- 1Parasitology Unit, Department of Veterinary Pathology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 2Nakhon Ratchasima Zoo, The Zoological Park Organization, Nakhon Ratchasima, Thailand
- 3Biomarkers in Animals Parasitology Research Unit, Chulalongkorn University, Bangkok, Thailand
Brugia malayi and B. pahangi, potential zoonotic pathogens transmitted by mosquitoes, are believed to primarily infect dogs and cats as reservoir hosts. Although previous studies have indicated nematode infections in lions, particularly in zoo environments where human contact with these reservoirs is possible, limited documentation exists regarding Brugia sp. infections in lions in Thailand. This study aims to investigate a case of Brugia infection in a lion from a zoo in Thailand. The blood sample was collected and examined from a female lion, using staining methods to morphologically identify microfilaria at the genus level. Subsequently, the PCR was employed targeting specific genes, including mitochondrial 12S rDNA, 18S rDNA, cytochrome oxidase I (COI) and Wolbachia surface protein (wsp), to confirm the species of the filarial nematode parasite. The genetic sequencing results revealed a high similarity (99–100%) to B. malayi for the 12S rDNA, 18S rDNA, COI and wsp genes. Phylogenetic analysis based on nucleotide sequences from the 12S rDNA, 18S rDNA, COI and wsp genes showed that the sequences from this study belong to different clusters. This marks the inaugural documentation of molecular identification of Brugia infection in a lion, signifying that lions could function as reservoirs for this parasite and present a potential public health risk in the region. Our research underscores the effectiveness of molecular techniques and phylogenetic analysis in discerning and comprehending the evolution of filarial parasites. Additionally, it emphasizes the significance of these methods in enhancing the diagnosis, control, and prevention of zoonotic filarial nematode infections.
1 Introduction
Brugia spp., nematodes of the Onchocercidae family, have a global presence in infecting the lymphatic system of mammals. Lymphatic disease, commonly referred to as elephantiasis, is regarded as the primary manifestation of filariasis (1). Among Brugia species of medical importance are Brugia malayi and Brugia timori, causative agents of lymphatic filariasis in south and southeast Asia (2, 3). Brugia malayi can also naturally infect mammals such as monkeys and cats (4–6). Other filarial species, such as B. pahangi, have been reported to have associations with domestic animals (7, 8). Brugia pahangi, a closely related species of B. malayi, is a lymphatic filarial worm of mammals, particularly of cats, dogs and wild carnivores (9, 10). Although there have been reports of the presence of this parasite’s microfilaria in human blood samples, it is not currently identified as a cause of human disease in its natural environment (11, 12).
Filarial nematodes have previously been reported in different species of wild or captive felids, such as Dirofilaria immitis has been found in Bengal tiger (Panthera tigris) (13, 14), snow leopard (Uncia uncia) (15), clouded leopard (Neofelis nebulosa) (16), African lion (Panthera leo) (17), leopard (Panthera pardus pardus) (18). Dirofilaria striata has been reported in Florida panthers (Felis concolor coryi) and the bobcat (Lynx rufus) (19). Genet cats (Genetta tigrina) were infected with D. repens in East Africa (20). To date, there have been no documented cases of lymphatic filariasis resulting from Brugia sp. infection in lions globally, including Thailand. Nevertheless, the result showed an instance of Brugia sp. infection in a captive lion from a private zoo in Thailand. Molecular characterization of the infection was achieved through the analysis of the 12S rDNA, 18S rDNA, COI, and wsp genes.
2 Materials and methods
2.1 Sample collection
On October 27, 2022, a routine health examination was conducted on two lions (Panthera leo) at a private zoo. Both lions, a 2-year-old male and a 2-year-old female, weighing 90 kg each, were anesthetized for the examination. The lions had a body condition score of 3. Anesthesia was induced using a combination of 1 mg/kg xylazine (X-LAZINE, Thailand) and 4 mg/kg ketamine (Hameln Pharma, United Kingdom). Subsequently, blood samples were drawn from the femoral vein using EDTA tubes. These samples were then sent to a private standard laboratory center (Nakhon Ratchasima Province) to determine the presence of microfilariae through Giemsa staining (21) and Acid phosphatase staining (22). A blood sample was subjected to testing using an FIV/FeLV test kit (IDEXX, United States) and a CPV/CCV test kit (IDEXX, United States). The results revealed that a female lion tested positive for Brugian filariasis. The remaining EDTA blood from the Brugian filariasis-positive lion was preserved at −20°C for subsequent DNA extraction.
2.2 DNA extraction and molecular assay
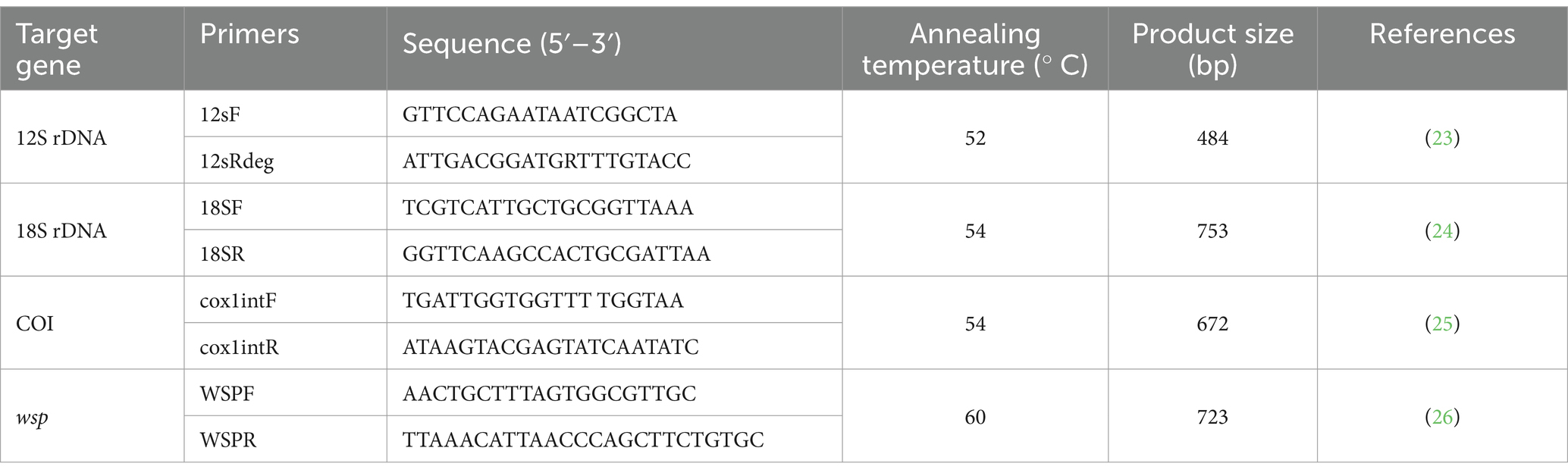
For molecular identification, NucleoSpin® Blood (MACHEREY-NAGEL, Germany) was used to extract filarial nematode DNA according to the manufacturer’s specifications. The DNA was used as a template for the PCRs with the GoTaq® Green Master Mix (Promega Corporation, United States), amplifying a section of the 12S rDNA, 18S rDNA, COI, and wsp genes. The cycling conditions for PCR and specific details of primer sequences were employed as outlined in Table 1. All PCR amplifications included D. immitis adult worm DNA as an amplification control, and nuclease-free water served as a no-template control. The amplified PCR products were stained with RedSafe™ Nucleic Acid Staining Solution (INtRON Biotechnology, Korea) and checked in 1.5% agarose gel. Further amplified DNA product was excised from the gel and purified using a PCR clean-up gel extraction kit (NucleoSpin® Gel and PCR Clean-up, MACHEREY-NAGEL, Germany) as per the manufacturer’s instructions. Purified PCR products were carried out in both directions and confirmed by barcode taq (BT) sequencing. DNA sequences obtained in the study were identified using BLAST with sequences available in the GenBank database. The 12S rDNA, 18S rDNA, COI and wsp sequences obtained from this study were aligned along with the reference sequences retrieved from NCBI using CLUSTALW. Finally, the phylogenetic analysis was performed using MEGA X software (27). The maximum likelihood algorithm was used as the best fit model with 1,000 bootstrap replicates (28) and the nucleotide distance was calculated using the p-distance method (29).
3 Results
3.1 Parasite identification
A 2-year-old female lion blood sample was brought to Vet Central Lab for screening of microfilariae. The Giemsa blood smear taken from the positive case showed the presence of sheathed microfilariae. These sheathed microfilariae had a pink sheath, with a head-space at the front and two clear tail nuclei at the back (Figure 1A). Histochemical staining was performed to confirm species identification by acid phosphatase activity in microfilariae. When the sheathed microfilariae were stained with acid phosphatase, a pattern of four points staining emerged, indicating acid phosphatase activity at the amphid (AM), excretory pore (EP), anal pore (AP), and phasmid (PM) (Figure 1B). When it comes to B. malayi microfilaria, bright red points were seen, which were easily visible even under low magnification.
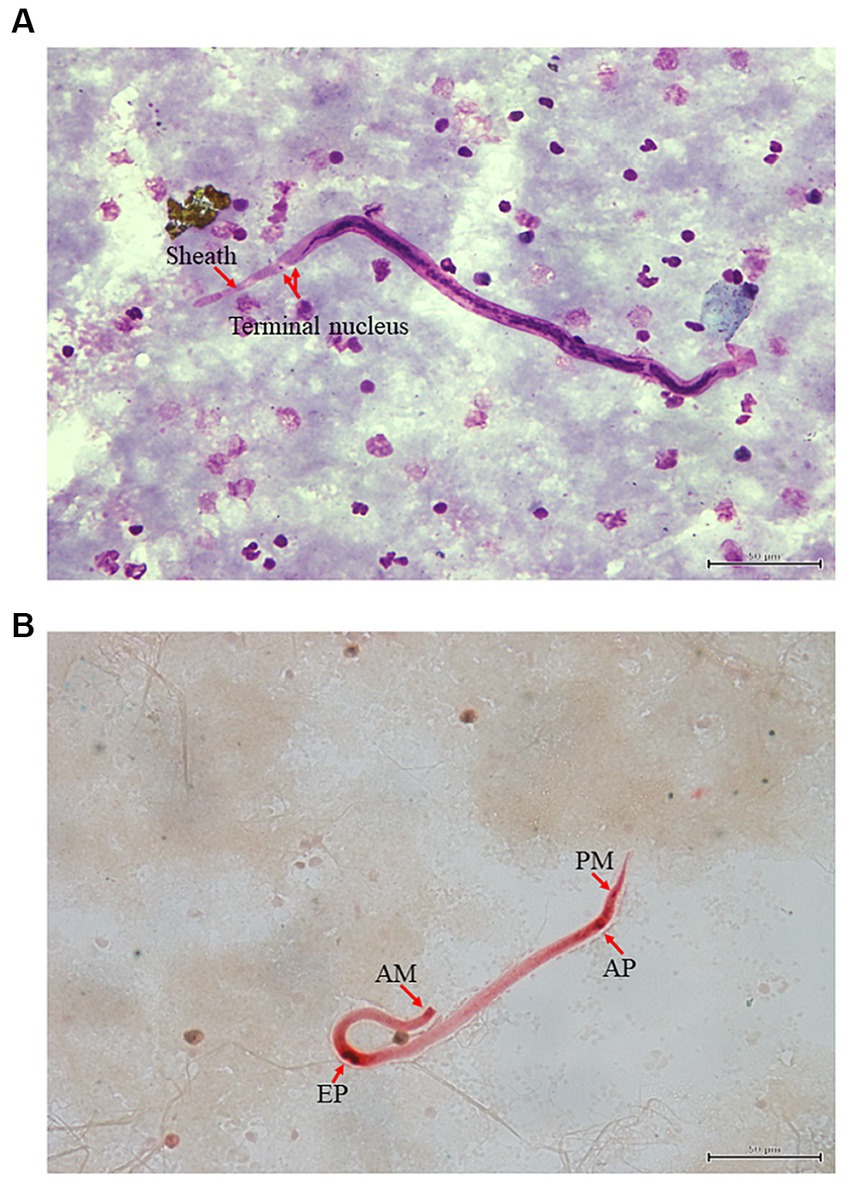
Figure 1. Microscopic examination using Giemsa and Acid phosphatase straining (A,B): Giemsa staining of microfilaria infection from lion (Panthera leo) in Thailand. Red arrow indicates the presence of sheathed microfilaria (A). Brugia microfilaria showing acid phosphatase activity at the amphid (AM), excretory pore (EP), anal pore (AP) and phasmid (PM) (B).
3.2 Similarity analysis
The primers used for PCR amplification successfully detected filarial nematode DNA in the blood samples. The Brugia sp. was specifically identified through PCR products of various sizes, including 484 bp (12S rDNA gene), 753 bp (18S rDNA gene), 672 bp (COI gene) and 723 bp (wsp gene). These PCR products were then sequenced and analyzed using BLAST. The BLAST results demonstrated that the 12S rDNA, 18S rDNA, COI and wsp sequences shared a high level of identity (99–100%) with B. malayi. The nucleotide sequence data mentioned in the paper have been deposited in GenBank, as indicated in Supplementary Table S1. Comparative analysis of the sequences revealed a high sequence similarity percentage (99.7–100%, 95.2–100%, 99.5–100% and 99.4–100%) for the 12S rDNA, 18S rDNA, COI and wsp sequences, respectively (Supplementary Tables S2–S5).
3.3 Phylogenetic analysis
The genetic relationships among B. malayi isolates can be more accurately determined by representing them in a phylogenetic tree. In our study, the Maximum Likelihood (ML) was used to construct these trees. By aligning the sequences of the B. malayi 12S rDNA gene obtained in our study with other sequences from GenBank, divided the resulting phylogenetic tree into 14 clusters. Notably, a B. malayi sequence identified in our study was placed in cluster 1 along with sequences from the UK, USA, and Thailand (Figure 2). In the phylogenetic tree of the 18S rDNA gene, the sequences were classified into 20 clusters within the phylogram. Interestingly, a B. malayi sequence from a lion in Thailand was assigned to cluster 9 and showed genetic similarities with sequences from the UK and USA, indicating the genetic variability of the B. malayi 18S rDNA sequence in Thailand (Figure 3). The COI gene sequences were also analyzed, which were grouped into 8 clusters. A B. malayi COI gene sequence from a lion in Thailand was classified into cluster 1, alongside other B. malayi COI sequences from Thailand, Vietnam, and the USA (Supplementary Figure S1). Furthermore, the phylogenetic analysis of the wsp gene in Wolbachia endosymbionts revealed clear separation into 18 clusters. Within the cluster associated with Brugia sp., a more detailed subdivision into 9 subclusters was observed (Supplementary Figure S2). The B. malayi sequence identified in our investigation was classified within the 1st cluster, exhibiting genetic affinities with sequences originating from the USA.
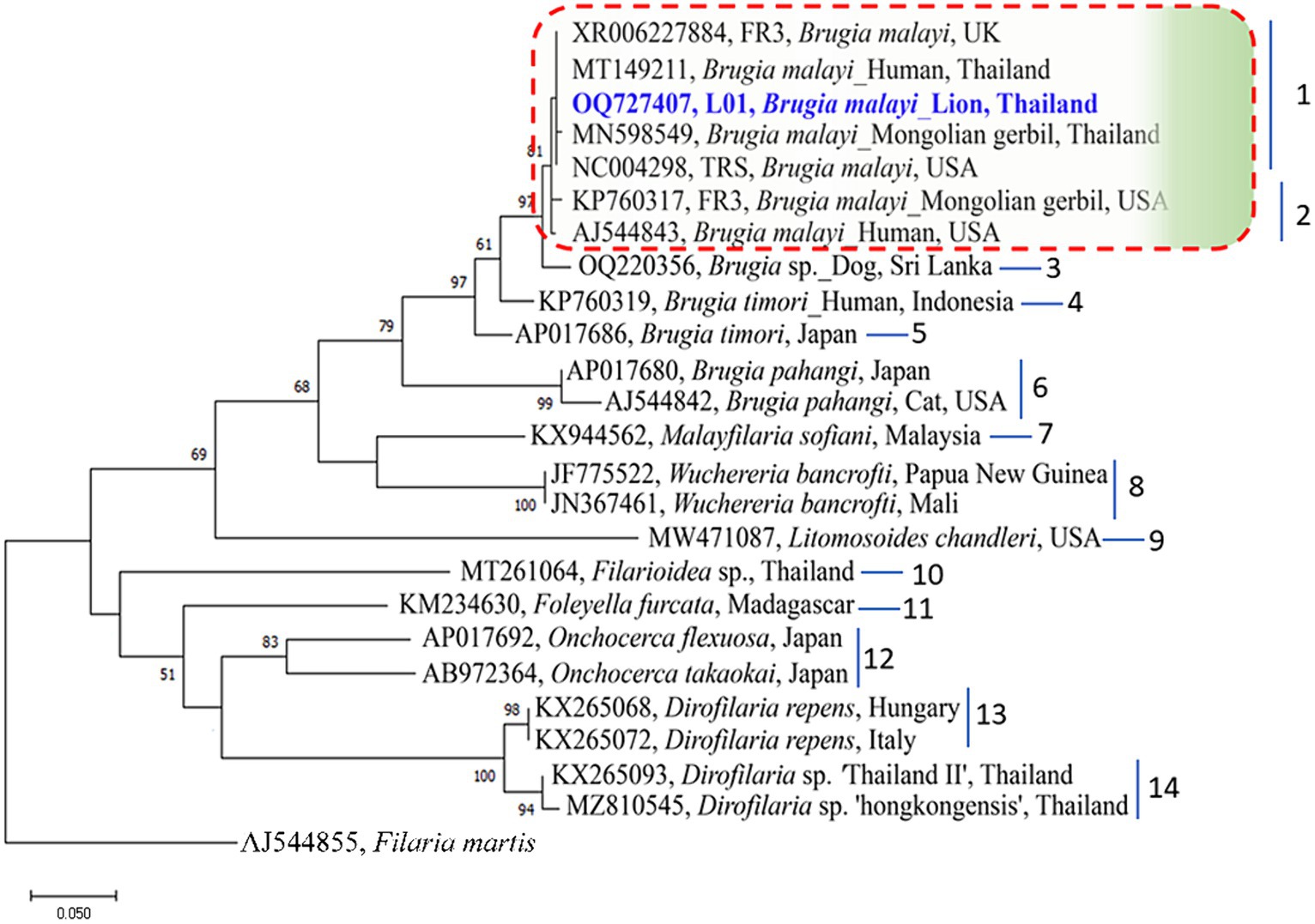
Figure 2. Phylogenetic tree based on an alignment of 12S rDNA gene sequences. Bootstrap confidence values (1,000 replicates) are shown as percentages. Values less than 50% are omitted. The units for the scale bar are substitutions per site. NCBI accession numbers are included. The 12S rDNA sequence of B. malayi from lion in Thailand, with taxon name shown in blue color and bold font, was generated as part of this study. Letters to the right of the bracketed branches denote the clusters. Filaria martis was used as out group.
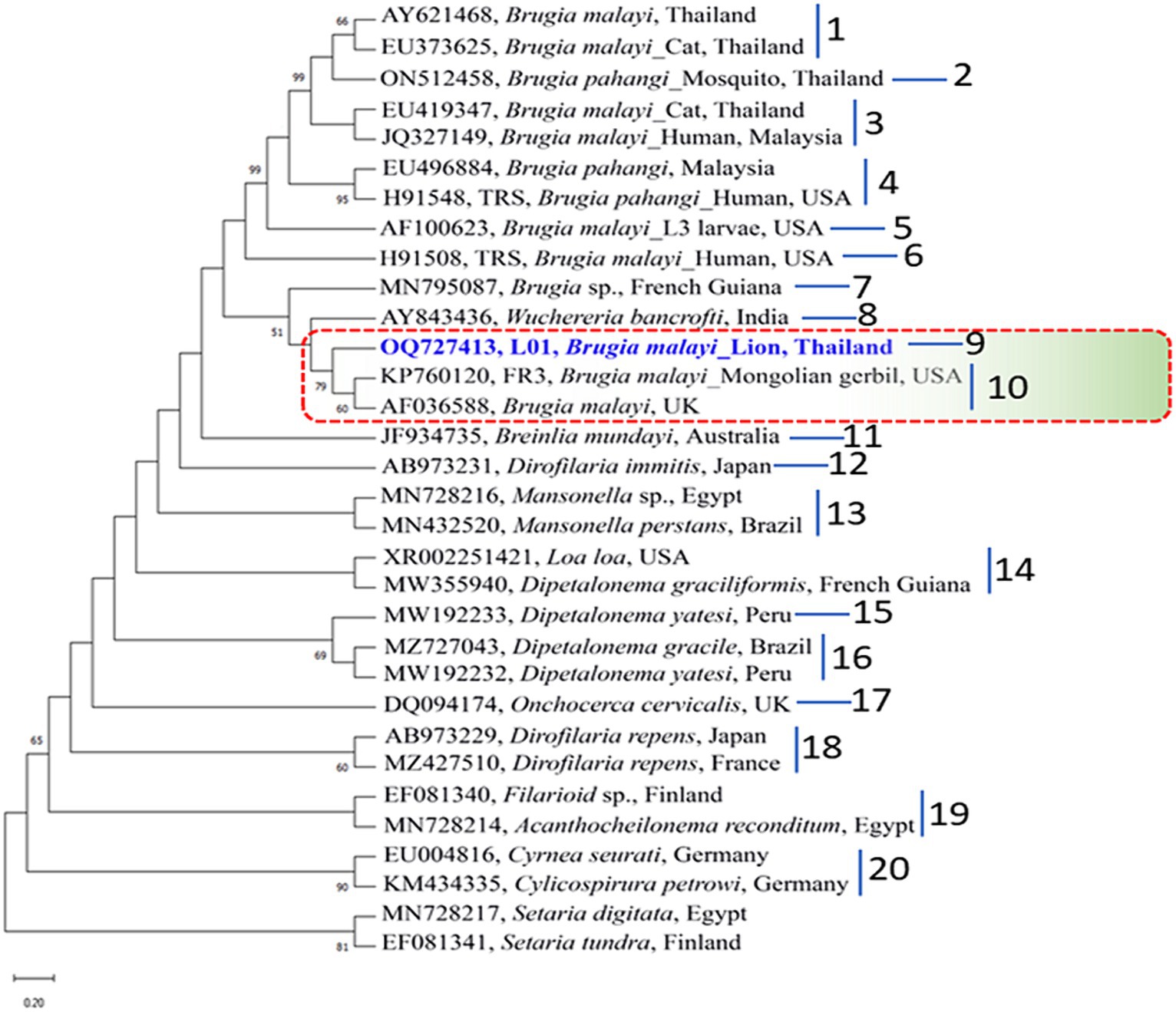
Figure 3. Phylogenetic tree based on an alignment of 18S rDNA gene sequences. Bootstrap confidence values (1,000 replicates) are shown as percentages. Values less than 50% are omitted. The units for the scale bar are substitutions per site. NCBI accession numbers are included. The 18S rDNA sequence of B. malayi from lion in Thailand, with taxon name shown in blue color and bold font, was generated as part of this study. Letters to the right of the bracketed branches denote the clusters. Setaria digitata and S. tundra were used as out group.
4 Discussion
Presently, there is a lack of literature on Brugia infection in zoos and wildlife animals in Thailand. However, to the best of our knowledge, this study is the inaugural one to present both morphological and molecular characterization of Brugia sp. infection in captive zoo lions in Thailand. Morphological assessment is employed to identify potential causes, and subsequent molecular characterization aids in pinpointing the most likely parasitic source. Knowledge of lion parasites mainly from zoo lions and mostly focused on intestinal parasites (30, 31). Limited information exists regarding the prevalence of parasites in free-ranging lions (32). Parasites in lions have been documented in various regions, including Tanzania, Ethiopia, South Africa, the USA, Italy, India, and Malaysia (16, 18, 19, 31–34) with a few notable exceptions where a particular group of parasites has been studied, such as blood parasites (35–39), intestinal parasites (32, 40) or where the results of veterinary examinations in a National Park (33) have been reported. A previous study reported that Dirofilaria sp. is a common parasite of lions in the Kruger Park (33), while our study found microfilaria of Brugia sp. infected in lion. The result of this study similar to result reported by Zahedi et al. (16) who found microfilaria of Brugia in a clouded leopard (Neofelis nebulosa). However, the study found a mixed infection of B. pahangi and D. immitis, which differs from our study that reported the infection of B. malayi.
The criteria for morphological identification of microfilariae include their size, sheath, and nuclear column (41). Filarial infection with multiple species and morphological alterations of microfilariae are not easily differentiated morphologically even by trained persons (42). Histochemical staining to detect acid phosphatase activity can overcome most of these problems (43). Our result showed the lion microfilariae are sheathed with four points staining pattern with acid phosphatase activity at the amphid (AM), excretory pore (EP), anal pore (AP) and phasmid (PM). Moreover, B. malayi microfilaria had bright red points that were distinctly visible even under low power. Species identification based on morphology also requires analysis of the adult-stage filaria. Unfortunately, our study did not find adult-stage filaria in lions. Nevertheless, the prevalence of microfilaria in lions should be noted and sampled for further analysis.
Routinely, diagnosis is carried out through microscopic examination of the morphology in microfilariae isolated from blood. It is known that B. malayi and B. pahangi are very similar morphologically. However, species identification by Giemsa staining is not sufficient (43). Although acid phosphatase staining is effective it is not reproducible and the procedure is complicated (44). Molecular methods based on species-specific PCRs are simple and easy to perform and have been introduced for discrimination of Brugia sp. (6, 44). Molecular diagnostic methods, based on the amplification of parasite DNA by PCR methods have the advantage of being more sensitive in detecting parasites than the usual microscopy methods, especially in case of low mf densities, as well as increased certainty in the identification of the species or even strain level. This study applied molecular phylogenetic analysis of 12S rDNA, 18S rDNA, COI, and wsp nucleotide sequences for confirmation of Brugia sp. in a single lion reservoir. In the tree analysis, the phylogenetic tree based on 12S rDNA, 18S rDNA and COI gene sequences were grouped with B. malayi sequences from other animals and other countries such as Thailand, Vietnam and USA. Notably, a B. malayi isolate obtained from a lion in a Thai zoo exhibited close genetic proximity to isolates from humans and dogs, suggesting a potential reservoir host. While suspicions of B. malayi presence in dogs have persisted concerning lymphatic filariasis, reports of B. malayi infection in lions were previously nonexistent. Our findings provide evidence for the potential role of lions as reservoir hosts for B. malayi. Moreover, there have been reports of B. malayi infection in humans in southern Thailand, particularly in regions near the Thai-Malaysian border. Studies have identified Mansonia sp. as a significant carrier of infective Brugia larvae in these areas, suggesting its role as one of the primary vectors for B. malayi in southern Thailand (45, 46). However, conclusive determination of the lion’s reservoir capacity in endemic areas necessitates further on-site investigations involving animals in the vicinity of the zoo.
The primer set COXI-int-F and COXI-int-R employed in our study functioned as universal primers, capable of amplifying the COI region from 11 diverse species of blood and tissue filariae, including B. malayi, B. pahangi, D. immitis, D. repens, Wuchereria bancrofti, and Onchocerca spp. (25). In a 2019 study by Satjawongvanit et al., the detection of filarial nematode DNA in blood samples from domestic dogs in the Bangkok Metropolitan Region, Thailand, was accomplished using COI and internal transcribed spacer 1 (ITS1) gene-based PCR. Based on partial nucleotide sequences of the COI gene (~690 bp), they identified three species in domestic dogs: D. immitis (57.89%), B. pahangi (22.81%), and B. malayi (7.02%). The study emphasized the utility of the COI gene as a valuable marker for distinguishing between D. immitis, B. pahangi, and B. malayi, making COI-based PCR a suitable tool for detecting filarial nematode infections in dogs (47). Additionally, mitochondrial 12S rDNA primers were employed to construct the phylogeny. Although both the 12S rDNA and COI gene were deemed useful markers for detecting filarial nematodes in our study, Gaillard et al. (48) reported that the phylogenetic analyses and amplification of the 12S rDNA gene demonstrated less discriminating power compared to the COI fragment.
The phylogenetic trees of wsp gene for Wolbachia endosymbiont were distinctly separated into 2 groups B. malayi and B. pahangi. However, the results of wsp showed little differences in nucleotide sequences between the two Brugia species with is differs from previous reports by Bazzocchi et al. (49) who showed that the sequences of B. malayi and B. pahangi were grouped in wsp phylogeny. However, all the phylogenetic relationships which are unquestioned for the host nematodes are matched by the Wolbachia phylogeny based on wsp this indicated that the Wolbachia-filaria association is stable and species-specific.
5 Conclusion
To the authors’ knowledge, this case represents the initial occurrence of Brugia infection in a captive lion from Thailand. The nematode was identified through both morphological and molecular methods, revealing a B. malayi infection, which has been documented in domestic animals within Thailand. Nonetheless, our findings bring attention to the insufficient understanding concerning the diversity of Brugia species and the interactions between hosts and parasites in wildlife animals from Thailand. This knowledge gap has potential implications in the fields of veterinary medicine and public health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The requirement of ethical approval was waived by Biosafety Committee of Chulalongkorn University, Faculty of Veterinary Science (IBC 2231033) for the studies involving animals because this study was a part of the routine health check at a private farm, for which ethical approval was not obligatory. All procedures conducted during the study adhered to the relevant guidelines and regulations. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was not obtained from the owners for the participation of their animals in this study because this was a part of the routine health check at Nakhon Ratchasima Zoo, Zoological Park Organization of Thailand, for which ethical approval was not obligatory.
Author contributions
WJ: Conceptualization, Investigation, Methodology, Validation, Writing – original draft. PK: Investigation, Methodology, Visualization, Writing – original draft. AP: Investigation, Resources, Visualization, Writing – original draft. PT: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported the Thailand Science Research and Innovation Fund, Chulalongkorn University (FOODF67310022), Thailand and the Second Century Fund (C2F), Chulalongkorn University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1376208/full#supplementary-material
SUPPLEMENTARY FIGURE 1. | Phylogenetic tree based on an alignment of COI gene sequences. Bootstrap confidence values (1,000 replicates) are shown as percentages. Values less than 50% are omitted. The units for the scale bar are substitutions per site. NCBI accession numbers are included. The COI sequence of B. malayi from lion in Thailand, with taxon name shown in blue color and bold font, was generated as part of this study. Letters to the right of the bracketed branches denote the clusters. Setaria tundra and S. labiatopapilosa were used as out group.
SUPPLEMENTARY FIGURE 2. | Phylogenetic tree based on an alignment of wsp sequences. Bootstrap confidence values (1,000 replicates) are shown as percentages. Values less than 50% are omitted. The units for the scale bar are substitutions per site. NCBI accession numbers are included. The wsp sequence of B. malayi from lions in Thailand, with taxon name shown in blue color and bold font, was generated as part of this study. Letters to the right of the bracketed branches denote the clusters. Wolbachia endosymbiont of Aedes albopictus was used as out group.
References
1. Eberhard, ML. Zoonotic filariasis. Trop Med Infect Dis. (2006) 2:1189–203. doi: 10.1016/B978-0-443-06668-9.50106-X
2. Bizhani, N, Hafshejani, SH, Mohammadi, N, Rezaei, M, and Rokni, MB. Lymphatic filariasis in Asia: a systematic review and meta-analysis. Parasitol Res. (2021) 120:411–22. doi: 10.1007/s00436-020-06991-y
3. Dickson, BFR, Graves, PM, and McBride, WJ. Lymphatic filariasis in mainland Southeast Asia: a systematic review and meta-analysis of prevalence and disease burden. Trop Med Infect Dis. (2017) 2:32. doi: 10.3390/tropicalmed2030032
4. Edeson, JFB, and Wilson, T. The epidemiology of filariasis due to Wuchereria bancrofti and Brugia malayi. Annu Rev Entomol. (1964) 9:245–68. doi: 10.1146/annurev.en.09.010164.001333
5. Kanjanopas, K, Choochote, W, Jitpakdi, A, Suvannadabba, S, Loymak, S, Chungpivat, S, et al. Brugia malayi in a naturally infected cat from Narathiwat province, southern Thailand. Southeast Asian J Trop Med Public Health. (2001) 32:585–7.
6. Chansiri, K, Tejangkura, T, Kwaosak, P, Sarataphan, N, Phantana, S, and Sukhumsirichart, W. PCR based method for identification of zoonostic Brugia malayi microfilariae in domestic cats. Mol Cell Probes. (2002) 16:129–35. doi: 10.1006/mcpr.2001.0402
7. Nuchprayoon, S, Sangprakarn, S, Junpee, A, Nithiuthai, S, Chungpivat, S, and Poovorawan, Y. Differentiation of Brugia malayi and Brugia pahangi by PCR-RFLP of ITS1 and ITS2. Southeast Asian J Trop Med Public Health. (2003) 34:67–73.
8. Rawangchue, T, Sripirom, N, and Sungpradit, S. Surveillance of zoonotic Brugia pahangi in monastery cats, Samphran district, Nakhon Pathom, Thailand. Thai J Vet Med. (2022) 52:117–25. doi: 10.56808/2985-1130.3196
9. Denham, DA, and Mcgreevy, PB. Brugian filariasis: epidemiological and experimental studies. Adv Parasitol. (1977) 15:243–309. doi: 10.1016/S0065-308X(08)60530-8
10. Lau, Y, Lee, W, Xia, J, Zhang, G, Razali, R, Anwar, A, et al. Draft genome of Brugia pahangi: high similarity between B. Pahangi and B. Malayi. Parasit Vectors. (2015) 8:451. doi: 10.1186/s13071-015-1064-2
11. Palmieri, JR, Ratiwayanto, S, Masbar, S, Tirtokusumo, S, Rusch, J, and Marwoto, HA. Evidence of possible natural infections of man with Brugia pahangi in South Kalimantan (Borneo), Indonesia. Trop Geogr Med. (1985) 37:239–44.
12. Tan, LH, Fong, MY, Mahmud, R, Muslim, A, Lau, YL, and Kamarulzaman, A. Zoonotic Brugia pahangi filariasis in a suburbia of Kuala Lumpur City, Malaysia. Parasitol Int. (2011) 60:111–3. doi: 10.1016/j.parint.2010.09.010
13. Kennedy, S, and Patton, S. Heartworms in a Bengal Tiger (Panthera tigris). J Zoo Anim Med. (1981) 12:20–2. doi: 10.2307/20094502
14. Atkins, C, Moresco, A, and Litster, A. Prevalence of naturally occurring Dirofilaria immitis infection among nondomestic cats housed in an area in which heartworms are endemic. J Am Vet Med Assoc. (2005) 227:139–43. doi: 10.2460/javma.2005.227.139
15. Murata, K, Yanai, T, Agatsuma, T, and Uni, S. Dirofilaria immitis infection of a snow leopard (Uncia uncia) in a Japanese zoo with mitochondrial DNA analysis. J Vet Med Sci. (2003) 65:945–7. doi: 10.1292/jvms.65.945
16. Zahedi, M, Vellayan, S, Jeffery, J, and Krishnasamy, M. A case of double infection with Brugia pahangi Buckley and Edeson, 1956 and Dirofilaria immitis Leidy 1856, in a Malaysian clouded leopard, Neofelis nebulosa. Vet Parasitol. (1986) 21:135–7. doi: 10.1016/0304-4017(86)90154-8
17. Ruiz de Ybáñez, MR, Martínez-Carrasco, C, Martínez, JJ, Ortiz, JM, Attout, T, and Bain, O. Dirofilaria immitis in an African lion (Panthera leo). Vet Rec. (2006) 158:240–2. doi: 10.1136/vr.158.7.240
18. Mazzariol, S, Cassini, R, Voltan, L, Aresu, L, and Regalbono, AF. Heartworm (Dirofilaria immitis) infection in a leopard (Panthera pardus pardus) housed in a zoological park in North-Eastern Italy. Parasit Vectors. (2010) 3:25. doi: 10.1186/1756-3305-3-25
19. Lamm, MG, Roelke, ME, Greiner, EC, and Steible, CK. Microfilariae in the free-ranging Florida panther (Felis concolor coryi). J Helminthol Soc Wash. (1997) 64:137–41.
20. Nelson, GS, Heisch, RB, and Furlong, M. Studies in filariasis in East Africa. II. Filarial infections in man, animals, and mosquitoes on the Kenya coast. Trans R Soc Trop Med Hyg. (1962) 56:202–17. doi: 10.1016/0035-9203(62)90155-4
21. Rosenblatt, JE. Laboratory diagnosis of infections due to blood and tissue parasites. Clin Infect Dis. (2009) 49:1103–8. doi: 10.1086/605574
22. Chalifoux, L, and Hunt, RD. Histochemical differentiation of Dirofilaria immitis and Dipetalonema reconditum. J Am Vet Med Assoc. (1971) 158:601–5.
23. Casiraghi, M, Bain, O, Guerrero, R, Martin, C, Pocacqua, V, Gardner, SL, et al. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int J Parasitol. (2004) 34:191–203. doi: 10.1016/j.ijpara.2003.10.004
24. Laidoudi, Y, Ringot, D, Watier-Grillot, S, Davoust, B, and Mediannikov, O. A cardiac and subcutaneous canine dirofilariosis outbreak in a kennel in Central France. Parasite. (2019) 26:72. doi: 10.1051/parasite/2019073
25. Casiraghi, M, Anderson, T, Bandi, C, Bazzocchi, C, and Genchi, C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. (2001) 122:93–103. doi: 10.1017/S0031182000007149
26. Junsiri, W, Kamkong, P, Chinkangsadarn, T, Ouisuwan, S, and Taweethavonsawat, P. Molecular identification and genetic diversity of equine ocular setariasis in Thailand based on the COI, 12S rDNA, and ITS1 regions. Infect Genet Evol. (2023) 110:105425. doi: 10.1016/j.meegid.2023.105425
27. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, C. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
28. Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. (1985) 39:783–91. doi: 10.2307/2408678
29. Nei, M, and Kumar, S. Molecular evolution and phylogenetics. New York: Oxford University Press (2000).
30. Müller-Graf, CDM. A Coprological survey of intestinal parasites of wild lions (Panthera leo) in the Serengeti and the Ngorongoro crater, Tanzania, East Africa. J Parasitol. (1995) 81:812–4. doi: 10.2307/3283987
31. Dashe, D, and Berhanu, A. Study on gastrointestinal parasitism of wild animals in captivity at the zoological garden of Haramaya University, Ethiopia. Open J Vet Med. (2020) 10:173–84. doi: 10.4236/ojvm.2020.109015
32. Bjork, KE, Averbeck, GA, and Stromberg, BE. Parasites and parasite stages of free-ranging wild lions (Panthera leo) of northern Tanzania. J Zoo Wildl Med. (2000) 31:56–61. doi: 10.1638/1042-7260(2000)031[0056:PAPSOF]2.0.CO;2
33. Young, E. Some important parasitic and other diseases of lion, Panthera leo. In the Kruger1national park. J S Afr Vet Assoc. (1975) 46:181–3.
34. Pawar, RM, Lakshmikantan, U, Hasan, S, Poornachandar, A, and Shivaji, S. Detection and molecular characterization of ascarid nematode infection (Toxascaris leonina and Toxocara cati) in captive Asiatic lions (Panthera leo persica). Acta Parasitol. (2012) 57:67–73. doi: 10.2478/s11686-012-0012-y
35. Dennig, HK, and Brocklesby, DW. Babesia pantherae sp. nov., a piroplasm of the leopard (Panthera pardus). Parasitology. (1972) 64:525–32. doi: 10.1017/S0031182000045595
36. Averbeck, GA, Bjork, KE, Packer, C, and Herbst, L. Prevalence of hematozoans in lions (Panthera leo) and cheetah (Acinonyx jubatus) in Serengeti National Park and Ngorongoro crater, Tanzania. J Wildl Dis. (1990) 26:392–4. doi: 10.7589/0090-3558-26.3.392
37. Lopez-Rebollar, LM, Penzhorn, BL, de Waal, DT, and Lewis, BD. A possible new piroplasm in lions from the republic of South Africa. J Wildl Dis. (1999) 35:82–5. doi: 10.7589/0090-3558-35.1.82
38. Bosman, AM, Venter, EH, and Penzhorn, BL. Occurrence of Babesia felis and Babesia leo in various wild felid species and domestic cats in southern Africa, based on reverse line blot analysis. Vet Parasitol. (2007) 144:33–8. doi: 10.1016/j.vetpar.2006.09.025
39. Squarre, D, Nakamura, Y, Hayashida, K, Kawai, N, Chambaro, H, Namangala, B, et al. Investigation of the piroplasm diversity circulating in wildlife and cattle of the greater Kafue ecosystem, Zambia. Parasit Vectors. (2020) 13:599. doi: 10.1186/s13071-020-04475-7
40. Mukarati, NL, Vassilev, GD, Tagwireyi, WM, and Tavengwa, M. Occurrence, prevalence and intensity of internal parasite infections of African lions (Panthera leo) in enclosures at a recreation park in Zimbabwe. J Zoo Wildl Med. (2013) 44:686–93. doi: 10.1638/2012-0273R.1
41. Mathison, BA, Couturier, MR, and Pritt, BS. Diagnostic identification and differentiation of microfilariae. J Clin Microbiol. (2019) 57:e00706–19. doi: 10.1128/JCM.00706-19
42. Ravindran, R, Varghese, S, Nair, SN, Balan, VM, Lakshmanan, B, Ashruf, RM, et al. Canine filarial infections in a human Brugia malayi endemic area of India. Biomed Res Int. (2014) 2014:630160:1–9. doi: 10.1155/2014/630160
43. Yen, PK, and Mak, JW. Histochemical differentiation of Brugia, Wuchereria, Dirofilaria and Breinlia microfilariae. Ann Trop Med Parasitol. (1978) 72:157–62. doi: 10.1080/00034983.1978.11719298
44. Nuchprayoon, S, Junpee, A, Poovorawan, Y, and Scott, AL. Detection and differentiation of filarial parasites by universal primers and polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. (2005) 73:895–900. doi: 10.4269/ajtmh.2005.73.895
45. Meetham, P, Kumlert, R, Gopinath, D, Yongchaitrakul, S, Tootong, T, Rojanapanus, S, et al. Five years of post-validation surveillance of lymphatic filariasis in Thailand. Infect Dis Poverty. (2023) 12:113. doi: 10.1186/s40249-023-01158-0
46. Zielke, E, Hinz, E, and Sucharit, S. Lymphatic filariasis in Thailand: a review on distribution and transmission. Tropenmed Parasitol. (1993) 15:141–8.
47. Satjawongvanit, H, Phumee, A, Tiawsirisup, S, Sungpradit, S, Brownell, N, Siriyasatien, P, et al. Molecular analysis of canine filaria and its Wolbachia endosymbionts in domestic dogs collected from two animal university hospitals in Bangkok metropolitan region, Thailand. Pathogens. (2019) 8:114. doi: 10.3390/pathogens8030114
48. Gaillard, CM, Pion, SD, Hamou, H, Sirima, C, Bizet, C, Lemarcis, T, et al. Detection of DNA of filariae closely related to Mansonella perstans in faecal samples from wild non-human primates from Cameroon and Gabon. Parasit Vectors. (2020) 13:313. doi: 10.1186/s13071-020-04184-1
Keywords: Brugia malayi, lion, molecular analysis, PCR, Thailand
Citation: Junsiri W, Kamkong P, Phojun A and Taweethavonsawat P (2024) Unveiling zoonotic threats: molecular identification of Brugia sp. infection in a lion. Front. Vet. Sci. 11:1376208. doi: 10.3389/fvets.2024.1376208
Edited by:
Sirikachorn Tangkawattana, Khon Kaen University, ThailandReviewed by:
Georgiana Deak, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaZorica D. Dakić, University of Belgrade, Serbia
Copyright © 2024 Junsiri, Kamkong, Phojun and Taweethavonsawat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyanan Taweethavonsawat, UGl5YW5hbi5UQGNodWxhLmFjLnRo
 Witchuta Junsiri1
Witchuta Junsiri1 Piyanan Taweethavonsawat
Piyanan Taweethavonsawat