
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 30 May 2024
Sec. Veterinary Epidemiology and Economics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1362011
This article is part of the Research TopicPathogens at the interface of animals in close contact with humans: risks and benefits, with special regard to immunosuppressed peopleView all 7 articles
 Si Thu Hein1,2
Si Thu Hein1,2 Rangsiya Prathan2
Rangsiya Prathan2 Songsak Srisanga2
Songsak Srisanga2 Dudsadee Muenhor3
Dudsadee Muenhor3 Thidathip Wongsurawat4,5
Thidathip Wongsurawat4,5 Piroon Jenjaroenpun4,5
Piroon Jenjaroenpun4,5 Padet Tummaruk6
Padet Tummaruk6 Rungtip Chuanchuen2*
Rungtip Chuanchuen2*This study aims to investigate bacterial communities and antimicrobial resistance (AMR) in airborne dust from pig farms. Airborne dust, pig feces and feed were collected from nine pig farms in Thailand. Airborne dust samples were collected from upwind and downwind (25 meters from pig house), and inside (in the middle of the pig house) of the selected pig house. Pig feces and feed samples were individually collected from the pen floor and feed trough from the same pig house where airborne dust was collected. A direct total bacteria count on each sampling plate was conducted and averaged. The ESKAPE pathogens together with Escherichia coli, Salmonella, and Streptococcus were examined. A total of 163 bacterial isolates were collected and tested for MICs. Pooled bacteria from the inside airborne dust samples were analyzed using Metagenomic Sequencing. The highest bacterial concentration (1.9–11.2 × 103 CFU/m3) was found inside pig houses. Staphylococcus (n = 37) and Enterococcus (n = 36) were most frequent bacterial species. Salmonella (n = 3) were exclusively isolated from feed and feces. Target bacteria showed a variety of resistance phenotypes, and the same bacterial species with the same resistance phenotype were found in airborne dust, feed and fecal from each farm. Metagenomic Sequencing analysis revealed 1,652 bacterial species across all pig farms, of which the predominant bacterial phylum was Bacillota. One hundred fifty-nine AMR genes of 12 different antibiotic classes were identified, with aminoglycoside resistance genes (24%) being the most prevalent. A total of 251 different plasmids were discovered, and the same plasmid was detected in multiple farms. In conclusion, the phenotypic and metagenomic results demonstrated that airborne dust from pig farms contained a diverse array of bacterial species and genes encoding resistance to a range of clinically important antimicrobial agents, indicating the significant role in the spread of AMR bacterial pathogens with potential hazards to human health. Policy measurements to address AMR in airborne dust from livestock farms are mandatory.
Antimicrobial resistance (AMR) has been listed as one of the six global emerging environmental challenges by the United Nations Environment Programme (1). Transmission of AMR bacteria and their resistance determinants in environmental settings has been extensively researched in two primary habitats, aquaculture, and soils (2) e.g., lakes (3), soil with sewage and chicken manure (4), sediments, wastewater treatment plants (5), hospital wastewater (6). Prior studies have shown that ambient air contained a variety of bacteria and AMR determinants that may move long distances and across borders (7, 8). These place airborne AMR bacteria and their resistance determinants as an additional important route for the spread of AMR on a continental or global scale (8, 9), necessitating policies and regulation to reduce the spread of AMR through airborne dust.
WHO published a list of highly virulent and AMR bacterial pathogens as global targets for development of novel antibiotics, i.e., the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) that are also high priority organisms for AMR monitoring due to their ability to acquire high levels of resistance. The existence and spread of airborne AMR in ESKAPE pathogens in healthcare settings has been the subject of numerous research (10). The problem is far more extensive due to their expansion in animal farms and the environment (11). Transmission of AMR bacteria, including ESKAPE organisms, from animal farms to nearby communities through the air was demonstrated (12, 13).
The role of livestock farming, in particular pig farms, as a significant source and reservoir for AMR bacteria and determinants has been scientifically demonstrated (14). It is primarily caused by a high bacterial load and an increase in antimicrobial use at the farms, which creates a significant selective antimicrobial pressure and raises the risk of bacterial resistance development and spread. In addition to meat, soil and farm wastewater, airborne dust from pig farms has reportedly contained AMR bacteria and determinants, inevitably reaching the general population (7, 15). AMR bacteria-filled dust has the potential to become airborne and spread across the farms. It may also be released into the outside air by forced or natural ventilation, endangering neighboring people, farm animals, and the environment (16). To date, relatively few research has focused on AMR in ambient air, in comparison to food animals and their products. Despite the extensive antimicrobial use for a long time, little is still known about the bacterial community and AMR in airborne dust from livestock farms.
AMR studies typically rely on culture-based approaches, of which data on AMR phenotype and prevalence could be assessed. These approaches are usually time-consuming, laborious, and information limited. An advanced high-throughput technology and bioinformatic tool, Metagenomic sequencing methods, has proved the ways to overcome the limitations of the culture-based approach and to enhance the likelihood of finding AMR determinants, including novel resistance genes. It is anticipated that bacterial population and AMR traits in air samples from livestock farms would vary and be complex and therefore, culture-based methods and metagenomic sequencing analysis should be used in complementary for in-depth understanding.
The correlation between total levels of AMR and the use of antimicrobial agents was previously indicated (17). It was estimated that pigs consumed the highest antimicrobial quantity of 193 mg/PCU in 2017, accounting for 45% of the global increase in antimicrobial consumption from 2017 to 2030 (18). This is consistent with Thailand's 2020 One Health Report on Antimicrobial Consumption and AMR, which found that pigs consumed the highest amount of antibacterial agents in medicated feed (19). In addition, pig farm dust was previously shown to have higher levels of AMR bacteria and resistance genes in comparison to other livestock (20). Therefore, this study aimed to investigate bacterial communities and AMR in airborne pig farm dust.
This cross-sectional study was conducted in 9 pig farms located in Nakhon Sawan, Chainat, Supan Buri, Lopburi, and Saraburi of Thailand during the rainy season from late May to mid-October 2022 (Table 1). Samples were taken on a single visit to each farm. Due to the farm biosecurity, the selection of farms depended on the owners' willingness to participate in the study and the availability of veterinarians or animal health practitioners. All participating farms adopt a close house system with the number of pigs varying from 50 to 13,000. Three farms (Farm 4, Farm 6, and Farm 9) had large-scale commercial farming operations with between 11,000 and 13,000 pigs. Six farms were smaller scale farms, of which four farms had 2,000–5,000 pigs and another two farms had 50–200 pigs. Sampling was carried out at one pig house in each farm according to the owner of the farm's advice. For antimicrobial usage, amoxicillin was the most used antimicrobials by adding to feed either alone or in combination with tiamulin. Two farms (Farm 2 and Farm 3) declared using penicillin - streptomycin injection for treatment of ill pigs. Amoxicillin injection alone was used on one farm (Farm 2) for sick pigs. Information on the use of antimicrobial feed additives was unavailable.
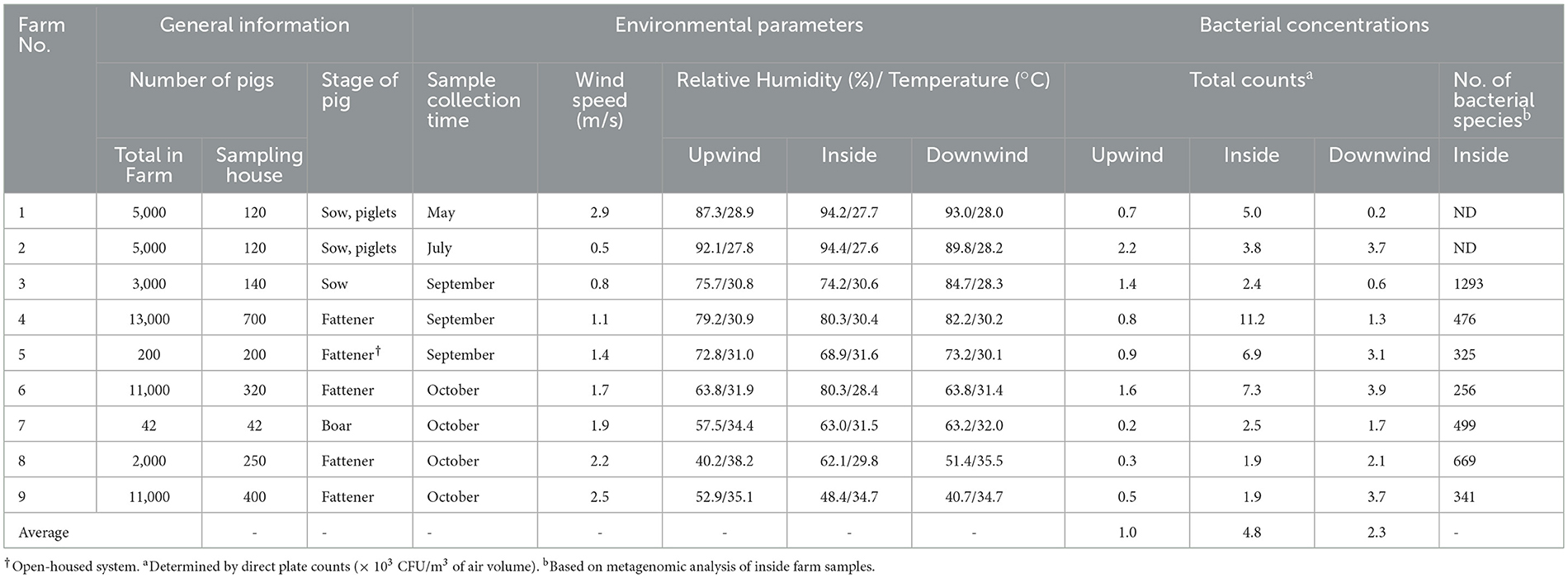
Table 1. General details of the participating pig farms, average environmental parameters, bacterial concentrations, and number of bacterial species in airborne dust at different sampling positions in pig farms (n = 9).
Airborne dust samples were collected using a BioStage® single-stage viable cascade impactor (SKC Inc., Eighty-Four, PA, USA) with 400 pores of 0.6 μm in size and a Quick Take 30 pump with a flow rate of 28.3 L/min. Tryptic soy agar (TSA, DifcoTM, MD, USA) in 100 mm × 15 mm petri dishes were placed inside the impactor for collecting dust. The impactor was positioned approximately 1.5 m above the ground, which is a person's average respiratory height (21). Airborne dust samples were taken from the selected pig house at three separate locations, inside (at the center of the pig house), upwind (25 m from the pig house), and downwind (25 m from the pig house). Sampling time was 2 min per plate. Three TSA plates were utilized to collect airborne dust samples at each location for a total of nine plates per farm.
The Kestrel 3000 Weather Meter (Nielsen-Kellermen, PA, USA) was used to record the environmental variables potentially affecting the composition and diversity of bacterial communities, including average wind speed (m/s), ambient air temperature (°C), and relative humidity (%) at each sampling site (Table 1).
Pig feces and feed samples at least 25 g of each were taken from the same pig house where airborne dust was collected. Fecal droppings were collected from the pen floor, and feed was collected from feeding trough. The samples were immediately placed in an ice box and transported to the laboratory within 6 h of collection.
Total bacteria were directly counted on TSA plates after a 24-h incubation at 37°C (22). The average colony-forming units per cubic meter of air (CFU/m3) were calculated from the counting results of three plates at each sampling position. Bioaerosol concentrations were estimated using the following formula (23).
Bioaerosol concentration (CFU/m3) = Number of colonies / (Adjusted flow rate of the sampling machine × Sampling duration in minutes).
The pooled bacteria from all three TSA plates obtained from each sampling position, pig feed samples (25 g each) and feces samples (25 g each) were separately pre-enriched in Buffer Peptone Water (BPW) and incubated at 37°C for 24 h. Then, target bacterial species were isolated and identified using previously published protocols, including the ESKAPE bacteria (i.e., Enterococcus species (24, 25), Staphylococcus species (26), Klebsiella species (27, 28), Acinetobacter (29), Pseudomonas species (30), Enterobacter species (27, 31), Escherichia coli (24, 32), Salmonella (33), and Streptococcus species (34, 35). One loopful of bacteria pooled from the three agar plates was steaked on selective media, Klebsiella species, Enterobacter species and Escherichia coli, MacConkey agar (DifcoTM & BBLTM); Enterococcus species, Slanetz & Bartley agar (OXOID®, Hampshire, UK); Staphylococcus species, mannitol salt agar (BBLTM, MD, USA); Acinetobacter species, CHROMagar™ Acinetobacter (CHROMagar, Paris, France); Pseudomonas species, Pseudomonas Agar Base (DifcoTM, MI, USA); Salmonella, Xylose Lysine Deoxycholate (XLD) agar (DifcoTM) and Streptococcus species, Columbia agar supplemented with 5% sheep blood agar (DifcoTM) and incubated at 37°C for 18–24 h.
Typical colonies of E. coli from MacConkey agar were streaked on Eosin Methylene Blue (EMB) agar (DifcoTM) and confirmed by Indole test. Typical Salmonella colonies from XLD plates were confirmed by growth in Triple sugar iron agar (TSI) (DifcoTM) and Motility Indole Lysine (MIL) agar (DifcoTM). Enterococcus species, Klebsiella species, Enterobacter species, and Streptococcus species were confirmed by PCR using the following primer sets; Enterococcus species, EN-1 5′-TACTGACAAACCATTCATGATG-3′ and EN-2 5′-AACTTCGTCACCAACGCGAAC-3′; Klebsiella species, KL-1 5′-CGCGTACTATACGCCATGAACGTA-3′ and KL-2 5′-ACCGTTGATCACTTCGGTCAGG-3′; Enterobacter species, Ent-1 5′-GGCAAAGCTCAACCCGGAGGTATTCT-3′ and Ent-2 5′-CAAAGAAAGATAATAATTTCACGGTTAGTC-3′ and Streptococcus species, C-1 5′-GCGTGCCTAATACATGCAA-3′ and C-2 5′-TACAACGCAGGTCCATCT-3′.
A single purified colony of each bacterial species was collected from each sampling position in each pig house and stored as a 20% glycerol stock at −80°C for further analysis.
A total of 163 bacterial isolates (n = 163) were collected (Table 4) and tested for their antimicrobial susceptibilities by broth microdilution method using the SensititreTM Complete Automated AST System (Thermo Fisher Scientific, MA, USA). Different SensititreTM MIC plates were used for different bacteria as follows: EUVSEC2, ASSECAF, and ASSECB for E. coli, Salmonella, Klebsiella species, and Enterobacter species; GNX2F for Acinetobacter and Pseudomonas species; EUST2 for Staphylococcus species and STP67 for Streptococcus species. All antibiotic plates were purchased from Trek Diagnostic Systems, West Sussex, UK. Two-fold agar dilution method was used for the susceptibility testing of Enterococcus species (36). The CLSI interpretive criteria was used for defining bacterial isolates as resistant or susceptible (36). E. coli ATCC 25922, P. aeruginosa ATCC 27853, S. aureus ATCC 29213 and E. faecalis ATCC 29212 served as quality control.
A loopful of the pooled airborne dust samples was taken directly from the pooled three TSA plates collected at the inside position of each pig house (n = 7) before the enrichment for isolation of target bacterial species to extract genomic DNA using ZymoBIOMICSTM DNA Miniprep Kit (Zymo Research, Irvine, CA, USA) following the manufacturer's instructions. The genomic DNA from Farm 1 and 2 samples were of poor quality and unusable for Metagenomic Sequencing. The concentration and quality of the extracted DNA were measured using a NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific). The quality and degradation were additionally evaluated by running 1 μl of the genomic DNA on 0.8% agarose gel electrophoresis stained with RedSafe nucleic acid staining solution (Thermo Fisher Scientific). The genomic DNA was submitted for Metagenomic Sequencing at Siriraj Long-read Lab (Si-LoL), Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand, using an Oxford Nanopore sequencing platform. The Metagenomic Sequencing results were analyzed as previously described (37). Briefly, the output fastq files were uploaded to BugSeq workflow v20.07.1 (https://bugseq.com) for metagenomic classification. The reads were quality controlled with fastp v20.07.1, a FASTQ data pre-processing tool, using a minimum average read quality of Phred 7, a minimum read length of 100 bp, and the default low complexity filter. The reads were then mapped with minimap2 v2.17 to identify consensus with all microbes in the NCBI nt database. The obtained results were visualized in MultiQC reporting tool. AMR genes were identified by aligning the reads against the Resfinder (http://genomicepidemiology.org) with minimap2. Analysis from BugSeq outputs and visualizations were performed in Pavian R package v1.2.0 in RStudio (R version 4.1.0) (https://www.r-project.org/). Kraken 2 was used to analyze bacterial species and visualize the Pavian R package results.
The highest average total bacterial counts were observed inside the pig houses (4.8 × 103 CFU/m3, 1.9–11.9 × 103 CFU/m3) (Table 1), followed by the downwind positions (2.3 × 103 CFU/m3, 0.2–3.9 × 103 CFU/m3) and the upwind positions (1.0 × 103 CFU/m3, 0.3–2.2 × 103 CFU/m3). The inside pig houses had higher bacterial loads in most farms, except for Farms 8 and 9, where the downwind concentrations were higher. The bacterial concentration inside the pig house at Farm 4 was highest (11.2 × 103 CFU/m3), while that of Farm 7 was lowest (2.5 × 103 CFU/m3). Only Farm 2 had comparable concentrations at the inside and downwind.
Using conventional-standard methods, the abundance and distribution of the ESKAPE species, E. coli, Salmonella and Streptococcus species were determined (Table 2). Staphylococcus (n = 37) and Enterococcus species (n = 36) were the most frequently detected bacterial species in all samples. Of all five Klebsiella isolates, two were obtained from airborne samples, while the others were isolated from feed samples. Salmonella species (n = 3) were found only in feed and feces but not in the airborne dust samples. Only one Pseudomonas isolate, which came from the feed sample, was obtained.
Overall, the bacterial isolates obtained from pig farm environment in this study exhibited various AMR phenotype and rates (Table 3). The number of target bacterial species varied greatly amongst the farms, therefore the comparison between farms is not appropriate.
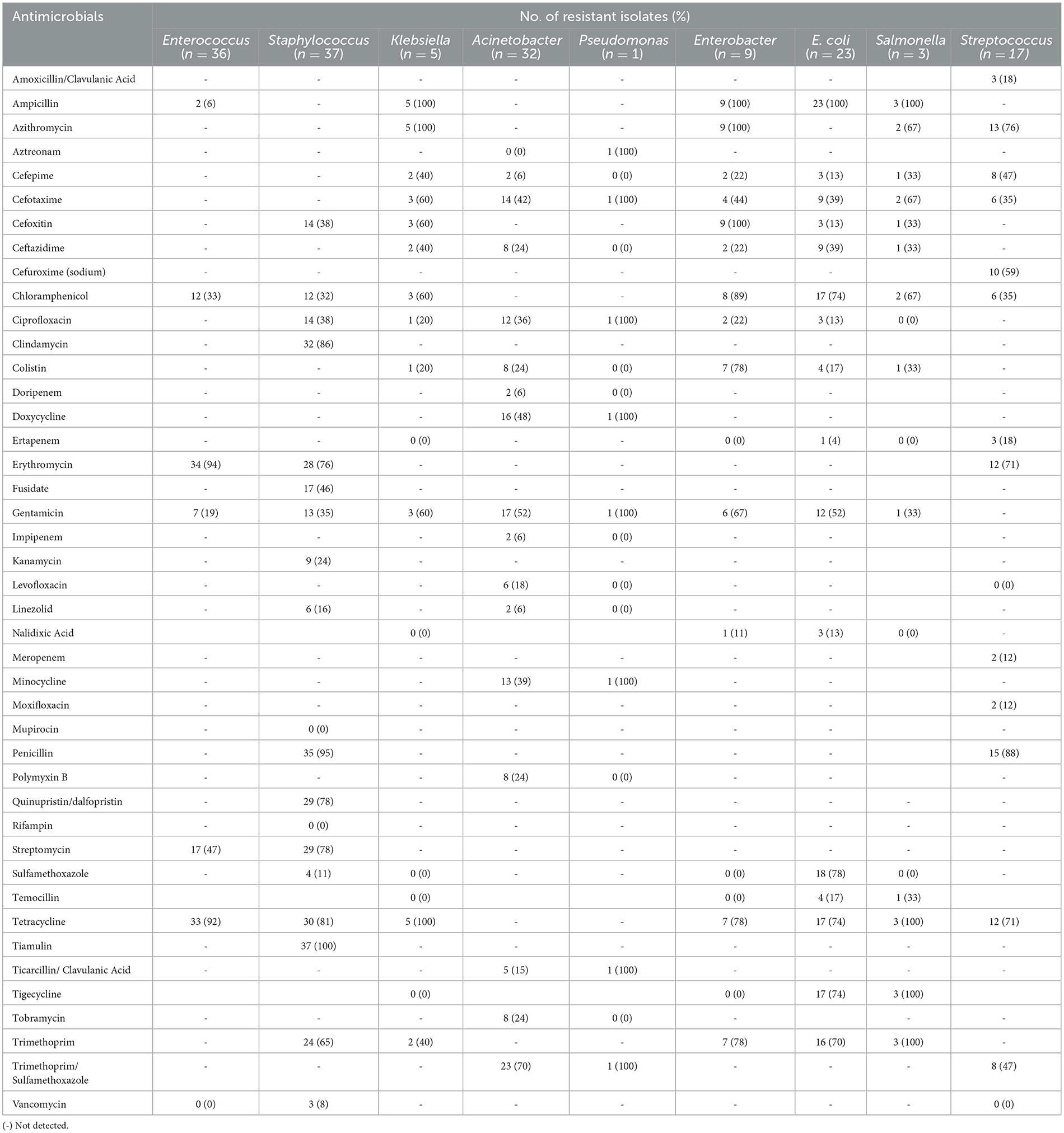
Table 3. Antimicrobial resistance rates (%) of bacterial species isolated from airborne dust, feed, and feces from the pig farms.
High resistance rates to erythromycin (94%) and tetracycline (92%) were observed for the Enterococcus isolates from airborne dust, feed, and feces (n = 36). The Staphylococcus isolates (n = 37) showed high resistance rates to tiamulin (100%), penicillin (95%), and clindamycin (86%). Notably, Staphylococcus isolates collected from airborne dust displayed resistance to a wider range of antibiotics compared to those from feed and feces samples.
All the E. coli isolates (n = 23) were resistant to ampicillin, while displayed high resistance rates to most antimicrobials tested. All three Salmonella isolates were resistant to ampicillin, tetracycline, tigecycline and trimethoprim. They also exhibited resistance to cefotaxime, cefoxitin but none were resistant to ciprofloxacin. The Streptococcus isolates (n = 17) showed high resistance rates to penicillin (88%), azithromycin (76%), erythromycin (71%), and tetracycline (71%). All Klebsiella species (n = 5) obtained from airborne dust and feed samples were resistant to ampicillin, azithromycin, and tetracycline, while some were resistant to trimethoprim/sulfamethoxazole (16/5, 70%) and gentamicin (12/5, 52%).
The only one isolate of Pseudomonas species, obtained from a feed sample of Farm 5, was resistant to a wide range of antimicrobials, including aztreonam, cefotaxime, and ciprofloxacin. Enterobacter isolates (n = 9) demonstrated resistance to most antimicrobials, including ampicillin, azithromycin, cefoxitin, chloramphenicol, and gentamicin, with high resistance rates ranging from 89% to 100%. Resistance to cefepime, cefotaxime, and ceftazidime was also observed at a lower frequency.
The bacteria isolate with various AMR phenotypes widely distributed among farms (Table 4). For each farm, the same bacterial species with the same resistance phenotype were found in airborne dust, feed, and fecal samples, for example, Enterococcus resistant to ampicillin, azithromycin, cefotaxime, ceftazidime, chloramphenicol, gentamicin, tigecycline and trimethoprim were found in airborne dust, feed and fecal samples obtained from Farm 3.
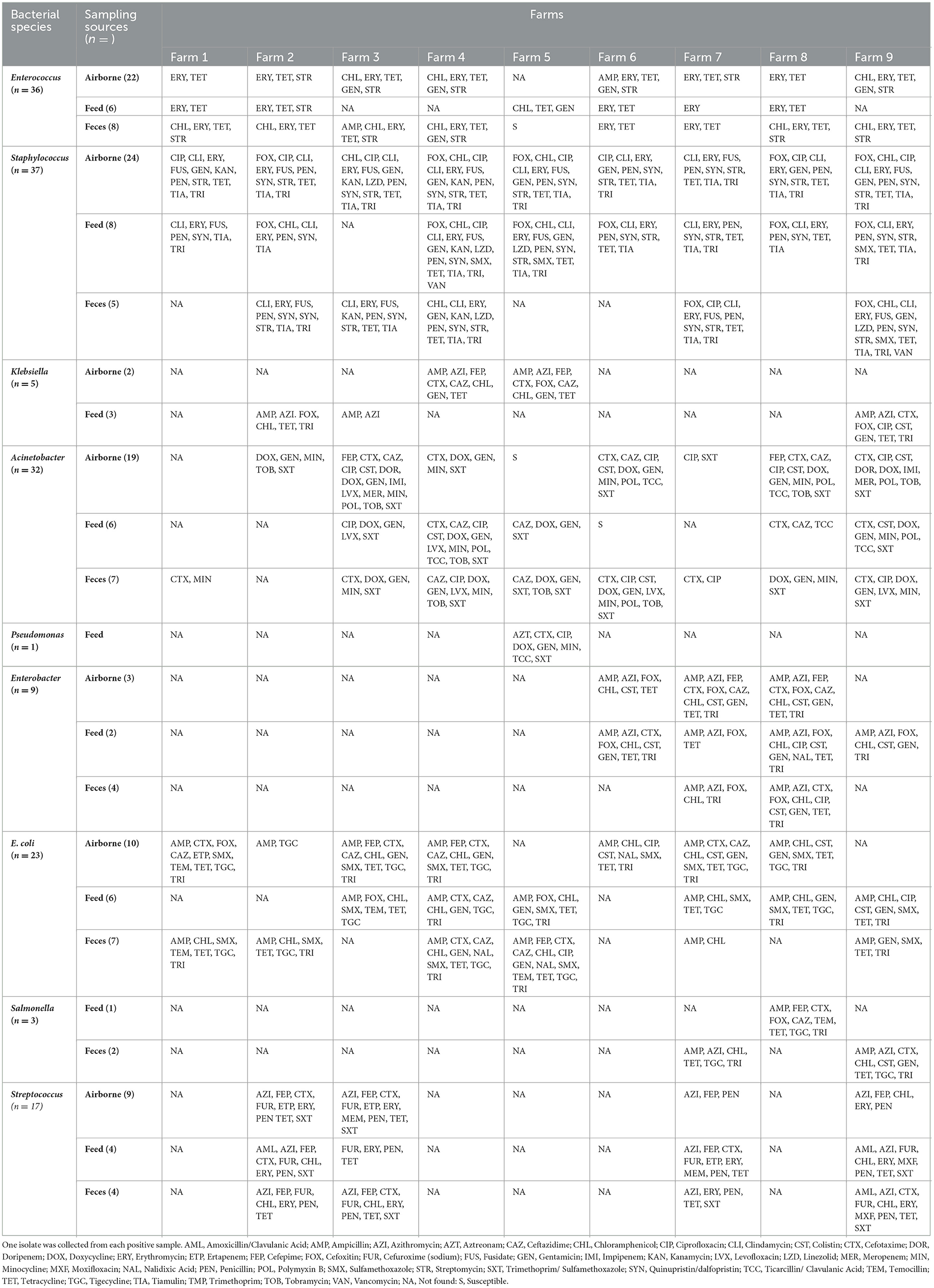
Table 4. Phenotypic characteristics of antimicrobial resistance in target bacterial species isolated from airborne dust, feed, and feces in pig farms.
Based on the Kraken 2 analysis, a total number of 1,652 bacterial species were found in all seven pig farms (Supplementary Table S1), of which Staphylococcus chromogenes was the most common bacterial species, followed by Mammaliicoccus sciuri and Staphylococcus haemolyticus. The Sankey visualization revealed that the predominant bacterial phyla observed inside pig house were Bacillota and Pseudomonadota (Figure 1). The Burkholderales order was discovered in Farms 4, 6, and 8. The relative proportions of phyla and genus of bacterial community compositions present in airborne dust from all pig farms are shown in Figure 2. The Bacillota phylum occupied the highest proportion of all farms (19.7–73.7%), of which Farm 9 had the highest percentage (73.7%). Staphylococcus was the most detected genus (50.8%). This bacterial pathogen was identified in all farms, except for Farm 4 where Acinetobacter was predominant. The number of bacterial species identified in each farm varied from 1,293 in Farm 3 to 325 in Farm 5 (Table 1).
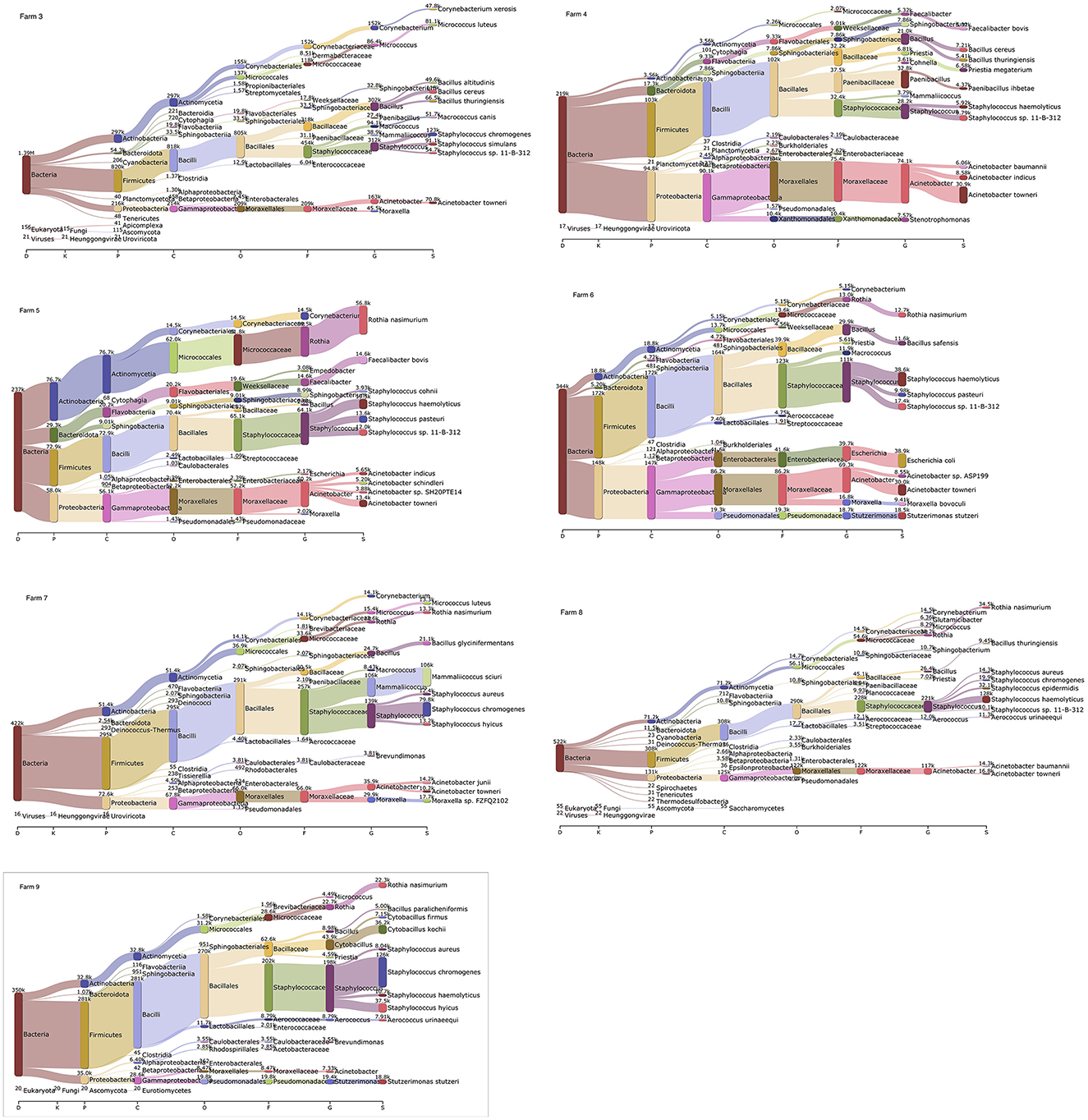
Figure 1. Sankey visualization from the Kraken 2 analysis of airborne bacterial communities inside pig farms (n = 7). It is shown in different taxonomy levels. The higher the portion for each phylum, the higher read counts. The number indicates the read counts. D, Domain; K, Kingdom; P, Phylum; C, Class; O, Order; F, Family; G, Genus; S, Species.
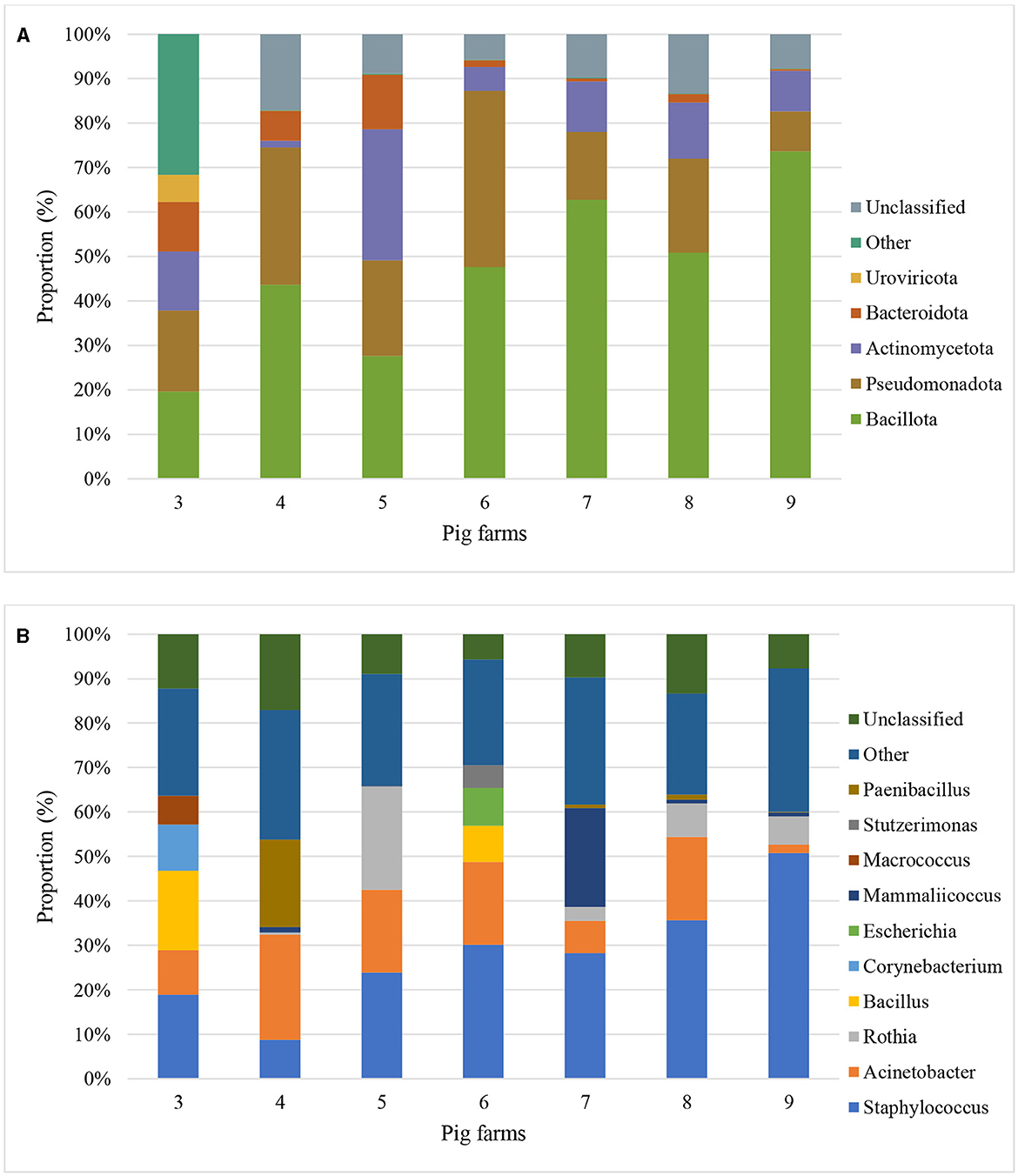
Figure 2. Metagenomic classifications of bacterial community compositions at Phylum level (A) and Genus level (B) of airborne dust inside seven pig farms by Proportion (percentage of sequencing reads that align or map to a specific phyla and genus to the total number of reads) of top 5 phyla and genus.
A hundred-fifty nine distinct AMR genes of 12 different antibiotic classes were identified in airborne dust from all pig farms (Table 5), including genes encoding resistance to aminoglycosides (24%), tetracycline (17%), macrolides (16%), β-lactams (13%), folate pathway antagonist (9%), lincomycin (7%), amphenicol (8%), as well as quinolone, polymyxin, glycoprotein, fosfomycin and fusidic acid at 1–2% of each (Figure 3).
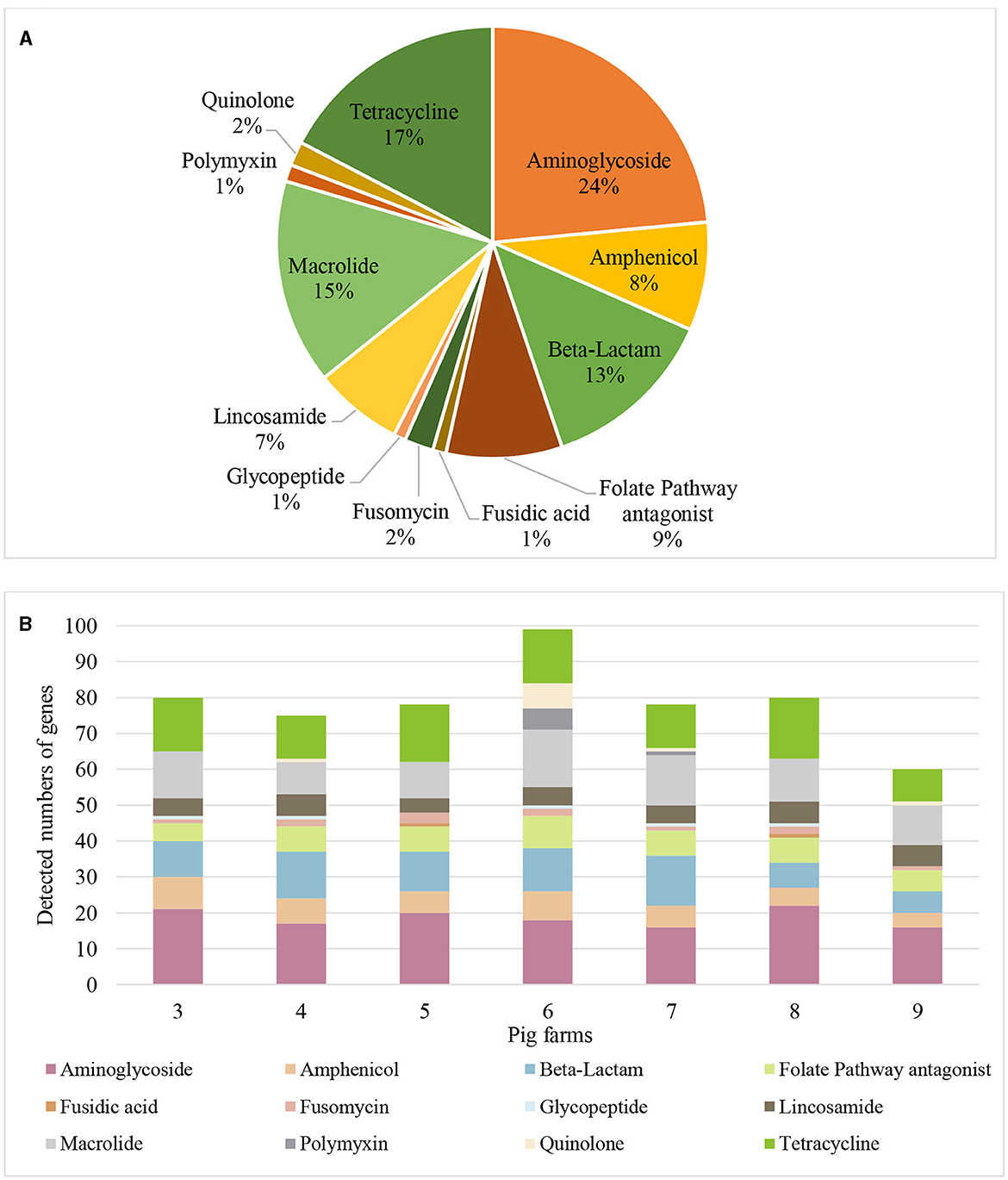
Figure 3. Antimicrobial resistance genes found in airborne dust inside pig farms (n = 7) by metagenomic sequencing. (A) Proportions of the number of detected genes for each antibiotic class. Proportion represents the percentage of number of detected genes for each antibiotic class to the total number of detected genes of resistance genes of different antibiotic classes from airborne dust collected inside the 7 pig farms. (B) Resistance gene profile in airborne dust inside the 7 pig farms. The stacked bar chart represents the number of detected resistance genes in each antibiotic class in each pig farm. Each colored segment within the bars corresponds to a specific resistance gene.
Seventy-four resistance genes were identified in ESKAPE species, of which Staphylococcus species (32 resistance genes) and Acinetobacter species (25 resistance genes) carried most AMR genes. The mcr 6.1 gene was detected in Klebsiella species, while Pseudomonas and Enterobacter species did not host any resistance genes as indicated by Bugseq metagenomic sequencing analysis data (Supplementary Table S3).
Several genes encoding resistance to clinically important antibiotics were determined, for example, ß-lactam resistance (e.g., blaCARB − 2, blaCARB − 4, blaOXA − 10, blaOXA − 164, blaOXA − 17, blaOXA − 209, blaOXA − 276, blaOXA − 284, blaOXA − 347, blaOXA − 360, blaOXA − 58) and colistin resistance (e.g., mcr-1.1, mcr-2.2, mcr-3.19, mcr-3.5, mcr-4.3, and mcr-6) (Table 5).
Two hundred-fifty-one plasmids were identified in all pig farms (Supplementary Table S4). Different numbers of plasmids were detected in different farms. The highest number of plasmids were detected in Farm 6 (n = 72), followed by Farm 3 (n = 56) and Farm 4 (n = 53). S. aureus (n = 48), A. baumannii (n = 19), and E. coli (n = 22) served as the most common hosts for the plasmids across all pig farms (Supplementary Table S4). No plasmids were identified in Pseudomonas and Enterobacter.
The same plasmid was detected in multiple farms, for example, pKKS49 (Accession no. NC_019149), and an unnamed3 plasmid (Accession no. CP027181) were found in Farms 3, 4, 6, 7 and 8. The predicted host of these two plasmids were S. aureus and A. baumannii, respectively. Another common plasmid is pSALNBL118 (Accession no. CP042023) with the predicted host of S. aureus that was found in Farms 3, 4, 5, 6 and 9. Several plasmids carried genes encoding resistance to clinically important antibiotics. For example, mcr-carrying plasmids were exclusively found in Farm 6 (n = 6), including pCHL5009T-102k-mcr3 (Accession no. CP032937), pEH_mcr4.3 (Accession no. CP038261), pMCR3_025943 (Accession no. CP027203), pSa4-CIP (Accession no. MG874042), pTR1 (Accession no. KJ187751), and pYY76–1–2 (Accession no. CP040929) (Supplementary Table S4). These plasmids were hosted by A. baumannii, E. coli, Salmonella spp., and Klebsiella pneumoniae.
One of the major findings in this study was the presence of various bacterial pathogens and plasmids in airborne dust within and around pig farms. The concentrations of culturable airborne bacteria in pig houses either average (4.8 103 CFU/m3) or individual farms (1.9–11.2 × 103 CFU/m3) in this study were comparable to a study conducted in farrowing, weaning, and fattening pig houses in China (38) but lower than previous studies in a nursery pig house in South Korea (1.34 × 105 CFU/m3) (39) and in pig confinement facility (1.8 × 104 CFU/m3) in the US (22). Thailand has a warm-humid tropical climate throughout the year and the sample collection was taken place in the rainy due to the expectation of high bacterial load inside the farm (23). The quantities of airborne microorganisms are affected by several factors e.g., different animal species, housing arrangements, management practices, and seasonal variations (40). However, the influence of these meteorological parameters was not pursued. The participating pig farms in this study implemented several farm management practices, including close house system with mechanical ventilation, routine cleaning and hygienic maintenance, biosecurity measures, age group separation in different houses, regular health monitoring and disease management systems, all of which helped to lower airborne bacterial counts. The Canadian Occupational Health and Safety Research Institute Robert Sauvé (IRSST) recommended that the concentration of total bacteria present during agricultural activity should not exceed 10,000 CFU/m3 in the air throughout an 8-h work period (41). The bacterial concentrations in all participating pig farms, except Farm 4 (11.2 × 103 CFU/m3), were lower than the suggested level. Farm 4 had the most total pigs (13,000 heads), as well as the most pigs (700 heads) in the sampling pen. Typically, larger pig populations in a confined space can result in higher microbial shedding, increased fecal matter and greater respiratory secretions, leading to an increased bacterial load in the air. These factors could explain the observation of the higher bacterial counts in airborne dust in Farm 4 that are greater than the advised level and those in other farms.
Staphylococcus has a remarkable ability to adapt their metabolic processes and composition to overcome obstacles, resulting in survival and persistence in an environment where many factors are unfavorable for growth and proliferation (42). Using either a culture-based approach or metagenomic sequencing analysis, Staphylococcus species were most prevalent in the airborne dust inside pig farms (Figure 1), in agreement with previous studies (43, 44). In contrast, Salmonella species was isolated only from fecal and feed samples, in agreement with previous studies conducted in pig buildings in the US (45) and Eastern Canada (46). The absence of Salmonella in airborne dust samples observed was possibly due to the Salmonella level being so low that they were undetectable or the culture media being inadequate for culturing airborne samples (46). The latter emphasizes the need for the development of more efficient methods to recover cultivable airborne Salmonella species.
It was previously demonstrated that Gram-positive bacteria predominated in airborne microbial contents whereas Gram-negative bacteria were in relatively low numbers (47). This is consistent with the current findings, which found that the most common species in all samples were Staphylococcus and Enterococcus species. This is likely because Gram-positive can produce spore that enhance their resistance to environmental stresses, leading to better growing in cultivation conditions (48). At the same time, Gram-negative bacteria tend to have short survival times in an airborne environment (49).
Pathogenic bacteria isolated from airborne dust in this study exhibited high AMR rates and most were resistant to multiple drugs (multidrug resistance, MDR), in agreement with previous studies (43, 50). The presence of MDR bacteria in the airborne dust raises a particular concern of the risk of occupational infections, and vice versa, there is a possibility that farm workers serve as asymptomatic reservoirs of MDR bacteria (50, 51). Airborne MDR bacteria have broader public health implications, as they can spread to the community and environment, including people who live close to pig farms.
Despite the limited number, the Acinetobacter and Pseudomonas isolates displayed resistance to various antimicrobials, e.g., aztreonam, cefotaxime, and ciprofloxacin. These two bacteria are opportunistic pathogens that may inflict diseases in people with impaired lung condition. Clindamycin is commonly used for the treatment of Staphylococcal and Streptococcal pneumonia, including community-acquired methicillin-resistant S. aureus (MRSA) infections (52, 53). Therefore, the presence of clindamycin-resistant Staphylococcus species in the airborne dust is of particular concern for public health. Taken together, the findings raise the alarm for the occupational and public health threat associated with airborne dust from pig farms and call for the development of effective measures to minimize the spread of AMR in farm environments.
Metagenomics provides a massive amount of data, yet a lot of it may be irrelevant or ambiguous. In this study, the combination of culture-dependent and -independent techniques was used. The total DNA for metagenomic sequencing analysis was prepared from the pooled bacteria, providing microbiome samples that mostly comprised bacteria DNA rather than other microorganisms. This combination was previously shown to improve understanding of human-associated microbial communities in relation to human health and disease (54). TSA, a non-selective media, contains nutrients required to support the growth of a wide range of bacteria and has been used for culturing bacteria from dust in several previous studies (22, 43, 55). The limitation was that TSA examined only culturable bacteria, resulting in the lack of non-culturable bacteria, fastidious bacteria with high nutrient requirements and bacteria with slow growth rate. The incubation conditions were aerobic; therefore, anaerobic bacteria are overlooked.
Metagenomic sequencing analysis in this study focused on the pooled bacteria collected inside each pig house to investigate the indoor microbial composition and its possible association with antimicrobial usage. Analyzing the microbiome composition of the outdoor environment of pig houses could provide insights into variations in microbial communities but the contributions of variables from other sources must be considered.
Bacillota was a major phylum in this study providing the evidence for the predominance of Gram-positive bacteria in airborne dust. This is in agreement with previous studies demonstrating the presence of the Bacillota in various atmospheric environments including animal farms (56) and animal feces served as a significant source of bacteria in this phylum (57). Therefore, the prolonged storage of manure in pits before its removal could be attributed to the higher bacteria level in the air within pig farms.
Indoor bioaerosols in food animal farms played a crucial role in the dissemination of AMR genes (12), in agreement with the observation of several genes encoding resistance to a wide range of antibiotic classes in this study (Supplementary Table S2). Aminoglycosides resistance encoding genes were at the highest abundance, in agreement with previous studies conducted in urban environment and pig farm (8, 58). A previous study showed that aminoglycoside resistance genes, such as aph(3′)-I, aadE, and aad were dominant in total suspended particulate samples collected from pig and chicken farms (58). The predominance of aadD, aadE, and aad(6) was previously observed in bioaerosols in chicken farms (59). High concentrations of aac6′-II, aadA1, aphA3, and strB in the air of various animal farms, e.g., pig, cattle, layer, and broiler farms were recently reported (13). These studies collectively underscore the importance of airborne dust from pig farms as a significant transmission pathway for AMR genes. Interestingly, no resistance genes were detected by Bugseq Metagenomic Sequencing Analysis in Pseudomonas and Enterobacter species, which are notorious for being MDR and containing multiple AMR genes (Supplementary Table S3). This may be attributed to low DNA content of these two species in airborne samples and the lack of certain AMR genes in the database. Further studies are warranted to explore the mechanisms and implications of AMR transmission through the atmosphere in livestock settings.
Previous studies showed that the global atmosphere is being polluted by AMR genes, in particular β-lactam resistance genes (7) and the occurrence of the genes in airborne particulate matter, dust, and human airways was positively correlated with Staphylococcus spp. (60). These agree with the metagenomic sequencing results in this study. A particular concern was the existence of blaOXA genes encoding OXA-type carbapenemases known to play a crucial role in carbapenem resistance in Acinetobacter and Pseudomonas species (61). The airborne transmission of these genes possibly provides the context for the clinical significance and potential treatment challenges in the clinical setting.
Colistin is one of the highest priorities critically important antibiotics but has been extensively used in pig farming. Colistin-resistant E. coli strains carrying plasmid-based mcr-1 were reported in food animals within China in 2016 (62). Since then, various mcr variants have been uncovered in livestock populations worldwide. The dissemination of colistin resistance gene including mcr-1, mcr-2 and mcr-3 genes were previously reported in Thai pig farms, of which mcr-3 was most common (63, 64). The Metagenomic Sequence analysis in this study revealed several plasmid-borne colistin resistance genes (e.g., mcr-1.1, mcr-2.2, mcr-3.19, mcr-3.5, mcr-4.3, and mcr-6). To our knowledge, mcr-4.3 and mcr-6 have never been reported in pig farms. No participating farms in this study explicitly mentioned the use of colistin. It is worth noting that colistin was frequently incorporated into medicated feed for suckling and nursery piglets in Thailand as a preventive measure against gastrointestinal tract infections. In Thai pig production, an estimated 40 tons of colistin were combined with medicated feed, of which 87.2% was designated for piglets (65). Extensive use of colistin may lead to the high colistin resistance in the pig production and can spread to the other farms via environmental contamination. The results underline the importance of further studies to comprehend the dynamics and implications of colistin resistance in the interconnected ecosystems of human, animal, and environmental health and to guide the development of effective strategies to preserve the efficacy of this critically important antibiotic.
Based on the metagenomic sequencing analysis, the same plasmids were detected in multiple farms, suggesting the spread of these plasmids among farms. It is well perceived that horizontal transfer of R plasmids plays a crucial role in the wide distribution of AMR. However, in vitro horizontal transmission of R plasmids was not pursued in this study.
The plasmids pSALNBL118, pKKS49, and an unnamed plasmid (Accession no. CP027181) were detected in up to 5 farms. pSALNBL118 is a phage like plasmid originated from S. aureus strain B3–4A isolated from beef liver (66) and might be important for horizontal gene transfer (67) and transmission of virulence factors. The plasmid pKKS49 carrying apmA encoding apramycin resistance, which were originated from an MRSA ST398 isolate obtained from a dust sample taken in a holding with breeding pigs in Portugal (68). Apramycin is frequently used for treatment and control of gastrointestinal tract infection in piglets in Thai pig farms (65), which can create selective pressure leading to resistant of this antibiotic (69). However, it was not disclosed if the antibiotic was used in the participating farm in this study. In addition, the unnamed plasmid carrying ant (2”)-Ia was commonly detected in gentamicin-resistant A. baumannii strain and should be further characterized due to the clinical importance of the pathogen.
The finding of mcr-harboring plasmids was limited to Farm 6, despite the farm not disclosing its use of the antibiotic. This suggests the possibility of contamination from external sources such as farm workers, contaminated feed, and water as well as the environmental contamination of AMR bacteria and genes, in addition to co-selection by other antibiotics. pCHL5009T-102k-mcr3 harboring mcr-3.5 and blaCTX − M55 was identified in E. coli. The plasmid was originally found to carry both mcr-1 and mcr-3 and isolated from E. coli in a patient in New Zealand who have experienced travel to Thailand (70). The observation of pCHL5009T-102k-mcr3 corresponded to previous studies that revealed the limited presence of mcr-1 and the predominance of mcr-3 in the E. coli isolates from healthy and sick pigs (64). Even though the reason underlying for the mcr-1 loss remains unclear, the co-localization of mcr and blaCTX − M55 on the same plasmid is an alarm for the distribution of bacterial pathogens resistant to last line antibiotics in the airborne dust in pig farm (71).
Additional limitations to this study are noted. Despite significant advancements in genomics, many resistance genes and bacterial species are still unidentified and unrepresented in databases. Furthermore, some bacteria with low biomass and resistance genes with low copies may not yield sufficient DNA copies for identifying by Metagenomic sequencing.
In conclusion, the results in this study demonstrated that the airborne dust around and within pig houses contained a wide array of microorganisms as well as AMR bacteria, posing potential risks to human, animal, and environmental health. The use of culture-based techniques in combination with metagenomic sequencing analysis enables the detection of a wider range of bacteria and their AMR genotype, providing valuable insights and providing crucial information for risk assessment, intervention planning, and informed decision-making to combat the spread of AMR. Specific guidelines to limit bioaerosol concentrations in livestock farm as well as to protect worker health are required. Future studies are suggested to examine the impact of AMR-contaminated airborne dust exposures in areas where animals are reared on the general public's health.
Metagenomic sequencing data were submitted to NCBI Sequence Read Archive (SRA) with accession numbers SRR25743046–SRR25743052 under the BioProject accession number PRJNA1006161.
SH: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing—original draft, Writing—review & editing. RP: Investigation, Writing—review & editing. SS: Investigation, Writing—review & editing. DM: Investigation, Methodology, Writing—review & editing. TW: Formal analysis, Software, Visualization, Writing—review & editing. PJ: Formal analysis, Software, Visualization, Writing—review & editing. PT: Investigation, Methodology, Writing—review & editing. RC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This Research is funded by the Thailand Science Research and Innovation Fund Chulalongkorn University [CU_FRB65_hea (88)_193_31_12]. It was also partially supported by the 90th year anniversary of Chulalongkorn University fund and National Research Council of Thailand (NRCT) Project ID N42A660897. SH is a recipient of Graduate Scholarship Programme for ASEAN and Non-ASEAN countries, Chulalongkorn University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1362011/full#supplementary-material
1. UNEP. Frontiers 2017 Emerging Issues of Environmental Concern. United Nations Environment Programme Nairobi (2017).
2. Wang J, Qin X, Guo J, Jia W, Wang Q, Zhang M, et al. Evidence of selective enrichment of bacterial assemblages and antibiotic resistant genes by microplastics in urban rivers. Water Res. (2020) 183:116113. doi: 10.1016/j.watres.2020.116113
3. D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D. Antibiotic resistance is ancient. Nature. (2011) 477:457–61. doi: 10.1038/nature10388
4. Chen Q. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int. (2016) 92:1–10. doi: 10.1016/j.envint.2016.03.026
5. Yang C, Zhang W, Liu R, Li Q, Li B, Wang S, et al. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ Sci Technol. (2011) 45:7408–15. doi: 10.1021/es2010545
6. Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A, et al. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. (2015) 69:234–42. doi: 10.1016/j.watres.2014.11.021
7. Li J, Cao J, Zhu Y-g, Chen Q-l, Shen F, Wu Y, et al. Global survey of antibiotic resistance genes in air. Environ Sci Technol. (2018) 52:10975–84. doi: 10.1021/acs.est.8b02204
8. Zhu G, Wang X, Yang T, Su J, Qin Y, Wang S, et al. Air pollution could drive global dissemination of antibiotic resistance genes. ISME J. (2021) 15:270–81. doi: 10.1038/s41396-020-00780-2
9. Gao M, Zhang X, Yue Y, Qiu T, Wang J, Wang X. Air path of antimicrobial resistance related genes from layer farms: emission inventory, atmospheric transport, and human exposure. J Hazard Mater. (2022) 430:128417. doi: 10.1016/j.jhazmat.2022.128417
10. Denissen J, Reyneke B, Waso-Reyneke M, Havenga B, Barnard T, Khan S, et al. Prevalence of eskape pathogens in the environment: antibiotic resistance status, community-acquired infection and risk to human health. J Hyg Environ. (2022) 244:114006. doi: 10.1016/j.ijheh.2022.114006
11. Muntean MM, Muntean A-A, Preda M, Manolescu LSC, Dragomirescu C, Popa M-I, et al. Phenotypic and genotypic detection methods for antimicrobial resistance in eskape pathogens. Exp Ther Med. (2022) 24:1–12. doi: 10.3892/etm.2022.11435
12. Bai H, He L-Y, Wu D-L, Gao F-Z, Zhang M, Zou H-Y, et al. Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ Int. (2022) 158:106927. doi: 10.1016/j.envint.2021.106927
13. Xin H, Gao M, Wang X, Qiu T, Guo Y, Zhang L. Animal farms are hot spots for airborne antimicrobial resistance. Sci Total Environ. (2022) 851:158050. doi: 10.1016/j.scitotenv.2022.158050
14. Song L, Wang C, Jiang G, Ma J, Li Y, Chen H, et al. Bioaerosol is an important transmission route of antibiotic resistance genes in pig farms. Environ Int. (2021) 154:106559. doi: 10.1016/j.envint.2021.106559
15. McEachran AD, Blackwell BR, Hanson JD, Wooten KJ, Mayer GD, Cox SB, et al. Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ Health Perspect. (2015) 123:337–43. doi: 10.1289/ehp.1408555
16. Casey JA, Kim BF, Larsen J, Price LB, Nachman KE. Industrial food animal production and community health. Curr Environ Health Rep. (2015) 2:259–71. doi: 10.1007/s40572-015-0061-0
17. Munk P, Knudsen BE, Lukjancenko O, Duarte ASR, Van Gompel L, Luiken RE, et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine european countries. Nat Microbiol. (2018) 3:898–908. doi: 10.1038/s41564-018-0192-9
18. Tiseo K, Huber L, Gilbert M, Robinson TP, Van Boeckel TP. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics. (2020) 9:918. doi: 10.3390/antibiotics9120918
19. Lekagul A, Kirivan S, Tansakul N, Krisanaphan C, Srinha J, Laoprasert T, et al. Antimicrobial consumption in food-producing animals in thailand between 2017 and 2019: the analysis of national importation and production data. PLoS ONE. (2023) 18:e0283819. doi: 10.1371/journal.pone.0283819
20. Luiken RE, Heederik DJ, Scherpenisse P, Van Gompel L, van Heijnsbergen E, Greve GD, et al. Determinants for antimicrobial resistance genes in farm dust on 333 poultry and pig farms in nine European countries. Environ Res. (2022) 208:112715. doi: 10.1016/j.envres.2022.112715
21. Chaivisit P, Fontana A, Galindo S, Strub C, Choosong T, Kantachote D. Airborne bacteria and fungi distribution characteristics in natural ventilation system of a university hospital in Thailand. Environ Asia. (2018) 11:2. doi: 10.14456/ea.2018.22
22. Green CF, Gibbs SG, Tarwater PM, Mota LC, Scarpino PV. Bacterial plume emanating from the air surrounding swine confinement operations. J Occup Environ Hyg. (2006) 3:9–15. doi: 10.1080/15459620500430615
23. Kumar P, Singh A, Singh R. Seasonal variation and size distribution in the airborne indoor microbial concentration of residential houses in Delhi and its impact on health. Aerobiologia. (2021) 37:719–32. doi: 10.1007/s10453-021-09718-3
24. EFSA. Report from the task force on zoonoses data collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia Coli and Enterococcus Spp. from food animals. EFSA J. (2008) 6:141r. doi: 10.2903/j.efsa.2008.141r
25. Ke D, Picard FoJ, Martineau F, Ménard C, Roy PH, Ouellette M, Bergeron MG. Development of a PCR assay for rapid detection of Enterococci. J Clin Microbiol. (1999) 37:3497–503. doi: 10.1128/JCM.37.11.3497-3503.1999
26. Sharp SE, Searcy C. Comparison of mannitol salt agar and blood agar plates for identification and susceptibility testing of Staphylococcus Aureus in specimens from cystic fibrosis patients. J Clin Microbiol. (2006) 44:4545–6. doi: 10.1128/JCM.01129-06
27. Dziri R, Klibi N, Alonso CA, Said LB, Bellaaj R, Slama KB, et al. Characterization of extended-spectrum B-lactamase (Esbl)-producing Klebsiella, Enterobacter, and Citrobacter obtained in environmental samples of a Tunisian hospital. Diagn Microbiol Infect Dis. (2016) 86:190–3. doi: 10.1016/j.diagmicrobio.2016.07.013
28. Younis A, Elbialy A, Abo Remila E, Ammar A. Molecular detection of genus Klebsiella and genotypic identification of Klebsiella Pneumoniae and Klebsiella Oxytoca by duplex polymerase chain reaction in poultry. Glob Vet. (2017) 18:234–41. doi: 10.5829/idosi.gv.2017.234.241
29. Anane A Y, Apalata T, Vasaikar S, Okuthe GE, Songca S. Prevalence and molecular analysis of multidrug-resistant Acinetobacter baumannii in the extra-hospital environment in Mthatha, South Africa. Braz J Infect Dis. (2020) 23:371–80. doi: 10.1016/j.bjid.2019.09.004
30. Ghane M, Azimi Z. Isolation, Identification and antimicrobial susceptibility of Pseudomonas Spp. isolated from hospital environment in Tonekabon, North of Iran. Appl Environ Microbiol. (2014) 2:97–101. doi: 10.12691/jaem-2-4-2
31. Asselin JAE, Bonasera JM, Beer SV. Pcr primers for detection of pantoea ananatis, Burkholderia Spp, and Enterobacter Sp from onion. Plant Dis. (2016) 100:836–46. doi: 10.1094/PDIS-08-15-0941-RE
32. Feng P, Weagant SD, Grant MA, Burkhardt W, Shellfish M, Water B. Bam: Enumeration of Escherichia Coli and the Coliform Bacteria. (2002), 1–13.
33. ISO. Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Detection of Salmonella Spp. Amendment 1: Anex D: Detection of Salmonella Spp. In Animal Faeces and in Environmental Samples from the Primary Production Stage. Geneva: International Standard Organization (2007).
34. Meiri-Bendek I, Lipkin E, Friedmann A, Leitner G, Saran A, Friedman S, et al. PCR-based method for the detectionof Streptococcus Agalactiae in milk. J Dairy Sci. (2002) 85:1717–23. doi: 10.3168/jds.S0022-0302(02)74245-8
35. Wyder AB, Boss R, Naskova J, Kaufmann T, Steiner A, Graber HU. Streptococcus Spp. and related bacteria: their identification and their pathogenic potential for chronic mastitis–a molecular approach. Res Vet Sci. (2011) 91:349–57. doi: 10.1016/j.rvsc.2010.09.006
36. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
37. Fan J, Huang S, Chorlton SD. Bugseq: a highly accurate cloud platform for long-read metagenomic analyses. BMC Bioinform. (2021) 22:1–12. doi: 10.1186/s12859-021-04089-5
38. Cui H, Zhang C, Liu J, Dong S, Zhao K, Chen L, et al. The distribution characteristics of aerosol bacteria in different types of pig houses. Animals. (2022) 12:1540. doi: 10.3390/ani12121540
39. Jo W-K, Kang J-H. Exposure levels of airborne bacteria and fungi in korean swine and poultry sheds. Arch Environ Occup Health. (2005) 60:140–6. doi: 10.3200/AEOH.60.3.140-146
40. Zhao Y, Aarnink AJ, De Jong MC, Groot Koerkamp PW. Airborne microorganisms from livestock production systems and their relation to dust. Crit Rev Environ Sci Technol. (2014) 44:1071–128. doi: 10.1080/10643389.2012.746064
41. Goyer N, Lavoie J, Lazure L, Marchand G. Bioaerosols in the Workplace: Evaluation, Control and Prevention Guide. London: Institut de recherche Robert-Sauvé en santé et en sécurité du travail (2001).
42. Onyango LA, Alreshidi MM. Adaptive metabolism in staphylococci: survival and persistence in environmental and clinical settings. J Patho. (2018) 2018:2632. doi: 10.1155/2018/1092632
43. Gibbs SG, Green CF, Tarwater PM, Mota LC, Mena KD, Scarpino PV. Isolation of antibiotic-resistant bacteria from the air plume downwind of a swine confined or concentrated animal feeding operation. Environ Health Perspect. (2006) 114:1032–7. doi: 10.1289/ehp.8910
44. Predicala BZ, Urban JE, Maghirang RG, Jerez SB, Goodband RD. Assessment of bioaerosols in swine barns by filtration and impaction. Curr Microbiol. (2002) 44:136–40. doi: 10.1007/s00284-001-0064-y
45. Elliott L, McCalla T, Deshazer J. Bacteria in the air of housed swine units. Appl Environ Microbiol. (1976) 32:270–3. doi: 10.1128/aem.32.2.270-273.1976
46. Pilote J, Létourneau V, Girard M, Duchaine C. Quantification of airborne dust, endotoxins, human pathogens and antibiotic and metal resistance genes in eastern canadian swine confinement buildings. Aerobiologia. (2019) 35:283–96. doi: 10.1007/s10453-019-09562-6
47. Cormier Y, Tremblay G, Meriaux A, Brochu G, Lavoie J. Airborne microbial contents in two types of swine confinement buildings in Quebec. Am Ind Hyg Assoc. (1990) 51:304–9. doi: 10.1202/0002-8894(1990)051<0304:AMCITT>2.0.CO;2
48. Ruiz-Gil T, Acuña JJ, Fujiyoshi S, Tanaka D, Noda J, Maruyama F, et al. Airborne bacterial communities of outdoor environments and their associated influencing factors. Environ Inter. (2020) 145:106156. doi: 10.1016/j.envint.2020.106156
49. Zucker BA, Trojan S, Müller W. Airborne gram-negative bacterial flora in animal houses. J Vet Med. (2000) 47:37–46. doi: 10.1046/j.1439-0450.2000.00308.x
50. Chapin A, Rule A, Gibson K, Buckley T, Schwab K. Airborne multidrug-resistant bacteria isolated from a concentrated swine feeding operation. Environ Health Perspect. (2005) 113:137–42. doi: 10.1289/ehp.7473
51. Just NA, Létourneau V, Kirychuk SP, Singh B, Duchaine C. Potentially pathogenic bacteria and antimicrobial resistance in bioaerosols from cage-housed and floor-housed poultry operations. Ann Occup Hyg. (2012) 56:440–9. doi: 10.1093/annhyg/mer105
52. Herigon JC, Hersh AL, Gerber JS, Zaoutis TE, Newland JG. Antibiotic management of Staphylococcus Aureus infections in US children's hospitals, 1999–2008. Pediatrics. (2010) 125:e1294–e300. doi: 10.1542/peds.2009-2867
53. Marcinak JF, Frank AL. Treatment of community-acquired methicillin-resistant Staphylococcus Aureus in children. Curr Opin Infect Dis. (2003) 16:265–9. doi: 10.1097/00001432-200306000-00014
54. Whelan FJ, Waddell B, Syed SA, Shekarriz S, Rabin HR, Parkins MD, et al. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat Microbio. (2020) 5:379–90. doi: 10.1038/s41564-019-0643-y
55. Vantarakis A, Paparrodopoulos S, Kokkinos P, Vantarakis G, Fragou K, Detorakis I. Impact on the quality of life when living close to a municipal wastewater treatment plant. J Environ Public Health. (2016) 2016:7023. doi: 10.1155/2016/8467023
56. de Groot GA, Geisen S, Wubs EJ, Meulenbroek L, Laros I, Snoek LB, et al. The aerobiome uncovered: multi-marker metabarcoding reveals potential drivers of turn-over in the full microbial community in the air. Environ Int. (2021) 154:106551. doi: 10.1016/j.envint.2021.106551
57. Gao X-L, Shao M-F, Wang Q, Wang L-T, Fang W-Y, Ouyang F, et al. Airborne microbial communities in the atmospheric environment of urban hospitals in China. J Hazard Mater. (2018) 349:10–7. doi: 10.1016/j.jhazmat.2018.01.043
58. Yang Y, Zhou R, Chen B, Zhang T, Hu L, Zou S. Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach. Chemosphere. (2018) 213:463–71. doi: 10.1016/j.chemosphere.2018.09.066
59. Yang F, Gao Y, Zhao H, Li J, Cheng X, Meng L, et al. Revealing the distribution characteristics of antibiotic resistance genes and bacterial communities in animal-aerosol-human in a chicken farm: from one-health perspective. Ecotoxicol Environ Saf. (2021) 224:112687. doi: 10.1016/j.ecoenv.2021.112687
60. Zhou Z-C, Liu Y, Lin Z-J, Shuai X-Y, Zhu L, Xu L, et al. Spread of antibiotic resistance genes and microbiota in airborne particulate matter, dust, and human airways in the urban hospital. Environ Int. (2021) 153:106501. doi: 10.1016/j.envint.2021.106501
61. Evans BA, Amyes SG. Oxa B-Lactamases. Clin Microbiol Rev. (2014) 27:241–63. doi: 10.1128/CMR.00117-13
62. Liu Y-Y, Wang Y, Walsh TR Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism Mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
63. Lay KK, Jeamsripong S, Sunn KP, Angkititrakul S, Prathan R, Srisanga S, et al. Colistin resistance and Esbl production in Salmonella and Escherichia Coli from Pigs and Pork in the Thailand, Cambodia, Lao Pdr, and Myanmar border area. Antibiotics. (2021) 10:657. doi: 10.3390/antibiotics10060657
64. Trongjit S, Assavacheep P, Samngamnim S, My TH, An VTT, Simjee S, et al. Plasmid-mediated colistin resistance and Esbl production in Escherichia Coli from clinically healthy and sick pigs. Sci Rep. (2022) 12:2466. doi: 10.1038/s41598-022-06415-0
65. Lekagul A, Tangcharoensathien V, Mills A, Rushton J, Yeung S. How Antibiotics Are Used in Pig Farming: A Mixed-Methods Study of Pig Farmers, Feed Mills and Veterinarians in Thailand. BMJ Glob Health. (2020) 5:e001918. doi: 10.1136/bmjgh-2019-001918
66. Karki AB, Neyaz L, Fakhr MK. Comparative genomics of plasmid-bearing Staphylococcus Aureus strains isolated from various retail meats. Front Microbiol. (2020) 11:574923. doi: 10.3389/fmicb.2020.574923
67. Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, et al. Diversity of prophages in dominant Staphylococcus Aureus clonal lineages. J bacteriol. (2009) 191:3462–8. doi: 10.1128/JB.01804-08
68. Kadlec K, Feßler AT, Couto N, Pomba CF, Schwarz S. Unusual small plasmids carrying the novel resistance genes Dfrk or Apma isolated from methicillin-resistant or-susceptible staphylococci. J Antimicrob Chemother. (2012) 67:2342–5. doi: 10.1093/jac/dks235
69. Luppi A. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag. (2017) 3:1–18. doi: 10.1186/s40813-017-0063-4
70. Creighton J, Anderson T, Howard J, Dyet K, Ren X, Freeman J. Co-Occurrence of Mcr-1 and Mcr-3 genes in a single Escherichia Coli in New Zealand. J Antimicrob Chemother. (2019) 74:3113–6. doi: 10.1093/jac/dkz311
Keywords: antimicrobial resistance, airborne dust, bacterial communities, metagenomic approach, pig farm
Citation: Hein ST, Prathan R, Srisanga S, Muenhor D, Wongsurawat T, Jenjaroenpun P, Tummaruk P and Chuanchuen R (2024) Metagenomic insights into isolable bacterial communities and antimicrobial resistance in airborne dust from pig farms. Front. Vet. Sci. 11:1362011. doi: 10.3389/fvets.2024.1362011
Received: 27 December 2023; Accepted: 30 April 2024;
Published: 30 May 2024.
Edited by:
Alda Natale, Experimental Zooprophylactic Institute of the Venezie (IZSVe), ItalyReviewed by:
Maria King, Texas A&M University, United StatesCopyright © 2024 Hein, Prathan, Srisanga, Muenhor, Wongsurawat, Jenjaroenpun, Tummaruk and Chuanchuen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rungtip Chuanchuen, Y2h1YW5jaHVlbi5yQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.