
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 17 June 2024
Sec. Livestock Genomics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1388339
This article is part of the Research Topic Rearing, Health, and Disease Management of Special Economic Animals: Paving the Way for a Sustainable Special Economy through Animal Agriculture View all 14 articles
Background: Natural cases of prion disease have not been reported in rabbits, and prior attempts to identify a prion conversion agent have been unsuccessful. However, recent applications of prion seed amplifying experimental techniques have sparked renewed interest in the potential susceptibility of rabbits to prion disease infections. Among several factors related to prion disease, polymorphisms within the prion-like protein gene (PRND), a member of the prion protein family, have been reported as significantly associated with disease susceptibility in various species. Therefore, our study aimed to investigate polymorphisms in the PRND gene of rabbits and analyze their genetic characteristics.
Methods: Genomic DNA was extracted from 207 rabbit samples to investigate leporine PRND polymorphisms. Subsequently, amplicon sequencing targeting the coding region of the leporine PRND gene was conducted. Additionally, linkage disequilibrium (LD) analysis was employed to assess the connection within and between loci. The impact of non-synonymous single nucleotide polymorphisms (SNPs) on the Doppel protein was evaluated using PolyPhen-2.
Results: We found nine novel SNPs in the leporine PRND gene: c.18A > G, c.76G > C, c.128C > T, c.146C > T, c.315A > G, c.488G > A, c.525G > C, c.544G > A, and c.579A > G. Notably, seven of these PRND SNPs, excluding c.525G > C and c.579A > G, exhibited strong LD values exceeding 0.3. In addition, LD analysis confirmed a robust link between PRNP SNP c.234C > T and PRND SNPs at c.525G > C and c.579A > G. Furthermore, according to PolyPhen-2 and SIFT analyses, the four non-synonymous SNPs were predicted to have deleterious effects on the function or structure of the Doppel protein. However, PANTHER and Missense3D did not indicate such effects.
Conclusion: In this paper, we have identified novel SNPs in the rabbit PRND gene and predicted their potential detrimental effects on protein function or structure through four non-synonymous SNPs. Additionally, we observed a genetic linkage between SNPs in the PRND and PRNP genes. These findings may provide insights into understanding the characteristics of rabbits as partially resistant species. To the best of our knowledge, this study is the first to genetically characterize PRND SNPs in rabbits.
Prion diseases are lethal neurodegenerative disorders characterized by aberrant misfolding of the cellular form of prion protein (PrPC) into the pathogenic, protease-resistant isoform of prion protein (PrPSc), resulting in their accumulation within the brain (1, 2). Host species exhibiting high susceptibility to prion diseases include humans (Creutzfeldt-Jakob disease, CJD), deer and elk (chronic wasting disease, CWD), cattle (bovine spongiform encephalopathy, BSE), and sheep and goats (scrapie), whereas dogs, horses, and chickens are resistant to prion diseases (3, 4). Occurrences of prion disease have not been reported in these resistant species in natural environments, and experimental infections have also failed to induce the disease (4, 5).
To date, there have been no reported cases of natural prion disease infection in rabbits (4, 6). Despite experimental attempts involving infection with the human kuru and CJD agents, as well as the scrapie agent isolated from sheep and mouse (Me7 strain), rabbits have proven to be resistant (7, 8). Further supporting this resistance, in vitro experiments demonstrated that rabbit PrPC did not convert to rabbit PrPSc in mouse neuroblastoma cells persistently infected with the mouse scrapie agent (RML strain) (9). Notably, a recent application of serial automated protein misfolding cyclic amplification (saPMCA) aimed to confirm protein misfolding and potential infection in rabbits, challenging the previous notion of rabbit resistance to prion diseases (10). The report indicated that rabbit brain homogenates, when amplified in vitro by saPMCA, exhibited resistance to proteinase K digestion. Upon injection into other rabbits, clinical symptoms emerged, leading to fatality. In brief, rabbits show prion resistance in natural transmission condition without extreme artificial replication techniques. Interestingly, rabbits exhibited a reduced propensity to transition to the β-structured state, which is believed to be associated with the mechanism of prion diseases, compared to susceptible species. However, they showed a higher propensity than dogs and horses (11). To further understand this partial resistance trait in rabbits, our investigation will focus on genetic factors associated with susceptibility to prion diseases.
Recent research has reported an association between the susceptibility to prion diseases and polymorphisms within the prion gene family (12–22). The prion protein gene family comprises the prion protein gene (PRNP), prion-like protein gene (PRND), prion-related protein gene (PRNT), and the shadow of prion protein gene (SPRN) (23). Among them, special attention is being given to PRND as a notable candidate gene. The PRND gene is located most closely to the PRNP gene, and the Doppel protein, encoded by PRND, shares biochemical and structural similarities with PrP (24, 25).
Previous studies propose an association between PRND polymorphisms and prion disease susceptibility in several species (26–30). In humans, significant differences in the frequencies of polymorphisms at codon 174 and the 3′ untranslated region (UTR) +28 of human PRND were observed in sporadic CJD patients compared to healthy controls (26, 27). In cattle, the genotype distributions of polymorphisms at codons 95 and 132 of the bovine PRND gene were significantly different between BSE-affected and healthy German Fleckvieh cattle (28). In sheep, linkage disequilibrium (LD) analysis revealed a significant linkage between the G allele of codon 26 of the ovine PRND gene and the ARR allele of the ovine PRNP gene, known to confer genetic resistance to scrapie (29). Even in goats, caprine PRND single nucleotide polymorphisms (SNPs) c.28 T > C, c.151A > G, and c.385G > C are strongly linked to caprine PRNP c.428A > G (H143R) (30). Consequently, the major homozygote genotype of caprine PRND SNPs is genetically associated with the caprine PRNP HH genotype, which is related to scrapie progression.
The exploration of polymorphisms in the PRND gene of prion disease-resistant species has yielded intriguing findings. In the canine PRND gene, four polymorphisms were identified, and researchers attempted to validate the association between PRNP and PRND through LD analysis (31). Despite the relatively shorter genetic distances in dogs compared to sheep and goats, no strong LD was observed between PRNP and PRND. In the equine PRND gene, SNPs were either absent or rare, depending on the breed (32, 33). Even in cases where they were rarely present, weak LD was confirmed between PRNP and PRND (33). Moreover, the PRND gene has not been identified in birds (34).
To date, studies on PRND polymorphisms in rabbits have not yet been performed, and this area is considered worth exploring. In this study, we investigated the genotype and allele frequencies of PRND polymorphisms in a group of 207 rabbits. In addition, we performed an LD analysis between PRNP and PRND to identify genetic linkage. Furthermore, we assessed the possible impact of non-synonymous SNPs on the structure and function of the Doppel protein using in silico prediction tools.
All 207 rabbit samples of hybrid breeding rabbits (New Zealand white and Flemish Giant FG) were provided by a slaughterhouse located in the Republic of Korea. Genomic DNA was isolated from 20 mg brain tissue following the manufacturer’s manuals using the Labopass Tissue Genomic DNA Isolation Kit (Cosmo Genetech Co., Ltd., Seoul). The overall experimental processes were approved by the Jeonbuk National University Institutional Animal Care and Use Committee (CBNU 2019-058). All experiments were performed in accordance with the Korea Experimental Animal Protection Act.
Primers were designed based on the leporine PRND gene sequence (Oryctolagus cuniculus) available in GenBank at the National Center for Biotechnology Information (NCBI) (Gene ID: 100347890). The gene-specific forward and reverse primer sequences were GGGTAGACCGGTTGGGAAAT and TGAGCACTGAAGCACTGAGG, respectively. Polymerase chain reaction (PCR) was performed targeting the coding region of the leporine PRND gene by an S-1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States). The PCR conditions followed the manual guide of BioFACT™ Taq DNA Polymerase (BioFACT Co., Ltd., Daejeon, Korea) with an annealing temperature of 65°C. The amplified products were purified using the FavorPrep™ GEL/PCR Purification Kit (Favorgen Biotech Corp., Kaohsiung, Taiwan), and then sequenced by an ABI PRISM 3730XL Analyzer (ABI, Foster City, CA, United States). These sequencing results for each sample were analyzed using Finch TV software (Geospiza Inc., Seattle, WA, United States).
The Hardy–Weinberg Equilibrium (HWE) test was conducted to assess the genotyping errors of the collected individual samples for this study. In HWE testing, a p-value lower than 0.05 indicates that the observed genotype or allele frequencies are not consistent with HWE (35, 36). The HWE test was conducted by the Michael H. Court’s calculator.
Additionally, LD analysis was performed to examine the statistical relationship between SNPs at each locus of the genes. LD, a measure of the correlation between two genetic loci (or SNPs), was assessed using the r2 value (37). The r2 value ranges from 0 to 1. Higher values indicate strong linkage between two genetic loci, suggesting the presence of associated genetic regions. Conversely, lower r2 values indicate that the variations at two genetic loci are independent or weakly correlated. The LD and haplotype distribution were estimated using the Haploview version 4.2 (Broad Institute, Cambridge, MA, United States).
PolyPhen-2 determines the effect of non-synonymous SNPs on the structure or function of a protein according to a position-specific independent counts (PSIC) score difference. The results are assigned as “probably damaging,” “possibly damaging” or “benign,” depending on the degree of risk. SIFT predicts the impact of amino acid substitutions on protein function based on sequence homology, assuming that alignment correlates well with evolution to maintain protein function. The SIFT score ranges from 0 to 1, where values below 0.05 are considered deleterious. PANTHER assesses the effect of non-synonymous SNPs on function using PANTHER-PSEP (position-specific evolutionary preservation). Positions conserved over longer periods are expected to have more detrimental effects. These effects are quantitatively scored as Pdel (probability of deleterious effect), and the results are classified as “probably damaging,” “possibly damaging,” and “probably benign.” Missense3D predicts structural changes resulting from deleterious variants that affect protein stability. It identifies structural damage through comprehensive analysis, which includes examining factors such as disruption of buried salt bridges and alterations in secondary structure.
To investigate the genetic linkage between PRNP and PRND SNPs, we conducted LD analysis between the loci of these two genes. Initially, we obtained the dataset of PRNP genotypes from rabbits, which included results from previously reported 203 samples. Subsequently, we performed preprocessing to match the PRNP genotype data with the individuals analyzed for PRND. Of the total genotype datasets examined, 201 matched, and these were used to analyze the LD scores between PRNP and PRND SNPs.
To investigate the leporine PRND polymorphisms, we analyzed DNA sequences targeting the open reading frame (ORF) of leporine PRND in 207 rabbits. The leporine PRND gene comprises two exons, with the ORF (537 bp) located in exon 2 (Figure 1A). PCR and sequencing were performed using a pair of primers designed in this study, and the sequencing results were identical to the leporine PRND gene registered in GenBank (Gene ID: 100347890). We found nine novel SNPs in leporine PRND: c.18A > G, c.76G > C, c.128C > T, c.146C > T, c.315A > G, c.488G > A, c.525G > C, c.544G > A, and c.579A > G (Figure 1B). Among them, four SNPs at c.76G > C (A26P), c.128C > T (T43M), c.146C > T (A49V), and c.488G > A (R163Q) are nonsynonymous SNPs within the ORF region, and two SNPs at c.544G > A and c.579A > G are located in the 3′ untranslated region (Figure 1A). Detailed information about the genotype and allele frequencies of the leporine PRND SNPs is described in Table 1.
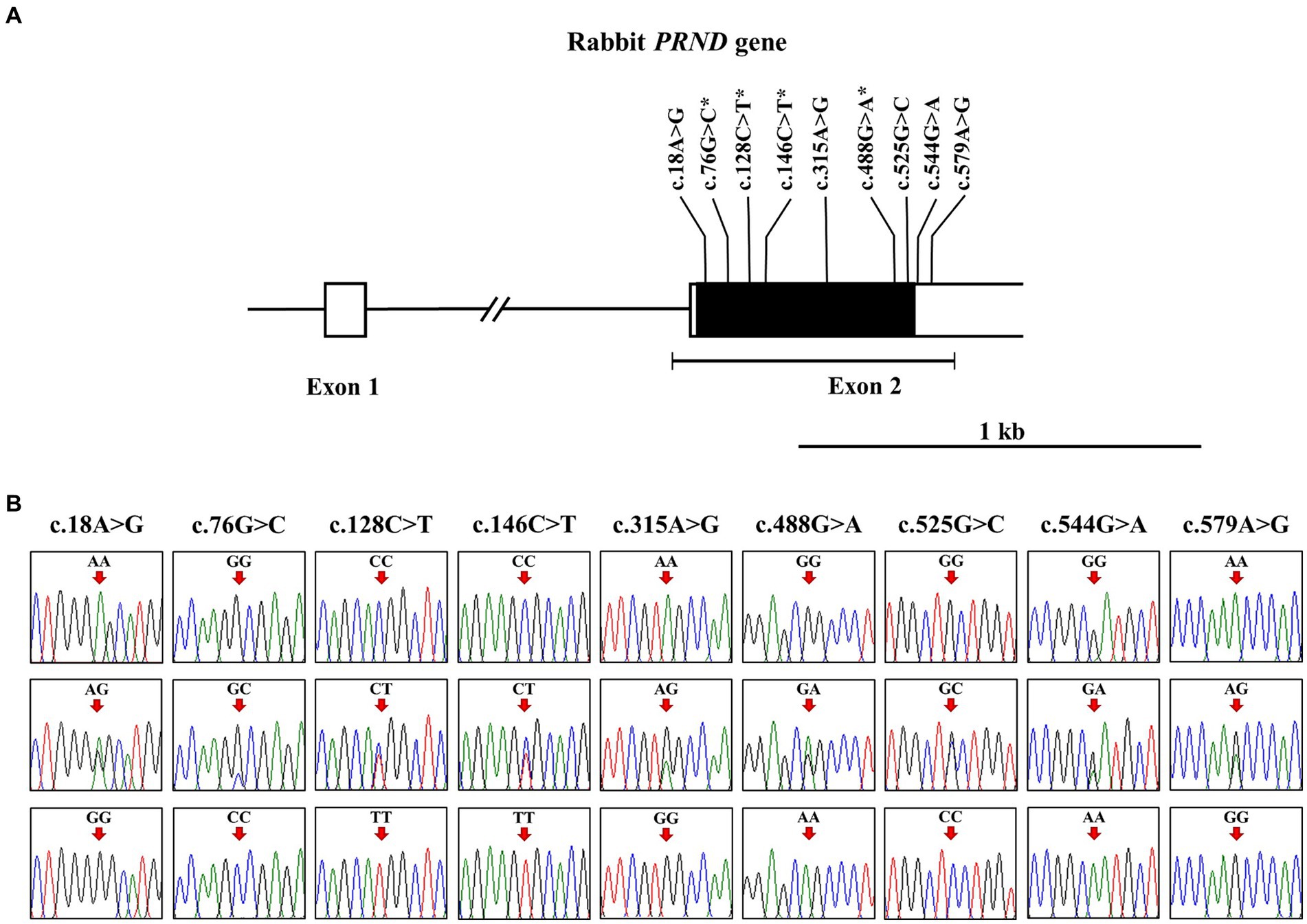
Figure 1. Identification of single nucleotide polymorphisms (SNPs) in the leporine prion-like protein gene (PRND). (A) The schematic diagram illustrates the genomic structure of the leporine PRND. The open reading frame (ORF) within exon 2 is represented by the black box, and the 5′ and 3′ untranslated regions (UTRs) from exon 1 to 2 are shown by the white boxes. The edged horizontal bar indicates the length of PCR products in this study. The positions of the polymorphisms identified in this study are shown in bold, with an asterisk denoting the non-synonymous SNP. (B) Electropherograms displaying nine novel SNPs discovered in the leporine PRND gene are presented. The electropherograms show three genotypes at c.18A > G, c.76G > C, c.128C > T, c.146C > T, c.315A > G, c.488G > A, c.525G > C, c.544G > A, and c.579A > G. The colors of the peaks represent each base of the DNA sequence as follows: green for adenine; red for thymine; blue for cytosine; black for guanine. Arrows indicate the position of the polymorphisms identified in this study. Upper panel, homozygote of the major allele; middle panel, heterozygote; lower panel, homozygote of the minor allele.
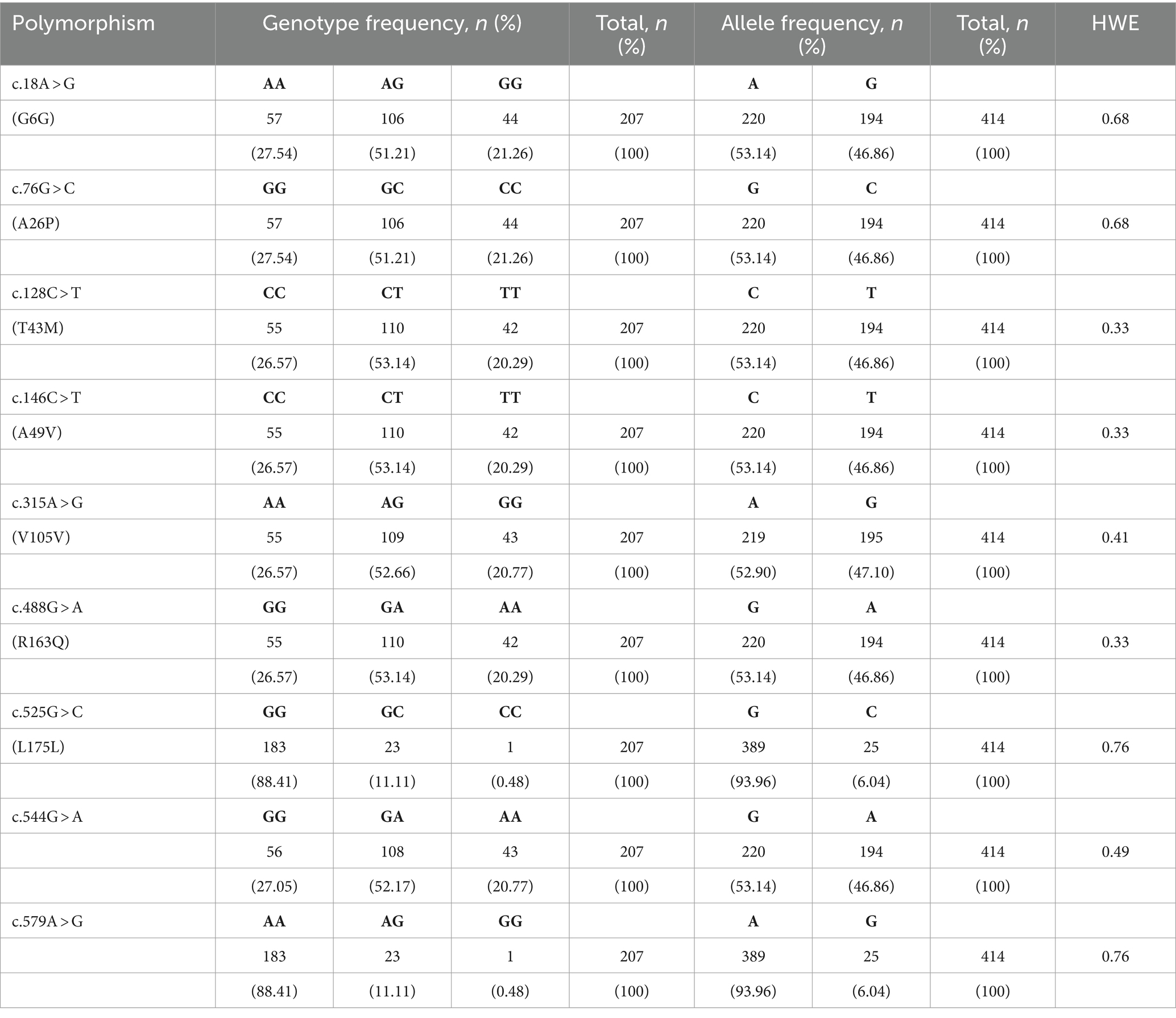
Table 1. Genotype and allele frequencies of the leporine prion-like protein gene (PRND) single nucleotide polymorphisms (SNPs).
We investigated the extent of LD among the leporine PRND SNPs by calculating the r2 values (Table 2). Seven of the PRND SNPs, excluding c.525G > C and c.579A > G, exhibited strong linkage with values greater than 0.3. In addition, we examined the haplotype frequency of the leporine PRND SNPs (Table 3). The most frequently observed haplotype was GCCCAGGGA (46.9%), followed by AGTTGAGAA (46.6%), AGCCAGCGG (6%), AGTTGAGGA (0.2%), and AGCCGGGAA (0.2%).
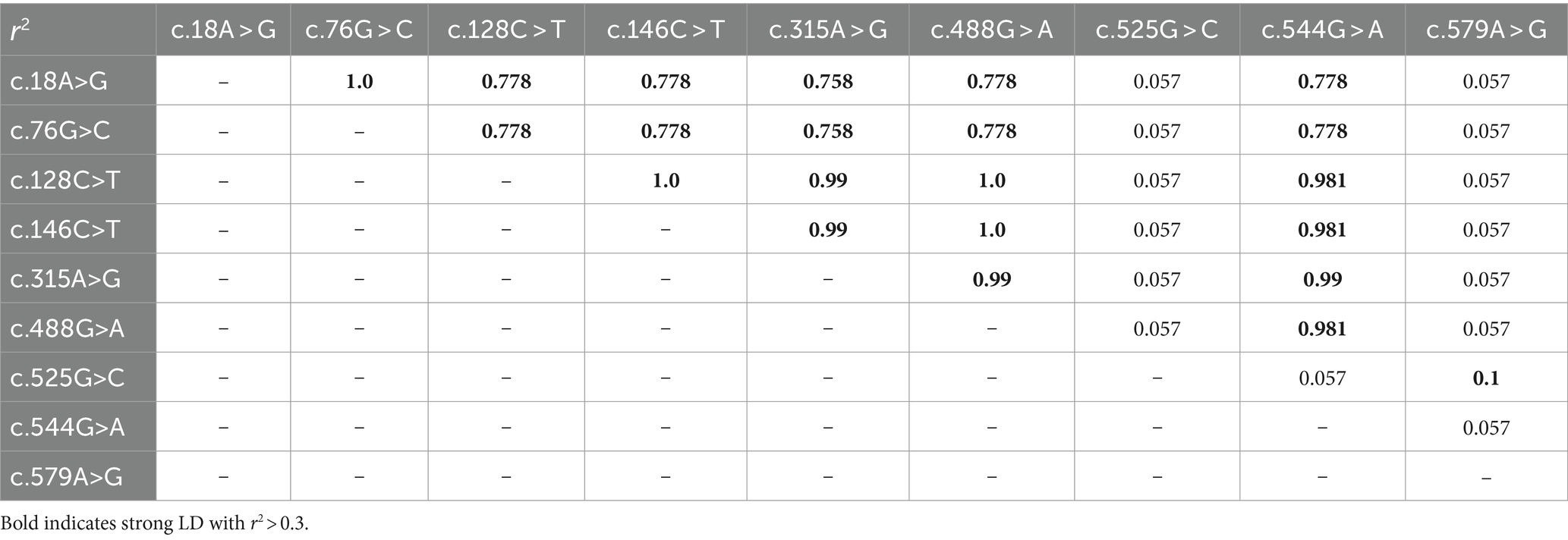
Table 2. Linkage disequilibrium (LD) scores of the leporine prion-like protein gene (PRND) single nucleotide polymorphisms (SNPs).
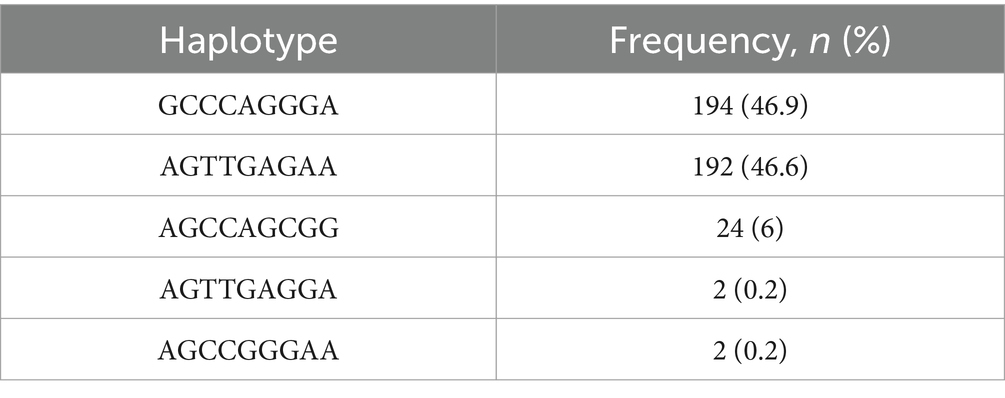
Table 3. Haplotype frequencies of the leporine prion-like protein gene (PRND) single nucleotide polymorphisms (SNPs).
We evaluated the potential effect of non-synonymous SNPs on leporine PRND using in silico prediction tools (Table 4). Both PolyPhen-2 and SIFT predicted that all four non-synonymous SNPs would have deleterious effects on protein function and structure. In Polyphen-2, the three SNPs at c.76G > C (A26P), c.128C > T (T43M), and c.488G > A (R163Q) were predicted as “Possibly damaging” with scores of 0.895, 0.924, and 0.816, respectively. Interestingly, the SNP at c.146C > T (A49V) was predicted as “Probably damaging” with a score of 0.991. In SIFT, all non-synonymous SNPs were predicted to impact protein function with a score of 0.00. However, both PANTHER and Missense3D classified all non-synonymous SNPs as benign.
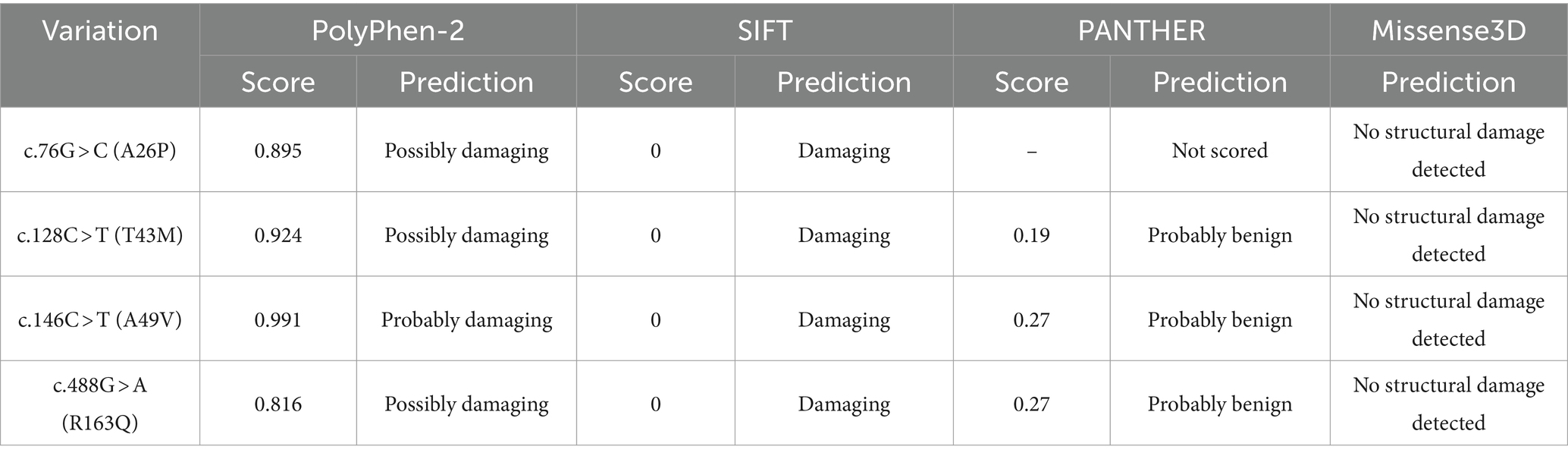
Table 4. In silico prediction of the functional effect of non-synonymous single nucleotide polymorphisms (SNPs) in the leporine prion-like protein gene (PRND).
A previous study reported a synonymous SNP (c.234C > T) identified in the ORF of the leporine PRNP gene (38). To investigate whether leporine PRND SNPs have strong genetic linkage with this leporine PRNP SNP, we performed LD analysis between PRNP and PRND SNPs using r2 values. LD scores were estimated in 201 animals after excluding 6 animals that did not have genotyping data for the PRNP gene. As shown in Table 5, PRND SNPs at c.525G > C and c.579A > G were strongly linked with PRNP SNP c.234C > T (r2 value of 0.912). The remaining seven PRND SNPs were weakly linked with r2 scores of less than 0.3.
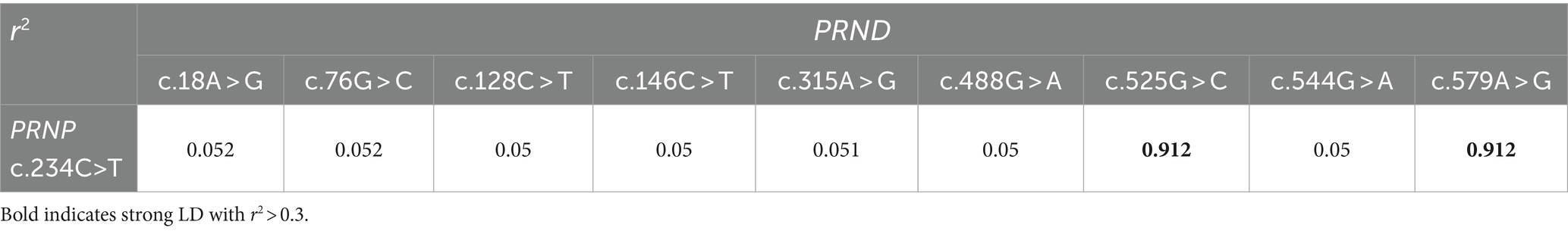
Table 5. Linkage disequilibrium (LD) scores between leporine prion protein gene (PRNP) and prion-like protein gene (PRND) single nucleotide polymorphisms (SNPs).
Since the pathological mechanisms of prion diseases remain elusive, investigating the genetic characteristics of prion disease-susceptible species is crucial for understanding this disease. However, exploring the genetic characteristic of resistant species offers a novel avenue to unravel the mysteries of prion diseases, providing valuable perspective (4). Previous research has suggested that resistance to prion diseases may be attributed to a specific structural stability unique to PrP in species resistant to such disease. For instance, the presence of an aspartic acid (Asp) residue at codon 163 of canine PrP enhances protein stability by forming an additional salt bridge compared to asparagine (Asn), the amino acid found at that position in prion disease-susceptible animal PrP (39, 40). This structural stabilization contributes to the resistance properties. Similarly, the Asp residue at codon 167 in equine PrP plays a crucial role in maintaining the well-defined structure of PrP, particularly within the β2–α2 loop, thereby contributing to disease resistance (41–44). While the amino acid structure of rabbit PrP shares similarities with that of prion disease-susceptible species, a detailed examination revealed that the β-sheet of rabbit PrP is shorter than that of human PrP (6). Notably, the β2–α2 loop of rabbit PrP spanning residues 165–175 exhibits a well-defined structure, contributing to the structural stability of rabbit PrP (11, 45). The V166 residue establishes hydrophobic contacts with the Y218 residue of helix 3 in rabbit PrP, potentially influencing the structural stability of the β2–α2 loop (11, 45). Furthermore, the S174 residue in rabbit PrP forms a robust hydrogen bond with the N171 residue, a key interaction believed to play a role in the observed resistance to prion susceptibility when serine replaces asparagine at codon 174 in mouse PrP (9, 11).
Given the influence of rabbit PrP-specific residues on structural stability, our attention may be directed towards exploring alterations in the rabbit PrP sequence and the potential influencing factors. In a recent study investigating polymorphisms within rabbit PRNP, a single SNP within the ORF region was identified. The study assessed the impact of substitutions in amino acids unique to rabbit compared to those conserved in other prion disease-susceptible species (38). While in silico analysis predicted benign effects on protein function and structure for amino acid substitutions in the unique residues of rabbit PrP (38), 3D structure predictions revealed weakened hydrogen bonds in residues such as S175, Q221, A226, and A230 of rabbit PrP resulting from these substitutions (38). However, the sensitivity to prion diseases cannot be fully explained by analyzing the sequence characteristics of PRNP alone. Recent studies have reported that the genetic profile of polymorphisms in the PRND gene, a member of the prion protein family, serves as an important cofactor associated with susceptibility to various types of prion diseases (12–22, 26–33, 46–53).
Recent case-controlled studies have suggested that susceptibility to prion diseases is associated with PRND polymorphisms at codon 174 and 3′ untranslated region (UTR) +28 in humans (12, 26, 27), codons 95 and 132 in cattle (28), codon 26 in sheep (29), and codon 10 in goats (54). Moreover, significant associations have been observed between polymorphisms of the PRNP and PRND genes in sheep and goats (29, 30). This suggests that PRND SNPs may indirectly contribute to susceptibility to prion diseases across various species. In our study, we investigated polymorphisms within the ORF region of the leporine PRND gene, which is located proximal to the PRNP gene and observed a strong linkage between PRNP (c.234C > T) and PRND (c.525G > C and c.579A > G) polymorphisms in rabbits. These results align with prion disease-susceptible species such as goats and sheep (strong LD between PRNP and PRND SNPs). Interestingly, weak LD values were observed for seven PRND SNPs (c.18A > G, c.76G > C, c.128C > T, c.146C > T, c.315A > G, c.488G > A, and c.544G > A). Particularly, weak linkage between PRNP and PRND SNPs was observed in dogs and horses, which can be interpreted as a characteristic of prion disease-resistant species. Although natural infections have not been reported in rabbits, they are considered partially resistant species with confirmed experimental infection potential. Therefore, further research is needed to explore various characteristics related to genetic diversity in rabbits. Additional research on the association of intergenic polymorphisms in rabbits, considering the observed weak LD in resistant species and strong LD in susceptible species, will require in depth exploration.
In addition, we identified several SNPs, and four non-synonymous SNPs showed a potent effect on leporine Doppel (Figure 1; Table 4). All non-synonymous SNPs were predicted to impact protein function by PolyPhen-2 and SIFT, whereas PANTHER and Missense3D did not indicate any effect. Since in silico prediction tools employ algorithms based on their specific criteria, further research is needed to validate these predictions using cellular or animal models in the future. Interestingly, since the Doppel protein is predominantly expressed in the testis, the impact induced by the PRND SNP may affect male fertility (55, 56). In the previous study, researchers investigated the influence of a single SNP, c.78G.A (A26A), identified in the PRND gene of sheep, on sperm reproductive ability (29, 57). Semen from sheep carrying the A allele exhibited a higher proportion of the F pattern in spermatozoa compared to those carrying the G allele. Additionally, there was an improvement in cleavage rate and enhanced embryo rates at 6 and 8 days. Consequently, the A allele at codon 26 of the PRND gene has been suggested to correlate with male reproductive performance. Although individuals carrying SNPs that may significantly impact reproductive capacity could have been culled from the breeding population, the degree of variation in reproductive ability may depend on the extent to which PRND SNPs affect the functional characteristics of Doppel protein. Therefore, further functional studies in natural rabbit populations, which lack the artificial selection present in the breeding population used in this study, are needed to elucidate the relationship between PRND polymorphism and reproductive capacity.
In this study, we investigated the polymorphism of the leporine PRND gene and analyzed its characteristics. The hybrid breeding rabbits we utilized are primarily consumed breeds in Korea. All individuals included in this study were sourced from rabbits slaughtered at regional abattoirs of the Korea Rabbit & Deer Farmers National Agricultural Cooperative Federation during sample collection. To determine whether rabbits from different breeds exhibit genotype distributions or novel polymorphisms distinct from those observed in this study, future genetic analysis studies on various rabbit breeds, particularly native breeds, will be necessary.
In the present study, we found nine novel SNPs in the leporine PRND gene, including four deleterious non-synonymous SNPs. In addition, we performed LD analysis between PRNP and PRND polymorphisms and found strong LD between PRNP (c.234C > T) and PRND SNPs (c.525G > C and c.579A > G). The remaining seven PRND SNPs (c.18A > G, c.76G > C, c.128C > T, c.146C > T, c.315A > G, c.488G > A, and c.544G > A) showed weak linkage with PRNP SNP. To the best of our knowledge, this study is the first to present a genetic characterization of PRND SNPs in rabbits.
The data presented in the study are deposited in the DRYAD repository (https://datadryad.org/stash/share/3gw51vGYS1FWIFzOBEC5JEfIhPxygvyC9eAgSh3SwKI).
The animal study was approved by the overall experimental processes were approved by the Jeonbuk National University Institutional Animal Care and Use Committee (CBNU 2019-058). All experiments were performed in accordance with the Korea Experimental Animal Protection Act. The study was conducted in accordance with the local legislation and institutional requirements.
M-JJ: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. Y-CK: Conceptualization, Formal analysis, Writing – review & editing. B-HJ: Conceptualization, Formal analysis, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (grant Nos. 2017R1A6A1A03015876 and 2021R1A6A3A01086488), National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (grant Nos. 2021R1A2C1013213 and 2022R1C1C2004792), and Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (grant No. 2021R1A6C101C369). M-JJ was supported by the BK21 Four Program in the Department of Bioactive Material Sciences, Jeonbuk National University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. Collins, SJ, Lawson, VA, and Masters, CL. Transmissible spongiform encephalopathies. Lancet. (2004) 363:51–61. doi: 10.1016/S0140-6736(03)15171-9
3. Mead, S, Lloyd, S, and Collinge, J. Genetic factors in mammalian prion diseases. Annu Rev Genet. (2019) 53:117–47. doi: 10.1146/annurev-genet-120213-092352
4. Qing, LL, Zhao, H, and Liu, LL. Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies. Dongwuxue Yanjiu. (2014) 35:436–45. doi: 10.13918/j.issn.2095-8137.2014.5.436
5. Moore, J, Hawkins, SA, Austin, AR, Konold, T, Green, RB, Blamire, IW, et al. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to the domestic chicken. BMC Res Notes. (2011) 4:501. doi: 10.1186/1756-0500-4-501
6. Myers, R, Cembran, A, and Fernandez-Funez, P. Insight from animals resistant to prion diseases: deciphering the genotype-Morphotype-phenotype code for the prion protein. Front Cell Neurosci. (2020) 14:254. doi: 10.3389/fncel.2020.00254
7. Barlow, RM, and Rennie, JC. The fate of Me7 scrapie infection in rats, guinea-pigs and rabbits. Res Vet Sci. (1976) 21:110–1. doi: 10.1016/S0034-5288(18)33406-4
8. Gibbs, CJ Jr, and Gajdusek, DC. Experimental subacute spongiform virus encephalopathies in Primates and other laboratory animals. Science. (1973) 182:67–8. doi: 10.1126/science.182.4107.67
9. Vorberg, I, Groschup, MH, Pfaff, E, and Priola, SA. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J Virol. (2003) 77:2003–9. doi: 10.1128/jvi.77.3.2003-2009.2003
10. Chianini, F, Fernandez-Borges, N, Vidal, E, Gibbard, L, Pintado, B, de Castro, J, et al. Rabbits are not resistant to prion infection. Proc Natl Acad Sci USA. (2012) 109:5080–5. doi: 10.1073/pnas.1120076109
11. Khan, MQ, Sweeting, B, Mulligan, VK, Arslan, PE, Cashman, NR, Pai, EF, et al. Prion disease susceptibility is affected by Beta-structure folding propensity and local side-chain interactions in Prp. Proc Natl Acad Sci USA. (2010) 107:19808–13. doi: 10.1073/pnas.1005267107
12. Peoc'h, K, Guerin, C, Brandel, JP, Launay, JM, and Laplanche, JL. First report of polymorphisms in the prion-like protein gene (PRND): implications for human prion diseases. Neurosci Lett. (2000) 286:144–8. doi: 10.1016/s0304-3940(00)01100-9
13. Won, SY, Kim, YC, Do, K, and Jeong, BH. The first report of genetic polymorphisms of the equine Sprn gene in outbred horses, Jeju and Halla horses. Animals (Basel). (2021) 11:2574. doi: 10.3390/ani11092574
14. Kim, YC, Kim, HH, Kim, AD, and Jeong, BH. Novel insertion/deletion polymorphisms and genetic features of the shadow of prion protein gene (Sprn) in dogs, a prion-resistant animal. Front Vet Sci. (2022) 9:942289. doi: 10.3389/fvets.2022.942289
15. Kim, YC, Kim, HH, Kim, K, Kim, AD, and Jeong, BH. Novel polymorphisms and genetic characteristics of the shadow of prion protein gene (Sprn) in cats, hosts of feline spongiform encephalopathy. Viruses. (2022) 14:981. doi: 10.3390/v14050981
16. Roh, IS, Kim, YC, Kim, HJ, Won, SY, Jeong, MJ, Hwang, JY, et al. Polymorphisms of the prion-related protein gene are strongly associated with Cervids' susceptibility to chronic wasting disease. Vet Rec. (2022) 190:e940. doi: 10.1002/vetr.940
17. Kim, YC, Kim, HH, and Jeong, BH. The first report of polymorphisms and genetic characteristics of the shadow of prion protein (Sprn) in prion disease-resistant animal, chickens. Front Vet Sci. (2022) 9:904305. doi: 10.3389/fvets.2022.904305
18. Jeong, BH, Kim, NH, Kim, JI, Carp, RI, and Kim, YS. Polymorphisms at codons 56 and 174 of the prion-like protein gene (PRND) are not associated with sporadic Creutzfeldt-Jakob disease. J Hum Genet. (2005) 50:311–4. doi: 10.1007/s10038-005-0254-8
19. Kim, YC, and Jeong, BH. First report of prion-related protein gene (Prnt) polymorphisms in cattle. Vet Rec. (2018) 182:717. doi: 10.1136/vr.104123
20. Roh, IS, Kim, YC, Kim, HJ, Won, SY, Jeong, MJ, Kang, HE, et al. Identification of the prion-related protein gene (Prnt) sequences in various species of the Cervidae Family. Mol Biol Rep. (2020) 47:6155–64. doi: 10.1007/s11033-020-05697-9
21. Kim, YC, and Jeong, BH. The first report of prion-related protein gene (Prnt) polymorphisms in goat. Acta Vet Hung. (2017) 65:291–300. doi: 10.1556/004.2017.028
22. Kim, YC, Kim, SK, and Jeong, BH. Scrapie susceptibility-associated Indel polymorphism of shadow of prion protein gene (Sprn) in Korean native black goats. Sci Rep. (2019) 9:15261. doi: 10.1038/s41598-019-51625-8
23. Jeong, BH, and Kim, YS. Genetic studies in human prion diseases. J Korean Med Sci. (2014) 29:623–32. doi: 10.3346/jkms.2014.29.5.623
24. Benvegnu, S, Franciotta, D, Sussman, J, Bachi, A, Zardini, E, Torreri, P, et al. Prion protein paralog Doppel protein interacts with Alpha-2-macroglobulin: a plausible mechanism for Doppel-mediated neurodegeneration. PLoS One. (2009) 4:e5968. doi: 10.1371/journal.pone.0005968
25. Moore, RC, Lee, IY, Silverman, GL, Harrison, PM, Strome, R, Heinrich, C, et al. Ataxia in prion protein (Prp)-deficient mice is associated with upregulation of the novel Prp-like protein Doppel. J Mol Biol. (1999) 292:797–817. doi: 10.1006/jmbi.1999.3108
26. Croes, EA, Alizadeh, BZ, Bertoli-Avella, AM, Rademaker, T, Vergeer-Drop, J, Dermaut, B, et al. Polymorphisms in the prion protein gene and in the Doppel gene increase susceptibility for Creutzfeldt-Jakob disease. Eur J Hum Genet. (2004) 12:389–94. doi: 10.1038/sj.ejhg.5201161
27. Jeong, BH, Kim, NH, Choi, EK, Lee, C, Song, YH, Kim, JI, et al. Polymorphism at 3' Utr +28 of the prion-like protein gene is associated with sporadic Creutzfeldt-Jakob disease. Eur J Hum Genet. (2005) 13:1094–7. doi: 10.1038/sj.ejhg.5201460
28. Kim, YC, and Jeong, BH. Bovine spongiform encephalopathy (Bse) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. J Dairy Res. (2018) 85:7–11. doi: 10.1017/S0022029917000814
29. Mesquita, P, Batista, M, Marques, MR, Santos, IC, Pimenta, J, Silva Pereira, M, et al. Prion-like Doppel gene polymorphisms and scrapie susceptibility in Portuguese sheep breeds. Anim Genet. (2010) 41:311–4. doi: 10.1111/j.1365-2052.2009.01992.x
30. Jeong, MJ, Kim, YC, and Jeong, BH. Prion-like protein gene (PRND) polymorphisms associated with scrapie susceptibility in Korean native black goats. PLoS One. (2018) 13:e0206209. doi: 10.1371/journal.pone.0206209
31. Won, SY, Kim, YC, Kim, K, Kim, AD, and Jeong, BH. The first report of polymorphisms and genetic features of the prion-like protein gene (PRND) in a prion disease-resistant animal, dog. Int J Mol Sci. (2019) 20:1404. doi: 10.3390/ijms20061404
32. Jeong, MJ, and Jeong, BH. No polymorphisms in the coding region of the prion-like protein gene in thoroughbred racehorses. Acta Vet Hung. (2019) 67:174–82. doi: 10.1556/004.2019.019
33. Won, SY, Kim, YC, Do, K, and Jeong, BH. Absence of strong genetic linkage disequilibrium between single nucleotide polymorphisms (Snps) in the prion protein gene (Prnp) and the prion-like protein gene (PRND) in the horse, a prion-resistant species. Genes (Basel). (2020) 11:518. doi: 10.3390/genes11050518
34. Premzl, M, and Gamulin, V. Comparative genomic analysis of prion genes. BMC Genomics. (2007) 8:1. doi: 10.1186/1471-2164-8-1
35. Hosking, L, Lumsden, S, Lewis, K, Yeo, A, McCarthy, L, Bansal, A, et al. Detection of genotyping errors by hardy-Weinberg equilibrium testing. Eur J Hum Genet. (2004) 12:395–9. doi: 10.1038/sj.ejhg.5201164
36. Gomes, I, Collins, A, Lonjou, C, Thomas, NS, Wilkinson, J, Watson, M, et al. Hardy-Weinberg quality control. Ann Hum Genet. (1999) 63:535–8. doi: 10.1046/j.1469-1809.1999.6360535.x
37. Gabriel, SB, Schaffner, SF, Nguyen, H, Moore, JM, Roy, J, Blumenstiel, B, et al. The structure of haplotype blocks in the human genome. Science. (2002) 296:2225–9. doi: 10.1126/science.1069424
38. Kim, DJ, Kim, YC, and Jeong, BH. First report of a novel polymorphism and genetic characteristics of the leporine prion protein (Prnp) gene. Front Vet Sci. (2023) 10:1229369. doi: 10.3389/fvets.2023.1229369
39. Fernandez-Borges, N, Parra, B, Vidal, E, Erana, H, Sanchez-Martin, MA, de Castro, J, et al. Unraveling the key to the resistance of canids to prion diseases. PLoS Pathog. (2017) 13:e1006716. doi: 10.1371/journal.ppat.1006716
40. Otero, A, Bolea, R, Hedman, C, Fernandez-Borges, N, Marin, B, Lopez-Perez, O, et al. An amino acid substitution found in animals with low susceptibility to prion diseases confers a protective dominant-negative effect in prion-infected transgenic mice. Mol Neurobiol. (2018) 55:6182–92. doi: 10.1007/s12035-017-0832-8
41. Sanchez-Garcia, J, and Fernandez-Funez, P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol Dis. (2018) 119:1–12. doi: 10.1016/j.nbd.2018.07.011
42. Perez, DR, Damberger, FF, and Wuthrich, K. Horse prion protein Nmr structure and comparisons with related variants of the mouse prion protein. J Mol Biol. (2010) 400:121–8. doi: 10.1016/j.jmb.2010.04.066
43. Kurt, TD, Bett, C, Fernandez-Borges, N, Joshi-Barr, S, Hornemann, S, Rulicke, T, et al. Prion transmission prevented by modifying the Beta2-Alpha2 loop structure of host Prpc. J Neurosci. (2014) 34:1022–7. doi: 10.1523/JNEUROSCI.4636-13.2014
44. Zhang, J. The structural stability of wild-type horse prion protein. J Biomol Struct Dyn. (2011) 29:369–77. doi: 10.1080/07391102.2011.10507391
45. Wen, Y, Li, J, Yao, W, Xiong, M, Hong, J, Peng, Y, et al. Unique structural characteristics of the rabbit prion protein. J Biol Chem. (2010) 285:31682–93. doi: 10.1074/jbc.M110.118844
46. Won, SY, Kim, YC, Kim, SK, and Jeong, BH. The first report of genetic and structural diversities in the SPRN gene in the horse, an animal resistant to prion disease. Genes (Basel). (2019) 11:39. doi: 10.3390/genes11010039
47. Lee, YR, Kim, YC, Won, SY, Jeong, MJ, Park, KJ, Park, HC, et al. Identification of a novel risk factor for chronic wasting disease (Cwd) in elk: S100g single nucleotide polymorphism (Snp) of the prion protein gene (Prnp). Vet Res. (2023) 54:48. doi: 10.1186/s13567-023-01177-7
48. Jo, WS, Kim, YC, Oem, JK, and Jeong, BH. First report of structural characteristics and polymorphisms of the prion protein gene in raccoon dogs: the possibility of prion disease-resistance. Front Vet Sci. (2022) 9:989352. doi: 10.3389/fvets.2022.989352
49. Kim, KH, Kim, YC, and Jeong, BH. Novel polymorphisms and genetic characteristics of the prion protein gene in pheasants. Front Vet Sci. (2022) 9:935476. doi: 10.3389/fvets.2022.935476
50. Kim, YC, Kim, SK, Won, SY, and Jeong, BH. Polymorphisms of shadow of prion protein gene (Sprn) in Korean native cattle (Hanwoo) and Holstein cattle. Sci Rep. (2020) 10:15272. doi: 10.1038/s41598-020-72225-x
51. Kim, Y, Kim, YC, and Jeong, BH. Novel single nucleotide polymorphisms (Snps) and genetic features of the prion protein gene (Prnp) in quail (Coturnix Japonica). Front Vet Sci. (2022) 9:870735. doi: 10.3389/fvets.2022.870735
52. Kim, HH, Kim, YC, Kim, K, Kim, AD, and Jeong, BH. Novel polymorphisms and genetic features of the prion protein gene (Prnp) in cats, hosts of feline spongiform encephalopathy. Genes (Basel). (2020) 12:13. doi: 10.3390/genes12010013
53. Jeong, MJ, Wang, Z, Zou, WQ, Kim, YC, and Jeong, BH. The first report of polymorphisms of the prion protein gene (Prnp) in Pekin ducks (Anas Platyrhynchos Domestica). Front Vet Sci. (2023) 10:1273050. doi: 10.3389/fvets.2023.1273050
54. Uboldi, C, Del Vecchio, I, Foti, MG, Azzalin, A, Paulis, M, Raimondi, E, et al. Prion-like Doppel gene (PRND) in the goat: genomic structure, Cdna, and polymorphisms. Mamm Genome. (2005) 16:963–71. doi: 10.1007/s00335-005-0084-1
55. Behrens, A, Genoud, N, Naumann, H, Rulicke, T, Janett, F, Heppner, FL, et al. Absence of the prion protein homologue Doppel causes male sterility. EMBO J. (2002) 21:3652–8. doi: 10.1093/emboj/cdf386
56. Paisley, D, Banks, S, Selfridge, J, McLennan, NF, Ritchie, AM, McEwan, C, et al. Male infertility and DNA damage in Doppel knockout and prion protein/Doppel double-knockout mice. Am J Pathol. (2004) 164:2279–88. doi: 10.1016/S0002-9440(10)63784-4
Keywords: prion, rabbit, Doppel, prion-like protein gene, PRND , polymorphism, SNP
Citation: Jeong M-J, Kim Y-C and Jeong B-H (2024) The first report of single nucleotide polymorphisms in the open reading frame of the prion-like protein gene in rabbits. Front. Vet. Sci. 11:1388339. doi: 10.3389/fvets.2024.1388339
Received: 15 March 2024; Accepted: 29 April 2024;
Published: 17 June 2024.
Edited by:
Jesus R. Requena, University of Santiago de Compostela, SpainReviewed by:
Christopher J. Silva, United States Department of Agriculture (USDA), United StatesCopyright © 2024 Jeong, Kim and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Hoon Jeong, YmhqZW9uZ0BqYm51LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.