
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 23 February 2024
Sec. Veterinary Infectious Diseases
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1325072
This article is part of the Research TopicInsights in Emerging or Re-emerging ZoonosesView all 14 articles
 Ana Abad-Fau1
Ana Abad-Fau1 Eloisa Sevilla1,2*
Eloisa Sevilla1,2* Ainara Oro1
Ainara Oro1 Inmaculada Martín-Burriel3
Inmaculada Martín-Burriel3 Bernardino Moreno1,2
Bernardino Moreno1,2 Mariano Morales1,4
Mariano Morales1,4 Rosa Bolea1,2
Rosa Bolea1,2Escherichia coli (E. coli) is a pathogen frequently isolated in cases of urinary tract infections (UTIs) in both humans and dogs and evidence exists that dogs are reservoirs for human infections. In addition, E. coli is associated to increasing antimicrobial resistance rates. This study focuses on the analysis of antimicrobial resistance and the presence of selected virulence genes in E. coli isolates from a Spanish dog population suffering from UTI. This collection of isolates showed an extremely high level of phenotypic resistance to 1st–3rd generation cephalosporins, followed by penicillins, fluoroquinolones and amphenicols. Apart from that, 13.46% of them were considered extended-spectrum beta-lactamase producers. An alarmingly high percentage (71.15%) of multidrug resistant isolates were also detected. There was a good correlation between the antimicrobial resistance genes found and the phenotypic resistance expressed. Most of the isolates were classified as extraintestinal pathogenic E. coli, and two others harbored virulence factors related to diarrheagenic pathotypes. A significant relationship between low antibiotic resistance and high virulence factor carriage was found, but the mechanisms behind it are still poorly understood. The detection of high antimicrobial resistance rates to first-choice treatments highlights the need of constant antimicrobial resistance surveillance, as well as continuous revision of therapeutic guidelines for canine UTI to adapt them to changes in antimicrobial resistance patterns.
Urinary tract infections are one the most common causes of primary care veterinary supervision in dogs and a treatment challenge due to their high recurrence and therapeutic implications. Escherichia coli is the most common bacterium isolated in UTIs in dogs and humans (1–3). In addition, E. coli bacteremia in humans (the most common cause of bacteremia in high-income countries) is caused by urinary tract infections in more than 50% of cases (4). Escherichia coli has also been associated with an increase in antimicrobial resistance (2, 5). Evidence suggests that dogs act as a reservoir of human infections with uropathogenic E. coli (UPEC) and are a source of spread of antimicrobial resistance (6).
Escherichia coli is classified into various pathotypes based on the presence of virulence factors. Uropathogenic E. coli is included within the group of extraintestinal pathogenic E. coli (ExPEC) and are characterized by specific virulence factor. Some of these virulence factors include P-fimbriae (papC), α-haemolysin (hlyA) and cytotoxic necrotizing factor type 1 (cnf1) (7). Eae, the gene that codifies for intimin and is associated with diarrheic strains, can also be found in uropathogenic strains (8). Other relevant E. coli virulent factors include Shiga toxins (Stx), also known as verotoxins and characteristic of Shiga toxin-producing E. coli (STEC), which are related to hemorrhagic diarrhea and hemolytic uremic syndrome, and have been described in several species, although natural infections are rarely described in dogs (9, 10). There is a potential zoonotic risk associated with the presence of these genetic elements in companion animals and other species (11, 12). Hybrid strains have gained recent attention, especially those that harbor several virulence factors traditionally associated with different pathotypes. These new types of strains are considered “heteropathogen” or hybrid, such as STEC/UPEC strains (2), and are considered as able to produce both outcomes, diarrhea, or UTI (13).
Virulence genes are encoded by plasmids, bacteriophages, or pathogenicity islands (PAI). Pathogenicity islands are mobile and unstable fragments of DNA present in pathogenic strains, but absent in the related non-pathogenic strains, which can be shared by horizontal transmission. PapC, hlyA and cnf1, among other virulence genes, are usually encoded simultaneously within PAIs in UPEC (14, 15). P-fimbriae, encoded by papC gene, plays an important role in kidney adherence and the inflammatory response (16). α-Haemolysin is a toxin known to produce renal injuries, and even though the mechanism is still unclear, cnf1 does not play a major role in the severity of the disease but it is usually associated with other virulence genes (15).
It has been previously demonstrated that some canine UPEC isolates are clonal with those isolated from humans, suggesting their zoonotic potential. It has also been proposed that dogs could act as a reservoir of this E. coli pathotype, hence the importance of the study of the potential implications of UTI in this animal species (17, 18).
In the last few decades there has been a rising concern about the increase in the number of E. coli isolates presenting a multidrug resistant (MDR) profile (19, 20). It has been described that the ownership of companion animals could be a risk factor in the spread of pathogenic E. coli strains between humans and pets, also favoring the dissemination of antimicrobial resistance in the community (21–23).
It is common to find antibiotic resistance in E. coli isolates from cases of UTI, which highlights the importance of monitoring the strain susceptibility to the antibiotic treatment, even if an experimental treatment has already been implemented. In fact, UPEC strains isolated from dogs have been described as MDR reservoirs in several countries (24, 25) and as carriers of extended-spectrum beta-lactamase (ESBL) genes (5). ESBL-producing E. coli have been previously found in cats and dogs, and human-dog co-carriage in the same household has also been demonstrated in fecal samples (26–28). In general, ESBL and the presence of other antibiotic resistance mechanisms can difficult the treatment of infectious diseases and, therefore, result in complicated chronic infections.
Although microbiological culture and susceptibility testing are recommended before any antimicrobial therapy is established, empiric treatment is frequently established and the most common recommendations to treat these infections in companion animals include amoxicillin (without clavulanic acid) and trimethoprim-sulfonamides as a first approach (29). Therefore, updated information about antimicrobial susceptibility patterns is highly needed.
The aim of this study was to determine the presence of E. coli in urine samples from a Spanish dog population presenting clinical signs of urinary tract infections and to characterize the isolates according to selected virulence factors and their antimicrobial resistance pattern, a research field scarcely investigated in Spain.
This study was conducted on a total of 52 E. coli isolates. This collection of isolates came from urine samples from dogs diagnosed with UTI. Samples were aseptically collected by cystocentesis as part of the daily activity of private veterinary practitioners in Zaragoza, Spain. The criteria followed to diagnose UTI were those used in everyday clinic, which include frequent urination, pain during urination, fever or vomiting, among others. The sampling period ranged from 2017 to 2019, and all urine samples were taken before any treatment was established. Mean age of the individuals was 8.97 years old (95% CI: 4.11–13.83%). In regard to the gender of these individuals, 21 of them were male and 31 were female.
Isolates were identified using VITEK® (bioMérieux, France) and those confirmed as E. coli were then stored at −20°C for further analysis.
DNA was extracted by boiling 3–5 colonies from pure cultures and then conventional PCR for the detection of virulence-related genes was performed. These genes included eae (intimin), Stx1 and Stx2 (Shiga toxins 1 and 2), papC (P-fimbriae), hlyA (α-haemolysin) and cnf1 (cytotoxic necrotizing factor type 1). Primers used in this study were those described in Table 1. PCR was performed in a BiometraTRIO 48 thermocycler (Analytik Jena, Germany), and PCR products were analyzed under UV light in 1.5% agarose gels stained with GelGreen® (Biotium, United States).
CECT 4783 strain was used as positive control for eae, Stx1 and Stx2 genes; C136b strain was the positive control for hlyA and cnf1 genes, kindly provided by Dr. J. A. Orden, University Complutense of Madrid, Spain. A canine strain previously isolated by our research group (Pe8 strain, GenBank accession number MK034302) was used as positive control for papC gene.
Escherichia coli isolates were classified in pathotypes according to the presence of the virulence factor genes analyzed. Enterohemorrhagic E. coli (EHEC) are described as those E. coli strains that harbor both intimin and Shiga toxins (12). When only one of these virulence factors was present, isolates were classified as enteropathogenic E. coli (EPEC) or STEC, respectively. If any of the other virulence factor genes analyzed were found, that is hlyA, PapC and/or cnf1, isolates were classified as extraintestinal pathogenic E. coli (ExPEC) (34).
Susceptibility to 74 different antimicrobials was determined using VITEK® (bioMérieux, France). The antimicrobial agents selected to test each isolate susceptibility depended on VITEK® guidelines, as clinically relevant antimicrobials recommended by VITEK® varied during the period in which the study was performed. Antimicrobials were classified in 12 categories: aminoglycosides, amphenicols, carbapenems, fluoroquinolones, nitrofurans, other β-lactams, penicillins, tetracyclines, sulfonamides, 1st–2nd generation cephalosporins, 3rd generation cephalosporins and 4th–5th generation cephalosporins, as shown in Table 2. All isolates were tested against at least one antibiotic of each category, except for 4th and 5th generation cephalosporins, which were added in the middle of the study. For those isolates having no information regarding 4th–5th generation cephalosporins, neither susceptibility nor resistance was included, and they were thus excluded from the prevalence analysis for that group. Resistance to a category of antimicrobials was defined as resistance to at least one of the agents in that category. MDR isolates were defined as those isolates with non-susceptibility to three or more antimicrobial categories (35).
Additionally, VITEK 2 ESBL test (bioMérieux) was used in these isolates for rapid detection of extended-spectrum β-lactamase (ESBL) production, which is based on simultaneous assessment of the inhibitory effects of cefepime, cefotaxime, and ceftazidime, alone and in the presence of clavulanate.
Breakpoints for the interpretation of minimal inhibitory concentration (MIC) results were applied according to the criteria stablished by bioMérieux for small animals (AST-GN97, bioMérieux, France), which include natural resistance and breakpoints from the Clinical and Laboratory Standards Institute (36).
Intermediate resistance category provides a flexible information in clinical practice. However, E. coli isolates have been previously found to harbor resistance genes (37) For this reason, when they had to be categorized into dichotomic variants they were assessed as resistant.
Those isolates showing the highest rate of phenotypic resistance, that is resistance to six or more antimicrobial categories, were selected for further characterization through whole genome sequencing (WGS). A total of ten E. coli isolates were cultured for 24 h in Nutrient Agar (Oxoid, United Kingdom) and DNA was then extracted using Wizard Genomic DNA Purification Kit (Promega, Madison, WI, United States). Quality parameters for DNA were checked both on Qubit 4 (Invitrogen) and gel electrophoresis. Genome sequencing was performed on an Illumina Miseq platform with a paired-end read length of 150 bp. Sequences were trimmed on Galaxy (Version 0.3.8.1) and assembled with Unicycler (Galaxy version 0.5.0 + Galaxy 1). All sequencing data have been submitted to NCBI Genome Database under BioProject PRJNA1031085, and individual accession numbers are the following: SAMN37924970 (isolate 258.883), SAMN37926527 (isolate 262.947), SAMN37926528 (isolate 263.715), SAMN37926529 (isolate 266.493), SAMN37926530 (isolate 267.252), SAMN37926531 (isolate 269.901), SAMN37926532 (isolate 271.550), SAMN37926533 (isolate 271.758), SAMN37926534 (isolate 271.811) and SAMN37926535 (isolate 271.960).
Antibiotic resistance genes, virulence factors, serotypes and sequence types (ST) were assigned to these sequenced genomes using tools that included ResFinder 4.1 (38–40), PathogenFinder 1.1 (41), VirulenceFinder 2.0 (39, 42), MLST 2.0 (E. coli #1 and #2) (39, 43–48), cgMLSTFinder 1.2 (42, 49), MGE v1.0.3 (39, 50–52) and SeroTypeFinder 2.0 (53). Visualization of the genomic data was carried out using Prok(see 54). A phylogenetic tree was created with Roary pipeline (55) based on Prokka annotation (56), and followed by use of IQ-TREE software (57).
Prevalence was calculated with 95% confidence intervals (CI). To test simple relationship between virulence factors and antibiotics, Fisher ‘s Exact Test was used, and the p-values determined, considering them statistically significant when value of p ≤0.05. Numeric values were calculated using Pearson’s coefficient. Isolates showing intermediate antibiotic resistance were considered as resistant for statistical comparisons. All the analyses and calculations were performed using R version 4.1.1 and RCommander 2.7–1.
According to the virulence factor analysis performed, the prevalence of the virulence-related genes was as follows: 1.92% for eae (95% CI: 0–5.66%), 1.92% for Stx2 (95% CI: 0–5.66%), 59.62% for papC (95% CI: 46.28–72.95%), 53.85% for hlyA (95% CI: 40.30–67.4%) and 32.69% for cnf1 (95% CI: 19.97–45.44%). However, Stx1 gene was not found in this study.
Regarding E. coli pathotype classification, 82.69% (95% CI: 79–87%) of isolates were classified as ExPEC, and around 20% (9/43) of them simultaneously harbored the three extraintestinal virulence factors analyzed. Additionally, 1.92% (95% CI: 0–5.66%) of isolates were defined as EPEC, and the same value was found for STEC. However, no EHEC isolates were detected. None of the virulence factors analyzed in this study were found in 13.46% (95%CI: 3–23%) of the isolates.
According to the antimicrobial resistance profiles observed, only one out of 52 (95% CI: 0–5.66%) E. coli isolates was susceptible to all the antimicrobials tested. Also, all the antimicrobial categories presented resistant isolates, although in a variable percentage.
According to antimicrobial resistance levels defined by the European Food Safety Authority (58), an extremely high resistance level was found for the categories of 1st–2nd and 3rd generation cephalosporins, followed by very high resistance to penicillins and fluoroquinolones. These isolates also displayed a high resistance level to amphenicols (Figure 1). A low resistance level was found in 5 out of the 12 categories: carbapenems, nitrofurans, other β-lactams, 4th–5th generation cephalosporins and aminoglycosides.
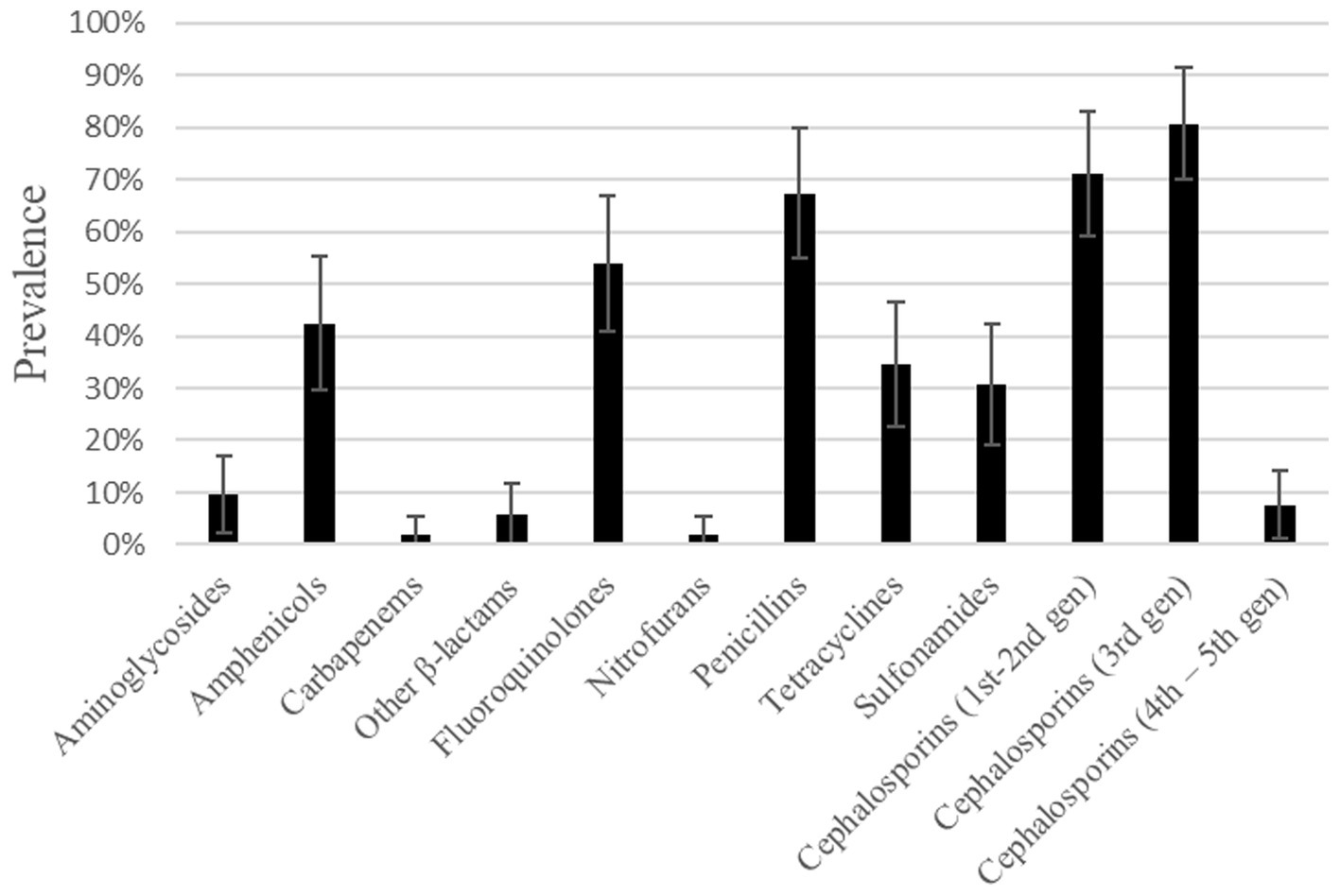
Figure 1. Prevalence of resistance to different antimicrobial categories found in E. coli isolates from dog urine.
Several isolates showed resistance to antibiotics which are considered critically important antimicrobials and are listed in category A (59). For example, three (out of 30) isolates were resistant to the β-lactam aztreonam, and there were several others found resistant to category A antibiotics from the penicillin group: three (out of 17) isolates were resistant to carbenicillin, 18 (out of 31) to ticarcillin, one (out of 33) to piperacillin and three (out of three) to mezlocillin. There was also one isolate showing intermediate resistance, and thus classified as resistant, to an agent from the carbapenem category (imipenem).
Apart from that, 13.46% (95% CI: 4.17–22.73%) of the isolates were considered ESBL-producers, and almost 60% (4/7) of them showed resistance to 9 or more out of the 12 antibiotic categories tested.
A total of 71.15% (95% CI: 58.84–83.46%) of the studied isolates were described as MDR.
Two main profiles of MDR, with a prevalence of 7.69% (95% CI: 0.45–14.93%) each of them, were observed. The isolates included in one of these profiles showed resistance to the following antimicrobial categories: penicillins, 1st–2nd and 3rd generation cephalosporins; while the other profile comprised those isolates resistant to amphenicols, fluoroquinolones, penicillins, and 1st–2nd and 3rd generation cephalosporins.
In silico molecular typing was performed in the sequenced genomes from those selected phenotypically resistant isolates (Table 3). Three different nomenclatures for sequence typing were assigned to each isolate according to Achtman’s MLST scheme, Pasteur MLST scheme and core genome (cg)-MLST.
Three of these isolates, that is 258.883, 262.947 and 263.715, shared the same serotype (O5H20), and the corresponding sequence type (Pasteur ST 901 / Achtman ST 6448) and core genome sequence type (cg-ST 174146), making this E. coli type the most prevalent one among the studied isolates. The rest of the isolates presented unique molecular types, although isolates 271.758 and 269.901 belonged to the same clonal complex (CC ST23) and were paired together in the phylogenetic tree (Figure 2). Annotated comparison of the isolates (Figure 3) showed no major missing regions.
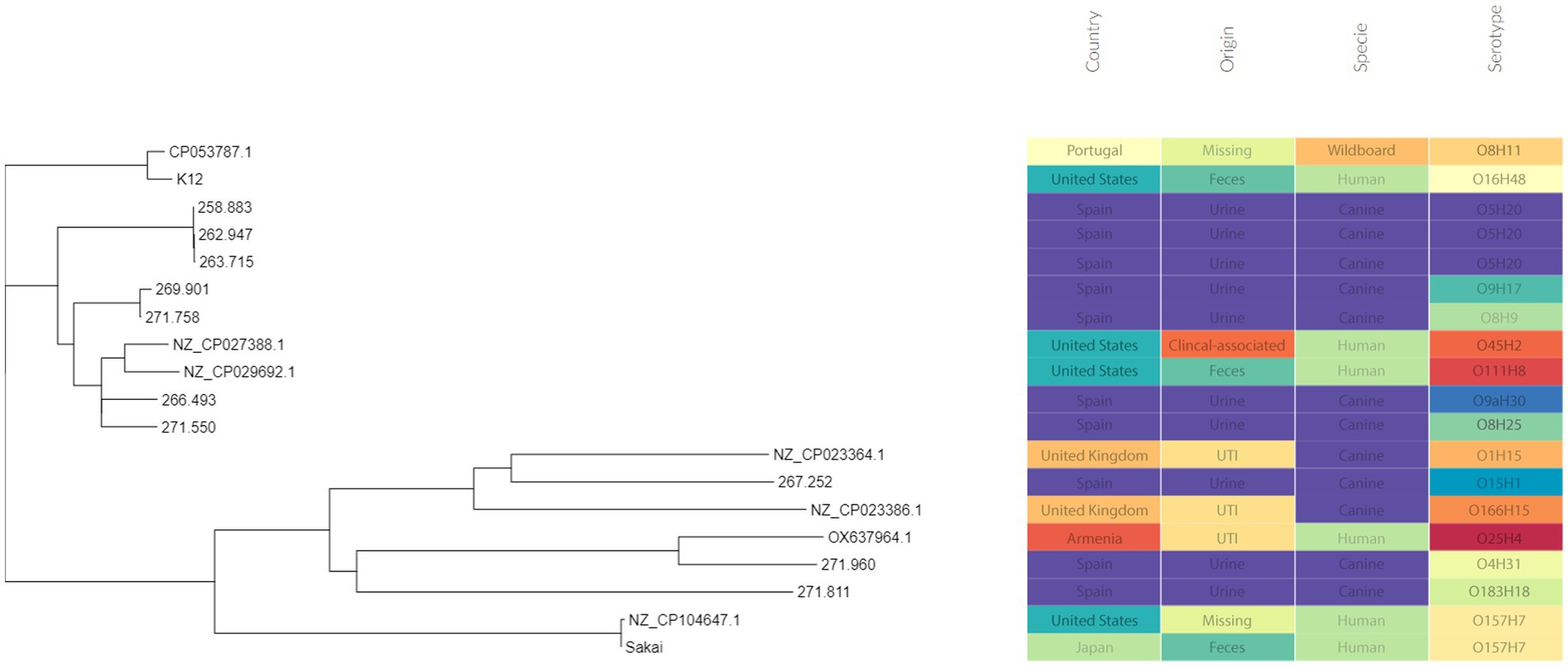
Figure 2. Phylogenetic tree including the sequenced E. coli isolates. Metadata was added using Phandango web application (60).
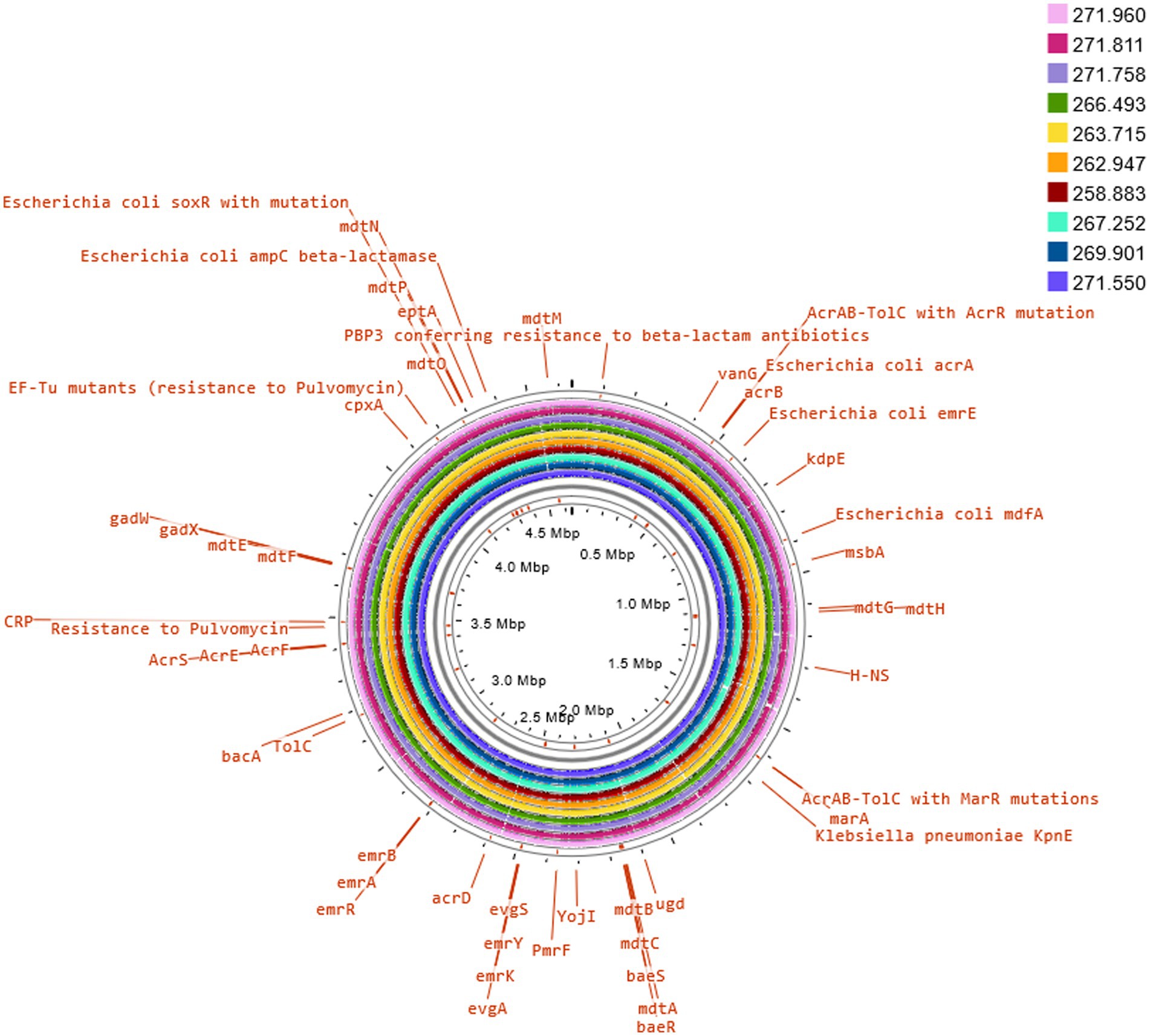
Figure 3. Comparison of E. coli K12 reference genome with sequenced isolates. The annotation of selected antimicrobial resistance genes was carried out on Proksee Server from the Stothard Research Group (University of Alberta, Canada) that uses BLAST analysis to illustrate conserved and missing genomic sequences (available online: https://proksee.ca/).
Several genes and mutations associated with resistance to different antimicrobial categories were detected in the sequenced isolates and are detailed in Table 4. In addition, some of these genes were associated to mobile genetic elements (MGE), which are described in Table 5.
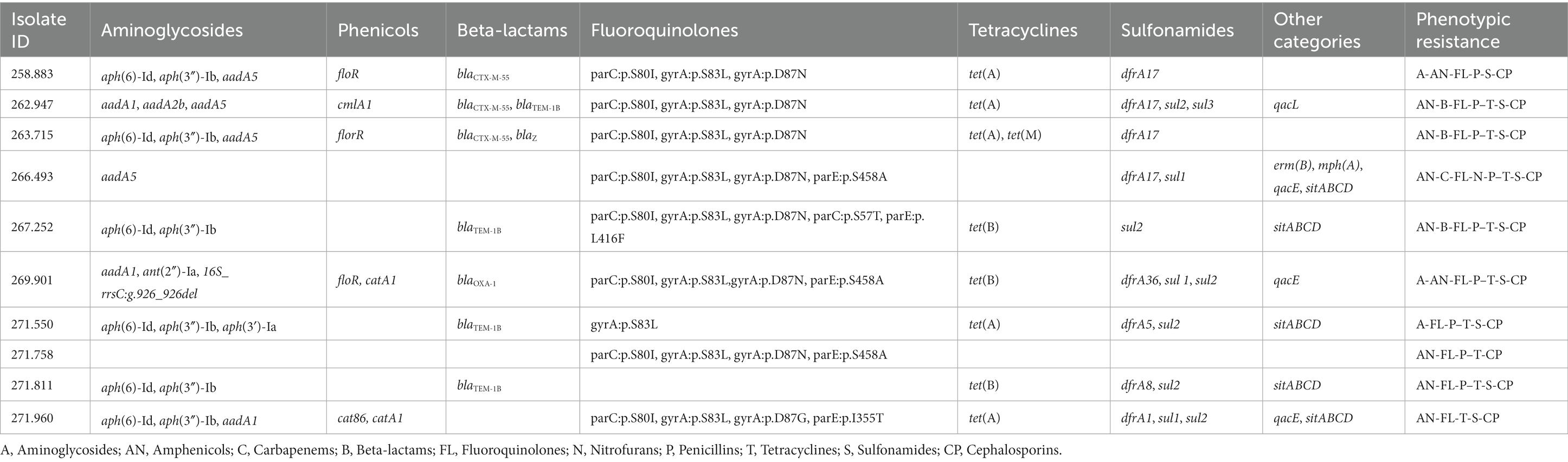
Table 4. Antimicrobial resistance genes and mutations found in the sequenced genomes of the selected E. coli isolates, as well as the corresponding phenotypic resistance pattern showed in the susceptibility testing assay.
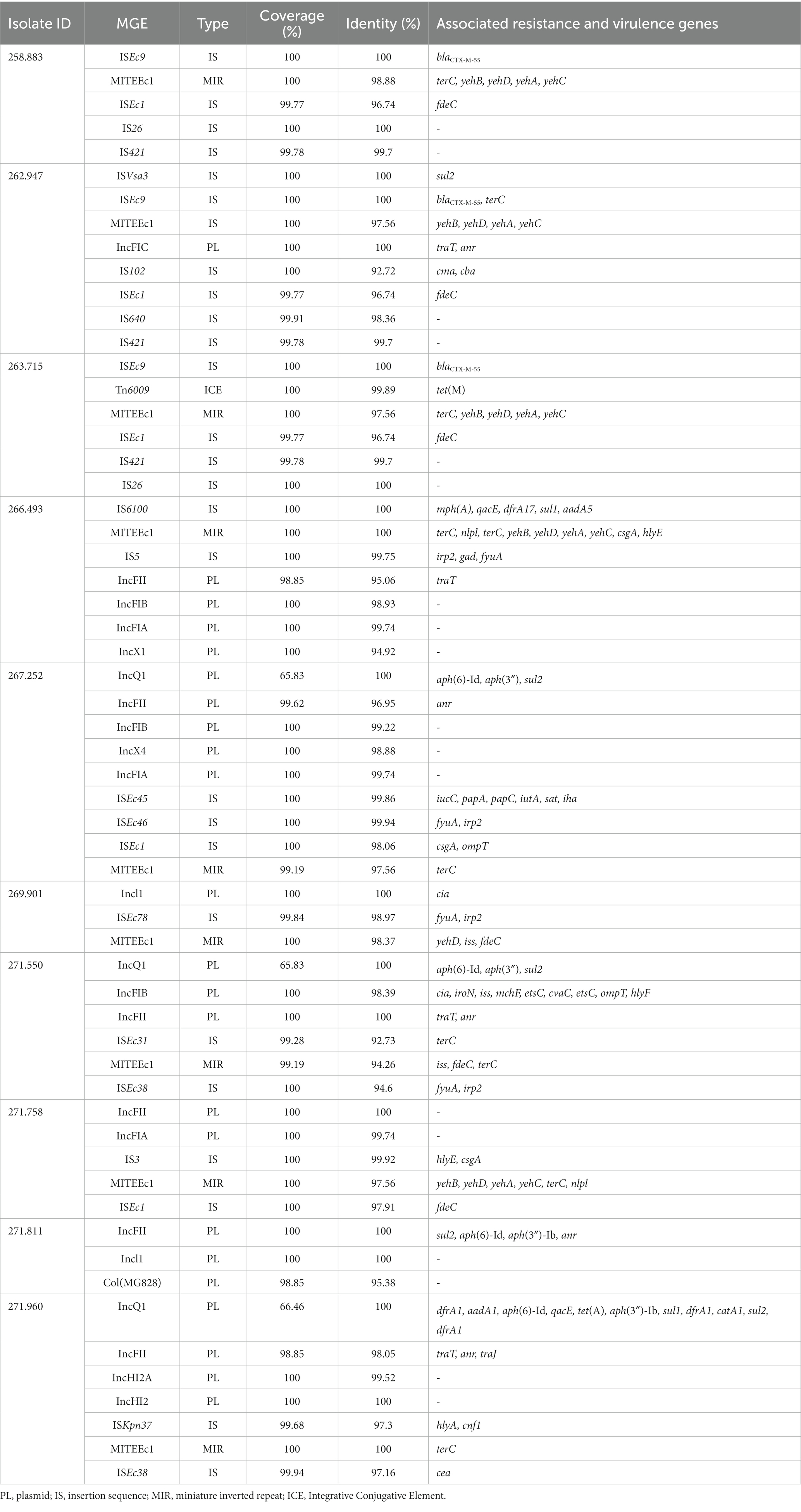
Table 5. Presence of mobile genetic elements (MGE) in sequenced isolates, and antibiotic resistance and associated virulence genes.
Interestingly, there were two MGE of particular interest due to its association with important resistance genes or a high number of them, that is IS6100 and ISEc9, which can be seen in Figure 4.
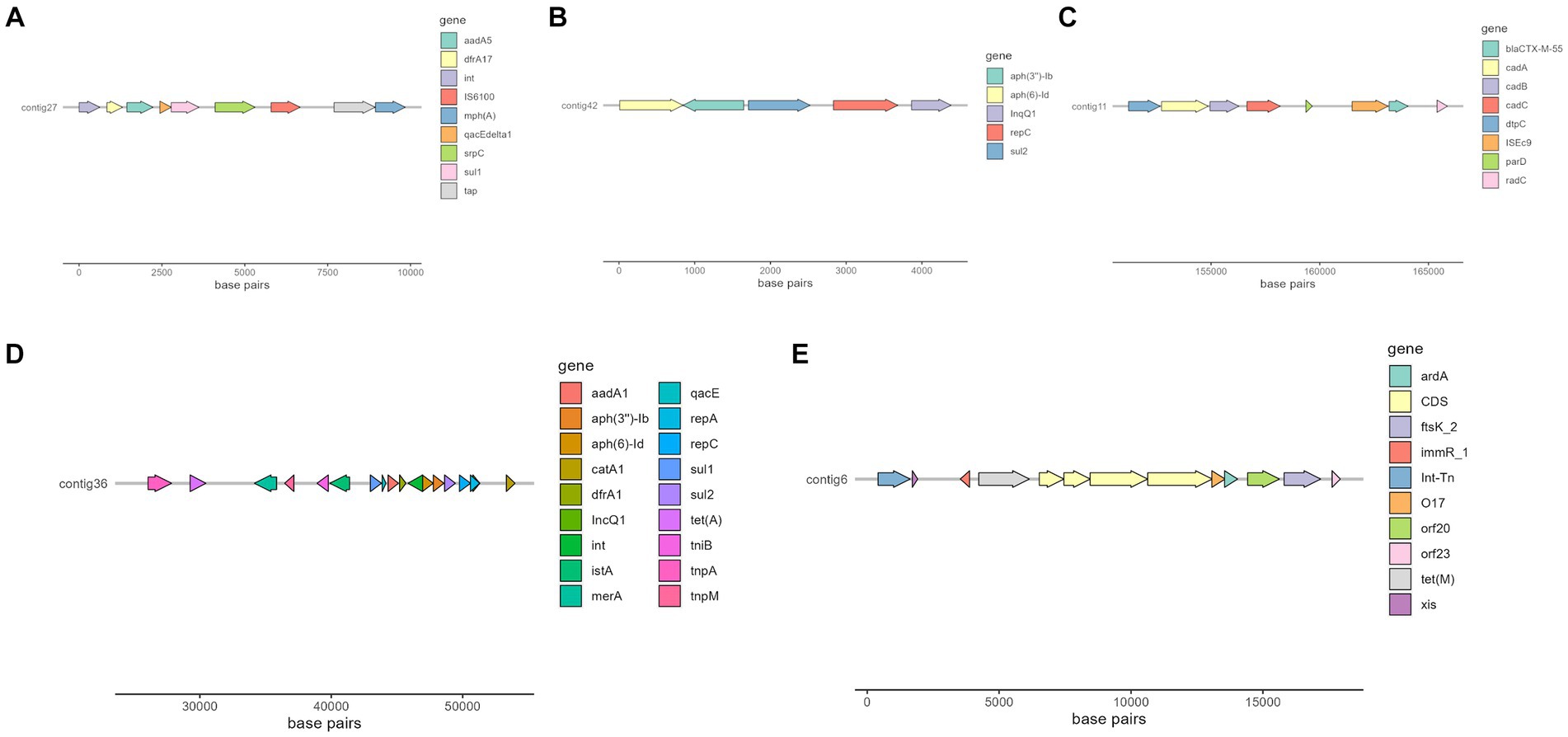
Figure 4. Selected contigs of different isolates containing antimicrobial resistance genes as well as MGE annotated with Prokka (56). (A) Organization of a fragment of contig 27 in isolate 266.493, which contains aadA5, dfrA17, qacE, sul1, and mphA genes. (B) Organization of contig 42 in isolate 269.901, which contains aph(6)-Id, aph(3″)-Ib and sul2 genes. (C) Organization of a fragment of contig 11 in isolate 258.883, which contains blaCTX-M-55 gene. (D) Organization of a fragment of contig 36 in isolate 271.960, which contains several resistance genes, such as aadA1, sul2 and tet(A). (E) Organization of a fragment of isolate 263.715, which contains tet(M) resistance gene.
When testing for simple relationships between phenotypic antimicrobial resistance and presence of virulence factors in these isolates, the carriage of cnf1 gene showed a significant association with resistance to several penicillins, as well as with the penicillin category itself (Value of p = 0.01). Cnf1 gene also showed a significant association (Value of p = 0.019) with MDR category. Apart from that, age of individuals was significantly associated with E. coli isolates showing resistance to various cephalosporins and to the 4th–5th generation cephalosporin category (Value of p = 0.034). Gender was associated with aminoglycoside resistant isolates (Value of p = 0.007).
ESBL production showed association with resistance to five out of the 12 antimicrobial categories tested (amphenicols, other β-lactams, 4th–5th generation cephalosporins, sulfonamides and fluoroquinolones). No significant relationship between the number of virulence-related genes and ESBL production was observed, however a negative relationship between the number of antimicrobial categories to which isolates showed resistance and number of virulence-related genes was found (Pearson coefficient = 0.33 value of – = 0.014).
This study has evaluated both phenotypic and genotypic antimicrobial resistance as well as the presence of selected virulence genes in E. coli isolates from Spanish dogs with UTI and has shown that dogs may be reservoirs of resistant uropathogenic strains of E. coli.
Comparing with results obtained in a previous study (61) in which samples were collected in a similar time and geographical location, although they had a different origin (feces), we found that E. coli isolates from urine were more susceptible to aminoglycosides than those obtained from dog feces (phenotypic resistance found in 9.62% vs. 40% of isolates). In general, the prevalence of antimicrobial resistances found are similar to those found in E. coli isolated from UTI patients (62, 63), except for the penicillin group, which was slightly higher in this study (67.31% vs. ~ 45–50%). Such high prevalence is also contrary to the decreasing trend of penicillin resistance in E. coli isolates in Europe (20). Some hypotheses for this phenomenon could be the trend of increase of antibiotic resistance year to year or the fact that more antibiotics from the penicillin group were studied in this work.
Escherichia coli aminoglycoside resistance was the only type of resistance linked to the gender of the animal, being found exclusively in male individuals. In fact, male gender has previously been associated with aminoglycoside resistance in Gram-negative bacteria (64).
When taking into account some of the antibiotics considered clinically important for human and animal health by the European Medicines Agency (59), it is worth mentioning that E. coli isolates showing resistance to several of these antibiotics were detected in this study, even to antibiotics from category A (“Avoid,” it includes antibiotics not authorized in veterinary medicine in the European Union), such as certain penicillins or carbapenems. Another relevant antibiotic category is category B (“Restrict”), which includes those listed as highest priority critically important antimicrobials (HP-CIAs) by the World Health Organization categorization, e.g., 3rd generation cephalosporins or fluoroquinolones. Indeed, as much as 80.77 and 53.85% of these isolates were considered resistant to 3rd generation cephalosporins and fluoroquinolones, respectively. The high amount of overall resistance found among all the categories could be biased by the fact that complicated UTI are more often requested for culture and antibiogram testing than simpler cases of UTI.
The treatment with amoxicillin or trimethoprim-sulfonamides as first-line agents is currently recommended for the management of bacterial UTI in dogs (29). However, considering these results, it seems that the use of these antimicrobials may be ineffective in a high percentage of cases, since 58.33% of isolates were found to be resistant to amoxicillin and 30.77% to trimethoprim-sulfonamides. Before suggesting any change in current guidelines for antibiotic treatment in canine UTI, it should be noted that E. coli is not the only pathogen responsible for UTI and that the data analyzed in this study might be overestimating the baseline resistance, mostly because of the selection of the patients. In any case, the use of antibiotic resistance testing as a routine allows not only the monitoring of the epidemiology of antibiotic resistance profiles but also the faster implementation of a treatment in case of failure of the empiric one.
It is worth mentioning the high percentage of MDR isolates found (71.15%). Among the MDR isolates found, more than 80% (30/37) were classified as ExPEC, and one of them corresponded to EPEC pathotype. This kind of strains possesses a potential zoonotic risk and can also serve as a reservoir of resistance genes (18), further contributing to the dissemination of antibiotic resistance and limiting the options for the treatment of infectious diseases in both humans and animals.
High antibiotic resistance has been associated with MGE in Enterobacteriaceae. Bacteria harboring these mobile elements can become a reservoir for antibiotic resistances and be transmitted then from pets to their owners or the environment. This phenomenon poses a serious health problem due to the spread of resistance and failure of current antibiotic treatments (65).
Of special interest is the presence of transposon Tn6009 carrying tet(M) gene in isolate 263.715. This element has been previously described in other Gram-negative bacteria, such as Enterococcus fecalis (66), and is associated with tetracycline resistance due to the presence of tet(M) (Figure 4E). This non-composite conjugative transposon is of clinical importance in Gram-positive bacteria and has a potential role in the dissemination of resistance (67, 68). Resistance to beta-lactams was encoded by blaCTX–M–55 gene in three isolates, which was located in a ISEc9 insertion sequence (IS1380-like). ISEc9 region has been previously associated with ESBL genes (69) and has been described in other bacteria such as Vibrio vulnificus (70). In all cases, the gene and the insertion sequence were 46 bp away. One of them (isolate 263.715) also harbored a blaZ gene, and another one (isolate 263.715) had a copy of blaTEM-1B. Beta-lactamase-encoding genes such as blaTEM-1B and blaOXA-1 were also identified in other isolates. For example, blaTEM-1B was present in two isolates that displayed no phenotypic resistance to beta-lactams (isolates 271.550 and 271.811), and in one that did (isolate 267.252). There was only one isolate (269.901) containing a blaOXA-1 gene. As expected, this isolate was resistant to several antibiotics in the penicillin group (ampicillin, amoxicillin + clavulanic acid, amoxicillin), and it was negative in the ESBL production test BlaOXA-1 has been found in ST131 or associated with other genes in plasmids (71). Despite this gene being originally described in MGE (72), we only identified IncI1 plasmid in this isolate, and it was not associated with any antibiotic resistance gene. Sulfonamide resistance genes (sul 1 or 2) were found in most of the sequenced genomes (7/10), and in five of them these genes were located in a MGE (IS6100 and IncQ1 for sul1, and ISVsa3, IncQ1 or IncFII for sul2). All these MGE also harbored other resistance genes, including those linked to streptomycin (aadA5, aph(6)-Id, aph(3″)-Ib and aadA1 in IS6100, IncFII and IncQ1) (73–76), trimethoprim (dfrA17 and dfrA1 in IS6100 and IncQ1) (77, 78), antiseptics (qacL and qacE in IS6100 and IncQ1, although all antiseptic resistance genes were incomplete) (79), erythromycin (mph(A) in IS6100) (80), doxycycline and tetracycline (tet(A) in IncQ1) (81) and chloramphenicol (catA1 in IncQ1) (82).
The ISVsa3 transposase was found in one of the sequenced isolates (262.947) and contained the sul2 gene, which has also been identified in other enteropathogens (83–85). Isolate 266.493 harbored dfrA17-aadA5-qacEdelta1-IS6100-mph(A)-sul1 integron structure, which is commonly identified in ExPEC pathotype (86).
Regarding the plasmids identified, IncFII plasmid has been previously documented in Spain as frequently linked to ESBL production (87). In this study, the plasmid was only identified in one isolate, although it was classified as non-ESBL producer.
Another plasmid identified was IncQ1, commonly found in E. coli and with ability to transfer between different bacterial species and strains, which facilitates the dissemination of antibiotic resistance in bacterial populations (88). This plasmid was detected in the genomes of three isolates (267.252, 271.550 and 271.960) and was found close to resistance genes linked to aminoglycoside resistance (aph(6)-Id and aph(3″)-Ib). All these three isolates were distant in the phylogenetic tree, which suggests that the plasmid has been likely acquired independently. One of these isolates harbored nine more resistance genes close to the detected plasmid (Figure 4D), indicating a potential hotspot for antibiotic resistance dissemination.
When studying antibiotic resistance genes in sequenced isolates, there was in general a consistent correlation between phenotypic and genetic resistance. However, there were two significant exceptions to this pattern. When examining aminoglycosides, several isolates exhibited susceptibility to this category despite carrying resistance genes related to both streptomycin and spectinomycin, which are included in this antimicrobial category. The second exception was observed with tetracycline, where the relationship between resistance genes and phenotypical resistance did not consistently align. This discrepancy in isolate 258.883 may be attributed to a nucleotide substitution at position 924 within the tet(A) gene (position 3,323, GenBank: AF534183.1), specifically transitioning from cytosine (C) to thymine (T). Because of this alteration, there is a shift in the protein composition from alanine (Ala) at position 118 to threonine (Thr). Although these are not the first E. coli isolates that harbor this gene mutation (89), to the authors knowledge our study is the first that associates this mutation in tet(A) to a failure in phenotypic response. Also, in more than 40% (4/9) of the isolates considered phenotypically resistant to amphenicols, no resistance gene associated with this antimicrobial category was found.
The phylogenetic tree (Figure 3) showed that canine isolates clustered together, except isolate 271.960 (ST 372), which was grouped with a human isolate (assembly reference OX637964.1) that belongs to ST131. ST 131 is one of the predominant sequence types within the ExPEC pathotype worldwide (71, 90). In fact, E. coli O25b:H4/ST131 was described as a prevalent clone in Spanish human population. In accordance with bibliography, this canine isolate was not associated with ESBL resistance and had a similar resistance profile to human strains (91, 92).
According to the virulence factors analyzed, most of the E. coli isolates found in urine samples were categorized in the ExPEC pathotype, as expected. The most frequently detected virulence factors were papC and hlyA, followed by cnf1. Cnf1 prevalence in these isolates was similar to that found in isolates from both dogs and humans, while papC and hlyA prevalence were higher in this study (18, 93–97). However, most of the sampled populations in these studies include healthy animals, which could lower the prevalence of E. coli virulence factors. The prevalence of these three virulence factors were higher in dog isolates than in those found in humans (95, 98–100). Some of these factors were found in MGE (Table 5), which highlights their potential of spread to other strains.
Almost 20% of these ExPEC isolates displayed in combination with the three extraintestinal virulence-related genes analyzed (papC, hlyA and cnf1), likely due to the presence of a PAI (101). This type of virulence factors are frequently found in E. coli strains causing extraintestinal disease in both humans and dogs, being thus this animal species a possible reservoir for the ExPEC pathotype (102).
Apart from that there was one Stx2-positive isolate, that did not harbor any other virulence factor studied. Shiga toxin 2 is believed to be associated with the development of HUS (11) and is better produced when it is found in combination with other strains or bacteria (103). However, there are also some descriptions of Shiga toxin-producing E. coli isolates associated with UTI cases, and it has been proposed that Shiga toxins can bind to receptors from urinary bladder epithelial cells and damage them (104, 105). Additionally, an EPEC isolate was also detected. It is not the first time that an eae-positive isolate has been found among UTI-associated strains, although its frequency seems to be quite low as well (8, 106, 107). The role of this gene product (i.e., intimin) in UTI pathogenesis is not fully understood and its significance remains to be studied (8).
Taking into consideration that fecal E. coli population might have a relationship with UTI pathogenesis (108), it may be suggested that certain diarrheagenic pathotypes also have potential to cause UTI, although uncommon. However, the role of these strains in UTI development and the molecular and pathogenic causes behind it are still poorly understood, and more research in this field is needed in order to comprehend the mechanisms and epidemiological causes. Nevertheless, the ability of such strains to cause an extraintestinal infection in the host is not only dependent on their virulence-related genes but also on risk factors such as age or immunosuppression (106).
It is also important to note that a wide variety of extraintestinal-associated virulence traits has been described in the literature. Thus, apart from these virulence genes typical of diarrheagenic strains, these two isolates might be also harboring some other extraintestinal virulence factors different from those analyzed in this study. In this regard, some E. coli strains have been recently classified as hybrids for harboring virulence factors usually associated with various pathotypes, e.g., STEC/UPEC strains (109). The genome plasticity of this microorganism promotes the exchange and combination of both intestinal and extraintestinal virulence determinants, resulting in an heteropathogenic potential (106, 107). The possible emergence of hybrid pathotypes not only in humans but also in animals should therefore be surveilled.
The finding of eight isolates (15.38%) considered neither intestinal nor extraintestinal pathogenic isolates could be explained by the fact that only a selection of virulence factors was tested. Thus, these E. coli might harbor other different virulence-related genes not analyzed in this study. However, another explanation could be that the causal agent for UTI in these dogs was different from E. coli, or even a non-infectious cause. It is also worth noting that the detection of virulence factor genes does not mean that they are phenotypically expressed, so the severity of the disease could not be only assessed with this information. Nevertheless, it is known that the severity of the disease is not caused by a single virulence factor but a combination of them (110, 111).
The most commonly isolated serotype in this study was O5H20. In this regard, O5:H(−) has been associated with STEC strains, and Shiga toxins have been also described in E. coli strains causing UTI (112, 113). However, these isolates did not harbor any Shiga toxin gene. The rest of E. coli serotypes are distributed along different STs and antibiotic resistance patterns, showing a heterogenic distribution.
Interestingly, low antibiotic resistance patterns were linked to a higher number of virulence factors. There is some literature (114, 115) that suggests a positive relationship between virulence factors and MDR. However, in isolates from this study only cnf1 carriage showed a significant association with MDR, while a high virulence factor carriage was associated with low resistance profiles. The reason for this mechanism is still unclear, but it is hypothesized that the acquisition of MDR is “sacrificed” in exchange for virulence factors, or that the low presence of virulence factors facilitates the acquisition of antibiotic resistance (116, 117). When analyzing correlation between all virulence factors found and the presence of antibiotic resistance genes in whole genome sequenced isolates, the relationship was non-significant (p = 0.14). However, there was a bias in selection of isolates, as only the more resistant ones were chosen.
Based on these data, a very high percentage of E. coli isolates found in urine samples from dogs suffering from UTI was considered MDR, the majority of them being classified as ExPEC. Phenotypic antimicrobial resistance to first-lines agents recommended in UTI management was also frequently observed, which could be associated with a treatment failure. Furthermore, several antimicrobial resistance genes, some of them contained in MGE, were identified in the genome of selected resistant isolates. The use of WGS could identify some of the genetic mechanisms underlying antimicrobial resistance, although there were a few discordances between phenotypic resistance and genes found. Combining both phenotypic and genetic data enhances our understanding of antibiotic resistance and improves treatment selection efficiency.
Overall, these findings are of concern for both animal and public health, since dogs could act as reservoirs of MDR pathogenic E. coli and contribute to the spread of antimicrobial resistance. Surveillance of antimicrobial resistance and revision of therapeutic guidelines should be therefore continuously addressed in clinical veterinary settings.
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/bioproject; PRJNA1031085.
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because samples were collected as part of the daily activity of private veterinary practitioners. Written informed consent was obtained from the owners for the participation of their animals in this study.
AA-F: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. ES: Data curation, Formal analysis, Supervision, Writing – original draft, Writing – review & editing. AO: Investigation, Writing – original draft. IM-B: Formal analysis, Supervision, Writing – review & editing. BM: Resources, Supervision, Writing – review & editing. MM: Conceptualization, Resources, Supervision. RB: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the project Microbiologia Veterinaria (No. 2019/1180, OTRI UNIZAR).
The authors would like to thank the staff of Albéitar Laboratories S. C. (Zaragoza, Spain) for their excellent technical assistance and providing some samples for the study. In addition, we would like to acknowledge the Microbiology Department of the Veterinary Faculty of Zaragoza, in which some techniques were performed.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Flores-Mireles, AL, Walker, JN, Caparon, M, and Hultgren, SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. (2015) 13:269–84. doi: 10.1038/nrmicro3432
2. Hall, JL, Holmes, MA, and Baines, SJ. Prevalence and antimicrobial resistance of canine urinary tract pathogens. Vet Rec. (2013) 173:549. doi: 10.1136/vr.101482
3. Teh, H. A review of the current concepts in canine urinary tract infections. Aust Vet J. (2022) 100:56–62. doi: 10.1111/avj.13127
4. Bonten, M, Johnson, JR, van den Biggelaar, AHJ, Georgalis, L, Geurtsen, J, de Palacios, PI, et al. Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin Infect Dis. (2021) 72:1211–9. doi: 10.1093/cid/ciaa210
5. Yousefi, A, and Torkan, S. Uropathogenic Escherichia coli in the urine samples of Iranian dogs: antimicrobial resistance pattern and distribution of antibiotic resistance genes. Biomed Res Int. (2017) 2017:1–10. doi: 10.1155/2017/4180490
6. Jacob, J, and Lorber, B. Diseases transmitted by Man's best friend: the dog. Microbiol Spectr. (2015) 3:15. doi: 10.1128/microbiolspec.IOL5-0002-2015
7. Landraud, L, Gibert, M, Popoff, MR, Boquet, P, and Gauthier, M. Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol Microbiol. (2003) 47:1653–67. doi: 10.1046/j.1365-2958.2003.03391.x
8. Abe, CM, Salvador, FA, Falsetti, IN, Vieira, MA, Blanco, J, Blanco, JE, et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol. (2008) 52:397–406. doi: 10.1111/j.1574-695X.2008.00388.x
9. de Brito, BG, Leite, DS, Linhares, RE, and Vidotto, MC. Virulence-associated factors of uropathogenic Escherichia coli strains isolated from pigs. Vet Microbiol. (1999) 65:123–32. doi: 10.1016/S0378-1135(98)00277-6
10. Dell'Orco, M, Bertazzolo, W, Pagliaro, L, Roccabianca, P, and Comazzi, S. Hemolytic-uremic syndrome in a dog. Vet Clin Pathol. (2005) 34:264–9. doi: 10.1111/j.1939-165X.2005.tb00052.x
11. Persson, S, Olsen, KE, Ethelberg, S, and Scheutz, F. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol. (2007) 45:2020–4. doi: 10.1128/JCM.02591-06
12. García, A, and Fox, JG. A one health perspective for defining and deciphering Escherichia coli pathogenic potential in multiple hosts. Comp Med. (2021) 71:3–45. doi: 10.30802/AALAS-CM-20-000054
13. Gati, NS, Temme, IJ, Middendorf-Bauchart, B, Kehl, A, Dobrindt, U, and Mellmann, A. Comparative phenotypic characterization of hybrid Shiga toxin-producing / uropathogenic Escherichia coli, canonical uropathogenic and Shiga toxin-producing Escherichia coli. Int J Med Microbiol. (2021) 311:151533. doi: 10.1016/j.ijmm.2021.151533
14. Lloyd, AL, Rasko, DA, and Mobley, HL. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. (2007) 189:3532–46. doi: 10.1128/JB.01744-06
15. Martin, HR, Taylor, NS, Buckley, EM, Marini, RP, Patterson, MM, and Fox, JG. Characterization of cytotoxic necrotizing factor 1-producing Escherichia coli strains from faeces of healthy macaques. J Med Microbiol. (2009) 58:1354–8. doi: 10.1099/jmm.0.012088-0
16. Lane, MC, and Mobley, HL. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. (2007) 72:19–25. doi: 10.1038/sj.ki.5002230
17. Johnson, JR, O'Bryan, TT, Low, DA, Ling, G, Delavari, P, Fasching, C, et al. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun. (2000) 68:3327–36. doi: 10.1128/IAI.68.6.3327-3336.2000
18. Johnson, JR, Stell, AL, and Delavari, P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect Immun. (2001) 69:1306–14. doi: 10.1128/IAI.69.3.1306-1314.2001
19. Silwedel, C, Vogel, U, Claus, H, Glaser, K, Speer, CP, and Wirbelauer, J. Outbreak of multidrug-resistant Escherichia coli sequence type 131 in a neonatal intensive care unit: efficient active surveillance prevented fatal outcome. J Hosp Infect. (2016) 93:181–6. doi: 10.1016/j.jhin.2016.02.014
20. ECDC. Antimicrobial resistance in the EU/EEA (EARS-net) - annual epidemiological report 2019: Stockholm: ECDC. (2020). Available at: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2019.
21. Ukah, UV, Glass, M, Avery, B, Daignault, D, Mulvey, MR, Reid-Smith, RJ, et al. Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiol Infect. (2018) 146:46–57. doi: 10.1017/S0950268817002680
22. Köck, R, Daniels-Haardt, I, Becker, K, Mellmann, A, Friedrich, AW, Mevius, D, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. (2018) 24:1241–50. doi: 10.1016/j.cmi.2018.04.004
23. Zechner, V, Sofka, D, Paulsen, P, and Hilbert, F. Antimicrobial resistance in Escherichia coli and resistance genes in Coliphages from a small animal clinic and in a patient dog with chronic urinary tract infection. Antibiotics (Basel). (2020) 9:652. doi: 10.3390/antibiotics9100652
24. Rzewuska, M, Czopowicz, M, Kizerwetter-Świda, M, Chrobak, D, Błaszczak, B, and Binek, M. Multidrug resistance in Escherichia coli strains isolated from infections in dogs and cats in Poland (2007-2013). ScientificWorldJournal. (2015) 2015:408205:1–8. doi: 10.1155/2015/408205
25. Yudhanto, S, Hung, CC, Maddox, CW, and Varga, C. Antimicrobial resistance in Bacteria isolated from canine urine samples submitted to a veterinary diagnostic laboratory, Illinois, United States. Front Vet Sci. (2022) 9:867784. doi: 10.3389/fvets.2022.867784
26. Costa, D, Poeta, P, Briñas, L, Sáenz, Y, Rodrigues, J, and Torres, C. Detection of CTX-M-1 and TEM-52 beta-lactamases in Escherichia coli strains from healthy pets in Portugal. J Antimicrob Chemother. (2004) 54:960–1. doi: 10.1093/jac/dkh444
27. O'Keefe, A, Hutton, TA, Schifferli, DM, and Rankin, SC. First detection of CTX-M and SHV extended-spectrum beta-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob Agents Chemother. (2010) 54:3489–92. doi: 10.1128/AAC.01701-09
28. van den Bunt, G, Fluit, AC, Spaninks, MP, Timmerman, AJ, Geurts, Y, Kant, A, et al. Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J Antimicrob Chemother. (2020) 75:342–50. doi: 10.1093/jac/dkz462
29. Weese, JS, Blondeau, J, Boothe, D, Guardabassi, LG, Gumley, N, Papich, M, et al. International society for companion animal infectious diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet J. (2019) 247:8–25. doi: 10.1016/j.tvjl.2019.02.008
30. Oswald, E, Schmidt, H, Morabito, S, Karch, H, Marchès, O, and Caprioli, A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. (2000) 68:64–71. doi: 10.1128/IAI.68.1.64-71.2000
31. Oh, KH, Kim, SB, Park, MS, and Cho, SH. Development of a one-step PCR assay with nine primer pairs for the detection of five diarrheagenic Escherichia coli types. J Microbiol Biotechnol. (2014) 24:862–8. doi: 10.4014/jmb.1312.12031
32. Blanco, M, Blanco, JE, Rodríguez, E, Abalia, I, Alonso, MP, and Blanco, J. Detection of virulence genes in uropathogenic Escherichia coli by polymerase chain reaction (PCR): comparison with results obtained using phenotypic methods. J Microbiol Methods. (1997) 31:37–43. doi: 10.1016/S0167-7012(97)00087-0
33. Yamamoto, S, Terai, A, Yuri, K, Kurazono, H, Takeda, Y, and Yoshida, O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. (1995) 12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x
34. Osugui, L, de Castro, AF, Iovine, R, Irino, K, and Carvalho, VM. Virulence genotypes, antibiotic resistance and the phylogenetic background of extraintestinal pathogenic Escherichia coli isolated from urinary tract infections of dogs and cats in Brazil. Vet Microbiol. (2014) 171:242–7. doi: 10.1016/j.vetmic.2014.03.027
35. Magiorakos, AP, Srinivasan, A, Carey, RB, Carmeli, Y, Falagas, ME, Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
36. CLSI. M100 performance standards for antimicrobial susceptibility testing a CLSI supplement for global application. Berwyn, PA: CLSI (2018).
37. Zhang, X, Zhang, Y, Wang, F, Wang, C, Chen, L, Liu, H, et al. Unravelling mechanisms of nitrofurantoin resistance and epidemiological characteristics among Escherichia coli clinical isolates. Int J Antimicrob Agents. (2018) 52:226–32. doi: 10.1016/j.ijantimicag.2018.04.021
38. Bortolaia, V, Kaas, RS, Ruppe, E, Roberts, MC, Schwarz, S, Cattoir, V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
39. Camacho, C, Coulouris, G, Avagyan, V, Ma, N, Papadopoulos, J, Bealer, K, et al. BLAST+: architecture and applications. BMC Bioinformatics. (2009) 10:421. doi: 10.1186/1471-2105-10-421
40. Zankari, E, Allesøe, R, Joensen, KG, Cavaco, LM, Lund, O, and Aarestrup, FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. (2017) 72:2764–8. doi: 10.1093/jac/dkx217
41. Cosentino, S, Voldby Larsen, M, Møller Aarestrup, F, and Lund, O. PathogenFinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS One. (2013) 8:e77302. doi: 10.1371/journal.pone.0077302
42. Clausen, PTLC, Aarestrup, FM, and Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. (2018) 19:307. doi: 10.1186/s12859-018-2336-6
43. Bartual, SG, Seifert, H, Hippler, C, Luzon, MA, Wisplinghoff, H, and Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. (2005) 43:4382–90. doi: 10.1128/JCM.43.9.4382-4390.2005
44. Griffiths, D, Fawley, W, Kachrimanidou, M, Bowden, R, Crook, DW, Fung, R, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. (2010) 48:770–8. doi: 10.1128/JCM.01796-09
45. Jaureguy, F, Landraud, L, Passet, V, Diancourt, L, Frapy, E, Guigon, G, et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics. (2008) 9:560. doi: 10.1186/1471-2164-9-560
46. Larsen, MV, Cosentino, S, Rasmussen, S, Friis, C, Hasman, H, Marvig, RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. (2012) 50:1355–61. doi: 10.1128/JCM.06094-11
47. Lemee, L, Dhalluin, A, Pestel-Caron, M, Lemeland, JF, and Pons, JL. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J Clin Microbiol. (2004) 42:2609–17. doi: 10.1128/JCM.42.6.2609-2617.2004
48. Wirth, T, Falush, D, Lan, R, Colles, F, Mensa, P, Wieler, LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. (2006) 60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x
49. Alikhan, NF, Zhou, Z, Sergeant, MJ, and Achtman, M. A genomic overview of the population structure of Salmonella. PLoS Genet. (2018) 14:e1007261. doi: 10.1371/journal.pgen.1007261
50. Carattoli, A, Zankari, E, García-Fernández, A, Voldby Larsen, M, Lund, O, Villa, L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
51. Joensen, KG, Scheutz, F, Lund, O, Hasman, H, Kaas, RS, Nielsen, EM, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. (2014) 52:1501–10. doi: 10.1128/JCM.03617-13
52. Zankari, E, Hasman, H, Cosentino, S, Vestergaard, M, Rasmussen, S, Lund, O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. (2012) 67:2640–4. doi: 10.1093/jac/dks261
53. Joensen, KG, Tetzschner, AM, Iguchi, A, Aarestrup, FM, and Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. (2015) 53:2410–26. doi: 10.1128/JCM.00008-15
54. Grant, JR, Enns, E, Marinier, E, Mandal, A, Herman, EK, Chen, CY, et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. (2023) 51:W484–92. doi: 10.1093/nar/gkad326
55. Page, AJ, Cummins, CA, Hunt, M, Wong, VK, Reuter, S, Holden, MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. (2015) 31:3691–3. doi: 10.1093/bioinformatics/btv421
56. Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
57. Nguyen, LT, Schmidt, HA, von Haeseler, A, and Minh, BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
58. EFSA, ECDC. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. (2020) 18:e06007. doi: 10.2903/j.efsa.2020.6007
59. EMA, CVMP, CHMP. Categorisation of antibiotics in the European Union. Report No.: Supplement_1. (2019). Available at: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf.
60. Hadfield, J, Croucher, NJ, Goater, RJ, Abudahab, K, Aanensen, DM, Harris, SR, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinfor. (2018) 34:292–293. doi: 10.1093/bioinformatics/btx610
61. Sevilla, E, Mainar-Jaime, RC, Moreno, B, Martín-Burriel, I, Morales, M, Andrés-Lasheras, S, et al. Antimicrobial resistance among canine enteric Escherichia coli isolates and prevalence of attaching-effacing and extraintestinal pathogenic virulence factors in Spain. Acta Vet Hung. (2020) 68:1–7. doi: 10.1556/004.2020.00013
62. Chang, SK, Lo, DY, Wei, HW, and Kuo, HC. Antimicrobial resistance of Escherichia coli isolates from canine urinary tract infections. J Vet Med Sci. (2015) 77:59–65. doi: 10.1292/jvms.13-0281
63. Yu, Z, Wang, Y, Chen, Y, Huang, M, Wang, Y, Shen, Z, et al. Antimicrobial resistance of bacterial pathogens isolated from canine urinary tract infections. Vet Microbiol. (2020) 241:108540. doi: 10.1016/j.vetmic.2019.108540
64. Richter, SE, Miller, L, Needleman, J, Uslan, DZ, Bell, D, Watson, K, et al. Risk factors for development of aminoglycoside resistance among gram-negative rods. Am J Health Syst Pharm. (2019) 76:1838–47. doi: 10.1093/ajhp/zxz201
65. Partridge, SR, Kwong, SM, Firth, N, and Jensen, SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. (2018) 31:17. doi: 10.1128/CMR.00088-17
66. Tóth, AG, Csabai, I, Judge, MF, Maróti, G, Becsei, Á, Spisák, S, et al. Mobile antimicrobial resistance genes in probiotics. Antibiotics (Basel). (2021) 10:287. doi: 10.3390/antibiotics10111287
67. Rice, LB. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother. (1998) 42:1871–7. doi: 10.1128/AAC.42.8.1871
68. Soge, OO, Beck, NK, White, TM, No, DB, and Roberts, MC. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J Antimicrob Chemother. (2008) 62:674–80. doi: 10.1093/jac/dkn255
69. Medugu, N, Tickler, IA, Duru, C, Egah, R, James, AO, Odili, V, et al. Phenotypic and molecular characterization of beta-lactam resistant multidrug-resistant Enterobacterales isolated from patients attending six hospitals in northern Nigeria. Sci Rep. (2023) 13:10306. doi: 10.1038/s41598-023-37621-z
70. Nakayama, T, Yamaguchi, T, Jinnai, M, Kumeda, Y, and Hase, A. ESBL-producing Vibrio vulnificus and V. alginolyticus harbour a plasmid encoding ISEc9 upstream of blaCTX-M-55 and qnrS2 isolated from imported seafood. Arch Microbiol. (2023) 205:569. doi: 10.1007/s00203-023-03569-x
71. Nicolas-Chanoine, MH, Bertrand, X, and Madec, JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. (2014) 27:543–74. doi: 10.1128/CMR.00125-13
72. Evans, BA, and Amyes, SG. OXA β-lactamases. Clin Microbiol Rev. (2014) 27:241–63. doi: 10.1128/CMR.00117-13
73. Sandvang, D. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob Agents Chemother. (1999) 43:3036–8. doi: 10.1128/AAC.43.12.3036
74. Meyer, R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. (2009) 62:57–70. doi: 10.1016/j.plasmid.2009.05.001
75. Scholz, P, Haring, V, Wittmann-Liebold, B, Ashman, K, Bagdasarian, M, and Scherzinger, E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. (1989) 75:271–88. doi: 10.1016/0378-1119(89)90273-4
76. Hollingshead, S, and Vapnek, D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. (1985) 13:17–30. doi: 10.1016/0147-619X(85)90052-6
77. Sandalli, C, Buruk, CK, Sancaktar, M, and Ozgumus, OB. Prevalence of integrons and a new dfrA17 variant in gram-negative bacilli which cause community-acquired infections. Microbiol Immunol. (2010) 54:164–9. doi: 10.1111/j.1348-0421.2010.00197.x
78. Sáenz, Y, Briñas, L, Domínguez, E, Ruiz, J, Zarazaga, M, Vila, J, et al. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. (2004) 48:3996–4001. doi: 10.1128/AAC.48.10.3996-4001.2004
79. Kazama, H, Hamashima, H, Sasatsu, M, and Arai, T. Characterization of the antiseptic-resistance gene qacE delta 1 isolated from clinical and environmental isolates of Vibrio parahaemolyticus and Vibrio cholerae non-O1. FEMS Microbiol Lett. (1999) 174:379–84.
80. Pawlowski, AC, Stogios, PJ, Koteva, K, Skarina, T, Evdokimova, E, Savchenko, A, et al. The evolution of substrate discrimination in macrolide antibiotic resistance enzymes. Nat Commun. (2018) 9:112. doi: 10.1038/s41467-017-02680-0
81. Roberts, MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. (2005) 245:195–203. doi: 10.1016/j.femsle.2005.02.034
82. Alton, NK, and Vapnek, D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. (1979) 282:864–9. doi: 10.1038/282864a0
83. Lewis, GL, Fenton, RJ, Moriyama, EN, Loy, JD, and Moxley, RA. Association of IS Vsa3 with multidrug resistance in Salmonella enterica isolates from cattle (Bos taurus). Microorganisms. (2023) 11:631. doi: 10.3390/microorganisms11030631
84. García, P, Guerra, B, Bances, M, Mendoza, MC, and Rodicio, MR. IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype. J Antimicrob Chemother. (2011) 66:543–9. doi: 10.1093/jac/dkq481
85. Zhou, S, Zhu, Y, Yan, Y, Wang, W, and Wang, Y. Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res. (2019) 161:610–20. doi: 10.1016/j.watres.2019.06.048
86. Jarocki, VM, Heß, S, Anantanawat, K, Berendonk, TU, and Djordjevic, SP. Multidrug-resistant lineage of Enterotoxigenic Escherichia coli ST182 with serotype O169:H41 in airline waste. Front Microbiol. (2021) 12:731050. doi: 10.3389/fmicb.2021.731050
87. Novais, A, Cantón, R, Moreira, R, Peixe, L, Baquero, F, and Coque, TM. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, −3, and −32) plasmids. Antimicrob Agents Chemother. (2007) 51:796–9. doi: 10.1128/AAC.01070-06
88. Rawlings, DE, and Tietze, E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. (2001) 65:481–96. doi: 10.1128/MMBR.65.4.481-496.2001
89. Vereecke, N, Van Hoorde, S, Sperling, D, Theuns, S, Devriendt, B, and Cox, E. Virotyping and genetic antimicrobial susceptibility testing of porcine ETEC/STEC strains and associated plasmid types. Front Microbiol. (2023) 14:312. doi: 10.3389/fmicb.2023.1139312
90. Bogema, DR, McKinnon, J, Liu, M, Hitchick, N, Miller, N, Venturini, C, et al. Whole-genome analysis of extraintestinal Escherichia coli sequence type 73 from a single hospital over a 2 year period identified different circulating clonal groups. Microb Genom. (2020) 6:255. doi: 10.1099/mgen.0.000255
91. López-Cerero, L, MeM, B, Serrano, L, Liró, J, Cisneros, JM, Rodríguez-Baño, J, et al. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enferm Infecc Microbiol Clin. (2013) 31:385–8. doi: 10.1016/j.eimc.2012.09.005
92. Dautzenberg, MJ, Haverkate, MR, Bonten, MJ, and Bootsma, MC. Epidemic potential of Escherichia coli ST131 and Klebsiella pneumoniae ST258: a systematic review and meta-analysis. BMJ Open. (2016) 6:e009971. doi: 10.1136/bmjopen-2015-009971
93. LeCuyer, TE, Byrne, BA, Daniels, JB, Diaz-Campos, DV, Hammac, GK, Miller, CB, et al. Population structure and antimicrobial resistance of canine Uropathogenic Escherichia coli. J Clin Microbiol. (2018) 56:18. doi: 10.1128/JCM.00788-18
94. Torkan, S, Bahadoranian, M, Khamesipour, F, and Anyanwu, M. Detection of virulence and antimicrobial resistance genes in Escherichia coli isolates from diarrhoiec dogs in Iran. Arch med vet. (2016) 48:181–90. doi: 10.4067/S0301-732X2016000200008
95. Tramuta, C, Robino, P, Nucera, D, Salvarani, S, Banche, G, Malabaila, A, et al. Molecular characterization and antimicrobial resistance of faecal and urinary Escherichia coli isolated from dogs and humans in Italy. Vet Ital. (2014) 50:23–30. doi: 10.12834/VetIt.1304.09
96. Siqueira, AK, Ribeiro, MG, DaS, L, Tiba, MR, Moura, C, Lopes, MD, et al. Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci. (2009) 86:206–10. doi: 10.1016/j.rvsc.2008.07.018
97. Yuri, K, Nakata, K, Katae, H, Tsukamoto, T, and Hasegawa, A. Serotypes and virulence factors of Escherichia coli strains isolated from dogs and cats. J Vet Med Sci. (1999) 61:37–40. doi: 10.1292/jvms.61.37
98. Blum, G, Falbo, V, Caprioli, A, and Hacker, J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. (1995) 126:189–95.
99. Landraud, L, Gauthier, M, Fosse, T, and Boquet, P. Frequency of Escherichia coli strains producing the cytotoxic necrotizing factor (CNF1) in nosocomial urinary tract infections. Lett Appl Microbiol. (2000) 30:213–6. doi: 10.1046/j.1472-765x.2000.00698.x
100. Johnson, JR, Delavari, P, Kuskowski, M, and Stell, AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. (2001) 183:78–88. doi: 10.1086/317656
101. Diard, M, Garry, L, Selva, M, Mosser, T, Denamur, E, and Matic, I. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol. (2010) 192:4885–93. doi: 10.1128/JB.00804-10
102. Johnson, JR, Stell, AL, Delavari, P, Murray, AC, Kuskowski, M, and Gaastra, W. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J Infect Dis. (2001) 183:897–906. doi: 10.1086/319263
103. Xiaoli, L, Figler, HM, Goswami Banerjee, K, Hayes, CS, and Dudley, EG. Non-pathogenic Escherichia coli enhance Stx2a production of E. coli O157:H7 through both bamA-dependent and independent mechanisms. Front Microbiol. (2018) 9:1325. doi: 10.3389/fmicb.2018.01325
104. Toval, F, Schiller, R, Meisen, I, Putze, J, Kouzel, IU, Zhang, W, et al. Characterization of urinary tract infection-associated Shiga toxin-producing Escherichia coli. Infect Immun. (2014) 82:4631–42. doi: 10.1128/IAI.01701-14
105. Beutin, L, Karch, H, Aleksić, S, Spencker, FB, and Rosenbaum, U. Occurrence of verotoxin (Shiga-like toxin) producing Escherichia coli in human urinary tract infection. Infection. (1994) 22:425. doi: 10.1007/BF01715504
106. Toval, F, Köhler, CD, Vogel, U, Wagenlehner, F, Mellmann, A, Fruth, A, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. (2014) 52:407–18. doi: 10.1128/JCM.02069-13
107. Valiatti, TB, Santos, FF, Santos, ACM, Nascimento, JAS, Silva, RM, Carvalho, E, et al. Genetic and virulence characteristics of a hybrid atypical Enteropathogenic and Uropathogenic Escherichia coli (aEPEC/UPEC) strain. Front Cell Infect Microbiol. (2020) 10:492. doi: 10.3389/fcimb.2020.00492
108. Moreno, E, Andreu, A, Pigrau, C, Kuskowski, MA, Johnson, JR, and Prats, G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. (2008) 46:2529–34. doi: 10.1128/JCM.00813-08
109. Gati, NS, Middendorf-Bauchart, B, Bletz, S, Dobrindt, U, and Mellmann, A. Origin and evolution of hybrid Shiga toxin-producing and Uropathogenic Escherichia coli strains of sequence type 141. J Clin Microbiol. (2019) 58:19. doi: 10.1128/JCM.01309-19
110. Brzuszkiewicz, E, Brüggemann, H, Liesegang, H, Emmerth, M, Olschläger, T, Nagy, G, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. (2006) 103:12879–84. doi: 10.1073/pnas.0603038103
111. Dale, AP, and Woodford, N. Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J Infect. (2015) 71:615–26. doi: 10.1016/j.jinf.2015.09.009
112. McLean, C, Bettelheim, KA, Kuzevski, A, Falconer, L, and Djordjevic, SP. Isolation of Escherichia coli O5: H −, possessing genes for Shiga toxin 1, intimin-β and enterohaemolysin, from an intestinal biopsy from an adult case of bloody diarrhoea: evidence for two distinct O5: H − pathotypes. J Med Microbiol. (2005) 54:605–7. doi: 10.1099/jmm.0.45938-0
113. Misra, RN. Serotyping and enterotoxigenicity of Escherichia coli isolated from urinary tract infections. Med J Armed Forces India. (1997) 53:83–6. doi: 10.1016/S0377-1237(17)30671-8
114. Karam, MRA, Habibi, M, and Bouzari, S. Relationships between virulence factors and antimicrobial resistance among Escherichia coli isolated from urinary tract infections and commensal isolates in Tehran, Iran. Osong Public Health Res Perspect. (2018) 9:217–24. doi: 10.24171/j.phrp.2018.9.5.02
115. Shah, C, Baral, R, Bartaula, B, and Shrestha, LB. Virulence factors of uropathogenic Escherichia coli (UPEC) and correlation with antimicrobial resistance. BMC Microbiol. (2019) 19:204. doi: 10.1186/s12866-019-1587-3
116. Wagner, S, Gally, DL, and Argyle, SA. Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet Microbiol. (2014) 169:171–8. doi: 10.1016/j.vetmic.2014.01.003
Keywords: dog, Escherichia coli, multidrug resistance, urinary tract infection, virulence factors
Citation: Abad-Fau A, Sevilla E, Oro A, Martín-Burriel I, Moreno B, Morales M and Bolea R (2024) Multidrug resistance in pathogenic Escherichia coli isolates from urinary tract infections in dogs, Spain. Front. Vet. Sci. 11:1325072. doi: 10.3389/fvets.2024.1325072
Received: 20 October 2023; Accepted: 12 February 2024;
Published: 23 February 2024.
Edited by:
Barbara Moroni, Experimental Zooprophylactic Institute for Piedmont, Liguria and Valle d'Aosta (IZSPLVA), ItalyReviewed by:
Yasmine Hasanine Tartor, Zagazig University, EgyptCopyright © 2024 Abad-Fau, Sevilla, Oro, Martín-Burriel, Moreno, Morales and Bolea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eloisa Sevilla, ZXNldmlsbHJAdW5pemFyLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.