- 1Liaocheng Research Institute of Donkey High-Efficiency Breeding and Ecological Feeding, Liaocheng University, Liaocheng, China
- 2Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
Omics methodologies, such as genomics, transcriptomics, proteomics, metabolomics, lipidomics and microbiomics, have revolutionized biological research by allowing comprehensive molecular analysis in livestock animals. However, despite being widely used in various animal species, research on donkeys has been notably scarce. China, renowned for its rich history in donkey husbandry, plays a pivotal role in their conservation and utilization. China boasts 24 distinct donkey breeds, necessitating conservation efforts, especially for smaller breeds facing extinction threats. So far, omics approaches have been employed in studies of donkey milk and meat, shedding light on their composition and quality. Similarly, omics methods have been utilized to explore the molecular basis associated with donkey growth, meat production, and quality traits. Omics analysis has also unraveled the critical role of donkey microbiota in health and nutrition, with gut microbiome studies revealing associations with factors such as pregnancy, age, transportation stress, and altitude. Furthermore, omics applications have addressed donkey health issues, including infectious diseases and reproductive problems. In addition, these applications have also provided insights into the improvement of donkey reproductive efficiency research. In conclusion, omics methodologies are essential for advancing knowledge about donkeys, their genetic diversity, and their applications across various domains. However, omics research in donkeys is still in its infancy, and there is a need for continued research to enhance donkey breeding, production, and welfare in China and beyond.
1 Introduction
In the field of biology, the term “omics” is commonly used to denote scientific disciplines focused on the comprehensive characterization of molecular components originating from various biological layers within living organisms. These layers encompass DNA, RNA, proteins, and metabolites, and their analysis is facilitated by high-throughput technologies (1–4). Recent advancements in both computational and experimental methodologies have substantially enhanced our capacity to profile multiple levels of cellular regulation, including the genome, transcriptome, epigenome, chromatin conformation, and metabolome, among other well-established “omics” (5–13). Consequently, these omics analyses have found widespread application in animal research in recent years (14, 15), as evidenced by the studies in cattle examining their role in production (16, 17), reproduction (18, 19), and metabolism/microbiome (20–22). Similarly, the contributions of omics techniques to understanding various traits in pigs (23–28), sheep, and goats (29) as well as chickens (30–32) have been comprehensively explored.
Furthermore, the utility of omics approaches extends to the authentication and assessment of various animal-derived products, including meat (33–40) and milk (41, 42). Surprisingly, despite these extensive investigations in multiple animal species, there is a notable absence of comprehensive reviews addressing the application of omics methodologies in donkey research. Therefore, this present review article aims to fill this gap by providing an overview of the genetic resources available for donkeys and examining the utilization of omics techniques in enhancing the productive and reproductive traits of donkeys in China. Additionally, this review delves into the role of omics applications in evaluating donkey-derived products, such as milk and meat.
2 Methodology for literature search
The central objective of this review article was to comprehensively investigate the evolving landscape of Omics applications within the domain of donkey research, with a particular emphasis on the Chinese context. This entailed conducting a systematic literature search to identify and assess pertinent studies, focusing exclusively on Chinese local donkey breeds. To ensure the selection of the most relevant literature, we devised stringent inclusion and exclusion criteria. The articles considered for inclusion were those published in recent years, which explored the utilization of Omics methodologies in livestock research, including pigs, cattle, sheep, goats, and poultry. Our review, however, honed in exclusively on studies related to donkeys, with a specific geographical focus on China. We meticulously chose a set of keywords that encapsulated the core themes of our review, including “donkey milk,” “donkey meat,” “donkey microbiome,” “donkey health,” “donkey reproduction,” and the overarching category of “omics.” Within the Omics umbrella, we included subdisciplines such as “genomics,” “transcriptomics,” “proteomics,” “metabolomics,” “microbiomics,” and “lipidomics.” Utilizing these keywords, we conducted an exhaustive search across reputable academic databases, including but not limited to PubMed, Scopus, Web of Science, and relevant academic journals. Following the initial search, the identified articles were subject to a rigorous screening process. Each article was assessed for relevance based on its title, abstract, and keywords. Only those articles meeting the criteria of investigating Omics applications in donkey research, particularly within the Chinese context, were retained. Relevant data from the selected articles were meticulously extracted and cataloged for subsequent analysis. This included information on the research objectives, methodologies employed, key findings, and any notable insights regarding the application of omics techniques in donkey research. The extracted data were synthesized, and patterns, trends, and advancements in the field of Omics applications in Chinese donkey research were critically evaluated. These insights were then organized and presented in a coherent manner within the review article.
3 Assessing the donkey welfare, distribution, and genetic resources in China
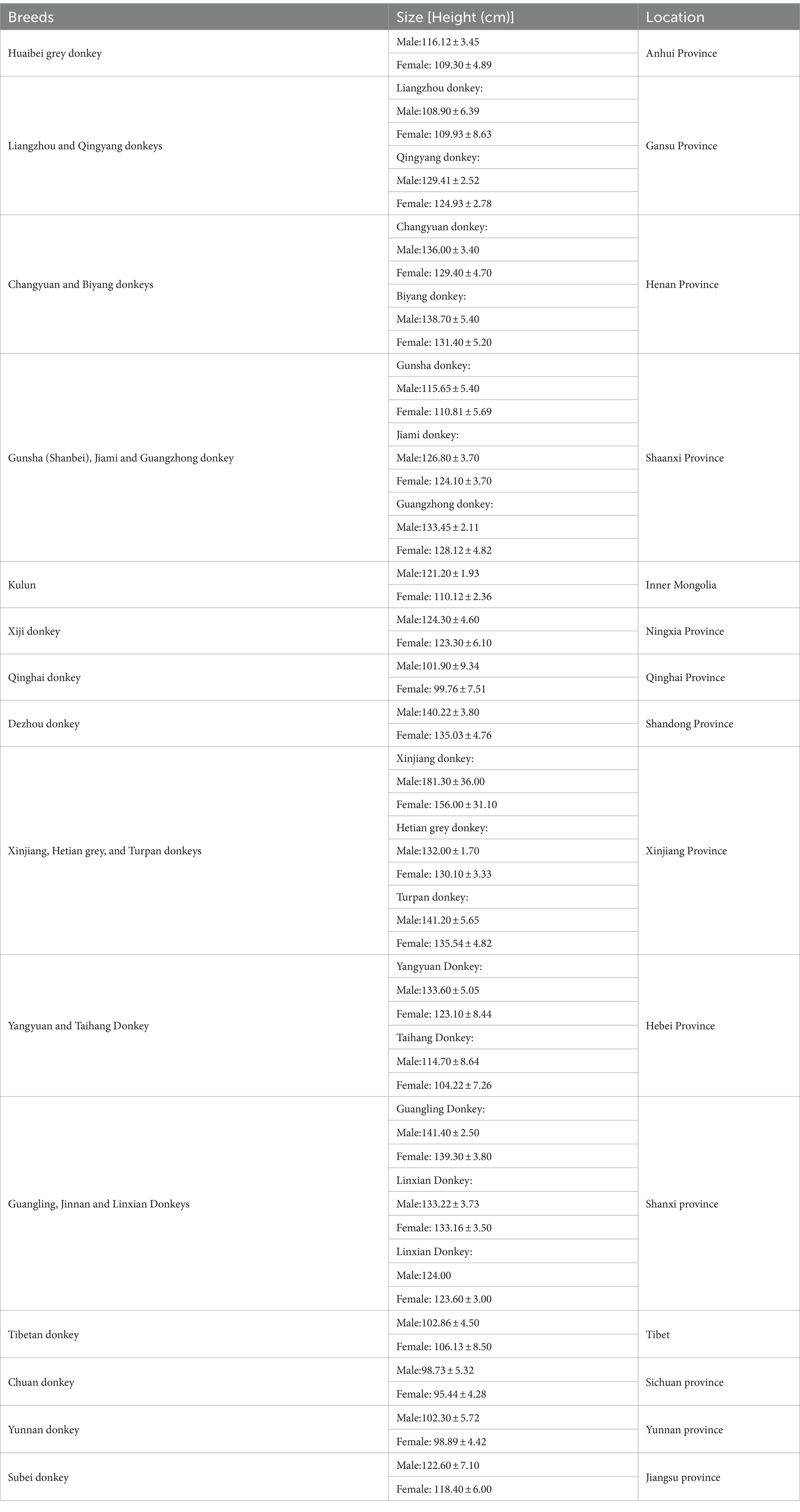
Donkey as a livestock animal has been ignored due to increased industrialization and mechanization because communities which previously relied on donkey traction now use motorized vehicles and machinery (43). In contrast, considerable attention has been given to the management and welfare and improvement of genetic resources and production of donkeys in China (44–49). According to our recent survey in different areas of China, we identified that there are well established and well-equipped donkey farms are existing in China (44). China realizes the importance of donkey as other livestock animals and still has reported 24 donkey breeds and number one in conservation of local donkey breeds [(48), https://zypc.nahs.org.cn/pzml/classify.html]. As per Food and Agricultural Organization report in 2020, the global donkey population was 50.45 million, of which 2.68 million are raised in China (47, 50). In China, donkeys are raised for milk, meat and hide production which might be another reason for the growing attention (44, 51–53).
The donkey is believed to have undergone a process of domestication approximately 5,000 years ago in Egypt, stemming from the African wild ass (54). This domestication event May have been prompted by environmental changes leading to a drier climate in the region. Previous research has suggested the possibility of dual domestication events, likely originating from the Nubian and Somali wild ass subspecies. These proposals are supported by patterns of mitochondrial DNA variation identified in both ancient and contemporary donkey populations (54–59). It is noteworthy that some extant subspecies of wild asses, as well as specific donkey breeds, face critical endangerment, leading to substantial conservation efforts (60). The pivotal contribution of genomics to the elucidation of the evolutionary history of equids has been comprehensively documented in a recently published studies (61–67). Consequently, researchers utilized the Chicago HiRise assembly technology to create a high-quality donkey genome assembly with sub-chromosomal scaffolds (60). This newly developed assembly has the potential to facilitate accurate assessments of heterozygosity in equine species beyond the horse, both at the genome-wide and local levels. Additionally, it aids in the detection of runs of homozygosity, which could be indicative of positive selection in domestic donkeys. Moreover, this advanced genome assembly enabled the identification of fine-scale chromosomal rearrangements between horses and donkeys, likely contributing to their divergence and eventual speciation (60). Recently, Liu et al. conducted genome-wide analyses using a novel donkey 40 K liquid SNP chip to study coat color diversity in the Chinese Dezhou donkey. However, SNP-Chip based for diversity purposes in donkey research is still in infancy (68).
China boasts a 4,000-year history of donkey husbandry and possesses abundant genetic resources in this domain (69–71). Animal genetic resources represent an indispensable facet of our genetic, economic, and cultural legacy, serving as a pivotal driver within the spheres of the economy, food production, regional identity, and ecosystem services (48, 72). Within this framework, it is crucial to recognize that local breeds, including those of donkeys, which May have lost their original purpose, are frequently confronting a dire existential threat. Consequently, there is an imperative need for concerted efforts aimed at their rehabilitation and reintegration into alternative economic utilization programs. Previous research has categorized Chinese donkey breeds into three regional branches: North China Plain, Loess Plateau, and Southwest China Plateau (73). The geographical distribution of these donkey breeds in China is primarily concentrated in Liaoning, Shanxi, Xinjiang, Inner Mongolia, Gansu, and Shandong Provinces (44, 48).
Recent research has identified a diverse array of donkey breeds within China’s genetic resources. These breeds encompass the Huaibei grey, Liangzhou, Qingyang, Changyuan, Biyang, Gunsha, Jiami, Guangzhong, Kulun, Xiji, Qinghai, Dezhou, Xinjiang, Hetian Grey, Turpan, Yangyuan, Taihang, Guangling, Jinnan, Linxian, Chuan, Xizang, Yunnan and Subei donkeys [(44, 74, 75), https://zypc.nahs.org.cn/pzml/classify.html]. These donkey breeds exhibit substantial variations in terms of body weight and size, with weight ranging from 130 to 260 kg and a height measuring between 110 cm to 130 cm. Morphologically, donkeys can be categorized into three main size groups: large-sized (Guanzhong, Hetian, Turpan, Changyuan, Jinnan, Guangling, and Dezhou donkeys), medium-sized (Jiami, Linxian, Biyang, Yangyuan and Qingyang donkeys), and small-sized (Kulun, Tibetan, Chuan, Subei, Huaibei, Xinjiang, Qinghai, Jiami Liangzhou, Taihang, Gunsha and Yunnan donkeys).1 These categories correspond to heights above 130 cm, between 115 cm and 125 cm, and below 110 cm, respectively (48). Among all donkey breeds, Dezhou donkey located in Shandong has extensively studied because of its heavy body, body height and length, predominantly black hair with a straight back and waist, arch of chest rib and round, firm hooves (44).
In China, traditional donkey breeding farms historically focused on a single local donkey breed. However, in a recent survey, 16 different local donkey breeds were identified. Notably, the Dezhou donkey currently dominates the Chinese donkey population, constituting over 57% of the total donkey population (44). This finding aligns with the findings of a prior study (48). In contrast, smaller-sized breeds such as Kulun, Qingyang, Xiji and Huaibei Grey donkeys constitute a significantly smaller proportion (44). This discrepancy is likely due to the relatively lower production value of meat and hide associated with these smaller breeds. Unfortunately, these smaller donkey breeds have faced indiscriminate slaughter in recent years, raising concerns about their potential extinction (57, 76). The details of donkey breeds and their location in China has been provided in Table 1. To preserve the genetic diversity of these smaller donkey breeds, it is crucial to establish dedicated breeding farms that are specifically designed for their conservation.
The breeds and their location information were obtained from following published sources [(44, 48, 49, 58, 73–82), https://zypc.nahs.org.cn/pzml/classify.html].
4 Utilization of omics approaches in donkey milk production research
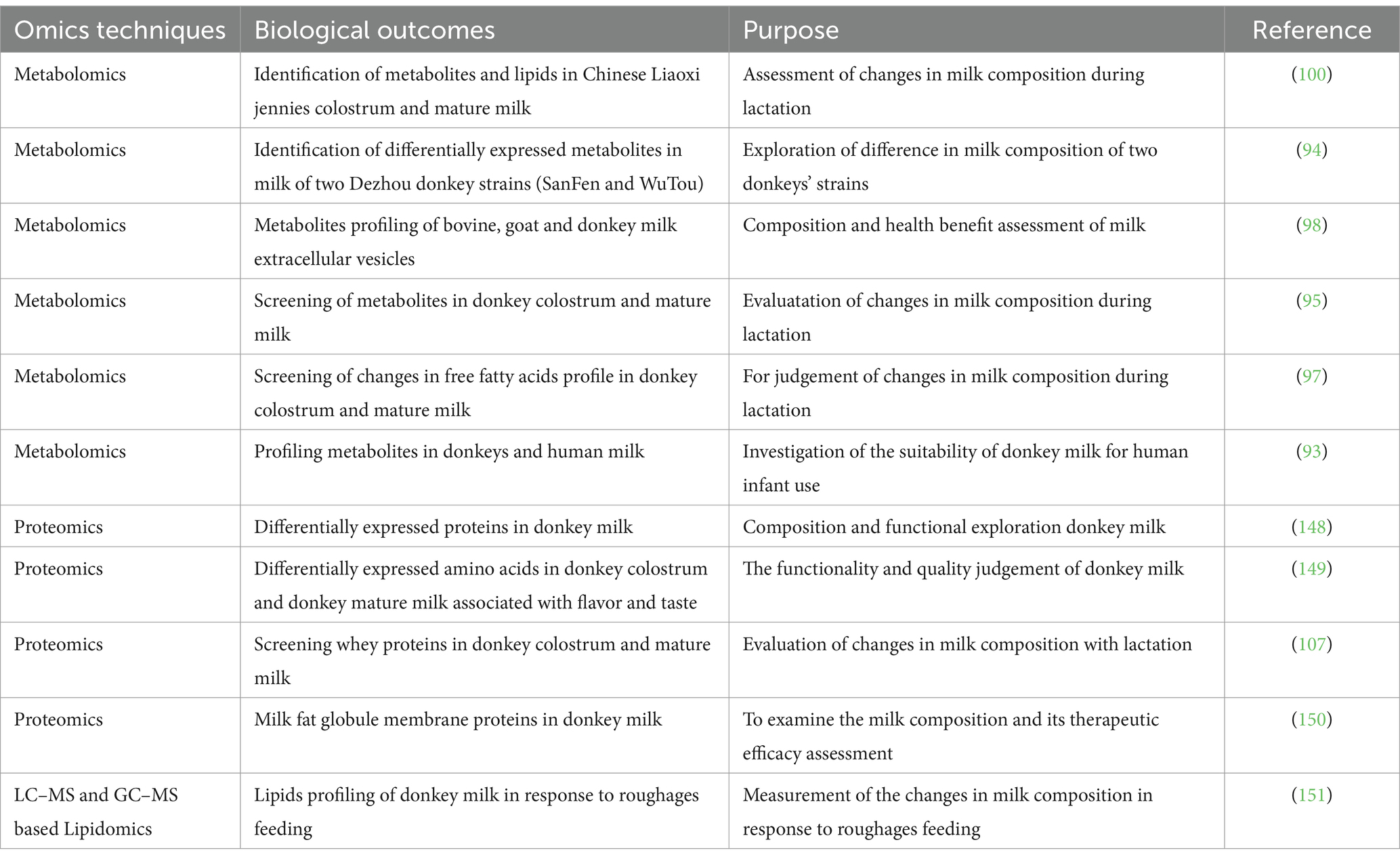
The research development on omics application in donkey milk research has been summarized in Table 2. New analytical technologies, with mass spectrometry being at the forefront, facilitate the generation of improved and innovative milk products based on the growing knowledge and understanding of milk bioactive compounds such as proteins, carbohydrates, lipids, and minerals, at global scale. The molecular understanding of biological milk function has emerged as a central theme in nutritional research (83–85). Mass spectrometry-based techniques enable the characterization of human and animal milk components not only in native fresh but also in processed milk. In recent years, the application of omics technologies has gained prominence in the field of donkey milk production research (86–92). Particularly, metabolomics, lipidomics, transcriptomics, and proteomics have played significant roles in advancing our understanding of various aspects of donkey milk and its potential applications. This paper provides a comprehensive overview of key findings and studies conducted in these areas.
Metabolomic research in donkey milk has primarily focused on comparing and characterizing metabolite profiles. Significant studies have been conducted in the field of metabolomics, including notable investigations such as the examination of metabolite profiles in both donkey milk and human milk through GC–MS analysis (93). Additionally, Li et al. (94) conducted a metabolomic comparison involving two distinct Dezhou donkey strains, SanFen and WuTou, employing LC–MS methodology. Furthermore, an extensive analysis of metabolites within donkey milk throughout various stages of lactation was performed using un-targeted metabolomics coupled with ultra-high-performance liquid tandem chromatography quadrupole time-of-flight mass spectrometry (95). These studies shed light on the chemical composition of donkey milk, which is essential for assessing its nutritional value, bioactive compounds, technical properties, and potential diagnostic applications (96). Accordingly, a study employed a metabolomic approach to identify differentially free fatty acids and related signaling pathways in donkey milk across various lactational stages (97). Moreover, mass spectroscopy (MS) coupled with Ultrahigh-performance liquid chromatography (UHPLC) technique has been utilized to screen the metabolites in bovine, goat and donkey milk to assess the anti-inflammatory and immunoregulatory properties of milk extracellular vesicles (98). Consistently, a study found lipids and metabolites in milk and colostrum through metabolomics analysis by using UHPLC and MS (99).
Lipidomics, a vital field within metabolomics, aims to elucidate the structures of lipid molecules. Several studies have examined various lipid subgroups in donkey milk using liquid chromatography–tandem mass spectrometry, including fatty acids, polar lipids, and glycolipids (100, 101). Moreover, investigations into the differences in lipid composition of donkey milk at different lactation stages have been conducted (97, 99, 102). Notably, a study has compared the lipid profiles of donkey milk with those of cow and human milk (102). These studies have contributed valuable insights into the lipid contents of donkey milk.
The proteomics applications have been extensively discussed in donkey milk research (103–105). Furthermore, proteomic analysis has been employed to study differentially expressed whey proteins in donkey milk (106). These proteins have been linked to processes such as protein processing in the endoplasmic reticulum, estrogen signaling, progesterone-mediated oocyte maturation, and the PI3K-Akt signaling pathway (107). Studies have also reported differentially expressed whey proteins in donkey colostrum and mature milk, with implications for signaling and antigen processing pathways (95, 107). In a study utilizing the Equine 670 k Chip, a study successfully identified genes (NUMB, ADCY8, and CA8) associated with milk production traits in Xinjiang Donkeys (108). Consistently, proteomic analysis revealed some key differentially expressed proteins that were involved in regulation of complement and coagulation cascades, staphylococcus aureus infection and AGE-RAGE signaling pathways in diabetic complications (51). In addition, these proteins have key role in promoting cell proliferation, enhancing antioxidant, immunoregulation, anti-inflammatory, and antibacterial effects, and enhancing skin moisture (51). In addition a study emphasized the significance of proteomics and peptidomics in comparing proteins and endogenous peptides in human, cow, and donkey milk (109). Additionally, transcriptomic screening of donkey mammary glands has been used to identify molecular factors associated with reduced susceptibility to mastitis (110). Another study has investigated molecular mechanisms regulating bioactive milk components in mammary glands through transcriptomic profiling (111). In summary, the utilization of omics methodologies, encompassing metabolomics, lipidomics, transcriptomics, and proteomics, has made substantial contributions to our comprehension of donkey milk’s characteristics. These applications have enabled us to gain insights into the composition of donkey milk, its alterations in composition throughout the lactation period, and the ability to distinguish donkey milk from other sources. Furthermore, omics techniques have furnished us with invaluable insights into the constituent components and inherent qualities of donkey milk, thus establishing a solid groundwork for further scientific exploration and the potential for groundbreaking advancements in this particular field.
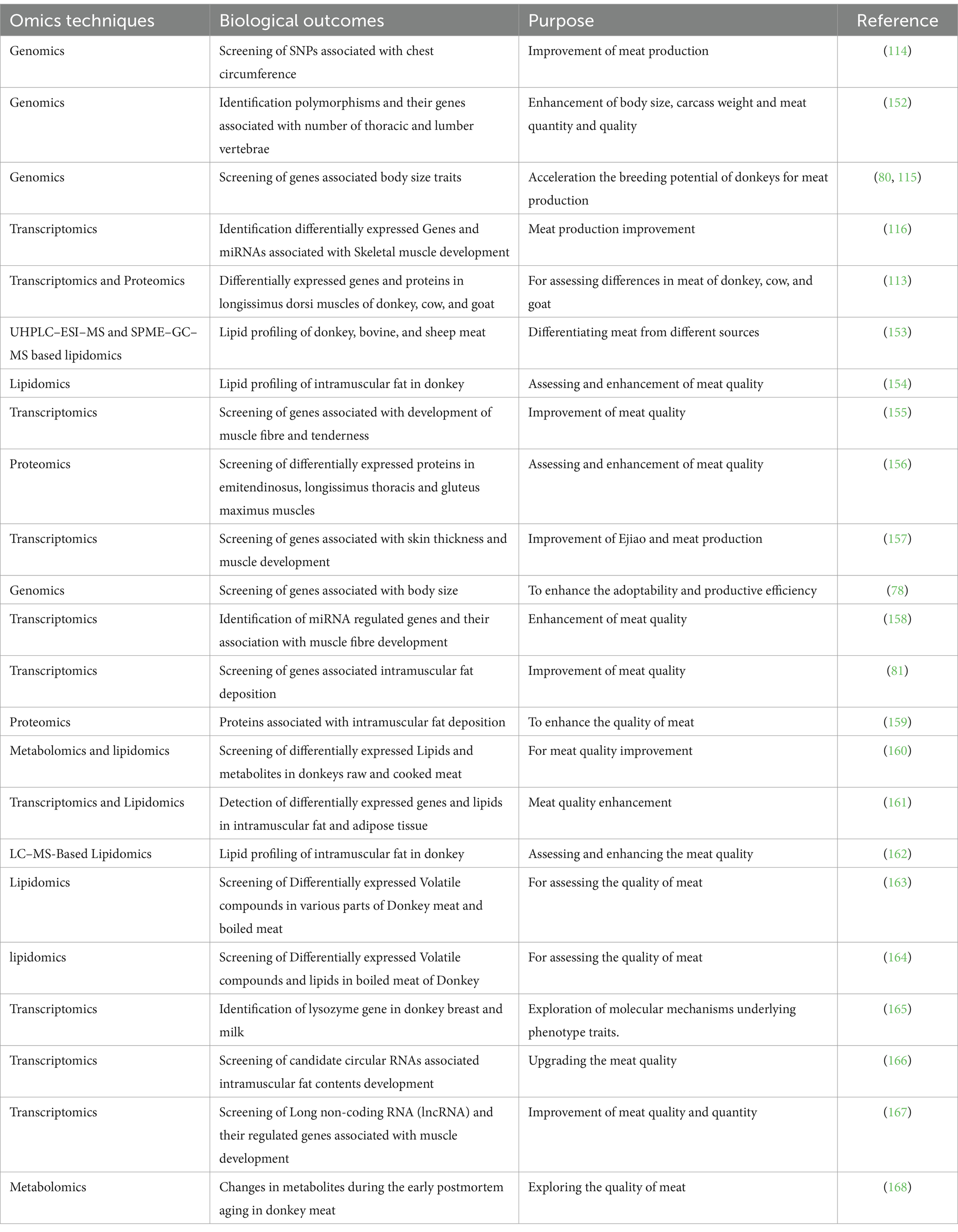
5 Utilization of omics approaches in donkey growth, meat production, and quality traits research
Recently a study conducted by Yu et al. (112) reported through transcriptomic screening several candidates Long non-coding RNAs (lncRNAs) that were involved in regulation of genes (DCN, ITM2A, MUSTN1, ARRDC2) associated with skeletal muscle development in donkeys (113). Consistently studies utilized genomic screening for polymorphisms and their genes that were associated with body size traits in Yangyuan donkeys (79) and chest circumference in Xinjiang Donkeys (114). In line with these studies, another study reported LCORL/NCAPG, FAM184B, TBX3, and IHH via Genomic screening which were associated with body height in Chinese 11 indigenous donkeys breeds (Biyang, Dezhou, Guangling, Hetian, Jiami, Kulun, Qingyang, Turpan, Tibetan, Xinjiang, and Yunnan) (115). While another study found eca-miR-1 regulated TMP3 gene via transcriptomic study, which is associated with skeletal muscle development (116). Besides, the omics methods have also been utilized to judge the quality of meat and changes in meat obtained from different sources of animals. The utilization of different omics techniques utilization in donkey growth and meat production and quality research have been summarized in Table 3.
6 Role of omics in donkey microbiota research
The gut microbiome of donkeys has garnered significant attention due to its pivotal role in donkey nutrition, as evidenced by recent studies (117, 118). A comparative investigation was conducted to assess the microbiota composition in Qinghai and Dezhou donkeys, revealing notable disparities. Specifically, it was observed that Qinghai Donkeys exhibited substantially higher flora diversity and richness in comparison to their Dezhou counterparts (119). While another study indicated that wild asses exhibit advantages over domestic donkeys in terms of dry matter digestion, gut microbial community composition, and function, it was also observed that wild asses possess a distinct intestinal flora adaptation for high altitudes on the Qinghai-Tibet plateau (120). This observation underscores the profound impact of gut microbiota on the adaptive evolution of donkeys. Utilizing advanced Omics techniques, multiple studies have consistently demonstrated a strong correlation between physiological variations and environmental changes with alterations in the gut microbiota of donkeys. These variations encompass diverse aspects such as pregnancy (121), age (122), transportation stress (122, 123) and altitude (118, 124, 125). Furthermore, these changes in gut microbiota composition and metabolite profiles have been shown to exert significant influence on maternal health, as well as the growth and development of the fetus (126–128). The relationship between feeding method and type of feed and the gut microbiota of weaned donkeys has been discussed in a recent study (129). Furthermore, the study by Huang et al. (130) found that supplementing donkeys with yeast polysaccharides significantly improved their gut microbiota and metabolites, which in turn were linked to enhanced immunity, better feed digestion, and improved growth in donkeys. Similarly, another study reported that high concentrate diet significantly improved the gut microbiota and metabolites following by enhanced of average daily gain and feed efficiency (131). By utilizing omics method, Zhang et al. (132) explored the differences in microbial diversity in small and large intestine and their impact on overall performance of donkeys (132). Recently our research team documented the dynamic changes in skin microbiota diversity and composition in donkeys of different ages and at different sites of the body (133). Collectively, these findings emphasize the imperative necessity of maintaining meticulous nutritional care and management practices for donkeys. Such measures are crucial not only for ensuring successful lactation but also for fostering optimal growth and health outcomes for both the donkeys and their foals. This scientific understanding underscores the significance of a comprehensive approach to donkey husbandry and underscores the importance of fostering a well-balanced gut microbiota for these animals’ overall well-being.
7 Omics application in donkey health research
Omics applications have gained prominence in the field of animal health (134), with recent attention being directed towards their utilization in donkey health research (135). Additionally, proteomic profiling in both neonatal and adult donkeys’ urine has proven valuable in assessing their health status (135). A multiomic approach has been employed to investigate the microbiota in the donkey hindgut, shedding light on its association with immunity and metabolism (117). Notably, whole genome sequencing has enabled the characterization of equine coronavirus obtained from donkeys with diarrhea in Shandong Province, China (136). In a study comparing jennies with and without reproductive issues, it was found that Streptococcus zooepidemicus isolates in jennies with reproductive problems exhibited a higher number of genes encoding virulence factors (137). Furthermore, a recent publication highlighted differentially expressed proteins in the serum of jennies with endometritis caused by E. coli, suggesting their potential as biomarkers for diagnosis (138). Despite these promising findings, it is evident that research in the realm of omics applications in donkey health remains limited, leaving ample room for further exploration and discovery in this domain.
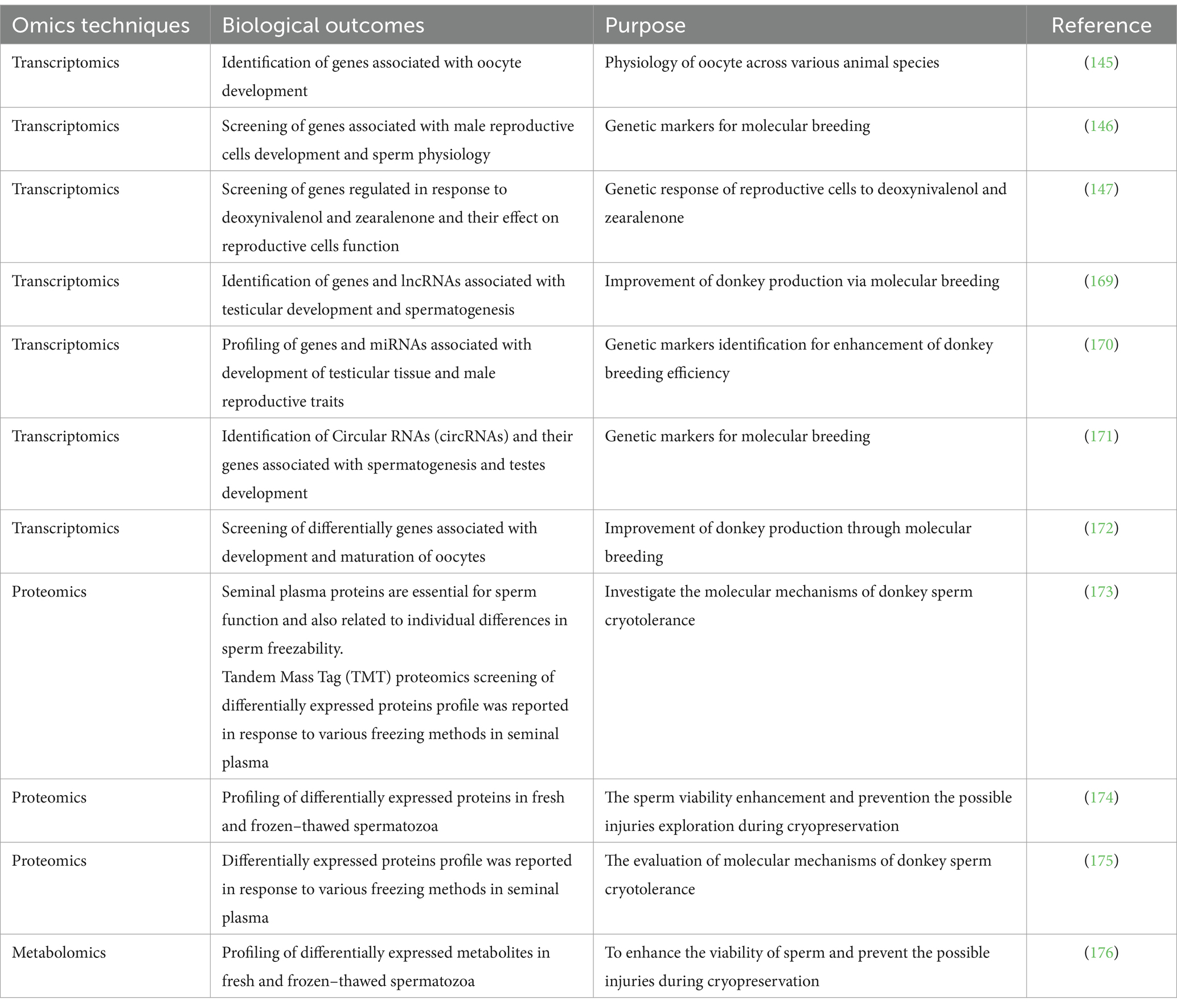
8 Omics applications in donkey reproductive research
Genetic selection and breeding are crucial tools for livestock improvement (139). Consistently, the prospective utilization of genomics, with particular emphasis on its applicability to equine diseases and fertility, has been comprehensively documented (140). The transcriptomic screening of granulosa cells in response to heat stress has been reported in our previous study (141, 142). Consistently, a study reported differentially expressed genes in Donkeys granulosa cells in response to vitamins A, D and E and micronutrients (143, 144). Furthermore, they observed that most of the differentially expressed genes were associated with steroidogenesis and follicular development (143). A study utilized transcriptomic approach and documented differentially expressed genes in donkey oocyte that were majorly associated with RNA metabolism and apoptosis (145). These findings revealed the uniqueness of donkey conclude that, compared to other species, donkey oocytes express a large number of genes related to RNA metabolism to maintain normal oocyte development during the period from germinal vesicle to metaphase II. Consistently, a study through integrative screening of miRNA and mRNA and found several genes and microRNAs associated with spermatogenesis (146). Deoxynivalenol and zearalenone, which are commonly found in feed products, exhibit serious negative effects on the reproductive systems of domestic animals. A recent study utilized transcriptomic approach to explore their negative effect on donkey endometrial cells by down regulating the androgen and estrogen secretion-linked genes and upregulating the cancer-promoting genes (147). These genes could be utilized for improvement of donkey breeding in future. For ease of reviewing, we have summarized the research development on omics application in donkey reproduction in Table 4.
9 Conclusion
Various omics technologies such as genomics, transcriptomics, metabolomics, lipidomics, and proteomics, have been used in different areas of donkey research. These include genetic resources, milk production, growth, meat quality, microbiota, health, and reproduction. However, it is important to note that the application of omics methods in donkey research is still in its early stages. There is significant potential for further exploration and discoveries in this field. Therefore, future research should concentrate on harnessing the potential of omics technologies to improve donkey health, productivity, and genetic conservation.
Author contributions
MK: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. WChe: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. XW: Data curation, Investigation, Software, Writing – review & editing. HL: Methodology, Conceptualization, Validation, Investigation, Visualization, Writing – review & editing. LW: Data curation, Investigation, Software, Writing – review & editing. BH: Data curation, Investigation, Methodology, Software, Writing – review & editing. XK: Data curation, Investigation, Software, Writing – review & editing. XL: Data curation, Methodology, Software, Writing – review & editing. ZZ: Data curation, Investigation, Software, Writing – review & editing. WCha: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – review & editing. AK: Data curation, Investigation, Methodology, Software, Writing – review & editing. YP: Data curation, Methodology, Conceptualization, Validation, Visualization, Investigation, Writing – review & editing. ChaW: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Shandong Province Modern Agricultural Technology System Donkey Industrial Innovation Team (grant number SDAIT-27), Livestock and Poultry Breeding Industry Project of the Ministry of Agriculture and Rural Affairs (grant number 19211162), Shandong Rural Revitalization Science and Technology Innovation Action Plan (Key Technology Innovation and Demonstration of Integrated Development of Dong-E Black Donkey Industry) (grant number 2021TZXD012), Open Project of Liaocheng University Animal Husbandry Discipline (grant number 319312101-14), Open Project of Shandong Collaborative Innovation Center for Donkey Industry Technology (grant number 3193308), Research on Donkey Pregnancy Improvement (grant number K20LC0901), and Liaocheng University Scientific Research Fund (grant number 318052025).
Acknowledgments
We express our sincere gratitude to the Liaocheng Research Institute of Donkey High-efficiency Breeding and Ecological Feeding, Liaocheng University for providing us financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Legarra, A, and Christensen, OF. Genomic evaluation methods to include intermediate correlated features such as high-throughput or omics phenotypes. JDS Commun. (2023) 4:55–60. doi: 10.3168/jdsc.2022-0276
2. Planell, N, Lagani, V, Sebastian-Leon, P, van der Kloet, F, Ewing, E, Karathanasis, N, et al. STATegra: multiomics data integration–a conceptual scheme with a bioinformatics pipeline. Front Genet. (2021) 12:620453. doi: 10.3389/fgene.2021.620453
3. Krassowski, M, Das, V, Sahu, SK, and Misra, BB. State of the field in multi-omics research: from computational needs to data mining and sharing. Front Genet. (2020) 11:610798. doi: 10.3389/fgene.2020.610798
4. Yugi, K, Kubota, H, Hatano, A, and Kuroda, S. Trans-omics: how to reconstruct biochemical networks across multiple “Omic” layers. Trends Biotechnol. (2016) 34:276–90. doi: 10.1016/j.tibtech.2015.12.013
5. Arias-Borrego, A, Callejón-Leblic, B, Collado, MC, Abril, N, and García-Barrera, T. Omics insights into the responses to dietary selenium. Proteomics. (2023) 23:e2300052. doi: 10.1002/pmic.202300052
6. Wang, X, Li, W, Feng, X, Li, J, Liu, GE, Fang, L, et al. Harnessing male germline epigenomics for the genetic improvement in cattle. J Anim Sci Biotechnol. (2023) 14:76. doi: 10.1186/s40104-023-00874-9
7. Shafi, A, Nguyen, T, Peyvandipour, A, Nguyen, H, and Draghici, SA. Multi-cohort and multi-omics meta-analysis framework to identify network-based gene signatures. Front Genet. (2019) 10:159. doi: 10.3389/fgene.2019.00159
8. Huang, S, Chaudhary, K, and Garmire, LX. More is better: recent progress in multi-omics data integration methods. Front Genet. (2017) 8:84. doi: 10.3389/fgene.2017.00084
9. Ramos, M, Schiffer, L, Re, A, Azhar, R, Basunia, A, Rodriguez, C, et al. Software for the integration of multiomics experiments in bioconductor. Cancer Res. (2017) 77:e39–42. doi: 10.1158/0008-5472.CAN-17-0344
10. Rohart, F, Gautier, B, Singh, A, and Lê Cao, KA. Mix omics: an R package for “omics” feature selection and multiple data integration. PLoS Comput Biol. (2017) 13:e1005752. doi: 10.1371/journal.pcbi.1005752
11. Gomez-Cabrero, D, Tarazona, S, Ferreirós-Vidal, I, Ramirez, RN, Company, C, Schmidt, A, et al. STATegra, a comprehensive multi-omics dataset of B-cell differentiation in mouse. Sci Data. (2019) 6:256. doi: 10.1038/s41597-019-0202-7
12. Gomez-Cabrero, D, Abugessaisa, I, Maier, D, Teschendorff, A, Merkenschlager, M, Gisel, A, et al. Data integration in the era of omics: current and future challenges. BMC Syst Biol. (2014) 8:I1. doi: 10.1186/1752-0509-8-S2-I1
13. González, I, Cao, KA, Davis, MJ, and Déjean, S. Visualising associations between paired "omics" data sets. BioData Min. (2012) 5:19. doi: 10.1186/1756-0381-5-19
14. Gong, Y, Li, Y, Liu, X, Ma, Y, and Jiang, L. A review of the pangenome: how it affects our understanding of genomic variation, selection and breeding in domestic animals? J Anim Sci Biotechnol. (2023) 14:1–9. doi: 10.1186/s40104-023-00860-1
15. Tan, X, He, Z, Fahey, AG, Zhao, G, Liu, R, and Wen, J. Research progress and applications of genome-wide association study in farm animals. Anim Res One Health. (2023) 1:56–77. doi: 10.1002/aro2.14
16. Behren, LE, König, S, and May, K. Genomic selection for dairy cattle behaviour considering novel traits in a changing technical production environment. Genes. (2023) 14:1933. doi: 10.3390/genes14101933
17. Kaur, H, Kaur, G, Gupta, T, Mittal, D, and Ali, SA. Integrating omics Technologies for a Comprehensive Understanding of the microbiome and its impact on cattle production. Biology. (2023) 12:1200. doi: 10.3390/biology12091200
18. Kertz, NC, Banerjee, P, Dyce, PW, and Diniz, WJ. Harnessing genomics and transcriptomics approaches to improve female fertility in beef cattle—a review. Animals. (2023) 13:3284. doi: 10.3390/ani13203284
19. Rabel, RC, Marchioretto, PV, Bangert, EA, Wilson, K, Milner, DJ, and Wheeler, MB. Pre-implantation bovine embryo evaluation—from optics to omics and beyond. Animals. (2023) 13:2102. doi: 10.3390/ani13132102
20. Ashokan, M, Rana, E, Sneha, K, Namith, C, Naveen Kumar, GS, Azharuddin, N, et al. Metabolomics—a powerful tool in livestock research. Anim Biotechnol. (2023) 34:3237–49. doi: 10.1080/10495398.2022.2128814
21. Lima, J, Ingabire, W, Roehe, R, and Dewhurst, RJ. Estimating microbial protein synthesis in the rumen—can “Omics” methods provide new insights into a long-standing question? Vet Sci. (2023) 10:679. doi: 10.3390/vetsci10120679
22. Dixit, S, Kumar, S, Sharma, R, Banakar, PS, Singh, M, Keshri, A, et al. Rumen multi-omics addressing diet–host–microbiome interplay in farm animals: a review. Anim Biotechnol. (2023) 34:3187–205. doi: 10.1080/10495398.2022.2078979
23. Fabrile, MP, Ghidini, S, Conter, M, Varrà, MO, Ianieri, A, and Zanardi, E. Filling gaps in animal welfare assessment through metabolomics. Front Vet Sci. (2023) 10:1129741. doi: 10.3389/fvets.2023.1129741
24. Jiang, YF, Wang, S, Wang, CL, Xu, RH, Wang, WW, Jiang, Y, et al. Pangenome obtained by long-read sequencing of 11 genomes reveal hidden functional structural variants in pigs. iScience. (2023) 26:106119. doi: 10.1016/j.isci.2023.106119
25. Lai, X, Zhang, Z, Zhang, Z, Liu, S, Bai, C, Chen, Z, et al. Integrated microbiome-metabolome-genome axis data of Laiwu and Lulai pigs. Sci Data. (2023) 10:280. doi: 10.1038/s41597-023-02191-2
26. Zeng, H, Zhang, W, Lin, Q, Gao, Y, Teng, J, Xu, Z, et al. Pig biobank: a valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. (2023) 52:gkad1080. doi: 10.1093/nar/gkad1080
27. Qiao, C, He, M, Wang, S, Jiang, X, Wang, F, Li, X, et al. Multi-omics analysis reveals substantial linkages between the oral-gut microbiomes and inflamm-aging molecules in elderly pigs. Front Microbiol. (2023) 14:14. doi: 10.3389/fmicb.2023.1250891
28. Kasper, C, Ribeiro, D, Almeida, AM, Larzul, C, Liaubet, L, and Murani, E. Omics application in animal science—a special emphasis on stress response and damaging behaviour in pigs. Genes. (2020) 11:920. doi: 10.3390/genes11080920
29. Wang, J, Fu, Y, Su, T, Wang, Y, Soladoye, OP, Huang, Y, et al. A role of multi-omics Technologies in Sheep and Goat Meats: Progress and way ahead. Food Secur. (2023) 12:4069. doi: 10.3390/foods12224069
30. Rice, ES, Alberdi, A, Alfieri, J, Athrey, G, Balacco, JR, Bardou, P, et al. A pangenome graph reference of 30 chicken genomes allows genotyping of large and complex structural variants. BMC Biol. (2023) 21:267. doi: 10.1186/s12915-023-01758-0
31. Urgessa, OE, and Woldesemayat, AA. OMICs approaches and technologies for understanding low-high feed efficiency traits in chicken: implication to breeding. Anim Biotechnol. (2023) 14:1–20. doi: 10.1080/10495398.2023.2187404
32. Su, G, Yu, C, Liang, S, Wang, W, and Wang, H. Multi-omics in food safety and authenticity in terms of food components. Food Chem. (2024) 437:137943. doi: 10.1016/j.foodchem.2023.137943
33. Herráiz-Gil, S, de Arriba, MD, Escámez, MJ, and León, C. Multi-Omic data integration in food science and analysis. Curr Opin Food Sci. (2023) 52:101049. doi: 10.1016/j.cofs.2023.101049
34. Dehau, T, Ducatelle, R, Van Immerseel, F, and Goossens, E. Omics technologies in poultry health and productivity-part 1: current use in poultry research. Avian Pathol. (2022) 51:407–17. doi: 10.1080/03079457.2022.2086447
35. Hwang, YH, Lee, EY, Lim, HT, and Joo, ST. Multi-omics approaches to improve meat quality and taste characteristics. Food Sci Anim Resour. (2023) 43:1067–86. doi: 10.5851/kosfa.2023.e63
36. Jia, W, Di, C, and Shi, L. Applications of lipidomics in goat meat products: biomarkers, structure, nutrition interface, and future perspectives. J Proteome. (2023) 270:104753. doi: 10.1016/j.jprot.2022.104753
37. Jia, W, Guo, A, Bian, W, Zhang, R, Wang, X, and Shi, L. Integrative deep learning framework predicts lipidomics-based investigation of preservatives on meat nutritional biomarkers and metabolic pathways. Crit Rev Food Sci Nutr. (2023):1–5. doi: 10.1080/10408398.2023.2295016
38. Mao, X, Bassey, AP, Sun, D, Yang, K, Shan, K, and Li, C. Overview of omics applications in elucidating the underlying mechanisms of biochemical and biological factors associated with meat safety and nutrition. J Proteome. (2023) 276:104840. doi: 10.1016/j.jprot.2023.104840
39. Ramanathan, R, Kiyimba, F, Suman, SP, and Mafi, GG. The potential of metabolomics in meat science: current applications, future trends, and challenges. J Proteome. (2023) 283–284:104926. doi: 10.1016/j.jprot.2023.104926
40. Zhang, R, Pavan, E, Ross, AB, Deb-Choudhury, S, Dixit, Y, Mungure, TE, et al. Molecular insights into quality and authentication of sheep meat from proteomics and metabolomics. J Proteome. (2023) 276:104836. doi: 10.1016/j.jprot.2023.104836
41. Jiang, N, Wu, R, Wu, C, Wang, R, Wu, J, and Shi, H. Multi-omics approaches to elucidate the role of interactions between microbial communities in cheese flavor and quality. Food Rev Int. (2023) 39:5446–58. doi: 10.1080/87559129.2022.2070199
42. Yuan, L, Dai, H, He, G, Yang, Z, and Jiao, X. Invited review: current perspectives for analyzing the dairy biofilms by integrated multi-omics. J Dairy Sci. (2023) 106:8181–92. doi: 10.3168/jds.2023-23306
43. Bennett, R, and Pfuderer, S. The potential for new donkey farming systems to supply the growing demand for hides. Animals. (2020) 10:718. doi: 10.3390/ani10040718
44. Zhang, Z, Huang, B, Wang, Y, Zhu, M, Liu, G, and Wang, C. A survey report on the donkey original breeding farms in China: current aspects and future prospective. Front Vet Sci. (2023) 10:1126138. doi: 10.3389/fvets.2023.1126138
45. Gao, R, Shi, L, Guo, W, Xu, Y, Jin, X, Yan, S, et al. Effects of housing and management systems on the growth, immunity, antioxidation, and related physiological and biochemical indicators of donkeys in cold weather. Animals. (2022) 12:2405. doi: 10.3390/ani12182405
46. Gao, WW, Zhang, S, Zhang, TH, Xiao, HD, Su, N, Tao, MF, et al. Prevalence and multilocus genotyping of Giardia duodenalis in donkeys in Shanxi Province, North China. Animals. (2023) 13:3771. doi: 10.3390/ani13243771
47. Deng, L, Shi, S, Li, J, Tang, C, Han, Y, and Xie, P. A survey of smallholder farms regarding demographics, health care, and management factors of donkeys in northeastern China. Front Vet Sci. (2021) 8:626622. doi: 10.3389/fvets.2021.626622
48. Seyiti, S, and Kelimu, A. Donkey industry in China: current aspects, suggestions, and future challenges. J Equine Vet. (2021) 102:103642. doi: 10.1016/j.jevs.2021.103642
49. Zeng, L, Dang, R, Dong, H, Li, F, Chen, H, and Lei, C. Genetic diversity and relationships of Chinese donkeys using microsatellite markers. Arch Anim Breed. (2019) 62:181–7. doi: 10.5194/aab-62-181-2019
50. FAOSTAT. Food and agricultural organization of the United Nations. In: FAO Statistical Database Website. (2020). Available at: http://www.fao.org/faostat/en/#data/QA (Accessed August 1, 2020).
51. Zhou, M, Huang, F, Du, X, Liu, G, and Wang, C. Analysis of the differentially expressed proteins in donkey Milk in different lactation stages. Food Secur. (2023) 12:4466. doi: 10.3390/foods12244466
52. Li, Y, Ma, Q, Liu, G, and Wang, C. Effects of donkey milk on oxidative stress and inflammatory response. J Food Biochem. (2022) 46:e13935. doi: 10.1111/jfbc.13935
53. Yue, Y, Li, L, Tong, M, Li, S, Zhao, Y, Guo, X, et al. Effect of varying dietary crude protein level on milk production, nutrient digestibility, and serum metabolites by lactating donkeys. Animals. (2022) 12:2066. doi: 10.3390/ani12162066
54. Kimura, B, Marshall, FB, Chen, S, Rosenbom, S, Moehlman, PD, Tuross, N, et al. Ancient DNA from Nubian and Somali wild ass provides insights into donkey ancestry and domestication. Proc R Soc B. (2011) 278:50–7. doi: 10.1098/rspb.2010.0708
55. Beja-Pereira, A, England, PR, Ferrand, N, Jordan, S, Bakhiet, AO, Abdalla, MA, et al. African origins of the domestic donkey. Science. (2004) 304:1781. doi: 10.1126/science.1096008
56. Rossel, S, Marshall, F, Peters, J, Pilgram, T, Adams, MD, and O'Connor, D. Domestication of the donkey: timing, processes, and indicators. Proc Natl Acad Sci USA. (2008) 105:3715–20. doi: 10.1073/pnas.0709692105
57. Wang, C, Li, H, Guo, Y, Huang, J, Sun, Y, Min, J, et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat Commun. (2020) 11:6014. doi: 10.1038/s41467-020-19813-7
58. Zeng, L, Liu, HQ, Tu, XL, Ji, CM, Gou, X, Esmailizadeh, A, et al. Genomes reveal selective sweeps in kiang and donkey for high-altitude adaptation. Zool Res. (2021) 42:450–60. doi: 10.24272/j.issn.2095-8137.2021.095
59. Liu, S, Su, J, Yang, Q, Sun, M, Wang, Z, Yu, J, et al. Genome-wide analyses based on a novel donkey 40K liquid chip reveal the gene responsible for coat color diversity in Chinese Dezhou donkey. Anim Genet. (2024) 55:140–6. doi: 10.1111/age.13379
60. Todd, ET, Tonasso-Calvière, L, Chauvey, L, Schiavinato, S, Fages, A, Seguin-Orlando, A, et al. The genomic history and global expansion of domestic donkeys. Science. (2022) 377:1172–80. doi: 10.1126/science.abo3503
61. Renaud, G, Petersen, B, Seguin-Orlando, A, Bertelsen, MF, Waller, A, Newton, R, et al. Improved de novo genomic assembly for the domestic donkey. Sci Adv. (2018) 4:4. doi: 10.1126/sciadv.aaq0392
62. Ahlawat, S, Sharma, U, Arora, R, Sharma, R, Chhabra, P, Singh, KV, et al. Mitogenomic phylogeny reveals the predominance of the Nubian lineage of African wild ass in Indian donkeys. Gene. (2023) 880:147627. doi: 10.1016/j.gene.2023.147627
63. Huang, B, Khan, MZ, Chai, W, Ullah, Q, and Wang, C. Exploring genetic markers: mitochondrial DNA and genomic screening for biodiversity and production traits in donkeys. Animals. (2023) 13:2725. doi: 10.3390/ani13172725
64. Liu, S, Su, J, Yang, Q, Sun, M, Wang, Z, Yu, J, et al. Genome-wide analyses based on a novel donkey 40K liquid chip reveal the gene responsible for coat color diversity in Chinese Dezhou donkey. Anim Genet. (2023) 54:104836.
65. Dong, H, Dong, Z, Wang, F, Wang, G, Luo, X, Lei, C, et al. Whole genome sequencing provides new insights into the genetic diversity and coat color of Asiatic wild ass and its hybrids. Front Genet. (2022) 13:818420. doi: 10.3389/fgene.2022.818420
66. Wang, Y, Hua, X, Shi, X, and Wang, C. Origin, evolution, and research development of donkeys. Genes. (2022) 13:1945. doi: 10.3390/genes13111945
67. Janečka, JE, Davis, BW, Ghosh, S, Paria, N, Das, PJ, Orlando, L, et al. Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes. Nat Commun. (2018) 9:2945. doi: 10.1038/s41467-018-05290-6
68. Bertolini, F, Scimone, C, Geraci, C, Schiavo, G, Utzeri, VJ, Chiofalo, V, et al. Next-generation semiconductor-based sequencing of the donkey (Equus asinus) genome provided comparative sequence data against the horse genome and a few million single nucleotide polymorphisms. PLoS One. (2015) 10:e0131925. doi: 10.1371/journal.pone.0131925
69. Wang, L, Sheng, G, Preick, M, Hu, S, Deng, T, Taron, UH, et al. Ancient mitogenomes provide new insights into the origin and early introduction of Chinese domestic donkeys. Front Genet. (2021) 12:759831. doi: 10.3389/fgene.2021.759831
70. Zhou, Z, Fan, Y, Wang, G, Lai, Z, Gao, Y, Wu, F, et al. Detection of selection signatures underlying production and adaptive traits based on whole-genome sequencing of six donkey populations. Animals. (2020) 10:1823. doi: 10.3390/ani10101823
71. Bai, D, Zhao, Y, Bei, L, Gerelchimeg, B, Zhang, X, and Dugar-Javiin, M. Progress in the whole genome of Equus by using high-throughput sequencing technologies. Hereditas. (2017) 39:974–83. doi: 10.16288/j.yczz.17-122
72. Ivanković, A, Šubara, G, Bittante, G, Šuran, E, Amalfitano, N, Aladrović, J, et al. Potential of endangered local donkey breeds in meat and Milk production. Animals. (2023) 13:2146. doi: 10.3390/ani13132146
73. Shen, J, Yu, J, Dai, X, Li, M, Wang, G, Chen, N, et al. Genomic analyses reveal distinct genetic architectures and selective pressures in Chinese donkeys. J Genet Genomics. (2021) 48:737–45. doi: 10.1016/j.jgg.2021.05.012
74. Chen, J, Zhang, S, Liu, S, Dong, J, Cao, Y, and Sun, Y. Single nucleotide polymorphisms (SNPs) and Indels identified from whole-genome Re-sequencing of four Chinese donkey breeds. Anim Biotechnol. (2023) 34:1828–39. doi: 10.1080/10495398.2022.2053145
75. Wang, T, Liu, Z, Shi, X, Zhang, Z, Li, Y, Huang, B, et al. An investigation of genetic diversity in three Dezhou donkey original breeding farms. Sci Rep. (2023) 13:11203. doi: 10.1038/s41598-023-38219-1
76. Lei, CZ, Ge, QL, Zhang, H, Liu, RY, Zhang, W, Jiang, YQ, et al. African maternal origin and genetic diversity of Chinese domestic donkeys. Asian Australas J Anim Sci. (2007) 20:645–52. doi: 10.5713/ajas.2007.645
77. Xia, J, Chang, L, Xu, D, Jia, Y, Ding, Y, Cao, C, et al. Next-generation sequencing of the complete Huaibei Grey donkey Mitogenome and Mitogenomic phylogeny of the Equidae Family. Animals. (2023) 13:531. doi: 10.3390/ani13030531
78. Wang, G, Wang, F, Pei, H, Li, M, Bai, F, Lei, C, et al. Genome-wide analysis reveals selection signatures for body size and drought adaptation in Liangzhou donkey. Genomics. (2022) 114:110476. doi: 10.1016/j.ygeno.2022.110476
79. Pengjia, B, Guo, X, Pei, J, Ding, X, Wu, X, Xiong, L, et al. Characterization of the complete mitochondrial genome of the Liangzhou donkey (Equus asinus). Mitochondrial DNA B. (2019) 4:1846–7. doi: 10.1080/23802359.2019.1613182
80. Song, S, Wang, S, Li, N, Chang, S, Dai, S, Guo, Y, et al. Genome-wide association study to identify SNPs and candidate genes associated with body size traits in donkeys. Front Genet. (2023) 14:1112377. doi: 10.3389/fgene.2023.1112377
81. Li, W, Qiu, L, Guan, J, Sun, Y, Zhao, J, and Du, M. Comparative transcriptome analysis of longissimus Dorsi tissues with different intramuscular fat contents from Guangling donkeys. BMC Genomics. (2022) 23:1–3. doi: 10.1186/s12864-022-08857-2
82. Yang, N. China's livestock and poultry genetic resources: horse, donkey and camel, Beijing, China: China Agricultural Press (2011).
83. Rocchetti, G, Galimberti, S, Callegari, ML, and Lucini, L. Metabolomics and proteomics approaches provide a better understanding of non-enzymatic Browning and Pink discoloration in dairy products: a mini review. Food Biosci. (2023) 56:103328. doi: 10.1016/j.fbio.2023.103328
84. Qin, C, Liu, L, Wang, Y, Leng, T, Zhu, M, Gan, B, et al. Advancement of omics techniques for chemical profile analysis and authentication of Milk. Trends Food Sci Technol. (2022) 127:114–28. doi: 10.1016/j.tifs.2022.06.001
85. Casado, B, Affolter, M, and Kussmann, M. OMICS-rooted studies of Milk proteins, oligosaccharides and lipids. J Proteome. (2009) 73:196–208. doi: 10.1016/j.jprot.2009.09.018
86. Guan, B, Zhang, Z, Cao, X, Yang, M, Chai, Y, Amantai, X, et al. Characterization and comparison site-specific N-glycosylation profiling of Milk fat globule membrane proteome in donkey and human colostrum and mature Milk. Food Chem. (2023) 419:136081. doi: 10.1016/j.foodchem.2023.136081
87. Auzino, B, Miranda, G, Henry, C, Krupova, Z, Martini, M, Salari, F, et al. Top-down proteomics based on LC-MS combined with cDNA sequencing to characterize multiple Proteoforms of Amiata donkey Milk proteins. Food Res Int. (2022) 160:111611. doi: 10.1016/j.foodres.2022.111611
88. Luoyizha, W, Zeng, B, Li, H, and Liao, X. A preliminary study of proteomic analysis on caseins and whey proteins in donkey Milk from Xinjiang and Shandong of China. eFood. (2021) 2:27–36. doi: 10.2991/efood.k.210222.001
89. Li, M, Zheng, K, Song, W, Yu, H, Zhang, X, Yue, X, et al. Quantitative analysis of differentially expressed Milk fat globule membrane proteins between donkey and bovine colostrum based on high-performance liquid chromatography with tandem mass spectrometry proteomics. J Dairy Sci. (2021) 104:12207–15. doi: 10.3168/jds.2021-20471
90. Proikakis, SC, Bouroutzika, EV, Anagnostopoulos, AK, and Tsangaris, GT. Proteomic data of Donkey's Milk. Data Brief. (2021) 39:107507. doi: 10.1016/j.dib.2021.107507
91. Piovesana, S, Capriotti, AL, Cavaliere, C, La Barbera, G, Samperi, R, Chiozzi, RZ, et al. Peptidome characterization and bioactivity analysis of donkey Milk. J Proteome. (2015) 119:21–9. doi: 10.1016/j.jprot.2015.01.020
92. Chianese, L, Calabrese, MG, Ferranti, P, Mauriello, R, Garro, G, De Simone, C, et al. Proteomic characterization of donkey Milk “Caseome”. J Chromatogr A. (2010) 1217:4834–40. doi: 10.1016/j.chroma.2010.05.017
93. Murgia, A, Scano, P, Contu, M, Ibba, I, Altea, M, Bussu, M, et al. Characterization of donkey Milk and metabolite profile comparison with human Milk and formula Milk. LWT. (2016) 74:427–33. doi: 10.1016/j.lwt.2016.07.070
94. Li, Y, Ma, QS, Zhou, MM, Zhang, ZW, Zhan, YD, Liu, GQ, et al. A metabolomics comparison in Milk from two Dezhou donkey strains. Eur Food Res Technol. (2022) 248:1267–75. doi: 10.1007/s00217-022-03962-8
95. Li, M, Kang, S, Zheng, Y, Shao, J, Zhao, H, An, Y, et al. Comparative metabolomics analysis of donkey colostrum and mature Milk using ultra-high-performance liquid tandem chromatography quadrupole time-of-flight mass spectrometry. J Dairy Sci. (2020) 103:992–1001. doi: 10.3168/jds.2019-17448
96. Tenori, L, Santucci, C, Meoni, G, Morrocchi, V, Matteucci, G, and Luchinat, C. NMR Metabolomic fingerprinting distinguishes Milk from different farms. Food Res Int. (2018) 113:131–9. doi: 10.1016/j.foodres.2018.06.066
97. Li, M, Li, W, Wu, J, Zheng, Y, Shao, J, Li, Q, et al. Quantitative Lipidomics reveals alterations in donkey Milk lipids according to lactation. Food Chem. (2020) 310:125866. doi: 10.1016/j.foodchem.2019.125866
98. Mecocci, S, Pietrucci, D, Milanesi, M, Pascucci, L, Filippi, S, Rosato, V, et al. Transcriptomic characterization of cow, donkey and goat Milk extracellular vesicles reveals their anti-inflammatory and immunomodulatory potential. Int J Mol Sci. (2021) 22:12759. doi: 10.3390/ijms222312759
99. Yang, Y, Lv, K, Zhang, Y, Wang, X, and Deng, L. Widely targeted metabolomics reveals dynamic alterations in colostrum, transitional, and mature milk of jennies. LWT. (2024) 200:116179. doi: 10.1016/j.lwt.2024.116179
100. Contarini, G, Pelizzola, V, Scurati, S, and Povolo, M. Polar lipid of donkey Milk fat: phospholipid, ceramide and cholesterol composition. J Food Compos Anal. (2017) 57:16–23. doi: 10.1016/j.jfca.2016.12.013
101. Martemucci, G, and D’Alessandro, AG. Fat content, energy value and fatty acid profile of donkey Milk during lactation and implications for human nutrition. Lipids Health Dis. (2012) 11:113. doi: 10.1186/1476-511X-11-113
102. Zhang, X, Li, H, Yang, L, Jiang, G, Ji, C, Zhang, Q, et al. Comparative Lipidomics profiling of donkey Milk with cow and human Milk by UHPLC-Q-Exactive Orbitrap mass spectrometry. J Food Compos Anal. (2021) 101:103988. doi: 10.1016/j.jfca.2021.103988
103. Li, M, Dong, Y, Li, W, Shen, X, Abdlla, R, Chen, J, et al. Characterization and comparison of whey proteomes from bovine and donkey colostrum and mature Milk. LWT. (2022) 158:113113. doi: 10.1016/j.lwt.2022.113113
104. Zhang, X, Jiang, G, Ji, C, Fan, Z, Ge, S, Li, H, et al. Comparative whey proteome profiling of donkey Milk with human and cow Milk. Front Nutr. (2022) 9:911454. doi: 10.3389/fnut.2022.911454
105. Li, M, Yu, H, Chen, J, Abdlla, R, Liu, A, Song, W, et al. Novel insights into whey protein differences between donkey and bovine Milk. Food Chem. (2021) 365:130397. doi: 10.1016/j.foodchem.2021.130397
106. Zhang, X, Li, H, Yu, J, Zhou, X, Ji, C, Wu, S, et al. Label-free-based comparative proteomic analysis of whey proteins between different Milk yields of Dezhou donkey. Biochem Biophys Res Commun. (2019) 508:237–42. doi: 10.1016/j.bbrc.2018.11.130
107. Li, W, Li, M, Cao, X, Han, H, Kong, F, and Yue, X. Comparative analysis of whey proteins in donkey colostrum and mature Milk using quantitative proteomics. Food Res Int. (2020) 127:108741. doi: 10.1016/j.foodres.2019.108741
108. He, HY, Liu, LL, Chen, B, Xiao, HX, and Liu, WJ. Study on lactation performance and development of KASP marker for Milk traits in Xinjiang donkey (Equus asinus). Anim Biotechnol. (2023) 34:2724–35. doi: 10.1080/10495398.2022.2114002
109. Ning, J, Yang, M, Liu, W, Luo, X, and Yue, X. Proteomics and Peptidomics as a tool to compare the proteins and endogenous peptides in human, cow, and donkey Milk. J Agric Food Chem. (2023) 71:16435–51. doi: 10.1021/acs.jafc.3c04534
110. Wang, X, Fei, Y, Shao, Y, Liao, Q, Meng, Q, Chen, R, et al. Transcriptome analysis reveals immune function-related mRNA expression in donkey mammary glands during four developmental stages. Comp Biochem Physiol Part D Genomics Proteomics. (2023) 49:101169–10. doi: 10.1016/j.cbd.2023.101169
111. Fei, Y, Gai, Y, Liao, Q, Zhang, L, Li, Z, Li, B, et al. An integrated analysis of lactation-related mi RNA and mRNA expression profiles in donkey mammary glands. Genes. (2022) 13:1637. doi: 10.3390/genes13091637
112. Yu, J, Yang, G, Li, S, Li, M, Ji, C, Liu, G, et al. Identification of Dezhou donkey muscle development-related genes and long non-coding RNA based on differential expression analysis. Anim Biotechnol. (2023) 34:2313–23. doi: 10.1080/10495398.2022.2088549
113. Sun, Y, Wang, Y, Li, Y, Li, H, Wang, C, and Zhang, Q. Comparative transcriptome and proteome analyses of the longissimus Dorsi muscle for explaining the difference between donkey meat and other meats. Anim Biotechnol. (2023) 34:3085–98. doi: 10.1080/10495398.2022.2134883
114. Liu, LL, Chen, B, Chen, SL, and Liu, WJ. A genome-wide association study of the chest circumference trait in Xinjiang donkeys based on whole-genome sequencing technology. Genes. (2023) 14:1081. doi: 10.3390/genes14051081
115. Liu, Y, Li, H, Wang, M, Zhang, X, Yang, L, Zhao, C, et al. Genetic architectures and selection signatures of body height in Chinese indigenous donkeys revealed by next-generation sequencing. Anim Genet. (2022) 53:487–97. doi: 10.1111/age.13211
116. Yang, G, Sun, M, Wang, Z, Hu, Q, Guo, J, Yu, J, et al. Comparative genomics identifies the evolutionarily conserved gene TPM3 as a target of eca-mi R-1 involved in the skeletal muscle development of donkeys. Int J Mol Sci. (2023) 24:15440. doi: 10.3390/ijms242015440
117. Li, Y, Ma, Q, Shi, X, Liu, G, and Wang, C. Integrated multi-omics reveals novel microbe-host lipid metabolism and immune interactions in the donkey hindgut. Front Immunol. (2022) 13:1003247. doi: 10.3389/fimmu.2022.1003247
118. Guo, R, Zhang, S, Chen, J, Shen, W, Zhang, G, Wang, J, et al. Comparison of gut microflora of donkeys in high and low altitude areas. Front Microbiol. (2022) 13:964799. doi: 10.3389/fmicb.2022.964799
119. Liu, H, Zhao, X, Han, X, Xu, S, Zhao, L, Hu, L, et al. Comparative study of gut microbiota in Tibetan wild asses (Equus kiang) and domestic donkeys (Equus asinus) on the Qinghai-Tibet plateau. PeerJ. (2020) 8:e9032. doi: 10.7717/peerj.9032
120. Li, Y, Ma, Q, Liu, G, Zhang, Z, Zhan, Y, Zhu, M, et al. Metabolic alternations during gestation in Dezhou donkeys and the link to the gut microbiota. Front Microbiol. (2022) 13:801976. doi: 10.3389/fmicb.2022.801976
121. Xing, J, Liu, G, Zhang, X, Bai, D, Yu, J, Li, L, et al. The composition and predictive function of the fecal microbiota differ between young and adult donkeys. Front Microbiol. (2020) 11:596394. doi: 10.3389/fmicb.2020.596394
122. Jiang, G, Zhang, X, Gao, W, Ji, C, Wang, Y, Feng, P, et al. Transport stress affects the fecal microbiota in healthy donkeys. J Vet Intern Med. (2021) 35:2449–57. doi: 10.1111/jvim.16235
123. Zhao, F, Jiang, G, Ji, C, Zhang, Z, Gao, W, Feng, P, et al. Effects of long-distance transportation on blood constituents and composition of the nasal microbiota in healthy donkeys. BMC Vet Res. (2020) 16:1–10. doi: 10.1186/s12917-020-02563-5
124. Guo, R, Zhang, W, Shen, W, Zhang, G, Xie, T, Li, L, et al. Analysis of gut microbiota in Chinese donkeys in different regions using metagenomic sequencing. BMC Genomics. (2023) 24:524. doi: 10.1186/s12864-023-09575-z
125. Zhang, H, Chen, Y, Wu, X, Shang, S, Chen, J, Yan, J, et al. Comparison of the gut microbiota in the Tibetan wild ass (Equus kiang) collected from high and low altitude. Pak J Zool. (2020) 52:2281. doi: 10.17582/journal.pjz/20180522070505
126. Zhang, Z, Huang, B, Gao, X, Shi, X, Wang, X, Wang, T, et al. Dynamic changes in fecal microbiota in donkey foals during weaning: from pre-weaning to post-weaning. Front Microbiol. (2023) 14:1105330. doi: 10.3389/fmicb.2023.1105330
127. Zhang, Z, Huang, B, Wang, Y, Zhan, Y, Zhu, M, and Wang, C. Dynamic alterations in the donkey fecal Bacteria community and metabolome characteristics during gestation. Front Microbiol. (2022) 13:927561. doi: 10.3389/fmicb.2022.927561
128. Zhang, Z, Huang, B, Wang, Y, Zhu, M, and Wang, C. Could weaning remodel the Oral microbiota composition in donkeys? An exploratory study. Animals. (2022) 12:2024. doi: 10.3390/ani12162024
129. Xie, L, Xing, J, Qi, X, Lu, T, Jin, Y, Akhtar, MF, et al. Effects of concentrate feeding sequence on growth performance, nutrient digestibility, VFA production, and fecal microbiota of weaned donkeys. Animals. (2023) 13:2893. doi: 10.3390/ani13182893
130. Huang, B, Khan, MZ, Chen, Y, Liang, H, Kou, X, Wang, X, et al. Yeast polysaccharide supplementation: impact on lactation, growth, immunity, and gut microbiota in Dezhou donkeys. Front Microbiol. (2023) 14:14. doi: 10.3389/fmicb.2023.1289371
131. Zhang, C, Zhang, C, Wang, Y, Du, M, Zhang, G, and Lee, Y. Dietary energy level impacts the performance of donkeys by manipulating the gut microbiome and metabolome. Front Vet Sci. (2021) 8:694357. doi: 10.3389/fvets.2021.694357
132. Zhang, R, Zhang, J, Dang, W, Irwin, DM, Wang, Z, and Zhang, S. Unveiling the biogeography and potential functions of the intestinal Digesta-and mucosa-associated microbiome of donkeys. Front Microbiol. (2020) 11:596882. doi: 10.3389/fmicb.2020.596882
133. Ma, Q, Yue, Y, Kou, X, Hou, W, Wang, M, Yang, X, et al. Dynamic distribution of skin microorganisms in donkeys at different ages and various sites of the body. Animals. (2023) 13:1566. doi: 10.3390/ani13091566
134. Leonardia, AM, Barbatoa, EM, Dall'Aglioa, C, and Antognonia, MT. Omics Technologies in Veterinary Medicine: literature review and perspectives in transfusion medicine. Transfus Med Hemother. (2023) 50:198–207. doi: 10.1159/000530870
135. Yu, F, Chen, Y, Liu, B, Wang, T, Ding, Z, Yi, Z, et al. DIA mass spectrometry characterizes urinary proteomics in neonatal and adult donkeys. Sci Rep. (2022) 12:22590. doi: 10.1038/s41598-022-27245-0
136. Qi, PF, Gao, XY, Ji, JK, Zhang, Y, Yang, SH, Cheng, KH, et al. Identification of a recombinant equine coronavirus in donkey, China. Emerg Microbes Infect. (2022) 11:1010–3. doi: 10.1080/22221751.2022.2056522
137. Dinis, PA, Carvalho, S, Marinho, C, Gonçalves, A, Sousa, M, Nóvoa, M, et al. Vaginal bacterial microbiota of an endangered donkey breed: a comparison between Miranda donkey breed (Equus asinus) jennies with and without reproductive problems. J Integr OMICS. (2016) 6:18–22. doi: 10.5584/jiomics.v6i1.193
138. Li, J, Zhao, Y, Mi, J, Yi, Z, Holyoak, GR, Wu, R, et al. Comparative proteome analysis of serum uncovers differential expression of proteins in donkeys (Equus Asinus) with endometritis caused by Escherichia Coli. J Equine Vet. (2023) 122:104221. doi: 10.1016/j.jevs.2023.104221
139. Khan, MZ, Chen, W, Huang, B, Liu, X, Wang, X, Liu, Y, et al. Advancements in genetic marker exploration for livestock vertebral traits with a focus on China. Animals. (2024) 14:594. doi: 10.3390/ani14040594
140. Chowdhary, BP, Paria, N, and Raudsepp, T. Potential applications of equine genomics in dissecting diseases and fertility. Anim Reprod Sci. (2008) 107:208–18. doi: 10.1016/j.anireprosci.2008.04.010
141. Khan, A, Dou, J, Wang, Y, Jiang, X, Khan, MZ, Luo, H, et al. Evaluation of heat stress effects on cellular and transcriptional adaptation of bovine granulosa cells. J Anim Sci Biotechnol. (2020) 11:1–20. doi: 10.1186/s40104-019-0408-8
142. Khan, MZ, Zhang, Z, Liu, L, Wang, D, Mi, S, Liu, X, et al. Folic acid supplementation regulates key immunity-associated genes and pathways during the periparturient period in dairy cows. Asian Australas J Anim Sci. (2020) 33:1507–19. doi: 10.5713/ajas.18.0852
143. Guo, Y, Dai, S, Wang, S, Song, S, and Zeng, S. Effects of feeding exogenous vitamins and microelements on follicular development and molecular function of granulosa cells in donkeys. J Equine Vet. (2023) 125:104804. doi: 10.1016/j.jevs.2023.104804
144. Guo, Y, Zhao, W, Li, N, Dai, S, Wu, H, Wu, Z, et al. Integration analysis of metabolome and transcriptome reveals the effect of exogenous supplementation with mixtures of vitamins ADE, zinc, and selenium on follicular growth and granulosa cells molecular metabolism in donkeys (Equus asinus). Front Vet Sci. (2022) 9:993426. doi: 10.3389/fvets.2022.993426
145. Zhang, FL, Zhang, SE, Sun, YJ, Wang, JJ, and Shen, W. Comparative transcriptomics uncover the uniqueness of oocyte development in the donkey. Front Genet. (2022) 13:839207. doi: 10.3389/fgene.2022.839207
146. Tian, F, Wang, J, Li, Y, Yang, C, Zhang, R, Wang, X, et al. Integrated analysis of mRNA and mi RNA in testis and cauda Epididymidis reveals candidate molecular markers associated with reproduction in Dezhou donkey. Livest Sci. (2020) 234:103885. doi: 10.1016/j.livsci.2019.103885
147. Song, JL, Sun, YJ, Liu, GQ, and Zhang, GL. Deoxynivalenol and Zearalenone: different mycotoxins with different toxic effects in donkey (Equus asinus) endometrial epithelial cells. Theriogenology. (2022) 179:162–76. doi: 10.1016/j.theriogenology.2021.11.021
148. Li, M, Zhu, Q, Hong, R, Feng, D, Liu, Y, and Yue, X. Comprehensive characterization of donkey Milk serum proteins. J Future Foods. (2022) 2:270–4. doi: 10.1016/j.jfutfo.2022.06.009
149. Ning, J, Chen, J, Liu, W, Chen, X, Luo, X, and Yue, X. Characterization and nutrition assessment of amino acids in different domains between donkey colostrum and mature milk. J Food Compos Anal. (2024) 132:106345. doi: 10.1016/j.jfca.2024.106345
150. Li, W, Li, M, Cao, X, Yang, M, Han, H, Kong, F, et al. Quantitative proteomic analysis of Milk fat globule membrane (MFGM) proteins from donkey colostrum and mature Milk. Food Funct. (2019) 10:4256–68. doi: 10.1039/C9FO00386J
151. Ren, W, Sun, M, Shi, X, Wang, T, Wang, Y, Wang, X, et al. Effects of roughage on the lipid and volatile-organic-compound profiles of donkey Milk. Food Secur. (2023) 12:2231. doi: 10.3390/foods12112231
152. Sun, Y, Li, YH, Zhao, CH, Jun, TE, Wang, YH, Wang, TQ, et al. Genome-wide association study for numbers of vertebrae in Dezhou donkey population reveals new candidate genes. J Integr Agric. (2023) 22:3159–69. doi: 10.1016/j.jia.2023.04.038
153. Man, L, Ren, W, Qin, H, Sun, M, Yuan, S, Zhu, M, et al. Characterization of the relationship between lipids and volatile compounds in donkey, bovine, and sheep meat by UHPLC–ESI–MS and SPME–GC–MS. LWT. (2023) 175:114426. doi: 10.1016/j.lwt.2023.114426
154. Ma, Q, Kou, X, Yang, Y, Yue, Y, Xing, W, Feng, X, et al. Comparison of lipids and volatile compounds in Dezhou donkey meat with high and low intramuscular fat content. Food Secur. (2023) 12:3269. doi: 10.3390/foods12173269
155. Chai, W, Qu, H, Ma, Q, Zhu, M, Li, M, Zhan, Y, et al. RNA-Seq analysis identifies differentially expressed genes in different types of donkey skeletal muscles. Anim Biotechnol. (2023) 34:1786–95. doi: 10.1080/10495398.2022.2050920
156. Chai, W, Xu, J, Qu, H, Ma, Q, Zhu, M, Li, M, et al. Differential proteomic analysis to identify potential biomarkers associated with quality traits of Dezhou donkey meat using a data-independent acquisition (DIA) strategy. LWT. (2022) 166:113792. doi: 10.1016/j.lwt.2022.113792
157. Wang, M, Li, H, Zhang, X, Yang, L, Liu, Y, Liu, S, et al. An analysis of skin thickness in the Dezhou donkey population and identification of candidate genes by RNA-Seq. Anim Genet. (2022) 53:368–79. doi: 10.1111/age.13196
158. Li, Y, Ma, Q, Shi, X, Yuan, W, Liu, G, and Wang, C. Comparative transcriptome analysis of slow-twitch and fast-twitch muscles in Dezhou donkeys. Genes. (2022) 13:1610. doi: 10.3390/genes13091610
159. Tan, X, He, Y, Qin, Y, Yan, Z, Chen, J, Zhao, R, et al. Comparative analysis of differentially abundant proteins between high and low intramuscular fat content groups in donkeys. Front Vet Sci. (2022) 9:951168. doi: 10.3389/fvets.2022.951168
160. Li, M, Ren, W, Chai, W, Zhu, M, Man, L, Zhan, Y, et al. Comparing the profiles of raw and cooked donkey meat by Metabonomics and Lipidomics assessment. Front Nutr. (2022) 9:851761. doi: 10.3389/fnut.2022.851761
161. Li, M, Zhu, M, Chai, W, Wang, Y, Song, Y, Liu, B, et al. Determination of the heterogeneity of intramuscular fat and visceral adipose tissue from Dezhou donkey by lipidomics and transcriptomics profiling. Front Nutr. (2021) 8:746684. doi: 10.3389/fnut.2021.746684
162. Li, M, Zhu, M, Chai, W, Wang, Y, Fan, D, Lv, M, et al. Determination of lipid profiles of Dezhou donkey meat using an LC-MS-based Lipidomics method. J Food Sci. (2021) 86:4511–21. doi: 10.1111/1750-3841.15917
163. Li, M, Sun, M, Ren, W, Man, L, Chai, W, Liu, G, et al. Characterization of volatile compounds in donkey meat by gas chromatography–ion mobility spectrometry (GC–IMS) combined with Chemometrics. Food Sci Anim Resour. (2024) 44:165–77. doi: 10.5851/kosfa.2023.e67
164. Li, M, Sun, L, Du, X, Ren, W, Man, L, Chai, W, et al. Characterization of lipids and volatile compounds in boiled donkey meat by lipidomics and volatilomics. J Food Sci. (2024) 89:3445–54. doi: 10.1111/1750-3841.17086
165. Wang, Y, Miao, X, Zhao, Z, Wang, Y, Li, S, and Wang, C. Transcriptome atlas of 16 donkey tissues. Front Genet. (2021) 12:682734. doi: 10.3389/fgene.2021.682734
166. Li, B, Feng, C, Zhu, S, Zhang, J, Irwin, DM, Zhang, X, et al. Identification of candidate circular RNAs underlying intramuscular fat content in the donkey. Front Genet. (2020) 11:587559. doi: 10.3389/fgene.2020.587559
167. Shi, T, Hu, W, Hou, H, Zhao, Z, Shang, M, and Zhang, L. Identification and comparative analysis of long non-coding RNA in the skeletal muscle of two Dezhou donkey strains. Genes. (2020) 11:508. doi: 10.3390/genes11050508
168. Chai, W, Wang, L, Li, T, Wang, T, Wang, X, Yan, M, et al. Liquid chromatography–mass spectrometry-based metabolomics reveals dynamic metabolite changes during early postmortem aging of donkey meat. Food Secur. (2024) 13:1466. doi: 10.3390/foods13101466
169. Yu, J, Wang, Z, Wang, F, Yang, G, Cheng, J, Ji, C, et al. Analysis of lncRNA and mRNA expression profiling in immature and mature DeZhou donkey (equine Taurus) testes. Reprod Domest Anim. (2023) 58:646–56. doi: 10.1111/rda.14330
170. Yu, M, Zhang, X, Yan, J, Guo, J, Zhang, F, Zhu, K, et al. Transcriptional specificity analysis of testis and epididymis tissues in donkey. Genes. (2022) 13:2339. doi: 10.3390/genes13122339
171. Sun, Y, Wang, Y, Li, Y, Akhtar, F, Wang, C, and Zhang, Q. Identification of circular RNAs of testis and caput epididymis and prediction of their potential functional roles in donkeys. Genes. (2022) 14:66. doi: 10.3390/genes14010066
172. Li, Z, Song, X, Yin, S, Yan, J, Lv, P, Shan, H, et al. Single-cell RNA-Seq revealed the gene expression pattern during the in vitro maturation of donkey oocytes. Genes. (2021) 12:1640. doi: 10.3390/genes12101640
173. Yu, J, Liu, H, Li, X, Ge, S, Zhao, X, Ji, C, et al. TMT-based comparative proteomic analysis of Dezhou donkey spermatozoa related to Freezability. J Proteome. (2023) 273:104793. doi: 10.1016/j.jprot.2022.104793
174. Liu, H, Yu, J, Li, M, Kang, S, Zhao, X, Yin, G, et al. Proteomic analysis of donkey sperm reveals changes in acrosome enzymes and redox regulation during cryopreservation. J Proteome. (2022) 267:104698. doi: 10.1016/j.jprot.2022.104698
175. Yu, J, Li, M, Ji, C, Li, X, Li, H, Liu, G, et al. Comparative proteomic analysis of seminal plasma proteins in relation to Freezability of Dezhou donkey semen. Anim Reprod Sci. (2021) 231:106794. doi: 10.1016/j.anireprosci.2021.106794
Keywords: omics application, donkey breeds, genetic resources, production traits, reproductive traits, microbiota, molecular breeding
Citation: Khan MZ, Chen W, Wang X, Liang H, Wei L, Huang B, Kou X, Liu X, Zhang Z, Chai W, Khan A, Peng Y and Wang C (2024) A review of genetic resources and trends of omics applications in donkey research: focus on China. Front. Vet. Sci. 11:1366128. doi: 10.3389/fvets.2024.1366128
Edited by:
Martino Cassandro, University of Padua, ItalyReviewed by:
Liang Deng, Shenyang Agricultural University, ChinaSharmila Ghosh, University of California, Davis, United States
Copyright © 2024 Khan, Chen, Wang, Liang, Wei, Huang, Kou, Liu, Zhang, Chai, Khan, Peng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changfa Wang, d2FuZ2NoYW5nZmFAbGN1LmVkdS5jbg==; Muhammad Zahoor Khan, emFob29yY2F1QGNhdS5lZHUuY24=
 Muhammad Zahoor Khan
Muhammad Zahoor Khan Wenting Chen1
Wenting Chen1 Xiaotong Liu
Xiaotong Liu Adnan Khan
Adnan Khan