
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 14 March 2024
Sec. Veterinary Infectious Diseases
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1378120
 Jun Zhang1,2,3
Jun Zhang1,2,3 Li Ma1,2,3
Li Ma1,2,3 Tuofan Li1,2,3
Tuofan Li1,2,3 Quan Xie1,2,3
Quan Xie1,2,3 Zhimin Wan1,2,3
Zhimin Wan1,2,3 Aijian Qin1,2,3,4
Aijian Qin1,2,3,4 Jianqiang Ye1,2,3,4
Jianqiang Ye1,2,3,4 Hongxia Shao1,2,3,4
Hongxia Shao1,2,3,4 Shengnan Wang1,2,3*
Shengnan Wang1,2,3*As an immunosuppressive disease virus, chicken infectious anemia virus (CIAV) mainly infects chickens, causing aplastic anemia and systemic lymphoid tissue atrophy. In recent years, the prevalence of CIAV in the poultry industry globally has caused huge economic losses. In this study, a total of 223 clinical samples, including anal swabs, tissues, blood, and vaccines, were collected from 19 broiler farms or breeding companies in Jiangsu province, with symptoms of significant anemia and immunosuppression during 2020–2022. Among them, 75 samples (75/223, 33.6%) were positive for CIAV in polymerase chain reaction (PCR) test, and 20 CIAV strains were successfully isolated. The phylogenetic trees of the 20 isolates and 42 CIAV strains deposited in GenBank formed four distinct groups (A-D). And the isolates mainly belonged to Group A but with high genetic diversity. Analysis for VP1 indicated that these isolates possess key characteristics of highly pathogenic strains. Meanwhile, VP2 and VP3 were much conserved with much fewer mutations compare to VP1. The above epidemiological study of CIAV provides novel insights into molecular characterization of CIAV and lays the foundation for developing efficient strategies for control of CIAV in China.
As an important immunosuppressive virus, chicken infectious anemia virus (CIAV) mainly causes aplastic anemia and systemic lymphoid tissue atrophy in chicks, while increasing susceptibility to other pathogens and decreasing immune response to vaccines (1–4). CIAV is classified into genus Gyrovirus, the family Anelloviridae by the International Committee on Taxonomy of Viruses (ICTV) (5). The single strand DNA of CIAV genome contains three overlapped open reading frames (ORFs) that encoding three viral proteins, namely VP1 (51.2 kDa), VP2 (24 kDa), and VP3 (13.6 kDa) (6). Among them, VP1 is the only capsid protein and the main immunogenic protein with neutralizing epitopes of CIAV (7). VP2 plays a role in viral assembly and has dual-specificity phosphatase activity, similar to Torque teno virus (TTV)-encoded ORF2 (8, 9). VP3 protein, also known as apoptin (10), can selectively mediate cell death (11, 12), and it is essential for viral replication (13, 14).
Since CIAV was first isolated and identified in Japan in 1979 (15), it has spread worldwide. Subsequently, CIAV was first reported and isolated from Heilongjiang province in China in 1992 (16). With the development of modern poultry industry globally, CIAV commercial attenuated vaccines are widely used in breeding flocks, but not in China. In China, CIAV infection and co-infection with other viruses, such as Marek’s disease virus (MDV), reticuloendotheliosis virus (REV), infectious bursal disease virus (IBDV), subgroup J avian leukosis virus (ALV-J), fowl adenovirus (FAdV), avian reovirus (ARV), H9N2 influenza virus (H9N2), and infectious bronchitis virus (IBV), are common in commercial chickens, which can aggravate the diseases (4, 17–21). Recent serological surveys of CIAV in chicken farms revealed high seropositivity rate in Zhejiang, Jiangsu, and Anhui provinces of China (22). Of note, CIAV can reduce the immune effect of vaccines and enhance the virulence of live attenuated vaccines, resulting in immune failure (4). CIAV could also enhance the pathogenicity of other pathogens and further induce high morbidity and mortality (4, 17–21). Thus, continuously following up on the incidence of CIAV infection in chicken flocks is extremely important for the prevention of CIAV.
In this study, we investigated the CIAV epidemiology of clinical samples from 19 broiler farms or breeding companies in Jiangsu province of China during 2020–2022. The PCR for CIAV showed that 75 samples (75/223, 33.6%) were positive and 20 CIAV strains were successfully isolated. Phylogenetic analysis for complete genome and amino acid sequences of VP1 revealed that all these 20 isolates belonged to highly virulent strains and could be clustered into four major groups (A-D). Our study provides novel insights into the epidemiology and efficient strategies for prevention and control of CIAV in China.
A total of 223 clinical samples (including anal swabs, tissues, blood, and vaccines) were received from 19 broiler farms or breeding companies in Jiangsu province (From Yancheng, Nantong, Taizhou, Changzhou, and Lianyungang Cities) during 2020–2022. These chickens showed anemia, thymus atrophy, and poor production performance, and PCR was mainly used to detect CIAV. The anal swabs and tissues samples were homogenized with phosphate-buffered saline (PBS), and supernatants were collected by centrifugation at 12000 rpm/min for 30 min at 4°C. Then, the collected supernatants were treated with 5 × penicillin and streptomycin for 45 min at 37°C and filtered through 0.22 μm filters for virus isolation. Blood and vaccine samples were directly inoculated into cells as shown below.
The genomic DNA was extracted from the clinical samples using a TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) and stored at-20°C. For the detection of CIAV, the PCR was performed with specific primers of VP3 gene listed in Table 1. The PCR reaction volume was 20 μL containing 10 μL of 2 × Taq Plus Master Mix II (Dye Plus) (Vazyme, Nanjing, China), 1 μL of each primer (10 μM), 7 μL of double-distilled water (ddH2O), and 1 μL of the DNA template. The PCR cycling conditions for the VP3 gene amplifications were as follows: 1 cycle of 95°C for 5 min, 35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, followed by a final extension step of 72°C for 10 min.

Table 1. Primers for PCR detection of CIAV VP3, linearization of pcDNA3.1, and complete genome amplification of CIAV.
The MDCC-MSB-1 cells (kept in our laboratory) were cultured at 37°C with 5% CO2 in the cell incubator. The supernatants of the PCR-positive samples were inoculated into MDCC-MSB-1 cells for virus isolation. The supernatants were collected at 3–4 days post-infection (dpi) and detected by PCR with the primers CIAV-VP3-F/R. To amplify the complete genome of the isolates, a pair of primers was designed for PCR amplification as listed in Table 1.
The pcDNA3.1 vector was linearized by PCR with primers listed in Table 1, and then the complete genome of the isolates with homologous arms was amplified with Phanta Super-Fidelity DNA Polymerase (Vazyme, China) and then cloned into pcDNA3.1 using a ClonExpress II One Step Cloning Kit (Vazyme, China). The PCR positive clones were sequenced by Genecfps (Wuxi, China) via sanger sequencing. The phylogenetic trees were constructed using neighbor-joining method using MEGA6.1 software with 1,000 bootstrap replicates based on the nucleotide sequences of the complete genome and amino acid sequences of VP1 of the isolates and the reference strains deposited in GenBank (Table 2). The deduced amino acid sequences of VP1, VP2, VP3, and untranslated regions (UTRs) of the isolates compared with the reference strains were also conducted using the Clustal W method in the MegAlign program by the Lasergene 7.0 software for molecular features.
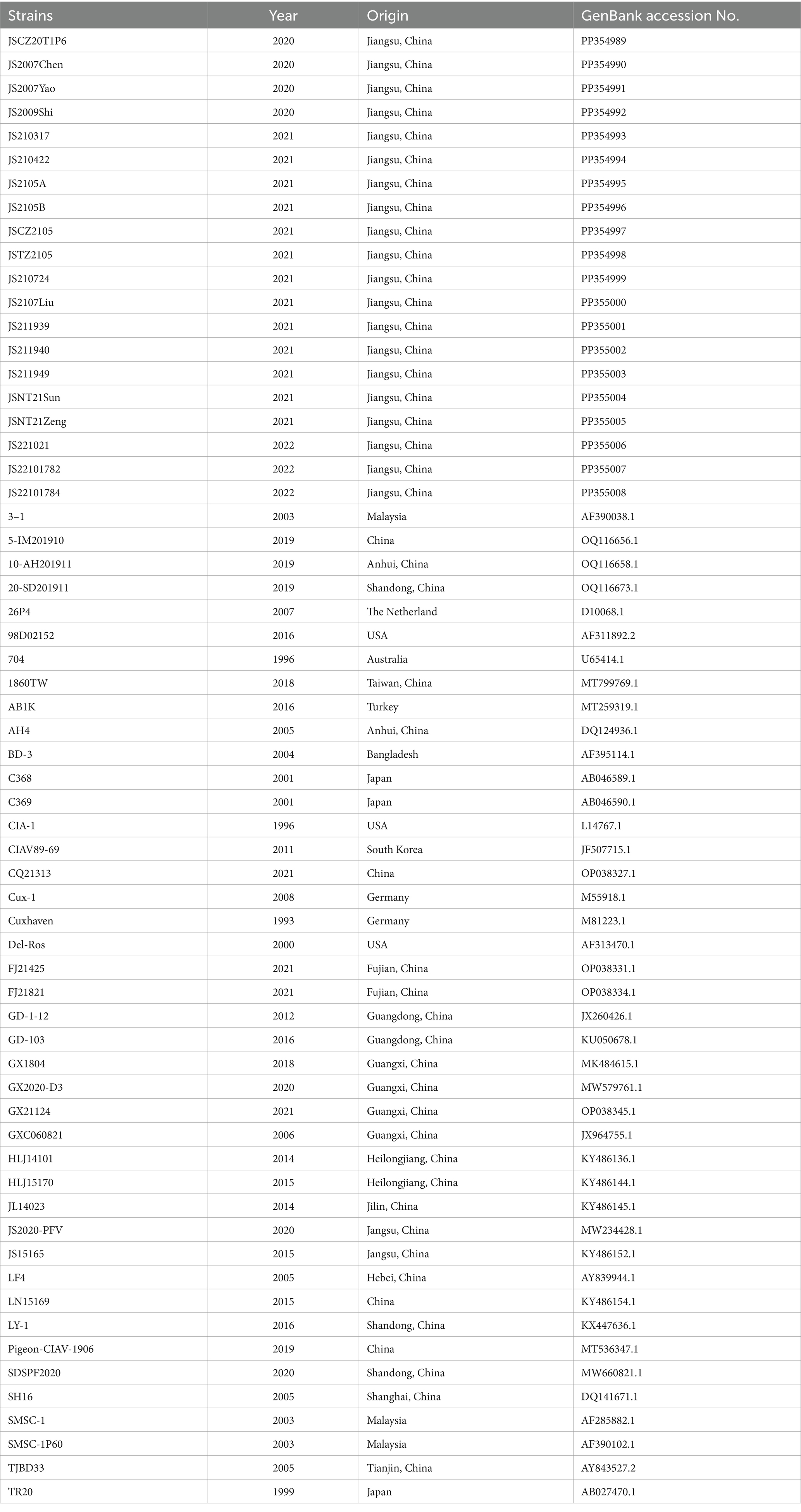
Table 2. The sequence information of 20 CIAV isolates and CIAV reference strains used in this study.
To investigate the epidemiology of CIAV in Jiangsu province of China, 223 clinical samples from 19 broiler flocks or breeding companies in Jiangsu province of China during 2020–2022 were tested. PCR result showed that 75 samples (75/223, 33.6%) were CIAV-positive. As shown in Figures 1, 36 clinical samples were CIAV-positive. These data demonstrated that CIAV was prevalent in Jiangsu province of China during 2020–2022. In addition, we also tested the co-infection status of these 36 samples, and the results showed that there are dual, triple, and quintuple co-infections of CIAV with ALV, FAdV, MDV, REV, and ARV (Supplementary Table S1). To isolate the clinical samples, 36 samples with strong CIAV-positive bands were inoculated into MDCC-MSB-1 cells for virus isolation, and 20 strains were successfully isolated for complete genome sequencing. The amplified complete genome of the 20 isolates with homologous arm of pcDNA3.1 were ligated with linearized pcDNA3.1 vector and further sequenced (Figure 2).
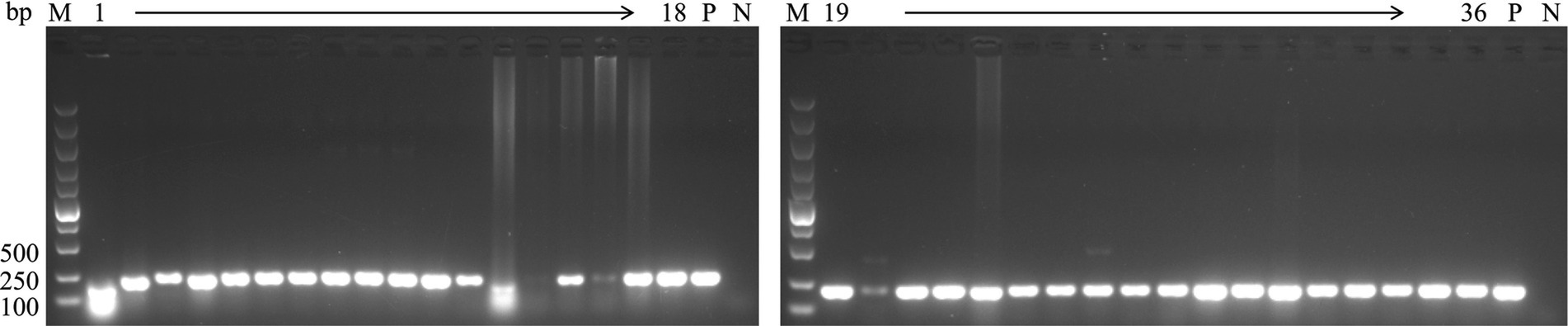
Figure 1. PCR detection for CIAV of clinical samples. The genomic DNA extracted from the clinical samples were detected by PCR using specific primers targeting VP3 gene of CIAV listed in Table 1. Lane M: Super DNA Marker; Lane P: positive control; Lane N: negative control; Lane 1–36: clinical samples with strong CIAV-positive in PCR detection.
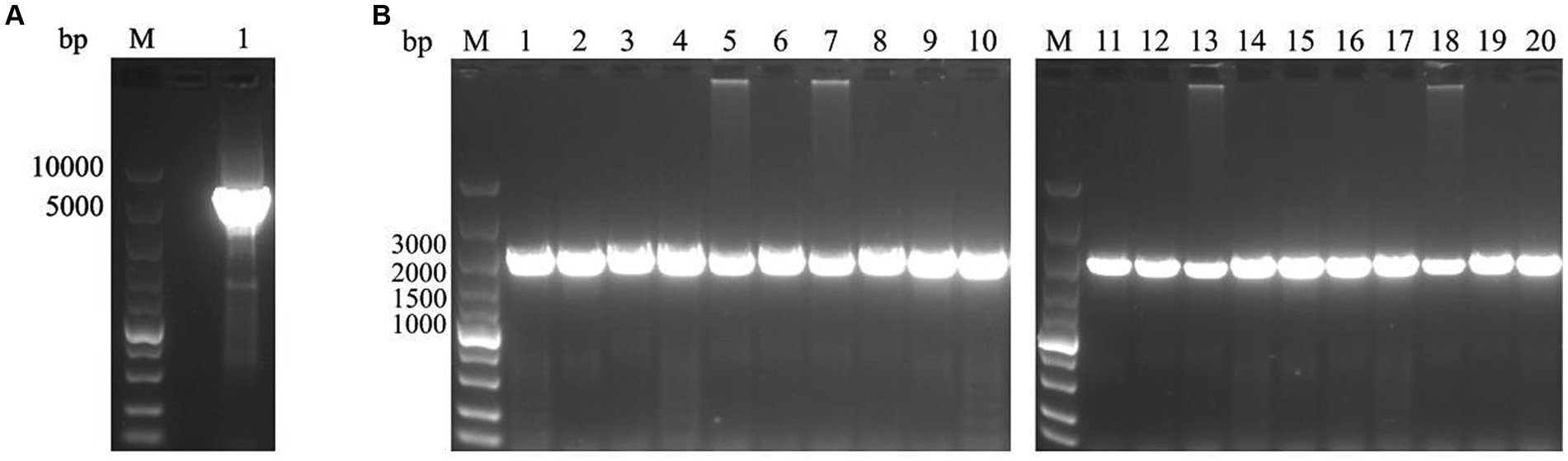
Figure 2. PCR amplification for linearization of pcDNA3.1 vector and the complete genome of 20 CIAV isolates. (A) The pcDNA3.1 vector was linearized by PCR with specific primers pcDNA3.1-F/R. lane M: Super DNA Marker; Lane 1: the linearized pcDNA3.1. (B) The complete genome of 20 isolates with homologous arm of pcDNA3.1 was amplified by PCR with specific primers pc-CIAV-F/R. lane M: Super DNA Marker; Lane 1–20: Total of 20 CIAV isolates from clinical samples.
Among the 20 CIAV isolates, the complete genome of the 17 isolates were 2,298 nucleotides in length, whereas three isolates had deletion or insertion in UTRs. The isolate JSNT21Sun (2,297 bp) had one nucleotide deletion, the isolate JS211949 (2,296 bp) had a deletion of two nucleotides and the isolate JSCZ2105 (2,299 bp) had one nucleotide insertion. The 20 isolates showed high nucleotide sequence identity with the three main vaccine strains, Cux-1 (96.5–98.0%, Germany), 26P4 (96.3–99.5%, The Netherland), and Del-Ros (97.9–98.6%, United States). The nucleotide similarity between the 20 isolates was high (96.3–100%), with the lowest 96.3% (JS2105B and JS2107Liu) and the highest 100% (JS2105A and JLTZ2105). Notably, the JS2105A was isolated from an attenuated live vaccine, and the JLTZ2105 sharing 100% nucleotide identity with the JS2105A was isolated from chicken liver tissue, suggesting that JLTZ2105 was derived from the vaccine contaminated with JS2105A. In addition, the deduced amino acid sequences of VP1 of the 20 isolates also shared high similarity with the three main vaccine strains Cux-1 (97.1–98.7%), 26P4 (97.6–99.6%), and Del-Ros (97.8–99.3%), and had high similarity (97.3–100%) with each other.
The phylogenetic trees were conducted based on the nucleotide sequences of the complete genome and the deduced amino acid sequences of VP1 of the 20 isolates and the 42 reference strains such as LF4 (China,), Cux-1, CIA (United States), and TR20 (Japan). As shown in Figure 3, the 62 strains were clustered into four major groups (A-D) based on the genome sequence. Fifteen of the 20 isolates belonged to Asian branch Group A and three (JS2105A, JS2105B, and JSTZ2105) of the 20 isolates belonged to Group B, the isolates JS221021 and JS2107Liu were clustered into Group C and Group D, respectively. As shown in Figure 4, similar to complete genome, the amino acid sequences of VP1of 62 strains were also clustered into four major groups (A-D). Among them, 16 of the 20 isolates belonged to Asian branch Group A, JS211949 and JSNT21Sun were clustered into Group B, and other two isolates (JS2107Liu and JS221021) belonged to Group D. The above data showed that the isolates in this study mainly belonged to Group A.
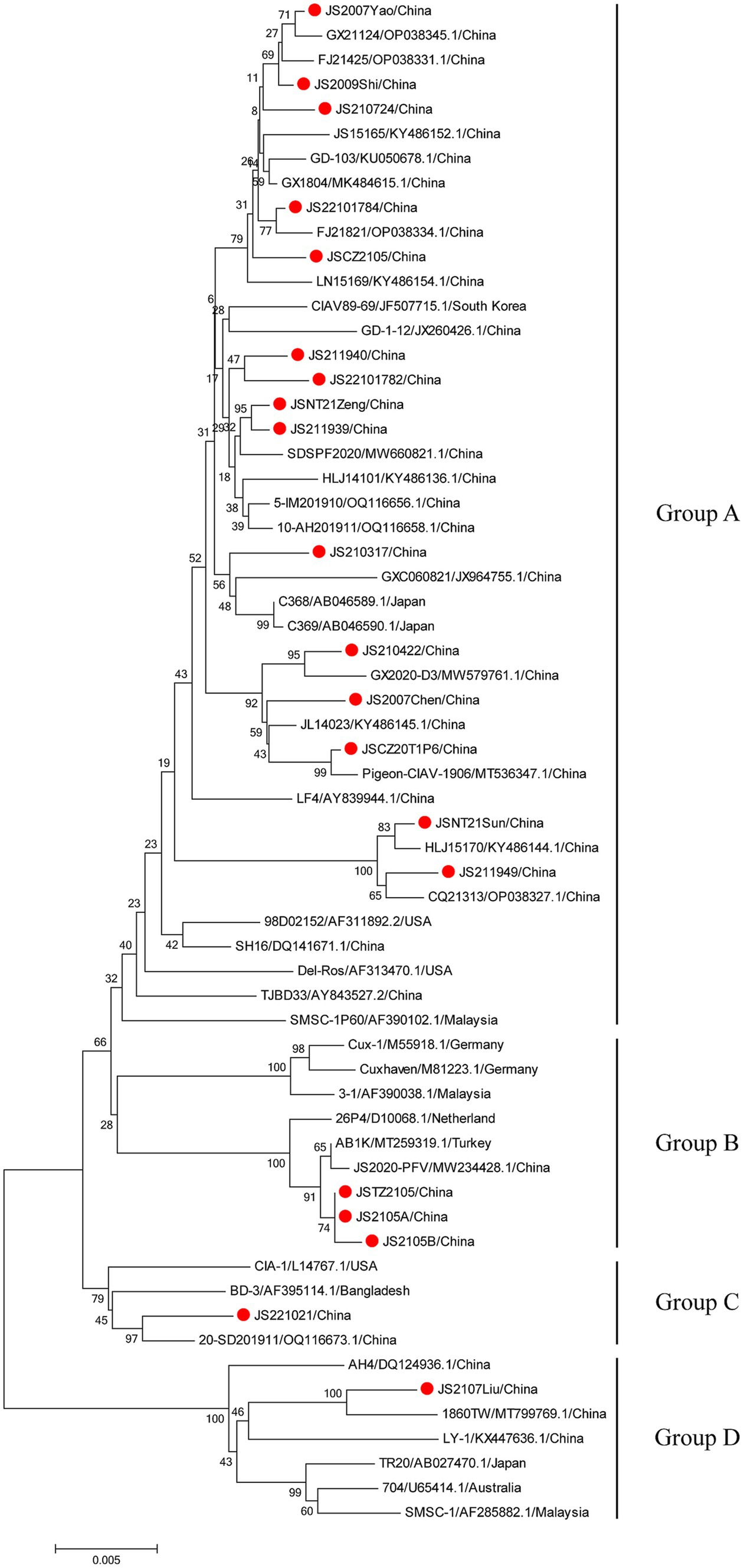
Figure 3. Phylogenetic tree analysis based on the nucleotide sequences of complete genome. The phylogenetic tree was constructed using the neighbor-joining method (1,000 bootstraps) with MEGA6. The 20 isolates in this study were indicated by the red closed circle.
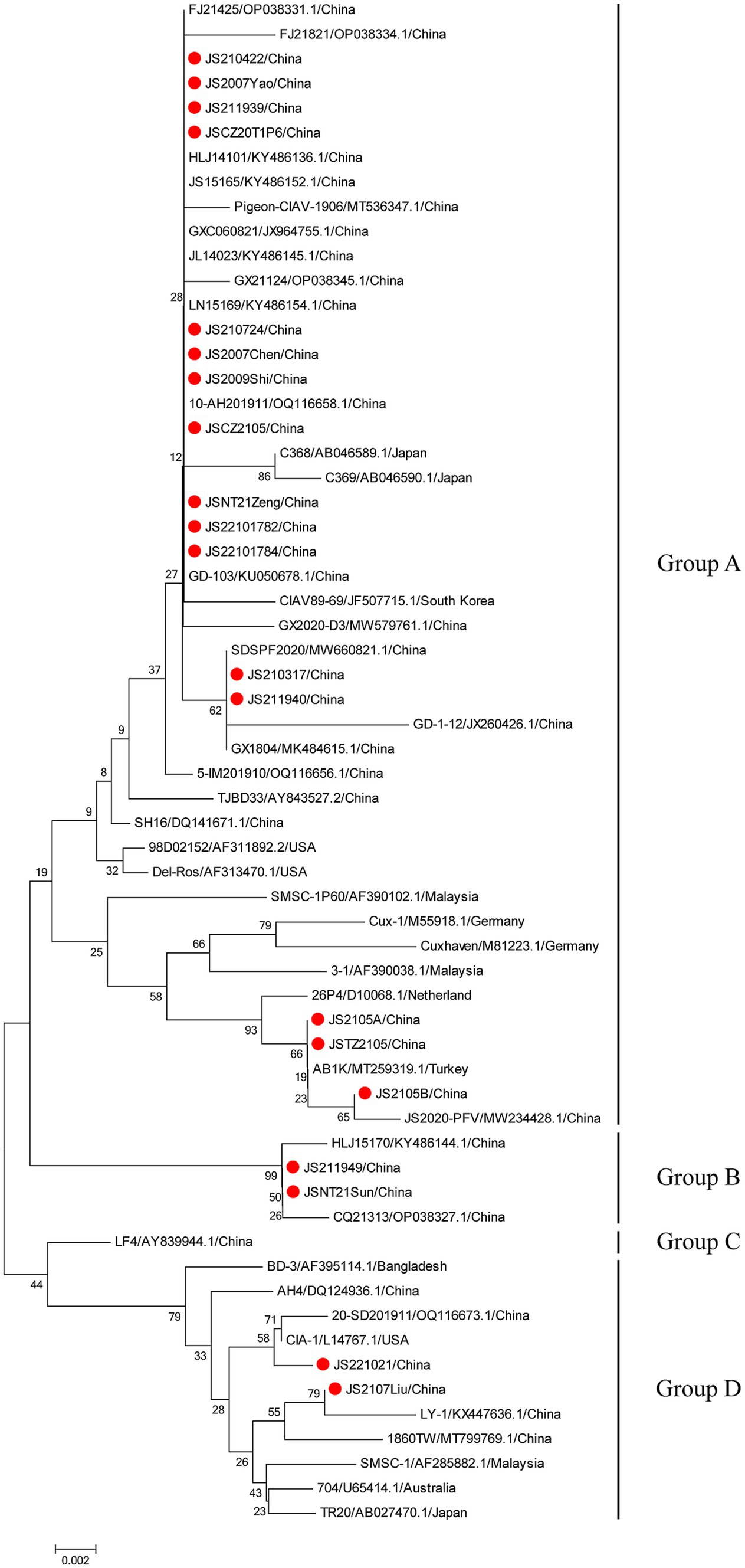
Figure 4. Phylogenetic tree analysis based on the amino acid sequences of VP1. The phylogenetic tree was constructed using the neighbor-joining method (1,000 bootstraps) with MEGA6. The 20 isolates in this study were indicated by the red closed circle.
In comparison with the 42 reference strains, the deduced amino acid sequences of VP1 of the 20 isolates in this study had 18 substitutions (V25I, V75I, T89K, G92D, M97L, L125I, K139Q, E144Q, V157M, E254G, S287T, S287A, A290P, Q294H, G370S, G370A, G370T, I376L, S413A, V436I, and S447T), and the mutation probability was 4%. As described in Figure 5, in the hypervariable region (aa 139–157) of VP1, JS2107Liu and JS221021 had two amino acid substitutions (K139Q and E144Q), which had an influence on the rate of replication or dissemination of infection in MDCC-MSB-1 cells (23). Seven isolates (JS210317, JS2105A, JS2105B, JSTZ2105, JS211940, JS211949, and JSNT21Sun) had one amino acid substitution (V157M). The known virulence-determining sites in VP1 of CIAV were shown in Table 3. The 20 isolates presented Q at amino acid sites 141 and 394, indicating all the isolates possessed sequence characteristics of highly virulent strains (24). In addition, at amino acid site 75, 18 isolates presented V instead of I as the majority virulent strains, except JS2107Liu and JS221021. At amino acid site 89, only JS2105B was K instead of T in the low virulent strains. Five isolates (JS2105A, JS2105B, JSTZ2105, JS2107Liu, and JS221021) presented I instead of L as the majority virulent strains shown at amino acid site 125. At amino acid sites 139 and 144, only JS2107Liu and JS221021 were Q, which is consist with the majority virulent strains, whereas the other 18 isolates were K or E. Seven isolates (JS210317, JS2105A, JS2105B, JSTZ2105, JS211940, JS211949, and JSNT21Sun) presented M instead of V as the low virulent strains at amino acid site 157. At amino acid site 287, six isolates (JS2105A, JS2105B, JSTZ2105, JS2107Liu, JS211949, and JSNT21Sun) presented T as the majority virulent strains, JS221021 had an A with rare reports, and the other 13 isolates presented S as the low virulent strains. Notably, amino acid site 370 in VP1 had three substitutions (G370S, G370A, and G370T). The substitutions of key amino acid sites in VP1 between the live attenuated vaccine sample JS2105A and reference strain AB1K were completely consistent, revealing a potential genetic evolutionary relationship.
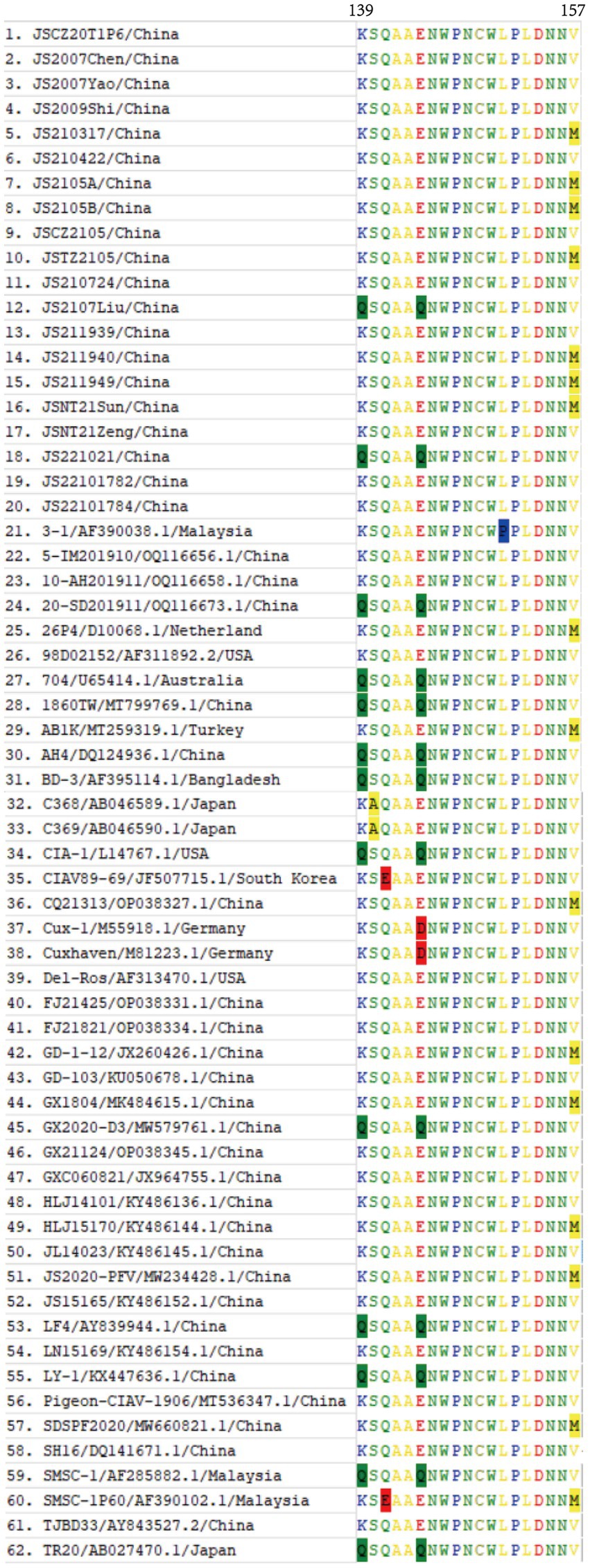
Figure 5. Amino acid alignment of the VP1 hypervariable region (aa 139–157) of 20 isolates and 42 CIAV reference strains available in GenBank. The substitutions of amino acids of VP1 occurring at sites 139, 140, 141, 144, 151, and 157 of 62 strains were highlighted with background color.
In addition, only two substitutions were found in VP2 (D169G: JS2105A, JS2105B, and JSTZ2105; G177A: JS2107Liu), with a mutation probability of 0.9%. Eight substitutions were detected in VP3 (L4P, P9Q, R23Q, P24L, L25S, V73A, S103N, and P112H), with a mutation probability of 6.6% (Table 4).
The UTR of CIAV is consists of approximately 300 centrally distributed nucleotides, containing more than a dozen conserved sequences known to be associated with replication and transcriptional regulation (25–28). In this study, the Clustal W method was used to analyze the nucleotide sequences of UTRs of 20 CIAV isolates, and the alignment result showed that the UTRs of the isolates contained DNA conserved regions with high G + C content (nucleotide homology 97.5–100%). Further analysis showed that the deletion and insertion in the UTRs of the isolates were not on the transcription factor binding motifs, and most of the motifs located in the UTRs were the same, such as Erthroid specific G-string, Polyadenylation signal, Core element of the SV40 enhancer, Lymphoid specific site, NF-AT2, SP1 site, and TATA box (Figure 6). However, there were also several single base mutations located in some motifs, such as NFκB+H2TF1 sites and GTII factor binding sites (Figure 6). Whether the mutations of these two motifs affect the binding of corresponding transcription factors and further have impacts on the replication of CIAV still need to be studied. In addition, transcription factor binding site analysis conducted by NSITE demonstrated that a tandem array consisting of four DR regions was found 1 nt upstream of the “CCAAT” box in the UTRs of all isolates (Figure 6). The ATF/CREB binding sites (ACGTCA) in the four DRs are similar to imperfect hormone response element half-sites (AGGTCA), and a cluster of GGTCA-like sequences was found downstream of the transcription initiation point (TSP) (Figure 6).
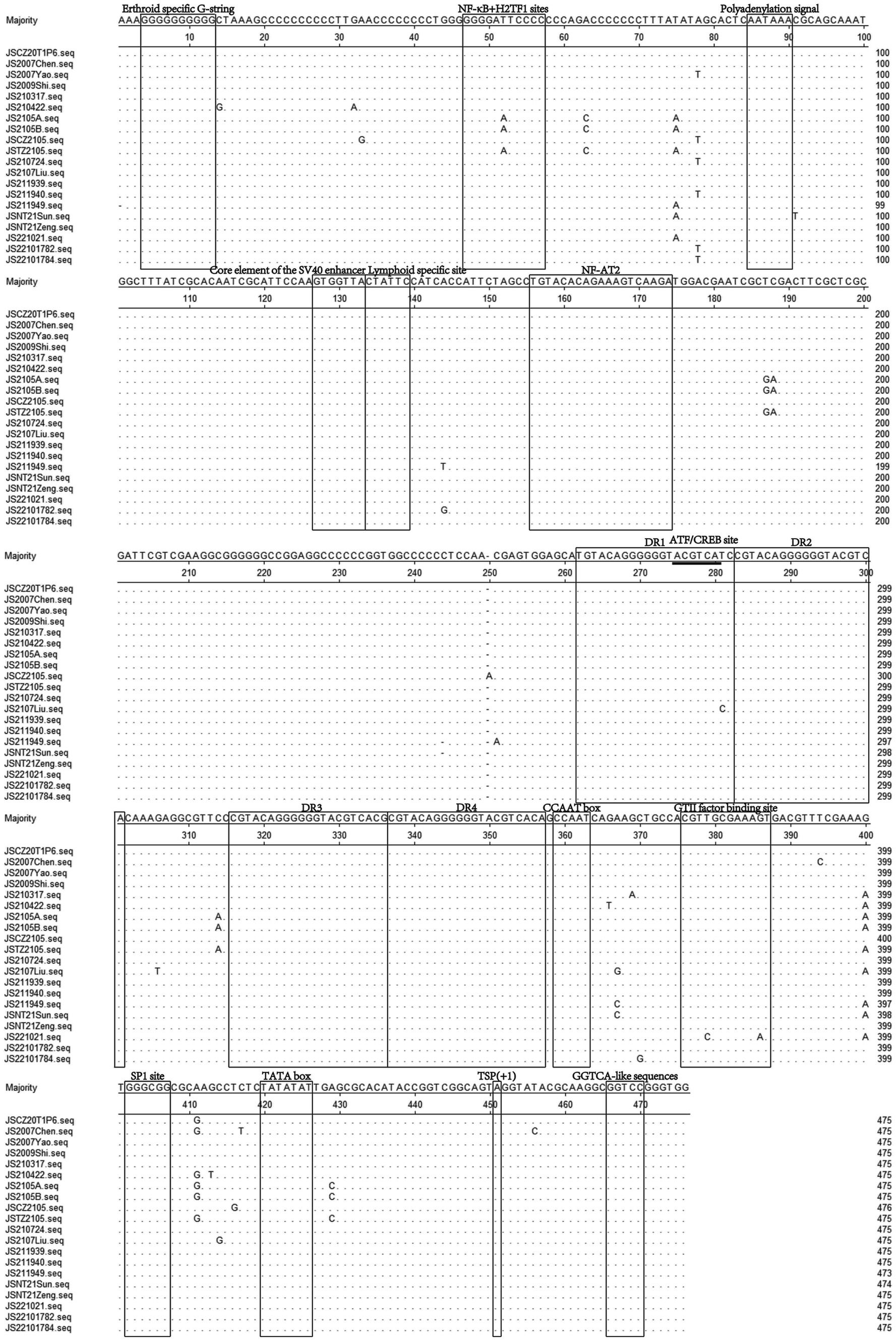
Figure 6. Sequence alignment of the UTRs nucleotide sequences of 20 CIAV isolates. The sequences in black frames are the motifs of transcriptional regulatory elements in this study. Conserved nucleotide sequences were indicated with dots and differences were indicated with letters.
Since Yuasa et al. first isolated CIAV in Japan in 1979 (15), CIAV has spread globally and caused huge economic losses to the poultry industry worldwide (25, 29). CIAV was first reported and isolated from Heilongjiang province in China in 1992 by Cui et al. (16). At present, from the perspective of the antigenicity of isolated strains worldwide, it is generally considered that there is only one serotype of CIAV (30, 31). However, the sequences of CIAV isolates in different regions were different, and the virulence changed greatly, which has become one of the important pathogens threatening the poultry industry worldwide (32).
In this study, we investigated CIAV infection in clinical samples from 19 broiler farms or breeding companies in Jiangsu province from 2020 to 2022 and compared the sequences of the obtained isolates with related reference strains. The investigation found that the positive rate of CIAV in 223 samples was 33.6% and 20 CIAV strains from different farms or company were successfully isolated. The nucleotide similarity of between the 20 isolates was 96.3–100%, whereas the identity of the complete genomic nucleotide sequence of CIAV isolated from the same farm or company were higher than 99%, except for JSNT21Sun and JSNT21Zeng (98.3%). Of note, although JSNT21Sun and JSNT21Zeng were clustered into Group A, their genetic evolutionary distance was higher when compared to other strains from the same field. Thus, it is necessary to be alert to the emergence of new recombinant viruses when strains with genetic distance exist in the same breeding farm. In addition, the co-infections of CIAV with other viruses, such as ALV-A/J/K, FAdV-4, MDV, and ARV, were also detected (Supplementary Table S1). These indicated that the co-infection of immunosuppressive pathogens still common in China, which should be taken more seriously. Currently, CIAV infection is generally prevented and controlled by vaccination in chickens, but no commercial vaccine is available in China (33). Thus, CIAV was very prevalent in China during the past years (34–37). Moreover, a study reported that a highly pathogenic CIAV strain was isolated from 2-month-old chickens for the first time in China, indicating that the epidemic CIAV strains have the potential to break age resistance with increasing pathogenicity (38).
As the only structural protein of CIAV, VP1 gene encodes the viral capsid and is the main immunogenic protein with neutralizing epitopes (7). VP1 is also the most variable protein in different CIAV strains (25, 39, 40). Moreover, the known variations in the hypervariable region of VP1 (139–151 aa) can impact on the protein structure prediction of VP1 (23, 41, 42). In this study, mutations were found in VP1 (Figure 4). Further research is needed to determine whether these mutations in VP1 could affect its antigenicity. VP2 is a non-structural protein in CIAV and acts as a scaffold protein to participate in the correct folding of VP1 during virion assembly (8, 43), assisting the production of neutralizing antibodies (44). Of note, VP2 is the most conserved protein in CIAV, with only two substitutions (D169G and G177A) in the isolates of this study. In addition, VP2 can also induce apoptosis (10), and has dual-specificity phosphatase activity, similar to TTV-encoded ORF2 (8, 9). Whether the two substitutions in VP2 impact its function still needs to be further studied. As another non-structural protein, VP3 is known as apoptin (10), which can selectively mediate cell death (11, 12), and it is essential for viral replication (13, 14). However, only eight substitutions (L4P, P9Q, R23Q, P24L, L25S, V73A, S103N, and P112H) were found in VP3 of the 20 isolates, which is much less than that of VP1. And the impact of these substitutions on the function of VP3 needs to be explored in the future. In addition, whether the mutations of NF-κB + H2TF1 sites and GTII factor binding sites in the UTRs of the isolates have impacts on the replication of CIAV should also be further studied.
A previous study found that amino acid site 394 of VP1 is a genetic determinant of pathogenicity in CIAV, and the mutation of Q to H can decrease the virulence of CIAV (24). Herein, all 20 isolates presented Q at amino acid site 394, indicating that all the isolates were highly likely to be highly pathogenic strains as well. Renshaw et al. (23) found that 139–151 aa is the hypervariable region in VP1, and the construction of various chimeric viruses of Cux-1 and CIA-1 proved that Q139 and/or Q144 impact the replication rate of CIAV and its infective ability in MDCC-MSB-1 cells. Meanwhile, the cell tropism of CIAV is related to amino acid sites 139, 144, and some upstream amino acid sites in VP1 (23). In this study, isolates JS2107Liu and JS221021 had Q at amino acid sites 139 and 144, representing the majority virulent strains, indicating that their replication in MDCC-MSB-1 cells or transmission in chickens may be different from the other 18 isolates. Notably, it is rarely reported that CIAV strains present A at amino acid site 287 (37), however, isolate JS221021 here had an A as well, which revealed that the mutation of rare amino acid site has a tendency to increase. All these demonstrate that the isolated strains with high sequence diversity got a high possibility of high pathogenicity.
It is noteworthy that the isolate JLTZ2105 sharing 100% nucleotide homology with JS2105A in chickens was derived from the attenuated vaccine contaminated with CIAV, which is consistent with the widespread presence of low pathogenic CIAV in SPF chickens in China reported in previous studies (34, 36). Since the replacements of key amino acid sites in VP1 between the vaccine sample JS2105A and reference strain AB1K were completely consistent. Therefore, the isolates JS2105A and JS2105B in this study may be mixed as contaminants in various vaccines to restore their virulence in the host (33). Of note, RDP4 software (RDP, GENECONV, Bootscan, MaxChi, Chimera, SiScan, and 3seq) and Simplot 3.51 software were used to assess the probability of genotype recombination of the 20 CIAV isolates and the 42 reference strains. However, the evaluation showed that there were potential recombination events in the reference strains (data not shown) instead of the 20 isolates, indicating that the variation of the isolates mainly due to natural mutations. Whether the genetic mutations in these isolates participate in the generation of new virus lineages needs to be further evaluated.
In summary, our study provided that the epidemiological data of CIAV in Jiangsu province of China during 2020–2022, which indicated a severe CIAV infection status in poultry industry. Notably, in clinical, it is necessary to pay more attention to the detection of CIAV in live vaccines to avoid CIAV infections caused by vaccination. Besides, the increasing isolation of CIAV strains with highly pathogenic characteristics is also worth our vigilance. Taken together, our study provides new insights into the epidemiology and effective strategies for the prevention and control of CIAV in China.
The data presented in the study are deposited in the GenBank repository, accession number PP354989-PP355008.
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
JZ: Investigation, Writing – original draft. LM: Investigation, Writing – original draft. TL: Data curation, Investigation, Writing – original draft, Writing – review & editing. QX: Investigation, Writing – review & editing. ZW: Data curation, Methodology, Writing – review & editing. AQ: Data curation, Writing – review & editing. JY: Data curation, Methodology, Writing – original draft, Writing – review & editing. HS: Funding acquisition, Methodology, Writing – review & editing. SW: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research & Development (R&D) Plan (2022YFD1800301), the National Natural Science Foundation of China (31872491), the Research Foundation for Talented Scholars in Yangzhou University and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1378120/full#supplementary-material
1. Rosenberger, JK, and Cloud, SS. Chicken anemia virus. Poult Sci. (1998) 77:1190–2. doi: 10.1093/ps/77.8.1190
2. Jeurissen, SH, Janse, ME, Van Roozelaar, DJ, Koch, G, and De Boer, GF. Susceptibility of thymocytes for infection by chicken anemia virus is related to pre-and posthatching development. Dev Immunol. (1992) 2:123–9. doi: 10.1155/1992/52484
3. Yuasa, N, Imai, K, and Nakamura, K. Pathogenicity of chicken anaemia agent in bursectomised chickens. Avian Pathol. (1998) 17:363–9. doi: 10.1080/03079458808436454
4. Zhang, J, Ma, L, Li, TF, Li, LY, Kan, QQ, Yao, XH, et al. Synergistic pathogenesis of chicken infectious anemia virus and J subgroup of avian leukosis virus. Poult Sci. (2021) 100:101468. doi: 10.1016/j.psj.2021.101468
5. Rosario, K, Breitbart, M, Harrach, B, Segales, J, Delwart, E, Biagini, P, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. (2017) 162:1447–63. doi: 10.1007/s00705-017-3247-y
6. Noteborn, MH, de Boer, GF, van Roozelaar, DJ, Karreman, C, Kranenburg, O, Vos, JG, et al. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. (1991) 65:3131–9. doi: 10.1128/JVI.65.6.3131-3139.1991
7. Noteborn, MH, Kranenburg, O, Zantema, A, Koch, G, de Boer, GF, and van der Eb, AJ. Transcription of the chicken anemia virus (CAV) genome and synthesis of its 52-kDa protein. Gene. (1992) 118:267–71. doi: 10.1016/0378-1119(92)90198-x
8. Noteborn, MH, Verschueren, CA, Koch, G, and Van der Eb, AJ. Simultaneous expression of recombinant baculovirus-encoded chicken anaemia virus (CAV) proteins VP1 and VP2 is required for formation of the CAV-specific neutralizing epitope. J Gen Virol. (1998) 79:3073–7. doi: 10.1099/0022-1317-79-12-3073
9. Peters, MA, Jackson, DC, Crabb, BS, and Browning, GF. Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J Biol Chem. (2002) 277:39566–73. doi: 10.1074/jbc.M201752200
10. Noteborn, MH, Todd, D, Verschueren, CA, de Gauw, HW, Curran, WL, Veldkamp, S, et al. A single chicken anemia virus protein induces apoptosis. J Virol. (1994) 68:346–51. doi: 10.1128/JVI.68.1.346-351.1994
11. Heilman, DW, Teodoro, JG, and Green, MR. Apoptin nucleocytoplasmic shuttling is required for cell type-specific localization, apoptosis, and recruitment of the anaphase-promoting complex/cyclosome to PML bodies. J Virol. (2006) 80:7535–45. doi: 10.1128/JVI.02741-05
12. Los, M, Panigrahi, S, Rashedi, I, Mandal, S, Stetefeld, J, Essmann, F, et al. Apoptin, a tumor-selective killer. Biochim Biophys Acta. (2009) 1793:1335–42. doi: 10.1016/j.bbamcr.2009.04.002
13. Kucharski, TJ, Ng, TF, Sharon, DM, Navid-Azarbaijani, P, Tavassoli, M, and Teodoro, JG. Activation of the chicken Anemia virus Apoptin protein by Chk1/2 phosphorylation is required for apoptotic activity and efficient viral replication. J Virol. (2016) 90:9433–45. doi: 10.1128/JVI.00936-16
14. Wang, YQ, Song, XQ, Gao, HL, Wang, XY, Hu, YH, Gao, YL, et al. C-terminal region of apoptin affects chicken anemia virus replication and virulence. Virol J. (2017) 14:38. doi: 10.1186/s12985-017-0713-9
15. Yuasa, N, Taniguchi, T, and Yoshida, I. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis. (1979) 23:366–85. doi: 10.2307/1589567
16. Cui, XL, Xing, GX, Wu, DL, Feng, JY, Li, F, and Qu, LX. Identification of chicken infectious anemia virus. Chin J Prev Vet Med. (1992) 6:3–5.
17. Miles, AM, Reddy, SM, and Morgan, RW. Coinfection of specific-pathogen-free chickens with Marek’s disease virus (MDV) and chicken infectious anemia virus: effect of MDV pathotype. Avian Dis. (2001) 45:9–18. doi: 10.2307/1593006
18. Zhang, YK, Cui, N, Han, N, Wu, JY, Cui, ZZ, and Su, S. Depression of Vaccinal immunity to Marek’s disease by infection with chicken infectious Anemia virus. Front Microbiol. (2017) 8:1863. doi: 10.3389/fmicb.2017.01863
19. Su, Q, Meng, FF, Li, Y, Zhang, YB, Zhang, ZJ, Cui, ZZ, et al. Chicken infectious anemia virus helps fowl adenovirus break the protection of maternal antibody and cause inclusion body hepatitis-hydropericardium syndrome in layers after using co-contaminated Newcastle disease virus-attenuated vaccine. Poult Sci. (2019) 98:621–8. doi: 10.3382/ps/pey153
20. Erfan, AM, Selim, AA, Helmy, SA, Eriksson, P, and Naguib, MM. Chicken anaemia virus enhances and prolongs subsequent avian influenza (H9N2) and infectious bronchitis viral infections. Vet Microbiol. (2019) 230:123–9. doi: 10.1016/j.vetmic.2019.01.024
21. Li, X, Zhang, KR, Pei, Y, Xue, J, Ruan, SF, and Zhang, GZ. Development and application of an MRT-qPCR assay for detecting coinfection of six vertically transmitted or immunosuppressive avian viruses. Front Microbiol. (2020) 11:1581. doi: 10.3389/fmicb.2020.01581
22. Zeng, FC, Li, WF, Ke, JH, Wen, F, Guo, JY, Zhang, XL, et al. Epidemiological characteristics and genetic diversity of chicken infectious anemia virus (CIAV) in Guangdong Province, China. Res Square. (2021) 2021:481. doi: 10.21203/rs.3.rs-385481/v1
23. Renshaw, RW, Soine, C, Weinkle, T, O’Connell, PH, Ohashi, K, Watson, S, et al. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J Virol. (1996) 70:8872–8. doi: 10.1128/JVI.70.12.8872-8878.1996
24. Yamaguchi, S, Imada, T, Kaji, N, Mase, M, Tsukamoto, K, Tanimura, N, et al. Identification of a genetic determinant of pathogenicity in chicken anaemia virus. J Gen Virol. (2001) 82:1233–8. doi: 10.1099/0022-1317-82-5-1233
25. Schat, KA. Chicken anemia virus. Curr Top Microbiol Immunol. (2009) 331:151–83. doi: 10.1007/978-3-540-70972-5_10
26. Noteborn, MH, Verschueren, CA, Zantema, A, Koch, G, and van der Eb, AJ. Identification of the promoter region of chicken anemia virus (CAV) containing a novel enhancer-like element. Gene. (1994) 150:313–8. doi: 10.1016/0378-1119(94)90444-8
27. Miller, MM, Jarosinski, KW, and Schat, KA. Negative modulation of the chicken infectious anemia virus promoter by COUP-TF1 and an E box-like element at the transcription start site binding deltaEF1. J Gen Virol. (2008) 89:2998–3003. doi: 10.1099/vir.0.2008/003103-0
28. Miller, MM, Jarosinski, KW, and Schat, KA. Positive and negative regulation of chicken anemia virus transcription. J Virol. (2005) 79:2859–68. doi: 10.1128/jvi.79.5.2859-2868.2005
29. Fatoba, A, and Adeleke, M. Chicken anemia virus: a deadly pathogen of poultry. Acta Virol. (2019) 63:19–25. doi: 10.4149/av_2019_110
30. McNulty, MS. Chicken anaemia agent: a review. Avian Pathol. (1991) 20:187–203. doi: 10.1080/03079459108418756
31. Noteborn, MH, and Koch, G. Chicken anaemia virus infection: molecular basis of pathogenicity. Avian Pathol. (1995) 24:11–31. doi: 10.1080/03079459508419046
32. Adair, BM. Immunopathogenesis of chicken anemia virus infection. Dev Comp Immunol. (2000) 24:247–55. doi: 10.1016/s0145-305x(99)00076-2
33. Varela, AP, Dos Santos, HF, Cibulski, SP, Scheffer, CM, Schmidt, C, Sales Lima, FE, et al. Chicken anemia virus and avian gyrovirus 2 as contaminants in poultry vaccines. Biologicals. (2014) 42:346–50. doi: 10.1016/j.biologicals.2014.08.002
34. Li, Y, Wang, YX, Fang, LC, Fu, JY, Cui, S, Zhao, YJ, et al. Genomic analysis of the chicken infectious Anemia virus in a specific pathogen-free chicken population in China. Biomed Res Int. (2016) 2016:4275718–5. doi: 10.1155/2016/4275718
35. Li, Y, Fang, LC, Cui, S, Fu, JY, Li, XH, Zhang, HM, et al. Genomic characterization of recent chicken Anemia virus isolates in China. Front Microbiol. (2017) 8:401. doi: 10.3389/fmicb.2017.00401
36. Li, Y, Hu, Y, Cui, S, Fu, J, Wang, Y, Cui, Z, et al. Molecular characterization of chicken infectious anemia virus from contaminated live-virus vaccines. Poult Sci. (2017) 96:1045–51. doi: 10.3382/ps/pew406
37. Yao, S, Tuo, TB, Gao, X, Han, CY, Yan, NN, Liu, AJ, et al. Molecular epidemiology of chicken anaemia virus in sick chickens in China from 2014 to 2015. PLoS One. (2019) 14:e0210696. doi: 10.1371/journal.pone.0210696
38. Fang, LC, Jia, HY, Hu, YH, Wang, YX, Cui, ZZ, Qi, LH, et al. Molecular characterization and pathogenicity study of a highly pathogenic strain of chicken anemia virus that emerged in China. Front Cell Infect Microbiol. (2023) 13:1171622. doi: 10.3389/fcimb.2023.1171622
39. Oluwayelu, DO, Todd, D, and Olaleye, OD. Sequence and phylogenetic analysis of chicken anaemia virus obtained from backyard and commercial chickens in Nigeria. Onderstepoort J Vet Res. (2008) 75:353–7. doi: 10.4102/ojvr.v75i4.111
40. Wang, D, Fan, W, Han, GZ, and He, CQ. The selection pressure analysis of chicken anemia virus structural protein gene VP1. Virus Genes. (2009) 38:259–62. doi: 10.1007/s11262-008-0316-z
41. Nogueira, EO, Ferreira, AJP, Martins Soares, R, Luiz Durigon, E, Lazzarin, S, and Brentano, L. Genome sequencing analysis of Brazilian chicken anemia virus isolates that lack MSB-1 cell culture tropism. Comp Immunol Microbiol Infect Dis. (2007) 30:81–96. doi: 10.1016/j.cimid.2006.11.001
42. van Santen, VL, Toro, H, and Hoerr, FJ. Biological characteristics of chicken anemia virus regenerated from clinical specimen by PCR. Avian Dis. (2007) 51:66–77. doi: 10.1637/0005-2086(2007)051[0066:BCOCAV]2.0.CO;2
43. McConnell, CD, Adair, BM, and McNulty, MS. Effects of chicken anemia virus on macrophage function in chickens. Avian Dis. (1993) 37:358–65. doi: 10.2307/1591659
Keywords: chicken infectious anemia virus (CIAV), isolation, complete genome, VP1, molecular characteristics
Citation: Zhang J, Ma L, Li T, Xie Q, Wan Z, Qin A, Ye J, Shao H and Wang S (2024) Isolation and genomic characterization of chicken infectious anemia virus in Jiangsu province of China during 2020–2022. Front. Vet. Sci. 11:1378120. doi: 10.3389/fvets.2024.1378120
Received: 29 January 2024; Accepted: 26 February 2024;
Published: 14 March 2024.
Edited by:
Chao-ting Xiao, Hunan University, ChinaReviewed by:
Jun Ji, Nanyang Normal University, ChinaCopyright © 2024 Zhang, Ma, Li, Xie, Wan, Qin, Ye, Shao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengnan Wang, d2FuZ3NoZW5nbmFuMTIzQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.