaving the large molecule AMP human RNase 7 (23). As expected, RNase 7 was cleaved with a clear cleavage product band remaining (<17.7 kDa) when incubated with the strain expressing pOmpT (such as strains E058 and E058ΔcompT). However, Strain E058ΔcompTΔpompT without c/pOmpT did not cleave RNase 7 with a clear RNase 7 protein band remaining (~17.7 kDa), indicating that pOmpT can also cleave RNase7 (Figure 6). Interestingly, RNase 7 was also cleaved by cOmpT (strain E058ΔpompT), which was shown to have no effect on RNase 7 in UPEC isolate cystitis 6 (23). Synthetic substrate digestion showed that residues 267 and 276 of c/pOmpT are involved in substrate recognition (Figure 6; Table 1), so we also checked the influence of changes in residues 267 and 276 on the proteolytic activity of c/pOmpT on RNase 7. The results showed that the replacement of residues 267 and 276 of cOmpT with that of pOmpT dramatically decreased the activity of cOmpT on RNase 7, and a clear band of intact RNase 7 was observed compared with that of cOmpT (Figure 6).
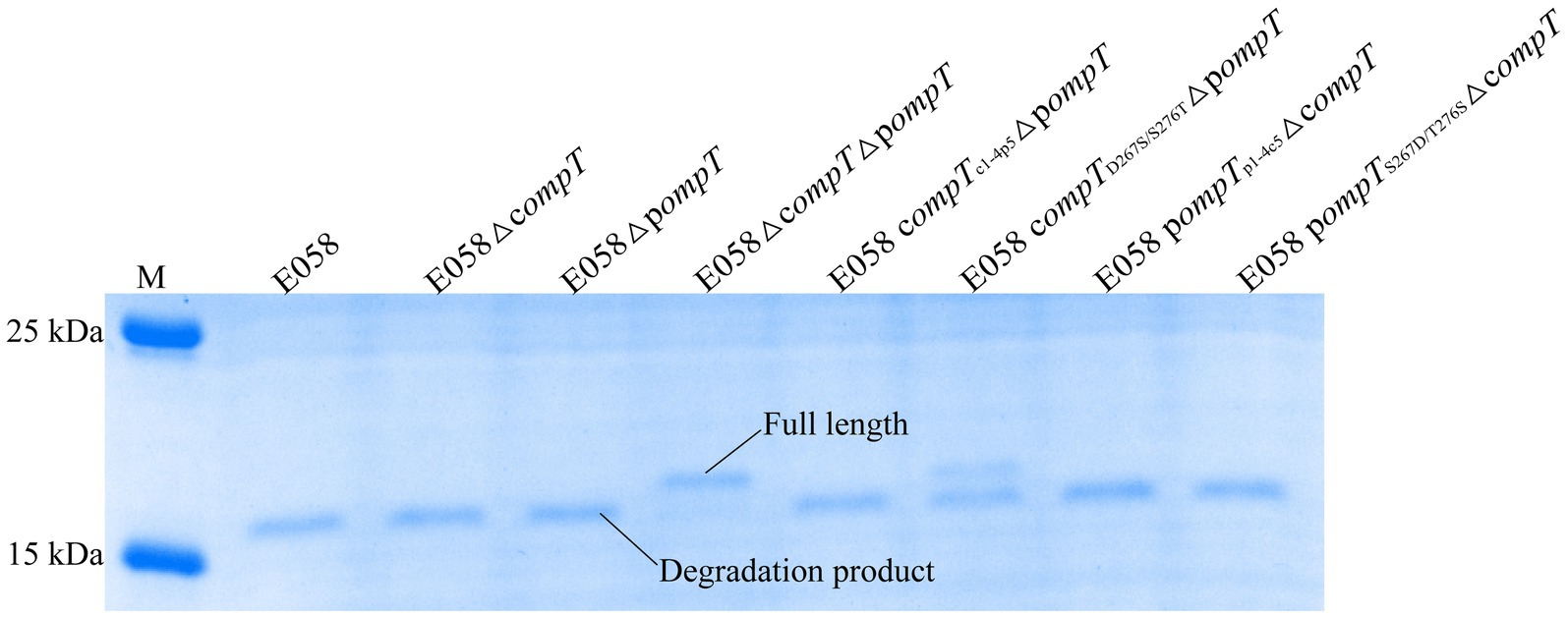
Figure 6. Changes of residues at positions 267 and 276 of pOmpT can also change its ability to cleave human RNase 7. RNase 7 was incubated with APEC E058 expressing cOmpT, pOmpT or their mutants for 15 min. 180 kDa Prestained Protein Marker (Vazyme, China) was used as the protein molecular size standard (lane M).
3.6 Residues 267 and 276 of cOmpT and pOmpT are closely related to APEC pathogenicity
The 50% lethal dose (LD50) in a chicken infection model was used to assess the pathogenicity of cOmpT, pOmpT, and their site-directed mutants in APEC E058. The LD50 for all strains is presented in Table 2 and Figure 7. The pathogenicity of single or double-gene deletion strains of cOmpT and pOmpT in APEC E058 was significantly reduced compared to the wild-type strain E058, by almost 5 (103.569/102.833, p < 0.05) (cOmpT single deletion), 112 (104.883/102.833, p < 0.01) (pOmpT single deletion), and 463 (105.499/102.833, p < 0.001) (cOmpT and pOmpT double deletion) times, respectively. This confirms the role of cOmpT and pOmpT as virulence factors in the pathogenicity of APEC E058. Interestingly, the LD50 of the mutant strain E058 compTD267SΔpompT was significantly lower, almost 80 times less than the single-gene deletion strain E058ΔpompT (106.784/104.883, p < 0.01), indicating that residue 267 of cOmpT contributes more to APEC pathogenicity than residue 276. Conversely, replacing residues 267 and 276 of pOmpT with those of cOmpT in compT gene deletion strains resulted in an LD50 higher than that of the single-gene deletion strain E058ΔcompT. The double substitution of residues 267 and 276 (102.741) exhibited the highest virulence, almost equivalent to the wild-type strain E058 (102.883/102.741, p > 0.05). Following this, the substitution of residue 267 (102.910) and residue 276 (103.193) occurred, respectively. This observation underscores a significant phenomenon: upon the deletion of cOmpT in E058, replacing only 2 residues of pOmpT resulted in its virulence matching that of the parental strain. Put differently, the substitution of two residues not only preserved the pathogenicity of pOmpT itself but also fully compensated for the virulence of cOmpT. These findings suggest that residues 267 and 276 of pOmpT also play an essential role in APEC virulence.
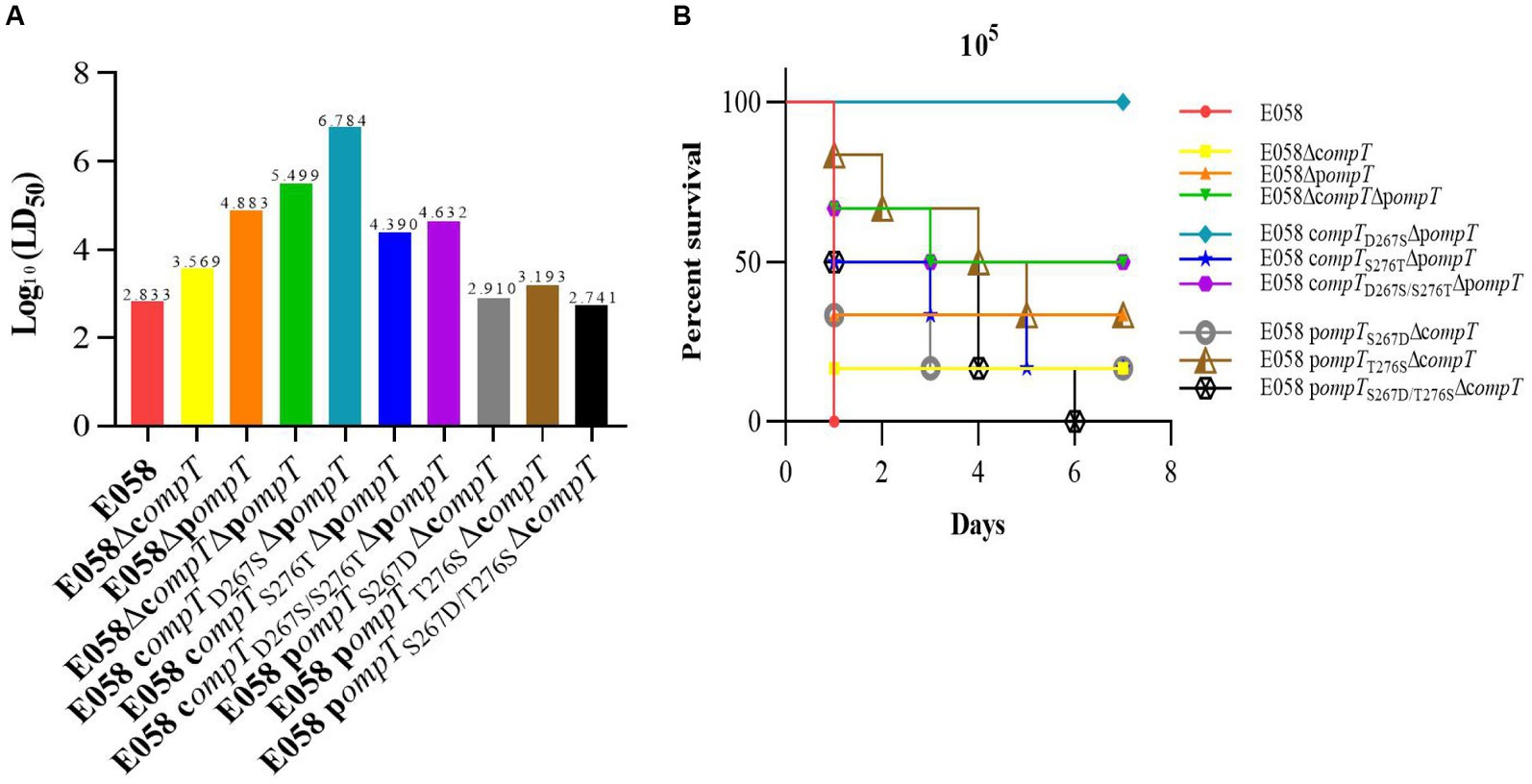
Figure 7. Residues 267 and 276 of cOmpT and pOmpT are closely related to APEC pathogenicity. (A) LD50 in a chicken infection model was used to assess the effect of cOmpT, pOmpT, and their site-directed mutants on the pathogenicity of APEC E058. The data of LD50 for all strains were presented in Table 2. (B) The survival of birds challenged with each strain at a dose of 105 CFU/bird was monitored for 7 days post-challenge based on the Kaplan Meyer survival curves.
4 Discussion
Omptins are key virulence factors involved in several gram-negative bacterial pathogens (14, 15, 17, 39). Numerous studies have suggested that OmpT-like proteins are present in intestinal or extraintestinal pathogenic E. coli strains that infect humans and animals and that they enhance bacterial virulence by participating in the cleavage of host AMPs (12, 13, 23, 41). Their contribution to E. coli virulence mainly depends on their own proteolytic activity and substrate specificity (53). Omptins in E. coli mainly includes OmpT, which is encoded on chromosomes, and OmpP/ArlC, which are encoded on episomal plasmids (23, 38, 39). Most pathogenic E. coli strains (e.g., EPEC, EHEC, UPEC) only harbor OmpT on their chromosomes. However, we discovered that the APEC E058 strain (O2 serotype), a member of the ExPECs, possesses the pompT on the ColV plasmid alongside the compT on the chromosome. Surprisingly, despite having only 76% amino acid sequence identity, both of these homologs are expressed on APEC E058’s outer membrane. Furthermore, the characterized active sites (including catalytic residues) and LPS-binding sites of cOmpT are conserved in pOmpT, with the exception of the two LPS binding sites. According to this study, APEC cOmpT and UPEC OmpT (from UPEC CFT073 and UPEC cystitis isolate 6) descended from the same ancestor, whereas pOmpT in APEC and ArlC in UPEC cystitis isolate 6 were homologs. OmpT can cleave protamine (35), which was confirmed by APEC E058. ArlC was discovered in the pathogenicity island on the plasmid of the AIEC NRG 857c strain and is involved in resistance to the human HDPs HD5, HBD2, and LL-37 (23). However, whether pOmpT or ArlC can resist protamine remains unclear. Here, the findings that pOmpT unable to cleave protamine fills the gap.
OmpT-like proteases exhibit variability in substrate specificity, indicating significant structural differences in the proteolytic active site enriched grooves between cOmpT and pOmpT despite their high amino acid sequence similarity (54). The only previously resolved structure of an E. coli OmpT-like subfamily protease was that of cOmpT, encoded on the chromosome. This structure featured a 10-chain antiparallel β-barrel, with its extracellular loop protruding above the lipid bilayer (24). Identified within this structural configuration were putative binding sites for LPS and the active sites (including the catalytic sites), located in the groove at the extracellular top of the vase-shaped β-barrel (24). Although software simulation results indicate that the overall structure of pOmpT closely resembles that of cOmpT, the true structure of pOmpT remains undiscovered. It has been found that omptins’ proteolytic activity and substrate specificity depend on residues within loops 1–5 (L1-L5), and mutations in these loops alter their activity (55). The analysis of cOmpT’s crystal structure confirms that the active sites are situated in the substrate-binding pocket region at the top of the extracellular loop (24). To investigate the resistance of protamine to pOmpT, we utilized the five extracellular loops of OmpT as entry points to examine the proteolytic activity and substrate specificity of pOmpT. The kinetics of protamine cleavage by loop-swap mutants indicated that L5 of cOmpT contributes to protamine cleavage activity. Furthermore, Asp267 and Ser276 in L5 of cOmpT were identified as the key residues that cleave protamine. Although the active sites and putative LPS-binding sites of cOmpT have been characterized, neither the 267th nor 276th residue is involved in these active sites. This result suggests that because of the characteristics of L5 and even its residues 267 and 276 of pOmpT, it loses the ability to resist protamine. Therefore, our findings are particularly important for supplementing critical information.
Escherichia coli OmpT was previously categorized as a serine protease, with residues Ser99 and His212 identified as typical active sites (56). Subsequent studies also classified OmpT as an aspartic protease (24), proposing a novel proteolytic mechanism involving a His212-Asp210 dyad and an Asp83-Asp85 pair that activates a putative nucleophilic water molecule, based on the crystal structure of E. coli OmpT (24). These active sites are fully conserved across the omptin family (24). This study investigates whether pOmpT operates via the same proteolytic mechanism as cOmpT and examines the impact of residues 267 and 276 on proteolytic activity. Sequence alignment analysis confirmed the conservation of these catalytic residues between the two homologs. Mutation of these four catalytic sites (Asp83, Asp85, Asp210 and His212) resulted in the complete abolition of the mutants’ proteolytic activity, demonstrating that neither residue 267 nor 276 interchange between cOmpT and pOmpT could restore the ability to cleave substrates, thereby indicating that these residues are not catalytic sites of OmpT. Earlier study supported the hypothesis that OmpT is a member of the serine protease family (57), since its proteolytic activity could be significantly inhibited by serine protease inhibitors such as diisopropylfluorophosphate (DFP) (27), phenylmethanesulfonyl fluoride (PMSF) (27), and aprotinin (49). Consistent with observations in cOmpT, the proteolytic activity of both wild-type pOmpT and its variant S267D/T276S were significantly inhibited by serine protease inhibitors (Aprotinin, PMSF, and Leupeptin), suggesting a shared catalytic mechanism between pOmpT and cOmpT. Subsequent analysis of the enzymatic activities of wild-type cOmpT, pOmpT, and their substitutions at residues 267 and 276 revealed a significantly lower proteolytic activity for pOmpT compared to cOmpT, aligning with pOmpT’s inability to cleave protamine. Substitution with either S276T or D267S/S276T significantly impaired the proteolytic activity of wild-type cOmpT; in contrast, replacement with S267D, T276S or S267D/T276S significantly enhanced the proteolytic activity of wild-type pOmpT. These results fully demonstrated that residues 267 and 276 of cOmpT and pOmpT are active sites that contribute to the proteolytic activity of both cOmpT and pOmpT, leading to different substrate cleavage specificities recognized by these two omptins. To elucidate the role of residues 267 and 276, the proteolytic kinetics of cOmpT, pOmpT, and their mutants involving these residues were examined using synthetic fluorescent peptides. Compared to the wild-type cOmpT, mutants displayed virtually unchanged catalytic constant (Kcat), values but the Km values were significantly increased: approximately 56% for the Asp267-to-Ser substitution [(16.7–10.7)/10.7], 344% for the Ser276-to-Thr substitution [(47.5–10.7)/10.7], and 281% for substitutions of both Asp267 and Ser276 [(40.8–10.7)/10.7]. Conversely, for pOmpT mutants relative to the wild-type pOmpT, the Km values were reduced by about 58% (Ser267-to-Asp substitution, [(66.7–28.1)/66.7]), 76% (Thr276-to-Ser substitution, [(66.7–15.7)/66.7]) and 68% (both Ser267 and Thr276 substitutions, [(66.7–21.5)/66.7]). This suggests that residues 267 and 276 in both cOmpT and pOmpT are involved in substrate binding rather than catalysis, similar to Tyr248 in carboxypeptidase A, which is involved in substrate binding as indicated by a Tyr248-to-Phe substitution that maintained the same Kcat value while increasing its Km value sixfold compared to the wild type (57). Thus, residues 267 and 276 in cOmpT and pOmpT likely serve as substrate binding sites, facilitating and promoting substrate cleavage by the catalytic center.
Protamine is a specific substrate for cOmpT but not for pOmpT; hence, we sought a specific substrate for pOmpT and investigated the mechanism underlying substrate cleavage specificity. ArlC in UPEC clinical isolates was reported to specifically cleave human AMP RNase 7 (23). However, it was observed in this study that both cOmpT and pOmpT from the APEC E058 strain could cleave human RNase 7. Substitutions of residues 267 and 276 between cOmpT and pOmpT also influenced their ability to cleave RNase 7, highlighting a unique feature of pOmpT from the APEC E058 strain: it does not cleave protamine but does cleave RNase 7, despite both being human AMPs. OmpT protease exhibits narrow cleavage specificity, preferring substrates at dibasic motifs (RR, RK, KK) (31, 32), a specificity determined by conserved residues Glu27 and Asp208 at the bottom of the deep S1 pocket and Asp97 in the shallower S1’ pocket (24). Despite the high conservation of these residues in pOmpT and cOmpT, molecular docking analysis suggests that residues 267 and 276 of cOmpT play a crucial role in substrate binding through interaction with arginine residues in protamine. This specificity variance between different omptins, driven by sequence variability in the outer loop (55), reveals differences in target substrate recognition by the 5-loop and its residues 267 and 276 of pOmpT, suggesting a structural specificity of pOmpT that prevents interaction with arginine residues in protamine, allowing for effective binding and cleavage of RNase 7 instead.
The evasion of APEC from host AMP killing effects remains a focal concern. OmpT, as a virulence factor, allows APEC to protect itself against the host. It is vital to understand whether the key sites determining substrate specificity influence APEC virulence. This study confirms that changes in the substrate-binding sites of cOmpT and pOmpT affect APEC infectivity, validating the hypothesis that OmpT-like proteases’ substrate specificity contributes to E. coli virulence. However, there was an unexplained outcome that substituting residue 267 on cOmpT with that of pOmpT in a pOmpT mutant (strain E058 compTD267SΔpompT, 106.784) was less pathogenic than the double mutant (strain E058ΔcompTΔpompT, 105.499), although the potential possibility of growth defect of this mutant had been ruled out in vitro. There were might some potential effects of residue 267 in cOmpT on other residues, even other virulence factors (viz. siderophores), leading to more attenuated virulence in the mutant. The findings were not only interesting, but also were worth making further study in future.
Given the WHO Reports in 2020 and 2021 highlighting the lack of effective treatments or preventions for bacterial infections due to antibiotic resistance, omptins could represent new targets for drug and vaccine development. Inhibiting or neutralizing these omptins is crucial in preventing septicemic bacterial infections (52, 53). Antimicrobial peptides, with their broad-spectrum antibacterial activity, low propensity for drug resistance development, and role in the innate immune system, have garnered significant interest (2, 58, 59). However, the natural composition of most antimicrobial peptides renders them susceptible to degradation by proteases like trypsin and pepsin in protease-rich physiological fluids (60). Therefore, antimicrobial peptides require modifications to resist enzymatic hydrolysis, enhancing their bactericidal efficacy for clinical application. Moreover, antimicrobial peptides also need to escape cleavage by bacterial omptins. For example, based on our findings, we can replace arginine at position 15 (R15) in protamine, which is readily bound to D267 in cOmpT, with the unnatural amino acid D-arginine, leading to escape from the recognition and binding of modified antimicrobial peptides by OmpT-like proteases. Since unnatural amino acids do not have protease recognition binding characteristics, their introduction into natural antimicrobial peptides can significantly reduce and even block protease degradation, ultimately improving antimicrobial activity (61).
In brief, we characterized the physiology of pOmpT, a newly discovered OmpT-like subfamily protease in pathogenic E. coli that causes intestinal and extraintestinal infections in both humans and poultry, and revealed its molecular mechanism involved in human AMP cleavage. Residues 267 and 276 were first characterized as substrate binding sites of omptins and play a critical role in the efficiency of AMP cleavage (62) and the pathogenicity of cOmpT and pOmpT. Importantly, both cOmpT and pOmpT of APEC, which were investigated in this study, are homologs of human ExPEC; in particular, cOmpT and pOmpT of avian origin confer APEC with the ability to cleave human AMPs, suggesting that APEC is a potential human pathogen (63). Our findings provide new insights for the development of antibacterial drugs that inhibit omptin activity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Yangzhou University (Chicken: SCXK(Su)2021-0027). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JL: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. LJ: Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. HW: Methodology, Resources, Writing – original draft, Writing – review & editing. JW: Methodology, Resources, Writing – original draft, Writing – review & editing. QG: Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. CH: Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. SG: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Key R&D Program of China (2021YFD1800404, 2017YFD0500203-2, 2017YFD0500705), the 111 Project (D18007), the National Natural Science Foundation of China (32373011, 31672553, 31972711), the National Program for High Technology Research and Development in China (2003 AA 222141), the Special Fund for Agroscientific Research in the Public Interest (201303044) to SG, and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Acknowledgments
Thanks very much bioRxiv for our study providing a preprint, you can access the preprint using the following link: https://www.biorxiv.org/content/10.1101/2022.06.17.496525v1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1410113/full#supplementary-material
Footnotes
1. ^http://www.glschina.com/en/profile.htm
3. ^https://swissmodel.expasy.org/
References
1. Samantha, G, and Le, MH. Resistance to antimicrobial peptides in gram-negative Bacteria. FEMS Microbiol Lett. (2012) 330:81–9. doi: 10.1111/j.1574-6968.2012.02528.x
2. Zasloff, M . Antimicrobial peptides of multicellular organisms. Nature. (2002) 415:389–95. doi: 10.1038/415389a
3. Hancock, R, and Sahl, HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. (2006) 24:1551–7. doi: 10.1038/nbt1267
4. Gallo, RL, and Hooper, LV. Epithelial antimicrobial Defence of the skin and intestine. Nat Rev Immunol. (2012) 12:503–16. doi: 10.1038/nri3228
5. Piers, KL, and Hancock, RE. The interaction of a recombinant Cecropin/Melittin hybrid peptide with the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. (1994) 12:951–8. doi: 10.1111/j.1365-2958.1994.tb01083.x
6. Zhang, L, Rozek, A, and Hancock, RE. Interaction of cationic antimicrobial peptides with model membranes. J Biol Chem. (2001) 276:35714–22. doi: 10.1074/jbc.M104925200
7. Brogden, KA . Antimicrobial peptides: pore formers or metabolic inhibitors in Bacteria? Nat Rev Microbiol. (2005) 3:238–50. doi: 10.1038/nrmicro1098
8. Hilchie, AL, Wuerth, K, and Hancock, RE. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol. (2013) 9:761–8. doi: 10.1038/nchembio.1393
9. Haiko, J, Suomalainen, M, Ojala, T, Lahteenmaki, K, and Korhonen, TK. Invited review: breaking barriers--attack on innate immune Defences by Omptin surface proteases of Enterobacterial pathogens. Innate Immun. (2009) 15:67–80. doi: 10.1177/1753425909102559
10. Kukkonen, M, and Korhonen, TK. The Omptin family of Enterobacterial surface proteases/Adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int J Med Microbiol. (2004) 294:7–14. doi: 10.1016/j.ijmm.2004.01.003
11. Le Sage, V, Zhu, L, Lepage, C, Portt, A, Viau, C, Daigle, F, et al. An outer membrane protease of the Omptin family prevents activation of the Citrobacter rodentium PhoPQ two-component system by antimicrobial peptides. Mol Microbiol. (2009) 74:98–111. doi: 10.1111/j.1365-2958.2009.06854.x
12. Thomassin, JL, Brannon, JR, Gibbs, BF, Gruenheid, S, and Le Moual, H. OmpT outer membrane proteases of Enterohemorrhagic and Enteropathogenic Escherichia coli contribute differently to the degradation of human Ll-37. Infect Immun. (2012) 80:483–92. doi: 10.1128/IAI.05674-11
13. Brannon, JR, Thomassin, JL, Desloges, I, Gruenheid, S, and Le Moual, H. Role of Uropathogenic Escherichia coli OmpT in the resistance against human cathelicidin Ll-37. FEMS Microbiol Lett. (2013) 345:64–71. doi: 10.1111/1574-6968.12185
14. Sodeinde, OA, Subrahmanyam, YV, Stark, K, Quan, T, Bao, Y, and Goguen, JD. A surface protease and the invasive character of plague. Science. (1992) 258:1004–7. doi: 10.1126/science.1439793
15. Lathem, WW, Price, PA, Miller, VL, and Goldman, WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. (2007) 315:509–13. doi: 10.1126/science.1137195
16. Korhonen, TK, Haiko, J, Laakkonen, L, Jarvinen, HM, and Westerlund-Wikstrom, B. Fibrinolytic and coagulative activities of Yersinia pestis. Front Cell Infect Microbiol. (2013) 3:35. doi: 10.3389/fcimb.2013.00035
17. Caulfield, AJ, Walker, ME, Gielda, LM, and Lathem, WW. The Pla protease of Yersinia pestis degrades Fas ligand to manipulate host cell death and inflammation. Cell Host Microbe. (2014) 15:424–34. doi: 10.1016/j.chom.2014.03.005
18. Franco, AA, Kothary, MH, Gopinath, G, Jarvis, KG, Grim, CJ, Hu, L, et al. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect Immun. (2011) 79:1578–87. doi: 10.1128/IAI.01165-10
19. Grodberg, J, and Dunn, JJ. OmpT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. (1988) 170:1245–53. doi: 10.1128/jb.170.3.1245-1253.1988
20. Sodeinde, OA, and Goguen, JD. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to OmpT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. (1989) 57:1517–23. doi: 10.1128/IAI.57.5.1517-1523.1989
21. Grodberg, J, and Dunn, JJ. Comparison of Escherichia Coli K-12 outer membrane protease OmpT and Salmonella typhimurium E protein. J Bacteriol. (1989) 171:2903–5. doi: 10.1128/jb.171.5.2903-2905.1989
22. Egile, C, D'Hauteville, H, Parsot, C, and Sansonetti, PJ. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. (2010) 23:1063–73. doi: 10.1046/j.1365-2958.1997.2871652.x
23. Desloges, I, Taylor, JA, Leclerc, JM, Brannon, JR, Portt, A, Spencer, JD, et al. Identification and characterization of OmpT-like proteases in Uropathogenic Escherichia coli clinical isolates. Microbiology. (2019) 8:e915. doi: 10.1002/mbo3.915
24. Vandeputte-Rutten, L, Kramer, RA, Kroon, J, Dekker, N, Egmond, MR, and Gros, P. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. (2001) 20:5033–9. doi: 10.1093/emboj/20.18.5033
25. Kramer, RA, Vandeputte-Rutten, L, Roon, G, Gros, P, Dekker, N, and Egmond, MR. Identification of essential acidic residues of outer membrane protease OmpT supports a novel active site. FEBS Lett. (2001) 505:426–30. doi: 10.1016/s0014-5793(01)02863-0
26. Sugimura, K, and Higashi, N. A novel outer-membrane-associated protease in Escherichia coli. J Bacteriol. (1988) 170:3650–4. doi: 10.1128/jb.170.8.3650-3654.1988
27. Sugimura, K, and Nishihara, T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: identity of protease VII and OmpT. J Bacteriol. (1988) 170:5625. doi: 10.1128/jb.170.12.5625-5632.1988
28. Yam, CH, Siu, WY, and Kaganovich, D. Cleavage of cyclin a at R70/R71 by the bacterial protease OmpT. Proc Natl Acad Sci USA. (2001) 98:497–501. doi: 10.1073/pnas.98.2.497
29. Gill, RT, Delisa, MP, Shiloach, M, Holoman, TR, and Bentley, WE. OmpT expression and activity increase in response to recombinant chloramphenicol acetyltransferase overexpression and heat shock in E. coli. J Mol Microbiol Biotechnol. (2000) 66:1304. doi: 10.1001/archgenpsychiatry.2009.156
30. Hui, CY, Guo, Y, He, QS, Liang, P, Wu, SC, Hong, C, et al. Escherichia coli outer membrane protease OmpT confers resistance to urinary cationic peptides. Microbiol Immunol. (2010) 54:452–9. doi: 10.1111/j.1348-0421.2010.00238.x
31. Dekker, N, Cox, RC, Kramer, RA, and Egmond, MR. Substrate specificity of the integral membrane protease OmpT determined by spatially addressed peptide libraries. Biochemistry US. (2001) 40:1694–701. doi: 10.1021/bi0014195
32. McCarter, JD, Stephens, D, Shoemaker, K, Rosenberg, S, Kirsch, JF, and Georgiou, G. Substrate specificity of the Escherichia coli outer membrane protease OmpT. J Bacteriol. (2004) 186:5919–25. doi: 10.1128/JB.186.17.5919-5925.2004
33. Varadarajan, N, Gam, J, Olsen, MJ, Georgiou, G, and Iverson, BL. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc Natl Acad Sci USA. (2005) 102:6855–60. doi: 10.1073/pnas.0500063102
34. Stathopoulos, C . Structural features, physiological roles, and biotechnological applications of the membrane proteases of the OmpT bacterial endopeptidase family: a Micro-review. Membr Cell Biol. (1998) 12:1–8.
35. Stumpe, S, Schmid, R, Stephens, DL, Georgiou, G, and Bakker, EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. (1998) 180:4002–6. doi: 10.1128/JB.180.15.4002-4006.1998
36. He, XL, Wang, Q, Peng, L, Qu, YR, Puthiyakunnon, S, Liu, XL, et al. Role of Uropathogenic Escherichia coli outer membrane protein T in pathogenesis of urinary tract infection. Pathog Dis. (2015) 73:ftv006. doi: 10.1093/femspd/ftv006
37. Thomassin, JL, Brannon, JR, Kaiser, J, Gruenheid, S, and Le Moual, H. Enterohemorrhagic and Enteropathogenic Escherichia coli evolved different strategies to resist antimicrobial peptides. Gut Microbes. (2012) 3:556–61. doi: 10.4161/gmic.21656
38. Hwang, BY, Varadarajan, N, Li, H, Rodriguez, S, Iverson, BL, and Georgiou, G. Substrate specificity of the Escherichia coli outer membrane protease OmpP. J Bacteriol. (2007) 189:522–30. doi: 10.1128/JB.01493-06
39. Mcphee, JB, Small, CL, Reid-Yu, SA, Brannon, JR, Moual, HL, and Coombes, BK. Host defense peptide resistance contributes to colonization and maximal intestinal pathology by Crohn's disease-associated adherent-invasive Escherichia coli. Infect Immun. (2014) 82:3383–93. doi: 10.1128/IAI.01888-14
40. Chen, J . Construction of compT, pompT gene knockout mutants of avian Pathogenic E. coli E058 and evaluation their pathogenicity. Yangzhou: Yangzhou University (2016).
41. Liu, J, Mu, X, Wang, X, Huan, H, Gao, Q, Chen, J, et al. Unexpected transcriptome pompT' contributes to the increased pathogenicity of a pompT mutant of avian pathogenic Escherichia coli. Vet Microbiol. (2019) 228:61–8. doi: 10.1016/j.vetmic.2018.11.011
42. Gao, S, Liu, X, Zhang, R, Jiao, X, Wen, Q, Wu, C, et al. The isolation and identification of pathogenic Escherichia coli isolates of chicken origin from some regions in China. Acta Veterinaria et Zootechnica Sinica. (1999) 30:164–71. doi: 10.3321/j.issn:0366-6964.1999.02.015
43. Xue, F, Yang, Z, Xu, Y, Wang, Y, Yu, T, Fan, M, et al. Development of Moloclonal antibodies against the cOmpT protein of avian pathogenic Escherichia coli and identification of epitopes recognized by Moloclonal antibodies. Acta Veterinaria et Zootechnica Sinica. (2020) 51:2270–80. doi: 10.11843/j.issn.0366-6964.2020.12.021
44. Nelson, DL, and Kennedy, EP. Magnesium transport in Escherichia coli: inhibition by cobaltous ion. J Biol Chem. (1971) 246:3042–9. doi: 10.1016/S0021-9258(18)62288-4
45. Jiang, Y, Chen, B, Duan, C, Sun, B, Yang, J, and Yang, S. Multigene editing in the Escherichia coli genome via the Crispr-Cas9 system. Appl Environ Microbiol. (2015) 81:2506–14. doi: 10.1128/AEM.04023-14
46. Koten, B, Simanski, M, Glaser, R, Podschun, R, Schroder, JM, and Harder, J. Rnase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One. (2009) 4:e6424. doi: 10.1371/journal.pone.0006424
47. Wang, H, Schwaderer, AL, Kline, J, Spencer, JD, Kline, D, and Hains, DS. Contribution of structural domains to the activity of ribonuclease 7 against Uropathogenic Bacteria. Antimicrob Agents Chemother. (2013) 57:766–74. doi: 10.1128/AAC.01378-12
48. Gao, S, Liu, X, and Zhang, R. An improved method for rapid isolation of the outer membrane proteins from Escherichia coli isolates of Chichen Origen. Microbiology China. (1996) 23:122–4.
49. Kramer, RA, Zandwijken, D, Egmond, MR, and Dekker, N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease OmpT. Eur J Biochem. (2000) 267:885–93. doi: 10.1046/j.1432-1327.2000.01073.x
50. Morris, GM, Huey, R, Lindstrom, W, Sanner, MF, Belew, RK, Goodsell, DS, et al. Autodock4 and Autodocktools4: automated docking with selective receptor flexibility. J Comput Chem. (2009) 30:2785–91. doi: 10.1002/jcc.21256
51. Zhang, S, Wang, Y, Han, L, Fu, X, Wang, S, Li, W, et al. Targeting N-terminal human maltase-Glucoamylase to unravel possible inhibitors using molecular docking, molecular dynamics simulations, and adaptive steered molecular dynamics simulations. Front Chem. (2021) 9:711242. doi: 10.3389/fchem.2021.711242
52. Brannon John, R, Burk David, L, Leclerc, J-M, Thomassin, J-L, Portt, A, Berghuis Albert, M, et al. Inhibition of outer membrane proteases of the Omptin family by Aprotinin. Infect Immun. (2015) 83:2300–11. doi: 10.1128/iai.00136-15
53. Hritonenko, V, and Stathopoulos, C. Omptin proteins: an expanding family of outer membrane proteases in gram-negative Enterobacteriaceae (review). Mol Membr Biol. (2007) 24:395–406. doi: 10.1080/09687680701443822
54. Brannon John, R, Thomassin, J-L, Gruenheid, S, and Le Moual, H. Antimicrobial peptide conformation as a structural determinant of Omptin protease specificity. J Bacteriol. (2015) 197:3583–91. doi: 10.1128/jb.00469-15
55. Kukkonen, M, Lähteenmäki, K, Suomalainen, M, Kalkkinen, N, Emödy, L, Lång, H, et al. Protein regions important for plasminogen activation and inactivation of Α2-Antiplasmin in the surface protease Pla of Yersinia pestis. Mol Microbiol. (2001) 40:1097–111. doi: 10.1046/j.1365-2958.2001.02451.x
56. Kramer, RA, Dekker, N, and Egmond, MR. Identification of active site serine and histidine residues in Escherichia coli outer membrane protease OmpT. FEBS Lett. (2000) 468:220–4. doi: 10.1016/S0014-5793(00)01231-X
57. Gardell, SJ, Craik, CS, Hilvert, D, Urdea, MS, and Rutter, WJ. Site-directed mutagenesis shows that tyrosine 248 of carboxypeptidase a does not play a crucial role in catalysis. Nature. (1985) 317:551–5. doi: 10.1038/317551a0
58. Lee, MO, Jang, H-J, Rengaraj, D, Yang, S-Y, Han, JY, Lamont, SJ, et al. Tissue expression and antibacterial activity of host defense peptides in chicken. BMC Vet Res. (2016) 12:231. doi: 10.1186/s12917-016-0866-6
59. Hancock, REW, Nijnik, A, and Philpott, DJ. Modulating immunity as a therapy for bacterial infections. Nat Rev Microbiol. (2012) 10:243–54. doi: 10.1038/nrmicro2745
60. Ong, ZY, Wiradharma, N, and Yang, YY. Strategies employed in the design and optimization of synthetic antimicrobial peptide Amphiphiles with enhanced therapeutic potentials. Adv Drug Deliv Rev. (2014) 78:28–45. doi: 10.1016/j.addr.2014.10.013
61. Ong, ZY, Cheng, J, Huang, Y, Xu, K, Ji, Z, Fan, W, et al. Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic Β-sheet forming peptide Amphiphiles. Biomaterials. (2014) 35:1315–25. doi: 10.1016/j.biomaterials.2013.10.053
62. Liu, JH, Ran, TT, Jiang, LY, Gao, QQ, Huan, CC, Wang, H, et al. Physiology and structure of pathogenic Escherichia coli pOmpT reveal two substrate-binding sites. bioRxiv. (2022). doi: 10.1101/2022.06.17.496525
63. Johnson, TJ, Wannemuehler, Y, Johnson, SJ, Stell, AL, Doetkott, C, Johnson, JR, et al. Comparison of Extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol. (2008) 74:7043–50. doi: 10.1128/AEM.01395-08
Keywords: avian pathogenic Escherichia coli, proteolytic activity, cleavage specificity, substrate affinity, substrate-binding site, virulence
Citation: Liu J, Jiang L, Wang H, Wu J, Gao Q, Huan C and Gao S (2024) Protamine cleavage specificity of the avian pathogen Escherichia coli OmpT reveals two substrate-binding sites related to virulence. Front. Vet. Sci. 11:1410113. doi: 10.3389/fvets.2024.1410113
Edited by:
Dirk Werling, Royal Veterinary College (RVC), United KingdomReviewed by:
Timothy James Wells, The University of Queensland, AustraliaKezong Qi, Anhui Agricultural University, China
Copyright © 2024 Liu, Jiang, Wang, Wu, Gao, Huan and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Gao, Z3NvbmdAeXp1LmVkdS5jbg==
 Juanhua Liu
Juanhua Liu Luyao Jiang
Luyao Jiang Hang Wang
Hang Wang Jiayan Wu
Jiayan Wu Qingqing Gao
Qingqing Gao Changchao Huan
Changchao Huan Song Gao
Song Gao