- 1Department of Veterinary Microbiology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 2Department of Pre-Clinic and Applied Animal Science, Faculty of Veterinary Science, Mahidol University, Nakhon Pathom, Thailand
- 3Siriraj Long-Read Lab (Si-LoL), Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 4Department of Microbiology, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
- 5School of Veterinary Medicine, Murdoch University, Perth, WA, Australia
- 6Center of Excellence in Diagnosis and Monitoring of Animal Pathogens (DMAP), Bangkok, Thailand
Introduction: Carbapenem and colistin-resistant Enterobacteriaceae, including Klebsiella pneumoniae, have become a growing global concern, posing a significant threat to public health. Currently, there is limited information about the genetic background of carbapenem and colistin-resistant K. pneumoniae isolates infecting humans and dogs in Thailand. This study aimed to characterize carbapenem and colistin-resistant genes in six resistant K. pneumoniae clinical isolates (three from humans and three from dogs) which differed in their pulse field gel electrophoresis profiles.
Methods: Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), antimicrobial susceptibility testing, and whole-genome sequencing were employed to identify and analyze the isolates.
Results and discussion: All six isolates were carbapenemase-producing K. pneumoniae isolates with chromosomally carried blaSHV, fosA, oqxA and oqxB genes, as well as nine to 21 virulence genes. The isolates belonged to five multilocus sequence types (STs): one isolate from a human and one from a dog belonged to ST16, with the other two human isolates being from ST340 and ST1269 and the other two dog isolates were ST147 and ST15. One human isolate and two dog isolates harbored the same blaOXA-232 gene on the ColKP3 plasmid, and one dog isolate carried the blaOXA-48 gene on the IncFII plasmid. Notably, one human isolate exhibited resistance to colistin mediated by the mcr-3.5 gene carried on the IncFII plasmid, which co-existed with resistance determinants to other antibiotics, including aminoglycosides and quinolones. In conclusion, this study provides a comprehensive characterization of both chromosome- and plasmid-mediated carbapenem and colistin resistance in a set of K. pneumoniae clinical isolates from unrelated humans and dogs in Thailand. The similarities and differences found contribute to our understanding of the potential widescale dissemination of these important resistance genes among clinical isolates from humans and animals, which in turn may contribute to outbreaks of emerging resistant clones in hospital settings.
Background
Klebsiella pneumoniae is designated as a critical pathogen of priority by the World Health Organization (WHO) (1), and it poses a significant threat to public health. This opportunistic bacterium commonly infects immunocompromised individuals in hospital settings, leading to various severe infections such as pneumonia, sepsis, urinary tract infections (UTIs), bacteremia, meningitis, and pyogenic liver abscesses (2). The emergence of resistance to antibiotics, particularly colistin and carbapenem, further complicates the treatment of K. pneumoniae infections in both human and animal populations.
The prevalence of carbapenem resistance in K. pneumoniae can vary depending on the geographical region and the specific population being studied. Generally, carbapenem-resistant K. pneumoniae (CRKP) strains have become a significant concern for both human and animal health (3–5). The prevalence of CRKP strains can be high in human healthcare setting in some regions, leading to increased morbidity and mortality rates among infected individuals (3). In animal settings, including in livestock and companion animals, CRKP isolates also have been reported, although the prevalence tends to be lower compared to in human populations (6, 7). Nevertheless, the presence of CRKP in animals raises concerns about potential transmission to humans through direct contact, environmental contamination, or foodborne routes (5). The One Health approach recognizes the interconnections between human, animal, and environmental health and emphasizes the importance of surveillance, prevention, and control measures to mitigate the spread of carbapenem resistance between different reservoirs (6).
Phenotypic methods are typically the first step in detecting carbapenem resistance. These methods involve performing antimicrobial susceptibility testing (AST) using disk diffusion or broth microdilution methods. Carbapenem-resistant strains exhibit reduced susceptibility or resistance to carbapenem antibiotics, such as imipenem, meropenem, or ertapenem (8). Carbapenemase enzymes are the most common mechanism of carbapenem resistance in K. pneumoniae. Several methods can detect carbapenemase production, including the Modified Hodge Test (MHT), Carba NP test, and Carba-R assay (9, 10). These tests detect the presence of carbapenemase enzymes by assessing the ability of the organism to hydrolyze carbapenem antibiotics. CRKP strains often possess various carbapenemase encoding genes, such as NDM-1, OXA variants, and KPC, which contribute to their resistant phenotype (11, 12).
Carbapenem resistance in K. pneumoniae is predominantly associated with the presence of carbapenemase genes carried on large, self-conjugative plasmids, although the blaOXA-232 gene (13) encoding resistance to carbapenems is commonly found on small ColE-like plasmids. Furthermore, the co-existence of multiple carbapenemase genes within a single isolate, such as blaNDM-1 and blaOXA-232 in sequence type (ST) 14, has been reported (14). Isolates of the globally prevalent K. pneumoniae ST16 also demonstrate diverse antimicrobial resistance profiles. For example, an outbreak of nosocomial infections caused by ST16 K. pneumoniae carrying blaCTX-M-15 occurred in Sweden and Denmark (4), while the co-occurrence of blaNDM-1 and blaOXA-232 in ST16 K. pneumoniae was identified in Italy (15). In a study in Thailand, the two STs that were most commonly found in K. pneumoniae from clinical affected humans were ST16 (n = 15) and ST231 (n = 14) (16).
The current study aimed to enhance understanding of the occurrence of antimicrobial resistance genes in K. pneumoniae isolates from clinically affected humans and dogs in hospital settings in Thailand. Specifically, we sought to elucidate the genetic mechanisms underlying carbapenem resistance in K. pneumoniae, assess the co-occurrence of other antimicrobial resistance genes, and compare the genetic characteristics of isolates from humans and dogs to help determine the possibility of transmission. Resistance to colistin was of interest as studies have revealed the presence of the mcr-3.5 gene in the IncFII plasmid type of pig-derived Escherichia coli isolates in Thailand and other countries (17–19). To date, the prevalence of K. pneumoniae in clinically affected companion animals in Thailand has not been elucidated. In this work, we outline the prevalence, characteristics, molecular typing, and whole genome sequence data for antibiotic resistant K. pneumoniae isolates from clinically affected companion dogs and humans in Hospital settings.
Materials and methods
Sample collection and identification
In 2020, 10 CRKP isolates from humans that met our inclusion requirements, which included being clinically resistant to the carbapenem drugs tested (imipenem, meropenem or ertapenem) were selected for study. They had been cultured from the urine of human patients with urinary tract infections (UTIs) at the King Chulalongkorn Memorial Hospital, Chulalongkorn University, Bangkok as part of routine diagnostic investigations. Another three isolates were collected from dogs with UTIs at the Small Animal Teaching Hospital, Faculty of Veterinary Science, Chulalongkorn University. All the isolates were obtained during the same month in 2020. The humans and dogs were all from different households. Isolates picked from MacConkey agar were identified as K. pneumoniae using IMViC biochemical tests and MALDI-TOF MS (Bruker-Daltonics, Bremen, Germany) (20). The isolates then were tested for antimicrobial susceptibility using the Vitek 2® (bioMérieux, France) and were subjected to Xba1-PFGE macrorestriction analysis using a previously described protocol (21). Briefly, the genomic DNA from the 13 CRKP isolates was digested with the restriction enzyme XbaI (Thermo Scientific). The Bio-Rad CHEF-DRIII system was used for gel electrophoresis, set with a 200 V field at an angle of 120° and run for 17–20 h: Salmonella serovar Braenderup H9812 DNA was incorporated as a standard control. Dendrograms were visualized by using the GeneTool program (Syngene, India) and analyzed with the GeneDirectory program (Syngene, India). Isolates showing different PFGE profiles then were subjected to multilocus sequence typing (MLST) by simplex PCR (Supplementary Table S1). Seven housekeeping genes including the beta-subunit of RNA polymerase B (rpoB), glyceraldehyde 3-phosphate dehydrogenase (gapA), malate dehydrogenase (mdh), phosphoglucose isomerase (pgi), phosphoporine E (phoE), translation initiation factor 2 (infB), and periplasmic energy transducer (tonB) were amplified (22). The MLST database at http://pubmlst.org/kpneumoniae was used to determine allele and sequence types (STs).
Three K. pneumoniae isolates from humans and three from dogs that were all resistant to carbapenem and appeared genetically distinct from each other were selected for whole genomic sequencing and additional analysis. The human isolates were designated KPH1, KPH3, and KPH4 and the dog isolates were KPA1, KPA2, and KPA3. There were no known connections between the six patients. The bacteria were preserved at −80°C in Tryptic Soy broth (Difco) containing 25% glycerol and were recovered on Tryptic Soy agar (Difco) containing 5% sheep blood.
Antimicrobial susceptibility testing
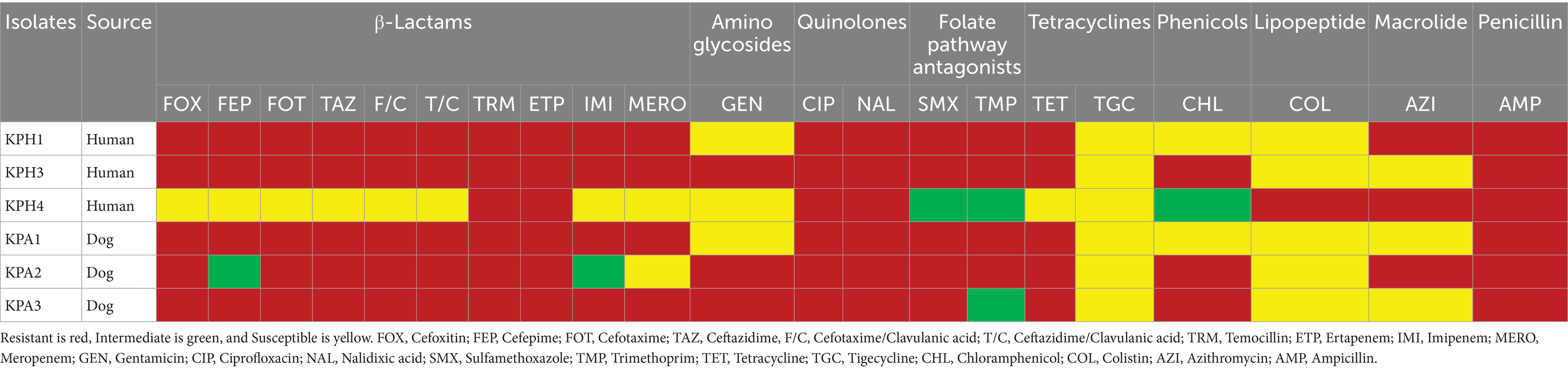
The six representative K. pneumoniae isolates were further tested for their carbapenem susceptibility using the Sensititre Complete Automated AST System (Thermo Scientific, United Kingdom). Following the Clinical and Laboratory Standards Institute (CLSI) standards VET01S (2023) (23), the inclusion criterion was non-susceptibility, which was characterized as resistance or intermediate phenotypes to at least one of the three carbapenems, imipenem, ertapenem, and meropenem. The CRKP isolates also were examined for their susceptibility to colistin, cefoxitin, cefepime, cefotaxime, ceftazidime, cefotaxime/clavulanic acid, ceftazidime/clavulanic acid, temocillin, gentamicin, ciprofloxacin, nalidixic acid, sulfamethoxazole, trimethoprim, tetracycline, tigecycline, chloramphenicol, azithromycin, and ampicillin. Isolates that were resistant to two or more antimicrobials were considered to be multi-drug resistant. The minimum inhibitory concentration (MIC) for colistin was determined by using the broth microdilution technique, with an MIC value of ≥4 μg/mL considered to indicate colistin resistance (23). Antimicrobial susceptibility testing for mcr positive K. pneumoniae isolates was performed by using the AST-GN 38 test kit in a Vitek2 apparatus (bioMérieux, France) (24). E. coli ATCC 25922 was used as the quality control strain for AST.
DNA preparation and whole genome sequencing
The six K. pneumoniae isolates were sub-cultured on Tryptic Soy agar at 37°C for 18 h. Genomic DNA was extracted from the isolates using the ZymoBIOMICS DNA Miniprep Kit following the manufacturer’s instructions, and a Qubit Fluorometer was used to assess the quantity of the extracted DNA. The genomic sequences of the isolates were obtained using the Illumina NovaSeq PE150 platform and MinION (Oxford Nanopore Technologies for long read sequencing).
Whole-genome analysis and bioinformatics
Paired-end reads were quality filtered to remove adapters and low-quality sequences with quality scores <30 using Trimmomatic v.0.36.5 (25). The associated bioinformatic studies were conducted on the European Galaxy server.1 Clean raw reads were assembled and analyzed with Unicycler hybrid assembly (Galaxy Version 0.4.8.0) using the default settings (26). Sequences were analyzed for species identification (KmerFinder 2.1), Multilocus Sequence Type (MLST 1.6), virulence factors (VirulenceFinder 1.2), antimicrobial resistance (ResFinder 2.1), plasmids (PlasmidFinder 1.2) and mobile elements (mobile element finders v1.0.3), using the Center for Genomic Epidemiology (CGE) pipeline (27). Using ABRicate on the Galaxy server, the presence of acquired antimicrobial resistance genes (ARGs) and K. pneumoniae virulence factors also were investigated (Galaxy Version 1.0.1). The CARD Resistance Gene Identifier (28) and ARG-ANNOT (Antibiotic Resistance Gene-ANNOTation) (29) databases were employed in this platform, while the VFDB databases were used for virulence genes (30). The NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) and Prokka (Prokaryotic genome annotation) (Galaxy Version 1.14.6) were used to annotate the genomes (31). Screening for biocide resistance genes using the antibacterial biocide and metal resistance gene databases ResDB and BacMet, was undertaken on the BacAnt server (32, 33).
Plasmid sequences were obtained using plasmid finder and annotated by Prokka (Galaxy Version 1.14.6) (31). PlasmidFinder and ResFinder, tools from the Center for Genomic Epidemiology (CGE),2 were used for predicting plasmids and genes associated with antimicrobial resistance, respectively. All the reference plasmid and other sequences used in the study were recovered from the NCBI database. VFDB, a tool on the Galaxy Europe server, was used for interpreting virulence genes (34). The pan-genome and single nucleotide polymorphisms (SNPs) were determined using Roary v. 3.13.0 (Galaxy Europe server) (35). For phylogenetic tree analysis, blaOXA-232 (in isolates KPH4, KPA1, and KPA2) was selected for multiple alignment of nucleotide sequences using MEGA11 software and the maximum likelihood method (100 bootstrap replicates). In addition, the whole genome sequences of the isolates carrying blaOXA-232 on the COlKP3 plasmid were compared to those of reference strains carrying blaOXA-232 obtained from Thailand (LC613148.1), Nepal (LC507653.1), Bangladesh (CP096175.1), China (CP097391.1), India (CP079128.1), the Netherlands (CP068856.1), Italy (MH523449.1), the USA (CP006802.1), South Korea (CP031737.1), Germany (CP091603.1), and the United Arab Emirates (MF774791.1).
Phylogenetic SNP tree analysis
Thirty-four complete genomes of KPC deposited in Genbank at NCBI were selected for comparative analysis. The core genes were analyzed in Roary version 3.13.0, then the find SNP site version 2.5.1 in the Galaxy website (see text footnote 1) was used to align single nucleotide polymorphism (SNP) sequences (36). A phylogenetic SNP tree was constructed in IQTree version 2.1.2 under maximum likelihood and visualized at the iTOL website3 (37). SNP distance matrix version 0.8.2 was used to compute distance in SNPs.
Plasmid conjugation assay
The six K. pneumoniae isolates (Donor strains) were examined for conjugative plasmid activity as previously described (38). Transfer frequencies were determined by dividing the number of transconjugants by the number of donor colonies (log of transconjugants on selective media/log of the donor). Colonies of the recipient E. coli J53 (Transconjugants) were selected on LB agar (Oxoid, England, United Kingdom) plates containing 200 μg/mL sodium azide and 1 μg/mL imipenem, while E. coli ATCC 25922 was used as a control strain. The transconjugant colonies were identified utilizing PCR. Bacterial DNA was extracted using the Thermo Scientific GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific). A multiplex-PCR for 11 acquired carbapenemase genes (blaIMP, blaVIM, blaSPM, blaGIM, blaSIM, blaKPC, blaNDM, blaAIM, blaDIM, blaBIC, and blaOXA-48) was undertaken on the extracted DNA following a previously published protocol (11).
Data availability statement
The whole-genome sequences of the K. pneumoniae isolates were deposited in the GenBank database with the following accession numbers: KPH1 (CP102546-48), KPH3 (CP102552-53 and CP102555), KPH4 (JAOAOE010000001, JAOAOE010000005, JAOAOE010000006, and JAOAOE010000009), KPA1 (CP101877-78, CP101880-81), KPA2 (CP102987-991), and KPA3 (CP102993-995).
Results
Antimicrobial susceptibility testing
Two human isolates (KPH1 and KPH3) and two dog isolates (KPA1 and KPA3) were resistant to all the antimicrobial drugs that were tested (Table 1; Supplementary Table S1). All six isolates were resistant to carbapenem (ertapenem), and four (KPH1, KPH3, KPA1, and KPA3) were resistant to imipenem and meropenem. Only KPH4 from a human showed co-resistance to colistin and ertapenem.
Genomic characterization of carbapenem resistant K. pneumoniae
The six CRKP isolates submitted for whole genome sequencing had a MIC of ≥2 μg/mL for ertapenem or a MIC of ≥4 μg/mL for imipenem or meropenem; they belonged to five different sequence types in MLST and all had different PFGE profiles. Their genome sizes ranged from 5 ~ 5.3 Mb. Information about the CRKP, including their STs, carbapenem-resistant Enterobacteriaceae (CRE) genes, and plasmid replicon type is presented in Table 2. All isolates were confirmed as CRKP by possessing NDM-1 (New Delhi metallo- β-lactamase) or OXA (oxacillinase) genes. KPH3 and KPH4 had both types of CRE genes, but with different OXA types. InFIA and IncFIB were common replicon types. A small (6,140 bp) ColKP3 plasmid was found in human isolate KPH4 and dog isolates KPA2.
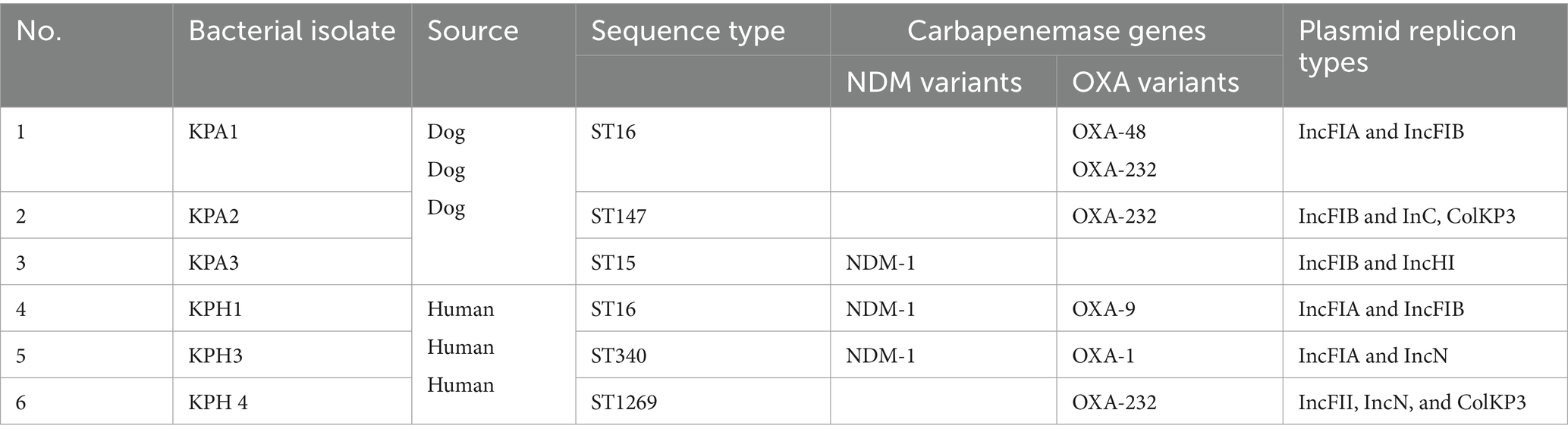
Table 2. Profiles of the six dog and human carbapenem resistant Klebsiella pneumoniae isolates detected by PCR.
All six isolates had a similar CG content percentage (57–57.2%), number of CDSs (5,247–5,458), number of rRNAs (24–25), and number of tRNAs (87–91). No CRE or CRISPR genes were present on the chromosome maps. The blaSHV family genes encoding beta-lactam resistance were present in all isolates, together with the fosA5 or fosA6 fosfomycin-resistance encoding genes. The oqxA and oqxB genes, encoding an RND-family multidrug efflux pump OqxAB that mediates quinolone resistance, also were identified on their chromosomes.
Between nine and 21 virulence genes were identified in the six CRKP isolates (Table 3). All six shared nine of these genes: ompA, entA, entB, fepC, yagV/ecpE, yagW/ecpD, yagX/ecpC, yagY/ecpB, and yagZ/ecpA. The virulence type and pattern of the 21-gene cluster contained by human isolate KPH1 and dog isolate KPA1 were identical. These isolates were both members of ST16 and had a similar resistance profile. The fepG gene was only found in dog isolate KPA2 (Table 3).
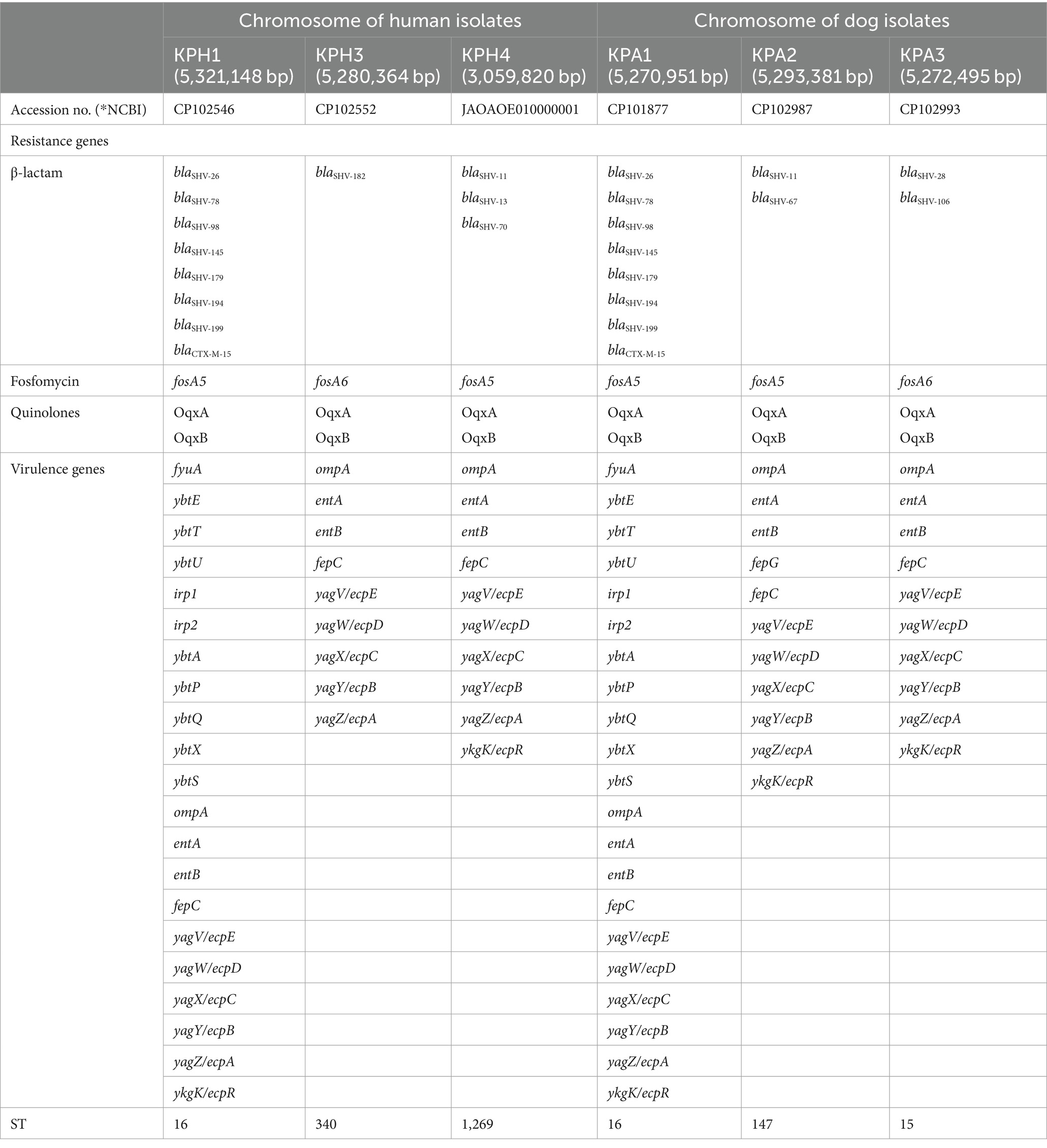
Table 3. Comparison of antimicrobial resistance genes and virulence genes on the chromosome of K. pneumoniae of human and dog origin.
Plasmid-mediated resistance genes
The CRKP isolates harbored between two to four plasmids, with sizes ranging from 188,688 to 6,141 base pairs (Table 4). Plasmids were designated by placing the letter p in front of the isolate name followed by an underscore and a numerical value (eg pKPH1_1). The replicon type of a 82,102 bp plasmid found in KPA2 was unidentified. The presence of numerous replicon types within a plasmid was observed in KPH1 and KPA1, which contained IncFIA, IncFIB, and IncFII; KPH2 contained IncFIA(HI1) and IncR; and KPA3 contained IncFIB(pQil), IncFII(K) and IncHI1B. In the human isolates, plasmids pKPH1_1, pKPH3_1, and ColKP3 plasmid pKPH4_3 were confirmed to contain blaOXA-1, blaOXA-9, and blaOXA-232, respectively. The gene blaNDM-1 encoding NDM was identified on plasmids pKPH1_2, pKPH3_2, and pKPA3_1 in two isolates from humans and one from a dog, respectively. The blaOXA-232 gene was identified on the plasmid pKPA2_4 and on the ColKP3 plasmid pKPA1_3, in the two respective dog isolates; ColKP3 also was found in human KPH4 and had the same size. Notably, blaOXA-48 was identified only in plasmid pKPA1_2. All the isolates contained gentamicin-resistance genes, and five of the six contained quinolone-resistance genes. Moreover, the mcr 3.5 gene was identified only on the IncFII plasmid in human pKPH4_1, while the IncFII plasmid in the dog isolates only contained blaOXA-48.
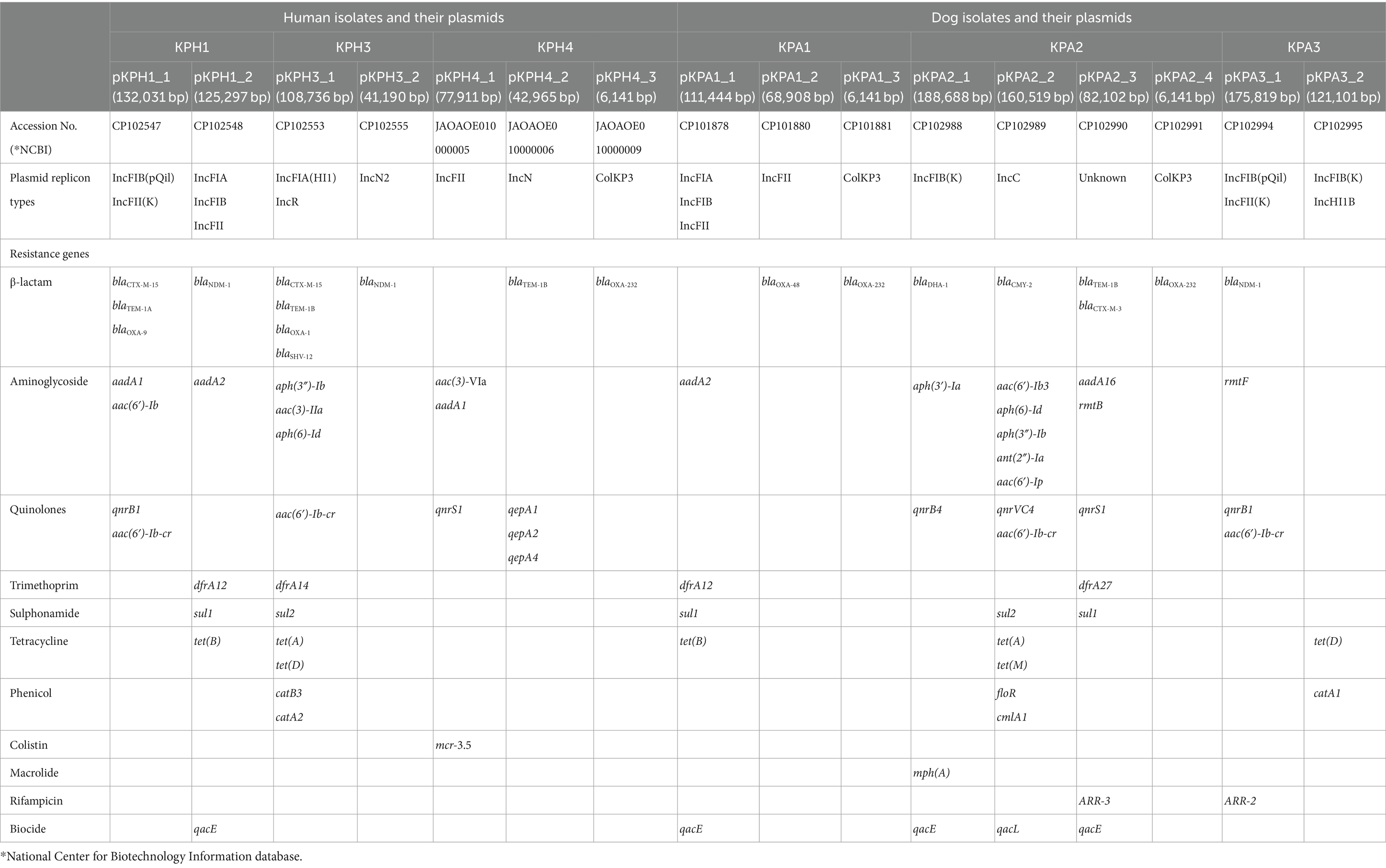
Table 4. Comparison of antimicrobial resistance genes and plasmid replicon types from K. pneumoniae of human and dog origin.
Genomic comparison and phylogenetic SNP tree analysis
For genomic comparisons and phylogenetic tree analysis the genome sequences of the three dog isolates and three human isolates were aligned with those of 34 K. pneumoniae carbapenemase containing (KPC) strains from Thailand and other countries. These showed different plasmids in relation to their ST, source, and country of origin (Figure 1). The Thai ColKP3 plasmids were very similar to each other, and they showed approximately 93 percent similarity to the nucleotide sequences of the plasmids from human strains possessing blaOXA-232 (Figure 2).
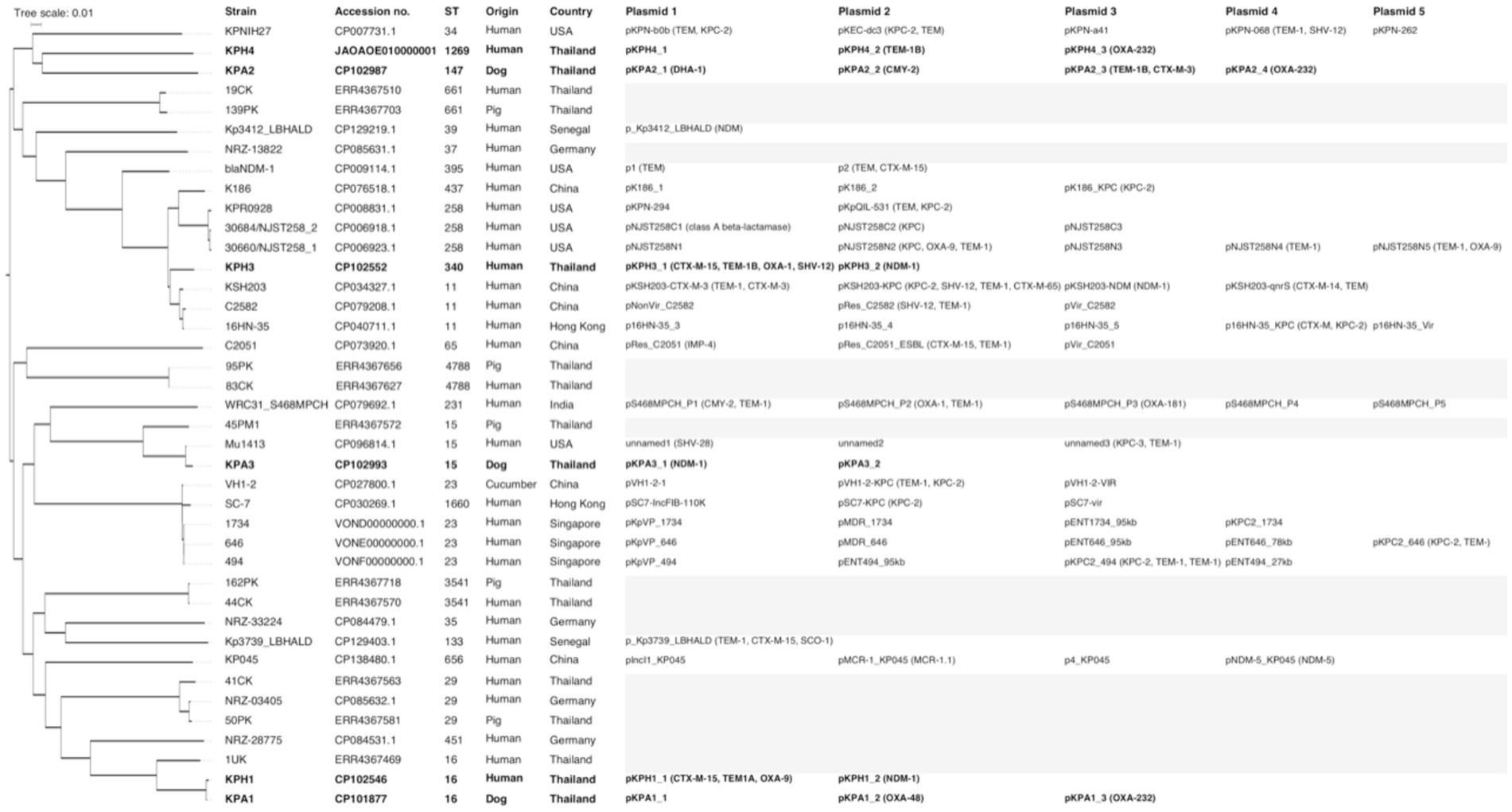
Figure 1. Phylogenetic single nucleotide polymorphisms (SNP) tree of 40 isolates of Klebsiella pneumoniae producing carbapenemase (KPC), including the six isolates from the current study (marked in bold), and 10 previous isolates from Thailand. Where available, the sequence type (ST), accession number, species and country of origin of the isolates, plasmid names, numbers, and carbapenem-resistance genes are presented. The gray highlight represents not identified or unavailable.
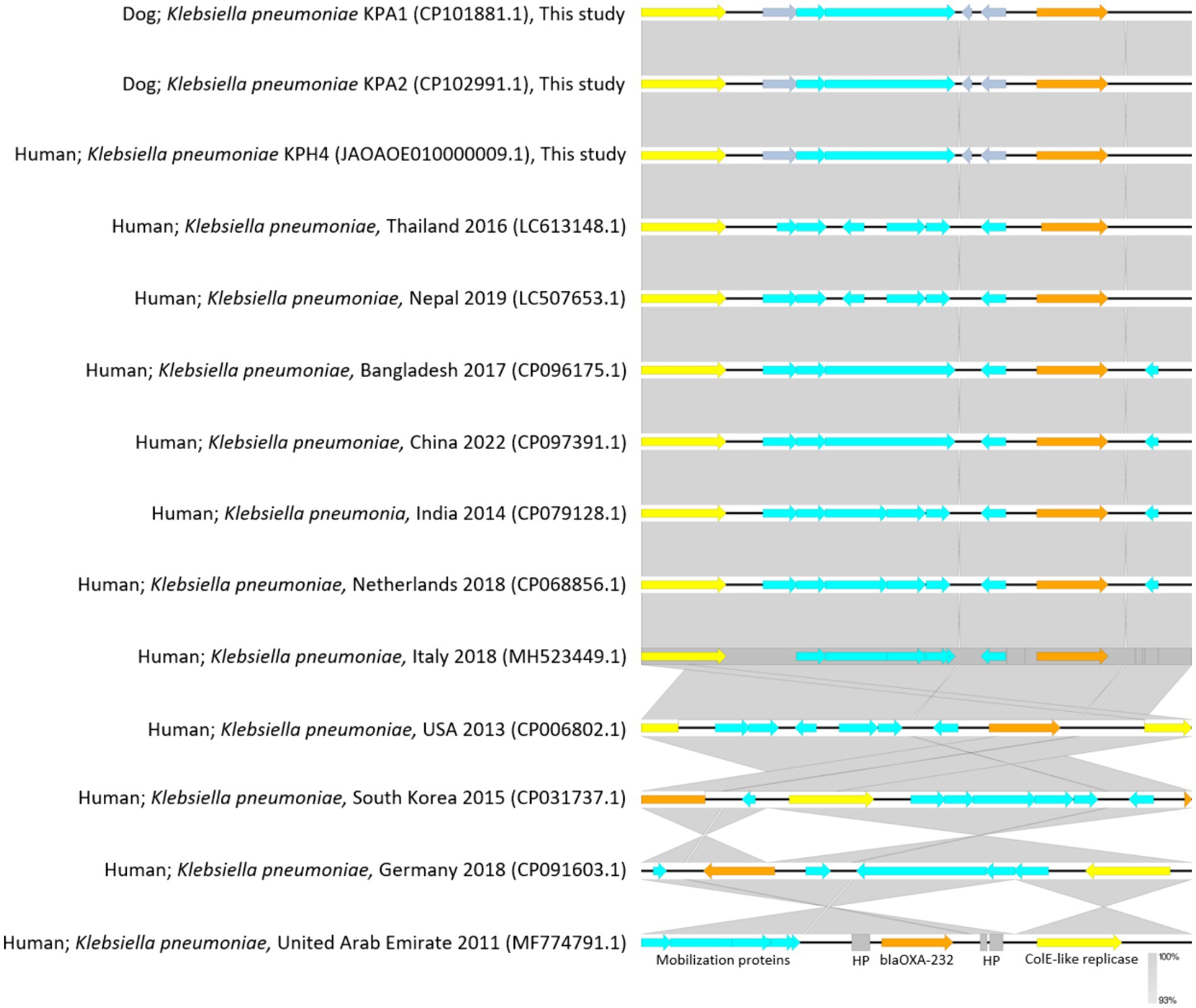
Figure 2. Plasmidome comparative analysis of sequenced regions of ColKP3 containing blaOXA-232 in Klebsiella pneumoniae from accession No. CP101881.1; dog Thailand 2020 (KPA1), No. CP102991.1; dog Thailand 2020 (KPA2), and No. JAOAOE010000009.1; human Thailand 2020 (KPH4) compared with K. pneumoniae from humans in each of the countries for blaOXA-232 (size; 6,141 bp) from No. LC613148.1; human Thailand 2016, No. LC507653.1; human Nepal 2019, No. CP096175.1; human Bangladesh 2017, No. CP097391.1; human China 2022, No. CP079128.1; human India 2014, No. CP068856.1; human the Netherlands 2018, No. MH523449.1; human Italy 2018, No. CP006802.1; human USA 2013, No. CP031737.1; human South Korea 2015, No. CP091603.1; human Germany 2018, and No. MF774791.1; human United Arab Emirates 2011. The gray area indicates the blast identities, and the percentage of identity is indicated in the legend. Open arrows represent coding sequences (blue for mobilization proteins, orange for blaOXA232, yellow for ColK-like replicase). The arrow size is proportional to the gene length. The image was generated using EasyFig with default parameters.
A comparison of the genetic environment of the mcr-3.5 cassette and blaOXA-48 from IncFII plasmids pKPH4_1 and pKPA1_2 in isolates pKPH4 and pKPA1 respectively, and the reference plasmid from a strain from a human urine sample (accession No. KU318420 from China 2013) is presented in Figure 3. The mcr-3.5 and blaOXA-48 genes were not in the core structure of the plasmid and were flanked by insertion sequence 6 (IS6), transposon 3 (Tn3) and other mobile genetic elements (Figure 3). The reference plasmid lacked these resistance genes.
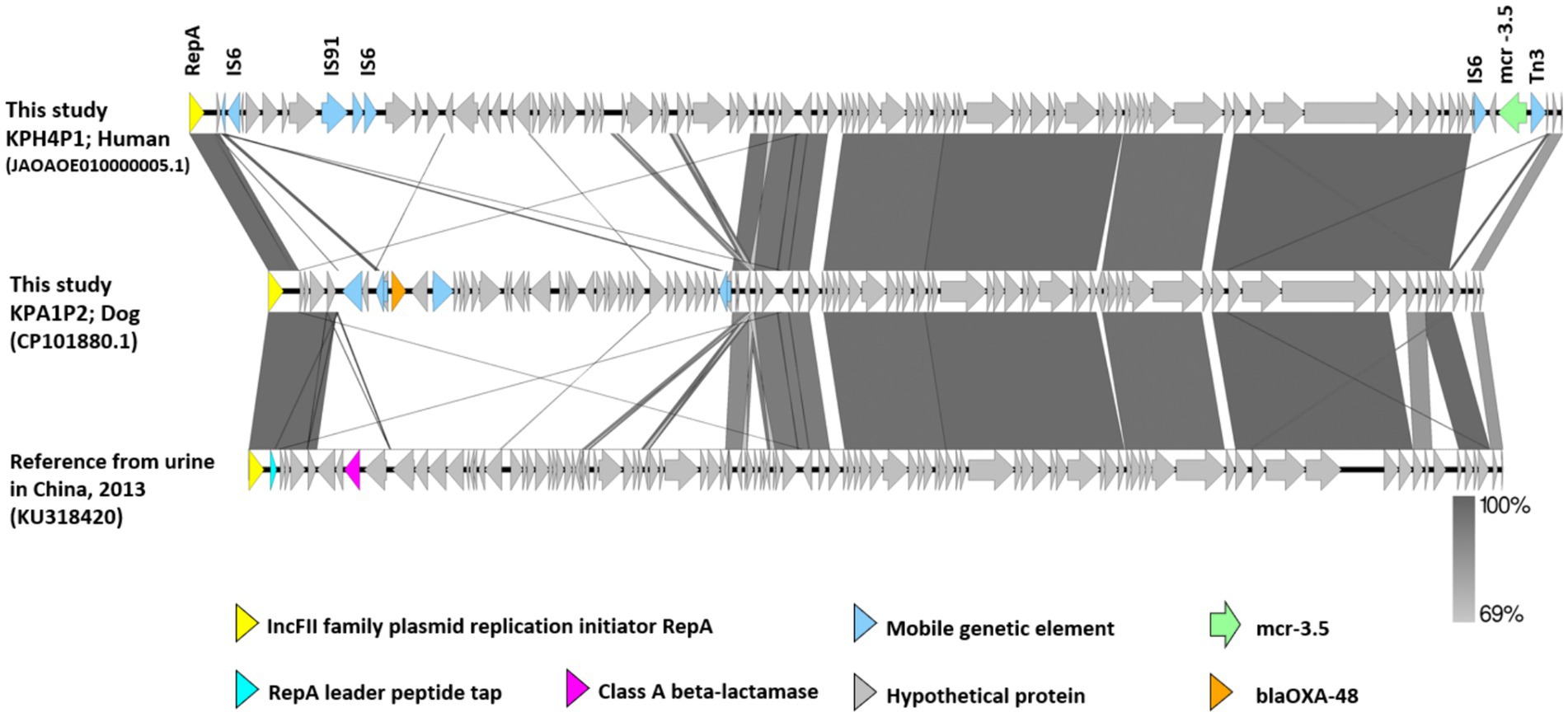
Figure 3. The genetic environment of mcr-3.5 and blaOXA-48 genes from carbapenem resistant K. pneumoniae isolates KPH4 and KPA1 containing an IncFII plasmid from this study compared with a reference plasmid. The gray area indicates the blast identities, and the percentage of identity is indicated in the legend. Open arrows represent coding sequences (green for mcr-3.5, orange for blaOXA48, purple for class A beta-lactamase and blue for mobile genetic elements). The arrow size is proportional to the gene length. The image was generated using EasyFig with default parameters. Accession no. JAOAOE010000005.1; human Thailand 2020 (KPH4P1) and accession no. CP101881.1; dog Thailand 2020 (KPA1P2) compared with a reference strain from a urine sample (IncFII; accession No. KU318420 from China 2013).
Conjugation of plasmids
The six transconjugants had conjugation rates ranging from 8 × 10−3 to 1.7 × 10−7 and were both genetically and phenotypically resistant to sodium azide and meropenem. KPA1 had the highest conjugation rate (Table 5).
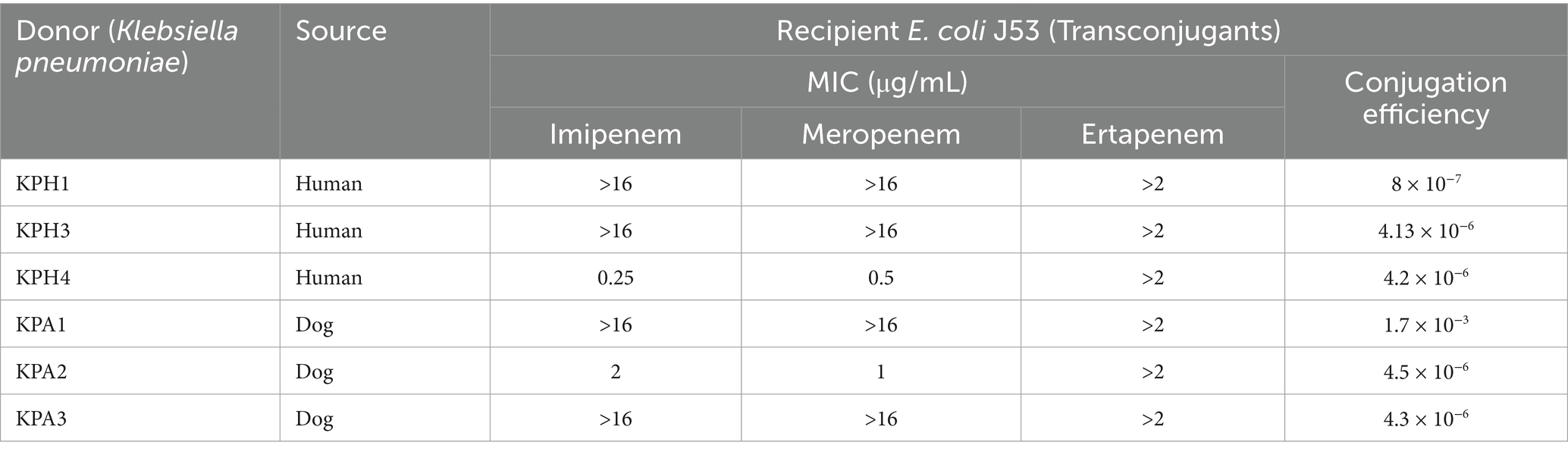
Table 5. Conjugation efficacy of E. coli J53 acquiring genes for carbapenem resistance from Klebsiella pneumoniae.
Discussion
The emergence and spread of carbapenem-resistant Klebsiella pneumoniae strains are of significant concern due to their limited treatment options and potential for causing severe infections (1). In this study, we characterized six CRKP isolates from humans and dogs with UTIs, focusing on their antimicrobial susceptibility, genomic features, and similarity of their plasmidome, focusing on plasmid-mediated resistance genes. The SNP tree indicated that KPH1 and KPA1 shared ST16 and were indistinguishable apart from their plasmid content (Figure 1). This suggests that these closely related isolates may be transmissible between humans and dogs. The other four isolates were dispersed across the SNP tree and were related to isolates from various sources and countries.
Antimicrobial susceptibility testing confirmed that all six isolates were phenotypically and genotypically resistant to ertapenem, with four showing additional resistance to imipenem and meropenem. The differential resistance pattern of K. pneumoniae to ertapenem compared to imipenem and meropenem can be attributed to several factors. Although all three antibiotics belong to the carbapenem class (39), there are subtle differences in their chemical structures and mechanisms of action, which can affect their effectiveness against resistant bacteria. K. pneumoniae can acquire various resistance mechanisms, including the production of carbapenemases that degrade or modify carbapenem antibiotics. Different carbapenemases may have varying affinities for different carbapenem drugs, leading to differences in resistance profiles (40). Importantly, none of the isolates exhibited tigecycline resistance, highlighting its potential as an alternative treatment option for CRKP infections (41).
Genomic characterization of the CRKP isolates revealed the presence of different carbapenemase genes, including NDM-1 and OXA variants. The presence of these genes indicates the potential for high-level carbapenem resistance in K. pneumoniae isolates and highlights the presence of specific resistance mechanisms that contribute to the spread and persistence of carbapenem resistance in isolates from animal and human patients (22).
The isolates shared common replicon types, such as InFIA and IncFIB, indicating the potential for plasmid-mediated horizontal gene transfer among them. The InFIA replicon type is associated with the plasmid family IncF, which is known for its broad host range and ability to transfer between different bacterial species. IncF plasmids often carry multiple resistance genes and are frequently found in clinical isolates of K. pneumoniae (27, 42, 43). In particular, InFIA and IFB plasmids have been associated with the dissemination of carbapenem resistance genes, such as blaNDM-1, in K. pneumoniae strains (22). Unfortunately, their identification in K. pneumoniae is a common finding, especially among multidrug-resistant strains and those carrying carbapenemase genes. Their presence in our isolates suggests the potential for horizontal gene transfer and the dissemination of resistance genes among bacterial populations in Thailand.
Notably, a small ColKP3 plasmid was found in both human and dog isolates, suggesting a potential reservoir for the dissemination of carbapenem resistance genes between humans and animals. The ColKP3 replicon plasmid has been implicated in the dissemination of carbapenemase encoding genes such as blaOXA-232 (44) that can hydrolyze carbapenems, leading to resistance. The presence of the ColKP3 replicon plasmid carrying carbapenemase genes could contribute to the spread of carbapenem resistance among K. pneumoniae strains. The ColKP3 replicon plasmid belongs to the group of small plasmids known as ColE-like plasmids (10) that are often self-transmissible and can replicate autonomously. Their small size facilitates their transfer between bacteria, allowing for the rapid dissemination of resistance genes (45).
Analysis of the chromosomes of the six isolates showed that they had similar characteristics, including comparable CG content, number of CDSs, rRNAs, and tRNAs. The presence of blaSHV genes encoding β-lactam resistance and fosA genes conferring fosfomycin resistance was observed on the chromosomes of all isolates. Additionally, the combination of oqxA and oqxB genes responsible for quinolone resistance was identified. This finding suggests that these resistance determinants are part of the inherent genetic makeup of the bacteria. In this case, their presence on the chromosome may not necessarily indicate the immediate potential for horizontal gene transfer. In clinical isolates the oqxA and oqxB genes generally locate on the chromosome and/or plasmids flanked by IS26-like elements, conferring low to intermediate resistance to quinoxalines, quinolones, tigecycline, and nitrofurantoin. These have the potential to co-spread with other antimicrobial resistance genes (blaCTX-M, rmtB and aac(6′)-Ib etc.) (46). The blaSHV genes encode beta-lactamase enzymes that can hydrolyze and inactivate beta-lactam antibiotics, including penicillins and some cephalosporins. The presence of blaSHV genes on the chromosome of K. pneumoniae strains may indicate the intrinsic resistance of these bacteria to beta-lactam antibiotics (47). However, the presence of plasmid-borne non-ESBL-encoding blaSHV is a risk factor for the development of an ESBL-positive phenotype, despite the same gene being present on the chromosome (4). This can impact the choice of antibiotics for treatment, as these strains may not respond well to certain drugs.
The ST16 clonal type of K. pneumoniae has proliferated globally and is commonly associated with multidrug-resistant strains carrying enzymes that produce NDM-1, CTX-M-15, and OXA-232 that cause resistance to crucial medicines (48). The spread of these antibiotic-resistant bacteria, particularly between animals and humans, poses a serious threat to public health since it reduces treatment options and can result in treatment failures (1). The two ST16 isolates from a human and a dog in this study had highly similar profiles of resistance and virulence genes on their chromosomes, including genes encoding ESBL-generating enzymes. On the other hand, blaNDM-1 and blaOXA-9 genes were identified on plasmids in the human isolates, whereas blaOXA-48 genes were found in the dog isolates. The identification of these specific resistance genes on plasmids in human and dog isolates suggests the involvement of mobile genetic elements in gene transmission. This finding emphasizes the potential for genetic exchange and the spread of resistance genes via mobile genetic components, but not chromosomal reliance (49).
The plasmid analysis revealed the presence of two to four plasmids in each CRKP isolate, with varying sizes and replicon types. Interestingly, several plasmids showed the coexistence of multiple replicon types, indicating the potential for diverse plasmid compatibility and dissemination mechanisms. The plasmids harbored various resistance genes, including blaOXA-1, blaOXA-9, blaOXA-48, blaOXA-232, and blaNDM-1. Prior to this study, examination of the genomic characteristics of multidrug-resistant ST11 K. pneumoniae isolates and their plasmids demonstrated the existence of several plasmids of various sizes and replicon types, indicating variable plasmid compatibility and dissemination mechanisms (50). In a previous study in Thailand, 12 of 15 K. pneumoniae ST16 isolates with chromosomal clonality contained both blaNDM-1 and blaOXA-232, indicating that these plasmids were widely distributed in K. pneumoniae (16). In comparison, all but one of 13 ST231 isolates in that study only harbored blaOXA-232 (16). Another study in clinically affected humans from Thailand also described K. pneumoniae ST16 as a clone that had virulence genes that were similar to those of other highly transmissible K. pneumoniae that carried blaNDM-1 and blaOXA-232 (51). When considering ST15, dog strain KPA3 in the current study showed resistance associated with plasmid pKPA3_1 (NDM-1) and pKPA3_2 (Figure 1). In another study in Thailand, pig strain 45 PM1 showed no plasmid-associated resistance determinants, whereas human strain Mu1413 isolated in the USA carried resistance determinants on plasmids unnamed1 (SHV-28), unnamed2, and unnamed3 (KPC-3, TEM-1) (52). For ST16, in the current study dog strain KPA1 carried resistance determinants on plasmids pKPA1_1, pKPA1_2 (OXA-48), and pKPA1_3 (OXA-232). Moreover, the closely related human strain KPH1 carried resistance determinants on plasmids pKPH1_1 (CTX-M-15, TEM1, OXA-9) and pKPH1_2 (NDM-1). In the earlier study in Thailand, human strain 1UK exhibited no resistance associated with the plasmids identified in other Thai isolates (52). These findings highlight the genetic relatedness and varied distribution of AMR genes among K. pneumoniae isolates, emphasizing the interconnectedness of human, animal, and environmental sources and transmission of resistance mechanisms across diverse hosts and environments.
Notably, the mcr-3.5 gene conferring colistin resistance was identified on an IncFII plasmid in one of the human isolates (KPH4), indicating the coexistence of multiple resistance mechanisms. The mcr-3.5 gene is a variant of the mobile colistin resistance gene (mcr) family (53). It encodes resistance to colistin, an antibiotic belonging to the polymyxin class. The mcr-3.5 gene was first identified in an Escherichia coli strain isolated from pig feces in China (54). It has been reported to be associated with IncFII plasmids, which are known to have a broad host range and can be transferred between different bacterial species (53). The presence of colistin resistance genes on IncFII plasmids supports the notion of multiple resistance mechanisms coexisting in K. pneumoniae strains. The findings will help guide the development of effective surveillance strategies and infection control measures, ultimately contributing to the preservation of public health and the improvement of patient outcomes.
In conclusion, this study offers valuable insights into the antimicrobial susceptibility, genomic characteristics, and plasmid-mediated resistance genes of CRKP isolates from unrelated humans and dogs with UTIs in Thailand. However, the limited number of isolates investigated in this study and the restricted clinical data rendered it difficult to determine whether the outbreak inside the animal and human hospital developed and evolved over time. Although human isolate KPH1 and dog isolate KPA1 both belonged to ST16 and were indistinguishable on the SNP tree, they had different plasmid types and had no known direct epidemiological connections. Nevertheless, the demonstration that human and dog isolates shared antimicrobial resistance and virulence genes on their chromosomes and had genes encoding NDM and OXA enzymes on plasmids that are identical to ColKP3 indicates the potential for interspecies transmission, and emphasizes the need for surveillance and control measures to prevent the spread of CRKP strains. Moreover, when the six isolates in the current study where compared to global isolates of K. pneumonia, these showed a variety of divergent genomes and plasmid types. There is a need for additional research into the precise mechanisms underlying the transmissibility and pathogenicity of CRKP strains and their potential effects on human and animal health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because all the isolates we used were obtained from the frozen stock of the Pathogen Bank in our institute. These were clinical isolates that were submitted for diagnostic purposes as a routine procedure. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
RP: Writing – review & editing, Writing – original draft, Validation, Software, Methodology, Investigation, Formal analysis, Data curation. NK: Writing – original draft, Resources, Methodology, Investigation, Formal analysis, Data curation. NPh: Writing – review & editing, Visualization, Methodology, Investigation, Formal analysis, Data curation. TW: Writing – review & editing, Validation, Software, Resources, Methodology. WN: Writing – review & editing, Resources, Methodology, Data curation. TC: Writing – review & editing, Resources, Methodology, Data curation. DH: Writing – review & editing, Supervision, Conceptualization. NPr: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Thailand Science Research and Innovation Fund Chulalongkorn University (CU_FRB65_hea (87)_192_31_11) and C2F Postdoctoral Scholarship, Chulalongkorn University. The open access fee for this article was paid by the Office of Research Affairs, Chulalongkorn University.
Acknowledgments
We are gratefully to the Pathogen Bank, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand for supporting work undertaken in this area.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1386496/full#supplementary-material
Footnotes
References
1. Tacconelli, E, Carrara, E, Savoldi, A, Harbarth, S, Mendelson, M, Monnet, DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. (2018) 18:318–27. doi: 10.1016/S1473-3099(17)30753-3
2. Paczosa, MK, and Mecsas, J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/MMBR.00078-15
3. Kumar, CK, Sands, K, Walsh, TR, O’Brien, S, Sharland, M, Lewnard, JA, et al. Global, regional, and national estimates of the impact of a maternal Klebsiella pneumoniae vaccine: a Bayesian modeling analysis. PLoS Med. (2023) 20:e1004239. doi: 10.1371/journal.pmed.1004239
4. Lester, CH, Olsen, SS, Jakobsen, L, Arpi, M, Fuursted, K, Hansen, DS, et al. Emergence of extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in Danish hospitals; this is in part explained by spread of two CTX-M-15 clones with multilocus sequence types 15 and 16 in Zealand. Int J Antimicrob Agents. (2011) 38:180–2. doi: 10.1016/j.ijantimicag.2011.03.018
5. Yen, NTP, Nhung, NT, Phu, DH, Dung, NTT, Van, NTB, Kiet, BT, et al. Prevalence of carbapenem resistance and its potential association with antimicrobial use in humans and animals in rural communities in Vietnam. JAC Antimicrob Resist. (2022) 4:dlac038. doi: 10.1093/jacamr/dlac038
6. Tesfa, T, Mitiku, H, Edae, M, and Assefa, N. Prevalence and incidence of carbapenem-resistant K. pneumoniae colonization: systematic review and meta-analysis. Syst Rev. (2022) 11:240. doi: 10.1186/s13643-022-02110-3
7. Kock, R, Daniels-Haardt, I, Becker, K, Mellmann, A, Friedrich, AW, Mevius, D, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. (2018) 24:1241–50. doi: 10.1016/j.cmi.2018.04.004
8. Oduyebo, OO, Falayi, OM, Oshun, P, and Ettu, AO. Phenotypic determination of carbapenemase producing enterobacteriaceae isolates from clinical specimens at a tertiary hospital in Lagos, Nigeria. Niger Postgrad Med J. (2015) 22:223–7. doi: 10.4103/1117-1936.173973
9. Nordmann, P, and Poirel, L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. (2013) 68:487–9. doi: 10.1093/jac/dks426
10. Chen, CY, Nguyen, LT, and Strobaugh, TP Jr. Sequence analysis and plasmid mobilization of a 6.6-kb kanamycin resistance plasmid, pSNC3-Kan, from a Salmonella enterica serotype Newport isolate. PLoS One. (2022) 17:e0268502. doi: 10.1371/journal.pone.0268502
11. Poirel, L, Walsh, TR, Cuvillier, V, and Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. (2011) 70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002
12. Anes, J, Hurley, D, Martins, M, and Fanning, S. Exploring the genome and phenotype of multi-drug resistant Klebsiella pneumoniae of clinical origin. Front Microbiol. (2017) 8:1913. doi: 10.3389/fmicb.2017.01913
13. Navon-Venezia, S, Kondratyeva, K, and Carattoli, A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. (2017) 41:252–75. doi: 10.1093/femsre/fux013
14. Doi, Y, Hazen, TH, Boitano, M, Tsai, YC, Clark, TA, Korlach, J, et al. Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob Agents Chemother. (2014) 58:5947–53. doi: 10.1128/AAC.03180-14
15. Avolio, M, Vignaroli, C, Crapis, M, and Camporese, A. Co-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016. Future Microbiol. (2017) 12:1119–22. doi: 10.2217/fmb-2017-0041
16. Boonyasiri, A, Elita, J, Lauren, MB, Chris, G, Kanokorn, L, Teerawit, T, et al. Genomic and clinical characterisation of multidrug-resistant carbapenemase producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC Infect Dis. (2021) 21:142. doi: 10.1186/s12879-021-05790-9
17. Kalova, A, Gelbicova, T, Overballe-Petersen, S, Litrup, E, and Karpiskova, R. Characterisation of Colistin -resistant Enterobacterales and Acinetobacter strains carrying mcr genes from Asian aquaculture products. Antibiotics. (2021) 10:838. doi: 10.3390/antibiotics10070838
18. Khine, NO, Wongsurawat, T, Jenjaroenpun, P, Hampson, DJ, and Prapasarakul, N. Comparative genomic analysis of Colistin resistant Escherichia coli isolated from pigs, a human and wastewater on colistin withdrawn pig farm. Sci Rep. (2023) 13:5124. doi: 10.1038/s41598-023-32406-w
19. Hamame, A, Davoust, B, Rolain, JM, and Diene, SM. Genomic characterisation of an mcr-1 and mcr-3-producing Escherichia coli strain isolated from pigs in France. J Glob Antimicrob Resist. (2022) 28:174–9. doi: 10.1016/j.jgar.2022.01.014
20. Singhal, N, Kumar, M, Kanaujia, PK, and Virdi, JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. (2015) 6:791. doi: 10.3389/fmicb.2015.00791
21. Li, J, Zhang, H, Ning, J, Sajid, A, Cheng, G, Yuan, Z, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. (2019) 8:44. doi: 10.1186/s13756-019-0489-3
22. Zhang, X, Chen, D, Xu, G, Huang, W, and Wang, X. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One. (2018) 13:e0194000. doi: 10.1371/journal.pone.0194000
23. CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals, VET01S. 6th ed. Wayne, PA: CLSI (2023).
24. Khine, NO, Lugsomya, K, Kaewgun, B, Honhanrob, L, Pairojrit, P, Jermprasert, S, et al. Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated Colistin resistance: mcr-1 and mcr-3 genes in contracted pig farms in Thailand. Front Vet Sci. (2020) 7:582899. doi: 10.3389/fvets.2020.582899
25. Bolger, AM, Lohse, M, and Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
26. Wick, RR, Judd, LM, Gorrie, CL, and Holt, KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. (2017) 13:e1005595. doi: 10.1371/journal.pcbi.1005595
27. Alba, P, Leekitcharoenphon, P, Franco, A, Feltrin, F, Ianzano, A, Caprioli, A, et al. Molecular epidemiology of mcr-encoded Colistin resistance in Enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoring. Front Microbiol. (2018) 9:1217. doi: 10.3389/fmicb.2018.01217
28. McArthur, AG, Waglechner, N, Nizam, F, Yan, A, Azad, MA, Baylay, AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. (2013) 57:3348–57. doi: 10.1128/AAC.00419-13
29. Gupta, SK, Padmanabhan, BR, Diene, SM, Lopez-Rojas, R, Kempf, M, Landraud, L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. (2014) 58:212–20. doi: 10.1128/AAC.01310-13
30. Chen, L, Zheng, D, Liu, B, Yang, J, and Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. (2016) 44:D694–7. doi: 10.1093/nar/gkv1239
31. Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
32. Hua, X, Liang, Q, Deng, M, He, J, Wang, M, Hong, W, et al. BacAnt: a combination annotation server for bacterial DNA sequences to identify antibiotic resistance genes, Integrons, and transposable elements. Front Microbiol. (2021) 12:649969. doi: 10.3389/fmicb.2021.649969
33. Pal, C, Bengtsson-Palme, J, Rensing, C, Kristiansson, E, and Larsson, DG. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. (2014) 42:D737–43. doi: 10.1093/nar/gkt1252
34. Liu, B, Zheng, D, Zhou, S, Chen, L, and Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. (2022) 50:D912–7. doi: 10.1093/nar/gkab1107
35. Page, AJ, Cummins, CA, Hunt, M, Wong, VK, Reuter, S, Holden, MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. (2015) 31:3691–3. doi: 10.1093/bioinformatics/btv421
36. Page, AJ, Taylor, B, Delaney, AJ, Soares, J, Seemann, T, Keane, JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. (2016) 2:e000056. doi: 10.1099/mgen.0.000056
37. Minh, BQ, Schmidt, HA, Chernomor, O, Schrempf, D, Woodhams, MD, von Haeseler, A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. (2020) 37:1530–4. doi: 10.1093/molbev/msaa015
38. Wang, M, Tran, JH, Jacoby, GA, Zhang, Y, Wang, F, and Hooper, DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. (2003) 47:2242–8. doi: 10.1128/AAC.47.7.2242-2248.2003
39. Alikhan, NF, Petty, NK, Ben Zakour, NL, and Beatson, SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. (2011) 12:402. doi: 10.1186/1471-2164-12-402
40. Aihara, M, Nishida, R, Akimoto, M, Gotoh, Y, Kiyosuke, M, Uchiumi, T, et al. Within-host evolution of a Klebsiella pneumoniae clone: selected mutations associated with the alteration of outer membrane protein expression conferred multidrug resistance. J Antimicrob Chemother. (2021) 76:362–9. doi: 10.1093/jac/dkaa439
41. Durante-Mangoni, E, Andini, R, and Zampino, R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect. (2019) 25:943–50. doi: 10.1016/j.cmi.2019.04.013
42. Ragupathi, NKD, Bakthavatchalam, YD, Mathur, P, Pragasam, AK, Walia, K, Ohri, VC, et al. Plasmid profiles among some ESKAPE pathogens in a tertiary care Centre in South India. Indian J Med Res. (2019) 149:222–31. doi: 10.4103/ijmr.IJMR_2098_17
43. Rimoldi, SG, Gentile, B, Pagani, C, Di Gregorio, A, Anselmo, A, Palozzi, AM, et al. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012-2014. BMC Infect Dis. (2017) 17:666. doi: 10.1186/s12879-017-2760-7
44. Shen, S, Han, R, Yin, D, Jiang, B, Ding, L, Guo, Y, et al. A Nationwide genomic study of clinical Klebsiella pneumoniae carrying Bla(OXA-232) and rmtF in China. Microbiol Spectr. (2023) 11:e0386322. doi: 10.1128/spectrum.03863-22
45. Li, X, Ma, W, Qin, Q, Liu, S, Ye, L, Yang, J, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol. (2019) 19:235. doi: 10.1186/s12866-019-1609-1
46. Fu, Y, Zhang, F, Zhang, W, Chen, X, Zhao, Y, Ma, J, et al. Differential expression of Bla(SHV) related to susceptibility to ampicillin in Klebsiella pneumoniae. Int J Antimicrob Agents. (2007) 29:344–7. doi: 10.1016/j.ijantimicag.2006.10.015
47. Hammond, DS, Harris, T, Bell, J, Turnidge, J, and Giffard, PM. Selection of SHV extended-spectrum-beta-lactamase-dependent cefotaxime and ceftazidime resistance in Klebsiella pneumoniae requires a plasmid-borne blaSHV gene. Antimicrob Agents Chemother. (2008) 52:441–5. doi: 10.1128/AAC.00359-07
48. Espinal, P, Nucleo, E, Caltagirone, M, Mattioni Marchetti, V, Fernandes, MR, Biscaro, V, et al. Genomics of Klebsiella pneumoniae ST16 producing NDM-1, CTX-M-15, and OXA-232. Clin Microbiol Infect. (2019) 25:385 e1–5. doi: 10.1016/j.cmi.2018.11.004
49. Gillings, MR, Paulsen, IT, and Tetu, SG. Genomics and the evolution of antibiotic resistance. Ann N Y Acad Sci. (2017) 1388:92–107. doi: 10.1111/nyas.13268
50. Mairi, A, Barraud, O, Muggeo, A, de Champs, C, and Touati, A. Genomic analysis of a multidrug-resistant Klebsiella pneumoniae ST11 strain recovered from barbary deer (Cervus elaphus barbarus) in Akfadou Forest. Algeria J Glob Antimicrob Resist. (2020) 22:515–8. doi: 10.1016/j.jgar.2020.04.027
51. Abe, R, Yukihiro, A, Dan, T, Noriko, S, Yo, S, Norihisa, Y, et al. Clonal dissemination of carbapenem-resistant Klebsiella pneumoniae ST16 co- producing NDM-1 and OXA-232 in Thailand. JAC Antimicrob Resist. (2022) 4:dlac084. doi: 10.1093/jacamr/dlac084
52. Leangapichart, T, Lunha, K, Jiwakanon, AS, Jarhult, JD, Magnusson, U, and Sunde, M. Characterization of Klebsiella pneumoniae complex isolates from pigs and humans in farms in Thailand: population genomic structure, antibiotic resistance and virulence genes. J Antimicrob Chemother. (2021) 76:2012–6. doi: 10.1093/jac/dkab118
53. Liu, L, Feng, Y, Zhang, X, McNally, A, and Zong, Z. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and Bla(NDM-5). Antimicrob Agents Chemother. (2017) 61:1–4. doi: 10.1128/AAC.01757-17
Keywords: Klebsiella pneumoniae, carbapenem resistance, colistin resistance, plasmid-mediated resistance, whole genome analysis
Citation: Pachanon R, Khine NO, Phumthanakorn N, Wongsurawat T, Niyomtham W, Chatsuwan T, Hampson DJ and Prapasarakul N (2024) Genomic characterization of carbapenem and colistin-resistant Klebsiella pneumoniae isolates from humans and dogs. Front. Vet. Sci. 11:1386496. doi: 10.3389/fvets.2024.1386496
Edited by:
Homer Pantua, BioAssets Corporation, PhilippinesReviewed by:
Rachel Amanda Hickman, Independent Researcher, Uppsala, SwedenAdriana Belas, Lusofona University, Portugal
Copyright © 2024 Pachanon, Khine, Phumthanakorn, Wongsurawat, Niyomtham, Chatsuwan, Hampson and Prapasarakul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuvee Prapasarakul, TnV2ZWUuUEBjaHVsYS5hYy50aA==
 Ruttana Pachanon1
Ruttana Pachanon1 Nwai Oo Khine
Nwai Oo Khine Thidathip Wongsurawat
Thidathip Wongsurawat Tanittha Chatsuwan
Tanittha Chatsuwan David J. Hampson
David J. Hampson Nuvee Prapasarakul
Nuvee Prapasarakul