- 1College of Life Sciences and Resources and Environment, Yichun University, Yichun, Jiangxi, China
- 2Shanxi Key Laboratory of Ecological Animal Science and Environmental Veterinary Medicine, College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, Shanxi, China
- 3College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, Shanxi, China
Orf is a zoonosis caused by the Orf virus (ORFV), which is endemic in goats, sheep, and wild ruminants worldwide. Orf infection is prevalent in China, with outbreaks reported in several provinces. Currently, there is limited information available regarding the characterization of ORFV strains in Jiangxi province. This study investigated an acute outbreak of Orf that occurred in 2021 in a goat herd in the Jiangxi province of China. Clinical signs in this case included lesions on the lips, nose, and inside the mouth. The presence of ORFV was confirmed from tissue samples by polymerase chain reaction (PCR). The nucleotide sequences of the B2L and F1L genes were fully sequenced and used to construct phylogenetic trees. The results of this investigation identified the ORFV JXxy2021 as the cause of the outbreak. The phylogenetic analysis revealed that the ORFV strain JXxy2021 had the highest similarity to the ORFV strains GO and FJ-SL from the neighboring province of Fujian. This suggests that JXxy2021 was likely transmitted from Fujian province. The results have provided valuable information on the genetic characteristics of JXxy2021 and the endemic situations of Orf in China.
1 Introduction
Orf, also known as sore mouth or contagious ecthyma, is a highly contagious zoonotic disease caused by the Orf virus (ORFV) (1). The disease usually manifests as scabbed sores on the lips, tongue, nostrils, and oral mucosa (2). The lesions are proliferative but self-limiting, and fatal outcomes are rare (3). However, the mortality of Orf can reach 90% in young animals, due to nutrient deficiency and secondary infection by pathogenic microorganisms (4). Orf primarily affects domestic small ruminants, such as sheep and goats, and occasionally wild small ruminants like deer and camels (5, 6). The disease can be transmitted to humans either through direct contact with infected animals or by consuming contaminated animal products (7–9). The case of human-to-human transmission of Orf has also been reported recently (10). Orf is widespread in most sheep- and goat-raising countries in the world (11). The global Orf endemic has caused significant economic losses for livestock farmers and poses a threat to public health as well (12).
Orf virus (ORFV), the etiologic agent of Orf, is the prototype member of the Parapoxvirus genus of the Poxviridae family (13). Other members of the genus include bovine papular stomatitis virus (BPSV), pseudocowpox virus (PCPV), and parapoxvirus of red deer in New Zealand (PVNZ) (14). The genome of ORFV is a linear double-strand DNA of about 138 kb with a G + C content as high as 64%, encoding 132 putative genes (15). The ORFV genome has a highly conserved core region (ORFs 009-111) and two variable terminal regions (ORFs 001-008, ORFs 112-132). The ORF011 (B2L) and ORF059 (F1L) genes, which are located in the core region of the ORFV genome, are highly conserved and commonly used for phylogenetic analysis of ORFV (16). Here we report the diagnosis and phylogenetic analysis of ORFV strain JXxy2021 in Jiangxi province of China. The full-length sequences of B2L and F1L genes were obtained. Multiple sequence alignments were performed based on the B2L and F1L genes. Phylogenetic trees were constructed to determine the genetic relationship of ORFV strain JXxy2021 with reference strains.
2 Materials and methods
2.1 Clinical cases and sampling
In 2021, a suspected outbreak of Orf occurred on a black goat farm in Jiangxi province, Southeast China. Following the first case, the goats on the farm were successively infected over an extended period. Tissue specimens were collected by scraping the crusts from affected areas, specifically the lips and nostrils, of goats with typical pathological changes consistent with Orf infection. The scab samples were stored at −80°C and analyzed further.
2.2 DNA extraction, PCR amplification, and sequencing
Pior to DNA extraction, the collected specimens were pre-treated as described below. Four samples of scab were collected from the goats exhibiting symptoms of the disease. A small quantity of each sample was selected and combined into a single sample. The mixed sample was then triturated in sterilized phosphate-buffered saline (PBS) using a mortar and pestle. The homogenized samples were centrifuged at 5000 r/min for 5 min at 4°C. The supernatant fluids were collected and used for downstream DNA extraction. Genomic DNA was extracted from 200 μL of viral stocks using the TIANamp RAN/DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions.
The polymerase chain reaction (PCR) was conducted to ascertain the presence of ORFV in the sample. Two pairs of primers targeting the entire open reading frame of the B2L and F1L genes of ORFV (Accession No. KX951407 and KX951408) were designed using the software Primer Premier 5. The sequences of the primers are as follows: B2L-F: 5′-atgtggccgttctcctccatc-3′, and B2L-R: 5′-ttaatttattggcttgcagaac-3′, F1L-F: 5′-atggatccacccgaaatcac-3′, and F1L-R: 5′-tcacacgatggccgtgacca-3′. The amplification reactions were performed in a 50 μL reaction volume consisting of 25 μL of 2 × Phanta Max Master Mix (Vazyme Biotechnology, Nanjing, China), 2 μL of each primer, 2 μL template, and 19 μL of distilled water. After pre-denaturation at 95°C for 3 min, 33 amplification cycles were carried out. The amplification process consisted of denaturation at 95°C for 15 s, annealing at 56°C for 15 s, and extension at 72°C for 60 s. This was followed by a final extension at 72°C for 5 min. The PCR products were analyzed on 1% agarose electrophoresis.
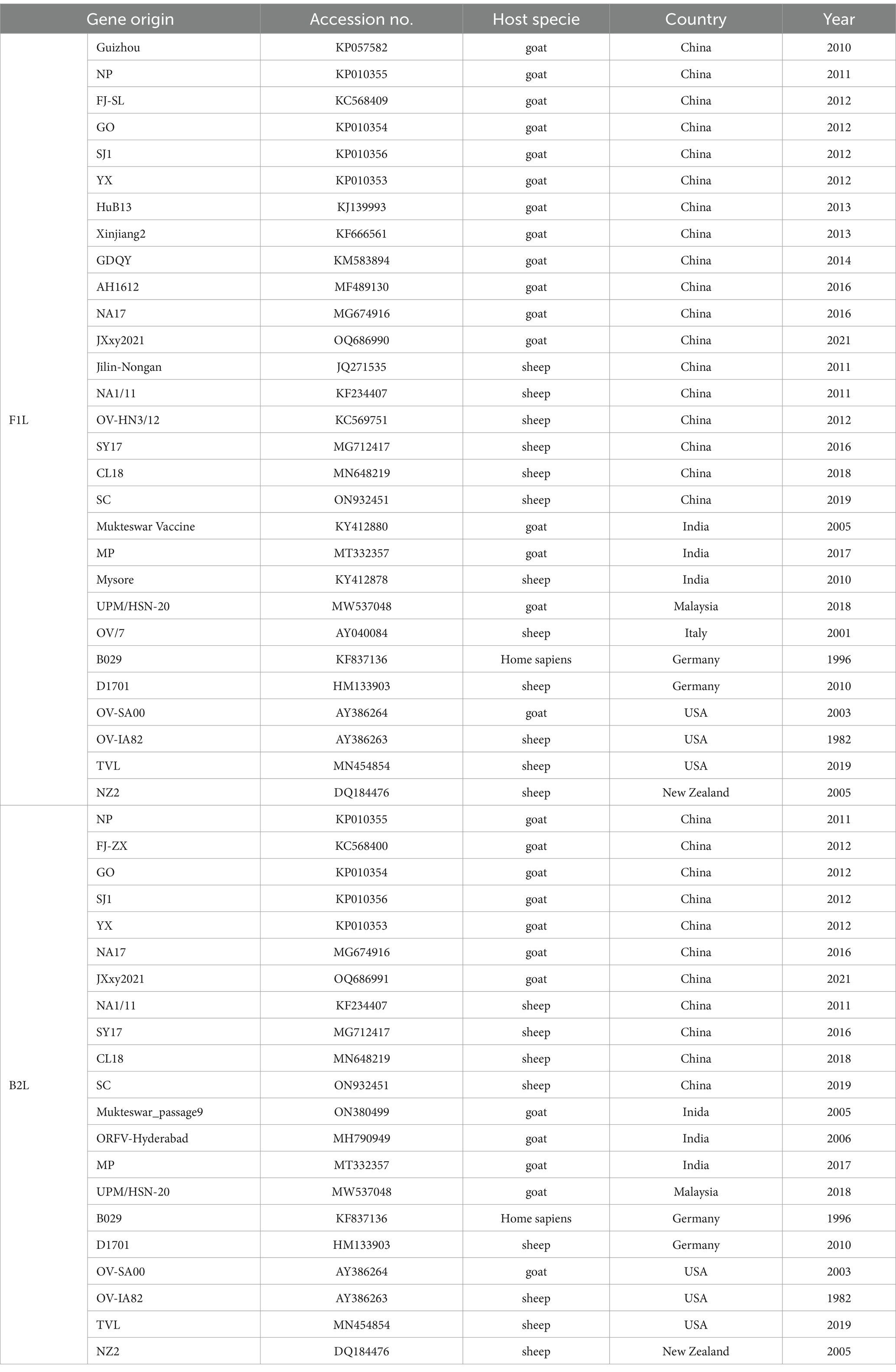
To get the full-length sequences of the B2L and F1L genes cloned in this study, the PCR products were purified using a Gel Extraction Kit (Thermo Fisher Scientific, Miami, USA) and were cloned into the TA/Blunt-Zero cloning Kit (Vazyme Biotechnology, Nanjing, China). The recombinant plasmids, TA/Blunt-Zero-B2L and TA/Blunt-Zero-F1L, were sequenced by Sangong Company (Shanghai, China). The sequences of B2L and F1L were deposited in the NCBI GenBank database under accession numbers OQ686991 and OQ686990, respectively.
2.3 Phylogenetic analysis
The ORFV strains in the NCBI GenBank that have the highest similarity to JXxy2021 were identified using the online BLAST program and were included in the phylogenetic analysis. The reference sequences were retrieved from the NCBI database, and the information for each sequence is listed in Table 1. The nucleotide sequences of B2L and F1L from ORFV strain JXxy2021 and the reference strains were used for phylogenetic analysis. Multiple sequence alignments were performed using the Clustal W method in DNASTAR software (v7.0). Phylogenetic trees were constructed by the neighbor-joining (NJ) method of MEGA 11 software, with 1,000 bootstrap test replications (17).
3 Results
3.1 Case presentation
In March of 2021, a suspected Orf outbreak was reported on a goat farm in Xinyu City, Jiangxi province (Figure 1). The goats that were suspected of having Orf first developed pimples and blisters on their lips, nostrils, gums, and tongue, which then led to ulceration (Figures 2A,B). This is consistent with the typical clinical manifestations of Orf (16). Over time, these sores tend to dry up and form a crust, but sores on the gums and tongue are more difficult to heal. Lesions around the lips, nostrils, tongue, and mouth can significantly impact a goat’s ability to feed, leading to a gradual decline in its physical and mental health. Based on these clinical signs, it was initially concluded that the goats were infected with the Orf virus (ORFV). The tissue samples were collected from the affected goats. The ORFV strain found in the samples was designated as JXxy2021 according to the time and location of the collection.
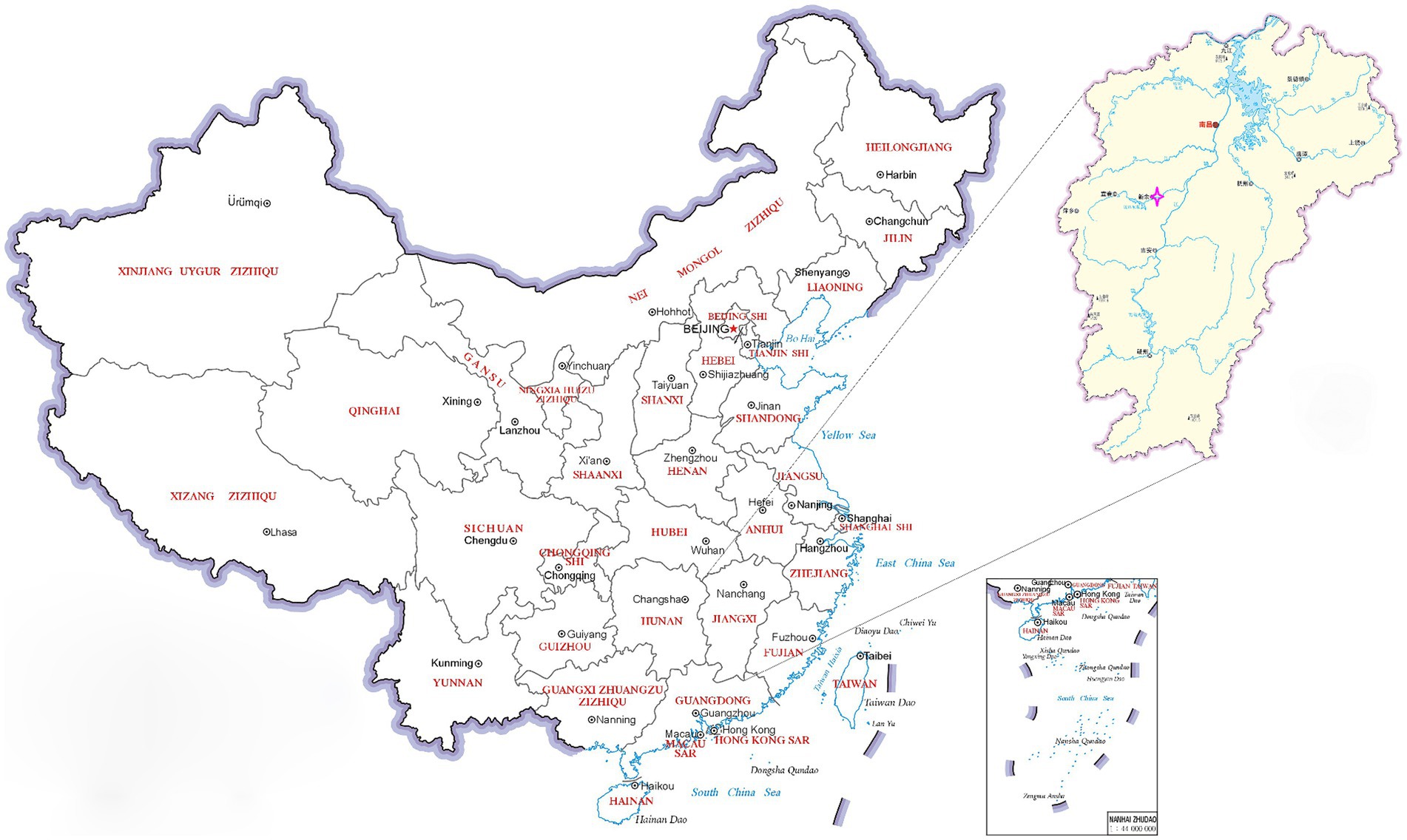
Figure 1. A map of China showing the sampling site. The pink four-pointed star  indicates the location of the outbreak of Orf. The map has been adapted from MAP WORLD (https://www.tianditu.gov.cn).
indicates the location of the outbreak of Orf. The map has been adapted from MAP WORLD (https://www.tianditu.gov.cn).
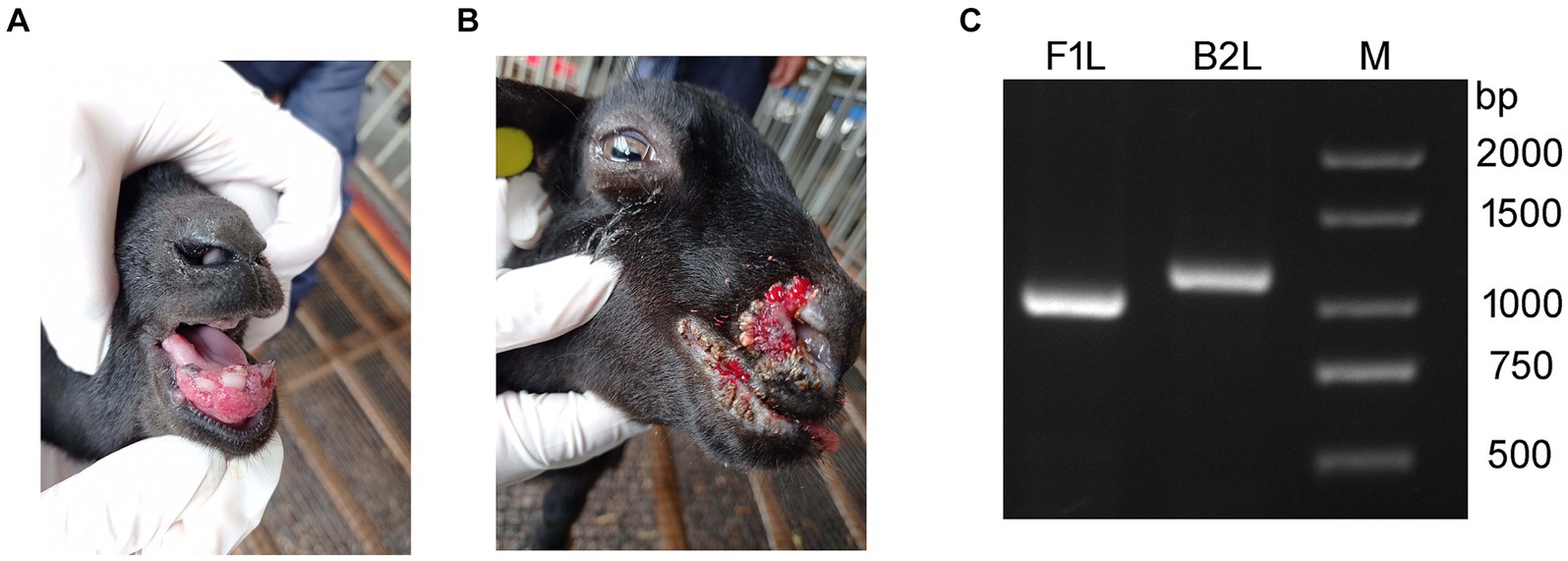
Figure 2. Representative clinical cases of ORFV infection in goat investigated using PCR. (A,B) Representative clinical cases of ORFV infection of goats. (A) Goat with severe lesions on the gum (Internal Orf). (B) Goat showing multiple nodular lesions on the lips and nostrils (External Orf). (C) PCR amplification of B2L and F1L genes using the DNA extracted from tissue samples. Lane M: DNA ladder.
3.2 Confirmatory PCR
To confirm the preliminary diagnostic results, PCR was used to detect ORFV-specific genes, B2L and F1L, from the DNA extracted from the tissue samples. The PCR-amplified products were subjected to agarose gel electrophoresis. Two bands corresponding to B2L and F1L were observed in the agarose gel between 1,000 and 1,500 bp, consistent with the expected gene sizes (Figure 2). The results indicated that ORFV was present in the samples collected from the sick goats. This is the first time an ORFV infection has been detected on this farm.
3.3 Sequence determination
Identifying the origin and genetic characteristics of ORFV JXxy2021 is crucial as no previous literature has reported the presence of ORFV in Jiangxi province. To address these issues, it is necessary to determine the sequence of B2L and F1L genes of JXxy2021. These two genes are the most commonly used targets for phylogenetic analysis of ORFV (18, 19). For the sake of obtaining the complete sequences of B2L and F1L genes, we cloned the PCR products into the commercial TA-cloning vectors for sequencing. The sequencing results showed that the B2L and F1L genes have a full length of 1,137 bp and 1,029 bp, respectively. The two cloned genes, B2L and F1L, can be read without interruption as there are no additional stop codons, except for the ends of the sequences. The deduced proteins of the B2L and F1L genes are 378 and 342 amino acids, respectively. Taken together, the molecular diagnosis and clinical symptoms suggest an ORFV infection on the farm in this study.
3.4 Phylogenetic analysis
To determine the phylogenetic relationship between JXxy2021 and other strains, we performed multiple sequence alignments of the B2L/F1L genes from JXxy2021 and reference strains. A phylogenetic tree was constructed by comparing the B2L nucleotide sequence with 20 reference strains obtained from the GenBank database. The result revealed that JXxy2021 shared the highest homology (98.8%) with the ORFV strain GO from Fujian province (Figure 3A; Supplementary Figure S1A). The phylogenetic tree, based on 29 F1L genes of various ORFV strains, indicated that the FJ-SL strain from Fujian province is genetically closely related to the JXxy2021 strain (Figure 3B), with a sequence identity of about 99.9% (Supplementary Figure S1B).
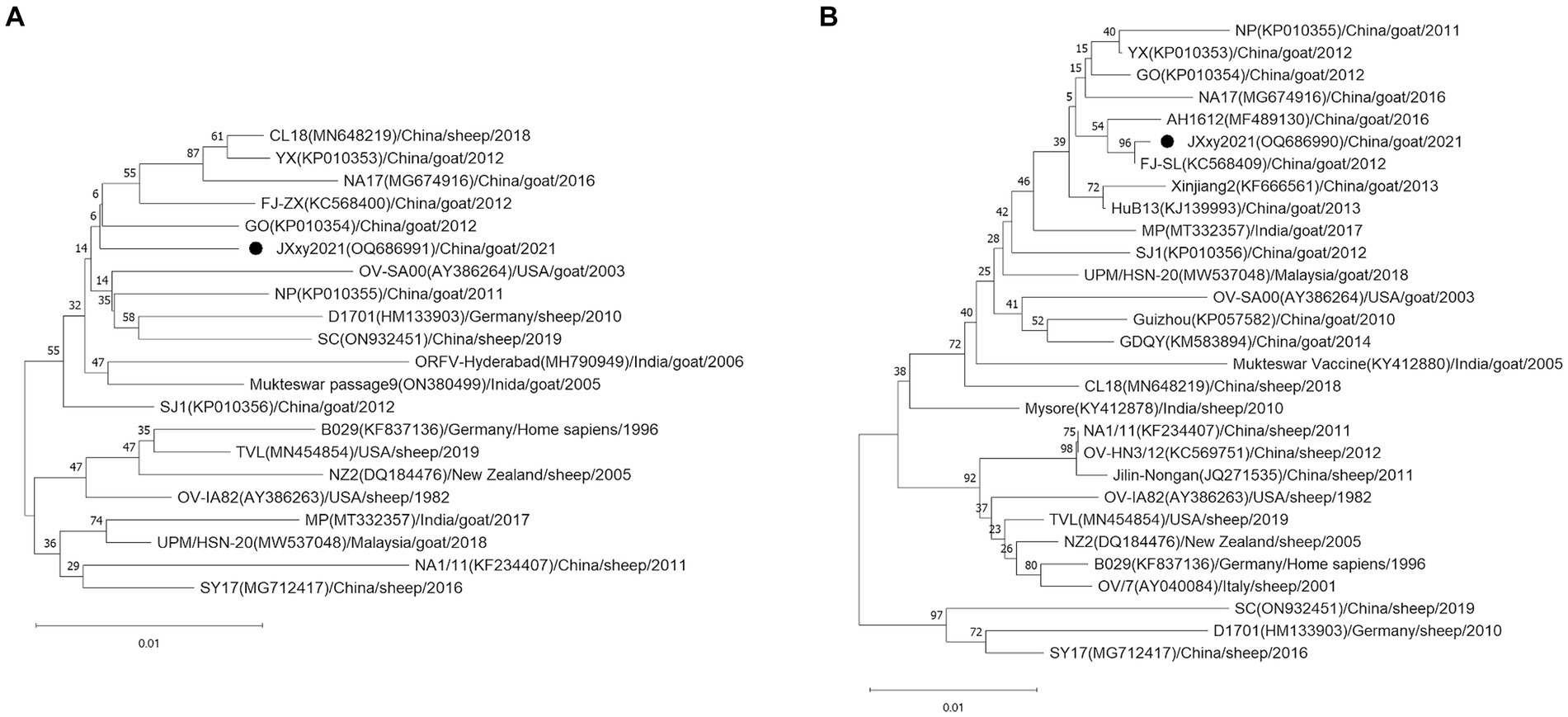
Figure 3. Phylogenetic trees based on the B2L (A) and F1L genes (B). The B2L and F1L sequences obtained from this study are indicated by the black point ●. Each strain is presented with a taxonomic name, including the corresponding GenBank accession number, host, year, and country of origin. The trees were constructed using the neighbor-joining (NJ) method of MEGA 11 software with 1,000 bootstrap test replications.
4 Discussion
Orf is a highly contagious zoonotic disease. The disease is common worldwide, especially in areas with high sheep or goat populations (20). China has the largest number of domestic small ruminants in the world (21). Orf is endemic in China, and several provinces have documented outbreaks of the disease (12, 22). The study reports an outbreak of Orf on a goat farm in Jiangxi province, Southeast China. The presence of Orf was detected through clinical signs and confirmed via molecular detection using PCR. In fact, the emergence of Orf on the farm occurred after breeding goats were introduced from the Sichuan province, Southwest China. It was initially hypothesized that Orf may have been caused by the introduction of breeding goats from outside. The ORFV strain SC was detected in a herd of sheep located in Sichuan province in 2017 (23). The nucleotide identity of the B2L and F1L genes between JXxy2021 and the SC strain is only 98.4 and 96.1% (Supplementary Figure S1), respectively. These values are not the highest among the results, indicating that the transmission of Orf from Sichuan province is less likely. Another possibility is that Orf was transmitted from the neighboring provinces to Jiangxi province. Jiangxi province shares borders with six provinces: Anhui, Hubei, Hunan, Guangdong, Fujian, and Zhejiang. Orf infection has been reported in four of the six provinces: Anhui, Hubei, Guangdong, and Fujian (22, 24–26). The phylogenetic trees based on the B2L and F1L genes indicate that the GO strain and FJ-SL strain have the highest homology to ORFV JXxy2021. Interestingly, GO and FJ-SL strains were discovered in Fujian province by the same research group (25, 27). The ORFV strains used for homology analysis in this study include those having the highest nucleotide sequence similarity to JXxy2021, which were identified through a BLAST search in the GenBank database. Based on the results obtained, JXxy2021 may have evolved from the ORFV strains found in the Fujian province. The findings indicate a potential cross-border transmission of ORFV due to close geographical proximity. However, the exact origin of the ORFV strain JXxy2021 remains unclear. Therefore, more epidemiological surveillance is needed in the future.
The B2L and F1L genes are regarded as epidemiologically relevant markers and are usually used in molecular epidemiological studies of ORFV (16, 25). The ORFV B2L gene encodes an envelope protein that is highly immunogenic and elicits a robust antibody response (28). The F1L protein is a key component of the microtubules found on the surface of ORFV and is responsible for stimulating the production of neutralizing antibodies in the host (29). Furthermore, the F1L protein is involved in the adsorption and invasion of host cells by the virus (30). However, the B2L and F1L genes are highly conserved in ORFV and may not accurately reflect the true genetic characteristics of different ORFV strains (31). Phylogenetic analysis based solely on conserved genes may overlook genetic heterogeneity among ORFV strains (32). Therefore, to accurately evaluate the genetic characteristics of ORFV strains, it is better to analyse more genes in addition to the B2L and F1L genes. However, the number of B2L and F1L genes of ORFV is the largest in the GenBank database. Currently, using the B2L or F1L genes as target genes for genetic evolutionary analysis of ORFV remains the dominant trend. Besides B2L and F1L genes, other genes, like ORF020 (VIR), GIF, vIL-10, ORF109, ORF110, ORF117, ORF119, ORF125, and ORF127 genes, have been utilized in the molecular epidemiological analysis of ORFV (6, 33–36). The ORF117, ORF119, ORF125, and ORF127 genes are located in the variable termini of the ORFV genome and are susceptible to mutation (37). Combining conserved and variable genes for phylogenetic analysis is more plausible than relying alone on conserved genes, which may introduce bias. This approach facilitates the acquisition of more comprehensive phylogenetic information on ORFV and enables better distinction between intraspecific strains (36).
In the view of some shepherds, Orf can be classified into two types: internal Orf and external Orf. If the lesions appear on the inside of the mouth, such as on the gums and tongue, it is considered as internal Orf. On the contrary, if Orf does not affect the inside of the oral cavity, it is referred to as external Orf. The healing process for lesions in the internal Orf is typically longer than that of the external Orf, resulting in a poorer outcome. The cases in this study (Figures 2A,B) happen to represent internal and external Orf, respectively. Similar situations can be observed in other cases and published literature (13, 24, 32). Currently, scholars may not yet be willing to accept the classification of Orf as internal or external based solely on clinical manifestations. The necessity and scientific validity of such a categorization should be discussed in the future.
This is the first report on the detection and phylogenetic analysis of an ORFV from Jiangxi province, Southeast China. This study identifies the genetic relatedness of JXxy2021 with other ORFV isolates from around the world. The results provide useful information on the endemic situations of Orf in China and may assist in preventing the transmission of Orf.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/nuccore/; OQ686990-OQ686991.
Ethics statement
The animal study was approved by the Animal Care and Ethics Committee of Yichun University (Permit No. JXSTUDKY2023130). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZZ: Funding acquisition, Writing – review & editing, Writing – original draft, Investigation, Conceptualization. XZ: Funding acquisition, Writing – original draft, Methodology, Investigation, Data curation. PM: Writing – original draft, Software, Methodology, Investigation. KF: Writing – original draft, Validation, Investigation. JG: Writing – original draft, Validation, Software, Methodology. ZY: Writing – original draft, Resources, Methodology, Data curation. TY: Writing – original draft, Resources, Funding acquisition. XX: Writing – original draft, Supervision, Data curation. WZ: Writing – original draft, Supervision, Resources, Funding acquisition. PL: Writing – review & editing, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32260880), Jiangxi Double Thousand Plan (jxsq2019101056), the Natural Science Foundation of Jiangxi Province (20202BABL205008, 20212BAB215032), the Science and Technology Research Program of Jiangxi Provincial Department of Education (GJJ190844, GJJ211630), and the Science and Technology Program of Xinyu City, Jiangxi Province, China (YKF2022-40, YKF2023-32).
Acknowledgments
The authors are grateful to Hong Zhang and Zhongping Zhang for facilitating the sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1389185/full#supplementary-material
SUPPLEMENTARY FIGURE S1 The percentages of identities and diversities of nucleotide sequences of the B2L (A) and F1L (B) gene among ORFV strains.
References
1. Santiago, L, Oliveira, D, Cardoso, JC, and Figueired, A. Human Orf: an under-recognized entity. Acta Dermatovenerol Croat. (2019) 27:280–1.31969245.
2. Hosamani, M, Scagliarini, A, Bhanuprakash, V, Mcinnes, CJ, and Singh, RK. Orf: an update on current research and future perspectives. Expert Rev Anti-Infect Ther. (2009) 7:879–93. doi: 10.1586/eri.09.64
3. Spyrou, V, and Valiakos, G. Orf virus infection in sheep or goats. Vet Microbiol. (2015) 181:178–82. doi: 10.1016/j.vetmic.2015.08.010
4. Bergqvist, C, Kurban, M, and Abbas, O. Orf virus infection. Rev Med Virol. (2017) 27:e1932. doi: 10.1002/rmv.1932
5. Alajlan, AM, and Alsubeeh, NA. Orf (Ecthyma Contagiosum) transmitted from a camel to a human: a case report. Am J Case Rep. (2020) 21:e927579. doi: 10.12659/ajcr.927579
6. Tryland, M, Beckmen, KB, Burek-Huntington, KA, Breines, EM, and Klein, J. Orf virus infection in Alaskan mountain goats, Dall's sheep, muskoxen, caribou and Sitka black-tailed deer. Acta Vet Scand. (2018) 60:12. doi: 10.1186/s13028-018-0366-8
7. Khan, Y, Currie, J, Miller, C, and Lawrie, D. Orf virus infection of the hand in a Scottish sheep farmer. A case report to increase awareness to avoid misdiagnosis. Case Reports Plast Surg Hand Surg. (2021) 9:26–9. doi: 10.1080/23320885.2021.2016057
8. Nougairede, A, Fossati, C, Salez, N, Cohen-Bacrie, S, Ninove, L, Michel, F, et al. Sheep-to-human transmission of Orf virus during Eid al-Adha religious practices France. Emerg Infect Dis. (2013) 19:102–5. doi: 10.3201/eid1901.120421
9. Macneil, A, Lederman, E, Reynolds, MG, Ragade, NJ, Talken, R, Friedman, D, et al. Diagnosis of bovine-associated parapoxvirus infections in humans: molecular and epidemiological evidence. Zoonoses Public Health. (2010) 57:e161–4. doi: 10.1111/j.1863-2378.2009.01317.x
10. Bouscarat, F, and Descamps, V. Wife to husband transmission of Ecthyma contagiosum (Orf). IDCases. (2017) 9:28–9. doi: 10.1016/j.idcr.2017.05.007
11. Lawan, Z, Bala, JA, Bukar, AM, Balakrishnan, KN, Mangga, HK, Abdullah, FFJ, et al. Contagious ecthyma: how serious is the disease worldwide? Anim Health Res Rev. (2021) 22:40–55. doi: 10.1017/s1466252320000018
12. Cheng, HY, Li, WJ, Li, XM, Fan, QL, Tang, XD, Liu, MJ, et al. Pathogenicity of blood orf virus isolates in the development of dairy goat contagious pustular dermatitis. Vet Microbiol. (2018) 219:178–82. doi: 10.1016/j.vetmic.2018.04.020
13. Chen, H, Li, W, Kuang, Z, Chen, D, Liao, X, Li, M, et al. The whole genomic analysis of orf virus strain HN3/12 isolated from Henan province, Central China. BMC Vet Res. (2017) 13:260. doi: 10.1186/s12917-017-1178-1
14. Scagliarini, A, Vaccari, F, Turrini, F, Bianchi, A, Cordioli, P, and Lavazza, A. Parapoxvirus infections of red deer, Italy. Emerg Infect Dis. (2011) 17:684–7. doi: 10.3201/eid1704.101454
15. Wang, X, Xiao, B, Zhang, J, Chen, D, Li, W, Li, M, et al. Identification and characterization of a cleavage site in the proteolysis of Orf virus 086 protein. Front Microbiol. (2016) 7:538. doi: 10.3389/fmicb.2016.00538
16. Zhao, K, Song, D, He, W, Lu, H, Zhang, B, Li, C, et al. Identification and phylogenetic analysis of an Orf virus isolated from an outbreak in sheep in the Jilin province of China. Vet Microbiol. (2010) 142:408–15. doi: 10.1016/j.vetmic.2009.10.006
17. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
18. Abrahao, JS, Borges, IA, Mazur, C, Lobato, ZI, Ferreira, PC, Bonjardim, CA, et al. Looking back: a genetic retrospective study of Brazilian Orf virus isolates. Vet Rec. (2012) 171:476. doi: 10.1136/vr.100634
19. Peralta, A, Robles, CA, Micheluod, JF, Rossanigo, CE, Martinez, A, Carosio, A, et al. Phylogenetic analysis of ORF viruses from five contagious Ecthyma outbreaks in Argentinian goats. Front Vet Sci. (2018) 5:5. doi: 10.3389/fvets.2018.00134
20. Kumar, R, Trivedi, RN, Bhatt, P, Khan, SUH, Khurana, SK, Tiwari, R, et al. Contagious pustular dermatitis (Orf disease) - epidemiology, diagnosis, control and public health concerns. Adv Animal Vet Sci. (2015) 3:649–76. doi: 10.14737/journal.aavs/2015/3.12.649.676
21. Mao, L, Li, W, Hao, F, Yang, L, Li, J, Sun, M, et al. Research Progress on emerging viral pathogens of small ruminants in China during the last decade. Viruses. (2022) 14:1288. doi: 10.3390/v14061288
22. Zhang, K, Shang, Y, Jin, Y, Wang, G, Zheng, H, He, J, et al. Diagnosis and phylogenetic analysis of Orf virus from goats in China: a case report. Virol J. (2010) 7:78. doi: 10.1186/1743-422X-7-78
23. Du, G, Wu, J, Zhang, C, Cao, X, Li, L, He, J, et al. The whole genomic analysis of the Orf virus strains ORFV-SC and ORFV-SC1 from the Sichuan province and their weak pathological response in rabbits. Funct Integr Genomics. (2023) 23:163. doi: 10.1007/s10142-023-01079-z
24. Wang, Y, Yang, K, Wang, YA, Yu, Z, Zhang, Q, Zhang, G, et al. Identification and phylogenetic analysis of an orf virus strain isolated in Anhui Province, east-Central China, in 2018. Acta Virol. (2019) 63:270–7. doi: 10.4149/av_2019_304
25. Chi, X, Zeng, X, Hao, W, Li, M, Li, W, Huang, X, et al. Heterogeneity among orf virus isolates from goats in Fujian Province, southern China. PLoS One. (2013) 8:e66958. doi: 10.1371/journal.pone.0066958
26. Duan, C, Liao, M, Wang, H, Luo, X, Shao, J, Xu, Y, et al. Identification, phylogenetic evolutionary analysis of GDQY orf virus isolated from Qingyuan City, Guangdong Province, southern China. Gene. (2015) 555:260–8. doi: 10.1016/j.gene.2014.11.016
27. Chi, X, Zeng, X, Li, W, Hao, W, Li, M, Huang, X, et al. Genome analysis of orf virus isolates from goats in the Fujian Province of southern China. Front Microbiol. (2015) 6:6. doi: 10.3389/fmicb.2015.01135
28. Sullivan, JT, Mercer, AA, Fleming, SB, and Robinson, AJ. Identification and characterization of an orf virus homologue of the vaccinia virus gene encoding the major envelope antigen p37K. Virology. (1994) 202:968–73. doi: 10.1006/viro.1994.1420
29. Scagliarini, A, Ciulli, S, Battilani, M, Jacoboni, I, Montesi, F, Casadio, R, et al. Characterisation of immunodominant protein encoded by the F1L gene of orf virus strains isolated in Italy. Arch Virol. (2002) 147:1989–95. doi: 10.1007/s00705-002-0850-2
30. Scagliarini, A, Gallina, L, Dal Pozzo, F, Battilani, M, Ciulli, S, and Prosperi, S. Heparin binding activity of orf virus F1L protein. Virus Res. (2004) 105:107–12. doi: 10.1016/j.virusres.2004.04.018
31. Wang, Y, Sun, S, Zhao, K, Du, L, Wang, X, He, W, et al. Orf virus DNA prime-protein boost strategy is superior to adenovirus-based vaccination in mice and sheep. Front Immunol. (2023) 14:1077938. doi: 10.3389/fimmu.2023.1077938
32. Li, S, Jing, T, Zhu, F, Chen, Y, Yao, X, Tang, X, et al. Genetic analysis of Orf virus (ORFV) strains isolated from goats in China: insights into epidemiological characteristics and evolutionary patterns. Virus Res. (2023) 334:199160. doi: 10.1016/j.virusres.2023.199160
33. Hosamani, M, Yadav, S, Kallesh, DJ, Mondal, B, Bhanuprakash, V, and Singh, RK. Isolation and characterization of an Indian ORF virus from goats. Zoonoses Public Health. (2007) 54:204–8. doi: 10.1111/j.1863-2378.2007.01046.x
34. Oem, JK, Chung, JY, Kim, YJ, Lee, KK, Kim, SH, Jung, BY, et al. Isolation and characterization of orf viruses from Korean black goats. J Vet Sci. (2013) 14:227–30. doi: 10.4142/jvs.2013.14.2.227
35. Zhang, K, Xiao, Y, Yu, M, Liu, J, Wang, Q, Tao, P, et al. Phylogenetic analysis of three orf virus strains isolated from different districts in Shandong Province, East China. J Vet Med Sci. (2015) 77:1639–45. doi: 10.1292/jvms.15-0368
36. Shimizu, K, Takaiwa, A, Takeshima, S-N, Okada, A, and Inoshima, Y. Genetic variability of 3′-proximal region of genomes of Orf viruses isolated from sheep and wild Japanese Serows (Capricornis crispus) in Japan. Front Vet Sci. (2020) 7:7. doi: 10.3389/fvets.2020.00188
Keywords: Orf virus, Jiangxi province, molecular detection, phylogenetic analysis, China, goat
Citation: Zhang Z, Zhang X, Meng P, Feng K, Gong J, Yang Z, Yang T, Xu X, Zheng W and Li P (2024) Molecular detection and phylogenetic analysis of Orf viruses from goats in Jiangxi province, China. Front. Vet. Sci. 11:1389185. doi: 10.3389/fvets.2024.1389185
Edited by:
Wen-Ping Guo, Chengde Medical University, ChinaReviewed by:
Chaoliang Leng, Nanyang Normal University, ChinaDomenico Galante, Experimental Zooprophylactic Institute of Puglia and Basilicata (IZSPB), Italy
Copyright © 2024 Zhang, Zhang, Meng, Feng, Gong, Yang, Yang, Xu, Zheng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengcheng Li, bGlwZW5nY2hlbmczMDVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Zhibang Zhang
Zhibang Zhang Xiaoyan Zhang
Xiaoyan Zhang Ping Meng1
Ping Meng1 Ziyin Yang
Ziyin Yang Pengcheng Li
Pengcheng Li