
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci., 31 January 2024
Sec. Veterinary Epidemiology and Economics
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1361788
This article is part of the Research TopicNontuberculous Mycobacterial Infections in Animals And Humans: Pathogenesis, Diagnosis, Prevention, Treatment, And EpidemiologyView all 13 articles
 Alberto Gomez-Buendia1,2
Alberto Gomez-Buendia1,2 Julio Alvarez1,2
Julio Alvarez1,2 Javier Bezos1,2
Javier Bezos1,2 Jorge Mourelo3
Jorge Mourelo3 Javier Amado4
Javier Amado4 Jose Luis Saez5
Jose Luis Saez5 Lucia de Juan1,2
Lucia de Juan1,2 Beatriz Romero1,2*
Beatriz Romero1,2*Non-tuberculous mycobacteria (NTM) are considered a relevant cause of non-specific reactions to the most widely applied bovine tuberculosis (bTB) test, the intradermal tuberculin test. In order to establish which NTM species might act as a potential source of such diagnostic interference, a collection of 373 isolates obtained from skin test positive cows from 359 officially tuberculosis-free (OTF) herds, culled in the framework of the bTB eradication campaign in Spain, were identified at the species level through PCR and Sanger sequencing of the 16S rDNA, hsp65 and rpoB genes. Of the 308 isolates for which a reliable identification was achieved, 32 different mycobacterial species were identified, with certain species being most represented: among M. avium complex members (n = 142, 46.1%), M. avium subsp. hominissuis (98; 69.0%) was the most abundant followed by M. avium subsp. avium (33, 23.2%), and M. intracellulare (7, 4.9%). Among non-MAC members (n = 166, 53.9%), M. nonchromogenicum (85; 27.6%) and M. bourgelatii (11; 5.6%) were the predominant species. In addition, mixed results were obtained in 53 isolates presenting up to 30 different genotypes, which could be indicative of new mycobacterial species. Our results represent a first step toward characterizing the diversity of NTM species that could interfere with official diagnostic tests for bTB eradication in Spain.
The genus Mycobacterium is very large, encompassing 196 different child taxa with validly published and correct names described (1). Mycobacteria can take on different roles: there are highly relevant animal and human pathogens such as M. bovis and M. caprae, members of the Mycobacterium tuberculosis complex (MTC) and the most common causative agents of bovine tuberculosis (bTB), while others like the non-tuberculous mycobacteria (NTM) are typically free-living microorganisms widely distributed in the environment. These NTM can be found in soil, water, dust, etc. (2), but in certain cases, usually linked to immunosuppressive processes, they can infect humans and animals and act as opportunistic pathogens (3). In cattle, infection with NTM has not been traditionally associated with the presence of clinical signs [with the notable exception of M. avium subsp. paratuberculosis (Map), the causative agent of paratuberculosis], although the presence of granulomatous lesions as a consequence of infection has been described (4–7).
Since the beginning of the establishment of bTB eradication programs in different regions of the world in the early 1900s, several countries have been successful in the eradication of the disease (8). In others, however, despite all the efforts made, the disease is still endemic (9). Failure of these programmes has been partly attributed to the limitations in the sensitivity and specificity of existing diagnostic tests. Among factors compromising the performance of these tests, NTMs have been repeatedly linked to the occurrence of non-specific reactions in the single and comparative intradermal tuberculin tests (SIT and CIT), the main diagnostic tools used as the basis of eradication programs worldwide (10).
Most studies evaluating the role of NTM in bTB diagnostic problems have focused on those produced by Map, showing that it can affect both specificity and sensitivity (11–14). Nevertheless, NTM other than Map, also known as atypical or environmental, can also impact the reliability of bTB diagnostic tests due to the occurrence of cross-reactions in TB-free animals (15, 16), while their impact on the sensitivity of the tests remains to be quantified. Several studies have identified which species may be most commonly associated with these cross-reactions on bTB diagnostic tests in different settings, with M. avium subspecies and M. nonchromogenicum being the most commonly retrieved mycobacteria from bovine samples (15, 17–19). However, not all samples evaluated in these studies were from test positive animals or officially tuberculosis-free (OTF) farms. For instance, in Spain a recent study describing the presence of NTM in animals included only three reactor cattle from OTF herds, with other isolates originating from herds in which the disease was present, thus making the role of these bacterial species on the occurrence of cross-reactions unclear (19).
Therefore, the objective of this study was to identify the species associated with non-specific reactions detected in the framework of the bTB eradication campaign in Spain in a large cohort (n = 373) of skin test-reactor cattle from OTF herds located in areas with different prevalence of the disease. This information can be useful for better understanding the primary species affecting the specificity of skin tests for bTB and to assess the geographical variability in their occurrence.
All NTM isolates available at the VISAVET Health Surveillance Center retrieved between 2011 and 2020 from skin test-positive cattle located in 359 OTF herds detected in the frame of the Spanish eradication program were evaluated. Isolates were either cultured directly at VISAVET from cattle samples collected at the abattoir and analyzed in the laboratory there or submitted by official veterinary laboratories located in two OTF regions (Asturias and Galicia, 72.1% of the herds), with the remaining sampled animals originating from different parts of the country (Figure 1). Veterinary laboratories submitted the isolates to VISAVET for identification once they had confirmed they did not belong to the M. tuberculosis complex except for the OTF region of Asturias, which mostly submitted isolates already identified as belonging to the M. avium complex (MAC).
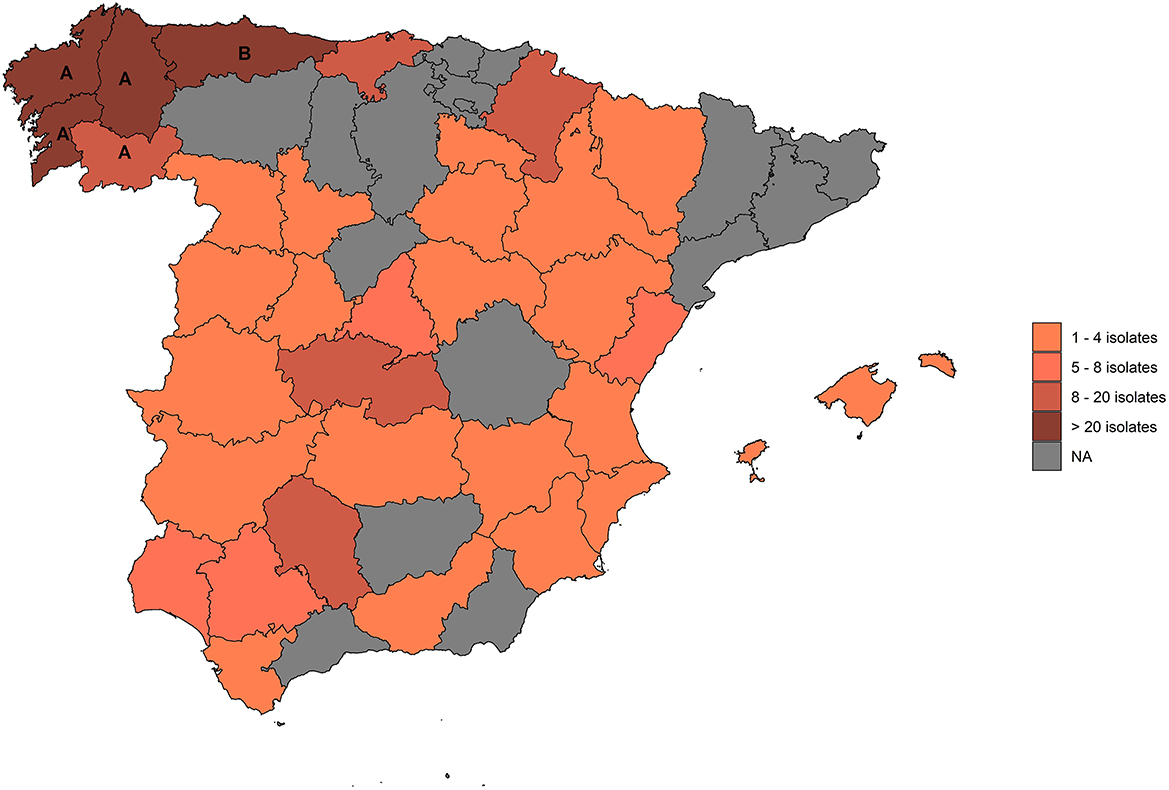
Figure 1. Provinces from which isolates included in this study originated. (A): Galicia; (B): Asturias.
According to the eradication program, when a skin test positive animal is identified in an OTF herd the animal must be culled within a maximum of 15 days and tissue samples are collected in the abattoir and submitted to an authorized laboratory in order to isolate or detect in the samples the presence of MTC members through bacteriology (20) and more recently direct PCR (21). Bacteriological analysis of these samples can occasionally lead to the identification of NTM species, which are identified as such in the laboratory according to the following protocol (Figure 2): once growth is observed, DNA is extracted and subjected to multiplex PCR for the detection of a Mycobacterium-specific DNA fragment (16S rDNA sequence) (22) and of a MTC-specific target (mpb70 gene) (22). If the presence of a MTC member is not detected, another PCR aiming at a MAC-specific 16S rDNA (22) is performed, followed in case of a positive result by two PCRs aiming at the IS901 and IS1245 sequences associated with M. avium subsp. avium (Maa) (23) and M. avium subsp. hominissuis (Mah) (24). The available NTM strain collection included all isolates identified as Mycobacterium spp. based on the detection of the 16S rDNA Mycobacterium fragment but not belonging to MTC based on the absence of the mpb70 gene that had been initially cultured at VISAVET or that were cultured in other authorized laboratories and later submitted to VISAVET for further identification. Isolates not belonging to MAC according to the methodology explained above were subjected to PCR amplification followed by Sanger sequencing of several targets: first, sequences of partial fragments of the 16S rDNA (22) and hsp65 (Telenti fragment) genes (25, 26) were generated. Forward and reverse sequences were curated using BioEdit software (27) and combined to yield a consensus sequence that was then screened using the NCBI Basic Local Alignment Search Tool (BLAST) to identify the bacterial species. An identification was considered reliable if sequence similarity and coverage with a target was >99 and 100%, respectively (28). If the 16S rDNA/hsp65 analysis yielded a non-reliable identification (due to low similarity/coverage and/or disagreement in the species identified through each target), amplification and sequencing of fragments of the rpoB gene (29) and then the 3′ region of the hsp65 gene (30) was conducted and analyzed as described above. Full workflow is shown in Figure 2. Prior to bacteriological culture, samples were evaluated for the presence of macroscopial lesions compatible with bTB (31).
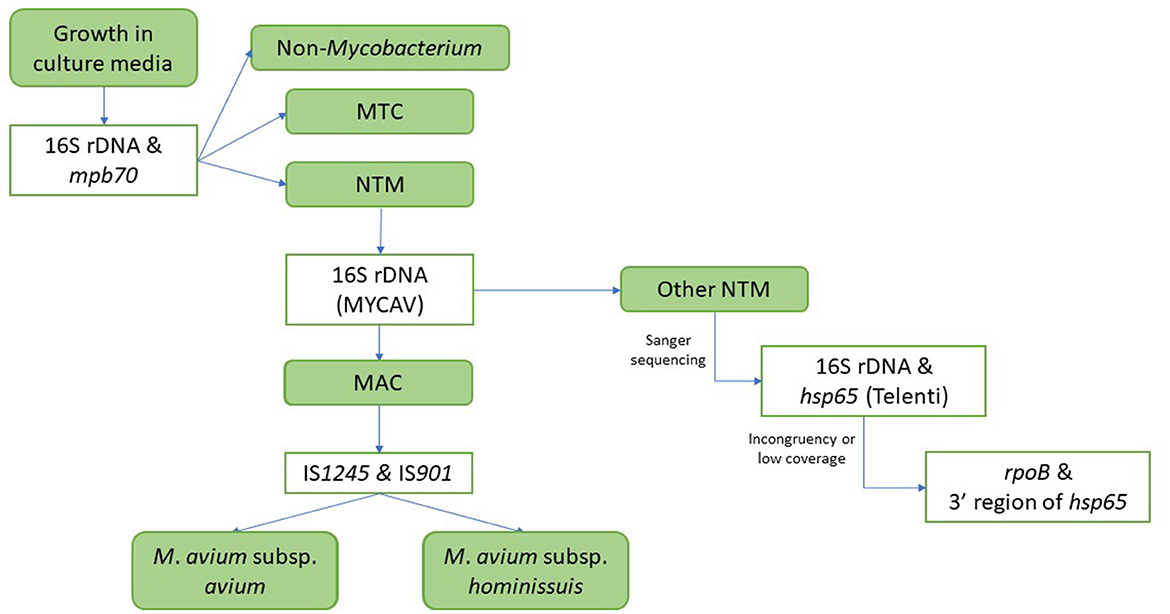
Figure 2. Workflow for the identification of isolates. MTC, Mycobacterium tuberculosis complex; NTM, non-tuberculous mycobacteria; MAC, Mycobacterium avium complex; MYCAV, MAC-specific 16S rDNA PCR.
All primers used are shown in Supplementary Table 1. Sequencing was performed by STABvida (Lisbon, Portugal).
Overall, 373 isolates recovered from positive cattle located in 359 OTF herds across 34 provinces (15 Autonomous Communities) were included in the study (Figure 1). Of these, 194 (54.0%) were beef herds, 150 (41.8%) were dairies, eight (2.2%) were fattening units, and seven (1.9%) were bullfighting herds. Of the 373 isolates, we were able to achieve a reliable identification in 308 (82.6%). Among these, 166 (53.9%) were classified as non-MAC NTM and 142 (46.1%) as MAC species. In the remaining 65 isolates (17.4% of all samples) a reliable identification was not achieved. Of them, 30 (46.2%) were classified as non-MAC NTM, 23 (35.4%) as MAC, and the 16S rDNA sequence revealed a bacteria not belonging to the Mycobacterium genus for the remaining 12 (18.5%) isolates.
Within the 166 non-MAC NTM isolates, 27 species were reliably identified (Table 1), being the most frequent species M. nonchromogenicum (n = 85; 51.2%), followed by M. bourgelatii (n = 11; 6.6%), M. shimoidei (8; 4.8%), M. kansasii (7; 4.2%) and M. intermedium (7; 4.2%). A reliable identification was not achieved in 30 isolates (15.3%), of which 8 and 16 did not yield a positive result for the rpoB and 3′ region of hsp65 genes, respectively (Table 2). Among these isolates, we identified 23 “genotypes” present in 1–5 isolates. A genotype is defined as distinct partial 16S rDNA and hsp65 (Telenti fragment), and, when available, rpoB and 3′ region of hsp65 sequences. Most of the non-MAC NTM were retrieved from Galicia (141/196). However, the diversity of genotypes was almost the same as in the non-OTF regions (29 and 28 genotypes, respectively).
For the 142 MAC isolates five species were identified (Mah, Maa, M. intracellulare, M. yongonense, and M. colombiense). Of these, M. avium subspecies accounted for 92.3% (n = 131) of the retrieved isolates, with Mah (98; 69.0%) being more represented than Maa (33; 23.2%). Most MAC isolates (63/165) originated from Asturias since this region submitted primarily cultures preliminarily identified as MAC, but 56 and 46 MAC isolates from Galicia and non-OTF regions, respectively, were also included. Regarding the predominant MAC species, Mah was recovered more frequently than Maa in all regions, and in the case of Galicia, M. intracellulare (4) was identified even more frequently than Maa (1).
Of the 23 isolates with non-reliable identification, we did not obtain a PCR amplicon for the rpoB and 3′ region of hsp65 genes in 11 and 20 isolates, respectively. Among these 23 isolates, seven genotypes, as previously defined, were identified in between 1 and 10 isolates each. The most common genotype was found in 10 isolates from five provinces in which a 99.8% similarity with the 16S rDNA sequence of M. colombiense, M. intracellulare, and M. bouchedurhonense coupled with a 97.4% similarity with the hsp65 M. scrofulaceum gene sequence was obtained. All genotypes for isolates with a non-reliable identification are shown in Table 2.
Finally, the 16S rDNA sequence revealed that 12 isolates did not belong to the Mycobacterium genus but instead to the following species: Prauserella rugosa (n = 3), Streptomyces hydrogenans (3), uncultured actinobacteria (3), Corynebacterium pseudotuberculosis (2), and Brevibacillus brevis (1).
Overall, macroscopical granulomatous bTB-like lesions were observed in lymph node samples from 20 animals (5.3%). From these samples, 11 different species were identified: Mah (n = 6), genotype 2 (2) (Table 2), M. nonchromogenicum (2), M. shimoidei (2), Maa (1), M. intracellulare (1), M. xenopi (1), Corynebacterium pseudotuberculosis (1), M. interjectum (1), M. yongonense, and genotype 7 (1) (Table 2).
Non-tuberculous mycobacteria are one of the limiting factors compromising diagnostic performance of bTB tests (15, 32). As the burden of bTB is decreasing in most countries in which eradication campaigns have been consistently applied, the need to maintain a high specificity is key to keeping the positive predictive value as high as possible, but without compromising the overall sensitivity of the surveillance system. The characterization of the factors affecting diagnostic specificity, such as the most prevalent NTM associated with non-specific reactions to bTB skin tests, can be useful to design strategies aiming at minimizing their impact. Here, we present a thorough characterization of a large panel of NTM isolates retrieved from skin test reactor cattle from OTF herds to assess their diversity in Spain.
A great percentage of the isolates included in the study belonged to MAC (specifically to the Mah, Maa, M. intracellulare, M. colombiense, and M. yongonense species, of which the first two were by far the most common), partly due to the fact that the second region from which a higher number of isolates were available (Asturias) provided mostly NTM preliminary identified as MAC (Table 1) (33). Nevertheless, the high frequency of isolation of MAC members from cattle here is in agreement with previous studies in which certain MAC members (Mah, Maa, Map, M. arosiense, M. bouchedurhonense, M. chimaera, M. colombiense, M. intracellulare, and M. vulneris) were also isolated from bovine samples and identified as a potential cause of non-specific reactions (15, 19, 34). The potential to elicit a cross-reacting immune response in bTB diagnostic tests by MAC members (and to some extent also by other NTMs) should be reduced by the use of the CIT, which also considers the response elicited by the avian protein purified derivative (PPD-A) obtained from a Maa isolate (35, 36), so diagnostic interference should be limited in areas where this test is used (33). This, however, can lead to a decrease in diagnostic sensitivity of between 14 and 44 percentage points according to some estimates (37), and therefore comparative tests should be used when the risk of tuberculosis is considered low. In our study, most of the animals (353/373) were reactors in the SIT (based only on the bovine PPD), and in fact, of the only 20 animals that were reactors in the CIT (all coming from a current OTF area), M. nonchromogenicum was isolated from 13 of them and only one MAC species (M. colombiense) was isolated, suggesting that the CIT could be compromised to a larger extent by certain mycobacterial species.
M. nonchromogenicum was the most frequently isolated non-MAC NTM. This species was the most common NTM retrieved in samples from cattle in Northern Ireland (23 of 48 animals) (18) and Hungary (30 of 104) (17), and the second most common in France (81 of 310) (15), although in these studies not all samples came from skin-positive animals or OTF herds. M. nonchromogenicum has also been described in cattle samples with lesions collected at abattoirs in Switzerland (6), South Africa (34) and Ethiopia (7, 38), as well as in milk samples from positive cows to the CIT in Brazil (39) and in the nasal mucosa of cattle (40). In another study carried out in Spain (19), including tissue samples from cattle and wild boar in the Basque country (from which no samples were included here), M. nonchromogenicum was isolated in three of 21 SIT positive cattle and nine wild boars. These results, coupled with our findings, demonstrate that M. nonchromogenicum can be found in reactor cattle from Northern regions in Spain, from which most isolates were retrieved, and could be a relevant contributor to the development of cross-reactions in the skin test. The ability of M. nonchromogenicum to elicit reactions after the inoculation of certain mycobacterial antigens (including PPD-A and PPD-B) was explored in an experimental infection model in guinea pigs, but the observed induced reactions were limited (16). Similarly, the ability of this species to cause false positive reactions in skin tests in cattle has not been demonstrated yet, since experimental studies to assess the cross-reaction potential of NTM have been only limited to M. kansasii (4, 41) and M. fortuitum (42).
Regarding the geographical distribution of non-MAC NTMs, 93% of the 85 M. nonchromogenicum isolates identified here were recovered from cattle from Galicia, representing 56% of all non-MAC NTMs available from this particular region. In contrast, no non-MAC NTM accounted for more than 10% of the isolates retrieved from non-OTF regions, suggesting a more heterogeneous distribution of NTM species. When considering MAC isolates, Mah was the most commonly found species, but its relative frequency compared with other MAC species varied between regions: in Galicia 85.4% of all MAC isolates with a reliable identification (41/48) were Mah and only one Maa isolate was retrieved. In contrast in Asturias 34 Mah and 25 Maa isolates were identified, accounting for 57.6 and 42.4% of the 59 MAC isolates with reliable identification, respectively, while in non-OTF regions Mah and Maa represented 65.7 and 20.0% of the 46 MAC isolates available from these regions, respectively. Although these differences may be pointing out at differences in the epidemiology of cross-reactions due to NTM in cattle between OTF and vs. non-OTF regions, the highly heterogeneous sampling efforts may have led to biases contributing to these results and thus these findings should be interpreted with care. Differences in the most prevalent NTM species associated with cross-reactions were not influenced by the production type, with M. nonchromogenicum being the most abundant in both beef and dairy herds in Galicia (42.3 and 38.4% of all isolates in each type, respectively). Similarly, the distribution of MAC members across different production type herds in Asturias was also similar, with Mah being isolated in 52.5 and 56.5% of the beef and dairy herds, respectively, and Maa in 40.0 and 34.8% of them. This suggests that the prevalence of the NTM may be more influenced by regional factors rather than linked to the type of production.
Interestingly, 53 isolates in our study could not be reliably assigned to a specific species. Among them, 10 isolates coming from beef herds exhibited identical sequences in the four sequenced genes (genotype 1, Table 2), which would constitute the fifth most common “species” in our study. These isolates had a similarity of 99.8% with M. colombiense/intracellulare/bouchedurhonense (MAC members) based on the 16S rDNA and of 100% with a sequence previously classified as belonging to MAC based on the rpoB gene (19). However, the closest match to their hsp65 short sequence was M. scrofulaceum (97.4% of similarity), while the highest similarity with a MAC member hsp65 sequence was only 95.7%, below the 97.3% threshold previously suggested for MAC members for this gene (33). This result highlights the importance of using multiple genetic markers to achieve a comprehensive identification of the species involved (28, 43). Further studies would be required to conclude on the nature of these isolates.
Our results demonstrate that multiple NTM species could be associated with the occurrence of non-specific reactions in the skin test in cattle in Spain. Nevertheless, the relative importance of MAC members (especially Mah and Maa) and M. nonchromogenicum, which constituted up to 69% of all isolates considered in the study, demonstrates that certain species are more likely to lead to diagnostic interferences in OTF herds in low prevalence/OTF areas of the country, in agreement with what has been described in other European (6, 15, 17–19) and non-European countries (34, 38, 39).
By including only isolates retrieved from positive animals in OTF herds (that maintained this status for at least 3 years after the reactor was found) we attempted to minimize the chance that animals could have also been infected with a MTC member. Further studies based on guinea pig and cattle experimental models will be conducted to confirm the ability of the characterized strains to induce non-specific reactions in non-bTB infected cattle, and to explore the ability of different antigens to discriminate the immune response induced in skin test and interferon-gamma assays. Altogether, this will contribute to limit the interference in the results of routine diagnostic tests, which can compromise the reliability of the herd status obtained and the non-desirable risk management measures that are necessary to implement in the context of an eradication programs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR642807—OR642833, OR648408—OR648468, OR671981—OR672046.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because no need. All the animals were slaughtered due to positive results to the official skin test following the Bovine Tuberculosis Eradication Program in Spain.
AG-B: Data curation, Formal analysis, Writing—original draft, Writing—review & editing. JAl: Data curation, Funding acquisition, Resources, Supervision, Writing—review & editing. JB: Funding acquisition, Writing—review & editing. JM: Conceptualization, Supervision, Writing—review & editing. JAm: Data curation, Resources, Writing—review & editing. JS: Investigation, Resources, Writing—review & editing. LdJ: Data curation, Funding acquisition, Writing—review & editing. BR: Data curation, Methodology, Supervision, Writing—review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AG-B holds a PhD fellowship (reference CT58/21-CT59/21) from the Universidad Complutense de Madrid and Banco Santander. This work was funded by the European Union Reference Laboratory for Bovine Tuberculosis, the Spanish Ministry of Agriculture, Fisheries and Food, and is a contribution to the project Integrated Strategies for Tuberculosis Control and Eradication in Spain (ERATUB) (RTI2018-096010-BC22, Ministerio de Ciencia, Innovación y Universidades, MICINN).
The authors would like to thank the collaboration and support of the EU-RL for Bovine Tuberculosis, the Spanish Ministry of Agriculture, Fisheries and Food, the Consellería do Medio Rural of the Xunta of Galicia, and to the Consejería de Medio Rural y Política Agraria of the Principado de Asturias. Also, we would like to thank Francisco Lozano, Nuria Moya, Tatiana Alende, Alexandra Gutiérrez, Cristina Viñolo, Daniela de la Cruz, and Laura Sánchez Martín from the Mycobacteria Unit of VISAVET Health Surveillance Center for their technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1361788/full#supplementary-material
1. Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. (2020) 70:5607–12. doi: 10.1099/ijsem.0.004332
2. Honda JR, Virdi R, Chan ED. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front Microbiol. (2018) 9:e02029. doi: 10.3389/fmicb.2018.02029
3. Primm TP, Falkinham JO. Nontuberculous mycobacteria. In: Reference Module in Biomedical Sciences. Elsevier (2014).
4. Waters WR, Whelan AO, Lyashchenko KP, Greenwald R, Palmer M V, Harris BN, et al. Immune responses in cattle inoculated with Mycobacterium bovis, Mycobacterium tuberculosis, or Mycobacterium kansasii. Clin Vacc Immunol. (2010) 17:247–52. doi: 10.1128/CVI.00442-09
5. Hernández-Jarguín AM, Martínez-Burnes J, Molina-Salinas GM, de la Cruz-Hernández NI, Palomares-Rangel JL, Mayagoitia AL, et al. Isolation and histopathological changes associated with non-tuberculous mycobacteria in lymph nodes condemned at a bovine slaughterhouse. Vet Sci. (2020) 7:1–10. doi: 10.3390/vetsci7040172
6. Ghielmetti G, Friedel U, Scherrer S, Sarno E, Landolt P, Dietz O, et al. Non-tuberculous Mycobacteria isolated from lymph nodes and faecal samples of healthy slaughtered cattle and the abattoir environment. Transbound Emerg Dis. (2018) 65:711–8. doi: 10.1111/tbed.12793
7. Nuru A, Zewude A, Mohammed T, Wondale B, Teshome L, Getahun M, et al. Nontuberculosis mycobacteria are the major causes of tuberculosis like lesions in cattle slaughtered at Bahir Dar Abattoir, northwestern Ethiopia. BMC Vet Res. (2017) 13:237. doi: 10.1186/s12917-017-1168-3
8. More SJ, Radunz B, Glanville RJ. Lessons learned during the successful eradication of bovine tuberculosis from Australia. Vet Rec. (2015) 177:224–32. doi: 10.1136/vr.103163
9. European Food Safety Authority (EFSA). The European Union One Health 2021 Zoonoses Report. EFSA J. (2022) 20:e8442. doi: 10.2903/j.efsa.2023.8442
10. WOAH. Mammalian tuberculosis (infection with Mycobacterium tuberculosis complex). In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. WOAH (2023).
11. Alvarez J, de Juan L, Bezos J, Romero B, Sáez JL, Reviriego Gordejo FJ, et al. Interference of paratuberculosis with the diagnosis of tuberculosis in a goat flock with a natural mixed infection. Vet Microbiol. (2008) 128:72–80. doi: 10.1016/j.vetmic.2007.08.034
12. Roupie V, Alonso-Velasco E, Van Der Heyden S, Holbert S, Duytschaever L, Berthon P, et al. Evaluation of mycobacteria-specific gamma interferon and antibody responses before and after a single intradermal skin test in cattle naturally exposed to M. avium subsp paratuberculosis and experimentally infected with M. bovis. Vet Immunol Immunopathol. (2018) 196:35–47. doi: 10.1016/j.vetimm.2017.12.007
13. Álvarez J, de Juan L, Bezos J, Romero B, Sáez JL, Marqués S, et al. Effect of paratuberculosis on the diagnosis of bovine tuberculosis in a cattle herd with a mixed infection using interferon-gamma detection assay. Vet Microbiol. (2009) 135:389–93. doi: 10.1016/j.vetmic.2008.09.060
14. Dunn JR, Kaneene JB, Grooms DL, Bolin SR, Bolin CA, Bruning-Fann CS. Effects of positive results for Mycobacterium avium subsp paratuberculosis as determined by microbial culture of feces or antibody ELISA on results of caudal fold tuberculin test and interferon-γ assay for tuberculosis in cattle. J Am Vet Med Assoc. (2005) 226:429–35. doi: 10.2460/javma.2005.226.429
15. Biet F, Boschiroli ML. Non-tuberculous mycobacterial infections of veterinary relevance. Res Vet Sci. (2014) 97:S69–77. doi: 10.1016/j.rvsc.2014.08.007
16. Fernández-Veiga L, Fuertes M, Geijo MV, Pérez de Val B, Vidal E, Michelet L, et al. Differences in skin test reactions to official and defined antigens in guinea pigs exposed to non-tuberculous and tuberculous bacteria. Sci Rep. (2023) 13:2936. doi: 10.1038/s41598-023-30147-4
17. Rónai Z, Eszterbauer E, Csivincsik, Guti CF, Dencso L, Jánosi S, et al. Detection of wide genetic diversity and several novel strains among non-avium nontuberculous mycobacteria isolated from farmed and wild animals in Hungary. J Appl Microbiol. (2016) 121:41–54. doi: 10.1111/jam.13152
18. Hughes MS, Ball NW, McCarroll J, Erskine M, Taylor MJ, Pollock JM, et al. Molecular analyses of mycobacteria other than the M. tuberculosis complex isolated from Northern Ireland cattle. Vet Microbiol. (2005) 108:101–12. doi: 10.1016/j.vetmic.2005.03.001
19. Varela-Castro L, Barral M, Arnal MC, Fernández de Luco D, Gortázar C, Garrido JM, et al. Beyond tuberculosis: diversity and implications of non-tuberculous mycobacteria at the wildlife–livestock interface. Transbound Emerg Dis. (2022) 69:e2978–93. doi: 10.1111/tbed.14649
20. Ministerio de Agricultura Pesca y Alimentación. Programa Nacional de Erradicación de Tuberculosis Bovina 2023 (Infección por el complejo Mycobacterium tuberculosis). Spain (2023).
21. Lorente-Leal V, Liandris E, Pacciarini M, Botelho A, Kenny K, Loyo B, et al. Direct PCR on tissue samples to detect Mycobacterium tuberculosis complex: an alternative to the bacteriological culture. J Clin Microbiol. (2021) 59:e01404-20. doi: 10.1128/JCM.01404-20
22. Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. Genome Res. (1992) 1:269–73. doi: 10.1101/gr.1.4.269
23. Kunze ZM, Portaels F, McFadden JJ. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J Clin Microbiol. (1992) 30:2366–72. doi: 10.1128/jcm.30.9.2366-2372.1992
24. Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. (1995) 33:304–7. doi: 10.1128/jcm.33.2.304-307.1995
25. Swanson DS, Kapur V, Stockbauer K, Pan X, Fronthingham R, Musser JM. Subspecific differentiation of Mycobacterium avium complex strains by automated sequencing of a region of the gene (hsp65) encoding a 65-Kilodalton heat shock protein. Int J Syst Bacteriol. (1997) 47:414–9. doi: 10.1099/00207713-47-2-414
26. Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. (1993) 31:175–8. doi: 10.1128/jcm.31.2.175-178.1993
27. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NTBioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. (1999) 41:95–8.
28. Tortoli E. Standard operating procedure for optimal identification of mycobacteria using 16S rRNA gene sequences. Stand Genomic Sci. (2010) 3:145–52. doi: 10.4056/sigs.932152
29. Adékambi T, Drancourt M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. (2004) 54:2095–105. doi: 10.1099/ijs.0.63094-0
30. Turenne CY, Semret M, Cousins D V, Collins DM, Behr MA. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J Clin Microbiol. (2006) 44:433–40. doi: 10.1128/JCM.44.2.433-440.2006
31. Gomez-Buendia A, Romero B, Bezos J, Lozano F, de Juan L, Alvarez J. Spoligotype-specific risk of finding lesions in tissues from cattle infected by Mycobacterium bovis. BMC Vet Res. (2021) 17:148. doi: 10.1186/s12917-021-02848-3
32. Jenkins AO, Gormley E, Gcebe N, Fosgate GT, Conan A, Aagaard C, et al. Cross reactive immune responses in cattle arising from exposure to Mycobacterium bovis and non-tuberculous mycobacteria. Prev Vet Med. (2018) 152:16–22. doi: 10.1016/j.prevetmed.2018.02.003
33. van Ingen J, Turenne CY, Tortoli E, Wallace Jr RJ, Brown-Elliott BA. A definition of the Mycobacterium avium complex for taxonomical and clinical purposes, a review. Int J Syst Evol Microbiol. (2018) 68:3666–77. doi: 10.1099/ijsem.0.003026
34. Gcebe N, Rutten V, Gey van Pittius NC, Michel A. Prevalence and distribution of non-tuberculous mycobacteria (NTM) in cattle, African buffaloes (syncerus caffer) and their environments in South Africa. Transbound Emerg Dis. (2013) 60:74–84. doi: 10.1111/tbed.12133
35. Gcebe N, Michel A, Gey van Pittius NC, Rutten V. Comparative genomics and proteomic analysis of four non-tuberculous Mycobacterium Species and Mycobacterium tuberculosis complex: occurrence of shared immunogenic proteins. Front Microbiol. (2016) 7:795. doi: 10.3389/fmicb.2016.00795
36. Infantes-Lorenzo JA, Moreno I, Risalde MDLÁ, Roy Á, Villar M, Romero B, et al. Proteomic characterisation of bovine and avian purified protein derivatives and identification of specific antigens for serodiagnosis of bovine tuberculosis. Clin Proteom. (2017) 17:36. doi: 10.1186/s12014-017-9171-z
37. Nuñez-Garcia J, Downs SH, Parry JE, Abernethy DA, Broughan JM, Cameron AR, et al. Meta-analyses of the sensitivity and specificity of ante-mortem and post-mortem diagnostic tests for bovine tuberculosis in the UK and Ireland. Prev Vet Med. (2018) 153:94–107. doi: 10.1016/j.prevetmed.2017.02.017
38. Berg S, Firdessa R, Habtamu M, Gadisa E, Mengistu A, Yamuah L, et al. The burden of mycobacterial disease in Ethiopian cattle: Implications for public health. PLoS ONE. (2009) 4:e5068. doi: 10.1371/journal.pone.0005068
39. Bolaños CAD, de Paula CL, Guerra ST, Franco MMJ, Ribeiro MG. Diagnosis of mycobacteria in bovine milk: an overview. Rev Inst Med Trop São Paulo. (2017) 59:1–13. doi: 10.1590/s1678-9946201759040
40. McCorry TP, McCormick CM, Hughes MS, Pollock JM, Neill SD. Mycobacterium nonchromogenicum in nasal mucus from cattle in a herd infected with bovine tuberculosis. Vet Microbiol. (2004) 99:281–5. doi: 10.1016/j.vetmic.2003.12.006
41. Waters WR, Palmer M V, Thacker TC, Payeur JB, Harris NB, Minion FC, et al. Immune responses to defined antigens of Mycobacterium bovis in cattle experimentally infected with Mycobacterium kansasii. Clin Vacc Immunol. (2006) 13:611–9. doi: 10.1128/CVI.00054-06
42. Michel AL. Mycobacterium fortuitum infection interference with Mycobacterium bovis diagnostics: natural infection cases and a pilot experimental infection. J Vet Diagn Investig. (2008) 20:501–3. doi: 10.1177/104063870802000415
Keywords: cattle, diagnosis, officially tuberculosis-free (OTF) herds, interference, non-tuberculous mycobacteria, tuberculosis, skin tests
Citation: Gomez-Buendia A, Alvarez J, Bezos J, Mourelo J, Amado J, Saez JL, de Juan L and Romero B (2024) Non-tuberculous mycobacteria: occurrence in skin test cattle reactors from official tuberculosis-free herds. Front. Vet. Sci. 11:1361788. doi: 10.3389/fvets.2024.1361788
Received: 26 December 2023; Accepted: 15 January 2024;
Published: 31 January 2024.
Edited by:
Cinzia Marianelli, National Institute of Health (ISS), ItalyReviewed by:
Marta Alonso-Hearn, Basque Research and Technology Alliance (BRTA), SpainCopyright © 2024 Gomez-Buendia, Alvarez, Bezos, Mourelo, Amado, Saez, de Juan and Romero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatriz Romero, YnJvbWVyb21AdmlzYXZldC51Y20uZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.