- 1College of Animal Science and Technology, Henan Agricultural University, Zhengzhou, China
- 2Institute of International Education, Henan Agricultural University, Zhengzhou, China
- 3College of Animal Science and Technology, Northeast Agricultural University, Harbin, China
- 4Henan Delin Biological Products Co., Ltd., Xinxiang, China
Fermented total mixed ration (FTMR) is an effective method of preserving high-moisture byproducts with higher aerobic stability after fermentation. FTMR has the potential to fulfill the daily nutritional requirements of cattle and enhance their production performance. The objective of this research was to examine the influence of FTMR on lactation performance, total tract apparent digestibility, fecal microbiota communities, and fermentation profiles in lactating dairy cows. A total of 12 cows were randomly assigned into two groups: the TMR group and the FTMR group. The TMR group was fed a total mixed ration (TMR) diet, and the FTMR group was fed an FTMR diet. The FTMR did not impact milk yield in dairy cows despite a decrease in dry matter intake, which increased the efficiency of the feed. In contrast to that in the TMR group, the milk fat content in the FTMR group was greater. The FTMR group showed greater digestibility of neutral detergent fiber (NDF), organic matter (OM), dry matter (DM), crude protein (CP), and acid detergent fiber (ADF) in the total digestive tract than did the TMR group. The FTMR increased the concentration of butyrate in the fecal matter and reduced the pH of the feces. The Chao1, ACE, and Shannon indices of the archaeal community in dairy cow feces were significantly higher in cow fed the FTMR compared to those fed the TMR. LefSe analysis revealed higher levels of Oscillospira, Lactobacillus, Prevotella, and Dehalobacterium in the feces of dairy cows fed the FTMR than in those fed the TMR. However, the abundances of Roseburia, rc4-4, Bulleidia and Sharpea exhibited the opposite trend. The abundances of Halobacteria, Halobacteriales, and Halobacteriaceae, which are biomarkers for distinguishing fecal archaea in the TMR from the FTMR, were substantially greater in the feces of dairy cows that consumed the TMR than in those that consumed the FTMR. Therefore, FTMR can improve the milk fat content, total tract apparent feed digestibility efficiency, and diversity of archaea in the feces. Additionally, this work provides a theoretical basis for the feasibility of FTMR feeding for dairy cows.
1 Introduction
One of the primary challenges confronting the cattle industry is the deficiency of high-quality protein and roughage resources in China (1). Despite the presence of abundant unconventional feed resources and crop residues in China, their digestion and utilization rates as feed resources remain low (2). Fermented total mixed ration (FTMR) technology represents a novel feeding approach developed from TMR modulation and processing technology (3). It effectively utilizes unconventional feed resources and crop residues as feed sources, minimizing waste, which helps mitigate feed resource shortages and reduce breeding costs (4). A high-quality diet with comprehensive, balanced nutrition and long-term storage is achieved by thoroughly mixing and fermenting various feed materials, including forage, concentrate, and essential minerals, with fermentation bacteria (5). After fermentation, the feed demonstrates high levels of crude protein, excellent palatability, and significant nutritive value (6, 7). FTMR can effectively utilize unconventional feed resources and crop residues as feed sources to reduce waste, thereby alleviating feed resource shortages and lowering breeding costs.
Due to the intricate conditions of anaerobic fermentation in FTMR, their microbial composition is highly diverse. Therefore, the microbial makeup of feed plays a critical role not only in fermentation quality but also in the aerobic stability of the feed (8). With advancements in microbial technology applied in the feed industry, the use of enzymes and bacteria has become a common strategy to enhance silage feed quality (9). Optimal combinations of enzymes and bacteria can effectively boost the fermentation of feed materials, improving both the quality and feeding efficiency (10–12). Cellulase breaks down cellulose, hemicellulose, and lignin in feed into accessible sugars that lactic acid bacteria can utilize, increasing their fermentation substrate, accelerating pH reduction, and enhancing silage fermentation quality (13, 14). Lactobacillus buchneri, a heterotypic fermentation bacterium, not only enhances aerobic stability and prolongs shelf life but also improves animal gut health and immunity, and increases milk production in cows (15).
In ruminants, the gastrointestinal microbes of dairy cows are pivotal for acquiring nutrients and energy (16, 17). Research has shown that diet largely determines the composition of bacteria in feces and the structure of the archaeal community in dairy cows, with the fecal microbiome serving as an indicator of hindgut microbial changes (18–20). Diets high in concentrates significantly decrease the populations of cellulolytic bacteria such as Ruminococcus, Fibrobacter, and Ruminiclostridium, as well as methanogens like Methanosarcina, Methanobrevibacter, and Methanosphaera (21). Al-Azzawi et al. demonstrated that adding powdered activated carbon to the diet reduces the abundance of Proteobacteria and Methanobrevibacter in feces (21). Additionally, hindgut archaea are responsible for methane production, which contributes significantly to global warming (22–24). However, limited information exists on the effects of co-fermentation TMR with Lactobacillus brucei and cellulase on fecal fermentation parameters and microbial diversity. This study aimed to evaluate and compare the impacts of total mixed rations (TMRs) and fermented total mixed rations (FTMR) on lactation performance, fecal fermentation parameters, and fecal microbiota in dairy cattle, providing valuable insights into the use of FTMR for dairy cow management.
2 Materials and methods
2.1 Animals and diets
All animal experimental procedures were approved by the Animal Care and Use Committee of Henan Agricultural University (Approval number: HENAU-2021-025).
Experimental dairy cows were obtained from Ruiya Dairy Farm located in Zhengzhou, Henan, China. A total of twelve parity 2 Holstein cows (average body weight = 616 ± 13.4 kg, average lactation period = 106 ± 7.55 days) were randomly assigned to either the total mixed ration (TMR, n = 6) or fermented total mixed ration (FTMR, n = 6) groups based on their daily milk yield and lactation period. Each group included 6 replicates (pens) with one cow per pen. The study was conducted over a period of 14 weeks, with the initial two weeks designated as an adaptation period.
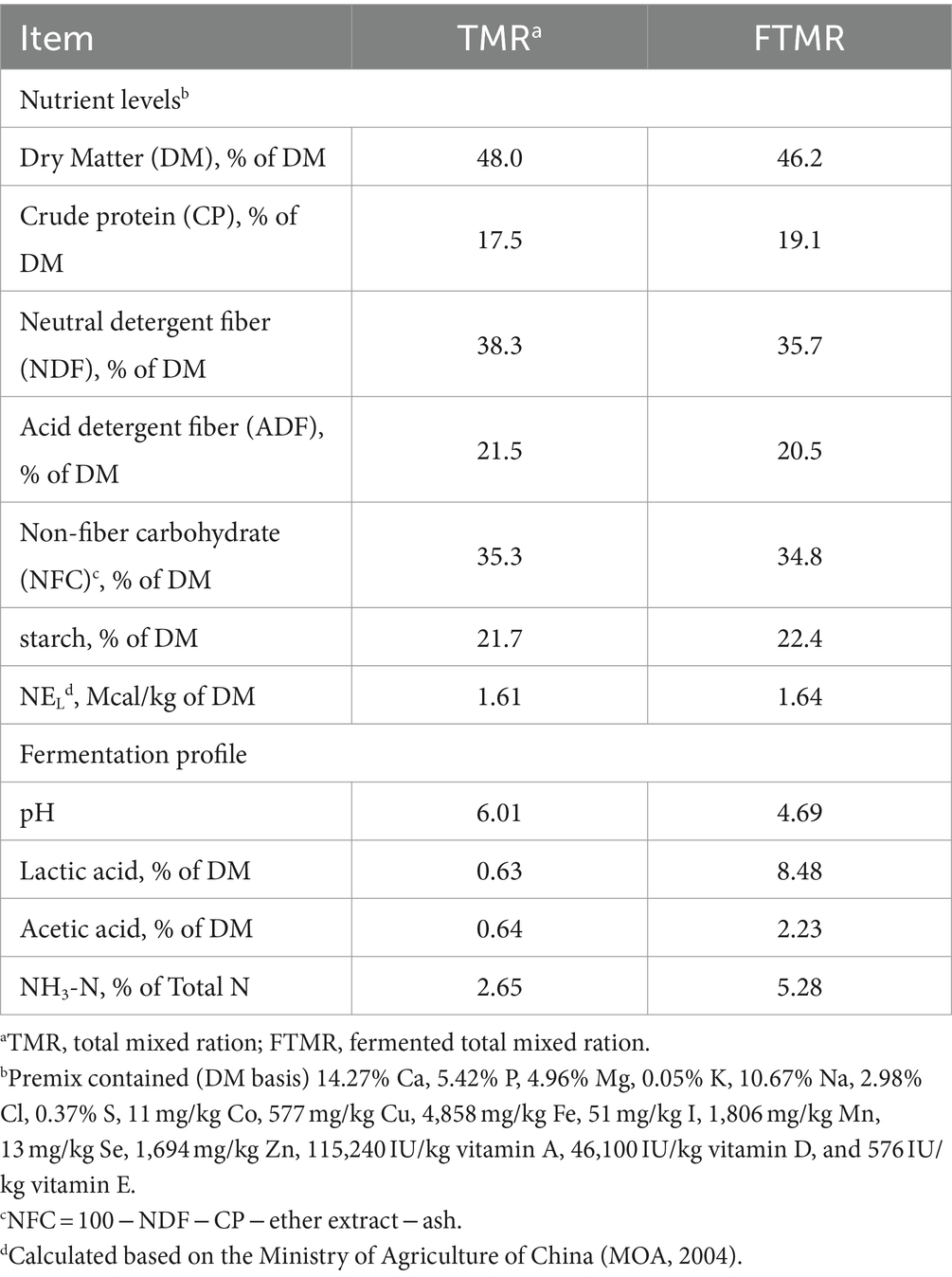
Both the TMR and FTMR diets consisted of 50% forage and 50% concentrate on a dry matter basis, formulated to meet nutritional requirements using the Cornell-Penn-Miner Dairy model version 3.08.01. The compositions of TMR and FTMR were identical (on a dry matter basis): alfalfa hay (11.80%), wet corn gluten feed (6.86%), corn stover (3.43%), corn silage (27.80%), ground corn (24.60%), soybean meal (9.80%), cottonseed meal (4.91%), DDGS (7.35%), expanded soybean (0.95%), and premix (2.50%). The moisture content was adjusted to 48.0%. Lactic acid bacteria (LAB) and cellulase were added to the FTMR. LAB included a blend of Lactobacillus plantarum (CGMCC 1.12934, obtained from the China General Microbiological Culture Collection Center) and Lactobacillus brucei (BNCC189797, obtained from Beina Bio, Beijing, China) in a 1:1 ratio, applied at 1 × 1011 cfu/g of fresh material. Cellulase (10,000 U/g, XS Biotechnology Co., Ltd., Beijing, China) was added at 10 g/kg of fresh material. Additives were dissolved in water and uniformly sprayed onto the mixture using a sprayer. The fermented mixture was packed using polyethylene stretch film with a compaction of 800 cm3 and fermented outdoors for 60 days at 16–30°C using a silage packer (Takakita MW1010H, Japan). Table 1 presents the nutrient compositions of both feeds. Cows were fed ad libitum twice daily at 12 h intervals (6:30 and 18:30) with free access to water in individual stall barns. Dairy cows were milked twice daily at 6:00 and 18:00.
2.2 Analysis of milk production and its components
At weeks 7 and 13 of the experiment, milk yield was recorded, and milk samples were collected over 6 consecutive days. Each day, at the Henan Dairy Herd Improvement Testing Center in Zhengzhou, China, a 50 mL aliquot of milk, proportional to the actual daily yield (morning and afternoon), was mixed using an automated near-infrared milk analyzer (MilkoScan, Foss Electric, Hillerød, Denmark). Potassium dichromate was added to preserve the milk samples, which were then stored at 4°C until analysis. Lactose, protein, fat, milk urea nitrogen (MUN), total solids (TS), and somatic cell count (SCC) were determined using infrared analysis methods described by Laporte and Paquin (25).
2.3 Fecal sample collection
During weeks 7 and 13 of the experiment, about 500 g of spot fecal samples were gathered from the cows’ rectums using sterile gloves. These samples were composited for each cow at both 6:00 and 18:00 over three consecutive days. To measure the pH, 5 g of fecal material was mixed with 250 mL of distilled water, and the pH was promptly measured using a portable pH meter (PHB-5, ShanghaiLeici, Shanghai, China).
One set of samples was used for total-tract apparent digestibility. Fecal samples were dried at 60°C in a forced-air oven and then ground using a 1 mm screen in a microplant grinding machine (FZ102, Taisite Instrument Co., Ltd., Tianjin, China). Subsequently, they were analyzed for dry matter (DM), crude protein (CP), ash, starch, neutral detergent fiber (NDF), acid detergent fiber (ADF), and indigestible NDF (iNDF). The other set of samples was used for fecal microorganism analysis. The samples were immediately frozen using liquid nitrogen. Equal amounts of frozen samples from each cow and time point were combined to ensure uniformity, using a sterile tap homogenizer (Shanghai Hannuo Ltd., Shanghai, China). These mixed samples were then stored at −80°C to minimize microbial activity, preparing them for subsequent DNA extraction.
2.4 Fecal sample analysis
Feed samples were analyzed for ash, dry matter (DM), crude protein (CP), and starch following procedures 942.05, 934.01, 976.05, and 982.30, respectively, as outlined by the Association of Official Analytical Chemists (AOAC, 1990). Neutral detergent fiber (NDF) and acid detergent fiber (ADF) concentrations were determined consecutively using an Ankom A200 fiber analyzer (Ankom Technology, Macedon, NY). Indigestible NDF (iNDF) served as an indirect marker, and total tract apparent nutrient digestibility was calculated accordingly. The iNDF marker was identified through in vitro analysis described by Goeser and Combs (26). For fecal samples, raw material was diluted 1:4 with distilled water to measure fecal pH using an Accumet AB150 pH meter (Fisher, Canada).
Fecal volatile fatty acid (VFA) content was determined using high-performance liquid chromatography (HPLC, Waters 600, Milford, Massachusetts, United States). Each sample preparation involved diluting 1 g of fecal material with 1 mL of water. Following this, 300 μL of an internal standard (4-methylvaleric acid, Sigma-Aldrich, St. Louis, MO) and 200 μL of 25% phosphoric acid were added, thoroughly mixed, and then centrifuged at 12,000 × g for 15 min at 4°C. The resulting supernatant was transferred to a new tube for analysis using HPLC, following the method detailed by Wang et al. (27).
2.5 Body condition score
During weeks 7 and 13 of the experiment, veterinarians assessed the body condition score (BCS) of the experimental cows shortly after the morning milking session. The assessment used a 5-point scale (1–5 points, with increments of 0.25) as outlined by Vasseur et al. (28).
2.6 Amplification of 16S rRNA and Illumina MiSeq sequencing
Genomic DNA extraction from fecal samples utilized the TIANamp Bacteria DNA Kit (TIANGEN, Peking, China) following the manufacturer’s protocol. Verification of DNA concentration and integrity was conducted using both agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Massachusetts, United States). The DNA was subsequently amplified in triplicate employing the Q5 High-Fidelity DNA Polymerase System (New England Biolabs (Beijing) LTD, Beijing, China). The V3–V4 region of the 16S rRNA gene for bacterial analysis was amplified using primers 338F (5′-ACTCCTRCGGGAGGCAGCAG-3′) and 806R (5′-GGACTACCVGGGTATCTAAT-3′) (23), while the 16S rRNA gene region V3–V4 for archaeal analysis was amplified using primers 524F (5′-TGYCAGCCGCCGCGGTAA-3′) and 958R (5′-YCCGGCGTTGAVTCCAATT-3′). PCR amplification consisted of an initial denaturation at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C, annealing at 60°C for bacteria/55°C for archaea, and elongation at 72°C, concluding with a final extension step at 72°C for 10 min. PCR products were analyzed by 2% agarose gel electrophoresis. Purification utilized the AxyPrep DNA Gel Extraction Kit (Axygen Bioscience, Union City, CA, United States), followed by quantification using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, Massachusetts, United States). Sequencing libraries were validated using an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), and their quantification was confirmed by an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and Promega QuantiFluor™-ST (Promega, Madison, WI, United States). Sequencing was performed by Frasergen Bioinformatics Technology Co., Ltd. (Wuhan, China) on an Illumina MiSeq platform (Illumina, Inc., San Diego, CA, United States), and the original RNA-seq data have been deposited into the NCBI Sequence Read Archive (SRA), accession number: PRJNA1119071.
2.7 Statistical and bioinformatics analyses
Processing of sequences from the MiSeq platform was performed using QIIME (version 1.8.0) (29). Reads meeting criteria of average quality score ≥ 25 and length between 220–250 nt were retained. Overlapping sequences (>10 bp overlap) were assembled using FLASH v1.2.7. To get high-quality clean tags, raw reads underwent specific filtering conditions via QIIME (v1.8.0) quality control: sequences ≤160 bp or with ≥8 bp homopolymers were excluded. Operational taxonomic units (OTUs) were clustered at 97% identity using UCLUST (30), and chimeric sequences were removed with USEARCH (v5.2.236, http://www.drive5.com/usearch/). The most prevalent sequence within each OTU (bacteria and archaea) was defined as the “representative sequence” and aligned against the SILVA bacterial database (version 119) (31), NCBI-nt protozoa database (32), Unite fungi ITS database (version 7.0) (33), and SILVA archaea database (31) using PyNAST (29) with standard parameters. Alpha diversity indices (ACE, Chao1, Shannon, Simpson) were computed by rarefied samples in QIIME to assess diversity and abundance of bacterial and archaeal communities. Principal component analysis (PCA) was utilized for beta diversity evaluation. Linear discriminant analysis effect size (LefSe) analysis on the Galaxy online platform (34) identified discriminative functional biomarkers, employing a size-effect threshold of 2.0 on the logarithmic LDA score to determine major abundant modules in the TMR and FTMR groups.
Independent-sample t-tests (for normally distributed data) or Mann–Whitney U tests (for nonnormally distributed data) were used to determine substantial differences in the relative abundance of the top 10 phyla and genera and in the alpha diversity indices between the two groups. p < 0.05 and p < 0.01 indicated statistically significant and extremely significant differences, respectively.
3 Results
3.1 Effect of TMR and FTMR on the lactation performance of dairy cows
Table 2 presents milk production and component findings in addition to feed intake. Dairy cows consuming the FTMR diet showed reduced DMI, increased milk fat concentration, higher ECM, and improved feed efficiency compared to those on the TMR diet. Significant differences in feed efficiency were observed between the TMR and FTMR groups.
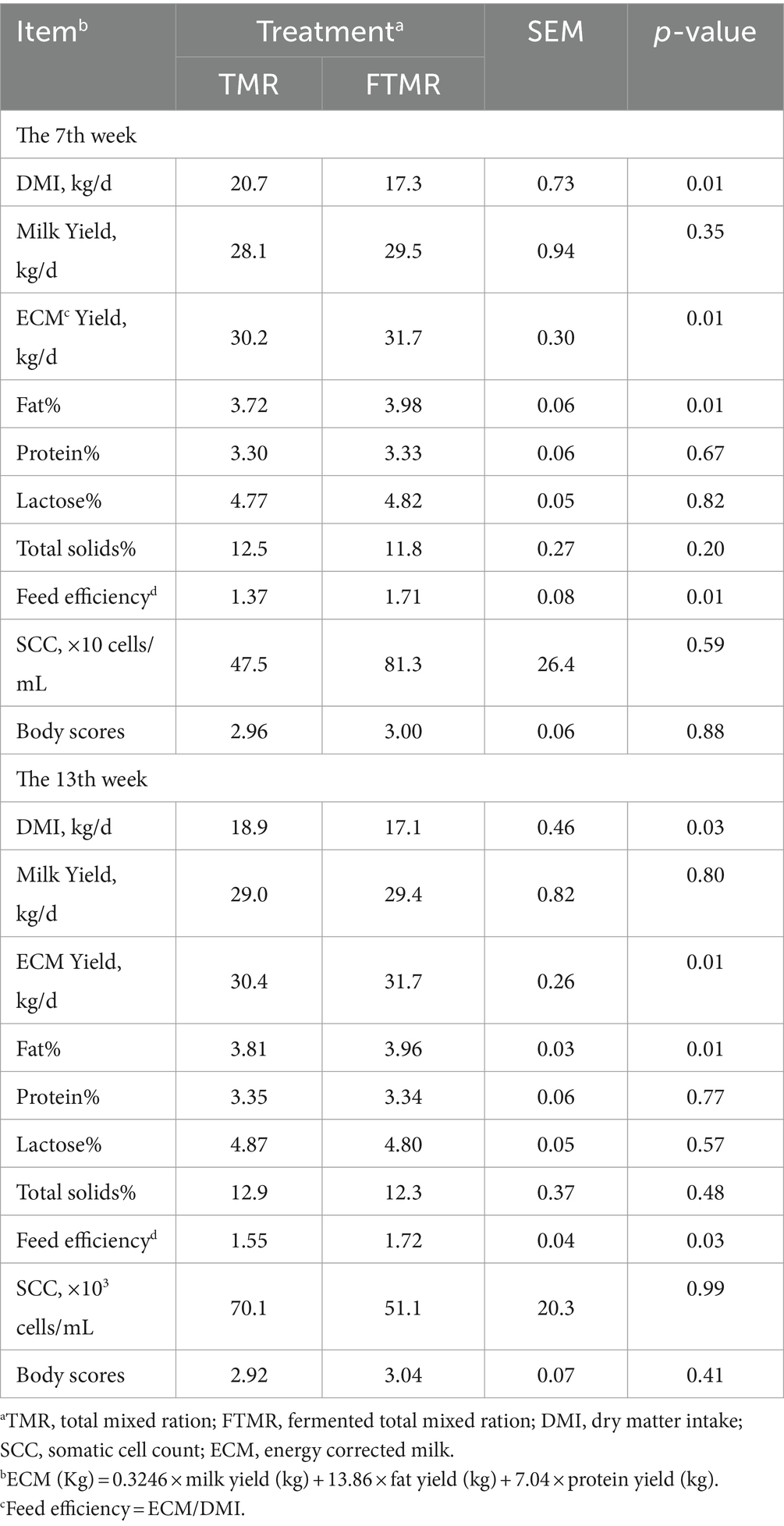
Table 2. Effects of feeding two treatment diets based on TMR and FTMR diets on lactation performance in dairy cows.
3.2 Effect of TMR and FTMR on the apparent total tract digestibility of dairy cow feces
Table 3 shows the apparent total tract nutrient digestibility of the two treatment diets, which was greater for the FTMR diet than for the TMR diet for DM, OM, CP, NDF and ADF.
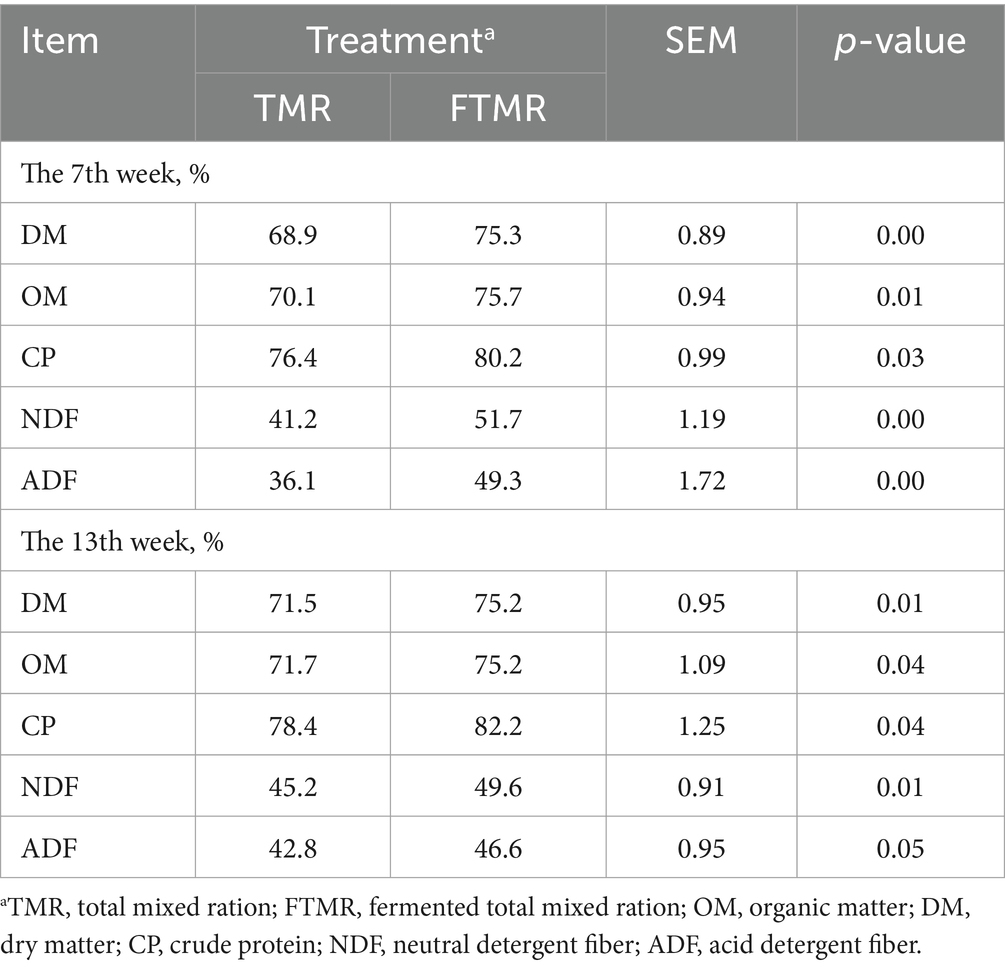
Table 3. Effects of feeding two treatment diets based on TMR and FTMR diets on total-tract apparent digestibility in dairy cows.
3.3 Effect of TMR and FTMR on the fecal VFA pattern and pH of dairy cows
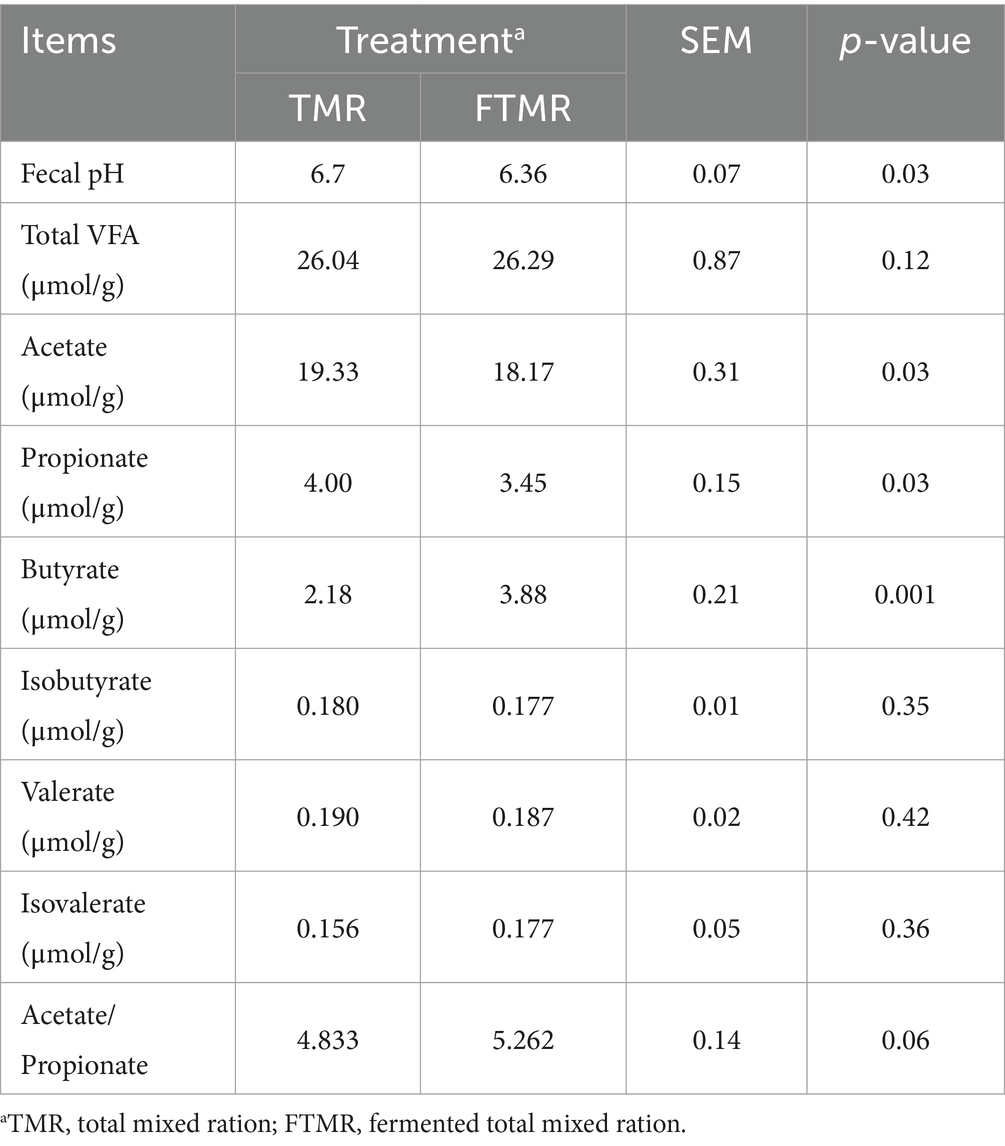
The fecal pH, acetate, and propionate were lower (p < 0.05) in the FTMR-treated samples than in the TMR-treated samples (Table 4). The content of butyrate in the feces of cows fed the FTMR was greater than that in the feces of cows fed the TMR. No significant effects were observed for fecal total VFA concentration, isobutyrate, valerate, isovalerate, and acetate/propionate ratio.
3.4 Analysis of 16S rRNA sequencing of bacteria and archaea in the feces of dairy cows fed the TMR and FTMR
Figure 1A displays bacterial species (OTUs) found in TMR and FTMR, and Figure 1C illustrates archaeal species (OTUs). Cows fed FTMR exhibited significantly more observed species than those fed TMR (p < 0.05).
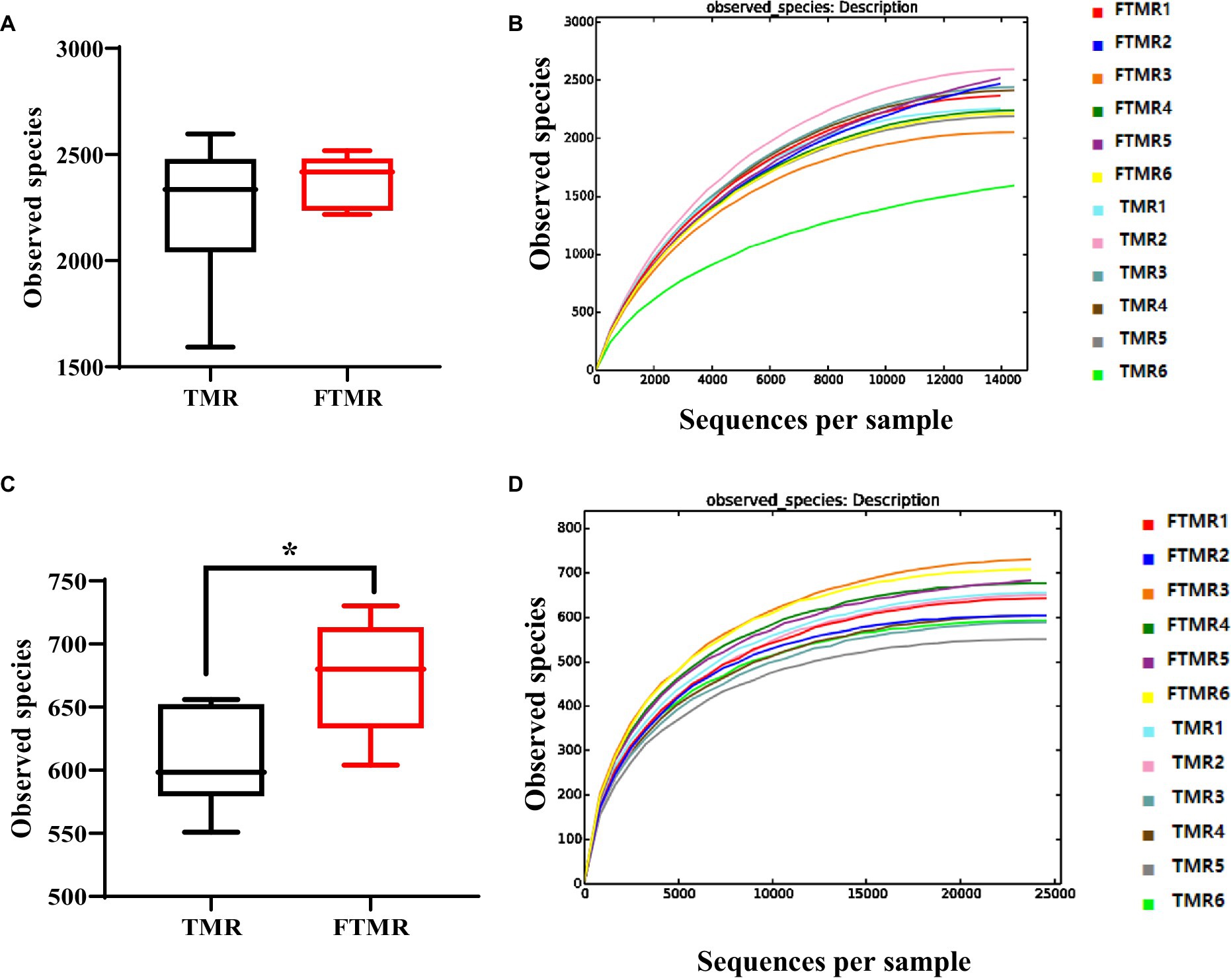
Figure 1. Boxplot and rarefaction curves of operational taxonomic units (OTUs). Bacterial (A) and archaea (C) boxplots represent the number of observed OTUs. The x-axis shows the observed species (OTUs), and the y-axis shows the relationship between TMR and FTMR. The OTU similarity threshold of 97% was considered. Boxes represent the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively), and the horizontal line inside the box defines the median. Whiskers represent the lowest and highest values within 1.5 times the IQR from the first and third quartiles. *p < 0.05 (Student’s t-test). Bacterial (B) and archaea (D) rarefaction curves of OTUs. The x-axis shows the number of valid sequences per sample, and the y-axis shows the observed species (OTUs). Each curve in the graph represents a different sample and is shown in a different color. As the sequencing depth increased, the number of OTUs also increased. Eventually, the curves began to plateau, indicating that as the number of extracted sequences increased, the number of OTUs detected decreased.
The sparsity curves of the OTUs identified in this study indicated that as sequencing depth increased, more species were detected. With higher numbers of sequences analyzed, the edges of the sparsity curves flattened, suggesting thorough coverage of the sequencing data (Figures 1B,D). Good’s coverage, which assesses how well samples are represented by sequencing, approached nearly 99%, indicating comprehensive detection of bacterial types in the samples.
3.5 Differences in bacterial and archaeal diversity between the feces of dairy cows fed the TMR and FTMR
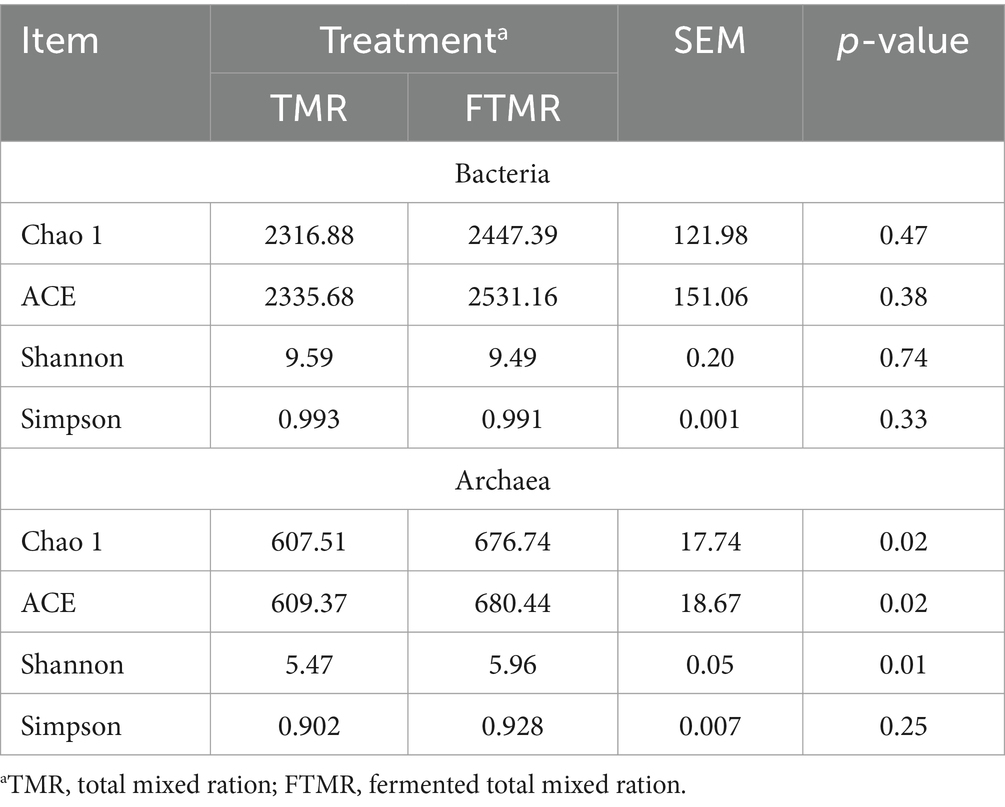
The alpha diversity indices commonly employed included abundance indices (Chao1 and ACE) and diversity indices (Simpson and Shannon). Analysis of fecal bacteria between the two groups indicated no statistically significant differences in the Chao1, ACE, Shannon, or Simpson indices, as indicated in Table 5 (p > 0.05). Moreover, FTMR significantly increased the fecal archaeal abundance indices (Chao and ACE indices) and diversity indices (Shannon index) (p < 0.05).
Beta diversity evaluations were used to analyze the similarities in community structure between the two groups. The NMDS analysis highlighted significant differences in fecal bacterial communities, clearly distinguishing between the TMR and FTMR groups at the OTU level. Similarly, the NMDS results showed substantial variability in fecal archaeal communities, with a considerable separation observed between the two groups, indicating distinct compositions of archaeal communities in the TMR and FTMR groups (See Figure 2).
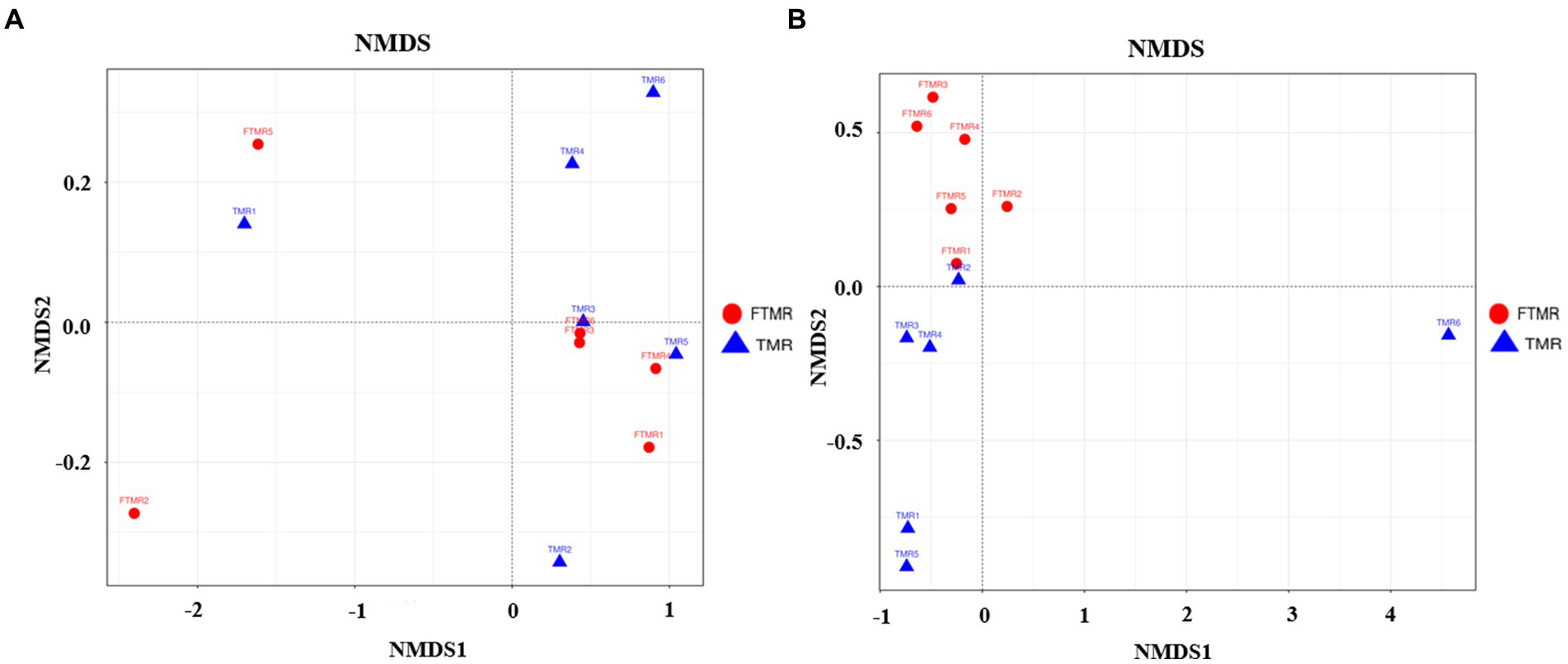
Figure 2. NMDS of fecal microbial communities. Weighted NMDS by fecal bacteria (A) and archaea (B). TMR, total mixed ration; FTMR, fermented total mixed ration.
3.6 Relative abundance and core microbiota of bacteria and archaea in the feces of dairy cows fed the TMR and FTMR
Figure 3 displays the distributions of the top 10 and 20 phyla and genera of fecal bacteria in the TMR and FTMR groups. Bacterial phyla with a relative abundance exceeding 1% were considered predominant. At the phylum level (Figure 3A), Firmicutes, Bacteroidetes, and Actinobacteria were prevalent in both groups. Among genera with a relative abundance exceeding 1% (Figure 3B), dominant bacteria included Unclassified_Ruminococcaceae, Unclassified_Bacteroidales, Unclassified_Clostridiales, Oscillospira, Unclassified_Peptostreptococcaceae, Unclassified_Lachnospiraceae, Bifidobacterium, Unclassified_Rikenellaceae, Unclassified_S24-7, Dorea, Unclassified_RF16, CF231, Clostridium, and Unclassified_Clostridiaceae. Oscillospira exhibited a notable difference between the TMR and FTMR groups (p = 0.01) (Figure 3E).
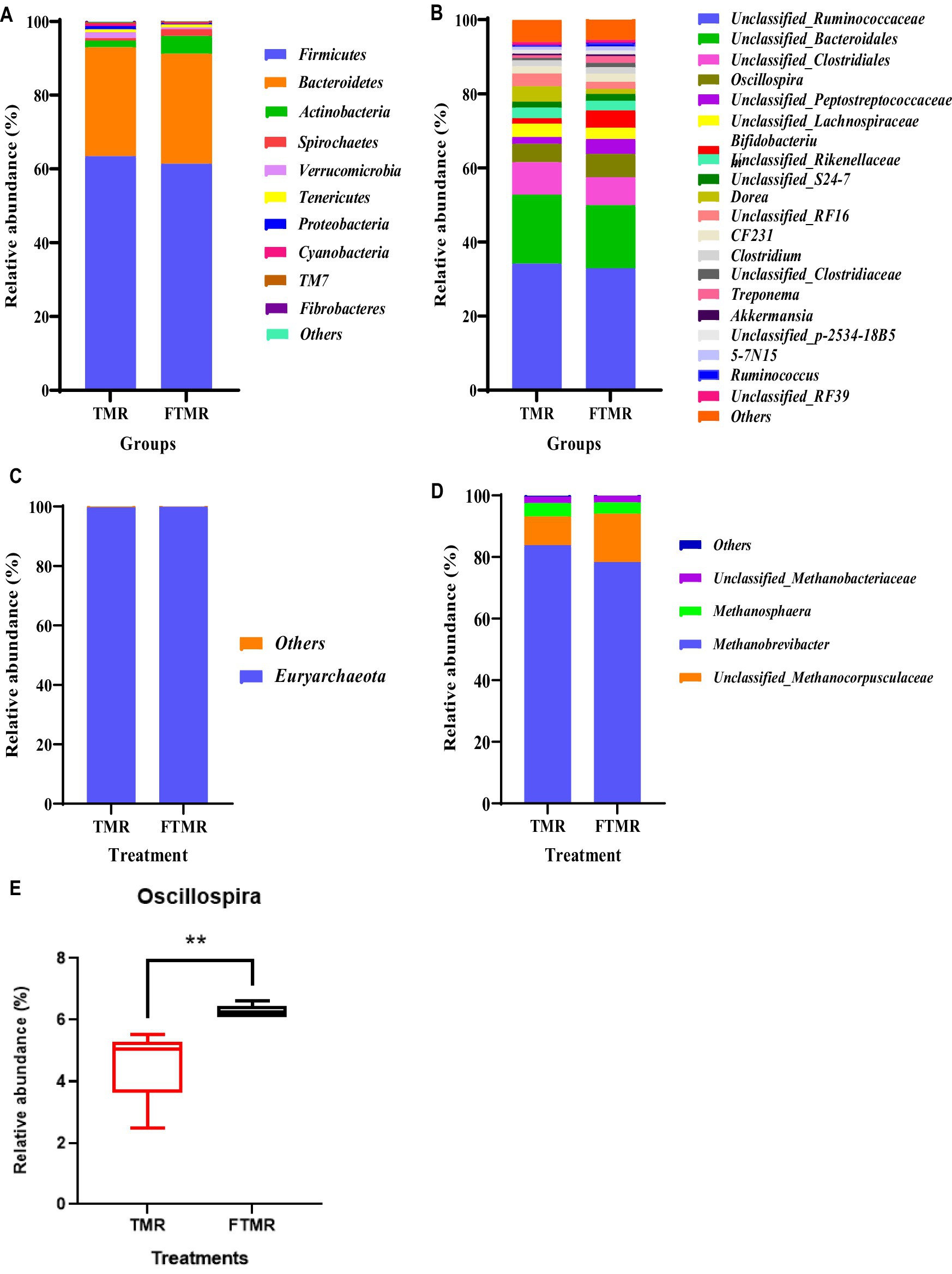
Figure 3. Relative fecal microbial abundances at phylum and genus levels. The abundances of phyla of bacteria (A) and archaea (C). The abundances of genera of bacteria (B) and archaea (D). Oscillospira genus (E) between both groups. Small box plots show the 25th, 50th and 75th percentiles, and whiskers show the extreme values of the data. TMR, total mixed ration; FTMR, fermented mixed ration.
Figure 3 presents the relative abundances of fecal archaea in the TMR and FTMR groups at both the phylum and genus levels. Archaeal phyla with a relative abundance exceeding 1% were classified as dominant. At the phylum level (Figure 3C), Euryarchaeota were the most prevalent archaea in both groups. Among archaeal genera with relative abundances exceeding 1% (Figure 3D), dominant archaea included Methanobrevibacter, Unclassified_Methanocorpusculaceae, Methanosphaera, and Unclassified_Methanobacteriaceae. Taxa shared between the TMR and FTMR groups were considered part of the core microbial community. The number of shared OTUs for bacteria (Figure 4A) was 3,762, and for archaea (Figure 4B), it was 947.
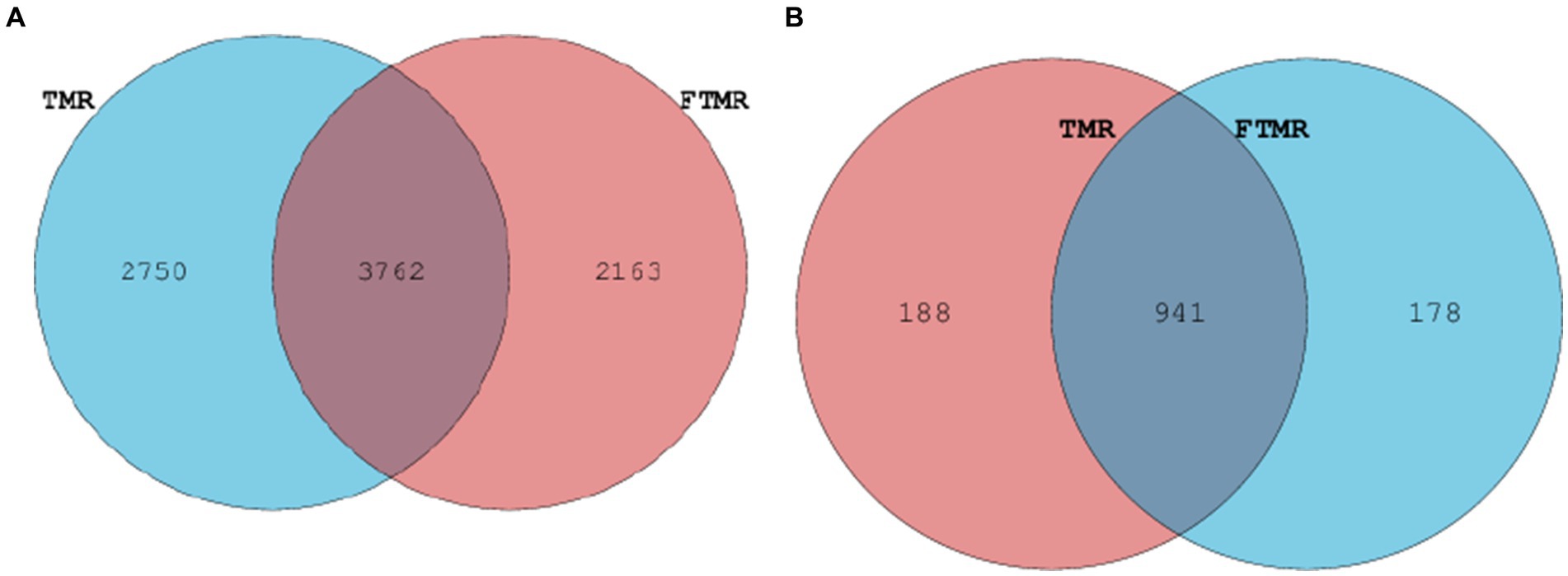
Figure 4. Venn diagram illustrating overlapping microbial OTUs at 3% dissimilarity level between both groups. Venn diagram of bacteria (A) and archaea (B) OTUs. TMR, total mixed ration; FTMR, fermented total mixed ration.
3.7 LDA effect size analysis between the TMR and FTMR groups
The enrichment module rankings were determined using LEfSe analysis. The cladogram (Figure 5A) visually confirmed differences in 16 bacterial taxa between the TMR and FTMR groups. Figure 5B, the LEfSe plot, displays the varying LDA scores of bacterial taxa between these groups. Significant biomarkers for bacteria included Roseburia, rc4-4, Bulleidia, and Sharpea at the genus level. Similarly, notable biomarkers for archaea between the groups were Lactobacillus, Prevotella, Dehalobacterium, and Oscillospira.
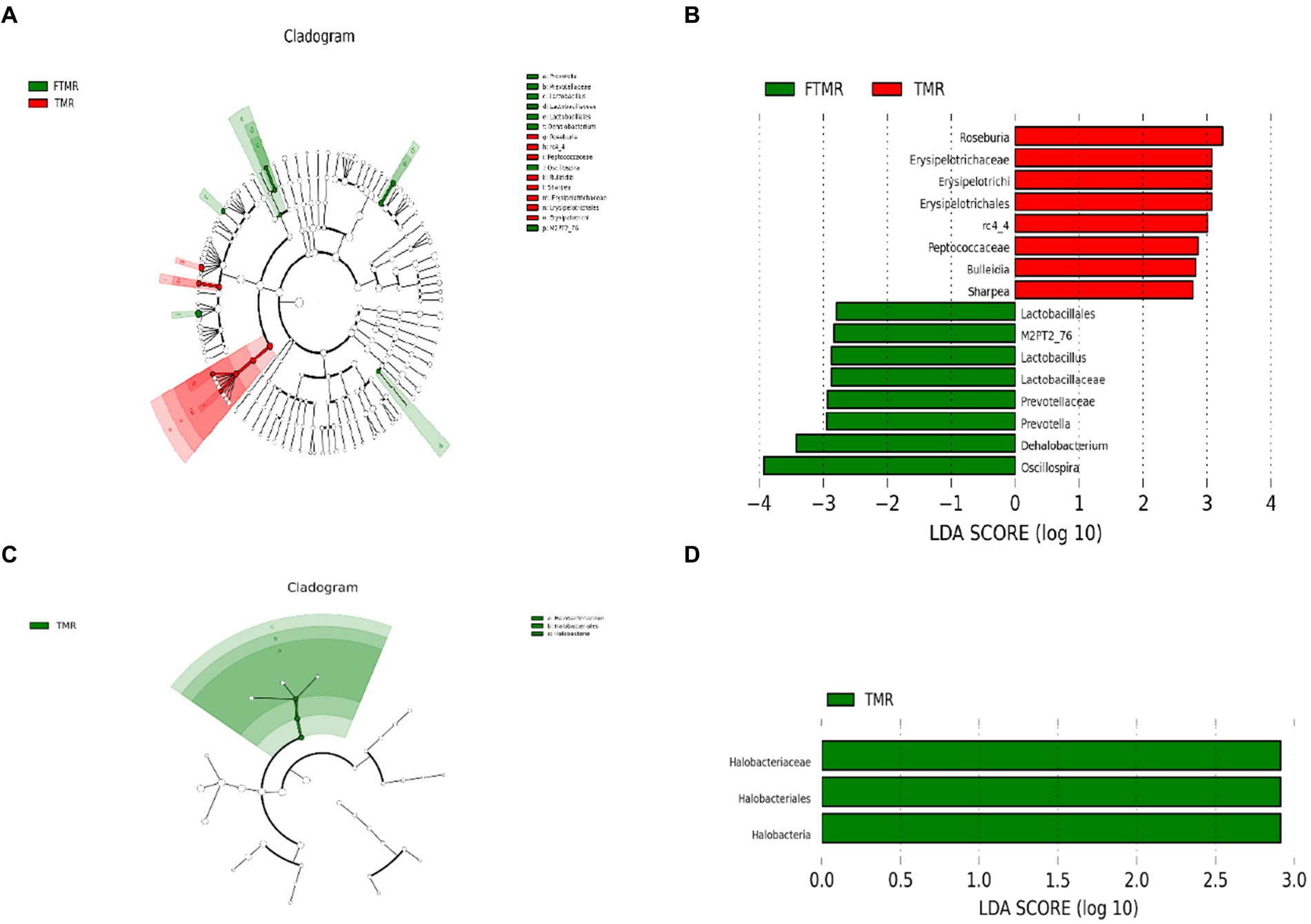
Figure 5. Effect size (LefSe) analysis for LDA. Cladogram diagram showing the microbial species with remarkable variation between TMR and FTMR groups. Cladogram diagram of bacteria (A) and archaea (C); species exhibiting remarkable differences with linear discriminant analysis (LDA) > 2.0, histograms of bacteria (B) and archaea (D). The length of the histogram denotes the LDA score, which contrasts the degree of impact by species exhibiting noteworthy differences between groups. TMR, total mixed diet; FTMR, fermented total mixed diet.
Figure 5C displays variations among the three archaeal taxa between the TMR and FTMR groups in a cladogram. In Figure 5D, the LEfSe analysis plot illustrates significant differences in LDA scores of archaeal taxa between these groups. Significant biomarkers distinguishing between the groups were evident at the class and order levels, specifically Halobacteria and Halobacteriales, respectively. Additionally, at the family level, Halobacteriaceae showed significant differences between the TMR and FTMR groups.
4 Discussion
4.1 Lactation performance, total tract apparent digestibility, fecal pH, and VFA of dairy cows
DMI is a crucial factor influencing dairy cow production performance (35). In this study, a decrease in DMI was observed in the FTMR group, while milk yield remained unaffected. This indicates that the FTMR diet demonstrates superior feed efficiency. The concentration of acetic acid in relation to total acids was negatively associated with silage intake (36). Previous research has shown that acetic acid concentrations exceeding 1.7% DM significantly reduce ruminant intake (37). Additionally, ammonia-N concentration in grass silage has been inversely correlated with dry matter intake in lactating dairy cows (38). As a result, the FTMR-fed group exhibited higher levels of organic acids and ammonia nitrogen, along with reduced dry matter intake compared to the TMR-fed group. However, FTMR had no impact on DMI in Ujumqin sheep, likely due to species differences and diet fermentation quality (39). Despite decreased DMI in the FTMR diet, milk yield was unaffected due to improved feed efficiency and increased ECM compared to the TMR diet. Previous research indicates that exogenous fibrolytic enzymes enhance fiber digestibility, thereby increasing available energy intake and improving feed efficiency (40). Compared to the TMR diet, FTMR demonstrated higher nutrient digestibility and feed efficiency, notably increasing DM digestibility (41). Cao et al. observed increased DM digestibility with fermented TMR in sheep compared to regular TMR consumption (39). Additionally, CP and NDF digestibility were enhanced in the FTMR diet, consistent with recent studies demonstrating improved CP and NDF digestibility in sheep fed FTMR, attributed to increased digestible CP and NDF levels (42). Enzyme levels have been reported to significantly impact total tract DM and NDF digestibility in a cubic manner (43), thereby equivalently elevating OM digestibility in cows fed the FTMR diet. Acetate is recognized as a primary precursor for milk fat synthesis in the mammary gland during lactation (44). Previous studies have shown that sheep fed FTMR exhibited increased acetate concentrations compared to those fed fresh TMR (39), contributing to increased milk fat production due to enhanced fiber digestibility from added fibrolytic enzymes (43).
Fermentation altered the carbohydrate composition in the FTMR, affecting hindgut fermentation patterns. Unlike the TMR group, the FTMR group showed a lower fecal pH. Adding starch (soluble carbohydrate) to dairy cow diets can significantly reduce fecal pH, potentially increasing nutrient bypassing in the rumen and accelerating passage rates (45). Hindgut microbes in dairy cows degrade 5 to 10% of carbohydrates, including starch, small particles escaping ruminal fermentation, and undigested rumen components (46, 47). After ensiling, the TMR exhibited reduced nonfibrous carbohydrate content, suggesting that FTMR increases concentrations of nonfibrous and soluble carbohydrates, speeds up feed passage rates, and boosts fermentable substances in the hindgut. Studies have reported that diets high in grains (rich in non-fiber carbohydrates) significantly increase fecal butyric acid levels and lower pH in cows (48), which is consistent with the current study’s findings. The FTMR group showed markedly higher fecal butyrate concentrations, corresponding with an increased relative abundance of key butyrate-producing bacteria like Ruminococcaceae, Oscillospira, and Bacteroidales. Acetate and propionate are crucial energy sources for ruminants. Research suggests that dietary fermentation promotes propionate fermentation and absorption in the rumen (49, 50), potentially influencing microbial populations producing these acids in both the rumen and hindgut.
4.2 Fecal bacterial community composition of dairy cows
The relative abundance histogram of phyla indicated that Firmicutes, along with Bacteroidetes, were the predominant fecal bacteria, consistent with prior studies (51–53). Previous research has linked the Firmicutes to Bacteroidetes ratio with energy extraction in humans and mice (54, 55). Our findings showed no change in the Firmicutes to Bacteroidetes ratio, suggesting that this ratio remains largely unaffected by ruminant diets (56). This could be attributed to the primary energy production occurring in the foregut rather than the hindgut (20).
In the feces, the dominant genera included Unclassified_Bacteroidales, Unclassified_Ruminococcaceae, Unclassified_Clostridiales, and Oscillospira. Previous studies have highlighted that Unclassified_Ruminococcaceae and Unclassified_Bacteroidales are integral parts of the rumen’s “core bacterial microbiome” (57). These genera play crucial roles in breaking down plant fibers within the gastrointestinal tract (57).
In this study, the abundance of Unclassified_Clostridiales, considered a core component of fecal bacteria, did not exhibit significant differences between the TMR and FTMR groups, which is consistent with prior research (58). This indicates that Unclassified_Clostridiales may contribute significantly to the fecal microbial ecosystem regardless of dietary differences. Further investigation involving the isolation and identification of representative strains of Unclassified_Clostridiales could provide deeper insights into their role in hindgut function (59).
Our findings revealed a significantly higher abundance of Oscillospira in the FTMR group compared to other groups at the genus level. The content of Oscillospira in the rumen of cattle fed a high-starch (soluble carbohydrates) diet increased (60). Zhao et al. suggested that fermentation increases the crystalline structure of starch grains, promoting the production of resistant starch (61). Therefore, the elevated levels of Oscillospira in the FTMR group may be attributed to the higher content of resistant starch in the hindgut of cows fed FTMR. Mackie et al. noted that Oscillospira is prevalent in the rumen of dairy cows and influenced by dietary factors (62), indicating its responsiveness to diet in the gastrointestinal tract. Oscillospira is known as a potential butyrate producer, contributing to intestinal health (63), suggesting that FTMR diets may enhance gastrointestinal health through increased Oscillospira abundance. Although Oscillospira has not been cultured in pure form (9), its potential as a probiotic for regulating the gastrointestinal tract of dairy cows merits consideration.
At the genus level, Roseburia, rc4-4, Bulleidia, and Sharpea were significantly more abundant in the TMR group compared to the FTMR group, consistent with findings by Kim et al. (58). This indicates that less common bacterial taxa are more responsive to dietary changes than those more commonly found. Roseburia species are known for their ability to break down dietary polysaccharides, producing butyrate (64), which correlates with our observation of higher butyric acid levels in the FTMR group than in the TMR group. The genus rc4-4 is associated with diets rich in fiber (65), suggesting that the TMR diet may have a higher fiber content in the hindgut compared to the FTMR diet. Sharpea plays a critical role in lactic acid production (66, 67), which increases during rumen acidosis in cows (68) and varies with diet composition in sheep (69). While lactic acid is not a primary product of normal rumen fermentation, diets high in carbohydrates and soluble sugars can lead to its accumulation (70). Hence, the higher abundance of Sharpea in the TMR group may be due to greater carbohydrate flow and soluble sugar levels from the rumen into the hindgut.
We identified four genera—Lactobacillus, Prevotella, Dehalobacterium, and Oscillospira—as distinguishing biomarkers between the FTMR and TMR groups. Lactobacillus is recognized for its probiotic role in ruminant gastrointestinal health and has been associated with reducing diarrhea rates in calves (71–73). Our findings showed that Lactobacillus constituted less than 1% of the fecal microbiota and was not considered a core community, consistent with Tang et al. (74). Its abundance varies with diet, transitioning from lactose dominance in younger animals to diets richer in complex carbohydrates as they mature (75–77). While Lactobacillus thrives in silage, its survival through the gastrointestinal tract, particularly in the rumen, is limited (50, 74). Nonetheless, our results indicated significantly higher levels of Lactobacillus in the feces of the FTMR group compared to the TMR group, suggesting silage could serve as a vehicle for probiotic delivery (50). Prevotella was abundant in both TMR and FTMR groups, consistent with its widespread presence in cow feces (56, 78, 79) and dominance in the rumen (57, 71, 80). Its prevalence in feces (81) varies with diet, such as higher levels in corn-based diets compared to those with wet distillers grains (82), indicating its diet-related presence in cattle feces (58, 83, 84). Prevotella species are known for their enzymatic capabilities in degrading proteins, starch, and hemicellulose to produce succinate and acetate (85–87). Dehalobacterium showed a higher abundance in FTMR feces compared to TMR, suggesting diet influences its presence (88). It has been linked to glucose-rich diets, although further research is needed on its role in the hindgut of dairy cows (89).
4.3 Fecal archaeal community composition of dairy cows
Our results showed that the archaeal Chao1 index in the FTMR group was markedly greater than that in the TMR group, confirming that the archaeal diversity in the FTMR group was markedly greater than that in the other groups. The predominant archaeal phylum identified in the feces was Euryarchaeota. Prior investigations have indicated that Euryarchaeota is the most prevalent archaeal bacterial phylum in the rumen (22, 85, 90), underscoring its critical function in the gastrointestinal system. It was further confirmed that Methanobrevibacter is the most prevalent archaeal bacterial genus in feces, aligning with previous studies (22, 91), which established their capability to generate CH4 from hydrogen and formic acid (91, 92). Nevertheless, no significant differences in Methanobrevibacter abundance were observed between the FTMR and TMR groups, implying that FTMR may not diminish methane production in the hindgut. Zhang et al. (93) noted that high-concentrate rations significantly decreased the abundances of Fibrobacter and Methanobrevibacter, demonstrating that an increase in these bacteria’s concentration in the diet could inhibit methane synthesis (94, 95). Conversely, this study did not alter the concentration-to-coarse ratio between TMR and FTMR. Moreover, a symbiotic interaction exists between bacteria and methanogens in the animal gut, and cellulolytic bacteria in the rumen of ruminants show a positive correlation with the number of methanogens (96). Notably, no significant alterations in the relative abundances of Fibrobacteres and Fibrobacter were detected in this study between the TMR and FTMR groups. Halobacteria, Halobacteriales, and Halobacteriaceae served as biomarkers to differentiate TMRs from FTMR, with their relative abundances being considerably higher in the TMR group than in the FTMR group, potentially due to variations in hindgut cell chloride content between the two groups (97).
5 Conclusion
In conclusion, FTMR was shown to increase milk fat content, improve nutrient digestibility, elevate the proportion of butyrate in fecal matter, and decrease fecal pH. These findings indicate that FTMR can increase hindgut fermentation. Additionally, FTMR significantly enhanced the diversity of fecal archaeal communities and increased the relative abundance of the genus Oscillospira, suggesting improved hindgut health compared to TMR. While Methanobrevibacter was found to be the most common archaeal genus in fecal samples, no significant disparities were observed between the TMR and FTMR groups, suggesting that FTMR may not have a significant impact on methane production in the hindgut. This study provides a theoretical basis for investigating the impact of FTMR on bacterial and archaeal communities in the hindgut, indicating that FTMR feeding can beneficially affect hindgut health in dairy cows.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA), accession number: PRJNA1119071.
Ethics statement
The animal study was approved by Animal Care Advisory Committee, Henan Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LW: Writing – original draft, Writing – review & editing, Conceptualization. SJ: Investigation, Software, Writing – review & editing. PW: Data curation, Writing – original draft. XL: Methodology, Writing – review & editing. CL: Formal analysis, Supervision, Writing – review & editing. SS: Formal analysis, Writing – review & editing. GZ: Project administration, Writing – review & editing. JC: Resources, Writing – review & editing. QY: Funding acquisition, Writing – review & editing. HZ: Resources, Visualization, Writing – original draft. QZ: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Research: Key Project of Science and Technology of Henan Province (232102110103), Natural Science Foundation of Henan (242300421573), Heilongjiang Province Natural Science Foundation for Joint Guide Project (LH2022C030), Key R&D Program Guidance Projects of Heilongjiang Province (GZ20230020), Henan Key Scientific and Technological Project (201111311100), Henan Key Research and Development Project (241111113700).
Acknowledgments
We would like to thank the staff at our laboratory for their ongoing assistance.
Conflict of interest
QZ was employed by Henan Delin Biological Products Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Li, Y, Wu, Q, Lv, J, Jia, X, Gao, J, Zhang, Y, et al. Associations of protein molecular structures with their nutrient supply and biodegradation characteristics in different byproducts of seed-used pumpkin. Animals. (2022) 12:956. doi: 10.3390/ani12080956
2. Saleem, AM, Ribeiro, GO, Sanderson, H, Alipour, D, Brand, T, Hünerberg, M, et al. Effect of exogenous fibrolytic enzymes and ammonia fiber expansion on the fermentation of wheat straw in an artificial rumen system (RUSITEC)1. J Anim Sci. (2019) 97:3535–49. doi: 10.1093/jas/skz224
3. Schingoethe, DJ . A 100-year review: Total mixed ration feeding of dairy cows. J Dairy Sci. (2017) 100:10143–50. doi: 10.3168/jds.2017-12967
4. Supapong, C, Cherdthong, A, Wanapat, M, Chanjula, P, and Uriyapongson, S. Effects of sulfur levels in fermented total mixed ration containing fresh cassava root on feed utilization, rumen characteristics, microbial protein synthesis, and blood metabolites in Thai native beef cattle. Animals. (2019) 9:261. doi: 10.3390/ani9050261
5. Zhao, J, Dong, Z, Chen, L, Wang, S, and Shao, T. The replacement of whole-plant corn with bamboo shoot shell on the fermentation quality, chemical composition, aerobic stability and in vitro digestibility of total mixed ration silage. Anim Feed Sci Tech. (2020) 259:114348. doi: 10.1016/j.anifeedsci.2019.114348
6. Weinberg, ZG, Chen, Y, Miron, D, Raviv, Y, Nahim, E, Bloch, A, et al. Preservation of total mixed rations for dairy cows in bales wrapped with polyethylene stretch film—a commercial scale experiment. Anim Feed Sci Tech. (2011) 164:125–9. doi: 10.1016/j.anifeedsci.2010.11.016
7. Xie, Y, Xu, S, Li, W, Wang, M, Wu, Z, Bao, J, et al. Effects of the application of Lactobacillus plantarum inoculant and potassium sorbate on the fermentation quality, in vitro digestibility and aerobic stability of total mixed ration silage based on alfalfa silage. Animals. (2020) 10:2229. doi: 10.3390/ani10122229
8. Chang, L, Mu, G, Wang, M, Zhao, T, Tuo, Y, Zhu, X, et al. Microbial diversity and quality-related physicochemical properties of spicy cabbage in northeastern China and their correlation analysis. Foods. (2022) 11:1511. doi: 10.3390/foods11101511
9. Liu, Q, Li, X, Desta, ST, Zhang, J-G, and Shao, T. Effects of Lactobacillus plantarum and fibrolytic enzyme on the fermentation quality and in vitro digestibility of total mixed rations silage including rape straw. J Integr Agr. (2016) 15:2087–96. doi: 10.1016/S2095-3119(15)61233-3
10. Kondo, M, Shimizu, K, Jayanegara, A, Mishima, T, Matsui, H, Karita, S, et al. Changes in nutrient composition and in vitro ruminal fermentation of total mixed ration silage stored at different temperatures and periods. J Sci Food Agr. (2016) 96:1175–80. doi: 10.1002/jsfa.7200
11. Wang, F, and Nishino, N. Resistance to aerobic deterioration of total mixed ration silage: effect of ration formulation, air infiltration and storage period on fermentation characteristics and aerobic stability. J Sci Food Agr. (2008) 88:133–40. doi: 10.1002/jsfa.3057
12. Gusmao, J, Danés, M, Casagrande, D, and Bernardes, T. Total mixed ration silage containing elephant grass for small-scale dairy farms. Grass and Forage Sci. (2018) 73:717–26. doi: 10.1111/gfs.12357
13. Bai, B, Qiu, R, Wang, Z, Liu, Y, Bao, J, Sun, L, et al. Effects of cellulase and lactic acid bacteria on ensiling performance and bacterial community of caragana korshinskii silage. Microorganisms. (2023) 11:337. doi: 10.3390/microorganisms11020337
14. Sun, Q, Gao, F, Yu, Z, Tao, Y, Zhao, S, and Cai, Y. Fermentation quality and chemical composition of shrub silage treated with lactic acid bacteria inoculants and cellulase additives. Anim Sci J. (2012) 83:305–9. doi: 10.1111/j.1740-0929.2011.00962.x
15. Arriola, KG, Vyas, D, Kim, D, Agarussi, MCN, Silva, VP, Flores, M, et al. Effect of Lactobacillus hilgardii, Lactobacillus buchneri, or their combination on the fermentation and nutritive value of sorghum silage and corn silage. J Dairy Sci. (2021) 104:9664–75. doi: 10.3168/jds.2020-19512
16. Xu, H, Huang, W, Hou, Q, Kwok, L-y, Sun, Z, Ma, H, et al. The effects of probiotics administration on the milk production, milk components and fecal bacteria microbiota of dairy cows. Sci Bulletin. (2017) 62:767–74. doi: 10.1016/j.scib.2017.04.019
17. Liu, J, Taft, DH, Maldonado-Gomez, MX, Johnson, D, Treiber, ML, Lemay, DG, et al. The fecal resistome of dairy cattle is associated with diet during nursing. Na Commun. (2019) 10:4406. doi: 10.1038/s41467-019-12111-x
18. Faulkner, MJ, Wenner, BA, Solden, LM, and Weiss, WP. Source of supplemental dietary copper, zinc, and manganese affects fecal microbial relative abundance in lactating dairy cows. J Dairy Sci. (2017) 100:1037–44. doi: 10.3168/jds.2016-11680
19. Welch, CB, Lourenco, JM, Krause, TR, Seidel, DS, Fluharty, FL, Pringle, TD, et al. Evaluation of the fecal bacterial communities of angus steers with divergent feed efficiencies across the lifespan from weaning to slaughter. Frontiers in Veterinary Science. (2021) 8:694. doi: 10.3389/fvets.2021.597405
20. Zhang, J, Shi, H, Wang, Y, Cao, Z, Yang, H, and Li, S. Effect of limit-fed diets with different forage to concentrate ratios on fecal bacterial and archaeal community composition in Holstein heifers. Front Microbiol. (2018) 9:976. doi: 10.3389/fmicb.2018.00976
21. Al-Azzawi, M, Bowtell, L, Hancock, K, and Preston, S. Addition of activated carbon into a cattle diet to mitigate GHG emissions and improve production. Sustain For. (2021) 13:8254. doi: 10.3390/su13158254
22. Kumar, S, Indugu, N, Vecchiarelli, B, and Pitta, DW. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front Microbiol. (2015) 6:781. doi: 10.3389/fmicb.2015.00781
23. Zhang, J, Shi, H, Wang, Y, Li, S, Cao, Z, Ji, S, et al. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front Microbiol. (2017) 8:2206. doi: 10.3389/fmicb.2017.02206
24. Ramin, M, Chagas, JC, Smidt, H, Exposito, RG, and Krizsan, SJ. Enteric and fecal methane emissions from dairy cows fed grass or corn silage diets supplemented with rapeseed oil. Animals. (2021) 11:1322. doi: 10.3390/ani11051322
25. Laporte, MF, and Paquin, P. Near-infrared analysis of fat, protein, and casein in cow’s milk. J Agric Food Chem. (1999) 47:2600–5. doi: 10.1021/jf980929r
26. Goeser, JP, and Combs, DK. An alternative method to assess 24-h ruminal in vitro neutral detergent fiber digestibility. J Dairy Sci. (2009) 92:3833–41. doi: 10.3168/jds.2008-1136
27. Wang, L, Zhang, G, Li, Y, and Zhang, Y. Effects of high forage/concentrate diet on volatile fatty acid production and the microorganisms involved in VFA production in cow rumen. Animals. (2020) 10:223. doi: 10.3390/ani10020223
28. Vasseur, E, Gibbons, J, Rushen, J, and de Passillé, AM. Development and implementation of a training program to ensure high repeatability of body condition scoring of dairy cows. J Dairy Sci. (2013) 96:4725–37. doi: 10.3168/jds.2012-6359
29. Caporaso, JG, Kuczynski, J, Stombaugh, J, Bittinger, K, Bushman, FD, Costello, EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
30. Edgar, RC . Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
31. Pruesse, E, Quast, C, Knittel, K, Fuchs, BM, Ludwig, WG, Peplies, J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. (2007) 35:7188–7196. doi: 10.1093/nar/gkm864
32. Wolf, MA, Sfriso, A, and Moro, I. Thermal pollution and settlement of new tropical alien species: The case of Grateloupia yinggehaiensis (Rhodophyta) in the Venice Lagoon. Estuar Coast Shelf S. (2014) 147:11–16. doi: 10.1016/j.ecss.2014.05.020
33. Kõljalg, U, Larsson, KH, Abarenkov, K, Nilsson, RH, Alexander, IJ, Eberhardt, U, et al. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. (2005) 166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x
34. Segata, N, Izard, J, Waldron, L, Gevers, D, Miropolsky, L, Garrett, WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
35. Ma, J, Sun, G, Shah, AM, Fan, X, Li, S, and Yu, X. Effects of different growth stages of amaranth silage on the rumen degradation of dairy cows. Animals. (2019) 9:793. doi: 10.3390/ani9100793
36. Wilkins, RJ, Hutchinson, KJ, Wilson, RF, and Harris, CE. The voluntary intake of silage by sheep: I. Interrelationships between silage composition and intake. JAS. (1971) 77:531–7. doi: 10.1017/S0021859600064613
37. Gerlach, K, Jobim, JLP, Nussio, CC, and Gustavo, L. A data analysis on the effect of acetic acid on dry matter intake in dairy cattle. Anim Feed Sci Technol. (2021) 272:114782. doi: 10.1016/j.anifeedsci.2020.114782
38. Huhtanen, P, Khalili, H, Nousiainen, JI, Rinne, M, Jaakkola, S, Heikkil, T, et al. Prediction of the relative intake potential of grass silage by dairy cows. Livest Prod Sci. (2002) 73:111–30. doi: 10.1016/S0301-6226(01)00279-2
39. Cao, Y, Zang, Y, Jiang, Z, Han, Y, Hou, JJ, Liu, H, et al. Fermentation quality and nutritive value of fresh and fermented total mixed rations containing Chinese wildrye or corn Stover. Grassl Sci. (2016) 62:213–23. doi: 10.1111/grs.12134
40. Tewoldebrhan, TA, Appuhamy, JADRN, Lee, JJ, Niu, M, Seo, S, Jeong, S, et al. Exogenous β-mannanase improves feed conversion efficiency and reduces somatic cell count in dairy cattle. J Dairy Sci. (2017) 100:244–52. doi: 10.3168/jds.2016-11017
41. Ferraretto, LF, Crump, PM, and Shaver, RD. Effect of cereal grain type and corn grain harvesting and processing methods on intake, digestion, and milk production by dairy cows through a meta-analysis. J Dairy Sci. (2013) 96:533–50. doi: 10.3168/jds.2012-5932
42. Cao, Y, Takahashi, T, Horiguchi, KI, Yoshida, N, and Cai, Y. Methane emissions from sheep fed fermented or non-fermented total mixed ration containing whole-crop rice and rice bran. Anim Feed Sci Tech. (2010) 157:72–8. doi: 10.1016/j.anifeedsci.2010.02.004
43. Refat, B, Christensen, DA, Mckinnon, JJ, Yang, W, Beattie, AD, Mcallister, TA, et al. Effect of fibrolytic enzymes on lactational performance, feeding behavior, and digestibility in high-producing dairy cows fed a barley silage–based diet. J Dairy Sci. (2018) 101:7971–9. doi: 10.3168/jds.2017-14203
44. Smith, SB . Contribution of the pentose cycle to lipogenesis in bovine adipose tissue. Arch Biochem Biophys. (1983) 221:46–56. doi: 10.1016/0003-9861(83)90120-0
45. Neubauer, V, Petri, RM, Humer, E, Kröger, I, Reisinger, N, Baumgartner, W, et al. Starch-rich diet induced rumen acidosis and hindgut dysbiosis in dairy cows of different lactations. Animals. (2020) 10:1727. doi: 10.3390/ani10101727
46. Gressley, TF, Hall, MB, and Armentano, LE. Ruminant nutrition symposium: productivity, digestion, and health responses to hindgut acidosis in ruminants. J Anim Sci. (2011) 89:1120–30. doi: 10.2527/jas.2010-3460
47. Lyu, J, Yang, Z, Wang, E, Liu, G, Wang, Y, Wang, W, et al. Possibility of using by-products with high NDF content to alter the fecal short chain fatty acid profiles, bacterial community, and digestibility of lactating dairy cows. Microorganisms. (2022) 10:1731. doi: 10.3390/microorganisms10091731
48. Mao, S, Zhang, R, Wang, D, and Zhu, W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet Res. (2012) 8:237. doi: 10.1186/1746-6148-8-237
49. Gastelen, S, Dijkstra, J, Gerrits, WJJ, Gilbert, MS, and Bannink, A. Short communication: quantifying postruminal starch fermentation in early-lactation Holstein-Friesian cows. Animal. (2023) 17:100974. doi: 10.1016/j.animal.2023.100974
50. Han, H, Ogata, Y, Yamamoto, Y, Nagao, S, and Nishino, N. Identification of lactic acid bacteria in the rumen and feces of dairy cows fed total mixed ration silage to assess the survival of silage bacteria in the gut. J Dairy Sci. (2014) 97:5754–62. doi: 10.3168/jds.2014-7968
51. Rice, WC, Galyean, ML, Cox, SB, Dowd, SE, and Cole, NA. Influence of wet distillers grains diets on beef cattle fecal bacterial community structure. BMC Microbiol. (2012) 12:25. doi: 10.1186/1471-2180-12-25
52. Li, S, Khafipour, E, Krause, DO, Kroeker, A, Rodriguez-Lecompte, JC, Gozho, GN, et al. Effects of subacute ruminal acidosis challenges on fermentation and endotoxins in the rumen and hindgut of dairy cows. J Dairy Sci. (2012) 95:294–303. doi: 10.3168/jds.2011-4447
53. Plaizier, JC, Li, S, Danscher, AM, Derakshani, H, Andersen, PH, and Khafipour, E. Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb Ecol. (2017) 74:485–95. doi: 10.1007/s00248-017-0940-z
54. Ley, RE, Turnbaugh, PJ, Klein, S, and Gordon, JI. Human gut microbes associated with obesity. Nature. (2006) 444:1022–3. doi: 10.1038/4441022a
55. Turnbaugh, PJ, Ley, RE, Mahowald, MA, Magrini, V, Mardis, ER, and Gordon, JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. doi: 10.1038/nature05414
56. Li, Y, Meng, Q, Zhou, B, and Zhou, Z. Effect of ensiled mulberry leaves and sun-dried mulberry fruit pomace on the fecal bacterial community composition in finishing steers. BMC Microbiol. (2017) 17:97. doi: 10.1186/s12866-017-1011-9
57. Henderson, G, Cox, F, Ganesh, S, Jonker, A, Young, W, and Janssen, PH. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. (2015) 5:1–15. doi: 10.1038/srep14567
58. Kim, M, Kim, J, Kuehn, LA, Bono, JL, Berry, ED, Kalchayanand, N, et al. Investigation of bacterial diversity in the feces of cattle fed different diets 1. J Anim Sci. (2014) 92:683–94. doi: 10.2527/jas.2013-6841
59. Kim, M, Morrison, M, and Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol Ecol. (2011) 76:49–63. doi: 10.1111/j.1574-6941.2010.01029.x
60. Myer, PR, Wells, JE, Smith, TP, Kuehn, LA, and Freetly, HC. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus. (2015) 4:454. doi: 10.1186/s40064-015-1201-6
61. Zhao, T, Li, X, Zhu, R, Ma, Z, Liu, L, Wang, X, et al. Effect of natural fermentation on the structure and physicochemical properties of wheat starch. Carbohydr Polym. (2019) 218:163–9. doi: 10.1016/j.carbpol.2019.04.061
62. Mackie, RI, Aminov, RI, Hu, W, Klieve, AV, Ouwerkerk, D, Sundset, MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. (2003) 69:6808–15. doi: 10.1128/AEM.69.11.6808-6815.2003
63. Walters, WA, Xu, Z, and Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. (2014) 588:4223–33. doi: 10.1016/j.febslet.2014.09.039
64. Rodriguez, J, Neyrinck, AM, Zhang, Z, Seethaler, B, Nazare, JA, Robles Sánchez, C, et al. Metabolite profiling reveals the interaction of chitin-glucan with the gut microbiota. Gut Microbes. (2020) 12:1810530. doi: 10.1080/19490976.2020.1810530
65. Zhong, Y, Nyman, M, and Fåk, F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res. (2015) 59:2066–76. doi: 10.1002/mnfr.201500187
66. Marketta, R, Marcia, F, Ilma, T, Tomasz, S, Ali-Reza, B, and Päivi, M. Effects of grass silage additive type and barley grain preservation method on rumen fermentation, microbial community and milk production of dairy cows. Agriculture. (2022) 12:1–18. doi: 10.3390/agriculture12020266
67. Lin, L, Xie, F, Sun, D, Liu, J, Zhu, W, and Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome. (2019) 7:83. doi: 10.1186/s40168-019-0701-y
68. Plaizier, JC, Li, S, Tun, HM, and Khafipour, E. Nutritional models of experimentally-induced subacute ruminal acidosis (SARA) differ in their impact on rumen and hindgut bacterial communities in dairy cows. Front Microbiol. (2017) 7:2128. doi: 10.3389/fmicb.2016.02128
69. Kamke, J, Kittelmann, S, Soni, P, Li, Y, Tavendale, M, Ganesh, S, et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome. (2016) 4:56. doi: 10.1186/s40168-016-0201-2
70. Dewanckele, L, Toral, PG, Vlaeminck, B, and Fievez, V. Invited review: role of rumen biohydrogenation intermediates and rumen microbes in diet-induced milk fat depression: an update. J Dairy Sci. (2020) 103:7655–81. doi: 10.3168/jds.2019-17662
71. Koike, S, Ueno, M, Ashida, N, Imabayashi, T, and Kobayashi, Y. Effect of Bacillus subtilis C-3102 supplementation in milk replacer on growth and rumen microbiota in preweaned calves. Anim Sci J. (2021) 92:e13580. doi: 10.1111/asj.13580
72. Timmerman, HM, Mulder, L, Everts, H, Van Espen, D, Van Der Wal, E, Klaassen, G, et al. Health and growth of veal calves fed milk replacers with or without probiotics. J Dairy Sci. (2005) 88:2154–65. doi: 10.3168/jds.S0022-0302(05)72891-5
73. Nocek, J, and Kautz, W. Direct-fed microbial supplementation on ruminal digestion, health, and performance of pre-and postpartum dairy cattle. J Dairy Sci. (2006) 89:260–6. doi: 10.3168/jds.S0022-0302(06)72090-2
74. Tang, MT, Han, H, Yu, Z, Tsuruta, T, and Nishino, N. Variability, stability, and resilience of fecal microbiota in dairy cows fed whole crop corn silage. Appl Microbiol Biotechnol. (2017) 101:6355–64. doi: 10.1007/s00253-017-8348-8
75. Hennessy, ML, Indugu, N, Vecchiarelli, B, Bender, J, Pappalardo, C, Leibstein, M, et al. Temporal changes in the fecal bacterial community in Holstein dairy calves from birth through the transition to a solid diet. PLoS ONE. (2020) 15:e0238882. doi: 10.1371/journal.pone.0238882
76. Gomez, D, Arroyo, L, Costa, M, Viel, L, and Weese, J. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J Vet Intern Med. (2017) 31:928–39. doi: 10.1111/jvim.14695
77. Yang, Q, Huang, X, Wang, P, Yan, Z, Sun, W, Zhao, S, et al. Longitudinal development of the gut microbiota in healthy and diarrheic piglets induced by age-related dietary changes. Microbiology. (2019) 8:e923. doi: 10.1002/mbo3.923
78. Dowd, SE, Callaway, TR, Wolcott, RD, Sun, Y, McKeehan, T, Hagevoort, RG, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. (2008) 8:125–8. doi: 10.1186/1471-2180-8-125
79. Albonico, F, Barelli, C, Albanese, D, Manica, M, Partel, E, Rosso, F, et al. Raw milk and fecal microbiota of commercial alpine dairy cows varies with herd, fat content and diet. PLoS One. (2020) 15:e0237262. doi: 10.1371/journal.pone.0237262
80. Xue, M, Sun, H, Wu, X, Guan, L, and Liu, J. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J Dairy Sci. (2019) 102:5031–41. doi: 10.3168/jds.2018-15974
81. Liu, J, Ml, Z, Ry, Z, Wy, Z, and Sy, M. Comparative studies of the composition of bacterial microbiota associated with the ruminal content, ruminal epithelium and in the faeces of lactating dairy cows. Microb Biotechnol. (2016) 9:257–68. doi: 10.1111/1751-7915.12345
82. Durso, L, Wells, J, Harhay, G, Rice, W, Kuehn, L, Bono, J, et al. Comparison of bacterial communities in faeces of beef cattle fed diets containing corn and wet distillers’ grain with solubles. Lett Appl Microbiol. (2012) 55:109–14. doi: 10.1111/j.1472-765X.2012.03265.x
83. Shanks, OC, Kelty, CA, Archibeque, S, Jenkins, M, Newton, RJ, McLellan, SL, et al. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microb. (2011) 77:2992–3001. doi: 10.1128/AEM.02988-10
84. Durso, LM, Harhay, GP, Smith, TP, Bono, JL, DeSantis, TZ, Harhay, DM, et al. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microb. (2010) 76:4858–62. doi: 10.1128/AEM.00207-10
85. Danielsson, R, Dicksved, J, Sun, L, Gonda, H, Müller, B, Schnürer, A, et al. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front Microbiol. (2017) 8:226. doi: 10.3389/fmicb.2017.00226
86. Stevenson, DM, and Weimer, PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Environ Microb. (2007) 75:165–74. doi: 10.1007/s00253-006-0802-y
87. Cotta, MA . Interaction of ruminal bacteria in the production and utilization of maltooligosaccharides from starch. Appl Environ Microb. (1992) 58:48–54. doi: 10.1128/aem.58.1.48-54.1992
88. Lau, SKP, Teng, JLL, Chiu, TH, Chan, E, Tsang, AKL, Panagiotou, G, et al. Differential microbial communities of omnivorous and herbivorous cattle in southern China. Comput Struct Biotechnol J. (2018) 16:54–60. doi: 10.1016/j.csbj.2018.02.004
89. O’Connor, A, Quizon, PM, Albright, JE, Lin, FT, and Bennett, BJ. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm Genome. (2014) 25:583–99. doi: 10.1007/s00335-014-9540-0
90. Zhou, M, Hernandez-Sanabria, E, and Guan, LL. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl Environ Microbiol. (2009) 75:6524–33. doi: 10.1128/AEM.02815-08
91. Holman, DB, Hao, X, Topp, E, Yang, HE, and Alexander, TW. Effect of co-composting cattle manure with construction and demolition waste on the archaeal, bacterial, and fungal microbiota, and on antimicrobial resistance determinants. PLoS One. (2016) 11:e0157539. doi: 10.1371/journal.pone.0157539
92. Zhang, T, Mu, Y, Zhang, D, Lin, X, Wang, Z, Hou, Q, et al. Determination of microbiological characteristics in the digestive tract of different ruminant species. Microbiology Open. (2018) 8:e00769. doi: 10.1002/mbo3.769
93. Zhang, R, Liu, J, Jiang, L, Wang, X, and Mao, S. The remodeling effects of high-concentrate diets on microbial composition and function in the hindgut of dairy cows. Front Nutr. (2021) 8:809406. doi: 10.3389/fnut.2021.809406
94. Mc Geough, E, O’kiely, P, Hart, K, Moloney, A, Boland, T, and Kenny, D. Methane emissions, feed intake, performance, digestibility, and rumen fermentation of finishing beef cattle offered whole-crop wheat silages differing in grain content. J Anim Sci. (2010) 88:2703–16. doi: 10.2527/jas.2009-2750
95. Knapp, JR, Laur, G, Vadas, PA, Weiss, WP, and Tricarico, JM. Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J Dairy Sci. (2014) 97:3231–61. doi: 10.3168/jds.2013-7234
96. Morvan, B, Bonnemoy, F, Fonty, G, and Gouet, P. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr Microbiol. (1996) 32:129–33. doi: 10.1007/s002849900023
Keywords: fermented total mixed ration, dairy cows, digestibility, fecal microbiota, fecal fermentation pattern
Citation: Wang L, Jin S, Wang P, Li X, Liu C, Sun S, Zhang G, Chang J, Yin Q, Zhang H and Zhu Q (2024) Fermented total mixed ration enhances nutrient digestibility and modulates the milk components and fecal microbial community in lactating Holstein dairy cows. Front. Vet. Sci. 11:1408348. doi: 10.3389/fvets.2024.1408348
Edited by:
Izhar Hyder Qazi, Shaheed Benazir Bhutto University of Veterinary & Animal Sciences, PakistanReviewed by:
Yu Pi, Chinese Academy of Agricultural Sciences, ChinaRuangyote Pilajun, Ubon Ratchathani University, Thailand
Copyright © 2024 Wang, Jin, Wang, Li, Liu, Sun, Zhang, Chang, Yin, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqiang Yin, cXF5MTk2NEBoZW5hdS5lZHUuY24=; Haiyang Zhang, ZGFoYWkwODA2NzAyQDEyNi5jb20=
 Lijun Wang
Lijun Wang Sanjun Jin
Sanjun Jin Ping Wang
Ping Wang Xinxin Li1
Xinxin Li1 Juan Chang
Juan Chang Qingqiang Yin
Qingqiang Yin