- 1Department of Microbiological Sciences, North Dakota State University, Fargo, ND, United States
- 2Lacombe Research and Development Centre, Agriculture and Agri-Food Canada, Lacombe, AB, Canada
- 3Department of Biological Sciences, North Dakota State University, Fargo, ND, United States
Introduction: The rise in antibiotic resistant pathogens associated with bovine respiratory disease (BRD) poses a serious challenge, particularly to the beef feedlot industry, as they currently depend on antibiotics to prevent BRD to mitigate the financial burden (approx. $1 billion annual loss) inflicted by BRD-associated high mortality and morbidity in feedlot cattle. Thus, there is an impetus need for the development of antimicrobial alternative strategies against BRD. This study aimed to screen and select candidate essential oils (EOs) for the development of an intranasal EO spray that can inhibit BRD pathogens and promote microbiota-mediated respiratory health.
Methods: The effects of selected EOs (ajowan, cinnamon leaf, citronella, grapefruit, fennel, and thyme) on a bovine nasopharyngeal microbiota culture were evaluated using 16S rRNA gene sequencing. The microbiota culture was enriched by incubating nasopharyngeal swabs obtained from finishing beef heifers in brain heart infusion broth with and without EOs (0.025%, v/v). These EOs were then also evaluated for their immunomodulatory effects on bovine turbinate (BT) cells by analyzing the concentrations of 15 cytokines and chemokines in cell culture after 24 h incubation. The crystal violet assay was done to assess the antibiofilm activity of EOs against Escherichia coli UMN026 strain. Finally, 15 EOs were screened for their antiviral activity against the bovine viral diarrhea virus 1 (BVDV-1) using BT cells and a fluorescence-based method.
Results: Ajowan, fennel, and thyme resulted in a moderate reduction of overall nasopharyngeal microbiota growth with significant alterations of both alpha and beta diversity, and the relative abundance of predominant bacterial families (e.g., increasing Enterobacteriaceae and decreasing Moraxellaceae) compared to the control (p < 0.05). Co-incubation of BT cells with selected EOs resulted in minimal alterations in cytokine and chemokine levels (p > 0.05). Ajowan, thyme, fennel, and cinnamon leaf exhibited antibiofilm activity at concentrations of 0.025 and 0.05%. Reduction of BVDV-1 replication in BT cells was observed with thyme (strong), and ajowan and citronella (moderate) at 0.0125% concentration.
Discussion: Accordingly, ajowan, thyme, fennel, cinnamon leaf, and citronella EOs were selected for further development as an intranasal EO spray to prevent and control of BRD pathogens in feedlot cattle.
1 Introduction
Bovine respiratory disease (BRD) is a complex infectious process resulting from the interplay of various factors, including environmental conditions, the immune status of the animal, and the presence of bacterial and viral agents (1). The primary bacterial pathogens associated with BRD are Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis (2). In addition to bacteria, viruses also play a significant role in BRD, including bovine herpesvirus-1 (BHV-1), bovine viral diarrhea virus (BVDV), bovine respiratory syncytial virus (BRSV), parainfluenza-3 (PI-3), and bovine respiratory coronavirus (BRCV) (3).
Feedlot cattle, particularly those newly feedlot arrivals are susceptible to BRD due to the numerous stress factors, such as weaning, transportation, changes in diet, contact with other animals, and concurrent diseases (4). These stressors can weaken the immune response of the cattle, making them more susceptible to viral infections and disrupting respiratory microbiota homeostasis. This disruption can then predispose animals to pneumonia due to the translocation of pathogenic bacteria from the upper to lower respiratory tract (1, 5). Despite extensive prevention and treatment efforts, including vaccination and antibiotic administration, BRD morbidity and mortality rates persist or even increased, which is partially due to the rise in antimicrobial resistance (AMR) (4, 6–8). Recent research shows that more than 50% of bovine respiratory pathogens harbor AMR levels that can exceed 50% and can often display multidrug resistance (9–12), which limits antibiotic effectiveness, particularly against multidrug-resistant strains. Therefore, manipulating the microbiota to restore respiratory homeostasis presents a promising approach for enhancing respiratory resilience against BRD (13).
Essential oils (EOs) are aromatic compounds extracted from plants and can exhibit antimicrobial activity against bacteria and viruses (14–16). These characteristics make them potential natural antibiotic alternatives. Developing antimicrobials that can inhibit pathogen growth without collateral damage to the commensal microbiota is an alternative approach to mitigating infectious diseases. The EOs of ajowan (AJO), cinnamon leaf (CIN), citronella (CIT), fennel (FEN), and thyme (THY), among others, were previously characterized in vitro for their antimicrobial activity against the BRD-associated bacterial pathogens M. haemolytica, P. multocida, and H. somni (17–19). Studies also suggest that EOs may modify antimicrobial resistance, potentially reverting multidrug-resistant bacteria to a susceptible state when used alongside antibiotics (20–23). Moreover, EOs have been shown to possess antiviral activity against other respiratory viral pathogens (24–26). For example, an EO blend consisting of three EOs inhibited influenza A (H1N1) and herpes simplex virus 1 as well as the bacterial pathogens (methicillin-resistant Staphylococcus aureus, Streptococcus pneumoniae, and Klebsiella pneumoniae) in vitro, suggesting potential for treating influenza and post-influenza bacterial pneumonia (27). Additionally, some EOs have been reported to have antifungal (27), antibiofilm (28, 29), and immunomodulatory properties (30). All of which makes EO-based strategy as an appealing antimicrobial alternative approach to mitigate BRD in feedlot cattle. The objective of this study was to screen and select candidate EOs for development of an intranasal EO spray against bacterial and viral pathogens associated with BRD in feedlot cattle, as an alternative to antibiotics. The screening criteria included the effect of EOs on bovine nasopharyngeal swab (NS) microbiota culture, antiviral activity against bovine respiratory viral pathogens, antibiofilm activity against Escherichia coli, and immunomodulatory activities.
2 Materials and methods
We previously identified minimum inhibitory concentrations (MICs) and minimum bactericidal concentrations of 15 EOs against M. haemolytica, P. multocida, and H. somni (18). Among those 15 EOs, five EOs were selected for this study and these selected EOs include AJO, THY, FEN (MIC ≤ 0.025%), CIN, and CIT (MIC ≤ 0.05%). The selected EOs displayed greater inhibition against all three BRD pathogens as compared to the remaining 9 EOs. In the present study, we further evaluated these five selected EOs for their antimicrobial, antiviral, immunomodulatory, and antibiofilm activities (Figure 1). Of note, we have previously tested 15 EOs including those EOs studied in the present study for their cytotoxicity on BT cells using a lawn assay, and none of these EOs exhibited any cytotoxicity within the tested range of concentrations (0.0125–0.4%, v/v) (18).
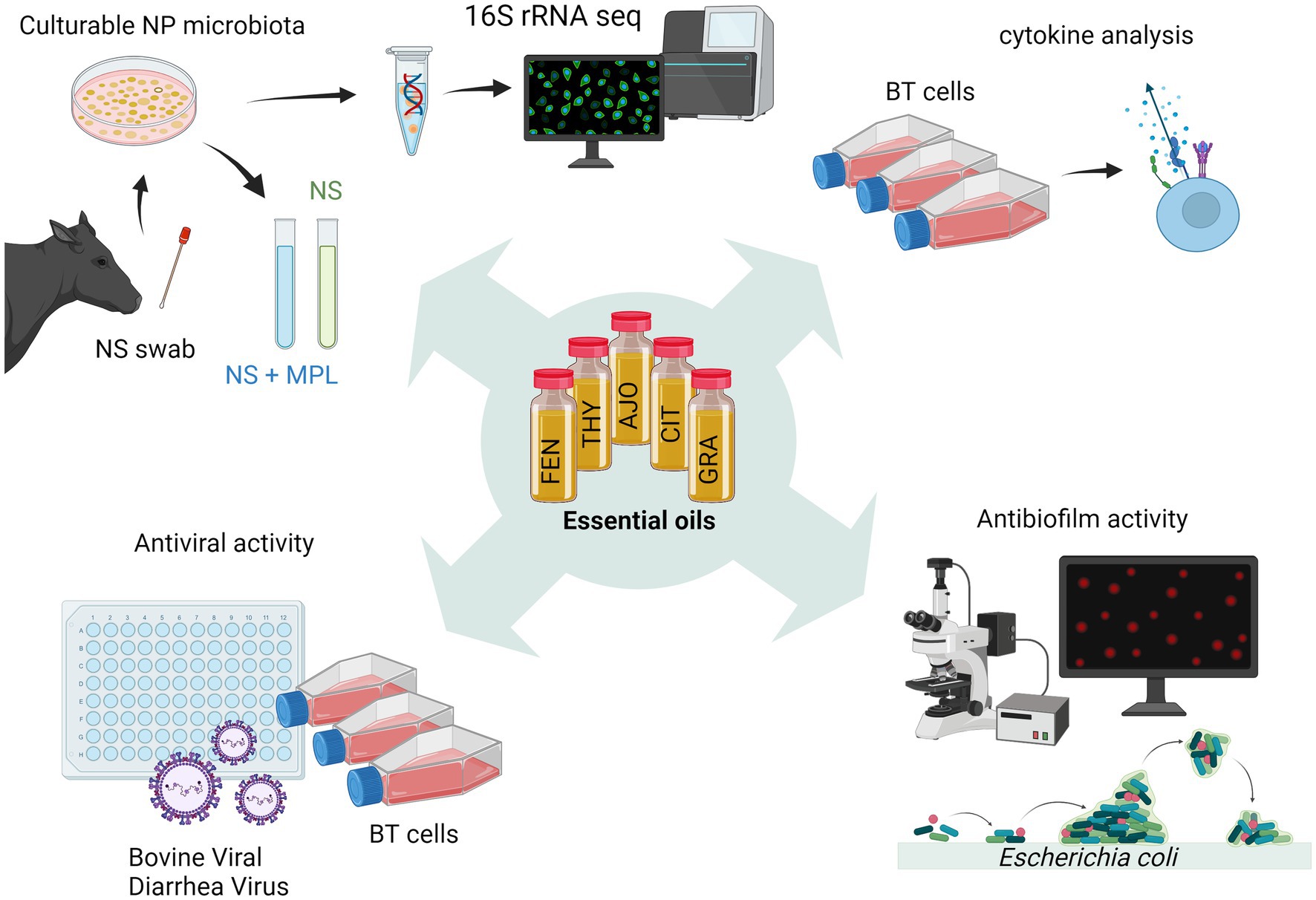
Figure 1. Schematic workflow diagram illustrating the process of screening essential oils (EOs) in vitro (created with BioRender.com).
2.1 Antimicrobial activity of EOs against BRD-associated bacterial pathogens
First, we evaluated the effects of AJO, THY, FEN, CIN, and grapefruit (GRA) EOs on the growth of completely different M. haemolytica and P. multocida strains (obtained from the Veterinary Diagnostic Lab, North Dakota State University, Fargo, ND, United States) than those strains used in our previous study (18), as well as on the overall growth of NS microbiota using the broth macrodilution method as described previously (18). Of note, the GRA EO was included in this study as a negative control as it did not inhibit the growth of M. haemolytica at the maximum tested concentration of 0.4% (18). While the CIT EO was selected for this study, and was further characterized for its immune modulation, antibiofilm and antiviral activities described below, this EO was not included in the present antimicrobial activity tests as it displayed similar antimicrobial activity against BRD pathogens and the commensal bacteria as with CIN EO (18) plus the limited budget availability for 16S rRNA gene sequencing.
For the evaluation of antimicrobial activity against M. haemolytica and P. multocida, 25 μL of each EO was diluted in 975 μL dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO, United States). Then a 50 μL aliquot of EO dissolved in DMSO was added to 5 mL brain heart infusion broth (BHI; BD, Franklin Lakes, NJ, United States) followed by inoculation with 50 μL of each of the 18 h culture of M. haemolytica or P. multocida containing 1 to 2 × 108 colony forming units (CFU) per ml. The mixture was incubated aerobically at 37°C with agitation at 200 rpm for 24 h. Controls included DMSO (1%, v/v) without EOs and a negative control with no DMSO or EO added. After incubation, the optical density (OD600) of each bacterial culture was measured.
2.2 Effects of EOs on the culture-enriched bovine nasopharyngeal microbiota
To investigate the impact of EOs on the overall growth of the culture-enriched bovine NS microbiota in vitro, NS swab samples collected from a cohort of crossbred finishing beef heifers (n = 31; initial BW = 494 ± 10 (SE) kg) were used. Animal housing, handling, and feeding conditions were described elsewhere (31). Deep nasopharyngeal swabs were collected from the right nostril of each heifer using a long, guarded swab with a rayon tip (27 cm long, MW 124, Medical Wire & Equipment, Corsham, United Kingdom) as described previously (31). Upon collection, NS swabs were stored in 1 mL BHI broth containing 20% glycerol, flash frozen with dry ice and stored at −80°C. From these NS swab samples stored in BHI/glycerol, 5 samples were randomly selected and vortexed vigorously for 1 min to release and dispense microbial cells attached to the swab. Then, 200 μL of the media containing NS microbial cells from each swab were pooled. One hundred microliters of the pooled sample were then inoculated into 900 μL BHI broth and incubated at 37°C with agitation at 200 rpm for approximately 3 h until the cell density reached an OD600 of 0.2. From this 3 h culture, 100 μL was inoculated into 5 mL BHI broth containing 0.025% of each EO (AJO, THY, FEN, CIN, and GRA) or DMSO or no EO (negative controls) and incubated for 24 h under the same conditions as described above. To test whether the presence of the NS commensal microbiota would influence the antimicrobial activity of EOs against M. haemolytica and P. multocida or a mixture of both species, 50 μL of an overnight (18 h) culture was added to 5 mL BHI containing NS microbiota culture and an EO and incubated for 24 h. A 50 μL aliquot of an overnight (18 h) culture of Lactobacillus fermentum (ATCC-9338, American Type Culture Collection, Manassas, VA, United States) culture was also added along to the M. haemolytica or P. multocida cultures. Of note, Lactobacillus spp. present in the nasopharynx of beef cattle are beneficial and can inhibit colonization of respiratory pathogens and induce positive modulation of nasopharyngeal microbiota in feedlot cattle (13, 32, 33). Therefore, L. fermentum was added as a representative of the beneficial bacteria, and to identify whether EOs could inhibit the beneficial bacteria in the presence of nasopharyngeal microbiota. After incubation, the overall cell growth of each NS microbiota culture was measured at an OD of 600 nm, and 1 mL of each 24 h culture was taken and stored at −80°C for genomic DNA extraction.
2.3 Genomic DNA extraction from nasopharyngeal microbiota cultures
Genomic DNA was extracted from the NS microbiota culture using the Qiagen DNeasy Blood and Tissue kit (Qiagen Inc., Germantown, MD, United States) according to the manufacturer’s instructions with modifications as outlined in our previous paper (34). Briefly, 100 μL of a 24 h NS microbiota culture was pelleted by centrifugation (13,000 × g for 5 min), and the supernatant was removed, and then the pellet was resuspended in 180 μL of lysis buffer (20 mM Tris–HCl, pH 8.0, 2 mM sodium EDTA, and 1.2% Triton X-100) containing 300 U/mL of mutanolysin and 20 mg/mL of lysozyme. The resuspended pellet in lysis buffer was vortexed and incubated at 37°C for 1 h with agitation at 800 rpm. Then, 25 μL of proteinase K and 200 μL of buffer AL (provided in the kit, no ethanol added) were added, and the mixture was incubated at 56°C for 30 min with shaking at 800 rpm. Silica/zirconia beads (0.1 mm) were then added to the tubes (approximately 400 mg) and samples were mechanically lysed at 6.0 m/s for 20 s in a FastPrep-24 classic bead beater (MP Biomedicals, Irvine, CA, United States). Samples were then centrifuged at 13,000 × g for 5 min and 400 μL of the supernatant was transferred to the columns provided by the kit and from this step onward the instructions from the kit were followed.
2.4 16S rRNA gene sequencing and analysis
The 16S rRNA gene amplification and sequencing were carried out by Molecular Research LP (MRDNA; Shallowater, TX, United States) using the 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers targeting the V4 hypervariable region, with reaction preparation and cycling conditions as described previously (35). Briefly, a 30-cycle PCR amplification was performed using the HotStarTaq Plus Master Mix Kit (Qiagen Inc., Germantown, MD, United States) and PCR products were checked for intensity and correct band size on a 2% agarose gel electrophoresis. Amplicon concentrations was normalized, indexed, pooled together, and then purified using AMPure XP beads (Beckman Coulter, Brea, CA, United States) for an Illumina DNA library preparation (Illumina Inc., San Diego, CA, United States). Sequencing was carried out in an Illumina MiSeq instrument using the MiSeq reagent kit v3 (2 × 300 bp) following the manufacturer’s instructions.
Sequences were quality filtered and processed with the DADA2 v.1.18 package (36) in R v. 4.3.1. Forward and reverse reads without primer sequences were initially trimmed to 220 bp, denoised, merged with a minimum overlap of 100 bp, and chimeras removed. The SILVA SSU database release 138.1 (37) was used to assign taxonomy to these amplicon sequence variants (ASVs) with the naïve Bayesian RDP classifier (38). Any ASVs classified as either chloroplasts, mitochondria, or eukaryota, as well as those present in the negative extraction control samples were removed from the analysis, as they were considered as contaminants. Samples were randomly subsampled to 171,000 reads prior to the calculation of richness (number of ASVs) and diversity indices (Shannon and inverse Simpson) and the Bray-Curtis dissimilarities with Phyloseq v. 1.46.0 (39) and vegan v. 2.5–7 (40) in R.
2.5 Immunomodulation effect of EOs on bovine turbinate cells
The selected six EOs (AJO, CIN, CIT, FEN, GRA, and THY) were evaluated for their immunomodulatory effects on BT cells. The BT cells (ATCC-1390; American Type Culture Collection) were seeded onto 6-well flat-bottom tissue culture plates at 1 × 105 cells per well and incubated using the standard culture conditions until a complete cell monolayer was achieved. The BT cell culture conditions are described previously (32). Briefly, the cell monolayer was washed with Dulbecco’s Modified Eagle Medium (DMEM, ATCC, Manassas, VA, United States), and then incubated with 2 mL of cell culture media supplemented with 0.025% EO (v/v) for 24 h. Ten microliters of each EO were diluted in 90 μL DMSO and a 25 μL aliquot of the EO dissolved in DMSO was added into 75 μL of DMEM to obtain a 2.5% EO stock solution. From each 2.5% EO stock solution, 20 μL was added to 2 mL of cell culture media. Negative controls included DMSO (0.2%, v/v) without EO and a negative control with no DMSO or EO added. After 24 h incubation, 1 mL of culture media was stored at −80°C for cytokine analysis. This experiment was repeated on three different days with three different passages of cells and each time had 2 replicates for each of EO. A total of 15 cytokines and chemokines were quantified using the Bovine Cytokine 15-Plex Discovery Assay® (Millipore Sigma, Burlington, Massachusetts, United States) on the Luminex™ 200 instrument (Eve Technologies Corp, Calgary, Alberta, Canada) according to the manufacturer’s instructions. The 15-plex assay consisted of interferon gamma (IFN-γ), interleukins (IL) IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IL-17A, and IL-36RA, interferon gamma inducible protein-10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammation proteins (MIP) MIP-1α and MIP-1β, tumor necrosis alpha (TNFα), and vascular endothelial growth factor-A (VEGF-A). Multiplexing allowed for the simultaneous detection of the cytokines and chemokines at a sensitivity range of 0.05 to 66.51 pg./mL (Millipore Sigma MILLIPLEX® MAP protocol).
2.6 Antibiofilm activity of EOs against Escherichia coli
The same selected EOs used for the immunomodulatory effects on BT cells (AJO, CIN, CIT, FEN, GRA, and THY) were also evaluated for their antibiofilm activity against E. coli strain UMN026 using a crystal violet (CV) biofilm assay (41). UMN026 is a clinical uropathogenic E. coli strain that displayed relatively strong biofilm forming capacity (P. Bergholz et al., unpublished data). For the biofilm assay, a single colony of E. coli UMN026 was inoculated into 200 μL of lysogeny broth (LB; BD, Franklin Lakes, NJ, United States) in a 96-well plate and incubated at 37°C for 8 h. Two microliters of this 8 h culture was then transferred to a well containing 198 μL glucose defined minimal media (GDMM) supplemented with 0.5% casamino acids (CAA; VWR, Radnor, PA, United States), prepared as described elsewhere (42), and the plate was then sealed and incubated overnight at 37°C. On the following day, 2 μL of the overnight culture was transferred to 198 μL GDMM supplemented with 0.5% CAA, incubated at 37°C for 8 h, and then 2 μL of 8 h culture was transferred to 198 μL GDMM +0.5% CAA and incubated overnight. On day 3, 2 μL of the overnight culture was transferred to 198 μL GDMM +0.5% CAA containing 0.025% or 0.05% EOs and incubated at 37°C for 48 h, to allow for biofilm formation. For EO stock solutions, 2.5% of EO stock solution was prepared by adding 25 μL of 10% EO dissolved in DMSO to 75 μL LB media. From the 2.5% EO stock solution, 2 or 4 μL was added to each well. For negative controls, DMSO (0.2%) without EO and with LB broth only was added.
After 48 h incubation, the media from the plate was removed and the well was washed using 200 μL of 1X phosphate buffered saline (PBS; Corning, Corning, NY, United States). This was repeated three times. The plate was then air dried at room temperature for 1 h. Then, 200 μL of 0.1% CV was added to the wells and incubated at room temperature for 15 min. The CV was removed by washing the wells with PBS three times, and the plate was air dried at room temperature for 1 h. Two hundred microliters of freshly prepared 80:20 ethanol:acetone solution was added to the wells and incubated at room temperature for 15 min. After incubation, 150 μL of dye solution was transferred to a new 96-well plate and the OD was measured at 580 nm using a Cytation 5 Cell Imaging Multi-Mode Reader (BioTeK, Winooski, VT, United States). For blanks, 150 μL of pure 80:20 ethanol:acetone solution was used.
2.7 Antiviral activity of EOs against bovine respiratory viruses
The main viral agents involved in BRD in feedlot cattle are BHV-1, BRSV, BVDV, and PI-3. Given that BVDV is often used for coinfection with BRD bacterial pathogens to experimentally induce BRD in cattle (43, 44), BVDV-1 was used as a model BRD viral pathogen for this in vitro screening. The 6 EOs (AJO, CIN, CIT, FEN, GRA, and THY) plus 9 more EOs [black pepper, carrot seed, eucalyptus, ginger grass, lavender, niaouli, rosemary, sandalwood, tangerine, and tea tree; further details on the origin of these EOs are provided in our previous publication (18)] that have been tested against BRD bacterial pathogens in our previous study were evaluated for their antiviral activity against BVDV-1 (ATCC-VR534; American Type Culture Collection) infection of BT cells (ATCC-1390) using a fluorescence-based assay. BT cell culturing was performed as described in our previous papers (18, 32), with minor modifications. Briefly, the BT cells were seeded onto 96-well flat-bottom tissue culture plates at 1 × 103 cells per well and incubated in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Oakville, ON, Canada) supplemented with 10% fetal horse serum (ATCC) and 1X antibiotic/antimycotic solution (Penicillin–Streptomycin-Amphotericin B; Hyclone Laboratories Inc., Logan, UT, United States) at 37°C with 5% CO2 until 85–90% confluency was obtained. The cell monolayer was then washed three times with Hank’s Balanced Salt Solution (HBSS; Cytiva Hyclone, Marlborough, MA, United States).
Stock BVDV-1 cultures were prepared by infecting BT cell monolayers at 85–90% confluence. Following incubation for 72 h, the infected flasks were freeze-thawed three times to release cell associated virus. Stock BVSV-1 titers were determined by the standard tissue culture infective dose (TCID50) assay. Briefly, BT cells were seeded in a 96-well plate. When the BT cells reached 80–90% confluency in the wells, a serial dilutions of the stock BVDV-1 virus ranging from −1 to −4 was added to the BT monolayers in the wells in a volume of 100 μL, in 4 replicates per dilution, and incubated at 37°C. After 3 h of incubation, 100 μL of DMEM with 5% fetal horse serum and 1 X antibiotic/antimycotic were added to each well and incubated for at least 72 h. The plates were observed under a light microscope for any visible CPE on the BT cell sheet. The titer of the stock culture was determined and calculated using the Reed-Muench formula (45). The culture was resuspended to a titer of 1.0 × 104 TCID50/mL in DMEM to perform the antiviral activity assay with the 15 EOs.
The antiviral activity assay was assessed as a reduction in BVDV-1 replication in the presence of EO’s, when compared to a control without EO treatment. Essentially as described before with little modification (46–48). Briefly, after BT cells seeded into 96-well plates became 80–90% confluent, the growth culture media was removed from each well and washed three times with Hank’s balanced salt solution (HBSS) (ATCC). Next, 200 μL of 1 × 104 TCID50/ml BVDV-1 in DMEM was mixed EO to obtain at a final concentration of EO at 0.025% or 0.0125% and added to respective wells. After 3 h of incubation, fetal horse serum was added to a final concentration of 5% (v/v) to each well and incubated at 37°C with the presence of 5% CO2 for another 45 h. After 48 h incubation, the culture media was removed, and washed with HBSS and fixed with a 1:1 mixture of methanol and acetone. An immunofluorescence assay was carried out on the fixed cell sheet with a BVDV fluorescent antibody conjugate (243-FA. 1801, NVSL, Ames, IA, United States), and with 6-diamidino-2-phenylindole (DAPI; Life Technologies, Carlsbad, CA, United States). Apple-green fluorescence observed under a fluorescence microscope was indicative of BVDV-1 replication. For viral positive controls, 1 × 104 TCID50/ml BVDV-1 in DMEM without EO or with DMSO, which was the solvent for EO, and for negative control DMEM with 5% fetal horse serum and without BVDV-1, without EO were used. The intensity of the apple-green fluorescence observed under the microscope was compared between EO-treated and control samples.
2.8 Statistical analysis
The effect of EO treatment on the culture-enriched NS microbiota was assessed using the Bray-Curtis dissimilarities and PERMANOVA (adonis2 function) with vegan in R. Pairwise comparisons of the Bray-Curtis dissimilarities between different EO treatment groups were done using the pairwise Adonis v. 0.4 R package with the Benjamini-Hochberg procedure used to correct p-values for multiple comparisons. The effect of EO treatment on alpha diversity indices (number of observed ASVs, Shannon diversity index) and the relative abundance of phyla, family, and genera of culture-enriched NS microbiota, as well as on cytokine and chemokine concentrations, and biofilm cell counts were determined using the generalized linear mixed model estimation procedure (PROC GLIMMIX) LSMEANS statement (ver. 9.4, SAS Institute Inc., Cary, NC, United States). The Shapiro–Wilk test was used to determine whether a dataset follows a normal distribution. Significance was considered at p < 0.05.
3 Results
3.1 Antimicrobial activity of EOs against BRD-associated bacterial pathogens
The AJO, CIN, FEN, and THY EOs significantly inhibited the growth of M. haemolytica, P. multocida, and L. fermentum when compared to GRA and the negative controls, both in pure culture and in combination with the culture-enriched NS microbiota (p ≤ 0.05), based on the OD600 measurements (Figure 2).
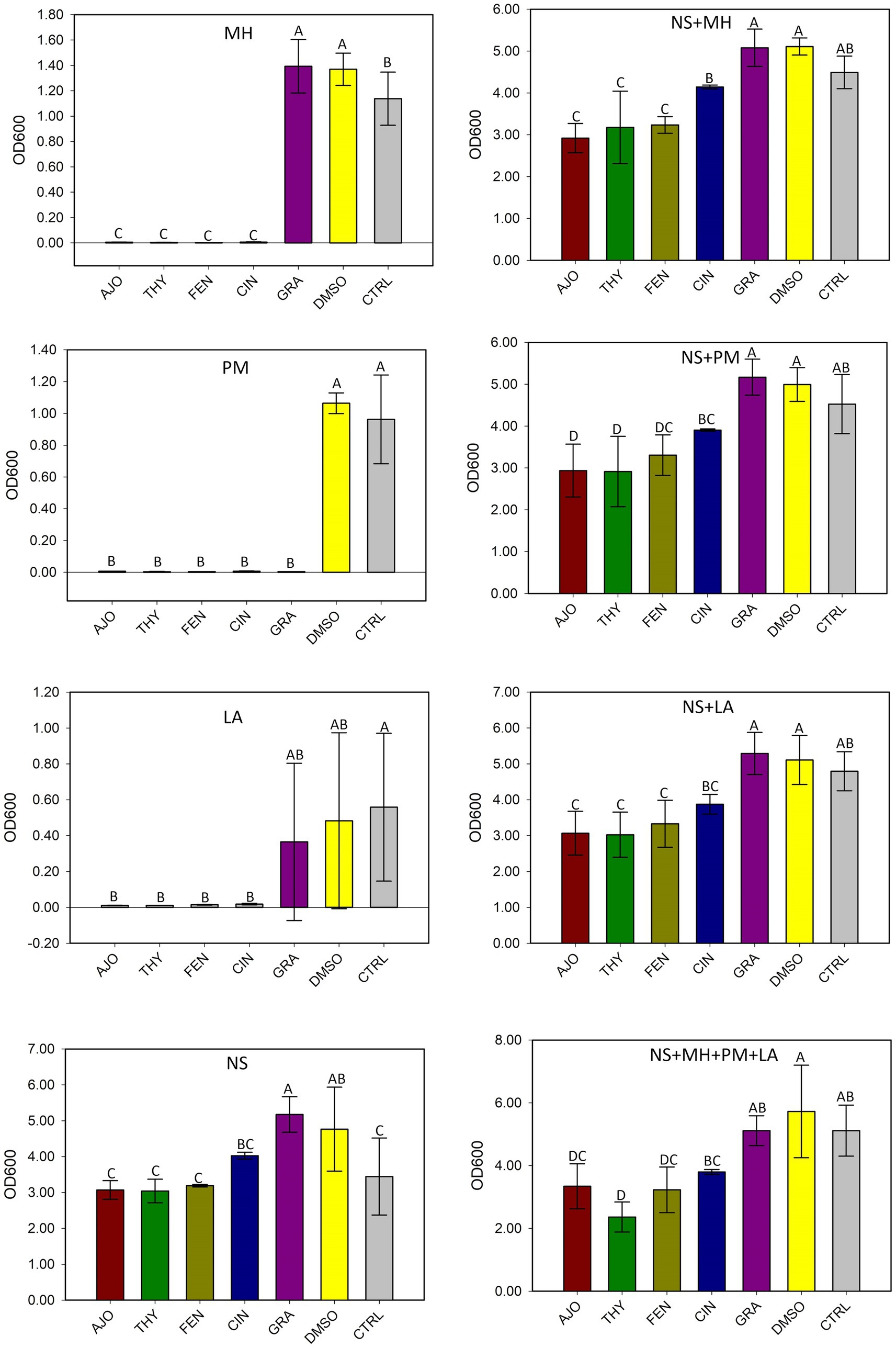
Figure 2. The effects of essential oils on the growth of Mannheimia haemolytica (MH), Pasteurella multocida (PM), Lactobacillus fermentum (LA), and the culture-enriched bovine nasopharyngeal microbiota alone (NS) or with presence of these bacteria. NS with one of the three strains (NS + MH, NS + PM, or NS + LA). NS plus all three bacterial inoculums combined (NS + MH + PM + LA). The values are the means from three replicates. Different uppercase letters indicate mean values differ (p < 0.05). The vertical bars indicate standard error of the mean.
3.2 Effects of EOs on the culture-enriched bovine NS microbiota
3.2.1 16S rRNA gene sequencing and analysis
After processing and quality filtering, the average number of sequences per sample was 265,699 ± 60,006 (SD) (n = 32 samples). These sequencing reads were taxonomically assigned into 14 bacterial phyla, 94 families, and 149 genera.
3.2.2 Microbial community structure, diversity, and composition
The microbial composition of the culture-enriched NS microbiota was similar between NS microbiota-only (NS) and NS with M. haemolytica, P. multocida, and L. fermentum added (MPL) samples. Although samples that were treated with the AJO, FEN, and THY clustered together and were well separated from samples that were treated with CIN, GRA, DMSO, and CTRL (Figure 3), the statistical significant difference based on PERMANOVA was not established due to the replicates in each treatment was being small (sample size = 2).
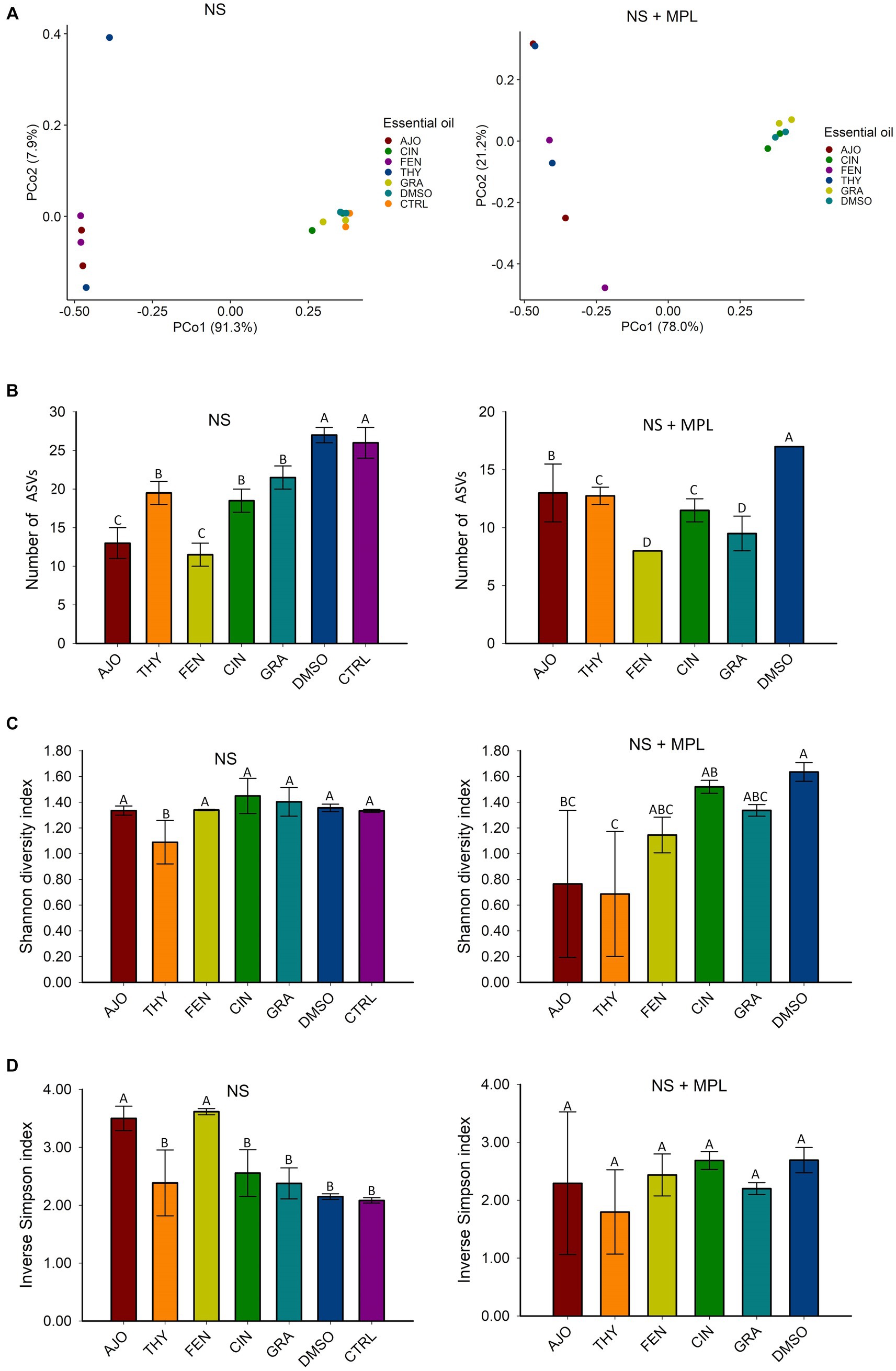
Figure 3. The effects of essential oils (EOs) on the beta and alpha diversities of the culture-enriched bovine nasopharyngeal swab microbiota (NS) along or with presence of Mannheimia haemolytica, Pasteurella multocida, and Lactobacillus fermentum (NS + MPL). Principle coordinates analysis (PCoA) plot of the Bray Curtis dissimilarities (A), and Alpha diversity indices (B–D) for the enriched-NS microbiota. The values (B–D) are the means from two replicates. Different uppercase letters indicate mean values differ (p < 0.05). The vertical bars indicate standard error of the mean.
The richness of the culture-enriched NS microbiota (number of observed ASVs) was significantly different between the EO and control groups. For the NS samples, the AJO-and FEN-treated samples had the lowest number of observed ASVs, followed by CIN, GRA, and THY. Samples that were not exposed to EOs such as DMSO and control samples had the greatest richness (p < 0.05). For NS + MPL (Culture-enriched NS microbiota containing additional M. haemolytica, P. multocida, and L. fermentum inoculum) samples, the overall number of ASVs was lower than the NS samples alone, and samples treated with the FEN and GRA EOs had the lowest observed number of ASVs, followed by THY, CIN, AJO, and the DMSO control (p < 0.05; Figure 3).
The diversity of the NS samples as determined by the Shannon and inverse Simpson indices, did not differ between any of the EOs and the control samples, except for THY, which had a higher Shannon diversity index value (p < 0.05) than the others (Figure 3). Additionally, samples treated with AJO and FEN had higher inverse Simpson index value than all other groups (p < 0.05), including the control samples (Figure 3). Similarly, the diversity of the culture-enriched NS + MPL microbiota was similar between all the tested EOs and the control samples, except for AJO and THY, which had lower Shannon diversity index values than the remaining samples (p < 0.05; Figure 3).
The most relatively abundant phyla in the culture-enriched NS microbiota were Proteobacteria (99.59%), followed by Firmicutes (0.31%), and Bacteroidota (0.07%). The effects of EOs on the relative abundance of these three phyla were detected (Figure 4). None of the EOs tested on the NS microbiota either with (NS + MPL) or without (NS) M. haemolytica, P. multocida, and L. fermentum added had a significant effect on the relative abundance of Proteobacteria. Moreover, all EOs tested significantly reduced the relative abundance of Firmicutes and Bacteroidota in the NS microbiota when compared to DMSO or the negative control samples (p < 0.05). However, when M. haemolytica, P. multocida, and L. fermentum were added to the NS microbiota, FEN did not reduce the relative abundance of Firmicutes when compared to the DMSO control, as AJO, CIN, GRA, and THY did (Figure 4).
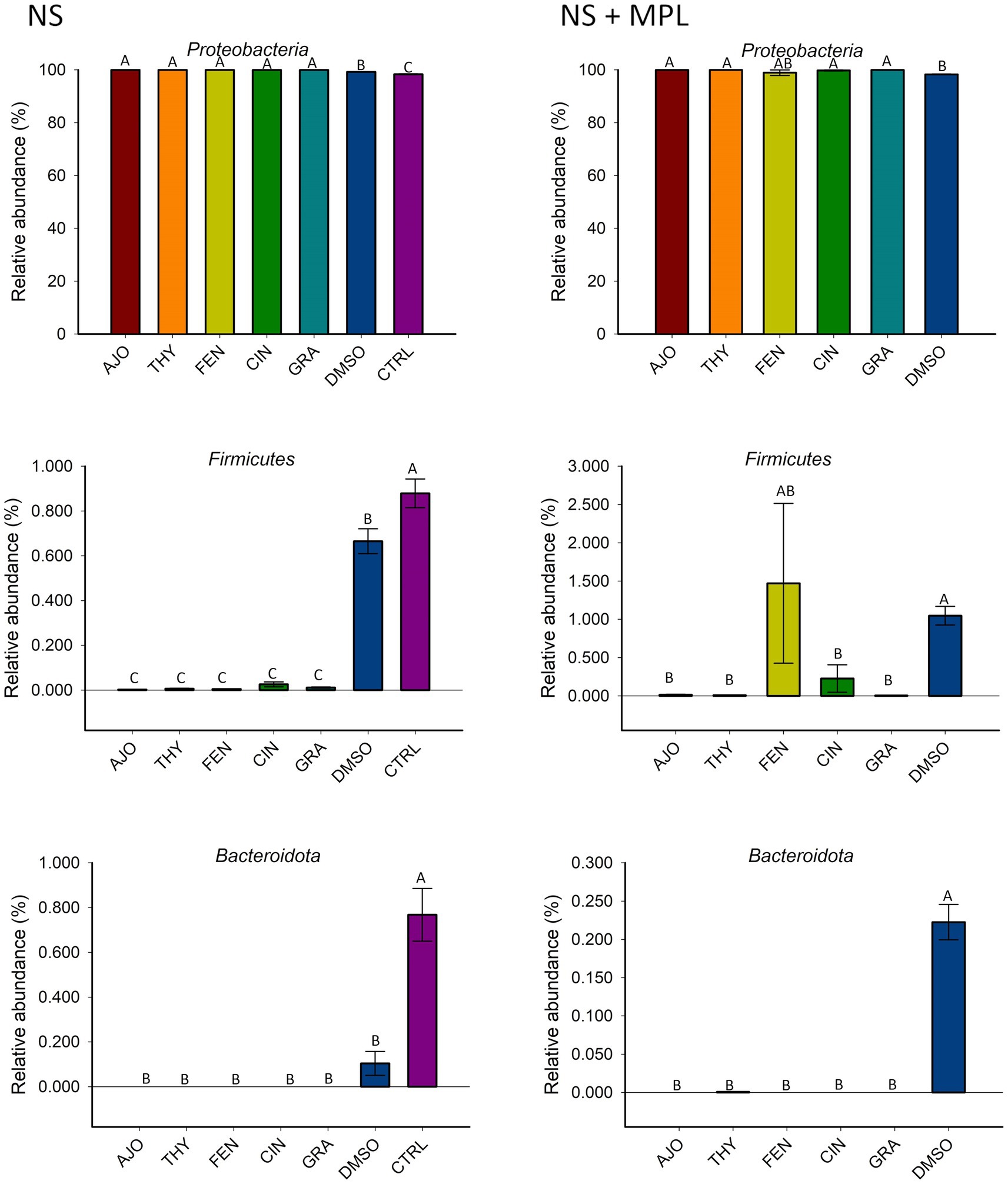
Figure 4. The effects of essential oils on relative abundance of the most predominant bacterial phyla in the culture-enriched bovine nasopharyngeal microbiota. The values are the means from two replicates. Different uppercase letters indicate significantly different means (p < 0.05).
The most relatively abundant families in the culture-enriched NS microbiota were Enterobacteriaceae (56.9%), Moraxellaceae (42.8%), Enterococcaceae (0.21%), Dysgonomonadaceae (0.13%), Bacilaceae (0.01%), Lactobaciliaceae (0.005%), and Pasteurellaceae (0.003%). The effects of EOs on the relative abundance of bacterial families were similar between NS and NS + MPL samples. Samples treated with AJO, FEN, and THY were dominated by Enterobacteriaceae (>99.97%), while the most abundant families in samples treated with CIN, GRA, and DMSO were Moraxellaceae (65.7, 77.8, and 77.9%, respectively), followed by Enterobacteriaceae (34.2, 22.2%. and 21.4%, respectively; Figure 5).
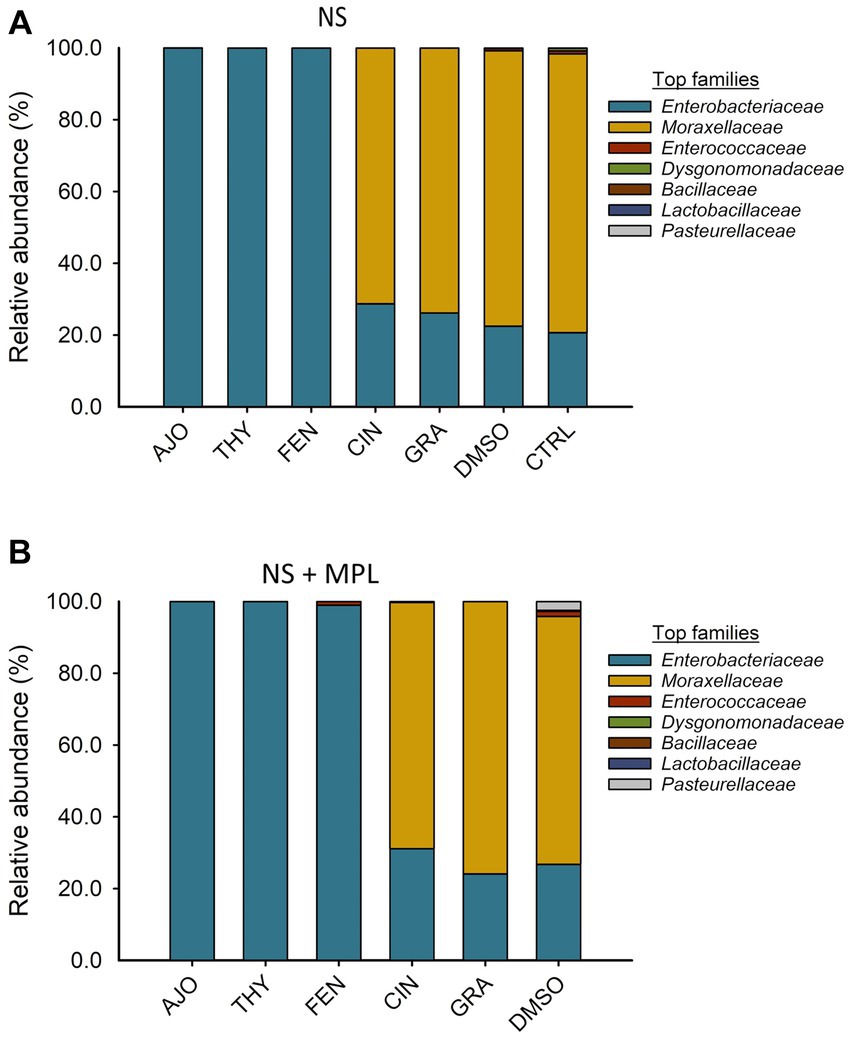
Figure 5. The effects of essential oils on relative abundance of the mostpredominant bacterial families present in the culture-enriched bovine nasopharyngeal microbiota with NS (A) or without NS + MPL (B) added Mannheimia haemolytica, Pasteurella multocida, and Lactobacillus fermentum. The values are the means from two replicates.
The most relatively abundant genera present in the culture-enriched NS microbiota (NS vs. NS + MPL) were Acinetobacter (42.8% vs. 35.6%), Escherichia-Shigella (25.5% vs. 41.5%), Klebsiella (18.9% vs. 12.0%), Enterococcus (0.21% vs. 0.45%), Citrobacter (0.15% vs. 0.16%), Dysgonomonas (0.13% vs. 0.04%), Mannheimia (0.01% vs. 0.05%), and Pasteurella (< 0.001% vs. 0.37%). The AJO, THY, and FEN EOs reduced the relative abundance of Acinetobacter (p < 0.05) and increased the relative abundance of Escherichia-Shigella and Klebsiella (p < 0.05) in comparison with the negative control samples (Figure 6). All EOs reduced the relative abundance of Enterococcus and Dysgonomonas (p < 0.05). Only FEN and CIN reduced the relative abundance of Citrobacter (p < 0.05), while AJO, FEN, GRA, and DMSO completely reduced the relative abundance of Mannheimia (p < 0.05), when compared to the control sample, in NS culture only. For the NS + MPL culture-enriched microbiota, all EOs reduced the relative abundance of Pasteurella and Mannheimia (p < 0.05) when compared to the DMSO control (Figure 7).
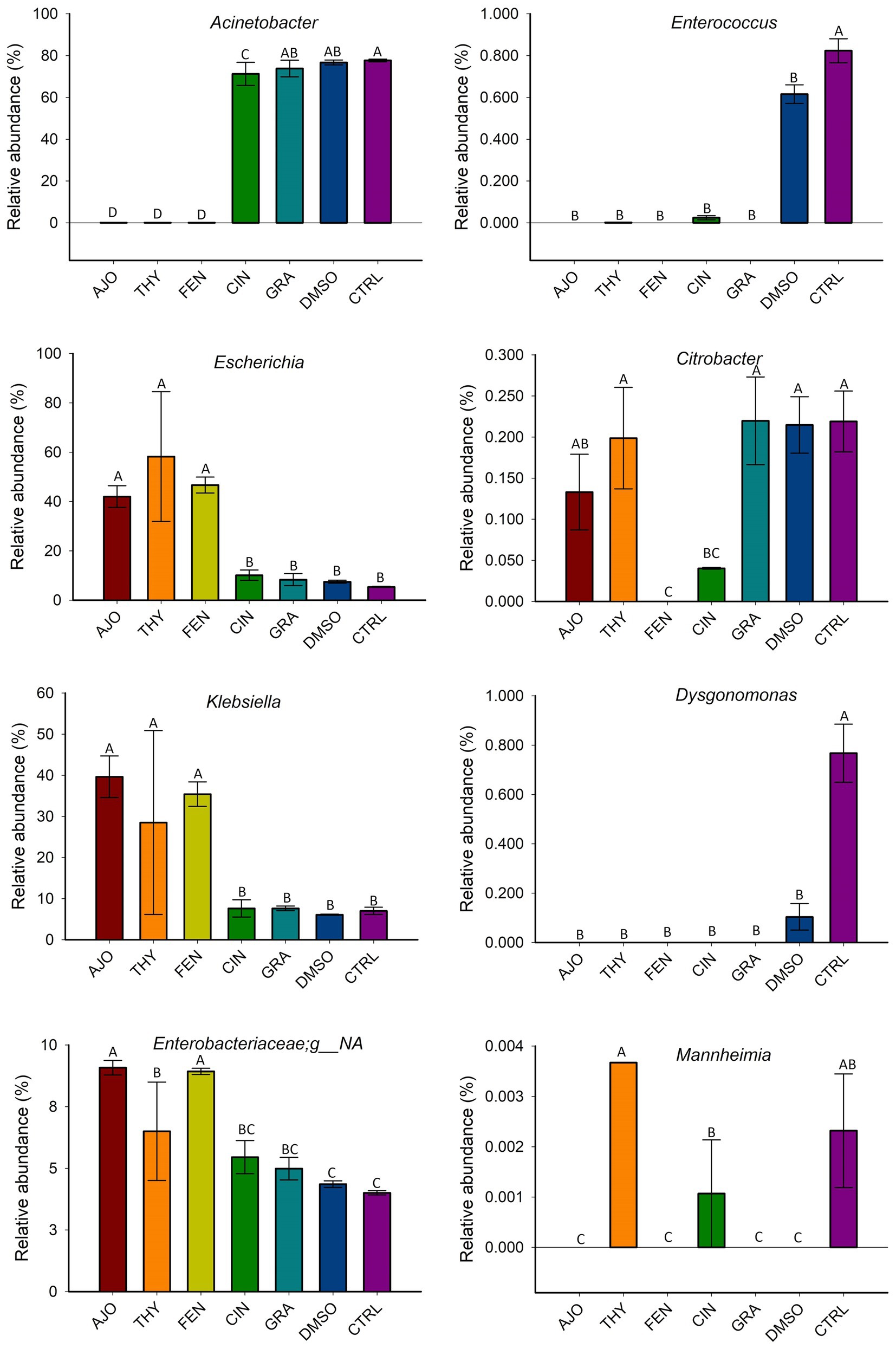
Figure 6. The effect of essential oils on the relative abundance of the most relatively abundant bacterial genera present in the culture-enriched bovine nasopharyngeal microbiota (NS). The values are the means from two replicates. Different uppercase letters indicate significantly different means (p < 0.05).
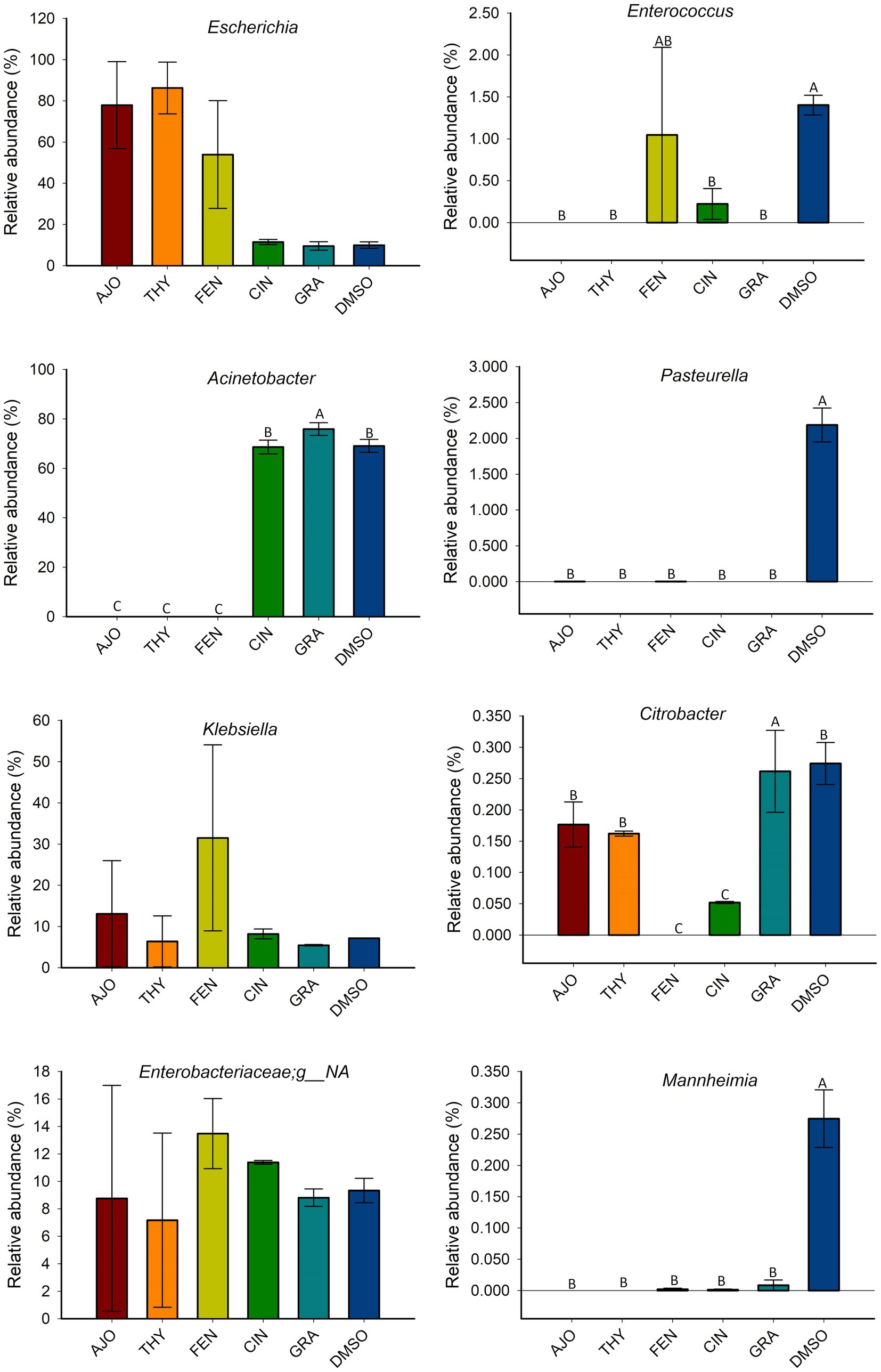
Figure 7. The effects of essential oils on relative abundance of the most predominant bacterial genera present in the culture-enriched bovine nasopharyngeal microbiota with Mannheimia haemolytica, Pasteurella multocida, and Lactobacillus fermentum added (NS + MPL). The values are the means from two replicates. Different uppercase letters indicate significantly different means (p < 0.05).
3.3 Immunomodulatory effects of EOs on BT cells
The effects of co-incubation of BT cells with one of the selected EOs (AJO, THY, CIN, CIT, and GRA) for 24 h on the production of cytokine and chemokines from BT cell monolayers are presented in Figure 8. The concentrations of IFN-γ, IL-1α, IL-1β, IL-4, IL-17A, MIP-1α, MIP-1β, and TNF-α did not differ between any of the EOs tested and control groups (DMSO and CTRL) (p > 0.05). The concentrations of IL-36R and IP-10 were lower in BT cells co-cultured with CIN as compared to BT cells co-cultured without EO (CTRL) (p < 0.05). The concentration of MCP-1 was reduced in BT cells in response to co-culturing with CIN and GRA compared to the control group (p < 0.05). The impact of EOs on IL-6, IL-8, and VEGF-A concentrations were not established as there were significantly larger variations between replicates. Overall, co-incubation of EO for 24 h incubation resulted in minimal immune stimulation in BT cell monolayers.
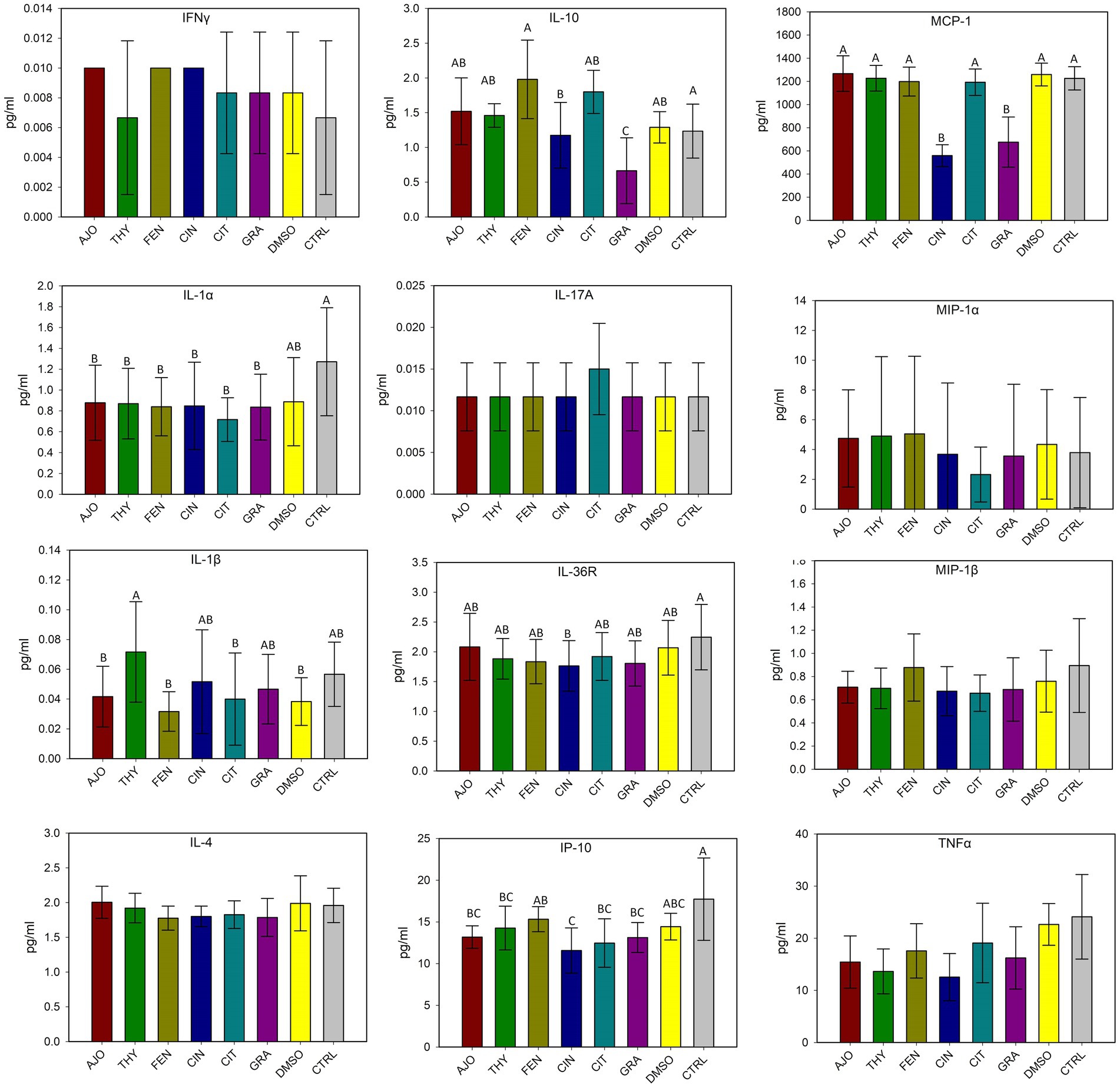
Figure 8. Evaluation of the effects of essential oils on cytokine production in bovine turbinate cell lines. Different uppercase letters indicate significantly different means (p < 0.05). The values are the means obtained from three independent experiments performed on different days. Different uppercase letters indicate significantly different means (p < 0.05).
3.4 Antibiofilm activity of EOs against Escherichia coli
With the use of CV assay, biofilms produced by E. coli were quantified after 48 h of incubation. Only AJO, THY, and GRA at a final concentration of 0.025% (v/v), significantly reduced (p < 0.05) biofilm formation by E. coli as compared to the DMSO and the negative control samples (Figure 9A). When the final concentration of the EOs was increased to 0.05% (v/v), all 6 EOs tested were able to significantly reduce (p < 0.05) the biofilm-forming capacity of E. coli (Figure 9). However, the most significant reduction of biofilm formation by E. coli was observed with AJO, THY, and CIN (Figure 9B).
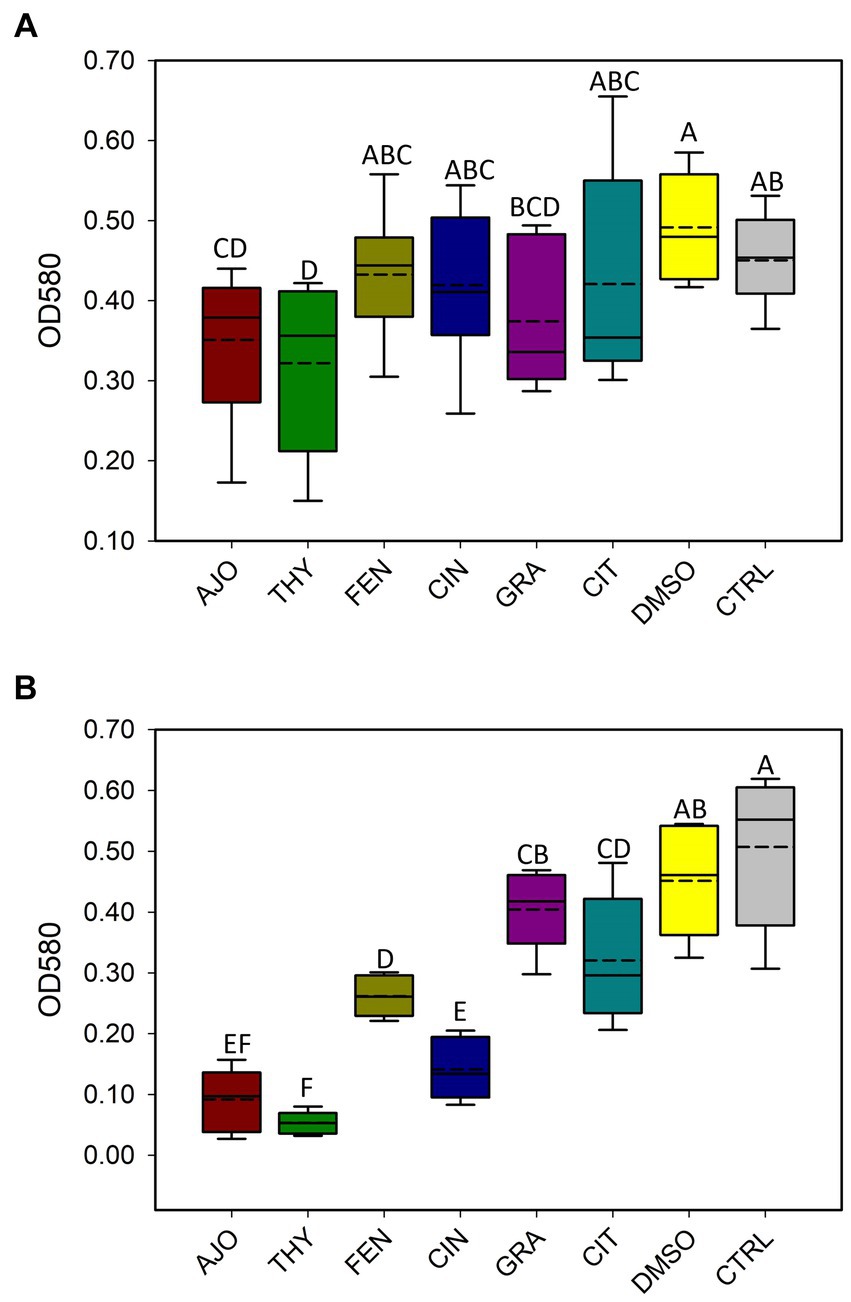
Figure 9. Antibiofilm activities of selected essential oils against Escherichia coli strain UMN026 at concentrations of (A) 0.025% and (B) 0.05% (v/v) based on optical density (OD) at 580 nm. The values are the means from four replicates. Different uppercase letters indicate significantly different means (p < 0.05).
3.5 Antiviral activity of EOs against the bovine respiratory viral pathogens
Bovine turbinate cells infected with BVDV-1 showed a CPE when observed under the microscope, while BT cells treated with the EOs did not and maintained a normal cytology morphology (Figures 10A,B). The EOs reduced the replication of BVDV-1 when observed through a fluorescence microscope. The fluorescein isothiocyanate (FITC) tagged anti-BVDV antibody fluorescence intensity, which represents viral replication, was the brightest in the viral positive control sample (no EOs) where the virus replicated freely in BT cells. However, the sample that received an EO in addition to the virus, had diminished fluorescence intensity, indicating a reduction in BVDV-1 replication (Figures 10C–E). Among the 15 tested EOs, the inhibitory effects on BVDV-1 replication on BT cell monolayer was observed with THY, AJO, and CIT.
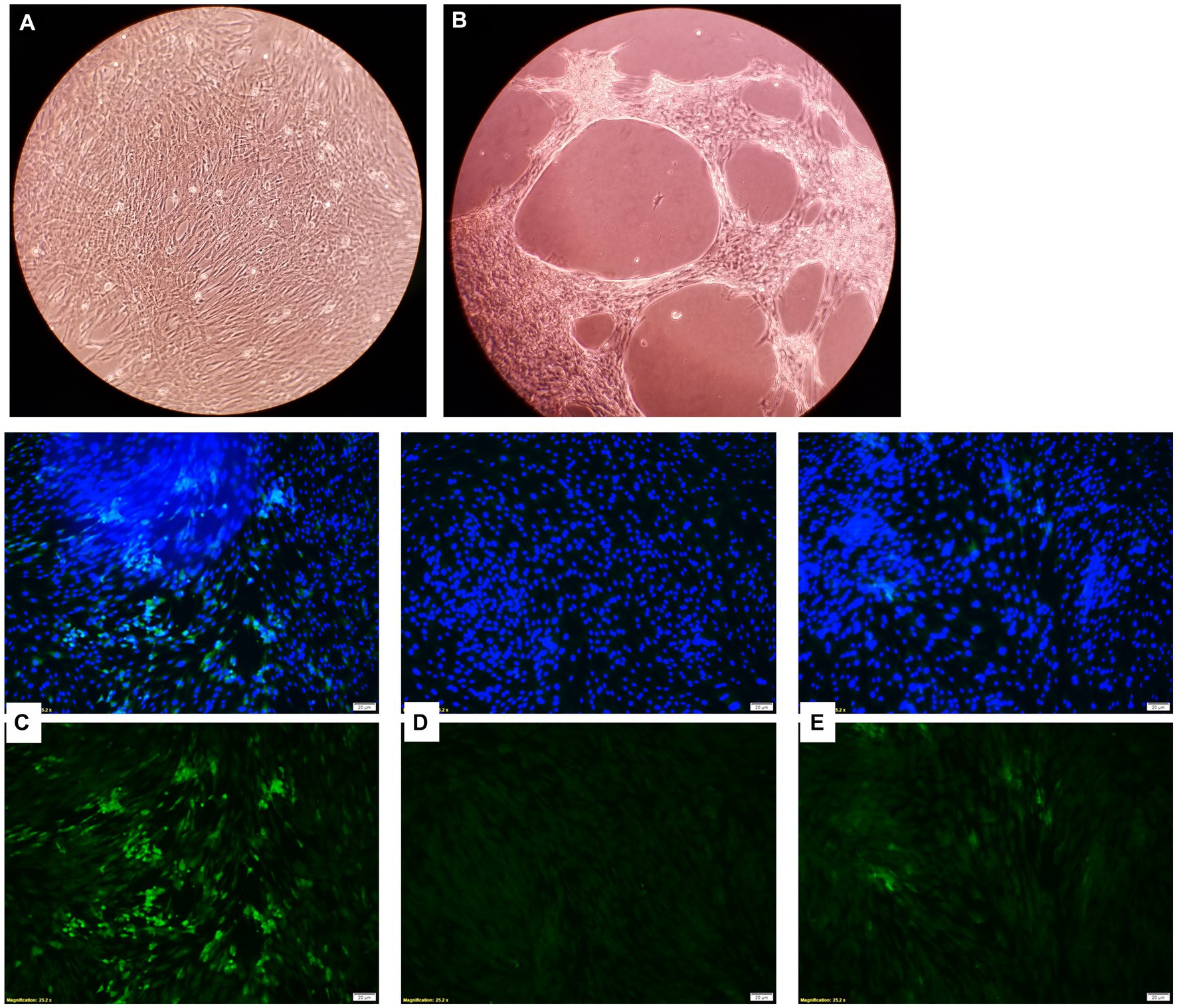
Figure 10. Microscopic images of bovine turbinate cells without (A) and with (B) bovine viral diarrhea virus 1 infection; Inhibition of bovine viral diarrhea virus 1 (BVDV-1) replication by essential oils. Green fluorescence is indicative of viral replication stained with FITC tagged anti-BVDV antibody and the blue fluorescence indicates the nuclei of the bovine turbinate (BT) cells stained with DAPI. (C) Showing the maximum viral replication on the BT cells for virus control (without EO); (D) Showing no viral replication on BT cells incubated DMEM control (No virus added); (E) Showing a minimal viral replication on BT cells incubated with THY EO (0.025%).
4 Discussion
The main components of EOs can vary greatly (49) and therefore it is important to determine the exact chemical composition of individual EOs evaluated in vitro for reproducibility and precision. The composition of the EOs evaluated in this study was previously determined via mass spectroscopy–gas chromatography (18). Most EOs often have 1 to 3 major components (>20%) that mainly contribute to the antimicrobial activity observed with the respective EOs. As such, we selected 5 different EOs that displayed relative strong antimicrobial activities against bovine respiratory pathogens, and that they contain different main chemical components that could provide broader spectrum of antimicrobial activities against BRD pathogens when these EOs applied together. The 5 EOs tested in the present study include AJO and THY (both of which were made up of three main components such as thymol, γ-terpinene, and p-cymene), CIN (84% eugenol), CIT (38% citronellal and 23% geraniol) and FEN (78% anethole) (18). Thymol has been shown to have antimicrobial activity against many bacterial pathogens including foodborne S. aureus (50), E. coli and Clostridium perfringens (51), as well as clinical isolates of Proteus mirabilis and Pseudomonas aeruginosa (52), and spoilage bacteria such as Leuconostoc citreum (53). Along with thymol, carvacrol, trans-anethole, and 1,8 cineole EO components have been investigated for their potential use as antibacterial agents or as adjuvants for the antibiotics against M. haemolytica and P. multocida (54). While no reports on testing other pure EO components against bovine respiratory bacterial pathogens, EO components anethole (55), eugenol (56), geraniol (57), and citronellal (58) have been shown promising antibacterial activities against several human respiratory pathogens. Because of these antibacterial components, the growth of M. haemolytica and P. multocida strains were significantly inhibited by the selected AJO, CIN, FEN, and THY EOs (0.025%), as they did with different country and feedlot origin M. haemolytica and P. multocida strains tested previously (18). Overall growth of culture-enriched NS microbiota determined by OD600 measurement was reduced at a moderate level by AJO, THY, and FEN under both NS microbiota cultured alone or co-cultured together with M. haemolytica, P. multocida and L. fermentum. These results observed in culture tubes suggest that intranasal inoculation of these EOs could not only inhibit BRD bacterial pathogens but also could influence overall growth of the microbiota residing within the nasopharynx in cattle.
To further identify the impact of EOs on community structure, microbial richness, diversity, and composition of the enriched-NS microbiota, we performed 16S rRNA gene sequencing on the NS microbiota culture samples harvested at the end of 24 h incubation. Beta-diversity of the enriched-NS microbiota alone or in the presence of MPL inoculums were impacted by AJO, THY, and FEN, as shown in the distinctive clustering of these samples from DMS control, and CIN and GRA treated samples (Figure 3A). All 5 EOs (AJO, THY, FEN, CIN, and GRA) tested reduced species richness in enriched-NS microbiota, with AJO and FEN being the strongest reducers. THY was the only EO that resulted in significant alterations of community diversity, and the diversity was reduced by this EO. Overall, the addition of M. haemolytica, P. multocida, and L. fermentum inoculums to NS microbiota culture induced some degree of fluctuations of the EO impact on alpha diversity indices, but the significant impact of the EOs retained in NS-MPL microbiota culture. Lower microbial richness and diversity of the respiratory microbiota has been reported in cattle that developed BRD (59, 60). However, microbial richness and diversity of healthy feedlot calves were reduced in response to intranasal bacterial therapeutics comprised of bovine nasopharyngeal origin Lactobacillus spp. (33) as compared to the intranasal administration of saline (Control). Antibiotic tulathromycin injection, on the other hand, increased both species richness and diversity in those calves as compared to calves received intranasal bacterial therapeutics or saline (Control). The direct comparison of the impact of bacterial therapeutics and antibiotic tulathromycin suggests that reduced microbial richness and diversity of bovine upper respiratory tract may have positive association with respiratory health. As such, EO-induced microbial richness reduction in enriched-NS microbiota is most likely to be associated with positive impact on nasopharyngeal microbiota. However, this warrants further testing in vivo.
The cultured-enriched NS microbiota was mainly composed of Proteobacteria, which accounted for 99.6% of the total sequencing reads. Other phyla with lower abundance detected were Firmicutes, Mycoplasmatota, Actinobacteria, and Bacteroidota. Treatment with EOs resulted in increased abundance of Proteobacteria and reduced Firmicutes and Bacteroidota abundance in both enriched-NS microbiota alone or enriched-NS microbiota along with MPL. At family level, AJO, THY, and FEN EOs significantly reduced abundance of Moraxellaceae while promoting the growth of Enterobacteriaceae. An increase in Moraxellaceae abundance has been associated with a predisposition to BRD in cattle (57). At genus level, EO specific effect on bacterial genera was observed. Overall, the effects among the three EOs (AJO, THY, and FEN) were the same on those genera affected by EO such as Acinetobacter was completely suppressed, while Escherichia and Klebsiella were enriched by these three EOs. Citrobacter was inhibited by FEN and CIT EOs. The genera Mannheimia, Pasteurella, Enterococcus and Dysgonomonas were completely diminished by all the five EOs (AJO, THY, FEN, CIN, and GRA) tested. The variations in bacterial cellular structure and EO chemical composition could be attributed to the difference in the susceptibility of these genera to EO treatments. The effectiveness of EO on bacteria varies between Gram-positive and Gram-negative bacteria (61), and those tested EOs displayed different MICs against BRD associated pathogens and commensal bacteria such as Lactobacillus, Bacillus, and Staphylococcus spp. isolated from the nasopharynx of feedlot cattle (18). The EO specific effects on certain bacterial genera observed in this study highlights that using combination of different EOs could enhance the modulatory effects on the upper respiratory microbiota in cattle as each EO has different bacterial targets. Another important finding from these 16S rRNA gene sequencing results is that the tested EOs were able to inhibit the abundance of BRD-associated genera Mannheimia and Pasteurella in the presence of culturable nasopharyngeal microbiota. Antibiotic tolerance of a bacteria is modulated by the metabolic cross-feeding interactions between different bacteria (62). Antibiotic concentrations required to inhibit the target bacteria differ when bacteria are grown in the presence of other bacteria than in monocultures (63). Commensal bacteria could influence the virulence of opportunistic pathogens (64). Since antibiotic tolerance of a bacteria, and virulence of a pathogen are influenced by a metabolic interdependence of different bacterial species in a community, it was thus important to evaluate whether EOs can still inhibit the growth of BRD pathogens in the presence of cultivable nasopharyngeal microbiota. To the best of our knowledge, this is the first study to characterize the culturable bovine respiratory microbiota using 16S rRNA gene sequencing. The nasopharyngeal swabs were cultured in relatively less-selective media BHI, which may have limited the growth of certain bacteria that do not thrive in this media. The mean observed total ASVs in NS microbiota culture that was not treated with EO was about 30, which is less than the number of ASVs observed in nasal microbiota of newborn calves (about 150 ASVs) (65), and the ASVs (about 450 ASVs) observed from the direct sequencing of the original nasopharyngeal swabs (31) that were used in this present study.
The concentrations of most cytokines evaluated were not significantly different between EO and control groups, suggesting that EOs may have limited immune stimulatory effects on BT cell monolayers. This is an important finding as it indicates that EOs applied to upper respiratory tract would not trigger a disproportional immune response. Whether immunomodulation is beneficial depends on the intended use of the stimulant tested. As the EOs explored in this study are intended for use as an intranasal cocktail spray to mitigate BRD pathogen growth and colonization, it is preferable that they do not result in an exacerbated inflammatory response, as this can lead to tissue damage and predispose or intensify infection by opportunistic pathogens. Other EOs and their main components, such as eucalyptol (60), eugenol, and cinnamaldehyde (66), have had their immunomodulatory and anti-inflammatory properties demonstrated at lower doses. However, the applied EO dosage is critical, as higher concentrations of EOs can be cytotoxic, hepatotoxic, and/or nephrotoxic (67), highlighting the importance of demonstrating their safety in vitro before testing them in an in vivo study.
Among the 6 EOs tested, AJO, THY, and GRA reduced biofilm formation by E. coli strain UMN026 at 0.025% concentration, whereas CIN, CIT, FEN, and GRA displayed antibiofilm activity against E. coli when the concentration was 0.05%. The similar effect of AJO and THY on the biofilm formation may be expected due to their similar chemical composition, as they both have thymol, γ-terpinene, and p-cymene among their main components (18). Thus, thymol could be a potent antibiofilm compound as other studies that have used thymol alone (68) or in synergistic combination with other antimicrobial agents (69) have reported a similar effect on biofilm formation. In addition to E. coli, other studies have observed antibiofilm activity of EOs against S. aureus (70), oral cariogenic bacteria (71), and foodborne pathogens (72, 73) in vitro, showing the potential EOs have against persistent pathogenic biofilm formers in diverse environments. E. coli UMN026 was selected for this biofilm assay because bacterial BRD-associated pathogens do not normally produce biofilms; however, the NS microbiome contains biofilm-forming species, and these bacteria can influence resistance or susceptibility to the development of clinical BRD. The capacity to form biofilms by members of the respiratory tract microbiota is associated with chronic infections (74) and biofilms offer a protection barrier to these species against other bacteria occurring in the same environment. Consequently, these biofilms can also protect the host mucosa while preserving the commensal microbial diversity and persistence within the biofilms (75). There are several important human respiratory pathogens that use biofilms to persist in the respiratory tract environment and cause disease, such as P. aeruginosa (76). Biofilms are therefore an important bacterial defense mechanism to evade the host immune system in the respiratory tract. Neutrophils are ineffective against persistent biofilm formers (77) which also confers protection from antibiotics (78). Therefore, it was important to test the antibiofilm capacity of the selected EOs, as disrupting biofilms is an important characteristic of any substance used in the nasopharynx for BRD prevention or treatment.
Reduction of BVDV1 viral replication on BT cells was observed with THY (strong), and AJO and CIT (moderate) (0.0125%). The antiviral capacity of some EOs or EO component such as Ocimum basilicum and Salvia officinalis (79) has been demonstrated against BVDV (80, 81) and other important respiratory viral pathogens, such as influenza viruses (24, 25, 82) and respiratory syncytial virus (80) have been shown to be inhibited by EOs in vitro. Additionally, studies using the active compounds of EOs against clinically relevant viruses demonstrated that thymol, carvacrol, p-cymene, components of AJO and THY, and limonene, and citronellal, the major component of CIT, have antiviral activity against herpes simplex virus type 1 (83) and type 2 (84), as well as dengue virus (85) in vitro. The mechanism of action of EOs on viral cells is not completely elucidated but is speculated to be due to their lipophilic properties, which allow them to cross the lipid bilayer and disrupt the envelope in enveloped viruses, leading to viral inactivation. A few studies have shown antiviral activity of EOs and their constituents when tested on enteroviruses and their surrogates, although with some limitations, like the incubation temperatures (86, 87). Antiviral drugs often target viral polymerases and replication, and since EOs and their components most likely affect the envelope and capsid of viruses, this means that EOs pose minimal risk for inducing antiviral drug resistance in clinical settings.
There are a couple limitations to be acknowledged in the present study. First, using BHI to culture nasopharyngal swab associated microbiota may have limited the growth of certain bacteria that do not thrive in this media, despite BHI being non-selective medium. In addition, culturing conditions such as temperature and atmosphere, and freeze/thawing cycles could contribute to the recovery of the lower species richness in the enriched-NS microbiota. Future studies should further evaluate the impact of EOs on culturable NS microbiota in vitro using different culturing media, growth conditions, prolonged incubation time, and larger sample size. The second limitation is associated with antiviral assay. Due to the logistic, resources and time constraints, we were unable to proceed and enumerate the reduction of BVDV1 viral cells after treatment with the EO. Therefore, the results on antiviral activities of EOs presented in the present study were only qualitative and should be interpreted cautiously, and future quantitative assay based studies should be conducted to further confirm our results and identify the extent to which these EOs can inhibit BRD viral pathogens.
5 Conclusion
The EOs AJO, THY, FEN, and CIN inhibited the growth of BRD-associated pathogens M. haemolytica and P. multocida both in individual cultures and in the presence of culture-enriched NS microbiota. EOs of AJO, THY, FEN, and CIN displayed significant modulation of community structure, species richness and composition of enriched NS-microbiota. Co-culturing BT cells with AJO, THY, FEN, CIN, CIT, or GRA had minimal effect on cytokine and chemokine release from BT cells. AJO, THY, FEN, and CIN EOs demonstrated antibiofilm activity against E. coli UMN026. BVDV-1 viral replication in BT cell monolayer was inhibited by THY (strong), and AJO and CIT (moderate). Overall, the results of this in vitro study suggest that THY, AJO, CIN, CIT, FEN EOs could be used as an intranasal EO spray to modulate nasopharyngeal microbiota and mitigate BRD pathogens in feedlot cattle as an antibiotic alternative.
Data availability statement
The raw sequencing data presented in the study can be found at the NCBI Sequence Read Archive under BioProject accession PRJNA1040875 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1040875). Other data that support the findings of this study are presented within the paper.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
SA: Conceptualization, Data curation, Writing – original draft, Writing – review & editing, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization. GM: Formal analysis, Visualization, Writing – original draft, Writing – review & editing. AR: Methodology, Writing – review & editing. DH: Data curation, Formal analysis, Software, Writing – review & editing. KS: Methodology, Writing – review & editing. LK: Methodology, Writing – review & editing. SR: Methodology, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by ND EPSCoR STEM grants program, and as part of a start-up funding from ND Agriculture Experimentation Station for S.A.
Acknowledgments
The authors would like to thank Dr. Birgit Pruess at the North Dakota State University for providing the E. coli strain and Dr. Kelli Maddock at the Veterinary Diagnostic Laboratory at North Dakota State University for supplying the Mannheimia haemolytica and Pasteurella multocida isolates.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Taylor, JD, Fulton, RW, Lehenbauer, TW, Step, DL, and Confer, AW. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can Vet J. (2010) 51:1095–102.
2. Griffin, D, Chengappa, MM, Kuszak, J, and McVey, DS. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract. (2010) 26:381–94. doi: 10.1016/j.cvfa.2010.04.004
3. Cusack, P, McMeniman, N, and Lean, I. The medicine and epidemiology of bovine respiratory disease in feedlots. Aust Vet J. (2003) 81:480–7. doi: 10.1111/j.1751-0813.2003.tb13367.x
4. Smith, RA, Step, DL, and Woolums, AR. Bovine respiratory disease: looking back and looking forward, what do we see? Vet Clin N Am Food Anim Pract. (2020) 36:239–51. doi: 10.1016/j.cvfa.2020.03.009
5. Zeineldin, M, Lowe, J, and Aldridge, B. Contribution of the mucosal microbiota to bovine respiratory health. Trends Microbiol. (2019) 27:753–70. doi: 10.1016/j.tim.2019.04.005
6. Hilton, WM. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim Health Res Rev. (2014) 15:120–2. doi: 10.1017/S1466252314000115
7. Edwards, TA. Control methods for bovine respiratory disease for feedlot cattle. Vet Clin N Am Food Anim Pract. (2010) 26:273–84. doi: 10.1016/j.cvfa.2010.03.005
8. Vogel, GJ, Bokenkroger, CD, Rutten-Ramos, SC, and Bargen, JL. A Retrospective evaluation of animal mortality in US feedlots: Rate, timing, and cause of death. Bovine Practitioner. (2015) 49:113–23. doi: 10.21423/bovine-vol49no2p113-123
9. Klima, CL, Zaheer, R, Cook, SR, Booker, CW, Hendrick, S, Alexander, TW, et al. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol. (2014) 52:438–48. doi: 10.1128/JCM.02485-13
10. Anholt, RM, Klima, C, Allan, N, Matheson-Bird, H, Schatz, C, Ajitkumar, P, et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front Vet Sci. (2017) 4:207. doi: 10.3389/fvets.2017.00207
11. Snyder, E, Credille, B, Berghaus, R, and Giguère, S. Prevalence of multi drug antimicrobial resistance in isolated from high-risk stocker cattle at arrival and two weeks after processing. J Anim Sci. (2017) 95:1124–31. doi: 10.2527/JAS.2016.1110
12. Timsit, E, Hallewell, J, Booker, C, Tison, N, Amat, S, and Alexander, TW. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet Microbiol. (2017) 208:118–25. doi: 10.1016/j.vetmic.2017.07.013
13. Amat, S, Alexander, TW, Holman, DB, Schwinghamer, T, and Timsit, E. intranasal bacterial therapeutics reduce colonization by the respiratory pathogen Mannheimia haemolytica in dairy calves. mSystems. (2020) 5:e00629-19. doi: 10.1128/mSystems.00629-19
14. Burt, S. Essential oils: their antibacterial properties and potential applications in foods—a review. Int J Food Microbiol. (2004) 94:223–53. doi: 10.1016/j.ijfoodmicro.2004.03.022
15. Hammer, KA, Carson, CF, and Riley, TV. Antimicrobial activity of essential oils and other plant extracts. J Appl Microbiol. (1999) 86:985–90. doi: 10.1046/j.1365-2672.1999.00780.x
16. Yap, PSX, Yiap, BC, Ping, HC, and Lim, SHE. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol J. (2014) 8:6–14. doi: 10.2174/1874285801408010006
17. Amat, S, Subramanian, S, Timsit, E, and Alexander, TW. Probiotic bacteria inhibit the bovine respiratory pathogen Mannheimia haemolytica serotype 1 in vitro. Lett Appl Microbiol. (2017) 64:343–9. doi: 10.1111/LAM.12723
18. Amat, S, Baines, D, Timsit, E, Hallewell, J, and Alexander, TW. Essential oils inhibit the bovine respiratory pathogens Mannheimia haemolytica, Pasteurella multocida and Histophilus somni and have limited effects on commensal bacteria and turbinate cells in vitro. J Appl Microbiol. (2019) 126:1668–82. doi: 10.1111/JAM.14238
19. Bismarck, D, Becker, J, Müller, E, Becher, V, Nau, L, and Mayer, P. Screening of antimicrobial activity of essential oils against bovine respiratory pathogens – focusing on Pasteurella multocida. Planta Med. (2022) 88:274–81. doi: 10.1055/a-1726-9291
20. Yap, PSX, Lim, SHE, Hu, CP, and Yiap, BC. Combination of essential oils and antibiotics reduce antibiotic resistance in plasmid-conferred multidrug resistant bacteria. Phytomedicine. (2013) 20:710–3. doi: 10.1016/J.PHYMED.2013.02.013
21. Knezevic, P, Aleksic, V, Simin, N, Svircev, E, Petrovic, A, and Mimica-Dukic, N. Antimicrobial activity of Eucalyptus camaldulensis essential oils and their interactions with conventional antimicrobial agents against multi-drug resistant Acinetobacter baumannii. J Ethnopharmacol. (2016) 178:125–36. doi: 10.1016/j.jep.2015.12.008
22. Basavegowda, N, Patra, JK, and Baek, K-H. Essential oils and Mono/bi/tri-metallic nanocomposites as alternative sources of antimicrobial agents to combat multidrug-resistant pathogenic microorganisms: an overview. Molecules. (2020) 25:1058. doi: 10.3390/molecules25051058
23. Ragno, R, Papa, R, Patsilinakos, A, Vrenna, G, Garzoli, S, Tuccio, V, et al. Essential oils against bacterial isolates from cystic fibrosis patients by means of antimicrobial and unsupervised machine learning approaches. Sci Rep. (2020) 10:2653. doi: 10.1038/s41598-020-59553-8
24. Wu, S, Patel, KB, Booth, LJ, Metcalf, JP, Lin, H-K, and Wu, W. Protective essential oil attenuates influenza virus infection: An in vitro study in MDCK cells. BMC Complement Altern Med. (2010) 10:69. doi: 10.1186/1472-6882-10-69
25. Choi, HJ. Chemical constituents of essential oils possessing anti-influenza A/WS/33 virus activity. Osong Public Health Res Perspect. (2018) 9:348–53. doi: 10.24171/J.PHRP.2018.9.6.09
26. Asif, M, Saleem, M, Saadullah, M, Yaseen, HS, and al Zarzour, R. COVID-19 and therapy with essential oils having antiviral, anti-inflammatory, and immunomodulatory properties. Inflammopharmacol. (2020) 28:1153–61. doi: 10.1007/s10787-020-00744-0
27. Brochot, A, Guilbot, A, Haddioui, L, and Roques, C. Antibacterial, antifungal, and antiviral effects of three essential oil blends. MicrobiologyOpen. (2017) 6:e00459. doi: 10.1002/mbo3.459
28. Gómez-Sequeda, N, Cáceres, M, Stashenko, EE, Hidalgo, W, and Ortiz, C. Antimicrobial and antibiofilm activities of essential oils against Escherichia coli O157:H7 and methicillin-resistant Staphylococcus aureus (MRSA). Antibiotics. (2020) 9:730. doi: 10.3390/antibiotics9110730
29. Liu, T, Wang, J, Gong, X, Wu, X, Liu, L, and Chi, F. Rosemary and tea tree essential oils exert antibiofilm activities in vitro against staphylococcus aureus and Escherichia coli. J Food Prot. (2020) 83:1261–7. doi: 10.4315/0362-028X.JFP-19-337
30. Sandner, G, Heckmann, M, and Weghuber, J. Immunomodulatory activities of selected essential oils. Biomol Ther. (2020) 10:1139. doi: 10.3390/BIOM10081139
31. Winders, TM, Holman, DB, Schmidt, KN, Luecke, SM, Smith, DJ, Neville, BW, et al. Feeding hempseed cake alters the bovine gut, respiratory and reproductive microbiota. Sci Rep. (2023) 13:8121. doi: 10.1038/s41598-023-35241-1
32. Amat, S, Timsit, E, Baines, D, Yanke, J, and Alexander, TW. Development of bacterial therapeutics against the bovine respiratory pathogen Mannheimia haemolytica. Appl Environ Microbiol. (2019) 85:e01359-19. doi: 10.1128/AEM.01359-19
33. Amat, S, Timsit, E, Workentine, M, Schwinghamer, T, Van Der Meer, F, Guo, Y, et al. A single intranasal dose of bacterial therapeutics to calves confers longitudinal modulation of the nasopharyngeal microbiota: a pilot study. Microbial systems. (2023) 8:e01016–22. doi: 10.1128/msystems.01016-22
34. Amat, S, Holman, DB, Schmidt, K, Menezes, ACB, Baumgaertner, F, Winders, T, et al. The nasopharyngeal, ruminal, and vaginal microbiota and the core taxa shared across these microbiomes in virgin yearling heifers exposed to divergent in utero nutrition during their first trimester of gestation and in pregnant beef heifers in response to mineral supplementation. Microorganisms. (2021) 9:2011. doi: 10.3390/microorganisms9102011
35. Santiago-Rodriguez, TM, Fornaciari, G, Luciani, S, Dowd, SE, Toranzos, GA, Marota, I, et al. Taxonomic and predicted metabolic profiles of the human gut microbiome in pre-Columbian mummies. FEMS Microbiol Ecol. (2016) 92:fiw182. doi: 10.1093/femsec/fiw182
36. Callahan, BJ, McMurdie, PJ, Rosen, MJ, Han, AW, Johnson, AJA, and Holmes, SP. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13: 13:581–583. doi: 10.1038/nmeth.3869
37. Quast, C, Pruesse, E, Yilmaz, P, Gerken, J, Schweer, T, Yarza, P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
38. Wang, Q, Garrity, GM, Tiedje, JM, and Cole, JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
39. McMurdie, PJ, and Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
40. Oskanen, J, Kindt, R, and O’Hara, R. vegan: Community Ecology Package. (2012).Available at: https://cir.nii.ac.jp/crid/1571980076147214336 (Accessed May 27, 2023).
41. O’Toole, GA. Microtiter dish biofilm formation assay. J Vis Exp. (2011) 47:2437. doi: 10.3791/2437
42. Neidhardt, FC, Bloch, PL, and Smith, DF. Culture medium for enterobacteria. J Bacteriol. (1974) 119:736–47. doi: 10.1128/jb.119.3.736-747.1974
43. Shahriar, FM, Clark, EG, Janzen, E, West, K, and Wobeser, G. Coinfection with bovine viral diarrhea virus and Mycoplasma bovis in feedlot cattle with chronic pneumonia. Can Vet J. (2002) 43:863–8.
44. Bürgi, N, Josi, C, Bürki, S, Schweizer, M, and Pilo, P. Mycoplasma bovis co-infection with bovine viral diarrhea virus in bovine macrophages. Vet Res. (2018) 49:2. doi: 10.1186/s13567-017-0499-1
45. Reed, LJ, and Muench, H. A simple method of estimating fifty per cent endpoints12. Am J Epidemiol. (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
46. Bonifaz, LC, Arzate, S, and Moreno, J. Endogenous and exogenous forms of the same antigen are processed from different pools to bind MHC class II molecules in endocytic compartments. Eur J Immunol. (1999) 29:119–31. doi: 10.1002/(SICI)1521-4141(199901)29:01<119::AID-IMMU119>3.0.CO;2-O
47. Rakibuzzaman, AGM, Kolyvushko, O, Singh, G, Nara, P, Piñeyro, P, Leclerc, E, et al. Targeted alteration of antibody-based immunodominance enhances the heterosubtypic immunity of an experimental PCV2 vaccine. Vaccine. (2020) 8:506. doi: 10.3390/vaccines8030506
48. Rakibuzzaman, A, Piñeyro, P, Pillatzki, A, and Ramamoorthy, S. Harnessing the Genetic plasticity of porcine circovirus type 2 to target suicidal replication. Viruses. (2021) 13:1676. doi: 10.3390/v13091676
49. Teixeira, B, Marques, A, Ramos, C, Serrano, C, Matos, O, Neng, NR, et al. Chemical composition and bioactivity of different oregano (Origanum vulgare) extracts and essential oil. J Sci Food Agric. (2013) 93:2707–14. doi: 10.1002/jsfa.6089
50. Rúa, J, del Valle, P, de Arriaga, D, Fernández-Álvarez, L, and García-Armesto, MR. Combination of carvacrol and thymol: antimicrobial activity against staphylococcus aureus and antioxidant activity. Foodborne Pathog Dis. (2019) 16:622–9. doi: 10.1089/fpd.2018.2594
51. Sepahvand, S, Amiri, S, Radi, M, and Akhavan, H-R. Antimicrobial activity of thymol and thymol-nanoemulsion against three food-borne pathogens inoculated in a sausage model. Food Bioprocess Technol. (2021) 14:1936–45. doi: 10.1007/s11947-021-02689-w
52. Sim, JXF, Khazandi, M, Chan, WY, Trott, DJ, and Deo, P. Antimicrobial activity of thyme oil, oregano oil, thymol and carvacrol against sensitive and resistant microbial isolates from dogs with otitis externa. Vet Dermatol. (2019) 30:524–e159. doi: 10.1111/vde.12794
53. Lee, S, Kim, H, Beuchat, LR, Kim, Y, and Ryu, J-H. Synergistic antimicrobial activity of oregano and thyme thymol essential oils against Leuconostoc citreum in a laboratory medium and tomato juice. Food Microbiol. (2020) 90:103489:103489. doi: 10.1016/j.fm.2020.103489
54. Kissels, W, Wu, X, and Santos, RR. Short communication: Interaction of the isomers carvacrol and thymol with the antibiotics doxycycline and tilmicosin: In vitro effects against pathogenic bacteria commonly found in the respiratory tract of calves. J Dairy Sci. (2017) 100:970–4. doi: 10.3168/JDS.2016-11536
55. Kwiatkowski, P, Pruss, A, Masiuk, H, Mnichowska-Polanowska, M, Kaczmarek, M, Giedrys-Kalemba, S, et al. The effect of fennel essential oil and trans-anethole on antibacterial activity of mupirocin against Staphylococcus aureus isolated from asymptomatic carriers. Postepy Dermatol Alergol. (2019) 36:308–14. doi: 10.5114/ada.2018.76425
56. Yadav, MK, Chae, S-W, Im, GJ, Chung, J-W, and Song, J-J. Eugenol: a phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PLoS One. (2015) 10:e0119564. doi: 10.1371/journal.pone.0119564
57. Mączka, W, Wińska, K, and Grabarczyk, M. One hundred faces of geraniol. Mol Ther. (2020) 25:3303. doi: 10.3390/molecules25143303
58. Ács, K, Balázs, VL, Kocsis, B, Bencsik, T, Böszörményi, A, and Horváth, G. Antibacterial activity evaluation of selected essential oils in liquid and vapor phase on respiratory tract pathogens. BMC Complement Altern Med. (2018) 18:227. doi: 10.1186/s12906-018-2291-9
59. Holman, DB, McAllister, TA, Topp, E, Wright, ADG, and Alexander, TW. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet Microbiol. (2015) 180:90–5. doi: 10.1016/J.VETMIC.2015.07.031
60. Yin, C, Liu, B, Wang, P, Li, X, Li, Y, Zheng, X, et al. Eucalyptol alleviates inflammation and pain responses in a mouse model of gout arthritis. Br J Pharmacol. (2020) 177:2042–57. doi: 10.1111/bph.14967
61. Prabuseenivasan, S, Jayakumar, M, and Ignacimuthu, S. In vitro antibacterial activity of some plant essential oils. BMC Complement Altern Med. (2006) 6:39. doi: 10.1186/1472-6882-6-39
62. Adamowicz, EM, Flynn, J, Hunter, RC, and Harcombe, WR. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. (2018) 12:2723–35. doi: 10.1038/s41396-018-0212-z
63. Adamowicz, EM, Muza, M, Chacón, JM, and Harcombe, WR. Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica. PLoS Pathog. (2020) 16:e1008700. doi: 10.1371/journal.ppat.1008700
64. Stacy, A, Fleming, D, Lamont, RJ, Rumbaugh, KP, and Whiteley, M. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. M bio. (2016) 7:e00782–16. doi: 10.1128/mBio.00782-16
65. Luecke, SM, Holman, DB, Schmidt, KN, Gzyl, KE, Hurlbert, JL, Menezes, ACB, et al. Whole-body microbiota of newborn calves and their response to prenatal vitamin and mineral supplementation. Front Microbiol. (2023) 14:1207601. doi: 10.3389/fmicb.2023.1207601
66. Mateen, S, Rehman, MT, Shahzad, S, Naeem, SS, Faizy, AF, Khan, AQ, et al. Anti-oxidant and anti-inflammatory effects of cinnamaldehyde and eugenol on mononuclear cells of rheumatoid arthritis patients. Eur J Pharmacol. (2019) 852:14–24. doi: 10.1016/j.ejphar.2019.02.031
67. Janes, SEJ, Price, CSG, and Thomas, D. Essential oil poisoning: N-acetylcysteine for eugenol-induced hepatic failure and analysis of a national database. Eur J Pediatr. (2005) 164:520–2. doi: 10.1007/s00431-005-1692-1
68. Čabarkapa, I, Čolović, R, Đuragić, O, Popović, S, Kokić, B, Milanov, D, et al. Anti-biofilm activities of essential oils rich in carvacrol and thymol against Salmonella Enteritidis. Biofouling. (2019) 35:361–75. doi: 10.1080/08927014.2019.1610169
69. Hossain, MI, Rahaman Mizan, MF, Toushik, SH, Roy, PK, Jahid, IK, Park, SH, et al. Antibiofilm effect of nisin alone and combined with food-grade oil components (thymol and eugenol) against Listeria monocytogenes cocktail culture on food and food-contact surfaces. Food Control. (2022) 135:108796. doi: 10.1016/j.foodcont.2021.108796
70. Budzyńska, A, Wieckowska-Szakiel, M, Sadowska, B, Kalemba, D, and Rózalska, B. Antibiofilm activity of selected plant essential oils and their major components. Pol J Microbiol. (2011) 60:35–41. doi: 10.33073/pjm-2011-005
71. de Oliveira Carvalho, I, Purgato, GA, Píccolo, MS, Pizziolo, VR, Coelho, RR, Diaz-Muñoz, G, et al. In vitro anticariogenic and antibiofilm activities of toothpastes formulated with essential oils. Arch Oral Biol. (2020) 117:104834. doi: 10.1016/j.archoralbio.2020.104834
72. Snoussi, M, Dehmani, A, Noumi, E, Flamini, G, and Papetti, A. Chemical composition and antibiofilm activity of Petroselinum crispum and Ocimum basilicum essential oils against Vibrio spp. strains. Microb Pathog. (2016) 90:13–21. doi: 10.1016/j.micpath.2015.11.004
73. Mouhoub, A, Guendouz, A, Belkamel, A, el Alaoui Talibi, Z, Ibnsouda Koraichi, S, el Modafar, C, et al. Assessment of the antioxidant, antimicrobial and antibiofilm activities of essential oils for potential application of active chitosan films in food preservation. World J Microbiol Biotechnol. (2022) 38:179. doi: 10.1007/s11274-022-03363-9
74. Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS. (2013) 121:1–58. doi: 10.1111/apm.12099
75. Burmølle, M, Thomsen, TR, Fazli, M, Dige, I, Christensen, L, Homøe, P, et al. Biofilms in chronic infections - a matter of opportunity - monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol. (2010) 59:324–36. doi: 10.1111/j.1574-695X.2010.00714.x
76. Parsek, MR, and Tolker-Nielsen, T. Pattern formation in Pseudomonas aeruginosa biofilms. Curr Opin Microbiol. (2008) 11:560–6. doi: 10.1016/j.mib.2008.09.015
77. Yonker, LM, Cigana, C, Hurley, BP, and Bragonzi, A. Host-pathogen interplay in the respiratory environment of cystic fibrosis. J Cyst Fibros. (2015) 14:431–9. doi: 10.1016/j.jcf.2015.02.008
78. Drenkard, E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. (2003) 5:1213–9. doi: 10.1016/j.micinf.2003.08.009
79. Madeddu, S, Marongiu, A, Sanna, G, Zannella, C, Falconieri, D, Porcedda, S, et al. Bovine viral diarrhea virus (BVDV): a preliminary study on antiviral properties of some aromatic and medicinal plants. Pathogens. (2021) 10:403. doi: 10.3390/pathogens10040403
80. Pilau, MR, Alves, SH, Weiblen, R, Arenhart, S, Cueto, AP, and Lovato, LT. Antiviral activity of the Lippia graveolens (Mexican oregano) essential oil and its main compound carvacrol against human and animal viruses. Braz J Microbiol. (2011) 42:1616–24. doi: 10.1590/S1517-83822011000400049
81. Kubiça, TF, Alves, SH, Weiblen, R, and Lovato, LT. In vitro inhibition of the bovine viral diarrhoea virus by the essential oil of Ocimum basilicum (basil) and monoterpenes. Braz J Microbiol. (2014) 45:209–14. doi: 10.1590/S1517-83822014005000030
82. Vimalanathan, S, and Hudson, J. Anti-influenza virus activity of essential oils and vapors. Am J Essent Oils Nat Products. (2014) 2:47–53.
83. Sharifi-Rad, J, Salehi, B, Schnitzler, P, Ayatollahi, SA, Kobarfard, F, Fathi, M, et al. Susceptibility of herpes simplex virus type 1 to monoterpenes thymol, carvacrol, p-cymene and essential oils of Sinapis arvensis L., Lallemantia royleana Benth. and Pulicaria vulgaris Gaertn. Cell Mol Biol (Noisy-le-Grand). (2017) 63:42–7. doi: 10.14715/cmb/2017.63.8.10
84. Allahverdiyev, A, Duran, N, Ozguven, M, and Koltas, S. Antiviral activity of the volatile oils of Melissa officinalis L. against Herpes simplex virus type-2. Phytomedicine. (2004) 11:657–61. doi: 10.1016/j.phymed.2003.07.014
85. Nagy, MM, Al-Mahdy, DA, Abd El Aziz, OM, Kandil, AM, Tantawy, MA, and El Alfy, TSM. Chemical composition and antiviral activity of essential oils from Citrus reshni hort. ex Tanaka (Cleopatra mandarin) cultivated in Egypt. J Essent Oil Bearing Plants. (2018) 21:264–72. doi: 10.1080/0972060X.2018.1436986
86. Gilling, D, Kitajima, M, Torrey, JR, and Bright, KR. Antiviral efficacy and mechanisms of action of oregano essential oil and its primary component carvacrol against murine norovirus. J Appl Microbiol. (2014) 116:1149–63. doi: 10.1111/jam.12453
Keywords: bovine respiratory pathogens, essential oil, antimicrobial, antiviral and antibiofilm activities, immunomodulation, nasopharyngeal microbiota
Citation: Amat S, Magossi G, Rakibuzzaman A, Holman DB, Schmidt KN, Kosel L and Ramamoorthy S (2024) Screening and selection of essential oils for an intranasal spray against bovine respiratory pathogens based on antimicrobial, antiviral, immunomodulatory, and antibiofilm activities. Front. Vet. Sci. 11:1360398. doi: 10.3389/fvets.2024.1360398
Edited by:
Ambreen Ashar, North Carolina State University, United StatesReviewed by:
Mihaela Niculae, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaZeeshan Ahmad Bhutta, Chungbuk National University, Republic of Korea
Copyright © 2024 Amat, Magossi, Rakibuzzaman, Holman, Schmidt, Kosel and Ramamoorthy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samat Amat, c2FtYXQuYW1hdEBuZHN1LmVkdQ==
 Samat Amat
Samat Amat Gabriela Magossi
Gabriela Magossi AGM Rakibuzzaman1
AGM Rakibuzzaman1 Devin B. Holman
Devin B. Holman Kaycie N. Schmidt
Kaycie N. Schmidt Luke Kosel
Luke Kosel Sheela Ramamoorthy
Sheela Ramamoorthy