
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 14 May 2024
Sec. Livestock Genomics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1399548
Background: Prion diseases in mammals are caused by the structural conversion of the natural prion protein (PrPC) to a pathogenic isoform, the “scrapie form of prion protein (PrPSc).” Several studies reported that the shadow of prion protein (Sho), encoded by the shadow of prion protein gene (SPRN), is involved in prion disease development by accelerating the conformational conversion of PrPC to PrPSc. Until now, genetic polymorphisms of the SPRN gene and the protein structure of Sho related to fragility to prion disease have not been investigated in pheasants, which are a species of poultry.
Methods: Here, we identified the SPRN gene sequence by polymerase chain reaction (PCR) and compared the SPRN gene and Sho protein sequences among various prion disease-susceptible and -resistant species to identify the distinctive genetic features of pheasant Sho using Clustal Omega. In addition, we investigated genetic polymorphisms of the SPRN gene in pheasants and analyzed genotype, allele, and haplotype frequencies, as well as linkage disequilibrium among the genetic polymorphisms. Furthermore, we used in silico programs, namely Mutpred2, MUpro and AMYCO, to investigate the effect of non-synonymous single nucleotide polymorphisms (SNPs). Finally, the predicted secondary and tertiary structures of Sho proteins from various species were analyzed by Alphafold2.
Results: In the present study, we reported pheasant SPRN gene sequences for the first time and identified a total of 14 novel SNPs, including 7 non-synonymous and 4 synonymous SNPs. In addition, the pheasant Sho protein sequence showed 100% identity with the chicken Sho protein sequence. Furthermore, amino acid substitutions were predicted to affect the hydrogen bond distribution in the 3D structure of the pheasant Sho protein.
Conclusion: To the best of our knowledge, this is the first report of the genetic and structural features of the pheasant SPRN gene.
Prion diseases are fatal, infectious neurodegenerative disorders caused by the misfolding of a benign prion protein (PrPC) into an abnormal prion protein (PrPSc) (1). The exact process of converting PrPC to PrPSc remains unclear, but several factors that play crucial roles in the conversion process have been identified (1, 2). The ability to infect some species and not others is a remarkable prion characteristic (3). A wide variety of species, including various members of mammalian families such as cattle, goats, deer, cats and other primate families, are susceptible to prion diseases (4–8). However, horses, dogs, and chickens are resistant to prion diseases (9–11). Previous studies have also indicated that genetic characteristics of the SPRN gene are significantly different between prion disease-susceptible species (cattle, goats) (12, 13) and -resistant species (horses, dogs, and chickens) (9, 10, 14). Therefore, the investigation of the prion disease-related genes in prion-resistant species has been of particular interest as it provides insights into the determinants of susceptibility.
Among the prion protein family, there are members such as shadow of prion protein (Sho) and PrPC, which are mostly expressed in brain tissue (15, 16). These proteins have been reported to affect embryonic and mammary development (17). Previous studies suggest that the Sho protein, encoded by the shadow of prion protein gene (SPRN), interacts with PrPC (18). This interaction speeds up the conversion of PrPC to PrPSc (19). Since these two proteins interact and share similar characteristics, including protein expression profile and function (20, 21), it is essential to investigate the genetic characteristics of the SPRN gene to interpret the pathomechanism of prion diseases.
In the phylogenetic tree of the order Galliformes, chickens and pheasants have a close evolutionary relationship (22–24). Pheasants are members the genus Phasianus within the bird family (25). To date, prion infection has not been reported in birds and some other vertebrate species (26, 27). In our previous study, we reported 34 polymorphisms in the pheasant prion protein gene (PRNP), including 8 non-synonymous single nucleotide polymorphisms (SNPs) and 6 insertion/deletion polymorphisms (28). Furthermore, it has been reported that the pheasant PrPC has a 3D structure similar to the chicken PrPC; thus, pheasant PrPC is predicted to have relatively prion-resistant feature (28). Genetic polymorphisms in prion-related genes contribute significantly to interindividual variation, so they have been investigated as useful biomarkers in medicine, as well as in the study of pathology, pharmacology, epidemiology, and clinical immunology (29). In addition, polymorphisms of the SPRN gene in prion disease-susceptible species play a crucial role in determining their susceptibility to prion diseases (12, 30, 31). The SPRN polymorphisms, which contribute to expression level or protein function, show strong associations with the pathogenesis of prion diseases in different species (32, 33). However, the SPRN gene sequence, genetic polymorphisms, and structural features of Sho in pheasants have not yet been investigated.
In this study, we amplified the SPRN gene sequence by polymerase chain reaction (PCR) and performed multiple sequence alignment of pheasant SPRN gene and Sho protein sequences with those of prion disease-susceptible and -resistant species. In addition, we performed amplicon sequencing of SPRN to identify genetic polymorphisms and investigated the genotype, allele and haplotype frequencies as well as linkage disequilibrium (LD) of SNPs of pheasant SPRN gene. Furthermore, we evaluated the change of Sho by non-synonymous SNPs in the pheasant SPRN gene using in silico tools. Lastly, we predicted the secondary and 3D structure of Sho in several prion disease-susceptible and -resistant species using AlphaFold2.
Tissues of the cerebral cortex from pheasants (Phasianus colchicus) (n = 135) were collected from slaughterhouses in Korea and stored in a deep freezer (−80°C). The Institutional Animal Care and Use Committee (IACUC) at Jeonbuk National University (JBNU 2020-209) approved this study.
Genomic DNA was extracted from 20 mg cerebral cortex of 135 pheasants by a Bead Genomic DNA Prep kit (Biofact, Daejeon, Korea) according to the manufacturer’s instructions.
To amplify the pheasant SPRN gene, a PCR was performed with primers, designed based on the chicken SPRN gene (Gene ID: BN000836.1), namely SPRN-F (GTGCACTGCATGTGGTGAAGT) and SPRN-R (CGCATTGTCACCCAGCTTTA). Detailed information regarding the primers is described in Figure 1A. The 25 μL PCR mixture composed of 2.5 μL of 10× H-star Taq reaction buffer, 2.5 μL of 5× band helper, 1 μL of each 10 mM dNTP mix, 1 μL of each primer (10 μM), and 0.2 μL of H-star Taq DNA polymerase (BIOFACT, Daejeon, Korea). The following experimental conditions were used for PCR: 98°C for 15 min for denaturation; 40 cycles of 98°C for 20 s, 58°C for 40 s and 72°C for 1 min for denaturation, annealing and extension, respectively and 1 cycle of 72°C for 5 min for the final extension. The PCR products were purified using a FavorPrep GEL/PCR Purification Mini Kit (FAVORGEN, Pingtung City, Taiwan), and sequenced using an ABI 3730xl sequencer (ABI, Foster City, CA, USA). Genotyping was carried out using Finch TV software (Geospiza Inc., Seattle,WA, USA).
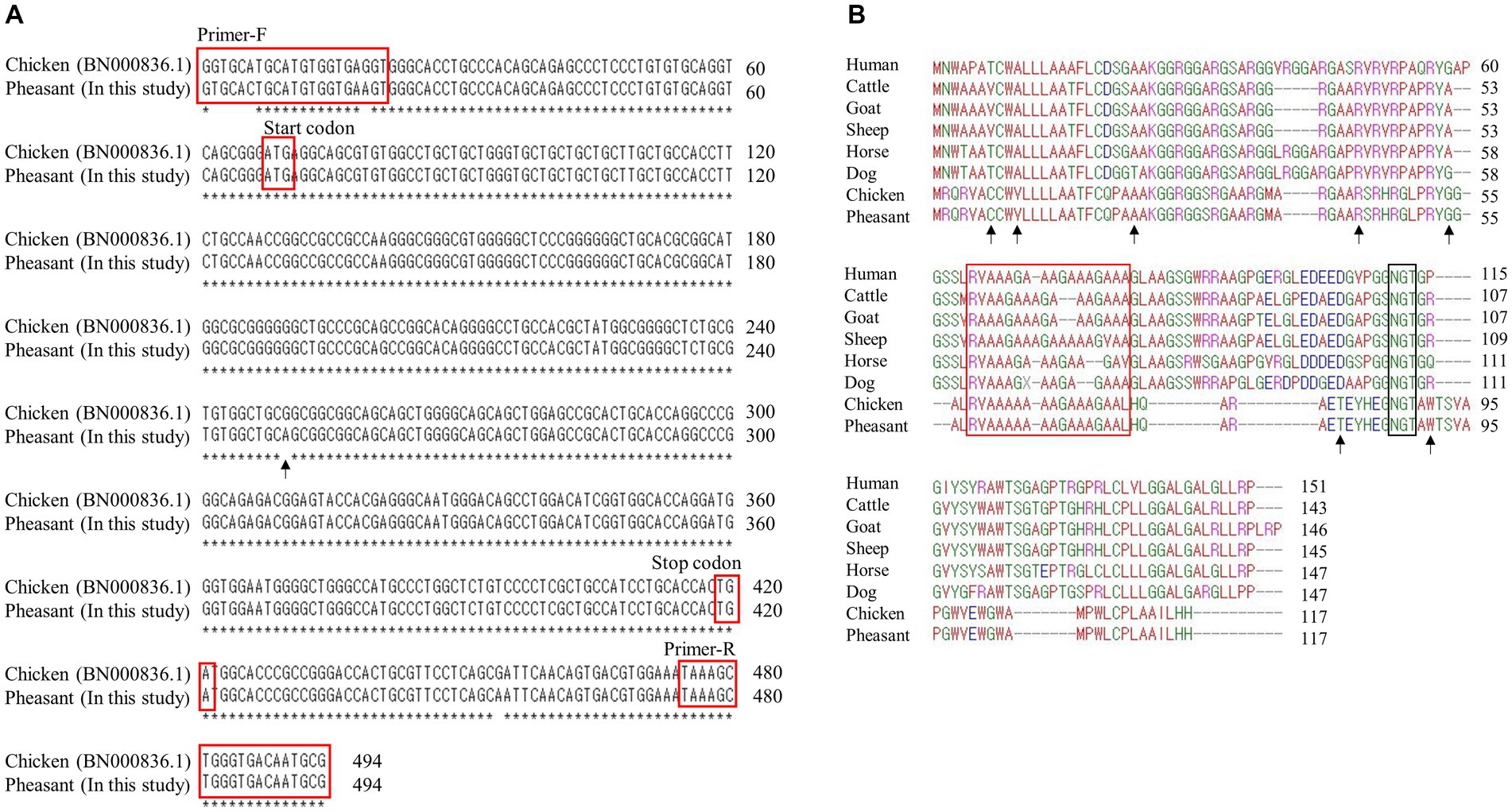
Figure 1. Identification of shadow of prion protein gene (SPRN) sequences in pheasants. (A) SPRN DNA sequence comparison between chicken and pheasant. The arrow indicates the one nucleotide (c.183) difference between chicken and pheasant. (B) Multiple sequence alignments of amino acid sequences of the shadow of prion protein (Sho) in various species including humans, cattle, goats, sheep, horses, dogs, chickens, and pheasants. Amino acid chemical properties are indicated by different colors; blue: acidic; red: small and hydrophobic; magenta: basic; green: hydroxyl, sulfhydryl, amine and glycine. The arrows denote the non-synonymous single nucleotide polymorphisms (SNPs) found in this study. The red box highlights the interaction region of Sho with prion protein (PrP) and the black box represents the NXT glycosylation motif of Sho.
To compare linkage between the SNPs of the pheasant’s PRNP and SPRN genes, data were obtained from our previous study (28). In brief, PCR was performed to amplify the pheasant PRNP gene with gene-specific primers, including PRNP-F (ATAAAGGAGGTGGGGAT GGG) and PRNP-R (CGTGGACACGATGTCATCTC). These primers were designed based on the pheasant PRNP gene (Gene ID: 116238382).
The amplicons of the SPRN gene in pheasants were analyzed by a web-based translation tool.1 Clustal Omega was used to align the amino acid sequences of Sho. The amino acid sequences of Sho from various species, humans (Homo sapiens, NP_001012526.2), cattle (Bos taurus, AAY83885.1), goats (Capra hircus, AGU17009.1), sheep (Ovis aries, NP_001156033.1), horses (Equus caballus, XP_023492126.1), dogs (Canis lupus familiaris, NC_051832), and chickens (Gallus gallus, CAJ43796.1), were obtained from the GenBank of the National Center for Biotechnology Information.
The impacts of each amino acid substitution were estimated using the Mutpred2,2 MUpro,3 and AMYCO in silico programs.4 MutPred2 is a web server designed to classify mutations as either disease-associated or neutral utilizing a machine-learning-based technique to estimate the molecular mechanism of the pathogenicity of an amino acid substitution. For a pathogenic mutation, the MutPred2 score is higher than 0.5 (34). MUpro can predict alterations in protein stability resulting from missense SNPs. If this tool score predicts a negative delta G (ΔG), it implies that the analyzed SNP may destabilize the protein (35). AMYCO program evaluates the impact of mutation on the aggregation propensity of prion-like domain (PrLD) in prion-like proteins (36).
The 3D structure of Pheasant SPRN was predicted by AlphaFold2, a tool reliant on machine learning.5 The confidence of the predicted structure was evaluated by the predicted local distance difference test (pLDDT) value, scaled from 0 to 100. The outcomes of the hydrogen bond alterations from the amino acid substitutions were predicted by the Swiss-PdbViewer (37).6
The LD and haplotype analyses were performed using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA). The Hardy–Weinberg Equilibrium (HWE) test was calculated by the chi-square test.
We first performed PCR to amplify the open reading frame (ORF) region of the pheasant SPRN gene. We designed SPRN gene-specific primers based on the SPRN gene sequences of the chicken (Gallus gallus) (Figure 1A). The PCR result revealed only one nucleotide difference (c.183) in the ORF of SPRN gene between the pheasant and chicken sequences.
Multiple sequence alignments of amino acid sequences of the Sho protein were performed among humans, cattle, goats, sheep, horses, dogs, chickens, and pheasants (Figure 1B). Pheasants and chickens share the same protein, despite differing by a single nucleotide (c.183). The Sho proteins of pheasants and chickens were the shortest among the eight species (117 amino acids). As in a previous study, although the Sho-PrP interaction regions (red box) and the NXT glycosylation motif (black box) were conserved among every species (14), the N-terminal and C-terminal regions exhibited significantly low sequence homology between mammals and pheasants.
We identified a total of 14 novel SNPs: 11 SNPs (c.18C > T (A6A), c.20G > A (C7Y), c.29 T > C (V10A), c.67G > A (A23T), c.131G > A (R44H), c.148C > T (L50L), c.160G > A (G54S), c.183A > G (A61A), c.219A > G (A73A), c.241A > G (T81A), c.271 T > C (W91R)) in the ORF region; 2 SNPs (c.-4C > T, c.-3G > A) upstream of the ORF of the SPRN gene; and 1 SNP (c.354 + 33A > G) downstream of the ORF of the SPRN gene (Figures 2A,B; Table 1). We found 7 non-synonymous and 4 synonymous SNPs. Detailed values of the genotype and allele frequencies of the SNPs in the pheasant SPRN gene are described in Table 1.
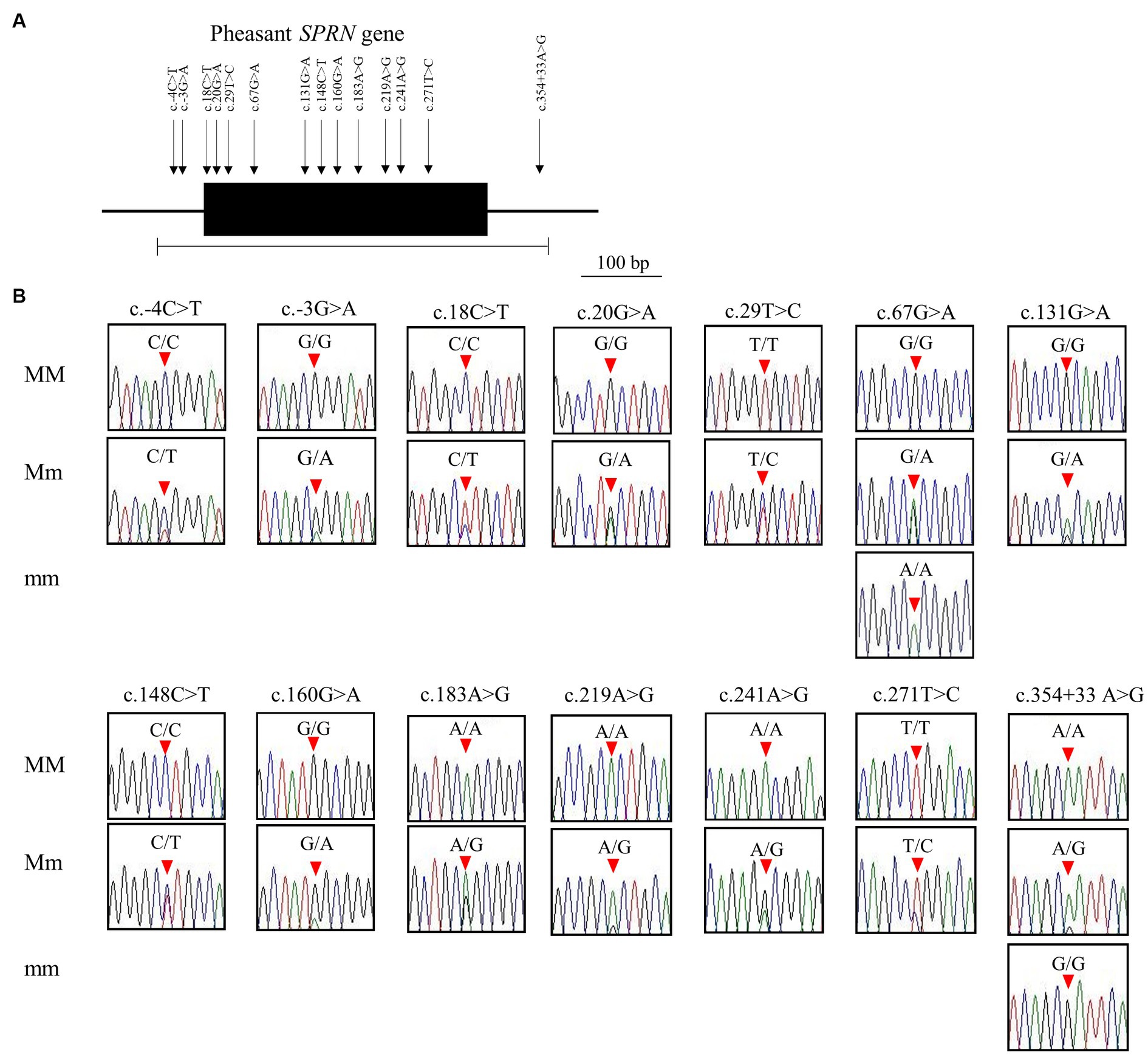
Figure 2. Identification of single nucleotide polymorphisms (SNPs) in pheasant shadow of prion protein gene (SPRN). (A) Gene map and polymorphisms identified in the SPRN in pheasant. The open reading frame (ORF) within exon is marked by a shaded block. Arrows indicate the 14 polymorphisms found in this study. The edged horizontal bar indicates the region sequenced. (B) Electropherograms of the 14 novel SNPs of the SPRN found in 135 pheasants. The peak colors indicate each base of the DNA sequence (green: adenine; red: thymine; blue: cytosine; black: guanine). The locations of the SNPs found in the present study are indicated by red arrowheads. MM indicates major homozygotes, Mm indicates heterozygotes, and mm indicates minor homozygotes.
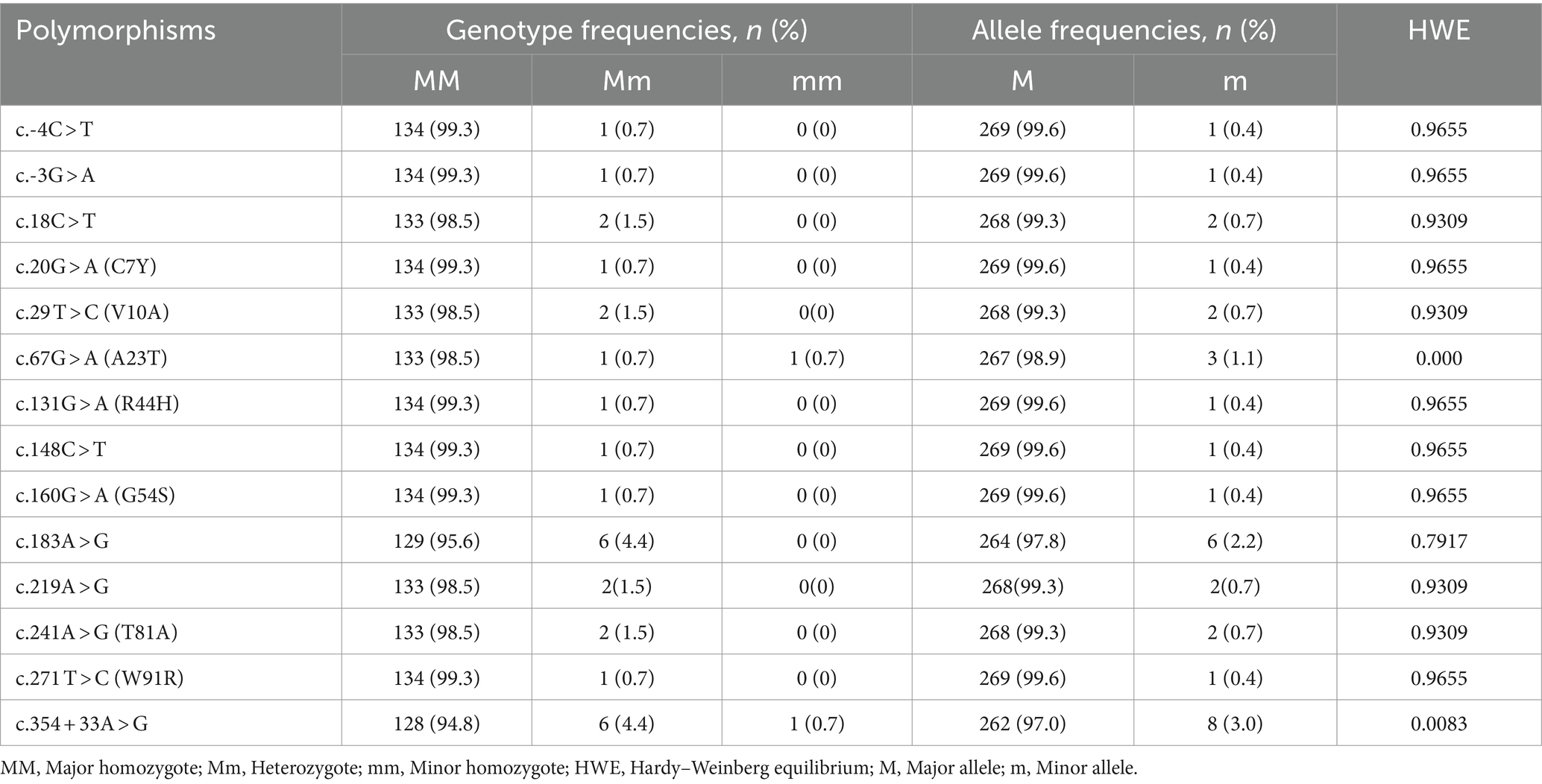
Table 1. Genotype and allele frequencies of shadow of prion protein gene (SPRN) polymorphisms in 135 pheasants.
We also analyzed the LD among all polymorphisms of the pheasant SPRN gene with their r2 values (Table 2). A total of 5 strong LDs (r2 > 0.333) were found. In addition, we investigated LD values between PRNP and SPRN polymorphisms. Strong LD (r2 > 0.333) was not observed between PRNP and SPRN polymorphisms in pheasants (Table 3). Furthermore, we conducted a haplotype analysis of all the polymorphisms found in the SPRN gene of the pheasant (Table 4). Three major haplotypes were identified in pheasants. The CGCGTGGCGAAATA (haplotype1, HT1) was most frequently observed (93.3%) in the pheasant SPRN gene, followed by CGCGTAGCGAAATA (haplotype2, HT2) (1.1%) and CGCGTGGCGAAATG (haplotype3, HT3) (1.1%).
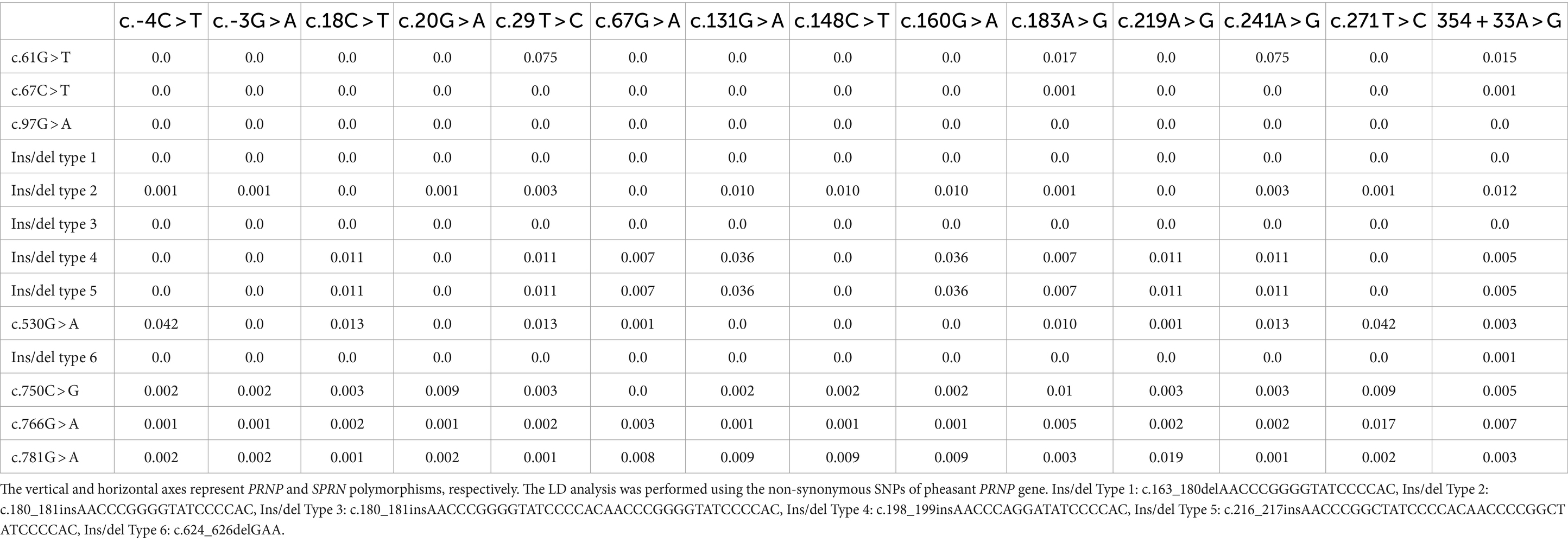
Table 3. Linkage disequilibrium (LD) analysis between polymorphisms in the PRNP and SPRN genes in pheasants.
To identify differences in the number of SNPs between pheasants and other species, we collected polymorphisms in the ORF of the SPRN gene from prion disease-susceptible (human, cattle, goat, sheep) and prion disease-resistant (horse, dog, chicken) species. Notably, the prion disease-susceptible species exhibited several genetic polymorphisms that lead to amino acid changes in the ORF of the SPRN gene. Three non-synonymous SNPs were reported in cattle and goats, and five non-synonymous SNPs were reported in sheep. However, only one synonymous SNP was identified in prion-resistant species, such as horses and chickens. In this study, we identified several genetic polymorphisms, including 7 non-synonymous and 4 synonymous SNPs, in pheasants (Figure 3).
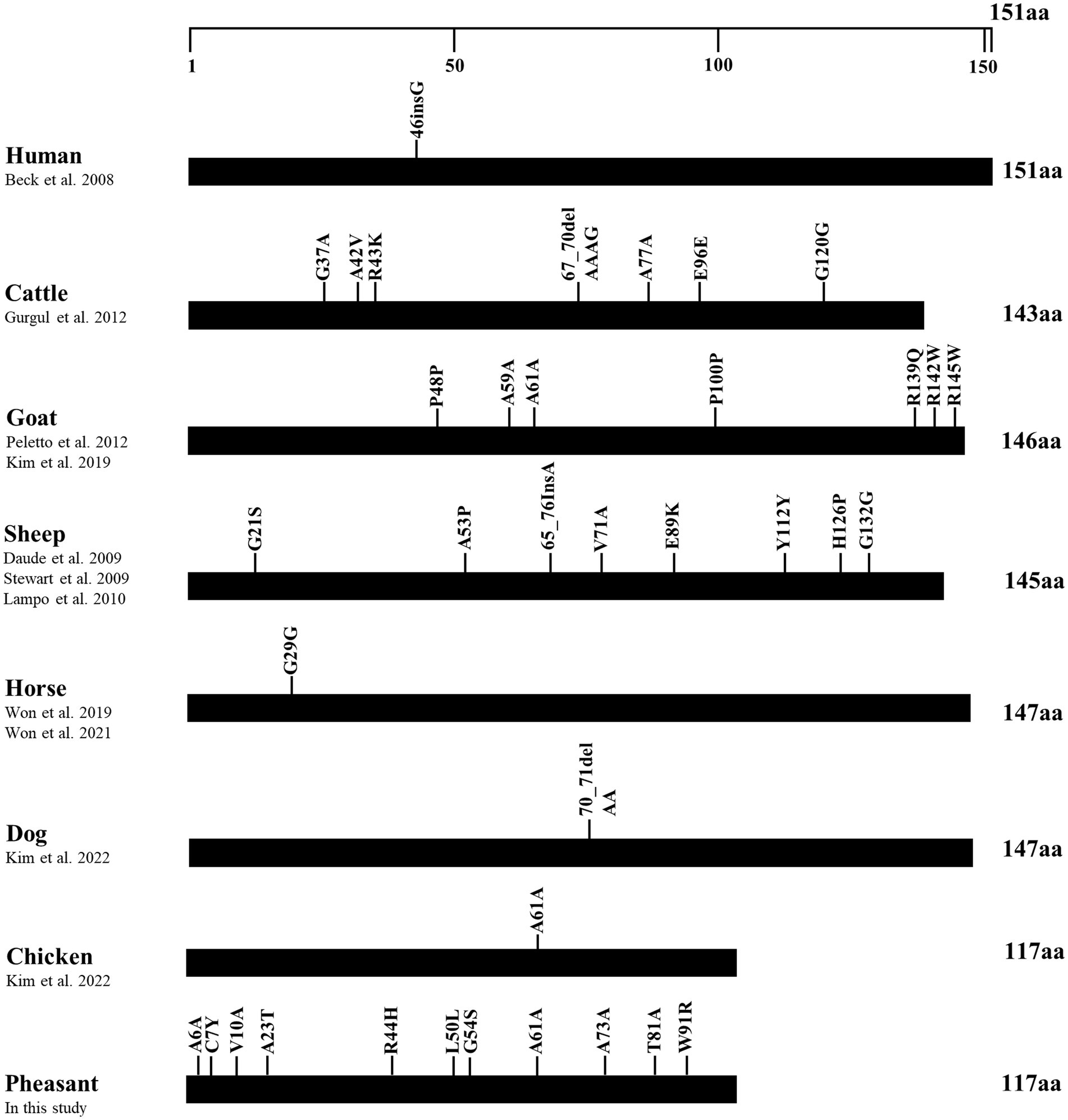
Figure 3. Distribution of genetic polymorphisms in the open reading frame (ORF) of the shadow of prion protein gene (SPRN) in various species. The figure shows previously reported genetic polymorphisms of the SPRN gene in a multitude of species such as humans, cattle, goats, sheep, horses, dogs, and chickens, as well as pheasants reported in this study. The edged horizontal bar signifies the length of the amino acids in the SPRN.
To analyze the functional and structural effects of non-synonymous SNPs in the pheasant Sho, we used MutPred2 and MUpro (Table 5). According to the MutPred2 analysis, the 7 non-synonymous SNPs had scores less than 0.5, indicating benign effects. However, the MUpro analysis gave these 7 non-synonymous SNPs scores below 0.0, indicating a decrease in protein stability (Table 5). We used AMYCO to analyze the effects of the 7 non-synonymous SNPs on the amyloid propensity of pheasant Sho. The pheasant Sho sequences containing the 7 non-synonymous SNPs had a score of 0.0, which is identical to that of the wild-type pheasant Sho (Table 5).
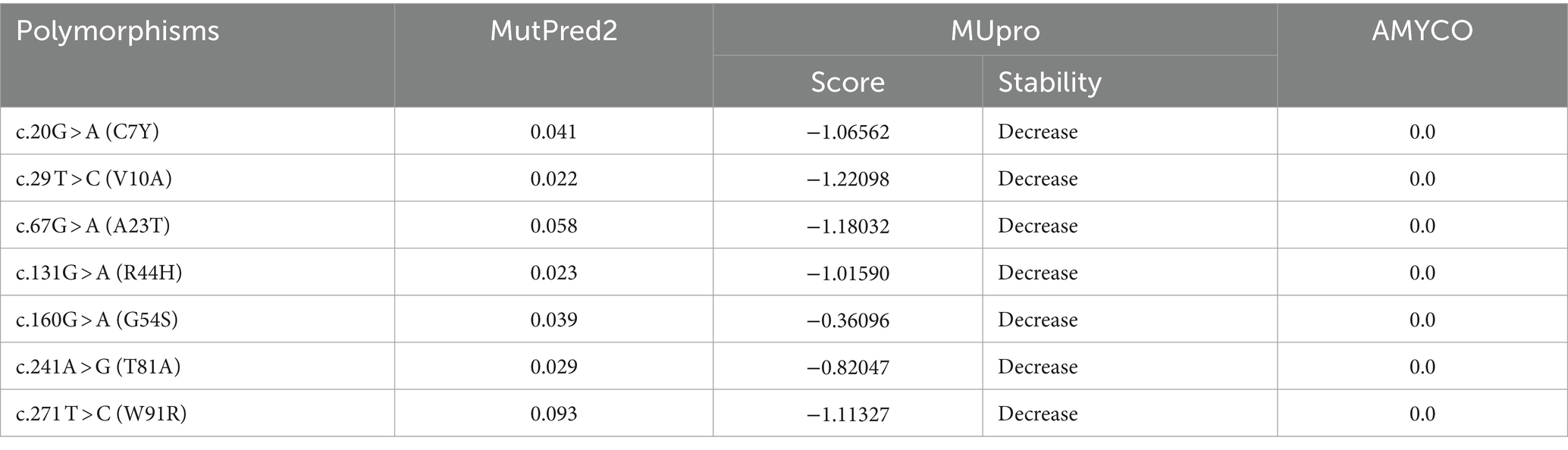
Table 5. In silico evaluation on effect of non-synonymous single nucleotide polymorphisms (SNPs) in the pheasant.
We used AlphaFold2 to analyze the 3D structures of Sho in eight species: humans, cattle, goats, sheep, horses, dogs, chickens, and pheasants. The structures of chicken and pheasant exhibited the same shape (Figure 4A). Two α-helices were predicted to be linked with the coil in Sho from all species except chicken and pheasant Sho. However, chicken and pheasant Sho proteins were predicted to have five α-helices (codons 3–22, 57–63, 70, 74–77 and 107–115) (Figure 4B). In addition, we predicted the 3D structure of the pheasant Sho carrying the CGCGTAGCGAAATA (haplotype2, HT2) and found it to be distinct from the wild type (Figure 5A). Pheasant Sho carrying the HT2 has three α-helices (codons 3–22, 102 and 106–115) (Figure 5B). Pheasant Sho carrying HT1 is the same as the wild-type, and HT3 is in the downstream of the ORF of the SPRN gene. HT3 has no effect on the structure of the Sho protein.
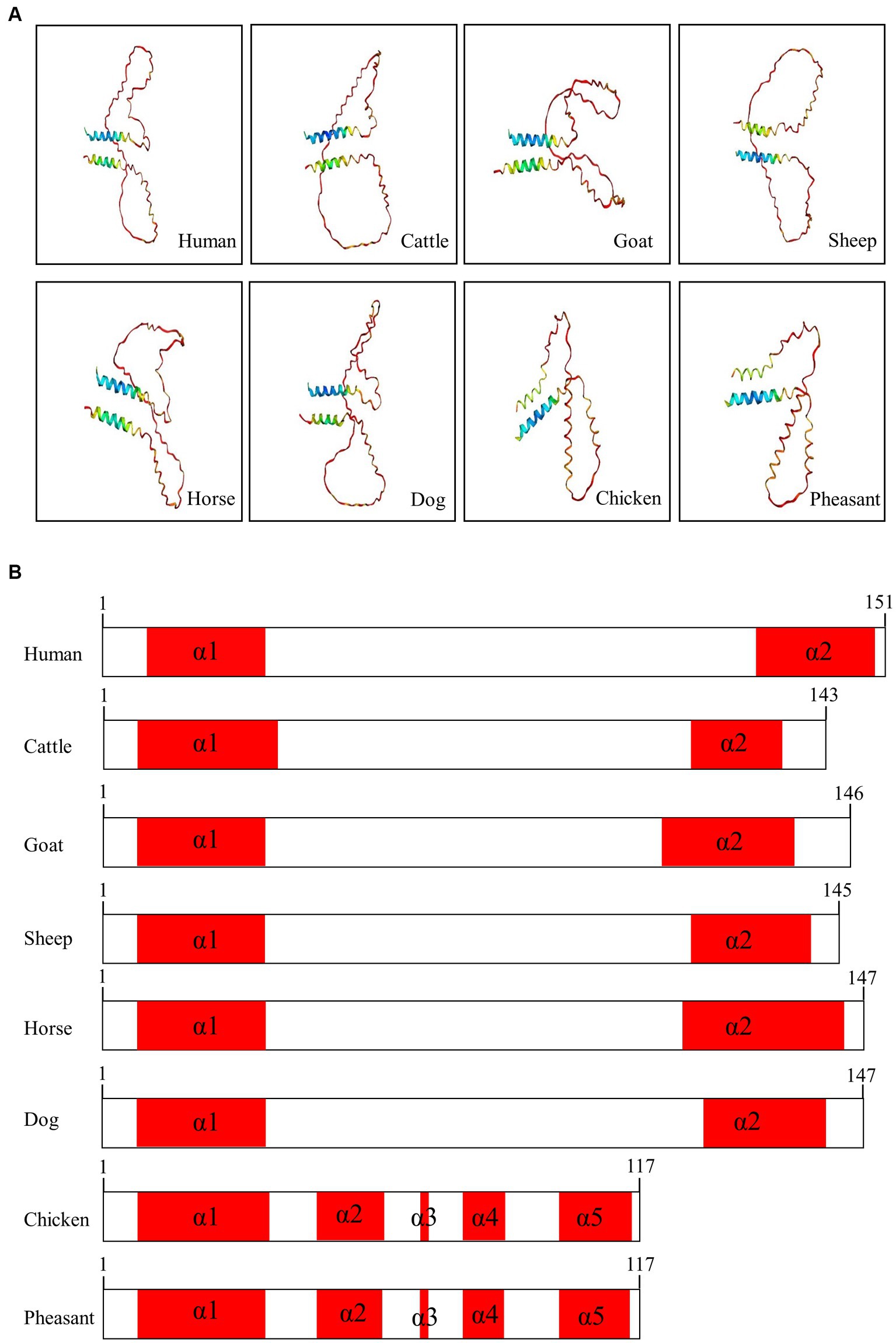
Figure 4. Prediction of 3D and secondary structures of the shadow of prion protein (Sho) in various species. (A) The 3D structures of human, cattle, goat, sheep, horse, dog, chicken, and pheasant Sho were analyzed by Alphafold2. (B) The secondary structures of human, cattle, goat, sheep, horse, dog, chicken, and pheasant Sho. In the structures, α-helices are represented in red, and coils are represented in white.
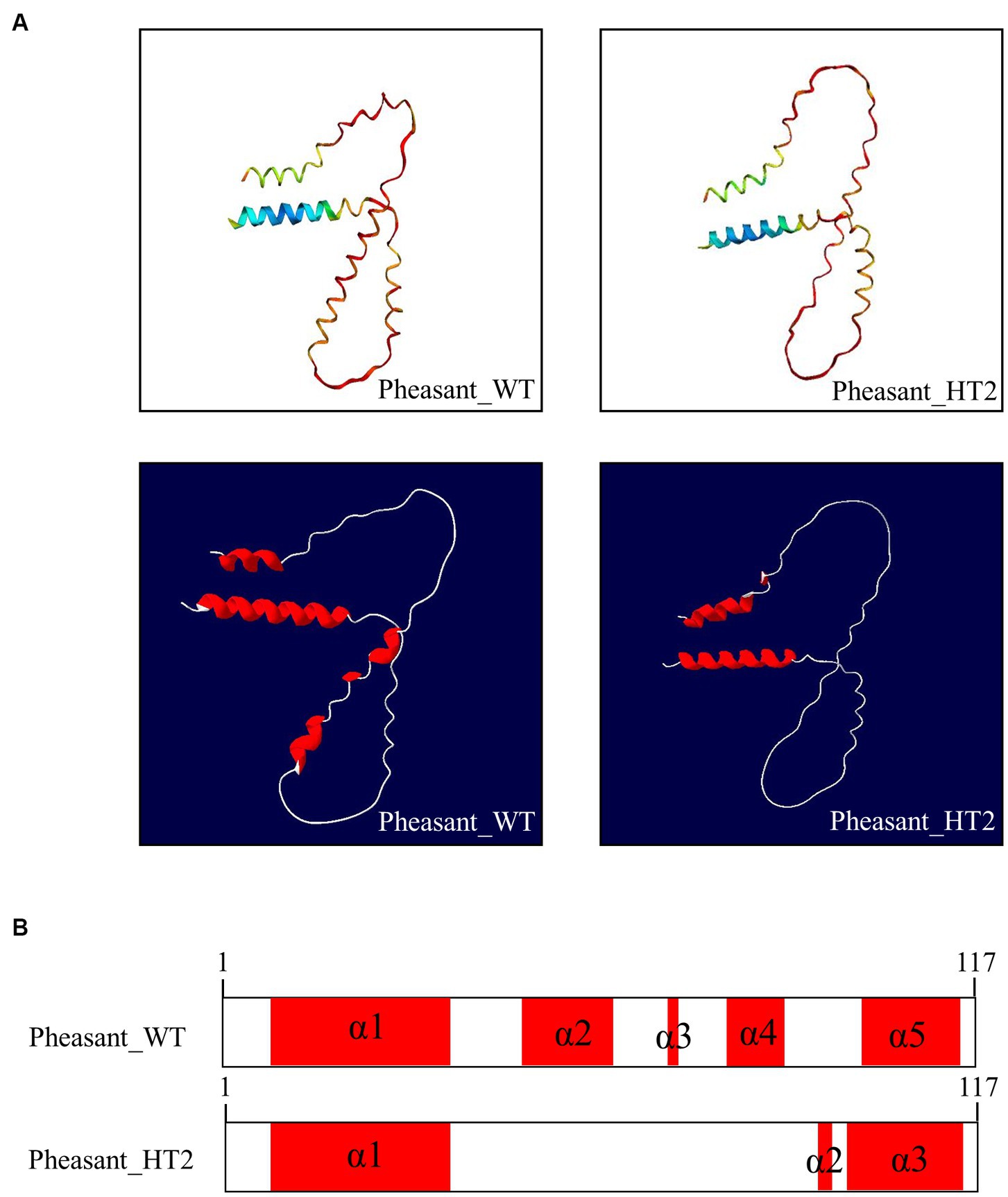
Figure 5. Prediction of 3D and secondary structures of the shadow of prion protein (Sho) in pheasants. (A) The 3D structures of wild-type and CGCGTAGCGAAATA (haplotype2, HT2) pheasant Sho. (B) The secondary structures of wild-type and CGCGTAGCGAAATA (haplotype2, HT2) pheasant Sho. In the structures, α-helices are represented in red, and coils are represented in white.
We explored the effects of 7 non-synonymous SNPs on the 3D structure of the pheasant Sho protein (Figure 6). First, the 3D structures of wild-type pheasant Sho were predicted by AlphaFold2. Then, the predicted structure was pictured using Swiss-PdbViewer, and the impact of 7 non-synonymous SNPs on pheasant Sho protein was analyzed. The C7 and Y7 alleles have three hydrogen bonds of the same length (Figure 6A). The V10 and A10 alleles have three hydrogen bonds of the same length (Figure 6B). Hydrogen bonds were absent in the A23 allele, but the T23 allele showed a hydrogen bond with Q20 (2.78 Å) (Figure 6C). Hydrogen bonds were absent in the R44, H44, G54 and S54 alleles (Figures 6D,E). The T81 allele showed a hydrogen bond with R78 (3.29 Å), but a hydrogen bond was absent in the A81 allele (Figure 6F). The W91 and R91 alleles also did not have hydrogen bonds (Figure 6G).
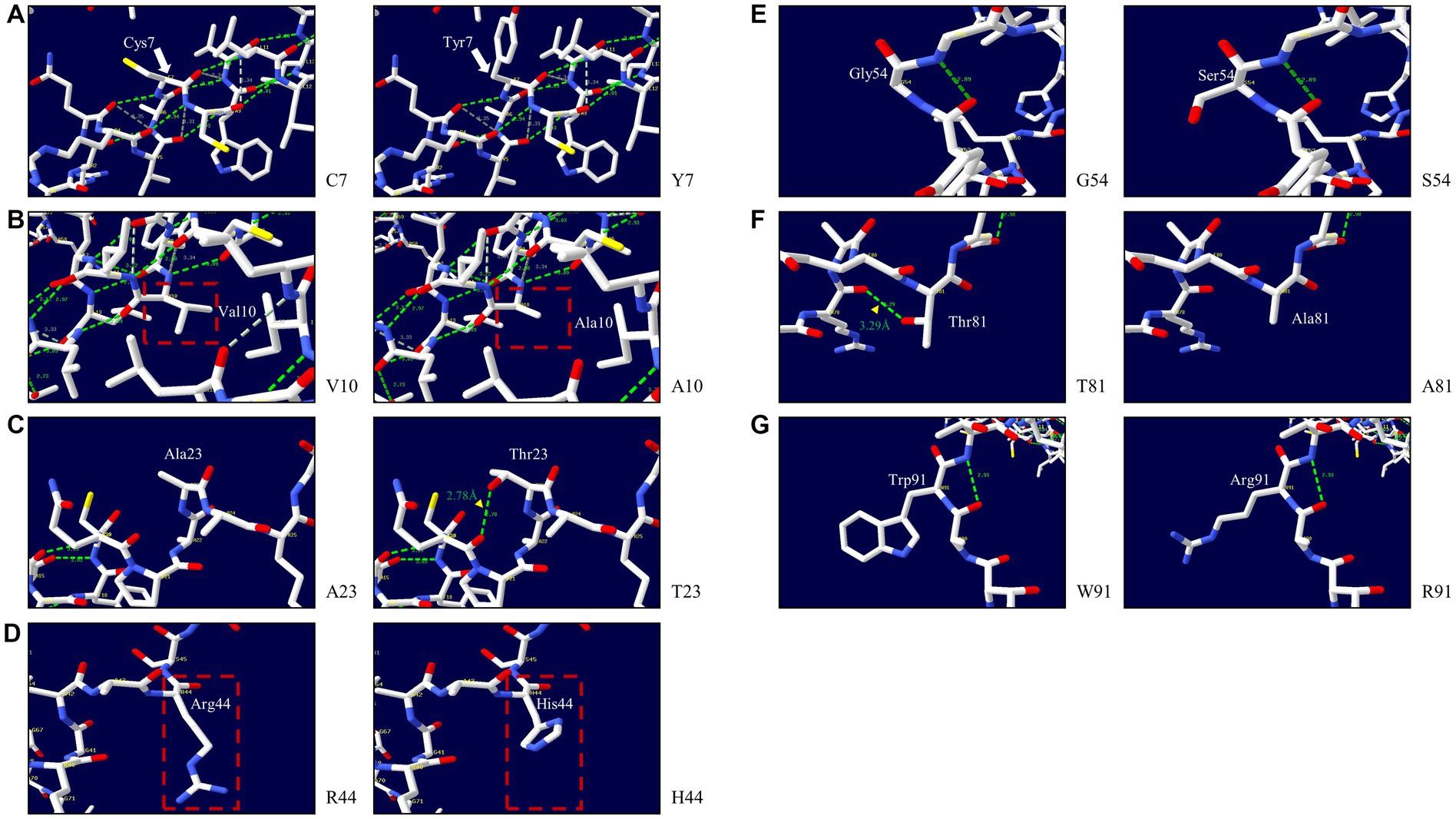
Figure 6. Prediction of the 3D structure and hydrogen bonds of the shadow of prion protein (Sho) in pheasants. (A) 3D structure of pheasant Sho carrying the C7 and Y7 alleles. (B) 3D structure of pheasant Sho carrying the V10 and A10 alleles. (C) 3D structure of pheasant Sho carrying the A23 and T23 alleles. (D) 3D structure of pheasant Sho carrying the R44 and H44 alleles. (E) 3D structure of pheasant Sho carrying the G54 and S54 alleles. (F) 3D structure of pheasant Sho carrying the T81 and A81 alleles. (G) 3D structure of pheasant Sho carrying the W91 and R91 alleles.
Considering that chickens are known to be resistant to prion diseases, it becomes crucial to investigate the genes associated with prion disease in pheasants. In the present study, we are the first to report pheasant SPRN gene sequences (Figure 1A). Chicken and pheasant have one nucleotide difference, but they share the same amino acid sequence. To identify the specific features of the pheasant Sho protein, we conducted a comparison of the amino acid sequences of the Sho from a variety of species and found that major components of Sho protein including prion interaction region and NXT glycosylation motif were conserved in pheasant like other prion-related species (Figure 1B).
In addition, we found 14 novel SNPs, including 7 non-synonymous and 4 synonymous SNPs, using amplicon sequencing (Figure 2). Previous studies have reported that prion disease-susceptible species, including cattle, goats, and sheep, have highly polymorphic SPRN genes (12, 13, 38, 39). In contrast, a prion disease-resistant species, horses and chickens have one polymorphism of the SPRN gene (9, 14, 40). Dogs, a prion disease-resistant animal, have one insertion/deletion polymorphism in SPRN gene (10). Interestingly, insertion/deletion polymorphisms have not been identified in the ORF of the pheasant SPRN gene. Although, phylogenetic analysis revealed that pheasants are closely related to chickens, a prion-resistant species (22–24), our results showed that pheasants have many polymorphisms. Further research is necessary to explore the SPRN gene in other species and to investigate the relationship between the number of SPRN gene polymorphisms and susceptibility to prion diseases. This is important because pathogenic mutations have a higher likelihood of occurring in SPRN genes that exhibit high polymorphism, as reported previously (10). Moreover, SPRN SNPs with low frequency have been discovered. Although the pheasant Sho sequence was identical to chicken Sho, the presence of a significant number of SNPs, albeit at a low frequency, can lead to structural abnormalities in the prion protein that encodes the prion-related protein. Therefore, further study is needed to comprehend how these characteristics could contribute to the progression of prion diseases and determine their impact on the prion pathomechanism.
To evaluate the impact of amino acid substitutions, we employed in silico programs (Table 5). The MutPred2 scores for all 7 non-synonymous SNPs were below 0.5, indicating no pathogenicity. We used MUpro to predict the effects of the SNPs on protein stability and found that a decrease in protein stability was associated with the 7 non-synonymous SNPs. Prion diseases are characterized by protein misfolding, and previous research has identified two important residues in PrP that promote stability in dogs and horses, animals known to be resistant to prion diseases (41). Those residues induce local changes that decrease the β-sheet content and enhance the structural stability of horse and dog PrPC, allowing the changes to spread to nearby regions (41). Further research is needed to understand how the decreased stability of the protein, in addition to its association with accelerated prion formation, can be interpreted in relation to the intrinsic functions of the Sho protein, such as embryo development. In addition, we employed AMYCO to estimate the impact of the mutations on the aggregation propensity of the PrLDs in prion-like proteins. Protein aggregation is a characteristic observed in various neurodegenerative disorders (42). The pheasant Sho sequences, when substituted with 7 non-synonymous SNPs, exhibited a score of 0.0, suggesting that the 7 non-synonymous SNPs do not have any amyloid-prone features.
We analyzed the 3D structures of Sho among various species predicted by AlphaFold2. Interestingly, with the exception of chicken and pheasant Sho, all species displayed a similar 3D structure, and none of the Sho proteins from any species exhibited a β-sheet (Figure 4). Additionally, we analyzed pheasant Sho according to the haplotype of pheasant Sho (Figure 5). By comparing the 3D structures of the pheasant Sho with both the wild-type allele and an HT2 variant, we observed a decrease in the number of α-helices in the pheasant Sho with HT2. The presence of HT1 and HT3 did not seem to impact the structure of the Sho protein. Since Sho plays a vital role in embryonic development, gaining insights into its structure can greatly contribute to comprehending its functional mechanisms (17, 43). Research is needed to investigate how structural changes in the haplotype affect the general function of Sho and its ability to interact with PrPC.
In this study, we first reported pheasant SPRN gene sequences and found 14 novel SNPs of the pheasant SPRN gene, including 7 non-synonymous and 4 synonymous in a total of 135 pheasants. In silico analysis, it was predicted that 7 non-synonymous SNPs induce decrease of protein stability. In addition, the secondary and tertiary structures of the wild-type pheasant Sho are identical to those of the chicken Sho. To the best of our knowledge, this is the first study on genetic polymorphisms of the pheasant SPRN gene.
The data presented in the study are deposited in the DRYAD repository https://datadryad.org/stash/share/9SErCsr_yxn1mmXQBsndJYntnvhR-sCJo-sjjnyumeE.
The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) at Jeonbuk National University (JBNU 2020-209). The study was conducted in accordance with the local legislation and institutional requirements.
D-IC: Conceptualization, Visualization, Writing – original draft. MZ: Formal analysis, Writing – review & editing. Y-CK: Formal analysis, Writing – review & editing. B-HJ: Conceptualization, Formal analysis, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2021R1A2C1013213, 2022R1C1C2004792). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876, 2021R1A6A3A01086488). This study was supported by Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (grant No. 2021R1A6C101C369).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. ^ https://web.expasy.org/translate/
2. ^ http://mutpred.mutdb.org/
3. ^ http://mupro.proteomics.ics.uci.edu/
4. ^ http://bioinf.uab.cat/amyco/
5. ^ https://colab.research.google.com/github/sokrypton/ColabFold/blob/v1.2.0/AlphaFold2.ipynb
2. Kim, YC, Lee, J, Lee, DW, and Jeong, BH. Large-scale lipidomic profiling identifies novel potential biomarkers for prion diseases and highlights lipid raft-related pathways. Vet Res. (2021) 52:105. doi: 10.1186/s13567-021-00975-1
3. Fernández-Borges, N, Parra, B, Vidal, E, Eraña, H, Sánchez-Martín, MA, de Castro, J, et al. Unraveling the key to the resistance of canids to prion diseases. PLoS Pathog. (2017) 13:e1006716. doi: 10.1371/journal.ppat.1006716
4. Wells, GA, Scott, AC, Johnson, CT, Gunning, RF, Hancock, RD, Jeffrey, M, et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. (1987) 121:419–20. doi: 10.1136/vr.121.18.419
5. Bons, N, Mestre-Frances, N, Belli, P, Cathala, F, Gajdusek, DC, and Brown, P. Natural and experimental oral infection of nonhuman primates by bovine spongiform encephalopathy agents. Proc Natl Acad Sci USA. (1999) 96:4046–51. doi: 10.1073/pnas.96.7.4046
6. Zeineldin, M, Cox-Struble, H, Camp, P, Farrell, D, Pritchard, R, Thacker, TC, et al. National Prevalence of caprine prion protein genetic variability at codons 146, 211, and 222 in goat herds in the United States. Vet Sci. (2023) 11:13. doi: 10.3390/vetsci11010013
7. Benestad, SL, and Telling, GC. Chronic wasting disease: an evolving prion disease of cervids. Handb Clin Neurol. (2018) 153:135–51. doi: 10.1016/b978-0-444-63945-5.00008-8
8. Iulini, B, Cantile, C, Mandara, MT, Maurella, C, Loria, GR, Castagnaro, M, et al. Neuropathology of italian cats in feline spongiform encephalopathy surveillance. Vet Pathol. (2008) 45:626–33. doi: 10.1354/vp.45-5-626
9. Won, SY, Kim, YC, Do, K, and Jeong, BH. The first report of genetic polymorphisms of the equine SPRN gene in outbred horses, Jeju and Halla horses. Animals. (2021) 11:2574. doi: 10.3390/ani11092574
10. Kim, YC, Kim, HH, Kim, AD, and Jeong, BH. Novel insertion/deletion polymorphisms and genetic features of the shadow of prion protein gene (SPRN) in dogs, a prion-resistant animal. Front Vet Sci. (2022) 9:942289. doi: 10.3389/fvets.2022.942289
11. Moore, J, Hawkins, SA, Austin, AR, Konold, T, Green, RB, Blamire, IW, et al. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to the domestic chicken. BMC Res Notes. (2011) 4:501. doi: 10.1186/1756-0500-4-501
12. Kim, YC, Kim, SK, Won, SY, and Jeong, BH. Polymorphisms of shadow of prion protein gene (SPRN) in Korean native cattle (Hanwoo) and Holstein cattle. Sci Rep. (2020) 10:15272. doi: 10.1038/s41598-020-72225-x
13. Kim, YC, Kim, SK, and Jeong, BH. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci Rep. (2019) 9:15261. doi: 10.1038/s41598-019-51625-8
14. Kim, YC, Kim, HH, and Jeong, BH. The first report of polymorphisms and genetic characteristics of the shadow of prion protein (SPRN) in prion disease-resistant animal, chickens. Front Vet Sci. (2022) 9:904305. doi: 10.3389/fvets.2022.904305
15. Onodera, T, Nishimura, T, Sugiura, K, and Sakudo, A. Function of prion protein and the family member, Shadoo. Curr Issues Mol Biol. (2020) 36:67–88. doi: 10.21775/cimb.036.067
16. Daude, N, and Westaway, D. Biological properties of the PrP-like Shadoo protein. Front Biosci. (2011) 16:1505–16. doi: 10.2741/3801
17. Passet, B, Castille, J, Makhzami, S, Truchet, S, Vaiman, A, Floriot, S, et al. The prion-like protein Shadoo is involved in mouse embryonic and mammary development and differentiation. Sci Rep. (2020) 10:6765. doi: 10.1038/s41598-020-63805-y
18. Premzl, M, Sangiorgio, L, Strumbo, B, Marshall Graves, JA, Simonic, T, and Gready, JE. Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene. (2003) 314:89–102. doi: 10.1016/s0378-1119(03)00707-8
19. Ciric, D, Richard, CA, Moudjou, M, Chapuis, J, Sibille, P, Daude, N, et al. Interaction between Shadoo and PrP affects the PrP-folding pathway. J Virol. (2015) 89:6287–93. doi: 10.1128/jvi.03429-14
20. Allais-Bonnet, A, and Pailhoux, E. Role of the prion protein family in the gonads. Front Cell Dev Biol. (2014) 2:56. doi: 10.3389/fcell.2014.00056
21. Makzhami, S, Passet, B, Halliez, S, Castille, J, Moazami-Goudarzi, K, Duchesne, A, et al. The prion protein family: a view from the placenta. Front Cell Dev Biol. (2014) 2:35. doi: 10.3389/fcell.2014.00035
22. Prum, RO, Berv, JS, Dornburg, A, Field, DJ, Townsend, JP, Lemmon, EM, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. (2015) 526:569–73. doi: 10.1038/nature15697
23. Helm-Bychowski, KM, and Wilson, AC. Rates of nuclear DNA evolution in pheasant-like birds: evidence from restriction maps. Proc Natl Acad Sci USA. (1986) 83:688–92. doi: 10.1073/pnas.83.3.688
24. van Raamsdonk, LWD, Prins, TW, Meijer, N, Scholtens, IMJ, Bremer, M, and de Jong, J. Bridging legal requirements and analytical methods: a review of monitoring opportunities of animal proteins in feed. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. (2019) 36:46–73. doi: 10.1080/19440049.2018.1543956
25. Ding, J, Jiang, T, Zhou, H, Yang, L, He, C, Xu, K, et al. The gut microbiota of pheasant lineages reflects their host genetic variation. Front Genet. (2020) 11:859. doi: 10.3389/fgene.2020.00859
26. Gibbs, CJ Jr, and Gajdusek, DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. (1973) 182:67–8. doi: 10.1126/science.182.4107.67
27. Barlow, RM, and Rennie, JC. The fate of ME7 scrapie infection in rats, guinea-pigs and rabbits. Res Vet Sci. (1976) 21:110–1. doi: 10.1016/S0034-5288(18)33406-4
28. Kim, KH, Kim, YC, and Jeong, BH. Novel polymorphisms and genetic characteristics of the prion protein gene in pheasants. Front Vet Sci. (2022) 9:935476. doi: 10.3389/fvets.2022.935476
29. Chiarella, P, Capone, P, and Sisto, R. Contribution of genetic polymorphisms in human health. Int J Environ Res Public Health. (2023) 20:912. doi: 10.3390/ijerph20020912
30. Kim, YC, Kim, HH, Kim, K, Kim, AD, and Jeong, BH. Novel polymorphisms and genetic characteristics of the shadow of prion protein gene (SPRN) in cats, hosts of feline spongiform encephalopathy. Viruses. (2022) 14:981. doi: 10.3390/v14050981
31. Gurgul, A, Polak, MP, Larska, M, and Słota, E. PRNP and SPRN genes polymorphism in atypical bovine spongiform encephalopathy cases diagnosed in polish cattle. J Appl Genet. (2012) 53:337–42. doi: 10.1007/s13353-012-0102-4
32. Beck, JA, Campbell, TA, Adamson, G, Poulter, M, Uphill, JB, Molou, E, et al. Association of a null allele of SPRN with variant Creutzfeldt-Jakob disease. J Med Genet. (2008) 45:813–7. doi: 10.1136/jmg.2008.061804
33. Peletto, S, Bertolini, S, Maniaci, MG, Colussi, S, Modesto, P, Biolatti, C, et al. Association of an indel polymorphism in the 3'UTR of the caprine SPRN gene with scrapie positivity in the central nervous system. J Gen Virol. (2012) 93:1620–3. doi: 10.1099/vir.0.041400-0
34. Choudhury, A, Mohammad, T, Anjum, F, Shafie, A, Singh, IK, Abdullaev, B, et al. Comparative analysis of web-based programs for single amino acid substitutions in proteins. PLoS One. (2022) 17:e0267084. doi: 10.1371/journal.pone.0267084
35. Kalmari, A, Hosseinzadeh Colagar, A, Heydari, M, and Arash, V. Missense polymorphisms potentially involved in mandibular prognathism. J Oral Biol Craniofac Res. (2023) 13:453–60. doi: 10.1016/j.jobcr.2023.05.007
36. Iglesias, V, Conchillo-Sole, O, Batlle, C, and Ventura, S. AMYCO: evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinform. (2019) 20:24. doi: 10.1186/s12859-019-2601-3
37. Guex, N, and Peitsch, MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. (1997) 18:2714–23. doi: 10.1002/elps.1150181505
38. Lampo, E, Van Poucke, M, Hugot, K, Hayes, H, Van Zeveren, A, and Peelman, LJ. Characterization of the genomic region containing the shadow of prion protein (SPRN) gene in sheep. BMC Genomics. (2007) 8:138. doi: 10.1186/1471-2164-8-138
39. Stewart, P, Shen, C, Zhao, D, and Goldmann, W. Genetic analysis of the SPRN gene in ruminants reveals polymorphisms in the alanine-rich segment of shadoo protein. JJ Gen Virol. (2009) 90:2575–2580. doi: 10.1099/vir.0.011494-0
40. Won, SY, Kim, YC, Kim, SK, and Jeong, BH. The first report of genetic and structural diversities in the sprn gene in the horse, an animal resistant to prion disease. Genes (Basel). (2019) 11:39. doi: 10.3390/genes11010039
41. Sanchez-Garcia, J, and Fernandez-Funez, P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol Dis. (2018) 119:1–12. doi: 10.1016/j.nbd.2018.07.011
42. Fassler, JS, Skuodas, S, Weeks, DL, and Phillips, BT. Protein aggregation and disaggregation in cells and development. J Mol Biol. (2021) 433:167215. doi: 10.1016/j.jmb.2021.167215
Keywords: pheasants, prion, SPRN, polymorphism, SNP
Citation: Choi D-I, Zayed M, Kim Y-C and Jeong B-H (2024) Novel polymorphisms and genetic studies of the shadow of prion protein gene (SPRN) in pheasants. Front. Vet. Sci. 11:1399548. doi: 10.3389/fvets.2024.1399548
Received: 12 March 2024; Accepted: 15 April 2024;
Published: 14 May 2024.
Edited by:
Sunday O. Peters, Berry College, United StatesReviewed by:
Christopher J. Silva, United States Department of Agriculture (USDA), United StatesCopyright © 2024 Choi, Zayed, Kim and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Hoon Jeong, YmhqZW9uZ0BqYm51LmFjLmty; Yong-Chan Kim, a3ljaEBhbnUuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.