- 1Department of Zoology, Abdul Wali Khan University Mardan, Khyber Pakhtunkhwa, Pakistan
- 2Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 3King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia
- 4Laboratory of Infectious Diseases, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
- 5Department of Emergency Medicine, Ditmanson Medical Foundation Chia-Yi Christian Hospital, Chiayi, Taiwan
- 6Department of Pathology, Ditmanson Medical Foundation Chia-Yi Christian Hospital, Chiayi, Taiwan
- 7Department of Cosmetic Science, Chia Nan University of Pharmacy and Science, Tainan, Taiwan
- 8Ph.D. Program in Translational Medicine, Rong Hsing Research Center for Translational Medicine, National Chung Hsing University, Taichung, Taiwan
- 9Department of Biotechnology and Bioindustry Sciences, College of Bioscience and Biotechnology, National Cheng Kung University, Tainan, Taiwan
Ticks pose significant threats to hosts by transmitting Borrelia spp., which are grouped into Lyme borreliae, relapsing fever borreliae (RF), and reptiles- and monotremes-associated borreliae. The RF borreliae encompass a group of Borrelia species predominantly transmitted by soft ticks, but some of its members can also be transmitted by hard ticks. Information on the detection and genetic characterization of tick-borne RF borreliae, including Borrelia theileri, is notably rare in Asia, particularly in Pakistan. Herein, we employed molecular techniques to detect borreliae in hard ticks collected from domestic animals in Khyber Pakhtunkhwa, Pakistan. Ticks were subjected to morphological analysis, followed by DNA extraction and PCR amplification of partial fragments of borrelial 16S rRNA and flaB genes. A total of 729 ticks were collected from 264 hosts, with Haemaphysalis cornupunctata (12.9%; 94/729) being the most prevalent, followed by Hyalomma anatolicum (11.7%; 85/729), Rhipicephalus microplus (10.0%; 73/729), Haemaphysalis kashmirensis (9.1%; 66/729), Haemaphysalis bispinosa (8.5%; 62/729), Rhipicephalus sanguineus (8%; 58/729), Haemaphysalis montgomeryi (6.2%; 45/729), Rhipicephalus turanicus (5.5%; 40/729), Hyalomma dromedarii and Ixodes kashmirensis (4.4%; 32/729 each), Rhipicephalus haemaphysaloides (4.1%; 30/729), Haemaphysalis sulcata and Hyalomma scupense (3.8%; 28/729 each), Haemaphysalis danieli (2.9%; 21/729), Hyalomma kumari (2.6%; 19/729), and Hyalomma isaaci (2.2%; 16/729). Based on 16S rRNA detection of Borrelia spp., only R. turanicus yielded positive results, resulting in an overall infection rate of 0.3% (2/160), while using flaB-based detection, four tick species including R. microplus, R. turanicus, Ha. sulcata, and Ha. cornupunctata showed positive results, yielding an overall infection rate of 6.9% (11/160). The amplified DNA fragments of borrelial 16S rRNA and flaB in R. turanicus from goats shared maximum identities of 100 and 99.40% with Borrelia theileri, respectively. Amplified borrelial flaB fragments in R. microplus from cows and sheep displayed 100% identity with B. theileri, while flaB fragments in Ha. cornupunctata and Ha. sulcata from goats revealed identities of 99.32 and 99.75% with undetermined RF Borrelia spp., respectively. Phylogenetic analysis revealed clustering of B. theileri from R. microplus and R. turanicus with the same species, while Borrelia spp. from Ha. cornupunctata and Ha. sulcata with undetermined RF Borrelia spp. Notably, this research marks the first documentation of B. theileri in R. turanicus and the identification of RF Borrelia spp. in Ha. cornupunctata and Ha. sulcata.
Introduction
Ticks are voracious blood feeders of all classes of terrestrial vertebrates (1, 2). They are distributed worldwide and they typically thrive more in warm and humid climates (1–4). Through various means, ticks can harm their vertebrate hosts, including the transmission of pathogens, such as Borrelia spp. (5–7).
Borrelia is a diverse genus of Gram-negative bacteria that act as obligatory parasites (8–10). Borrelia spp., alternate between arthropod vectors, including ticks, and vertebrate hosts such as domestic animals (8, 10). Due to their pathogenicity to vertebrate hosts, including humans, some Borrelia spp., are considered of significant global health concern, as they cause emerging and reemerging infectious diseases (11, 12). With approximately 42 known species, Borrelia spp., are divided into three main categories: Lyme borreliae (LB), relapsing fever borreliae (RF), and reptile-associated (REP) and monotreme associated borreliae (MON) (10, 11, 13–20). Owing to the diversity of spirochetes in the genus Borrelia, there have been proposals to split it into different genera (21, 22), however, this remains a controversial debate (10, 23).
Relapsing fever borreliae are endemic in temperate and tropical regions of the world, including Asia (8, 24). These agents are typically transmitted by soft ticks belonging to the genus Ornithodoros (25, 26). However, some members of RF borreliae including Borrelia theileri, Borrelia miyamotoi and Borrelia lonestari are primarily transmitted by hard ticks (26–29). With approximately 21 known species, RF borreliae are usually categorized into three groups: soft tick relapsing fever (STRF), hard ticks relapsing fever (HTRF), and louse-borne relapsing fever borreliae (6, 26). Relapsing fever borreliae are maintained in enzootic cycles covering birds and mammals (30). With the exception to Borrelia duttonii, which is originally associated with humans, other RF borreliae accidently infect humans.
Among the TBRF borreliae, B. theileri is the causative agent of bovine borreliosis in livestock including cows, goats and sheep (6, 31–34). This bacterium is worldwide distributed and transmitted by various hard tick species, mainly belonging to the genus Rhipicephalus (6, 26, 35).
The pathogenicity of B. theileri in domestic animals is known (36, 37), while there is a notable diversity and abundance of their vertebrate hosts and tick vectors in Pakistan (38–42). Concern exists regarding the negative impacts of this pathogen on the country’s livestock industry, though this impact has yet to be determined. In continuation with our previous studies, which detected REP Borrelia sp. and Borrelia anserina (43, 44) in ticks, the objective of this study was to genetically characterize RF borreliae in hard ticks infesting domestic animals.
Materials and methods
Study area
In toto, 15 districts of Khyber Pakhtunkhwa, including Abbottabad, Bajaur, Buner, Charsadda, Dir Lower, Dir Upper, Haripur, Malakand, Mardan, Mohmand, Nowshera, Peshawar, Shangla, Swabi, and Swat, were included in this study. Google Maps was utilized to determine the precise geographical coordinates of the collection points in the study area, and these information were organized in Microsoft Excel 2016. The land-cover map of the study area was created using ArcGIS version 10.3.1 (ESRI, Redlands, CA, United States; Figure 1).
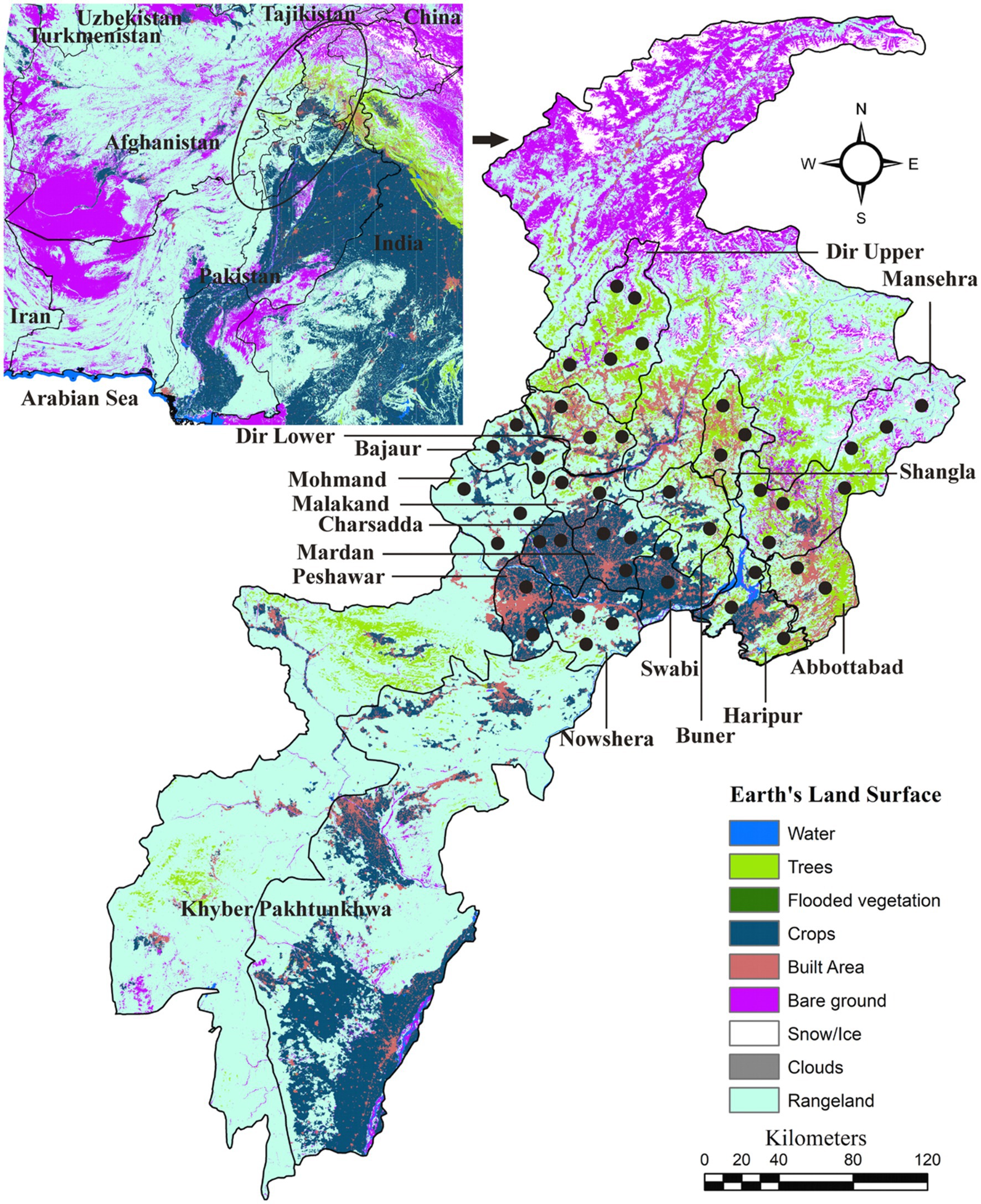
Figure 1. Land-use and land-cover based map showing locations (black circles) where domestic animals were screened for collecting ticks.
Tick collection, preservation, and identification
The study was conducted from March to September 2022, covering three seasons in Pakistan: spring (March–May), summer (June–August), and early autumn (September). Different domestic animals were screened for ticks in farms and grazing fields, when found, ticks were collected using tweezers. Essential information, including the host type, collection date, and collection site coordinates, was recorded during fieldwork. Before preservation in 70% ethanol, all collected ticks were washed with distilled water followed by 70% ethanol. Tick specimens were morphologically identified up to the species level using a stereomicroscope (Stemi 508, Zeiss, Germany) and standard taxonomic identification keys and morphological descriptions (45–51).
DNA extraction and polymerase chain reaction
A subset of 160 specimens, consisting of at least 1 female per tick species per district and 3 nymphs per tick species, underwent DNA extraction. Tick homogenization was performed individually using sterile scissors in 1.5 mL Eppendorf tubes. DNA extraction was carried out using the phenol-chloroform method with minor modifications (52). The extracted DNA was then hydrated by adding 20–30 μL of “nuclease-free” water, and its quantification was analyzed using a NanoDrop spectrophotometer (Nano-Q, Optizen, South Korea).
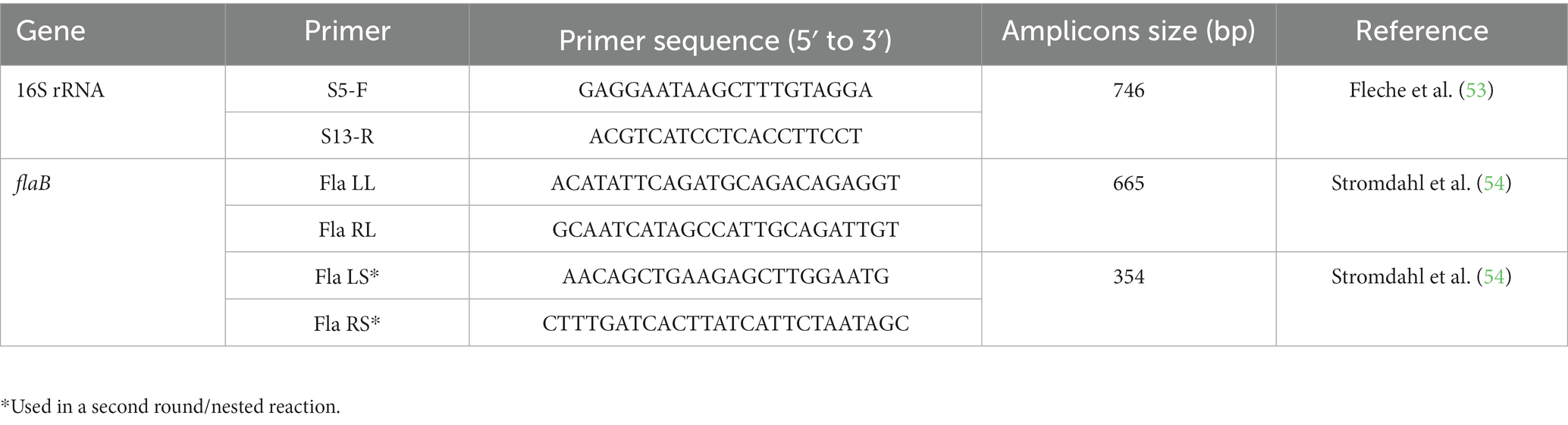
All extracted genomic DNA samples were subjected to a conventional PCR (BIOER, China). Partial fragments of borrelial 16S rRNA were amplified through standard PCR, while borrelial flaB amplification was achieved through nested-PCR. For 16S rRNA and the first round of flaB PCR, the reaction mixture had a total volume of 25 μL and included components: 2 μL of extracted DNA (50–100 ng/μL), 1 μL of each forward and reverse primer (Table 1) at a concentration of 10 pmol/μL, 8.5 μL of nuclease-free PCR water, and 12.5 μL of DreamTaq green MasterMix (2X). The second round of flaB PCR was carried out with 1 μL of the first round PCR product and 9.5 μL of PCR water. All PCRs were conducted under experimental conditions, as previously described (43). PCR water (nuclease-free) was used as a negative control, while DNA of Borrelia sp. from A. gervaisi ticks as a positive control. The amplified fragments were resolved on a 2% agarose gel, visualized using a Gel Documentation system (BioDoc-It™ Imaging Systems, Upland, CA, United States), and purified using the GeneClean II Kit (Qbiogene, Illkirch, France).
DNA sequencing and phylogenetic analysis
The amplicons were sent for DNA sequencing (Macrogen, Inc., Seoul, South Korea) using the Sanger sequencing method with an ABI 373XL system. The raw sequences obtained were then visualized and analyzed using SeqMan version 5.0 (DNASTAR, Inc., Madison, WI, United States) to obtain clean sequences. These clean sequences were subsequently subjected to BLAST (Basic Local Alignment Search Tool) analysis at the NCBI (National Center for Biotechnology Information) to identify the closest matches with sequences already deposited in the GenBank.
The obtained sequences, homologous sequences (downloaded from the BLAST results), and an appropriate outgroup were imported and aligned using the BioEdit alignment editor version 7.0.5 (55) with the ClustalW multiple alignment method (56). The tree topology was adapted from previous studies (10, 13, 16, 20, 43). The alignments were then used in Molecular Evolutionary Genetics Analysis (MEGA-X) (57) to construct phylogenetic trees, utilizing the Maximum Likelihood method with 1,000 bootstrap replicates.
Results
Tick and host description
Of the total collected specimens (729), Haemaphysalis ticks were the most abundant comprising 43.3% (316/729), followed by the Rhipicephalus ticks (27.6%, 201/729), Hyalomma (24.7%, 180/729) and Ixodes ticks (4.4%, 32/729). Within the genus Haemaphysalis, the most abundant species was Haemaphysalis cornupunctata (29.7%; 94/316) followed by Haemaphysalis kashmirensis (20.9%; 66/316), Haemaphysalis bispinosa (19.6%; 62/316), Haemaphysalis montgomeryi (14.2%; 45/316), Haemaphysalis sulcata (8.9%; 28/316), and Haemaphysalis danieli (6.6%; 21/316). Among Rhipicephalus ticks, the most abundant species was Rhipicephalus microplus (36.3%; 73/201) followed by Rhipicephalus sanguineus (28.9%; 58/201), Rhipicephalus turanicus (19.9%; 40/201) and Rhipicephalus haemaphysaloides (14.9%; 30/201). Among Hyalomma ticks, the most abundant species was Hyalomma anatolicum (42.3%; 85/180) followed by Hyalomma dromedarii (17.8%; 32/180), Hyalomma scupense (15.6%; 28/180), Hyalomma kumari (10.6%; 19/180) and Hyalomma isaaci (8.9%; 16/180). The genus Ixodes was represented by only one species, Ixodes kashmiricus (100%; 32/32). Of the total examined domestic hosts (264), the most abundant were goats (28%; 74/264) and sheep (27.3%; 72/264), followed by cows (18.6%; 49/264), dogs and horses (9.5%; 25/264 each), and camels (7.2%; 19/264). With an overall prevalence of infestation (48.5%; 129/266), the highest prevalence was found on goats (56.8%; 42/74) followed by sheep (51.4%; 37/72), dogs (48%; 12/25), cows (42.9%; 21/49), camels (38.1%; 8/19) and horses (36%; 9/25). With an overall tick burden of 2.8 ticks per examined host, the highest tick burden was noted on goats (3.6; 268/74), followed by sheep (3.2; 232/72), cows (2.6; 129/49), dogs (1.7; 43/25), camels (1.5; 28/19), and horses (1.1; 29/25). The details about the life stage, associated hosts, and collection site of each tick species are given in Table 2.
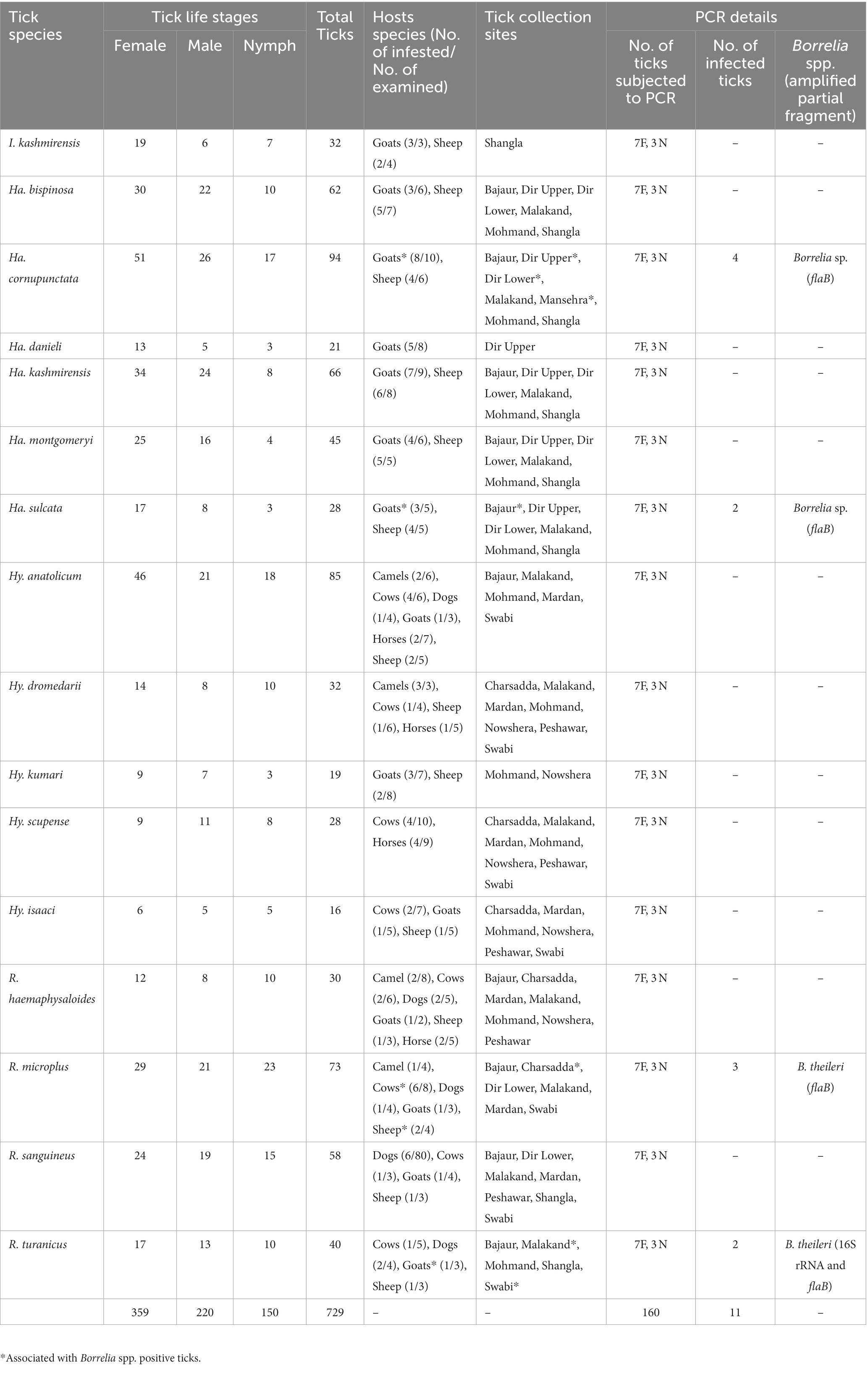
Table 2. Information related to tick species, their hosts and collection sites, as well as PCR results of associated Borrelia spp.
Sequences analyses
The partial fragments of borrelial flaB were amplified in 11 ticks, while partial fragments of borrelial 16S were amplified in 2 flaB-positive ticks. The attempts to amplify 16S rRNA in the remaining 9 flaB-positive ticks were unsuccessful. Overall, 4 sequences (1 forward and 1 reverse per positive tick sample) were obtained for 16S rRNA, while 22 longer sequences (1 forward and 1 reverse per positive tick sample) were obtained for flaB. A subset of sequences of 16S rRNA obtained from the genomic DNA of R. turanicus from goats and cattle were found to be identical, which resulted in consensus sequences of 644 bp. Similarly, all long sequences of flaB within each subset were identical in the following ways: (a) sequences obtained from R. microplus from sheep and cattle were identical, (b) sequences obtained from R. turanicus from goats were identical, (c) sequences obtained from Ha. cornupunctata from goats were identical, and sequences obtained from Ha. sulcata from goats were identical. These four subsets resulted in consensus sequences of 587, 522, 547, and 559 bp, respectively. Besides the long sequences for flaB, 11 short sequences (1 forward per positive tick sample) were obtained from their corresponding positive PCR samples.
Detection of Borrelia spp. in ticks
The consensus sequences of borrelial 16S rRNA obtained from genomic DNA of R. turanicus from goats shared a maximum identity of 100% with B. theileri. The long consensus sequences of borrelial flaB obtained from the same samples also showed a maximum identity of 99.40% with B. theileri. Other long consensus sequences of borrelial flaB obtained from tick’s genomic DNA showed their BLAST identities in the following ways: (a) Borrelia sp. detected in R. microplus from cows and sheep showed a maximum identity of 100% with B. theileri, (b) Borrelia sp. detected in Ha. cornupunctata from goats depicted a maximum identity of 99.32% with a Borrelia sp., and (c) Borrelia sp. detected in Ha. sulcata from goats displayed a maximum identity of 99.75% with Borrelia sp. Among the determined RF borreliae, all these sequences were found close to B. theileri followed by B. lonestari.
Considering 16S rRNA-based detection of Borrelia spp., only R. turanicus was positive, resulting in an overall infection rate of 0.3% (2/160). When considering flaB-based detection of Borrelia spp., four tick species, including R. microplus, R. turanicus, Ha. sulcata, and Ha. cornupunctata were positive, yielding an overall infection rate of 6.9% (11/160). Notably, within the overlapped region, the short and long sequences of flaB were identical, confirming the BLAST identities of each other. However, only long sequences were used in phylogenetic analyses. Table 2 gives details about Borrelia spp., including the associated hosts and the corresponding geographical sites.
The obtained sequences were submitted to GenBank under the following accession numbers: 16S rRNA OR561043 (B. theileri haplotype detected in R. turanicus from goats); and flaB OR574987 (B. theileri haplotype detected in R. turanicus from goats), OR574986 (B. theileri detected in R. microplus from cows and sheep), OR574984 (Borrelia sp. detected in Ha. cornupunctata from goats), OR574985 (Borrelia sp. detected in Ha. sulcata from goats).
Phylogenetic analysis
A phylogenetic tree was obtained based on 16S rRNA, in which B. theileri detected in R. turanicus from goats in the present study clustered with the same species from Egypt and Zambia (Figure 2A). Furthermore, this species appeared in a monophyletic group alongside B. lonestari and B. miyamotoi. Another phylogenetic tree was obtained based on flaB (Figure 2B), revealing the clustering of Borrelia spp. detected in the current study in the following manner. The haplotype of B. theileri found in R. microplus from sheep and cows, as well as another haplotype of B. theileri found in R. turanicus from goats clustered with the corresponding species from Brazil. The Borrelia sp. found in Ha. sulcata from goats clustered with undetermined Borrelia sp. from Brazil, while the Borrelia sp. found in Ha. cornupunctata from goats clustered with undetermined species from Portugal. Additionally, these species also formed a monophyletic group along with B. lonestari and B. miyamotoi.
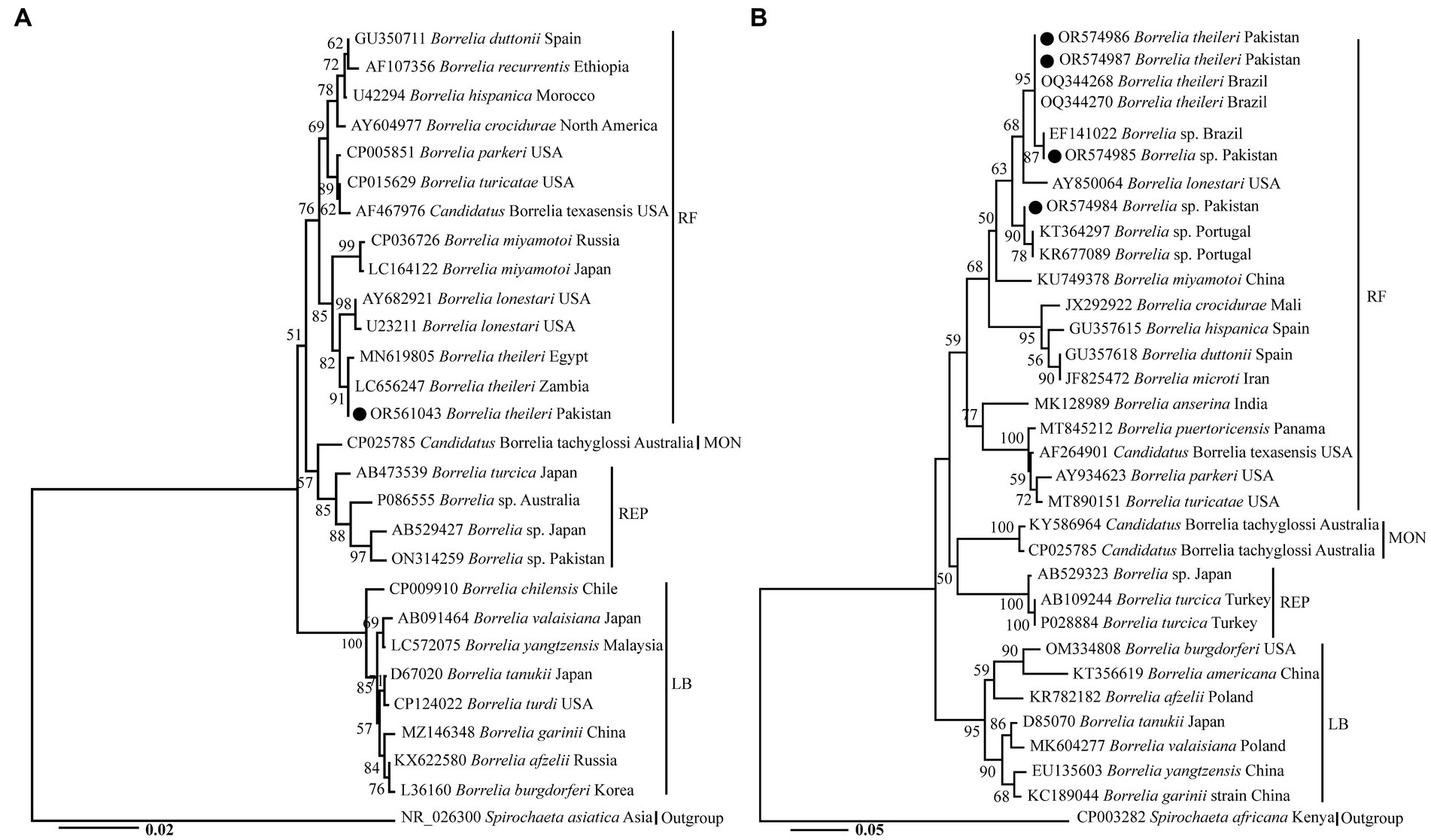
Figure 2. The phylogenetic trees were constructed for Borrelia spp. based on 16S rRNA (A) and flaB (B), with Spirochaeta asiatica, and Spirochaeta africana serving as the outgroups, respectively. The Neighbor-Joining method was utilized with 1,000 replicates for bootstrap analysis. GenBank accession numbers, species names, and geographic locations were assigned to all sequences. Borrelia spp. detected in this study are indicated by a black circle. RF, MON, REP, and LB represent relapsing fever, monotreme-associated, reptile-associated, and Lyme Borreliae, respectively.
Discussion
Compared to LB borreliae, RF borreliae have received less attention globally (26). Similarly, despite reported RF cases (58), the association of borreliae with ticks in in Asia in general and in Pakistan in particular is poorly known. Neglecting RF borreliae can have significant adverse consequences for both public and animal health. To address this knowledge gap, we conducted Borrelia spp. detection in 16/160 hard ticks collected from six different domestic animals across different geographical areas. In addition to detecting B. theileri in R. microplus, our study represents, to the best of our knowledge, the first report of B. theileri in R. turanicus, as well as the detection of Borrelia spp. in Ha. cornupunctata and Ha. sulcata.
In Pakistan, previous studies suggest that Haemaphysalis spp., Hyalomma spp., and Rhipicephalus spp. are commonly associated with domestic animals (38, 40, 42, 45, 59–63). Considering their distribution in previous studies, it can be inferred that Haemaphysalis spp. are prevalent in humid and vegetated areas, Hyalomma species prevalent in dry and desert areas, and Rhipicephalus spp. are abundant in humid and warm areas. Given that the current study area primarily consists of rangeland, cropland, and forested land surfaces, Haemaphysalis spp. and Rhipicephalus spp. were found the most abundant ticks in this study, in contrast to Hyalomma spp. Moreover, the higher abundance of Haemaphysalis and Rhipicephalus ticks could also be associated with their broader host range and greater number of main hosts.
Regarding HTRF borreliae, it is well studied that B. theileri is associated with the genus Rhipicephalus, especially R. microplus (26, 31). Along with R. microplus, the current study also detected this pathogen in R. turanicus, marking the earliest such finding. In contrast, the association of HTRF borreliae with the genus Haemaphysalis is poorly understood. This study and previous related reports (64, 65) suggest that there could be a considerable association between HTRF borreliae and Haemaphysalis ticks, which needs further investigation. Furthermore, apart from being found in hard ticks, previous studies have also detected B. theileri and other closely related undetermined Borrelia spp. in vertebrate hosts, including cattle, goats, and sheep (6, 31, 36). Given the known pathogenicity of B. theileri in domestic animals (26, 36, 37, 66), hard ticks could pose threats to domestic animals in the region. Several other factors in the studied region, including a high tick abundance, a large population of cattle and small ruminants, and their combined farming practices and unmonitored movements, could further exacerbate health threats.
Different analysis based on molecular data are considered powerful tools for the identification of biological species (67, 68). Furthermore, unlike the traditional systematic and taxonomy of TBRF borreliae, which was based on co-speciation of ticks and borreliae (26, 69), the advanced approach relies on molecular evidence (10, 26, 67–71). It is also studied that different molecular markers, including 16S rRNA and flaB are compatible in the case of identification and phylogenetic analysis of TBRF (67, 72). Therefore, 16S rRNA and flaB based molecular data was obtained for Borrelia spp. in the current study, which was subsequently subjected to phylogenetic analysis. The genetic variations in the detected Borrelia spp. could be associated with the difference in their tick hosts in general. Despite their mutual genetic variations, in the BLAST and phylogenetic analysis, these Borrelia spp. exhibited proximity to B. theileri, B. lonestari and B. miyamotoi. The observed closeness could be attributed to their shared niche, while most RF borreliae are associated with soft ticks, these are associated with hard ticks. Candidatus Borrelia texasensis, although considered to be transmitted by hard ticks (26, 73), did not cluster in the mentioned group.
Conclusion
This study not only confirmed the presence of B. theileri in R. microplus but also provided the first documented evidence of B. theileri in R. turanicus collected on cattle and sheep, along with the detection of RF Borrelia spp. in Ha. cornupunctata and Ha. sulcata collected on goats. The study contributes to the expansion of the geographical and tick host range of RF borreliae, which, in turn, could support future research efforts focusing on veterinary health. Further studies should be encouraged to investigate the ticks-borne bovine borreliosis in order to reduce potential risks in the region.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/; OR561043, OR574984-OR574987.
Ethics statement
The animal studies were approved by Ethical permission for conducting this study was granted under the reference number Dir/A&R/AWKUM/2018/1410 by the Advanced Studies and Research Board of Abdul Wali Khan University, Mardan (AWKUM). Oral permissions were obtained from the owners of the animals, and the animals were treated with care and handled gently during the tick collection process. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MK: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MA: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Writing – original draft, Writing – review & editing. ADA: Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. TT: Data curation, Formal analysis, Investigation, Software, Writing – review & editing. S-CC: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. C-CC: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. AIA: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Acknowledgments
The authors acknowledge the financial support provided by the Higher Education Commission (HEC), Pakistan, and Pakistan Science Foundation (PSF). The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia. We are grateful to Marcelo B. Labruna for revising an earlier version of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Guglielmone, AA, Petney, TN, and Robbins, RG. Ixodidae (Acari: Ixodoidea): descriptions and redescriptions of all known species from 1758 to December 31, 2019. Zootaxa. (2020) 4871:1–322. doi: 10.11646/zootaxa.4871.1.1
3. Anderson, JF. The natural history of ticks. Med Clin. (2002) 86:205–18. doi: 10.1016/S0025-7125(03)00083-X
4. Dantas-Torres, F. Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int J Parasitol: Parasites and Wildlife. (2015) 4:452–61. doi: 10.1016/j.ijppaw.2015.07.001
5. Boulanger, N, Boyer, P, Talagrand-Reboul, E, and Hansmann, Y. Ticks and tick-borne diseases. Med Mal Infect. (2019) 49:87–97. doi: 10.1016/j.medmal.2019.01.007
6. Faccini-Martínez, ÁA, Silva-Ramos, CR, Santodomingo, AM, Ramírez-Hernández, A, Costa, FB, Labruna, MB, et al. Historical overview and update on relapsing fever group Borrelia in Latin America. Parasit Vectors. (2022) 15:1–20. doi: 10.1186/s13071-022-05289-5
7. Muñoz-Leal, S, Faccini-Martínez, ÁA, Pérez-Torres, J, Chala-Quintero, SM, Herrera-Sepúlveda, MT, Cuervo, C, et al. Novel Borrelia genotypes in bats from the Macaregua cave, Colombia. Zoonoses Public Health. (2021) 68:12–8. doi: 10.1111/zph.12789
8. Dworkin, MS, Schwan, TG, Anderson, DE, and Borchardt, SM. Tick-borne relapsing fever. Infect Dis Clin. (2008) 22:449–68. doi: 10.1016/j.idc.2008.03.006
9. Lopez, JE, Krishnavahjala, A, Garcia, MN, and Bermudez, S. Tick-borne relapsing fever spirochetes in the Americas. Vet Sci. (2016) 3:16. doi: 10.3390/vetsci3030016
10. Margos, G., Fingerle, V., Cutler, S., Gofton, A., Stevenson, B., and Estrada-Peña, A., (2020). Controversies in bacterial taxonomy: The example of the genus Borrelia. Ticks and Tick-borne Diseases, 11, 101335.
11. Fesler, MC, Shah, JS, Middelveen, MJ, Du Cruz, I, Burrascano, JJ, and Stricker, RB. Lyme disease: diversity of Borrelia species in California and Mexico detected using a novel immunoblot assay. Dent Health. (2020) 8:097. doi: 10.3390/healthcare8020097
12. Ogden, NH, Artsob, H, Margos, G, Tsao, J, Sonenshine, DE, and Roe, M. Non-rickettsial tick-borne bacteria and the diseases they cause. Biolog Ticks. (2011) 2:278–312.
13. Binetruy, F, Garnier, S, Boulanger, N, Talagrand-Reboul, E, Loire, E, Faivre, B, et al. A novel Borrelia species, intermediate between Lyme disease and relapsing fever groups, in neotropical passerine-associated ticks. Sci Rep. (2020) 10:10596. doi: 10.1038/s41598-020-66828-7
14. Gupta, RS. Distinction between Borrelia and Borreliella is more robustly supported by molecular and phenotypic characteristics than all other neighbouring prokaryotic genera: response to Margos' et al. PloS One. (2019) 14:e0221397. doi: 10.1371/journal.pone.0221397
15. Kaenkan, W, Nooma, W, Chelong, IA, Baimai, V, Trinachartvanit, W, and Ahantarig, A. Reptile-associated Borrelia spp. in Amblyomma ticks, Thailand. Ticks Tick-borne Dis. (2020) 11:101315. doi: 10.1016/j.ttbdis.2019.101315
16. Margos, G, Gofton, A, Wibberg, D, Dangel, A, Marosevic, D, Loh, SM, et al. The genus Borrelia reloaded. PloS One. (2018) 13:e0208432. doi: 10.1371/journal.pone.0208432
17. Panetta, JL, Šíma, R, Calvani, NE, Hajdušek, O, Chandra, S, Panuccio, J, et al. Reptile-associated Borrelia species in the goanna tick (Bothriocroton undatum) from Sydney, Australia. Parasit Vectors. (2017) 10:1–13. doi: 10.1186/s13071-017-2579-5
18. Takano, A, Goka, K, Une, Y, Shimada, Y, Fujita, H, Shiino, T, et al. Isolation and characterization of a novel Borrelia group of tick-borne borreliae from imported reptiles and their associated ticks. Environ Microbiol. (2010) 12:134–46. doi: 10.1111/j.1462-2920.2009.02054.x
19. Margos, G, Wormser, GP, Schwartz, I, Markowicz, M, Henningsson, AJ, Lienhard, R, et al. Evidence of taxonomic bias in public databases: the example of the genus Borrelia. Ticks Tick-borne Dis. (2022) 13:101994. doi: 10.1016/j.ttbdis.2022.101994
20. Gofton, AW, Popa-Baez, A, Takano, A, Soennichsen, K, Michie, M, Short, M, et al. Characterisation and comparative genomics of three new Varanus-associated Borrelia spp. from Indonesia and Australia. Parasit Vectors. (2023) 16:1–15. doi: 10.1186/s13071-023-05937-4
21. Adeolu, M, and Gupta, RS. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. Nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek. (2014) 105:1049–72. doi: 10.1007/s10482-014-0164-x
22. Barbour, AG, and Gupta, RS. The family Borreliaceae (Spirochaetales), a diverse group in two genera of tick-borne spirochetes of mammals, birds, and reptiles. J Med Entomol. (2021) 58:1513–24. doi: 10.1093/jme/tjab055
23. Margos, G, Marosevic, D, Cutler, S, Derdakova, M, Diuk-Wasser, M, Emler, S, et al. There is inadequate evidence to support the division of the genus Borrelia. Int J Syst Evol Microbiol. (2017) 67:1081–4. doi: 10.1099/ijsem.0.001717
24. Jakab, Á, Kahlig, P, Kuenzli, E, and Neumayr, A. Tick borne relapsing fever-a systematic review and analysis of the literature. PLoS Negl Trop Dis. (2022) 16:e0010212. doi: 10.1371/journal.pntd.0010212
25. Cutler, SJ. Relapsing fever borreliae: a global review. Clin Lab Med. (2015) 35:847–65. doi: 10.1016/j.cll.2015.07.001
26. Talagrand-Reboul, E, Boyer, PH, Bergström, S, Vial, L, and Boulanger, N. Relapsing fevers: neglected tick-borne diseases. Front Cell Infect Microbiol. (2018) 8:98. doi: 10.3389/fcimb.2018.00098
27. Fukunaga, M, Okada, K, Nakao, M, Konishi, T, and Sato, Y. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int J Syst Evol Microbiol. (1996) 46:898–905. doi: 10.1099/00207713-46-4-898
28. Burkot, TR, Mullen, GR, Anderson, R, Schneider, BS, Happ, CM, and Zeidner, NS. Borrelia lonestari DNA in adult Amblyomma americanum ticks, Alabama. Emerg Infect Dis. (2001) 7:471–3. doi: 10.3201/eid0703.017323
29. Lambert, JS, Cook, MJ, Healy, JE, Murtagh, R, Avramovic, G, and Lee, SH. Metagenomic 16S rRNA gene sequencing survey of Borrelia species in Irish samples of Ixodes ricinus ticks. PloS One. (2019) 14:e0209881. doi: 10.1371/journal.pone.0209881
30. Schwan, TG, and Piesman, J. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg Infect Dis. (2002) 8:115–21. doi: 10.3201/eid0802.010198
31. McCoy, BN, Maïga, O, and Schwan, TG. Detection of Borrelia theileri in Rhipicephalus geigyi from Mali. Ticks Tick-borne Dis. (2014) 5:401–3. doi: 10.1016/j.ttbdis.2014.01.007
32. Morel, N, De Salvo, MN, Cicuttin, G, Rossner, V, Thompson, CS, Mangold, AJ, et al. The presence of Borrelia theileri in Argentina. Vet Parasitol: Regional Stud Reports. (2019) 17:100314. doi: 10.1016/j.vprsr.2019.100314
33. Qiu, Y, Squarre, D, Nakamura, Y, Lau, AC, Moonga, LC, Kawai, N, et al. Evidence of Borrelia theileri in wild and domestic animals in the Kafue ecosystem of Zambia. Microorganisms. (2021) 9:2405. doi: 10.3390/microorganisms9112405
34. Theiler, A. Spirillosis of cattle. J Compar Pathol Therapeutics. (1904) 17:47–55. doi: 10.1016/S0368-1742(04)80003-1
35. Elelu, N. Tick-borne relapsing fever as a potential veterinary medical problem. Vet Med Sci. (2018) 4:271–9. doi: 10.1002/vms3.108
36. Callow, LL. Observations on tick-transmitted spirochaetes of cattle in Australia and South Africa. Br Vet J. (1967) 123:492–7. doi: 10.1016/S0007-1935(17)39704-X
37. Paula, WVDF, Neves, LC, de Paula, LGF, Serpa, MCDA, de Oliveira, FP, Dantas-Torres, F, et al. First molecular detection of Borrelia theileri subclinical infection in a cow from Brazil. Vet Res Commun. (2023) 47:963–7. doi: 10.1007/s11259-022-10020-x
38. Ali, A, Shehla, S, Zahid, H, Ullah, F, Zeb, I, Ahmed, H, et al. Molecular survey and spatial distribution of Rickettsia spp. in ticks infesting free-ranging wild animals in Pakistan (2017–2021). Pathogens. (2022) 11:162. doi: 10.3390/pathogens11020162
39. Aneela, A, Almutairi, MM, Alouffi, A, Ahmed, H, Tanaka, T, da Silva Vaz, I, et al. Molecular detection of Rickettsia hoogstraalii in Hyalomma anatolicum and Haemaphysalis sulcata: updated knowledge on the epidemiology of tick-borne Rickettsia hoogstraalii. Vet Sci. (2023) 10:605. doi: 10.3390/vetsci10100605
40. Karim, S, Budachetri, K, Mukherjee, N, Williams, J, Kausar, A, Hassan, MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl Trop Dis. (2017) 11:e0005681. doi: 10.1371/journal.pntd.0005681
41. Ullah, S, Alouffi, A, Almutairi, MM, Islam, N, Rehman, G, Ul Islam, Z, et al. First report of Rickettsia conorii in Hyalomma kumari ticks. Animals. (2023) 13:1488. doi: 10.3390/ani13091488
42. Tila, H., Khan, M., Almutairi, M. M., Alouffi, A., Ahmed, H., Tanaka, T., et al., (2023). First report on detection of Hepatozoon ayorgbor in Rhipicephalus haemaphysaloides and Hepatozoon colubri in Haemaphysalis sulcata and Hyalomma anatolicum: risks of spillover of Hepatozoon spp. from wildlife to domestic animals. Frontiers in Veterinary Science, 10.
43. Khan, M, Islam, N, Khan, A, Islam, ZU, Muñoz-Leal, S, Labruna, MB, et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia sp. Ticks Tick-borne Dis. (2022) 13:102047. doi: 10.1016/j.ttbdis.2022.102047
44. Zahid, H, Alouffi, A, Almutairi, MM, Ateeq, M, Tanaka, T, Chang, SC, et al. Argas persicus and Carios vespertilionis ticks infesting ducks, domestic fowls and bats in Pakistan: first report on molecular survey and phylogenetic position of Borrelia anserina. Vet Sci. (2023) 10:628. doi: 10.3390/vetsci10100628
45. Alam, S, Khan, M, Alouffi, A, Almutairi, MM, Ullah, S, Numan, M, et al. Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms. (2022) 10:1663. doi: 10.3390/microorganisms10081663
46. Apanaskevich, DA. Differentiation of closely related species Hyalomma anatolicum and H. excavatum (Acari: Ixodidae) based on a study of all life cycle stages, throughout entire geographical range. Parazitologiia. (2003) 37:259–80.
47. Hoogstraal, H, and Kaiser, MN. Observations on Egyptian Hyalomma ticks (Ixodoidea, Ixodidae). 5. Biological notes and differences in identity of H. Anatolicum and its subspecies anatolicum Koch and excavatum Koch among Russian and other workers. Identity of H. lusitanicum Koch. Ann Entomol Soc Am. (1959) 52:243–61. doi: 10.1093/aesa/52.3.243
48. Kamran, K, Ali, A, Villagra, C, Siddiqui, S, Alouffi, AS, and Iqbal, A. A cross-sectional study of hard ticks (acari: ixodidae) on horse farms to assess the risk factors associated with tick-borne diseases. Zoonoses Public Health. (2021) 68:247–62. doi: 10.1111/zph.12809
49. Nosek, J, and Sixl, W. Central-European ticks (Ixodoidea). Mitt Abt Zool Landesmus Joanneum. (1972) 1:480.
50. Numan, M, Islam, N, Adnan, M, Zaman Safi, S, Chitimia-Dobler, L, Labruna, MB, et al. First genetic report of Ixodes kashmiricus and associated Rickettsia sp. Parasit Vectors. (2022) 15:1–12. doi: 10.1186/s13071-022-05509-y
51. Walker, JB, Keirans, JE, and Horak, IG. The genus Rhipicephalus (Acari, Ixodidae): A guide to the brown ticks of the world. Cambridge: Cambridge University Press (2000).
52. Sambrook, J, Fritsch, EF, and Maniatis, T. Molecular cloning: A laboratory manual. Ed. 2 ed. US: Cold Spring harbor Laboratory Press (1989).
53. Fleche, AL, Postic, D, Girardet, K, Peter, O, and Baranton, G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Evol Microbiol. (1997) 47:921–5. doi: 10.1099/00207713-47-4-921
54. Stromdahl, EY, Williamson, PC, Kollars, TM Jr, Evans, SR, Barry, RK, Vince, MA, et al. Evidence of Borrelia lonestari DNA in Amblyomma americanum (Acari: Ixodidae) removed from humans. J Clin Microbiol. (2003) 41:5557–62. doi: 10.1128/JCM.41.12.5557-5562.2003
55. Hall, T, Biosciences, I, and Carlsbad, C. BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
56. Thompson, JD, Higgins, DG, and Gibson, TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
57. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
58. Larsson, C, Andersson, M, and Bergström, S. Current issues in relapsing fever. Curr Opin Infect Dis. (2009) 22:443–9. doi: 10.1097/QCO.0b013e32832fb22b
59. Ahmad, I, Ullah, S, Alouffi, A, Almutairi, MM, Khan, M, Numan, M, et al. Description of male, redescription of female, host record, and phylogenetic position of Haemaphysalis danieli. Pathogens. (2022) 11:1495. doi: 10.3390/pathogens11121495
60. Ali, A, Numan, M, Ullah, S, Khan, M, and Kamran, K. Genetic characterization of Haemaphysalis (Rhipistoma) indica and Haemaphysalis (Segalia) montgomeryi ticks (Ixodoidea: Ixodidae). Ticks Tick-borne Dis. (2023) 14:102105. doi: 10.1016/j.ttbdis.2022.102105
61. Ali, A, Obaid, MK, Almutairi, M, Alouffi, A, Numan, M, Ullah, S, et al. Molecular detection of Coxiella spp. in ticks (Ixodidae and Argasidae) infesting domestic and wild animals: with notes on the epidemiology of tick-borne Coxiella burnetii in Asia. Front Microbiol. (2023) 14:1229950. doi: 10.3389/fmicb.2023.1229950
62. Khan, SM, Khan, M, Alouffi, A, Almutairi, MM, Numan, M, Ullah, S, et al. Phylogenetic position of Haemaphysalis kashmirensis and Haemaphysalis cornupunctata, with notes on Rickettsia spp. Gen. (2023) 14:360. doi: 10.3390/genes14020360
63. Shehla, S, Ullah, F, Alouffi, A, Almutairi, MM, Khan, Z, Tanaka, T, et al. Association of SFG Rickettsia massiliae and Candidatus Rickettsia shennongii with different hard ticks infesting livestock hosts. Pathogens. (2023) 12:1080. doi: 10.3390/pathogens12091080
64. Furuno, K, Lee, K, Itoh, Y, Suzuki, K, Yonemitsu, K, Kuwata, R, et al. Epidemiological study of relapsing fever borreliae detected in Haemaphysalis ticks and wild animals in the western part of Japan. PloS One. (2017) 12:e0174727. doi: 10.1371/journal.pone.0174727
65. Lee, K, Takano, A, Taylor, K, Sashika, M, Shimozuru, M, Konnai, S, et al. A relapsing fever group Borrelia sp. similar to Borrelia lonestari found among wild sika deer (Cervus nippon yesoensis) and Haemaphysalis spp. ticks in Hokkaido, Japan. Ticks Tick-borne Dis. (2014) 5:841–7. doi: 10.1016/j.ttbdis.2014.06.006
66. Bishop, GC. Borrelia theileri infection In: JAW Coetzer, GR Thomson, and RC Tustin, editors. Infectious diseases of livestock. Oxford: Oxford University Press (1994). 992–3.
67. Madden, T. The BLAST sequence analysis tool In: McEntyre, J, and Ostell, J, editors. The NCBI handbook. Bethesda, MD, USA: The NCBI Handbook. National Center for Biotechnology Information. (2003).
68. McGinnis, S, and Madden, TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. (2004) 32:W20–5. doi: 10.1093/nar/gkh435
69. Wang, G, and Schwartz, I. Genus II. Borrelia Swellengrebel 1907, 582AL In: DJ Brenner, NR Krieg, GM Garrity, and JT Staley, editors. Bergey's manual of systematic bacteriology, vol. 3. 2nd ed. New York, NY: Springer (2011). 484–98.
70. Maiden, MC, Van Rensburg, MJJ, Bray, JE, Earle, SG, Ford, SA, Jolley, KA, et al. Controversies in bacterial taxonomy: the example of the genus Borrelia. Ticks Tick-borne Dis. (2020) 11:101335. doi: 10.1016/j.ttbdis.2019.101335
71. Fingerle, V, Pritsch, M, Wächtler, M, Margos, G, Ruske, S, Jung, J, et al. "Candidatus Borrelia kalaharica" detected from a febrile traveller returning to Germany from vacation in southern Africa. PLoS Negl Trop Dis. (2016) 10:e0004559. doi: 10.1371/journal.pntd.0004559
72. Oshaghi, MA, Rafinejad, J, Choubdar, N, Piazak, N, Vatandoost, H, Telmadarraiy, Z, et al. Discrimination of relapsing fever Borrelia persica and Borrelia microtti by diagnostic species-specific primers and polymerase chain reaction–restriction fragment length polymorphism. Vector-Borne Zoonotic Dis. (2011) 11:201–7. doi: 10.1089/vbz.2009.0170
Keywords: hard ticks, Borrelia, Borrelia theileri, domestic animals, Pakistan
Citation: Khan M, Almutairi MM, Alouffi A, Tanaka T, Chang S-C, Chen C-C and Ali A (2023) Molecular evidence of Borrelia theileri and closely related Borrelia spp. in hard ticks infesting domestic animals. Front. Vet. Sci. 10:1297928. doi: 10.3389/fvets.2023.1297928
Edited by:
Münir Aktaş, Firat University, TürkiyeReviewed by:
Sara Savic, Scientific Veterinary Institute Novi Sad, SerbiaBenjamin Cull, University of Minnesota Twin Cities, United States
Copyright © 2023 Khan, Almutairi, Alouffi, Tanaka, Chang, Chen and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, dW9wX2FsaUB5YWhvby5jb20=; Shun-Chung Chang, MDc0NjBAY3ljaC5vcmcudHc=
 Mehran Khan
Mehran Khan Mashal M. Almutairi
Mashal M. Almutairi Abdulaziz Alouffi
Abdulaziz Alouffi Tetsuya Tanaka
Tetsuya Tanaka Shun-Chung Chang5*
Shun-Chung Chang5* Chien-Chin Chen
Chien-Chin Chen Abid Ali
Abid Ali