
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 12 March 2024
Sec. Veterinary Infectious Diseases
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1336663
 Yu Han1
Yu Han1 Kejia Lu1
Kejia Lu1 Riteng Zhang1
Riteng Zhang1 Xi Wei1
Xi Wei1 Hanwei Guo1
Hanwei Guo1 Lina Tong1,2
Lina Tong1,2 Xinglong Wang1
Xinglong Wang1 Sa Xiao1
Sa Xiao1 Haijin Liu1*
Haijin Liu1* Zengqi Yang1*
Zengqi Yang1*Bovine parainfluenza virus type 3 (BPIV3) is a key pathogen associated with bovine respiratory disease complex (BRDC). However, its specific pathogenesis mechanisms have not been fully elucidated. Reverse genetics provides a useful method for understanding the pathogenic mechanism of BPIV3. To ensure the functionality of the rescue platforms, we first constructed a minigenome (MG) system of BPIV3 utilizing a 5-plasmid system in this investigation. Then, a full-length infection clone of BPIV3 was obtained from the SX-2021 strain, and different methods were employed to identify the rescued virus. Additionally, we recovered a recombinant BPIV3 using the reverse genetics system that could express enhanced green fluorescence protein (eGFP). Through the growth curve assays, the replicate capability of rBPIV3-SX-EGFP was found to be similar to that of the parental virus. Subsequently, the rBPIV3-SX-EGFP was used to determine the antiviral activity of ribavirin. The results showed that ribavirin had an anti-BPIV3 effect in MDBK cells. In conclusion, the successful development of a reverse genetic system for the SX-2021 strain establishes a foundation for future studies on BPIV3, including investigations into its pathogenic mechanism, gene function, and antiviral screening properties.
BPIV3 is recognized as a significant pathogen contributing to BRDC, inducing a spectrum of mild-to-severe respiratory symptoms (1, 2). BPIV3 is a non-segmented, enveloped negative-strand RNA virus that is a member of the Respirovirus genus within the Paramyxovirdae family (2). In recent years, there has been an increasing number of BPIV3-induced respiratory illness outbreaks in cattle, resulting in substantial financial losses within the global livestock industry (3–5). BPIV3 has been classified into three genotypes, and the first report of BPIV3 genotype C as a new genotype was identified in China (6). Nevertheless, BPIV3c lacks a recognized therapy, and its pathogenic mechanism remains elusive.
To date, only limited studies on the reverse genetics of the BPIV3 genotype A have been reported (7, 8). Therefore, it is crucial to establish a reverse genetics system for BPIV3c to investigate pathogenic mechanisms and facilitate the development of a modified live virus vaccine. Mini-genome (MG) systems, also referred to as Mini replicon technology, are commonly utilized to explore the life cycle of viruses within the Paramyxoviridae family (9–11). Previously, reporter viruses have been effectively employed to examine the pathophysiology of paramyxoviruses and screen antiviral medicines with high sensitivity. Ribavirin, a broad-spectrum antiviral drug, is commonly used in antiviral tests (12).
In this study, we successfully developed the first reverse genetic system for BPIV3 genotype C using a 5-plasmid system. Furthermore, the rBPIV3-SX-EGFP was employed as a screening method for antivirals in vitro.
MDBK and BHK-21 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA), at 37°C with 5% CO2. The BPIV3 strain was isolated from a BPIV3-positive lung tissue sample, designated as BPIV3/SX/2021 (GenBank: ON804787). The pCMV and pCI-neo plasmids were used to construct the rBPIV3-SX cDNA clone and three helper plasmids, respectively. The pGAGGS-T7 plasmid, containing T7 RNA polymerase, was constructed in our laboratory. Polyclone antibodies against NP were prepared in our laboratory, and the specificity has been verified in our previous study (13). An anti-EGFP antibody (ab184601) was purchased from Abcam. HRP-conjugated Goat Anti-Mouse IgG was purchased from Invitrogen. FITC-labeled Goat Anti-Mouse IgG (H + L) was purchased from Beyotime.
A set of primers was designed to construct an MG system for BPIV3 based on the conserved genomic regions of the viral 5′ untranslated region (5′ UTR), EGFP, and the viral 3′ UTR. The three PCR products were assembled by overlap and inserted in the antisense orientation between the T7 promoter and terminator sequences, resulting in a construct named pT7-MG.
To construct a full-length cDNA clone of the SX-2021 strain, a set of primers was designed to amplify the viral RNA genome using reverse transcription-polymerase chain reaction (RT-PCR). The PCR products were sequenced by Sanger sequencing with three independent parallel tests. Six separate fragments (A to F) were assembled using unique restriction enzyme sites. A T7 promoter was inserted upstream of fragment A and three G residues preceding the T7 promoter sequence. Fragment F contained a 24-nucleotide (nt) partial HDV ribozyme (Supplementary Table S1). Additionally, a molecular marker for differentiating the rescued virus from the parental virus was created. The primers corresponding to fragments A and B were modified to eliminate the unnecessary NcoI site. The fragments of (A-C) and (D-F) were, respectively, inserted into the pCMV plasmid; pCMV (A-C) and pCMV (D-F) plasmids were digested by NcoI and RsrII. These two fragments were then linked, and the full-length BPIV3 cDNA was assembled, named pBPIV3-SX (Figure 1A). The NP, P, and L ORFs were separately subcloned into the pCI-neo plasmid to construct the three helper plasmids, pCI-NP, pCI-P, and pCI-L.
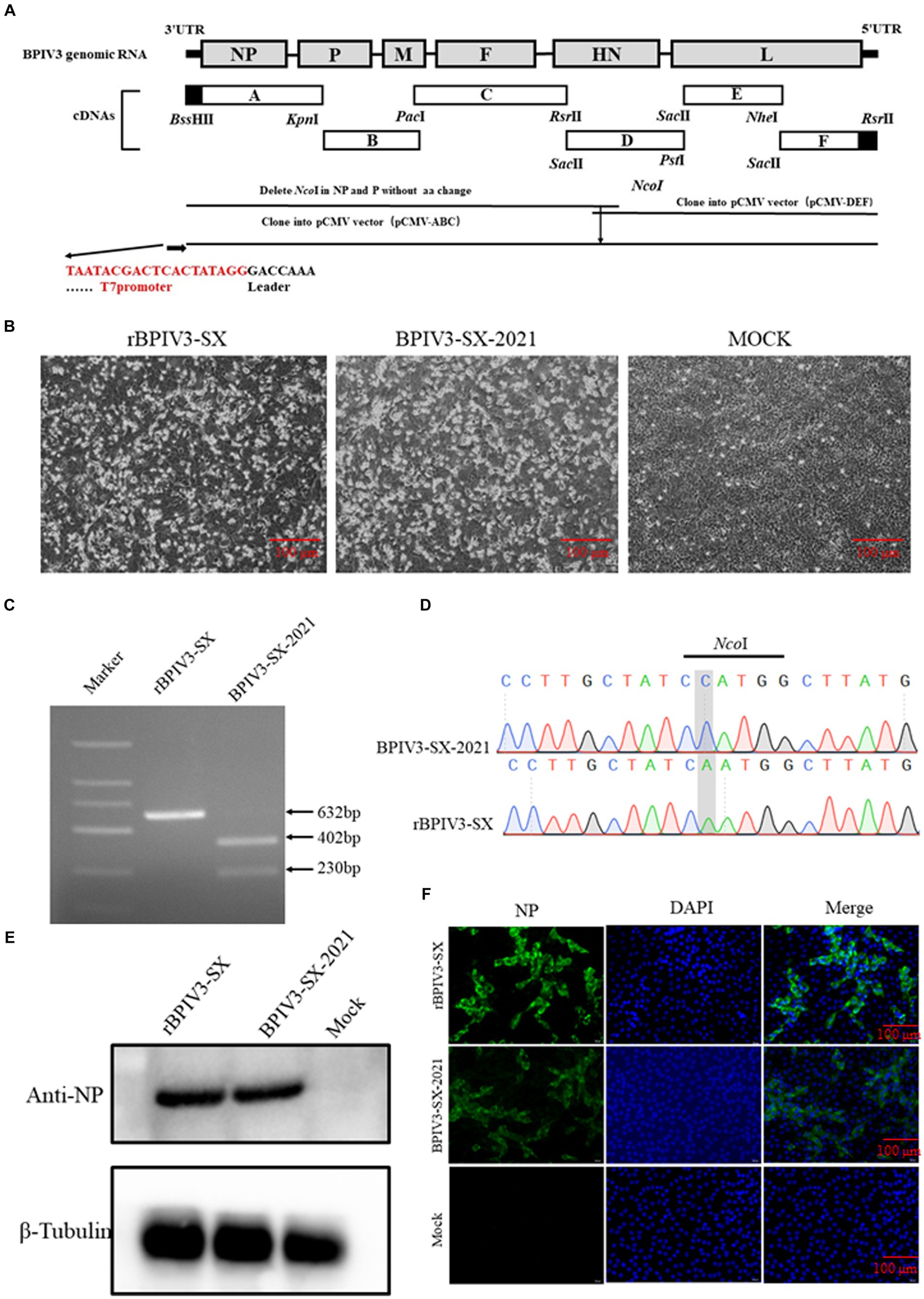
Figure 1. Identification of the rescued rBPIV3-SX virus assay. (A) Schematic representation showing the construction of pBPIV3-SX. (B) BPIV3-specific cytopathic effect (CPE) of the rescued virus. MDBK cell cultures were collected at 48 h after infection with the rescue and parental virus. (C) Identification of genetic marker in the rescued virus. The NcoI restriction enzyme site was mutated into the recombinant plasmid to create a genetic marker to distinguish the rescued virus from the parental virus. (D) The genetic marker mutation was identified by sequencing. (E) Expression of NP in rBPIV3-SX-infected cells. The MDBK cells infected with the rescue and parental virus were detected using Western blotting using anti-BPIV3 NP polyclonal antibody at 24 h. (F) Immunofluorescence analysis of the rescued virus. The MDBK cells were infected with the rescue and parental virus, and the infected cells were stained by anti-BPIV3 NP polyclonal antibody.
To construct a rescue virus containing eGFP, a plasmid containing the full-length sequence of BPIV3 was used, with a fusion sequence of eGFP. The fusion sequence, flanked by BssHII and PacI sites, was subcloned into the pBPIV3-SX, resulting in the plasmid named pBPIV3-SX-EGFP.
BHK-21 cells were seeded into a 12-well plate and transfected with a plasmid of the minigenome or full-length cDNA clone, along with pCI-NP, pCI-P, pCI-L, and pCAGGS-T7 at a ratio of 0.5 μg: 0.5 μg: 0.1 μg: 0.05 μg: 0.35 μg, using TurboFect™ reagent (Invitrogen, Carlsbad, CA). Replication of this minigenome was observed at various time points under a fluorescent microscope. After 60 h post-transfection with the full-length cDNA clone, the cell supernatant from BHK-21 cells was harvested and transferred to MDBK cells. The rescue viruses were harvested between 48 and 60 h post-infection when significant CPE was observed. Additionally, the rescue viruses underwent 20 serial passages in MDBK cells, using a multiplicity of infection (MOI) of 0.1. The virus was adsorbed onto confluent cell monolayers in 60-mm cell plates for 1 h at 37°C with frequent rocking. Following adsorption, the inoculum was removed, the cells were washed with DMEM, 4 mL of maintenance media was added, and the plates were incubated at 37°C. medium from each well was harvested at 72 hpi and stored in aliquots at −80°C. Following each passage, the viral harvest was quantified by TCID50 assay on MDBK cells and diluted to produce an MOI of 0.1 for each subsequent passage.
To determine the molecular marker (C341 to A341 mutation in the NcoI site) in the rescued virus, rBPIV3/SX, RT-PCR was performed on the genomic region containing the specific mutation. For this purpose, viral RNA was extracted from passage 5 of the rBPIV3-SX virus using the TRIzol reagent (Takara) and reverse transcribed into cDNA using a StarScript II RT Kit (Genstar, Beijing, China). RT-PCR was employed to amplify the relevant viral genomic region using the primer pair, Nco-F/Nco-R (Supplementary Table S1). The PCR product was gel-purified, digested with NcoI, and the DNA sequenced to confirm the presence of the C341 to A341 mutation.
MDBK cells were seeded into a 12-well plate and were infected with BPIV3-SX-2021, rBPIV3-SX, rBPIV3-SX-EGFP, or mock-infected at an MOI of 0.1. Cell lysates were harvested using 160 μL/well lysis buffer at 24 hpi, and the cell lysate was denatured at 95°C for 10 min in a 5x loading buffer (Bio-Rad, Hercules, CA). Proteins were analyzed on a 10% SDS-PAGE and then transferred to a PVDF membrane. The membrane was blocked with 5% skim milk for 2 h at room temperature. To identify the expression of BPIV3 NP or eGFP, the membrane was incubated overnight at 4°C with primary antibodies. The membrane was then washed three times with TBST, followed by incubation with secondary antibodies at room temperature for 1 h. Target proteins were analyzed using BeyoECL Plus (Beyotime, Beijing, China).
MDBK cells were seeded into a 24-well plate and infected with BPIV3-SX-2021, rBPIV3-SX, and rBPIV3-SX-EGFP at an MOI of 0.1 or mock-infected. At 24 hpi, cell monolayers were fixed with 4% paraformaldehyde at room temperature for 30 min and then blocked with PBS containing 1% bovine serum albumin (BSA) for 30 min and then permeabilized with 0.1% Triton X-100 at 5 min. Cell monolayers were incubated with pAb against NP for 1 h and then washed with PBS three times. Cell monolayers were further incubated for 1 h with a secondary antibody. After washing three times, the cells were analyzed under a fluorescent microscope (IX73; OLYMPUS, Tokyo, Japan).
MDBK cells were seeded into a 6-well plate and infected with BPIV3-SX-2021, rBPIV3-SX, and rBPIV3-SX-EGFP at an MOI of 0.1. The plates were incubated at 37°C for 1 h, and the cell culture supernatant was removed and replaced with DMEM supplemented containing 2% FBS. Cell culture supernatants were harvested at 12, 24, 36, 48, 60, and 72 hpi, and the viral titers were measured using the TCID50 assay on MDBK cells.
To determine the concentration of ribavirin-treated effect on MDBK cell viability, a CCK-8 assay was employed (Solarbio, CA1210). MDBK cells were seeded into a 96-well plate, and the supernatants were supplemented with DMEM containing ribavirin at various dosages. After incubating the ribavirin-treated cells for 24 h, the CCK-8 solution (10 μL/well) was added to each well. The plate was examined using a microplate reader at 450 nm after 4 h of incubation.
MDBK cells were seeded into a 12-well plate to assess the antiviral activities of ribavirin at various doses. The cells were infected with rBPIV3-SX-EGFP at an MOI of 1 for 1 h at 37°C. The supernatants were replaced and added to DMEM containing ribavirin at various doses. A fluorescent microscope was used to examine ribavirin-treated cells after 24 h. The supernatants were harvested for the viral titer determination using the TCID50 assay of rBPIV3-SX-EGFP.
In addition to reporter viruses, a minigenome system is a powerful tool for viral studies. Consequently, we constructed a minigenome system, with the 5′ and 3′ non-coding regions of the BPIV3 genome constituting the trailer and leader sequence. The genome structures of two genes were flanked by gene start (GS) and gene end (GE) sequences. The reporter plasmid contains the eGFP reporter gene inserted between the leader and trailer regions and is flanked by the T7 promoter and HDV ribozyme. The MG sequence of BPIV3 was schematically shown in Figure 2A. In addition to the MG plasmid, the three helper plasmids were used in the minigenome system (Figure 2B). The (pT7-MG) plasmid and pCAGGS-T7 plasmid, along with the three helper plasmids, were co-transfected into BHK-21 cells. The replication of this minigenome (MG) and eGFP activity was assessed at various time points. At 24 hpi, a few cells exhibited green fluorescence, with the proportion of such cells increasing over time. The eGFP activity increased substantially from 24 to 72 h post-transfection, indicating that our MG system replicated efficiently (Figure 2C).
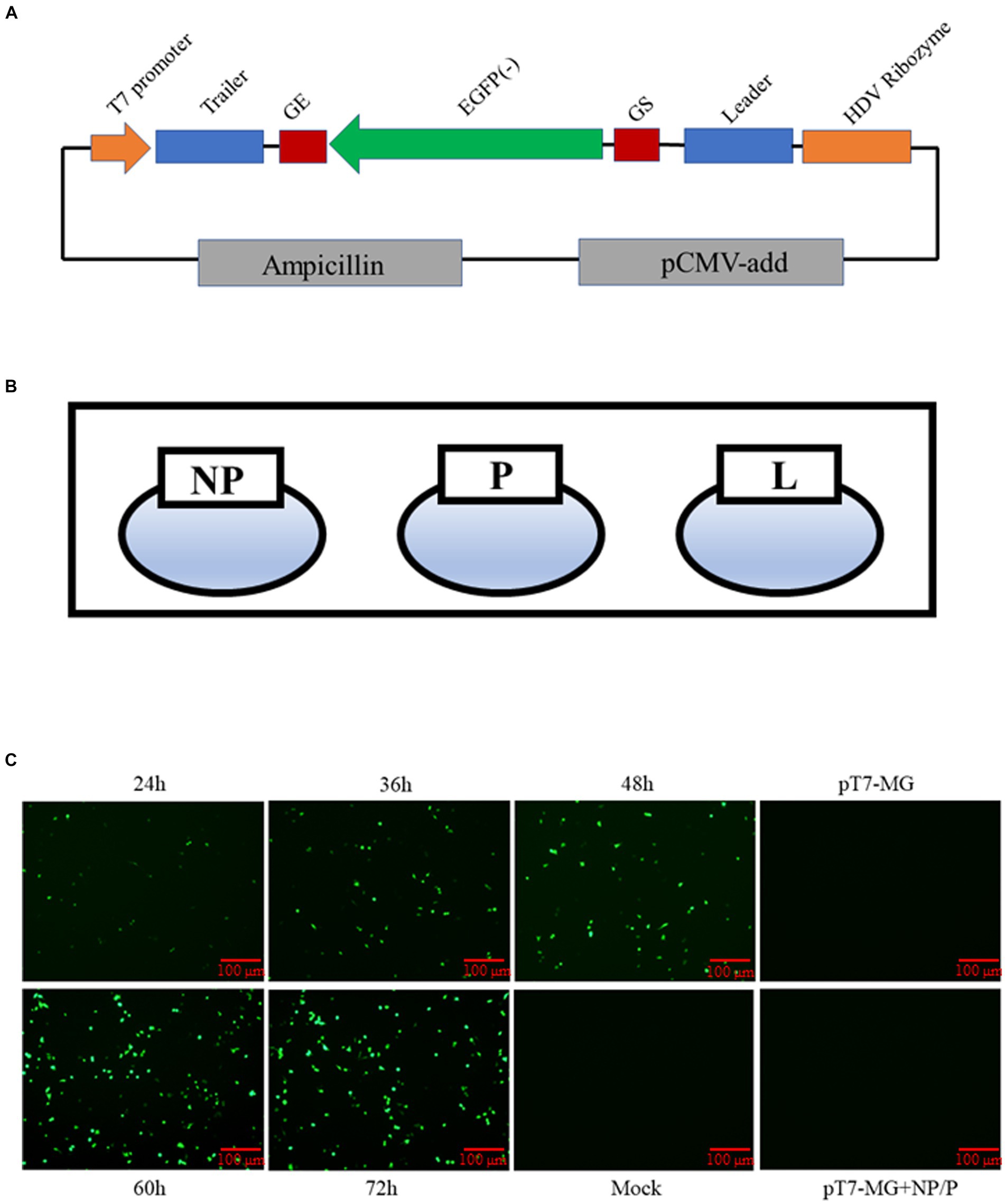
Figure 2. Construction of BPIV3 minigenome system. (A) Diagram showing the structure of the BPIV3 minigenome system. This reporter plasmid contains the eGFP reporter gene inserted between the leader and trailer regions and then flanked by the T7 promoter and HDV ribozyme. (B) Schematic map of the three helper plasmids for the rescue virus. (C) BHK-21 cells co-transfected with five plasmids at 24 h, 36 h, 48 h, 60 h, and 72 h. The expression levels of eGFP were determined under the fluorescence microscope.
To rescue the virus, the full-length infection clone plasmid of rBPIV3-SX was constructed and co-transfected into BHK-21 cells with the helper plasmids. After 72-h transfection, the supernatants from transfected cells were harvested and subsequently transferred onto the MDBK cells. Simultaneously, typical CPEs were observed in virus-infected cell monolayers at 48 h (Figure 1B). Total RNA was extracted from rBPIV3-SX or the parental virus, and the fragment containing the NcoI molecular marker in rBPIV3-SX was analyzed by RT-PCR using the primer Nco-F/R. PCR products were gel-purified and digested by NcoI. The parental virus produced a 402-bp and a 230-bp fragment following NcoI digestion, whereas the rescued virus could not achieve this (Figure 1C). Additionally, the PCR products were sequenced to confirm the presence of the NcoI site mutation (Figure 1D). The expression of NP protein in rBPIV3-SX infected cells was analyzed using the Western blot. The result showed the NP (69 kDa) protein was expressed in cells infected with the rBPIV3-SX (Figure 1E). BPIV3 NP expression was further assessed using a pAb against the NP protein by IFA. The virus-infected cell monolayer was visible in bright green fluorescence but not in the mock cells (Figure 1F).
Furthermore, we generated a full-length infectious clone of the BPIV3 containing an eGFP reporter. The P-M intergenic site has been demonstrated to be preferable for the optimal expression of foreign proteins (14). Using the rBPIV3 (SX-2021) cDNA clone as the backbone of the rBPIV3-SX-EGFP cDNA clone, an EGFP reporter gene was inserted between BPIV3 P and M genes of the rBPIV3-SX cDNA clone using overlapping technology (Figure 3A). To rescue the rBPIV3-SX-EGFP, we co-transfected the full-length plasmid and the helper plasmids into BHK-21 cells. As depicted in Figure 3B, to characterize the recombinant virus, the viral protein NP was confirmed using immunofluorescent microscopy, and the eGFP and NP proteins were expressed in rBPIV3-SX-EGFP virus-infected cells. To further determine the expression of NP and eGFP proteins using the Western blot, the results showed that the NP (69 kDa) and eGFP (27 kDa) proteins were expressed in cells infected with rBPIV3-SX-EGFP. However, only the NP protein was expressed in cells infected with the parental virus (Figure 3C). To assess the impact of eGFP expression on viral replication, the growth kinetics of rBPIV3-SX-EGFP were compared to those of rBPIV3-SX and the parental virus. The results have shown that all the viruses displayed a similar growth behavior during 72 h (Figure 3D). To evaluate the genetic stability of the recombinant virus, rBPIV3-SX-EGFP underwent 20 serial passages in MDBK cells. As shown in Figure 3E, eGFP was consistently expressed at a high level during passages.
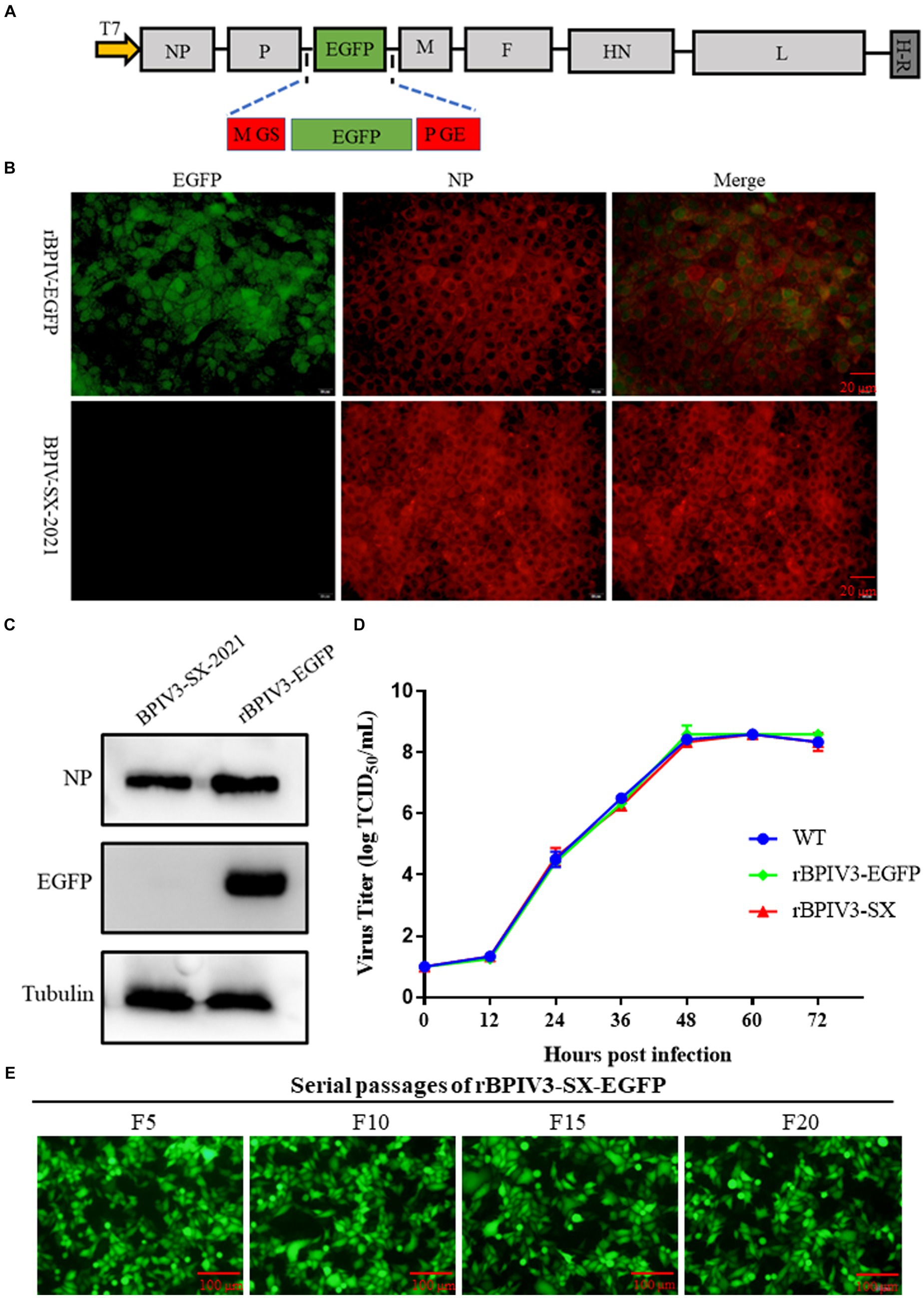
Figure 3. Construction, identification, and characterization of full-length BPIV3 reporter virus. (A) Diagram showing the structure of the full-length BPIV3 reporter virus. The H-R stands for HDV ribozyme. (B) Immunofluorescence analysis of the rescued virus. The MDBK cells were infected with rBPIV3-SX-EGFP and parental virus; the infected cells were stained by anti-BPIV3 NP polyclonal antibody. (C) Expression of NP and eGFP in rBPIV3-SX-EGFP-infected cells. The MDBK cells infected with rBPIV3-SX-EGFP and parental virus were detected using Western blotting using anti-BPIV3 NP polyclonal antibody and anti-EGFP monoclonal antibody at 24 h. (D) The growth kinetics of the rescued viruses. MDBK cells in six-well plates were infected with rBPIV3-SX, rBPIV3-SX-EGFP, and parental virus. The supernatant was harvested at 12, 24, 36, 48, 60, and 72 h and titrated on MDBK cells. (E) Serial passages of rBPIV3-SX-EGFP. The MDBK cells were infected with different passages of rBPIV3-SX-EGFP, and the expression of eGFP was determined under the fluorescence microscope.
Using the Cell Counting Kit 8 assay, we assessed the impact of ribavirin on MDBK cell viability. The results showed that ribavirin treatment did not affect the viability of the cells (Figure 4A). Ribavirin-treated cell monolayers were monitored at 24 hpi in a randomly selected field-of-view fluorescence microscope to assess the declining trend in eGFP expression. The results showed that the fraction of green cells gradually decreased with increasing drug concentration in the group treated with ribavirin (Figure 4B). At 24 hpi, all ribavirin-treated cell cultures were titrated using the TCID50 assay, and the viral titer steadily dropped with increasing ribavirin concentration, as observed under the fluorescence microscope (Figure 4C).
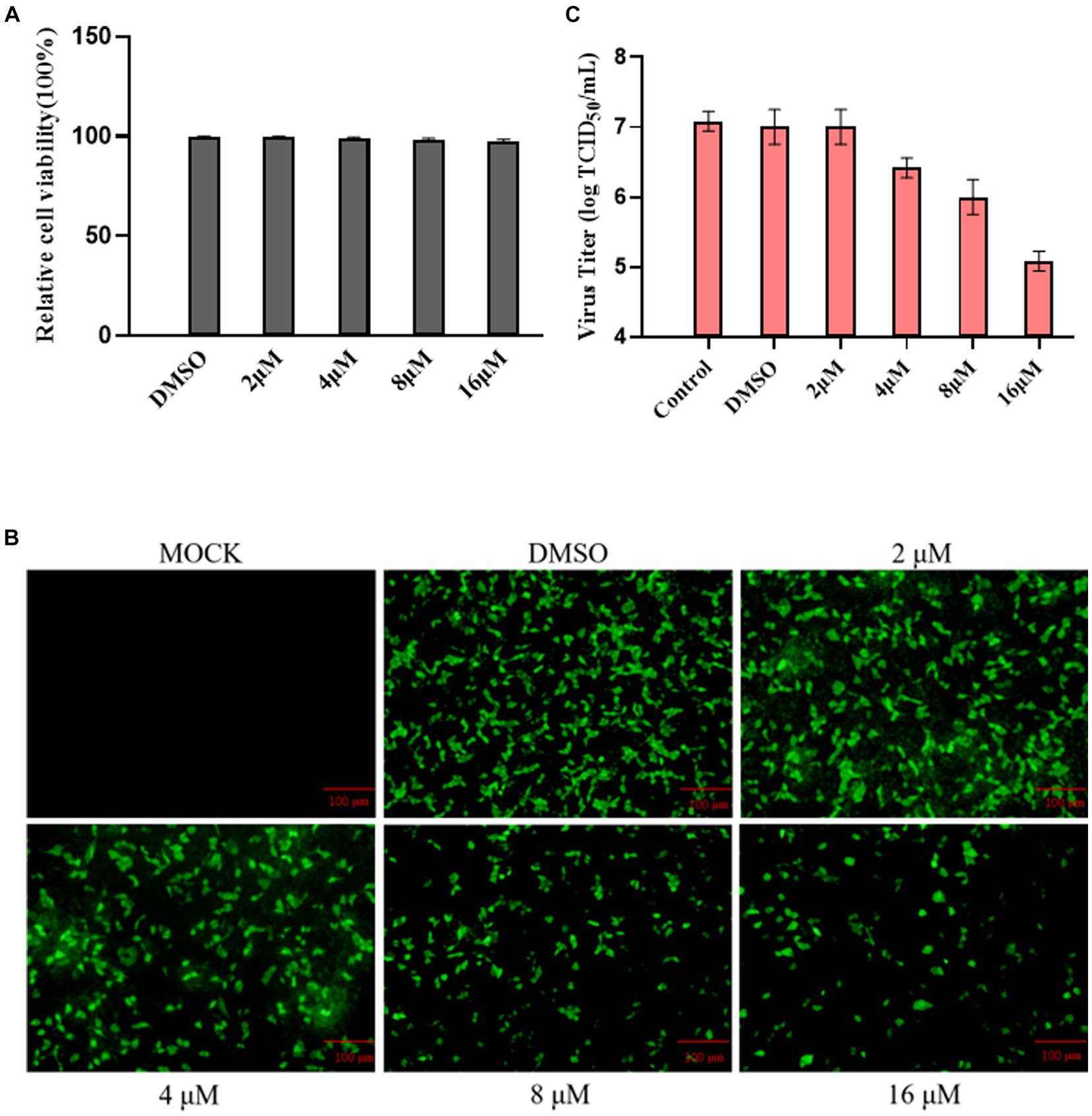
Figure 4. Ribavirin inhibitions of rescued rBPIV3-SX-EGFP replication. (A) The cytotoxic effect of ribavirin treatment on MDBK cells. The cells were treated with 0, 2, 4, 8, or 16 μM. Relative cell viability was determined using the CCK-8 assay. (B) Profiles of rBPIV3-SX-EGFP-expressed eGFP at 24 h after treatments separately with ribavirin at different concentrations. (C) The viral titer was detected with different concentrations of ribavirin treatment using the Reed–Muench assay.
BPIV3 infection primarily induces respiratory symptoms in both young and adult cattle, such as cough, fever, and nasal discharge, particularly in feedlot cattle (15, 16). Genotype C of BPIV3 as a potentially new genotype was isolated and identified in China in 2008 (6). Subsequently, BPIV3c has been identified in major cattle-producing countries, including the United States, Australia, Japan, and Korea. Its presence has led to significant economic losses in the global livestock industry (4, 5, 17).
Previous phylogenetic studies have revealed significant genetic diversity among different genotypes of BPIV3, with potential geographical and tempo ral ranges (6). Therefore, the construction of the reverse genetic systems for different viral strains could be useful for assessing the evolutionary and ecological assessments of BPIV3. The reverse genetics system for BPIV3 dates back to as early as 2000 when it was established as an RNA virus vector for expressing and distributing foreign antigens, contributing to disease prevention in humans (7). In this study, we first successfully built a reverse genetic system for the BPIV3c strain, SX-2021. To construct the reverse genetic system, we first developed a minigenome system (MG) to test the utility of the helper plasmids. Furthermore, the MG system is a powerful tool commonly utilized to study the life cycle of members of the Paramyxoviridae family. Moreover, the ratio of the helper plasmids has been shown to influence the efficiency of MG reporter gene expression (18). However, the MG system, being shorter than the full-length genome, has inherent limitations that need to be acknowledged. Therefore, rescuing a new recombinant virus has provided significant advantages for further studying the function of viral genes. To rescue the recombinant virus, we constructed a stable full-length cDNA clone carrying a genetic marker. This clone was then co-transfected with the infection clone into BHK-21 cells along with the helper plasmids. The rescued rBPIV3-SX and parental BPIV3 exhibited identical characteristics in the Western blot and immunofluorescence assays. To determine the genetic marker stability of the rescued rBPIV3-SX, we employed restriction enzyme analysis and Sanger sequencing.
A pivotal application of the reverse genetic system involves utilizing the viral backbone for expressing foreign genes (18). In a previous study with the BN-1 strain, a recombinant virus containing the eGFP reporter gene was constructed and inserted between the N and P genes (8). However, the reporter virus was only used to examine the tropism and pathogenesis of BPIV3 in hamsters. Previous studies on other paramyxoviruses have indicated that the expression of foreign proteins, with varying insertion sizes, could impact virus replication (19–21). In this study, we inserted reporter protein gene sequences between the P and M proteins in BPIV3, immediately fused with gene-start (GS) and gene-end (GE) sequences (14, 22). Upon comparing the replication of rBPIV3-SX-EGFP with wild-type parental BPIV3, our results demonstrated that rBPIV3-SX-EGFP exhibited similar growth kinetics to the parental virus. Additionally, the utilization of virus expression reporter genes serves as a powerful tool for screening and testing antiviral drugs (23–25). Ribavirin is a purine nucleoside derivative with broad-spectrum antiviral activity. In this study, we employed rBPIV3-SX-EGFP for drug cytotoxicity and viral infection tests to evaluate its potential in discovering novel antiviral drugs and conducting high-throughput drug screening for anti-BPIV3 drugs. These findings suggest that the rBPIV3-SX-EGFP holds promise for discovering novel antiviral drugs and conducting high-throughput drug screening for anti-BPIV3 drugs.
In summary, our success in developing both a minigenome system and a full-length genome for an emerging BPIV3c represents a significant achievement. Additionally, we successfully rescued a recombinant virus containing an eGFP reporter gene. This tool will be instrumental in conducting tests for antiviral drugs. We believe that the minigenome system, full-length genome, and the recombinant virus with an eGFP reporter gene can collectively contribute to further investigations into the biology, tissue tropism, and transmission dynamics of BPIV3.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary material.
YH: Writing – original draft. KL: Investigation, Writing – original draft. RZ: Software, Writing – original draft. XW: Validation, Writing – original draft. HG: Resources, Writing – original draft. LT: Investigation, Writing – original draft. XLW: Resources, Writing – original draft. SX: Data curation, Writing – original draft. HL: Formal analysis, Writing – original draft. ZY: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1336663/full#supplementary-material
1. Mahony, TJ , McCarthy, FM , Gravel, JL , West, L , and Young, PL . Construction and manipulation of an infectious clone of the bovine herpesvirus 1 genome maintained as a bacterial artificial chromosome. J Virol. (2002) 76:6660–8. doi: 10.1128/jvi.76.13.6660-6668
2. Horwood, PF , Gravel, JL , and Mahony, TJ . Identification of two distinct bovine parainfluenza virus type 3 genotypes. J Gen Virol. (2008) 89:1643–8. doi: 10.1099/vir.0.2008/000026-0
3. Konishi, M , Ohkura, T , Shimizu, M , Akiyama, M , Kameyama, K , and Takeuchi, K . Complete genome sequence of the first isolate of genotype C bovine parainfluenza virus type 3 in Japan. Genome Announc. (2014) 2:e01215–4. doi: 10.1128/genomeA.01215-14
4. Oem, JK , Lee, EY , Lee, KK , Kim, SH , Lee, MH , and Hyun, BH . Molecular characterization of a Korean bovine parainfluenza virus type 3 isolate. Vet Microbiol. (2013) 162:224–7. doi: 10.1016/j.vetmic.2012.10.013
5. Neill, JD , Ridpath, JF , and Valayudhan, BT . Identification and genome characterization of genotype B and genotype C bovine parainfluenza type 3 viruses isolated in the United States. BMC Vet Res. (2015) 11:112. doi: 10.1186/s12917-015-0431-8
6. Zhu, YM , Shi, HF , Gao, YR , Xin, JQ , Liu, NH , Xiang, WH, et al. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in China. Vet Microbiol. (2011) 149:446–51. doi: 10.1016/j.vetmic.2010.11.011
7. Haller, AA , Miller, T , Mitiku, M , and Coelingh, K . Expression of the surface glycoproteins of human parainfluenza virus type 3 by bovine parainfluenza virus type 3, a novel attenuated virus vaccine vector. J Virol. (2000) 74:11626–35. doi: 10.1128/jvi.74.24.11626-11635
8. Ohkura, T , Minakuchi, M , Sagai, M , Kokuho, T , Konishi, M , Kameyama, KI, et al. Infection of the upper respiratory tract of hamsters by the bovine parainfluenza virus type 3 BN-1 strain expressing enhanced green fluorescent protein. Virology. (2015) 476:134–40. doi: 10.1016/j.virol.2014.12.015
9. Minet, C , Yami, M , Egzabhier, B , Gil, P , Tangy, F , Brémont, M, et al. Sequence analysis of the large (L) polymerase gene and trailer of the peste des petits ruminants virus vaccine strain Nigeria 75/1: expression and use of the L protein in reverse genetics. Virus Res. (2009) 145:9–17. doi: 10.1016/j.virusres.2009.06.002
10. Iwasaki, M , Takeda, M , Shirogane, Y , Nakatsu, Y , Nakamura, T , and Yanagi, Y . The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol. (2009) 83:10374–83. doi: 10.1128/JVI.01056-09
11. Blondot, ML , Dubosclard, V , Fix, J , Lassoued, S , Aumont-Nicaise, M , Bontems, F, et al. Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein. PLoS Pathog. (2012) 8:e1002734. doi: 10.1371/journal.ppat.1002734
12. Graci, JD , and Cameron, CE . Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. (2006) 16:37–48. doi: 10.1002/rmv.483
13. Han, Y , Adam, FEA , Zhang, R , Gao, Y , Lei, J , Lu, K, et al. Isolation and identification of a genotype C bovine parainfluenza virus type 3 and its pathogenicity in albino Guinea pigs. Transbound Emerg Dis. (2023) 2023:1–10. doi: 10.1155/2023/8854528
14. Carnero, E , Li, W , Borderia, AV , Moltedo, B , Moran, T , and García-Sastre, A . Optimization of human immunodeficiency virus gag expression by Newcastle disease virus vectors for the induction of potent immune responses. J Virol. (2009) 83:584–97. doi: 10.1128/JVI.01443-08
15. Autio, T , Pohjanvirta, T , Holopainen, R , Rikula, U , Pentikäinen, J , Huovilainen, A, et al. Etiology of respiratory disease in non-vaccinated, non-medicated calves in rearing herds. Vet Microbiol. (2007) 119:256–65. doi: 10.1016/j.vetmic.2006.10.001
16. Snowder, GD , Van Vleck, LD , Cundiff, LV , Bennett, GL , Koohmaraie, M , and Dikeman, ME . Bovine respiratory disease in feedlot cattle: phenotypic, environmental, and genetic correlations with growth, carcass, and longissimus muscle palatability traits. J Anim Sci. (2007) 85:1885–92. doi: 10.2527/jas.2007-0008
17. Kumagai, A , Kanno, T , Kawauchi, K , Tanaka, K , Ishihara, R , and Hatama, S . Phylogenetic and antigenic analysis of bovine parainfluenza virus type 3 isolated in Japan between 2002 and 2019. Vet Microbiol. (2020) 247:108774. doi: 10.1016/j.vetmic.2020.108774
18. Su, J , Dou, Y , You, Y , and Cai, X . Application of minigenome technology in virology research of the Paramyxoviridae family. J Microbiol Immunol Infect. (2015) 48:123–9. doi: 10.1016/j.jmii.2014.02.008
19. Liang, B , Ngwuta, JO , Herbert, R , Swerczek, J , Dorward, DW , Amaro-Carambot, E, et al. Packaging and Prefusion stabilization separately and additively increase the quantity and quality of respiratory syncytial virus (RSV)-neutralizing antibodies induced by an RSV fusion protein expressed by a parainfluenza virus vector. J Virol. (2016) 90:10022–38. doi: 10.1128/JVI.01196-16
20. Liu, F , Li, L , Liu, Y , Sun, C , Liu, C , Wu, X, et al. Development of reverse genetics system for small ruminant morbillivirus: rescuing recombinant virus to express Echinococcus granulosus EG95 antigen. Virus Res. (2019) 261:50–5. doi: 10.1016/j.virusres.2018.12.008
21. Liu, F , Zhang, Y , Li, L , Zuo, Y , Sun, C , Xiaodong, W, et al. Rescue of eGFP-expressing small ruminant morbillivirus for identifying susceptibilities of eight mammalian cell lines to its infection. Virus Res. (2019) 261:60–4. doi: 10.1016/j.virusres.2018.12.011
22. Chen, C , Zhou, M , Yan, XG , Chen, YX , Cui, M , Chen, HC, et al. A recombinant canine distemper virus expressing interleukin-7 enhances humoral immunity. J Gen Virol. (2019) 100:602–15. doi: 10.1099/jgv.0.001247
23. Liu, F , Wang, Q , and Shan, H . Rescue of dual reporter-tagged parainfluenza virus 5 as tool for rapid screening of antivirals in vitro. Vet Microbiol. (2021) 259:109154. doi: 10.1016/j.vetmic.2021.109154
24. Ren, T , Zhou, L , Min, X , Sui, M , Zhi, X , Mo, Y, et al. Development of a recombinant reporter Getah virus for antiviral drug screening assays. Vet Microbiol. (2023) 281:109742. doi: 10.1016/j.vetmic.2023.109742
Keywords: bovine parainfluenza virus, reverse genetics system, virus rescue, antiviral drugs, ribavirin
Citation: Han Y, Lu K, Zhang R, Wei X, Guo H, Tong L, Wang X, Xiao S, Liu H and Yang Z (2024) Construction and characterization of a reverse genetics system of bovine parainfluenza virus type 3c as a tool for rapid screening of antivirals in vitro. Front. Vet. Sci. 11:1336663. doi: 10.3389/fvets.2024.1336663
Received: 11 November 2023; Accepted: 26 February 2024;
Published: 12 March 2024.
Edited by:
Kaori Sano, National Institute of Infectious Diseases (NIID), JapanReviewed by:
Kang Ouyang, Guangxi University, ChinaCopyright © 2024 Han, Lu, Zhang, Wei, Guo, Tong, Wang, Xiao, Liu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haijin Liu, bGl1aGFpamluQG53YWZ1LmVkdS5jbg==; Zengqi Yang, eXpxODE2MkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.