- 1Department of Zoology, Abdul Wali Khan University Mardan, Mardan, Khyber Pakhtunkhwa, Pakistan
- 2Department of Zoology, University of Loralai, Loralai, Balochistan, Pakistan
- 3Institute of Environmental and Occupational Health Sciences, Department of Public Health, College of Public Health, National Taiwan University, Taipei, Taiwan
- 4King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia
- 5Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 6Laboratory of Infectious Diseases, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
Alectorobius species are soft ticks primarily infesting birds, such as swallows, while Dermacentor species are hard ticks mainly infesting mammals, such as small ruminants. This study for the first time reported on the morphological and molecular bases of two tick species, namely A. coniceps and a Dermacentor sp. in Pakistan. The former species was examined in swallows’ nests in Khyber Pakhtunkhwa province, while the latter species was examined in small ruminants in Balochistan province. In total, 25 ticks were collected, with 14 ticks morphologically identified as A. coniceps (males = 9 and females = 5) and 11 ticks identified as Dermacentor sp. (males = 7 and females = 4). Following morphological identification, molecular identification was gained by obtaining 16S rDNA and cox1 sequences for these ticks. The BLAST results for the 16S rDNA and cox1 sequences from A. coniceps shared a maximum identity of 97.46% and 96.49% with the same species from Malta. The BLAST analysis of the 16S rDNA and cox1 sequences from Dermacentor sp. showed maximum identities of 98.42% and 97.45% with Dermacentor pavlovskyi from China. The phylogenetic analysis based on 16S rDNA and cox1 of A. coniceps showed a close evolutionary relationship with the same species. The case of Dermacentor sp., based on 16S DNA and cox1, indicated a close evolutionary relationship with Dermacentor pavlovskyi from China.
Introduction
Ticks are arthropods that fall under Arachnida and are further categorized into three families: hard ticks (Ixodidae), soft ticks (Argasidae), and Nuttalliellidae (1, 2). With medical significance, they are obligate ectoparasites of semi-terrestrial and terrestrial vertebrates (3–5). Usually, hard ticks take a single extended blood meal during each of their life stages, while soft ticks consume multiple brief blood meals during the nymphal and adult stages (6–8).
The genus Ornithodoros is recognized as the most diverse among soft ticks, comprising approximately 130 species (9–11). They have been documented on a wide variety of hosts, including amphibians, birds, mammals, and reptiles (12–16). Previously, this genus was believed to have seven subgenera, including Alectorobius, which encompassed ticks such as Ornithodoros (Alectorobius) coniceps (17, 18). However, Alectorobius was later reclassified as a distinct genus (19), and this reclassification is adopted in this study. These ticks are distributed worldwide primarily infesting birds and occasionally parasitize humans (6, 20). These parasites have been observed infesting pigeons, ruddy shelducks, swallows, swifts, sparrows, and chickens (6, 13, 20, 21). Throughout their lifespan, they may parasitize a single host or multiple hosts (6, 20). However, they exhibit nidicolous behavior, briefly attaching to their hosts and generally residing in their nests (6, 13, 22). Alectorobius coniceps is an ornithophilic species belonging to the mentioned genus, and there are limited morphologically based records of this species from the Oriental region (23).
With approximately 40 species, the Dermacentor is the fourth most diverse genus among hard ticks (24). Approximately half of its species are found in the Palearctic region, with the remaining species distributed across Afrotropical, Nearctic, and Oriental regions (24–26). They are primarily three-host ticks, with a prominent preference for mammals, including wild and domestic, and occasionally humans (24, 26, 27). The adult and nymph stages of these parasites have been observed on a range of larger mammals including pigs, deer, antelope, bison, elk, goats, sheep, cattle, camels, horses, and dogs, whereas their larval stages have been found infesting smaller hosts such as rodents and lagomorphs (24, 27). The subgenus Asiacentor is mainly found in Asia, and Dermacentor pavlovskyi is regarded as the type species for this subgenus (28–31). These ticks have primarily been found infesting small ruminants, such as goats and sheep (29, 31, 32). The known distribution of D. pavlovskyi includes Central Asia and China, which are parts of the Palearctic region (26, 29, 31, 32), and there have been no recorded data of this species in the Oriental region.
Currently, there is a lack of consensus on the systematic classification and taxonomy of argasid ticks, including the genus Alectorobius (18, 19). Similarly, although the current understanding recognizes seven subgenera within the genus Dermacentor (24, 33, 34), the systematics and taxonomy of this genus pose significant challenges (27, 35). In such circumstances, combining morphological and molecular studies on ticks, especially those that are poorly understood, could be crucial for clarifying their phylogenetic position (36–38). Moreover, understanding tick species from different geographic locations is important for shedding light on the evolutionary history of ticks. Although demonstrating characteristics of both the Palearctic and Oriental regions, the knowledge regarding the genera Alectorobius and Dermacentor is limited within Pakistan. No tick species from the Alectorobius genus has been reported, while only Dermacentor raskemensis and Dermacentor marginatus have been identified morphologically or molecularly within the Dermacentor genus in this country (39–43). To address this knowledge gap, in this study, we reported the occurrence and genetic characterization of Dermacentor and Alectorobius species in Pakistan using morphological and molecular approaches.
Materials and methods
Study area
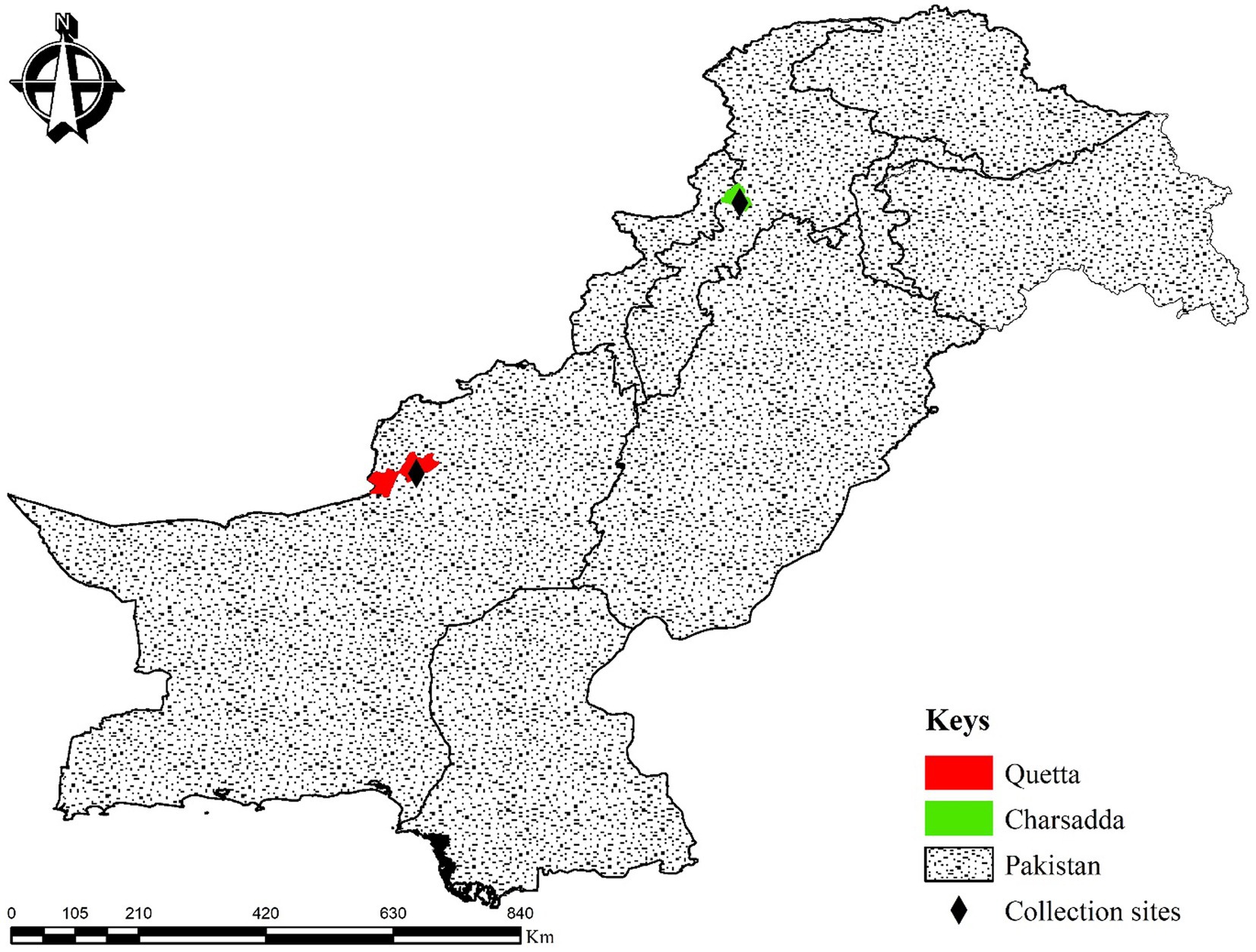
This study was carried out in the districts of Quetta (30°08′55.3″N, 66°57′42.1″E) and Charsadda (34°10′17.5″N, 71°45′31.7″E) in the provinces of Balochistan and Khyber Pakhtunkhwa, Pakistan, respectively. The geocoordinates of the collection areas were determined using the Global Positioning System (GPS), and the study map was designed using ArcGIS v. 10.3.1 (Figure 1).
Tick collection and preservation
Tick specimens were collected from sheep and nests of swift birds in 2023 from the districts of Quetta and Charsadda, respectively. In order to avoid any external damage to the specimens, the ticks were carefully detached from the host body and nests using tweezers. The specimens were rinsed in distilled water followed by 70% ethanol and subsequently preserved in 100% ethanol in 1.5 mL Eppendorf tubes.
Morphological identification of ticks
The collected specimens were morphologically identified using a stereo-zoom microscope (StereoBlue-euromex SB.1302-1, Arnhem, Netherlands) using standard morphological identification keys for Dermacentor spp. (28–30) and Alectorobius spp. (21, 44, 45).
DNA extraction and PCR
A total of 14 ticks including five Dermacentor spp. and nine Alectorobius spp. specimens selected for DNA extraction and were dried in an incubator for 30 min. With the use of sterilized scissors and a micro pestle, the specimens were homogenized in 200 μL of phosphate-buffered saline (PBS). The phenol-chloroform method was used to extract the genomic DNA (46), and 30 μL of “nuclease-free” PCR water was utilized to dilute the extracted DNA pellet. The genomic DNA was measured through NanoDrop (Nano-Q, Optizen, Daejeon, South Korea) and kept at −20°C for further experiments.
Conventional PCR (GE-96G, BIOER, Hangzhou, China) was used to amplify partial fragments of mitochondrial 16S rRNA and cox1 from the extracted genomic DNA of ticks (Table 1). Each PCR reaction mixture was performed in a total 25 μL volume—containing 1 μL each primer—forward and reverse (10 μM), 2 μL of genomic DNA template (100 ng/μL), 8.5 μL of “nuclease-free” PCR water, and 12.5 μL of DreamTaq green MasterMix (2x; Thermo Scientific, Waltham, MA, United States). A positive control (DNA of Argas persicus or Hyalomma anatolicum) and a negative control (PCR water that had been “nuclease-free” instead of DNA) were included in each PCR reaction. Lists of the primers used in the present study along with the thermocycler conditions are shown in Table 1.
The products of the PCR were electrophoresed on a 2% agarose gel and observed under ultraviolet light in a Gel Documentation System (RB Flash Digi, Robus Technologies, United Kingdom). The DNA Clean and Concentrator Kit (Zymo Research, Irvine, CA, United States) was used to purify the PCR-positive samples in accordance with the instructions provided by the manufacturer.
DNA sequencing and phylogenetic analysis
All amplified amplicons of cox1 and 16S rDNA partial fragments were sequenced bidirectionally (Macrogen Inc., Seoul, South Korea) using the Sanger sequencing method. The obtained sequences were cropped through SeqMan v. 5 (DNASTAR, Inc., Madison/WI, United States) to remove poor reading sequences and subjected to Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) at the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). After BLAST, high identity sequences were downloaded in FASTA format from the NCBI. The obtained sequences were aligned with the downloaded sequences using ClustalW multiple alignments in BioEdit Sequence Alignment Editor v. 7.0.5 (49). The phylogenetic trees were constructed individually for each gene sequence of the tick, using the maximum likelihood method with the Tamura-Nei model in Molecular Evolutionary Genetics Analysis (MEGA-X), with a bootstrapping value of 1,000 (50). The coding sequences were aligned using MUSCLE alignments (51).
Results
Ticks and their geographic origin
A total of 25 tick specimens were collected and morphologically identified into two distinct species, A. coniceps and Dermacentor sp., both of which were found in different areas. These comprised 14 out of 25 (56%; 9 males and 5 females) A. coniceps from three different nests of swift birds in the district of Charsadda, Khyber Pakhtunkhwa and 11 out of 25 (44%; 7 males and 4 females) Dermacentor sp. from sheep in the district of Quetta, Balochistan.
Morphology of Alectorobius coniceps
Female
They are broadly rounded posteriorly, obtusely angled anteriorly with a small, bluntly subtriangular hood, and the color is light to dark brownish to black. Approximately 10 setae and large central pores are found on the anterior labium, and irregularly divided longitudinal striations are found on the posterior labium. The coxal and supracoxal folds are conspicuous. The transverse part of the pre-anal groove is small. With comparatively larger disks adjacent to it, the posteromedian groove extends from the anus to the paired organ. The posterior paired organ can be found at approximately posterior 1 out of 7 to 1 out of 9 of the body. The anterior valve is small and finely striated in shape, whereas the posterior valve is wider and less finely striated in shape with coxa IV disks at each apex. The spiracular plates are large and positioned laterally to coxa IV. The capitulum in the camerostome is between the coxa I and the hood. Basis capituli ventral surface is pebbled and approximately two times as broad as long. Legs are narrow and long, and surfaces are pebbled. Tarsi are narrow, elongate, and abruptly tapering distally (Figure 2).

Figure 2. Male (A: dorsal and B: ventral) and female (C: dorsal and D: ventral) views of Alectorobius coniceps collected in this study from nests of swift birds in district Charsadda, KP.
Male
Except for sexual characteristics and size, male ticks are similar to female ticks. The integument around the genital operculum is finely and densely pebbled spiculate, and the posterior integument is transversely rugose. Nymphs resembled adults, except for the lack of external genitalia. Base capituli posthypostomal setae extending to the level of midpalpal segment 2 length. Legs have moderate length and humps (Figure 2).
Morphology of Dermacentor sp.
Female
The female tick’s body is elongated, oval, and brownish-red in color. Legs and capitulum are lighter. Except for punctations and grooves, the scutum is oval and whitish in color. Eyes are in front of the middle lateral border. The genital opening is level on coxae II and III. The spiracles are well developed. The capitulum is long and hairy with slightly developed whitish patterns dorsally. Cornua formed barely tubercles, and porose regions are subcircular. Palps are twice as long as they are wide; article I is small; article II and III are both well developed; and article II has a slight posterodorsal spine, while article IV is small and cylindrical. Coxa I has long, triangular, close-spaced, and parallel spurs with tapering or narrowly rounded tips; the external spur is usually nearly equal to or slightly shorter than the internal; and both the spurs are directed slightly posterolateral. Each part of coxae II and III has a sharp external spur and a smaller, broadly oval internal spur; coxa IV is enlarged and rounded with a narrow triangular external spur with a tapering apex. Legs are ornamented, hairy, and lack spines. The tarsi are slightly raised (Figure 3).
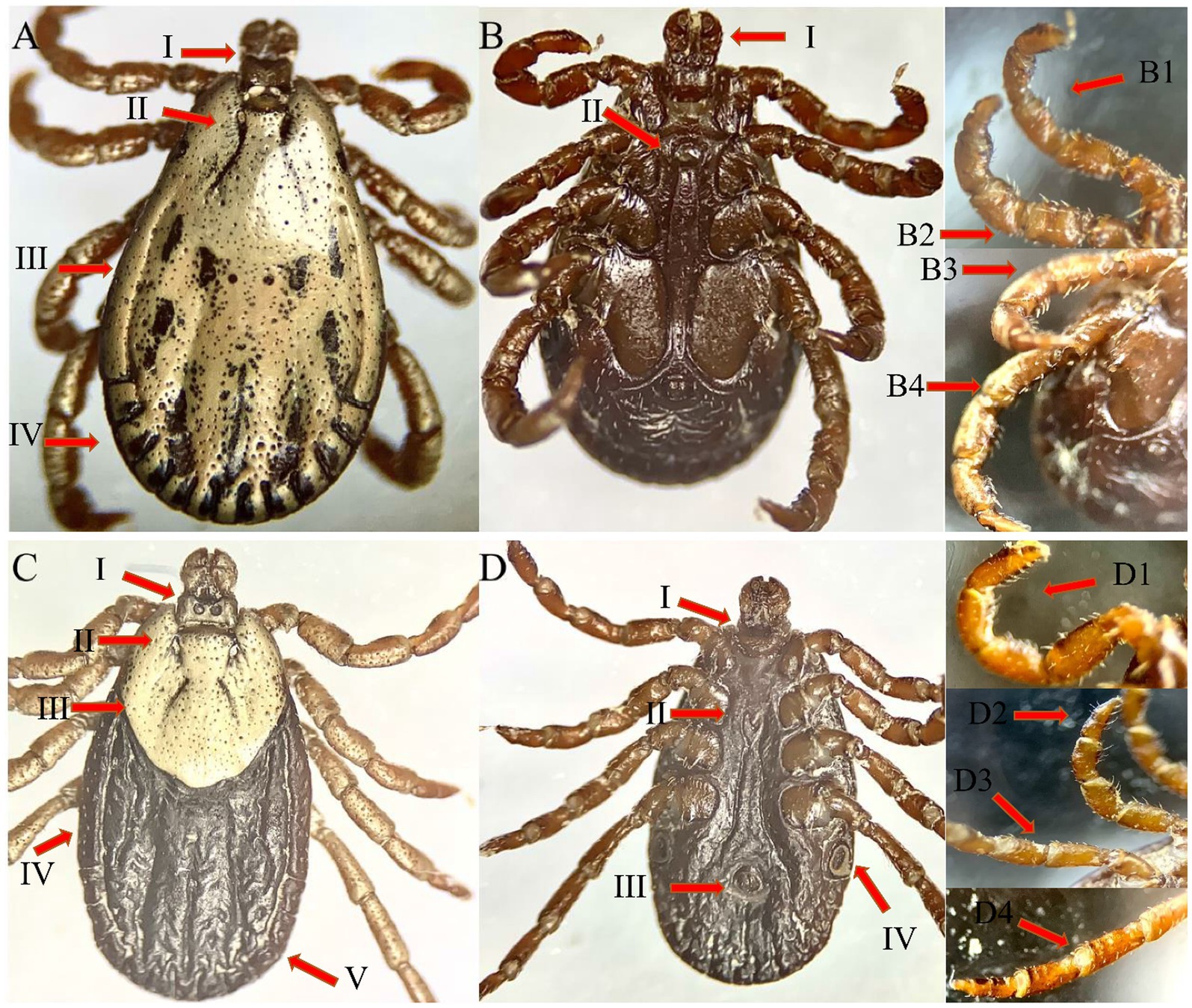
Figure 3. The male [A: dorsal—dorsally capitulum (I), cervical groove (II), Lateral groove (III), and festoon (IV), B: ventral—ventrally capitulum (I), and genital aperture, leg 1 (B1), leg 2 (B2), leg 3 (B3), leg 4 (B4)] and female [C: dorsal—dorsally capitulum (I), cervical groove (II), scutum (III), lateral groove (IV) and festoon (V), D: ventral—ventrally capitulum (I), genital aperture (II), anal groove (III) and spiracles (IV), leg 1 (D1), leg 2 (D2), leg 3 (D3), leg 4 (D4)] views of Dermacentor sp. collected in this study from district Quetta, Balochistan.
Male
They are oval and brownish-red in color. The scutum is partially curved, and the whole scutum is white except for the grooves and punctations. The cervical grooves are twisted outward, while the marginal grooves are deep. The outside festoons are wider than the inner ones. The punctations can be seen in some short hairs, some of which are dispersed. Eyes are on a level on coxa II. The genital aperture is parallel to coxa II, and the genital grooves extend almost parallel to coxa IV. They have thin and well-developed spiracles. The cornua is tapered to large spines. The palps are twice as long as the hypostome. Articles II and III are well developed, article II has a tiny spine postero-dorsally, article III is triangular dorsally, and article IV is small and cylindrical. Legs are similar to or resemble those of the female (Figure 3).
Molecular analysis
The BLAST analysis of the 16S rDNA sequence belonging to morphologically identified A. coniceps showed 97.46% maximum identity with the same species. In the phylogenetic tree, the 16S rDNA sequence for A. coniceps was clustered with the same species reported from Malta (MK946450) and grouped in a sister clade with Alectorobius capensis (KU946450) and Alectorobius sawaii (MK606017). The BLAST analysis of the 16S rDNA sequence for Dermacentor sp. showed 98.42% maximum identity with Dermacentor pavlovskyi, followed by 96.35% with Dermacentor marginatus, 95.79% with Dermacentor raskemensis, 95.60% with Dermacentor niveus, 94.23% with Dermacentor nuttalli, 94.20% with Dermacentor silvarum, and 93.70% with Dermacentor sinicus. Phylogenetically, the 16S rDNA sequence for Dermacentor sp. clustered with the D. pavlovskyi reported from China (OK493293-OK493294) and grouped in a sister clade with D. raskemensis, D. nuttalli, D. marginatus, D. silvarum, and D. sinicus (Figure 4). The obtained 16S rDNA sequence for A. coniceps and Dermacentor sp. were deposited to the GenBank under accession numbers: OR643824 and OR643821, respectively.
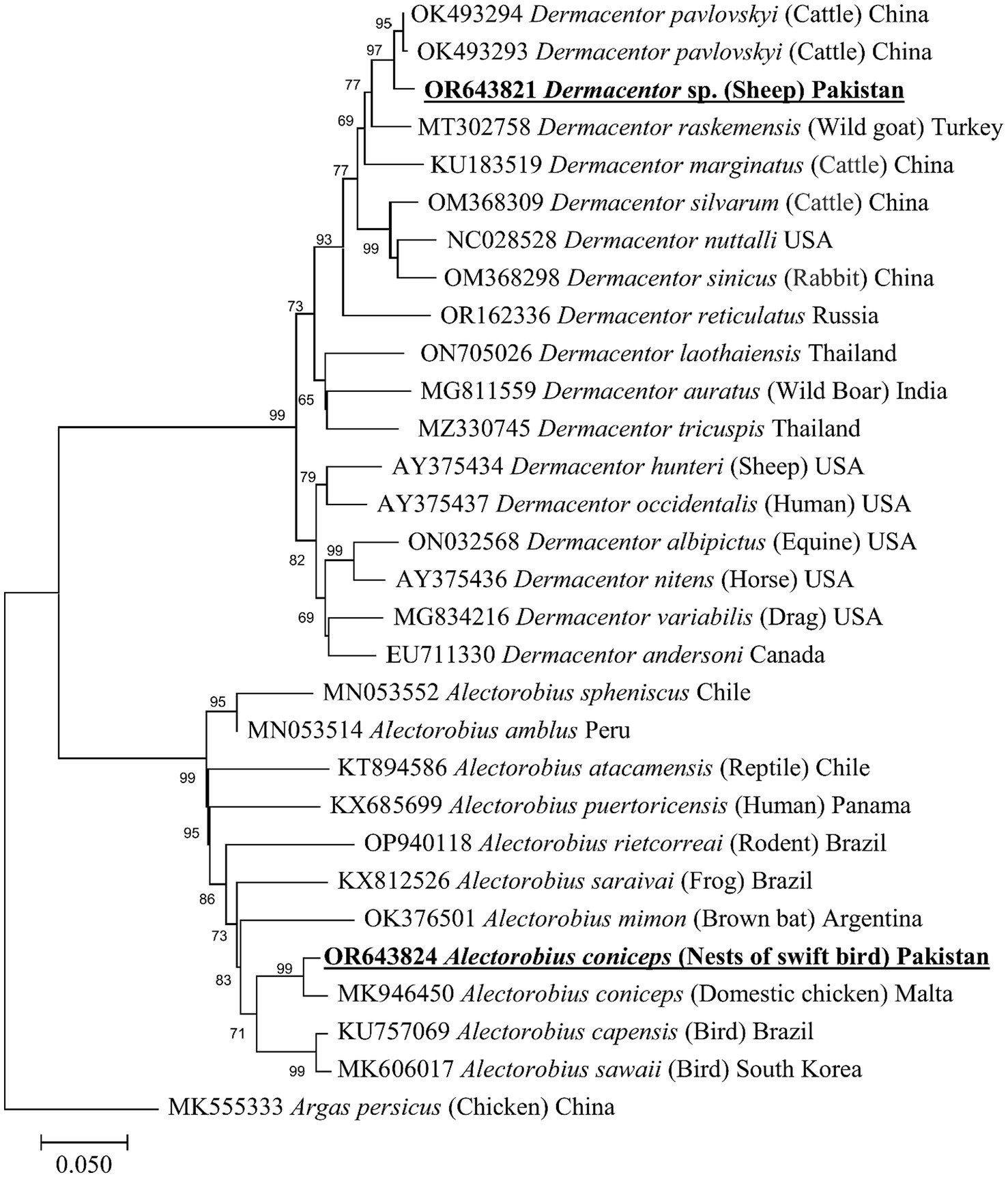
Figure 4. Maximum likelihood phylogenetic tree based on partial mitochondrial 16S ribosomal DNA sequences for Alectorobius spp. and Dermacentor spp. The 16S rDNA sequence of Argas persicus was used as an outgroup. The levels of bootstrap support (>65%) for phylogenetic groupings are given at each node; the accession numbers are followed by the species names, hosts, and locations (if applicable). The obtained sequences are shown in bold-underlined font.
The BLAST analysis of the mitochondrial cox1 partial sequence obtained for A. coniceps showed 96.49% maximum identity with the same species. In the phylogenetic tree, the cox1 sequence for A. coniceps clustered with the corresponding sequence was reported from Malta (MK946447). While the BLAST analysis of the obtained cox1 sequence for Dermacentor sp. showed 97.45% maximum identity with D. pavlovskyi followed by 93.85% with D. raskemensis, 92.09% with D. nuttalli, 91.92% with D. sinicus, and 91.39% with D. marginatus, D. silvarum, and D. niveus. In the phylogeny, the cox1 sequence for Dermacentor sp. clustered with the D. pavlovskyi species was reported from China (OK489456) and grouped in a sister clade with D. raskemensis, D. nuttalli, D. sinicus, D. marginatus, and D. silvarum (Figure 5). The obtained cox1 sequences for A. coniceps and Dermacentor sp. were deposited to the GenBank under accession numbers OR660126 and OR643819, respectively.
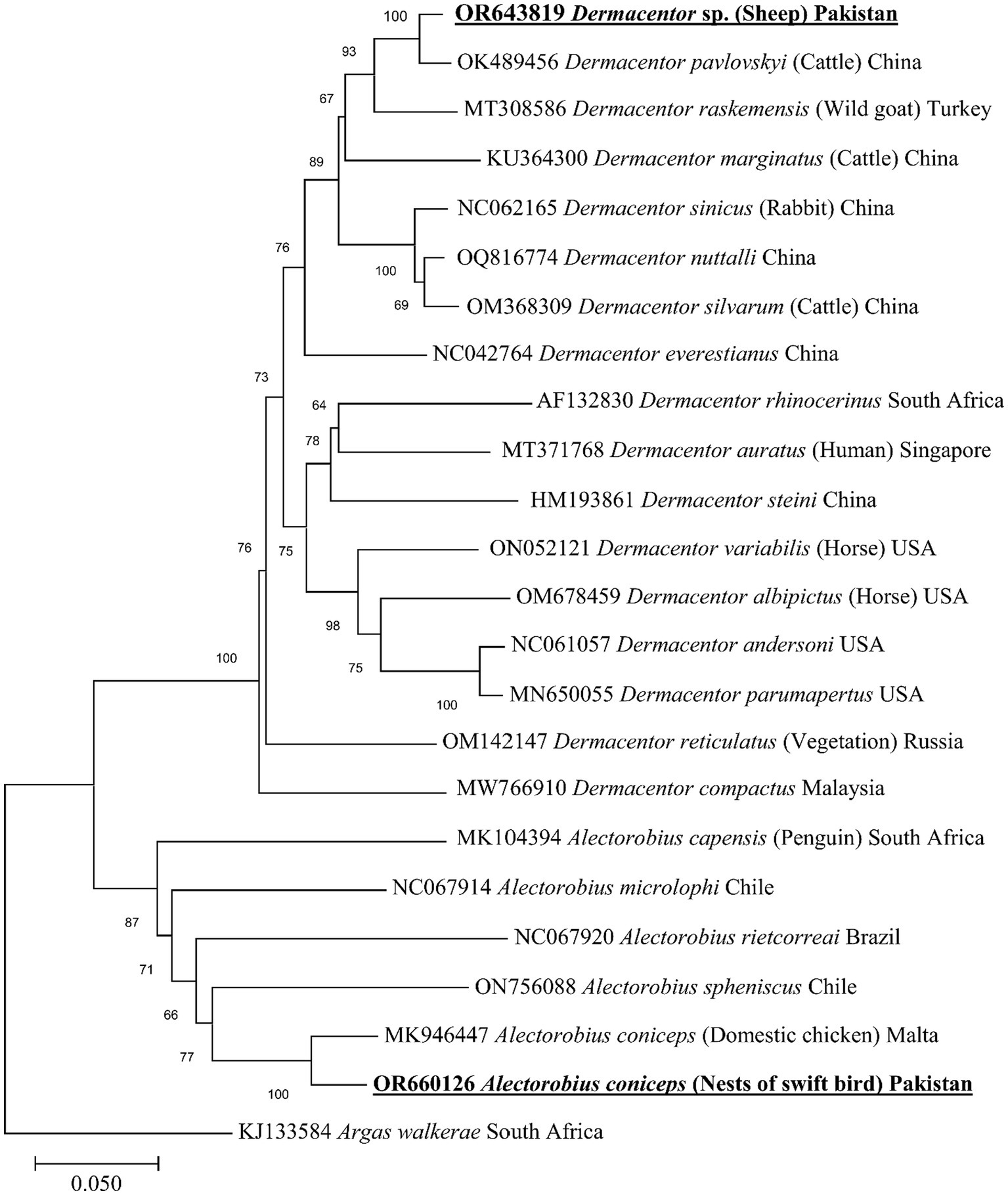
Figure 5. Maximum likelihood phylogenetic tree based on partial mitochondrial cox1 sequence for Alectorobius spp. and Dermacentor spp. The cox1 sequence of Argas walkerae was used as an outgroup. The levels of bootstrap support (>65%) for phylogenetic groupings are given at each node; the accession numbers are followed by the species names, hosts, and locations (if applicable). The obtained sequences are shown in bold-underlined font.
The sequences were used in the phylogenetic analysis for Alectorobius spp. (Tables 2, 3) and Dermacentor spp. (Tables 4, 5), and their identities with the species of the corresponding genus are shown in Table 2.
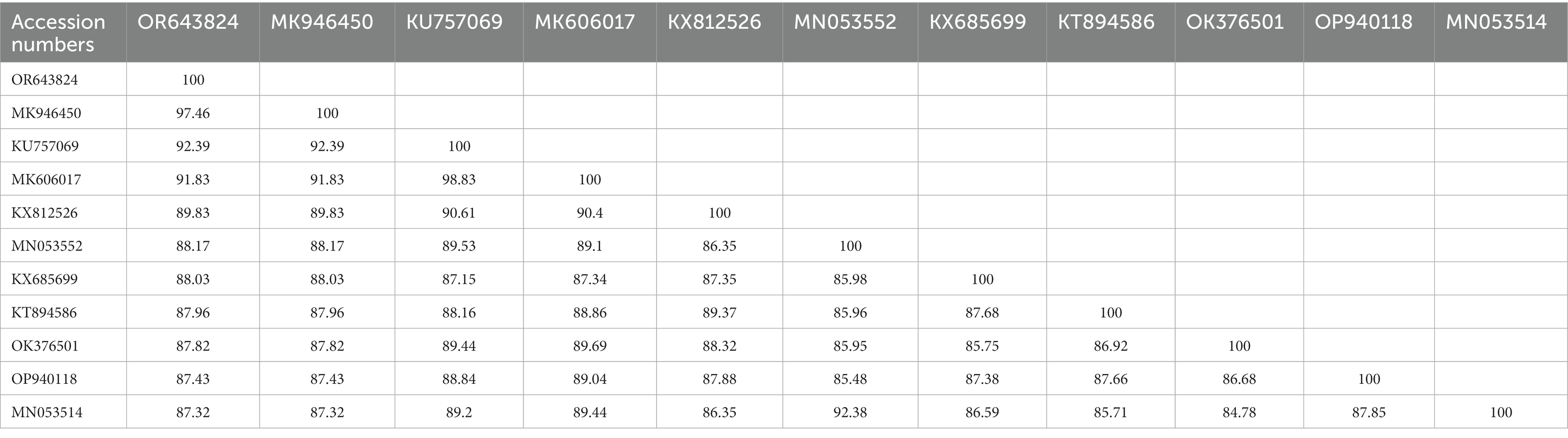
Table 2. Obtained 16S rDNA (OR643824; <460 bp) sequence identities with the diversity of Alectorobius species.
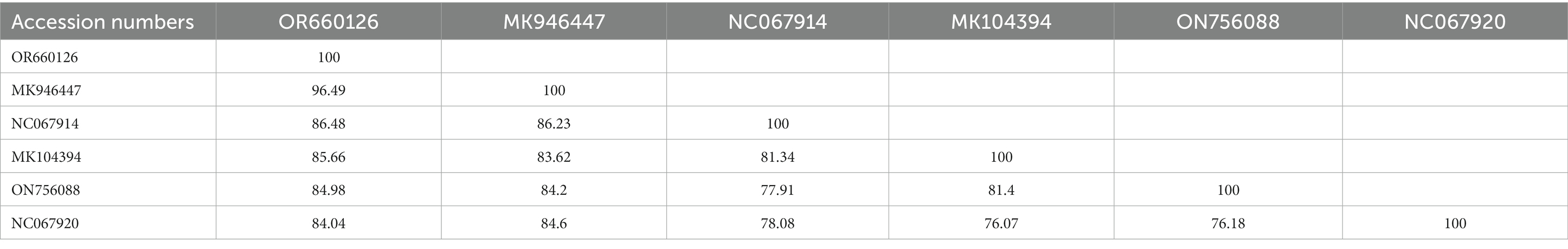
Table 3. Obtained cox1 (OR660126; <649 bp) sequence identities with the diversity of Alectorobius species.
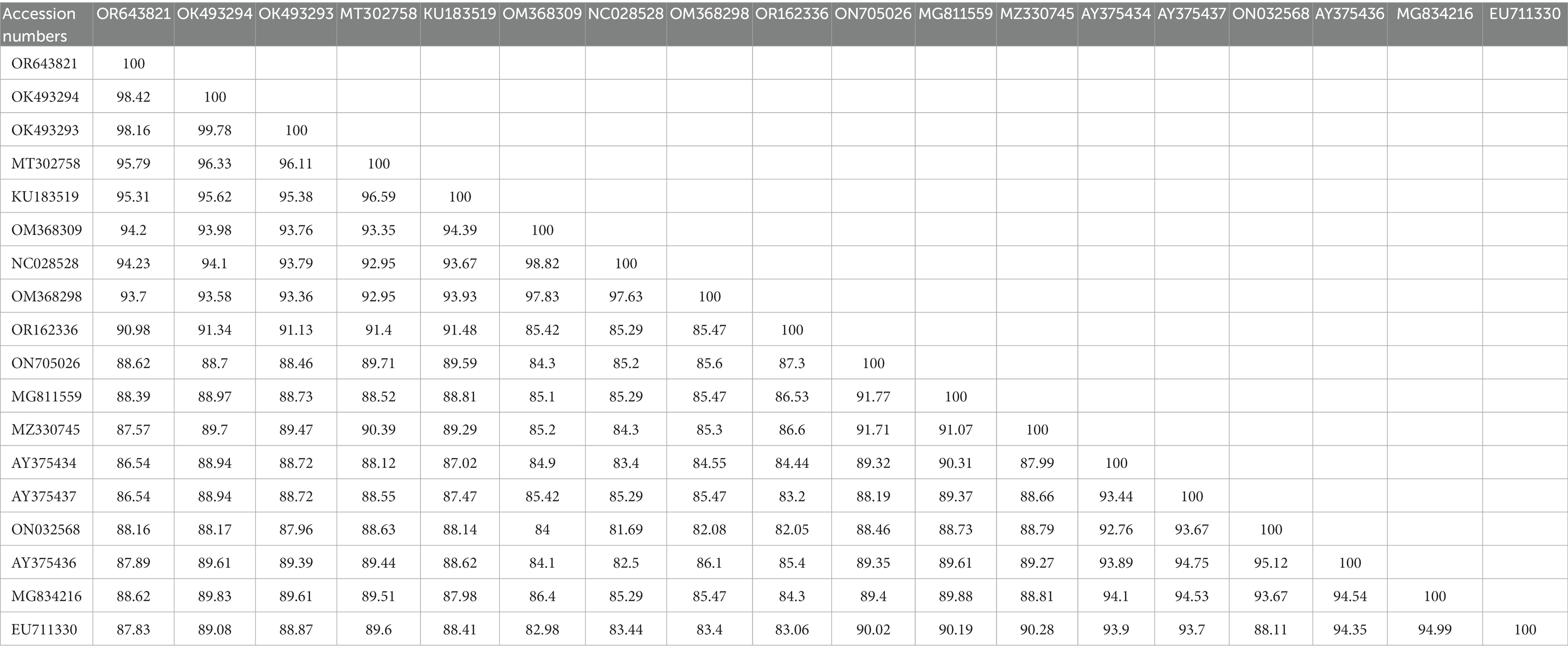
Table 4. Obtained 16S rDNA (OR643821; <460 bp) sequence identities with the diversity of Dermacentor species.
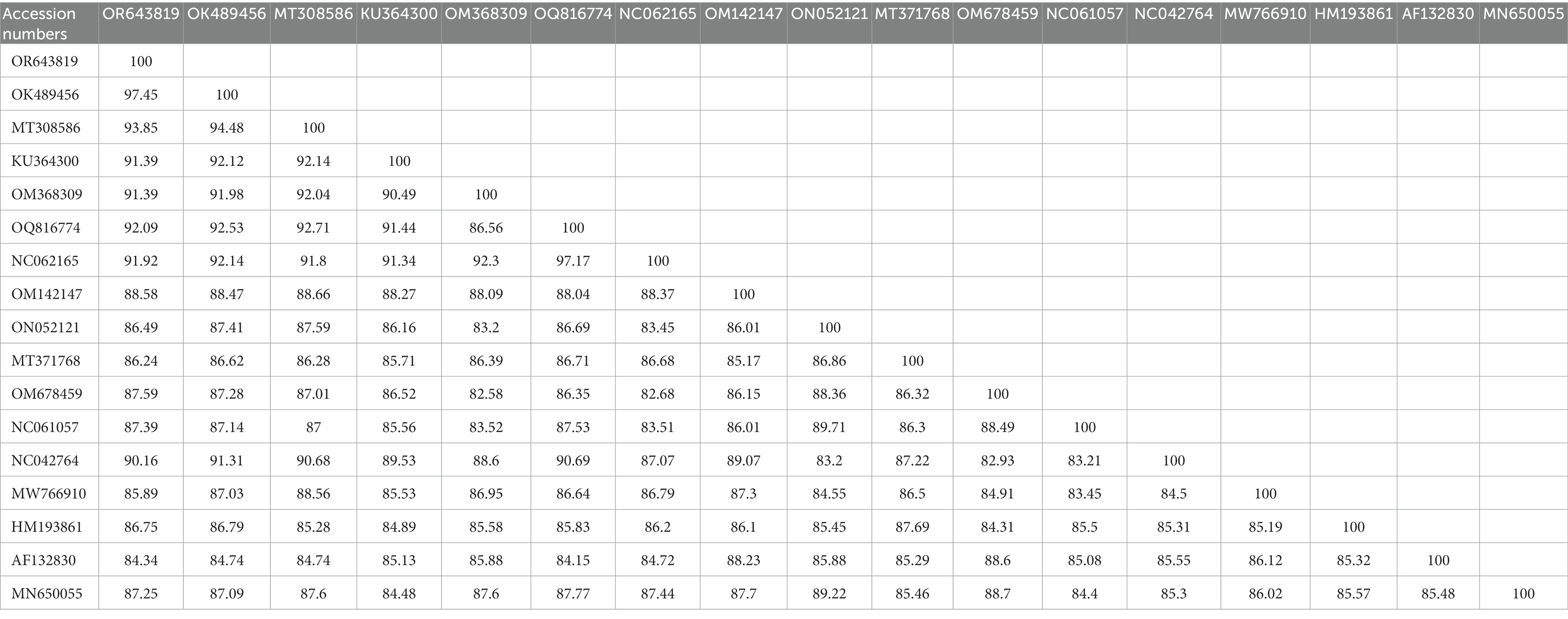
Table 5. Obtained cox1 (OR643819; <649 bp) sequence identities with the diversity of Dermacentor species.
Discussion
Ticks of the genus Alectorobius and Dermacentor have medical and economic importance worldwide (20, 34, 52). However, they are among the most neglected tick genera in the Oriental region, including Pakistan. Importantly, research in Pakistan has considerably focused on the exploration of ticks and tick-borne pathogens in the last half-decade (4, 5, 9, 39, 53–65). This study provides the first morphological and molecular record of A. coniceps and Dermacentor sp. from the Oriental region, including Pakistan.
Alectorobius spp. are distributed globally possibly due to their association with birds, facilitating an efficient distribution (6, 13, 20). Among Alectorobius ticks, A. coniceps has been collected from various locations such as caves, crevices, cliffs, ravines, nests, stables, wells, and lofts in both the Palearctic and Oriental regions (20, 21, 66). In this study, A. coniceps ticks were collected from swallows’ nests in Khyber Pakhtunkhwa, which is located at the junction of the Palearctic and Oriental regions. Moreover, the larval stages of Alectorobius ticks may feed on the same bird species, and therefore, they are relatively well understood from the Palearctic region (13, 21, 67). With this tendency, the adults and nymphs of A. coniceps were collected in this study. Future studies should also prioritize the investigation of larval stages from the Oriental region, as, to the best of our knowledge, this stage has not been described in the Oriental region.
Dermacentor spp. are believed to have evolved in central Asia, and this region exhibits the highest diversity of Dermacentor spp. (26, 68, 69). Among Dermacentor ticks, D. pavlovskyi has been documented infesting goats and sheep in mountainous regions, as reported in earlier studies (29, 31, 32). Similarly, the area (Balochistan), where Dermacentor sp., a closely related tick to D. pavlovskyi, was collected in the present study, is situated adjacent to the Palearctic region and has a mountainous terrain with an approximate elevation of 5,500 feet. Other than open areas, D. pavlovskyi ticks have been found on wild animals in nature reserves and national parks. For instance, these ticks were collected by the Republican Tropical Station in 1941–1955 in the Aksu-Dzhabagly nature reserve in Kazakhstan (31). Interestingly, the current study area is located near the renowned national park, “Hazarganji-Chiltan.” It could be assumed that D. pavlovskyi or closely related ticks have an affinity for wild goats and sheep in such protected areas from where they could invade domestic animals. The tendency of larval Dermacentor ticks to infest small animals has resulted in a limited understanding of this stage. Consequently, this study collected adult and nymphal stages of Dermacentor sp. ticks, and no larval stage of this species was collected.
Molecular-based analysis is pivotal for comprehending the debated systematics and taxonomy of tick species, including the genus Alectorobius and the genus Dermacentor (19, 37, 70, 71). Consequently, A. coniceps and Dermacentor sp. ticks of the current study were subjected to molecular-based analysis involving 16S rDNA and cox1 sequences. The analysis revealed that the A. coniceps ticks from this study displayed variations of 2.54 and 3.51% with their respective species from Malta, based on 16S rDNA and cox1 sequences, respectively. Similarly, Dermacentor sp. ticks exhibited variations of 1.68% and 2.58% with D. pavlovskyi from China as determined by 16S rDNA and cox1 sequences, respectively. At present, although ticks having this range of variations are considered a single species (72, 73, 4), Dermacentor sp. could not be validated as D. pavlovskyi due to some morphological variations. In contrast to D. pavlovskyi and Dermacentor montanus (28, 29), Dermacentor sp. examined in this study lacks prominent spines on their legs. In comparison to female ticks of D. montanus and D. pavlovskyi, Dermacentor sp. exhibited a genital aperture with a more wing-like shape. Furthermore, other morphological differences among these species were observed in punctuations, cervical grooves, and lateral grooves. However, further comprehensive studies such as mitochondrial genome sequencing of these ticks are essential to gain an accurate understanding of their genetic structure. Furthermore, these intraspecific variations can be attributed to various factors, including tick population size, ecology, and geographic isolation (37, 74). In addition to its closest evolutionary relationship with the same species, A. coniceps displayed proximity to A. capensis, confirming their classification into the same species complex (6, 13). Although the closest evolutionary relationship of Dermacentor sp. with D. pavlovskyi was observed, however, its closeness with other species of the same subgenus, such as D. montanus, could not be verified due to the lack of authentic genetic data in GenBank.
Conclusion
This study for the first time presented both morphological and molecular data on poorly known ticks, A. coniceps, and Dermacentor sp., closely related to D. pavlovskyi, from the junction of the Palearctic and Oriental regions in Pakistan. The geographic, morphological, and genetic data of these tick species may aid future studies on tick systematic and taxonomy. Furthermore, the study suggests that the study area, showcasing a combination of traits from two different zoogeographic regions, could harbor a notable diversity of ticks.
Data availability statement
The data presented in the study have been deposited in the GenBank repository with the following accession numbers: OR643824, OR643821, OR660126, and OR643819.
Ethics statement
The animal studies were approved by the Advance Studies Research Board (ASRB: Dir/A&R/AWKUM/2022/9396) Committee members of Abdul Wali Khan University, Mardan, KP, Pakistan, gave their approval for the proposed study. The owners of the animals gave their verbal consent for the observation and tick collections. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
AAli: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. MK: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. ZU: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. MN: Data curation, Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. K-HT: Data curation, Formal analysis, Validation, Writing – review & editing. AAlo: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. MA: Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. TT: Funding acquisition, Investigation, Methodology, Project administration, Writing – review & editing.
Acknowledgments
The authors acknowledge the financial support provided by the Higher Education Commission (HEC), Pakistan, and Pakistan Science Foundation (PSF). The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia. This work was supported by JSPS KAKENHI Grant Numbers JP20KK0154 and JP22H02522, and JSPS Bilateral Program Grant Numbers JPJSBP120206002, JPJSBP120219936, and JPJSBP120239937.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Guglielmone, AA , Robbins, RG , Apanaskevich, DA , Petney, TN , Estrada Peña, A , Horak, IG, et al. The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida) of the world: A list of valid species names. Exp Appl Acarol. (2010) 28:27. doi: 10.1023/a:1025381712339
2. Nava, S , Guglielmone, AA , and Mangold, AJ . An overview of systematics and evolution of ticks. Front Biosci. (2009) 14:2857–77. doi: 10.2741/3418
3. Boulanger, N , Boyer, P , Talagrand-Reboul, E , and Hansmann, Y . Ticks and tick-borne diseases. Med Mal Infect. (2019) 49:87–97. doi: 10.1016/j.medmal.2019.01.007
4. Khan, M , Islam, N , Khan, A , Islam, ZU , Muñoz-Leal, S , Labruna, MB, et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia sp. Ticks Tick Borne Dis. (2022) 13:102047. doi: 10.1016/j.ttbdis.2022.102047
5. Khan, M , Almutairi, MM , Alouffi, A , Tanaka, T , Chang, SC , Chen, CC, et al. Molecular evidence of Borrelia theileri and closely related Borrelia spp. in hard ticks infesting domestic animals. Front Vet Sci. (2023) 10:1297928. doi: 10.3389/fvets.2023.1297928
6. Dietrich, M , Gómez-Díaz, E , and McCoy, KD . Worldwide distribution and diversity of seabird ticks: implications for the ecology and epidemiology of tick-borne pathogens. Vector-Borne Zoonot Dis. (2011) 11:453–70. doi: 10.1089/vbz.2010.0009
7. Kahl, O . Hard ticks as vectors—some basic issues. Wien Klin Wochenschr. (2018) 130:479–83. doi: 10.1007/s00508-018-1360-x
8. Vial, L . Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite. (2009) 16:191–202. doi: 10.1051/parasite/2009163191
9. Ali, A , Numan, M , Khan, M , Aiman, O , Muñoz-Leal, S , Chitimia-Dobler, L, et al. Ornithodoros (Pavlovskyella) ticks associated with a Rickettsia sp. in Pakistan. Parasit Vectors. (2022a) 15:138. doi: 10.1186/s13071-022-05248-0
10. Bakkes, DK , De Klerk, D , Latif, AA , and Mans, BJ . Integrative taxonomy of Afrotropical Ornithodoros (Ornithodoros) (Acari: Ixodida: Argasidae). Ticks Tick Borne Dis. (2018) 9:1006–37. doi: 10.1016/j.ttbdis.2018.03.024
11. Vázquez-Guerrero, E , González-Quiroz, JL , Domínguez-López, ML , Kneubehl, AR , Krishnavajhala, A , Curtis, MW, et al. New records of Ornithodoros turicata (Ixodida: Argasidae) in rural and urban sites in the Mexican states of Aguascalientes and Zacatecas indicate the potential for tick-borne relapsing fever. Exp Appl Acarol. (2023) 91:99–110. doi: 10.1007/s10493-023-00830-2
12. Barros-Battesti, DM , Landulfo, GA , Luz, HR , Marcili, A , Onofrio, VC , and Famadas, KM . Ornithodoros faccinii n. sp. (Acari: Ixodida: Argasidae) parasitizing the frog Thoropa miliaris (Amphibia: Anura: cycloramphidae) in Brazil. Parasit Vectors. (2015) 8:1–11. doi: 10.1186/s13071-015-0877-3
13. Estrada-Peña, A , Kleinerman, G , and Baneth, G . Genus Ornithodoros Koch, 1844 In: Ticks of Europe and North Africa: A guide to species identification, 1st edn. Springer, Cham (2017). 41–3.
14. Muñoz-Leal, S , Venzal, JM , González-Acuña, D , Nava, S , Lopes, MG , Martins, TF, et al. A new species of Ornithodoros (Acari: Argasidae) from desert areas of northern Chile. Ticks Tick Borne Dis. (2016) 7:901–10. doi: 10.1016/j.ttbdis.2016.04.008
15. Venzal, JM , and Estrada-Peña, A . Larval feeding performance of two Neotropical Ornithodoros ticks (Acari: Argasidae) on reptiles. Exp Appl Acarol. (2006) 39:315–20. doi: 10.1007/s10493-006-9011-8
16. Zahid, H , Muñoz-Leal, S , Khan, MQ , Alouffi, AS , Labruna, MB , and Ali, A . Life cycle and genetic identification of Argas persicus infesting domestic fowl in Khyber Pakhtunkhwa, Pakistan. Front Vet Sci. (2021) 8:664731. doi: 10.3389/fvets.2021.664731
17. Clifford, CM , Kohls, GM , and Sonenshine, DE . The systematics of the subfamily Ornithodorinae (Acarina: Argasidae). I. The genera and subgenera. Ann Entomol Soc Am. (1964) 57:429–37. doi: 10.1093/aesa/57.4.429
18. Hoogstraal, H . Argasid and nuttalliellid ticks as parasites and vectors. Adv Parasitol. (1985) 24:135–238. doi: 10.1016/S0065-308X(08)60563-1
19. Mans, BJ , Kelava, S , Pienaar, R , Featherston, J , de Castro, MH , Quetglas, J, et al. Nuclear (18S-28S rRNA) and mitochondrial genome markers of Carios (Carios) vespertilionis (Argasidae) support Carios Latreille, 1796 as a lineage embedded in the Ornithodorinae: re-classification of the Carios sensu Klompen and Oliver (1993) clade into its respective subgenera. Ticks Tick Borne Dis. (2021) 12:101688. doi: 10.1016/j.ttbdis.2021.101688
20. Kleinerman, G , and Baneth, G . Ornithodoros (Alectorobius) coniceps (Canestrini, 1890) (figs. 14 and 15) In: Ticks of Europe and North Africa: a guide to species identification, 1st edn. Springer, Cham (2017). 51–4.
21. Hoogstraal, H , Clifford, CM , and Keirans, JE . The Ornithodoros (Alectorobius) capensis group (Acarina: Ixodoidea: Argasidae) of the Palearctic and oriental regions. O. (a.) coniceps identity, bird and mammal hosts, virus infections, and distribution in Europe, Africa, and Asia. J Parasitol. (1979) 65:395–407.
22. Gray, JS , Estrada-Peña, A , Vial, L , Sonenshine, DE , and Roe, RM . Ecology of nidicolous ticks. Biol Ticks. (2013) 2:39–60.
23. Hoogstraal, H . A small form of Ornithodoros (Alectorobius) coniceps (Canestrini, 1890) (Ixodoidea, Argasidae) from India and USSR with wild birds as hosts in India. Acarologia. (1962) 4:190–2.
24. Guglielmone, AA , Petney, TN , and Robbins, RG . Ixodidae (Acari: Ixodoidea): descriptions and redescriptions of all known species from 1758 to December 31, 2019. Zootaxa. (2020) 4871:1–322. doi: 10.11646/zootaxa.4871.1.1
25. Drehmann, M , Springer, A , Lindau, A , Fachet, K , Mai, S , Thoma, D, et al. The spatial distribution of Dermacentor ticks (Ixodidae) in Germany—evidence of a continuing spread of Dermacentor reticulatus. Front Vet Sci. (2020) 7:578220. doi: 10.3389/fvets.2020.578220
26. Filippova, N.A. , and Panova, I.V. Revision of the genus Dermacentor Koch of the Fauna of the USSR and adjoining territories (Ixodoidea, Ixodidae). Parazitolog Sb. (in Russian, NIH Library translation by T. Crump) Parazitol Sborn Zool Inst Akad Nauk SSSR (1990) 35:4955.
27. Apanaskevich, DA , and Apanaskevich, MA . Description of a new Dermacentor (Acari: Ixodidae) species from Thailand and Vietnam. J Med Entomol. (2015) 52:806–12. doi: 10.1093/jme/tjv067
28. Filippova, NA . Ixodid ticks of subfamily Amblyomminae. Nauka, St. Petersburg: Fauna of Russia and Neighbouring Countries (1997).
29. Gao, Z , Xu, X , Zhang, J , Xuan, Y , Bai, L , Chen, Z, et al. Morphological and molecular characterization of adult Dermacentor pavlovskyi Olenev, 1927 (Acari: Ixodidae). Syst Appl Acarol. (2022) 27:922–33. doi: 10.11158/saa.27.5.7
30. Olenev, NO . A new species of the genus Dermacentor (Ixodidae). Parasitology. (1927) 19:84–5. doi: 10.1017/S0031182000005540
31. Sayakova, ZZ , Sadovskaya, VP , Yeszhanov, AB , Meka-Mechenko, VG , Kunitsa, TN , Kulemin, MV, et al. Distribution of ticks of the genus Dermacentor Koch, 1844 (Ixodidae, Amblyomminae) in the south-eastern part of Kazakhstan. News Natl. Acad. Sci. Republic Kazakhstan. Biol Med Ser. (2019) 335:55–62. doi: 10.32014/2019.2519-1629.48
33. Chavatte, JM , and Octavia, S . The complete mitochondrial genome of Dermacentor (Indocentor) auratus (Acari, Ixodidae). Parasite. (2021) 28:6. doi: 10.1051/parasite/2021002
34. Yi, S , and Rong-Man, X . The genus Dermacentor and the subgenus Indocentor (Acari: Ixodidae) from China. Orient Insects. (2013) 47:155–68. doi: 10.1080/00305316.2013.811019
35. Perry, K.L. . Molecular phylogenetic relationships of north American Dermacentor ticks using mitochondrial gene sequences. Statesboro, GA: M.S. thesis, Georgia Southern University. (2014).
36. Burger, TD , Shao, R , Beati, L , Miller, H , and Barker, SC . Phylogenetic analysis of ticks (Acari: Ixodida) using mitochondrial genomes and nuclear rRNA genes indicates that the genus Amblyomma is polyphyletic. Mol Phylogenet Evol. (2012) 64:45–55. doi: 10.1016/j.ympev.2012.03.004
37. Mans, BJ . Paradigms in tick evolution. Trends Parasitol. (2023) 39:475–86. doi: 10.1016/j.pt.2023.03.011
38. Nava, S , Beati, L , Labruna, MB , Cáceres, AG , Mangold, AJ , and Guglielmone, AA . Reassessment of the taxonomic status of Amblyomma cajennense with the description of three new species, Amblyomma tonelliae n. sp., Amblyomma interandinum n. sp. and Amblyomma patinoi n. sp., and reinstatement of Amblyomma mixtum, and Amblyomma sculptum (Ixodida: Ixodidae). Ticks Tick Borne Dis. (2014) 5:252–76. doi: 10.1016/j.ttbdis.2013.11.004
39. Ahmad, I , Ullah, S , Alouffi, A , Almutairi, MM , Numan, M , Tanaka, T, et al. First molecular-based confirmation of Dermacentor marginatus and associated Rickettsia raoultii and Anaplasma marginale in the Hindu Kush Mountain range. Animals. (2023) 13:686. doi: 10.3390/ani13233686
40. Apanaskevich, DA . First description of the nymph and larva of Dermacentor raskemensis (Acari: Ixodidae), parasites of pikas and other small mammals in Central Asia. J Med Entomol. (2013) 50:959–64. doi: 10.1603/ME13051
41. Dhanda, V , Kulkarni, SM , and Pratt, P . Dermacentor raskemensis (Ixodoidea: Ixodidae), redescription and notes on ecology. J Parasitol. (1971) 57:1324–9. doi: 10.2307/3277993
42. Hoogstraal, H. , and Valdez, R . Fieldiana Zool. Ticks (Ixodoidea) from wild sheep and goats in Iran and medical and veterinary implications. (1980) 6:116.
43. McCarthy, VC . Ixodid ticks (Acarina, Ixodidae) of West Pakistan. College Park: University of Maryland (1967).
44. Estrada-Peña, A , Mihalca, AD , and Petney, TN . Ticks of Europe and North Africa: A guide to species identification. Basel: Springer International Publishing. (2018) 404.
45. Hoogstraal, H , Clifford, CM , Keirans, JE , Kaiser, MN , and Evans, DE . The Ornithodoros (Alectorobius) capensis group (Acarina: Ixodoidea: Argasidae) of the palearctic and oriental regions. O.(a.) maritimus: identity, marine bird hosts, virus infections, and distribution in western Europe and northwestern Africa. J Parasitol. (1976) 62:799–810.
46. Sambrook, J , Fritsch, EF , and Maniatis, T . Molecular cloning: a laboratory manual. 2nd ed Cold Spring Harbor Laboratory Press (1989).
47. Folmer, O , Hoeh, WR , Black, MB , and Vrijenhoek, RC . Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mar Environ Res. (1994) 3:294–9.
48. Mangold, AJ , Bargues, MD , and Mas-Coma, S . Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol Res. (1998) 84:478–84.
49. Hall, T , Biosciences, I , and Carlsbad, C . BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
50. Kumar, S , Stecher, G , Li, M , Knyaz, C , and Tamura, K . MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
51. Edgar, RC . MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
52. Manzano-Román, R , Díaz-Martín, V , de la Fuente, J , and Pérez-Sánchez, R . Soft ticks as pathogen vectors: distribution, surveillance and control. Parasitology. (2012) 7:125–62. doi: 10.5772/32521
53. Ahmad, I , Ullah, S , Alouffi, A , Almutairi, MM , Khan, M , Numan, M, et al. Description of male, Redescription of female, host record, and phylogenetic position of Haemaphysalis danieli. Pathogens. (2022) 11:1495. doi: 10.3390/pathogens11121495
54. Ali, A , Mulenga, A , and Vaz, IS . Editorial: tick and tick-borne pathogens: molecular and immune targets for control strategies. Front Physiol. (2020) 11:744. doi: 10.3389/fphys.2020.00744
55. Ali, A , Zahid, H , Zeb, I , Tufail, M , Khan, S , Haroon, M, et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit Vectors. (2021) 14:1–12. doi: 10.1186/s13071-021-04836-w
56. Aiman, O , Ullah, S , Chitimia-Dobler, L , Nijhof, AM , and Ali, A . First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick Borne Dis. (2022) 13:101899. doi: 10.1016/j.ttbdis.2022.101899
57. Ali, A , Shehla, S , Zahid, H , Ullah, F , Zeb, I , Ahmed, H, et al. Molecular survey and spatial distribution of Rickettsia spp. in ticks infesting free-ranging wild animals in Pakistan (2017–2021). Pathogens. (2022b) 11:162. doi: 10.3390/pathogens11020162
58. Ali, A , Numan, M , Ullah, S , Khan, M , and Kamran, K . Genetic characterization of Haemaphysalis (Rhipistoma) indica and Haemaphysalis (Segalia) montgomeryi ticks (Ixodoidea: Ixodidae). Ticks Tick Borne Dis. (2023) 14:102105. doi: 10.1016/j.ttbdis.2022.102105
59. Karim, S , Budachetri, K , Mukherjee, N , Williams, J , Kausar, A , Hassan, MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl Trop Dis. (2017) 11:e0005681. doi: 10.1371/journal.pntd.0005681
60. Numan, M , Islam, N , Adnan, M , Zaman Safi, S , Chitimia-Dobler, L , Labruna, MB, et al. First genetic report of Ixodes kashmiricus and associated Rickettsia sp. Parasit Vectors. (2022) 15:1–12. doi: 10.1186/s13071-022-05509-y
61. Numan, M , Alouffi, A , Almutairi, MM , Tanaka, T , Ahmed, H , Akbar, H, et al. First detection of Theileria sinensis-like and Anaplasma capra in Ixodes kashmiricus: with notes on cox1-based phylogenetic position and new locality records. Animals. (2023) 13:3232. doi: 10.3390/ani13203232
62. Tila, H , Khan, M , Almutairi, M , Alouffi, A , Ahmed, H , Tanaka, T, et al. First report on detection of Hepatozoon ayorgbor in Rhipicephalus haemaphysaloides and Hepatozoon colubri in Haemaphysalis sulcata and Hyalomma anatolicum: risks of spillover of Hepatozoon spp. from wildlife to domestic animals. Frontiers in veterinary. Science. (2023) 10:1255482. doi: 10.3389/fvets.2023.1255482
63. Ullah, S , Alouffi, A , Almutairi, MM , Islam, N , Rehman, G , Ul Islam, Z, et al. First report of Rickettsia conorii in Hyalomma kumari ticks. Animals. (2023) 13:1488. doi: 10.3390/ani13091488
64. Kamran, K , Ali, A , Villagra, C , Siddiqui, S , Alouffi, AS , and Iqbal, A . A cross‐sectional study of hard ticks (acari: ixodidae) on horse farms to assess the risk factors associated with tick‐borne diseases. Zoonoses and Public Health. (2021a) 68:247–62.
65. Kamran, K , Ali, A , Villagra, CA , Bazai, ZA , Iqbal, A , and Sajid, MS . Hyalomma anatolicum resistance against ivermectin and fipronil is associated with indiscriminate use of acaricides in southwestern Balochistan, Pakistan Parasitol Res. (2021b) 120:15–25.
66. Khoury, C , Bianchi, R , Massa, AA , Severini, F , Di Luca, M , and Toma, L . A noteworthy record of Ornithodoros (Alectorobius) coniceps (Ixodida: Argasidae) from Central Italy. Exp Appl Acarol. (2011) 54:205–9. doi: 10.1007/s10493-011-9429-5
67. Hornok, S , Grima, A , Takács, N , Szekeres, S , and Kontschán, J . First records and molecular-phylogenetic analyses of three tick species (Ixodes kaiseri, Hyalomma lusitanicum and Ornithodoros coniceps) from Malta. Ticks Tick Borne Dis. (2020) 11:101379. doi: 10.1016/j.ttbdis.2020.101379
68. Pomerantzev, BN . Geographic distribution of Ixodoidea ticks and structure of its fauna in Palearctic region. Ann Zool Inst. (1948) 7:132–48.
69. Rudakov, NV , Shpynov, SN , and Samoilenko, IE , Tankibaev, MA . Ecology and epidemiology of spotted fever group rickettsiae and new data from their study in Russia and Kazakhstan. Ann N Y Acad Sci. (2003) 990:12–24.
70. Barker, SC , and Murrell, A . Systematics and evolution of ticks with a list of valid genus and species names. Parasitology. (2004): 129:S15–S36.
71. Beati, L , and Klompen, H . Phylogeography of ticks (Acari: Ixodida). Annu Rev Entomol. (2019) 64:379–97.
72. Labruna, MB , Onofrio, VC , Beati, L , Arzua, M , Bertola, PB , Ribeiro, AF, et al., Redescription of the female, description of the male, and several new records of Amblyomma parkeri (Acari: Ixodidae), a South American tick species. Experimental and Applied Acarology. (2009) 49:243–60.
73. Lv, J. , Wu, S. , Zhang, Y. , Zhang, T. , Feng, C. , Jia, G, et al. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial Dna. (2014) 25:142–149.
Keywords: Alectorobius coniceps, Dermacentor sp., 16S rDNA, cox1, Pakistan
Citation: Ali A, Khan M, Ullah Z, Numan M, Tsai K-H, Alouffi A, Almutairi MM and Tanaka T (2024) First record of Alectorobius coniceps (Ixodoidea: Argasidae) and Dermacentor sp. (Ixodoidea: Ixodidae) in Pakistan. Front. Vet. Sci. 10:1326734. doi: 10.3389/fvets.2023.1326734
Edited by:
Alicia Rojas, University of Costa Rica, Costa RicaReviewed by:
Asadollah Hosseini-Chegeni, Lorestan University, IranThankGod Emmanuel Onyiche, University of Maiduguri, Nigeria
Copyright © 2024 Ali, Khan, Ullah, Numan, Tsai, Alouffi, Almutairi and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, dW9wX2FsaUB5YWhvby5jb20=; Tetsuya Tanaka, azYxOTk0MzFAa2FkYWkuanA=
 Abid Ali
Abid Ali Mehran Khan
Mehran Khan Zafar Ullah
Zafar Ullah Muhammad Numan
Muhammad Numan Kun-Hsien Tsai
Kun-Hsien Tsai Abdulaziz Alouffi
Abdulaziz Alouffi Mashal M. Almutairi
Mashal M. Almutairi Tetsuya Tanaka
Tetsuya Tanaka