
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 18 September 2023
Sec. Parasitology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1255482
This article is part of the Research Topic Innovative Approaches for Precise Identification and Control of Ticks and Tick-Borne Pathogens View all 10 articles
 Hadia Tila1
Hadia Tila1 Mehran Khan1
Mehran Khan1 Mashal M. Almutairi2
Mashal M. Almutairi2 Abdulaziz Alouffi3
Abdulaziz Alouffi3 Haroon Ahmed4
Haroon Ahmed4 Tetsuya Tanaka5
Tetsuya Tanaka5 Kun-Hsien Tsai6*
Kun-Hsien Tsai6* Abid Ali1*
Abid Ali1*This study aimed to detect Hepatozoon spp. in ticks infesting asymptomatic domestic animals and to provide insight into their potential spillover from wild to domestic animals. In total, 537 tick specimens were collected in Khyber Pakhtunkhwa, Pakistan, and morphologically identified. The most prevalent tick species was Haemaphysalis cornupunctata (69; 12.8%), followed by Haemaphysalis kashmirensis (62; 11.5%), Rhipicephalus microplus (58; 10.8%), Haemaphysalis montgomeryi (51; 9.5%), Rhipicephalus sanguineus (49; 9.1%), each Haemaphysalis bispinosa and Haemaphysalis sulcata (43; 8.0%), each Hyalomma anatolicum and Rhipicephalus turanicus (37; 6.9%), Rhipicephalus haemaphysaloides (33; 6.1%) Hyalomma scupense (30; 5.6%), and Hyalomma isaaci (25; 4.7%). The extracted DNA from a subset of each tick species was subjected to PCR to amplify 18S rRNA fragments of Hepatozoon spp. By BLAST analysis, the Hepatozoon sp. detected in Hy. anatolicum infesting cows and in Ha. sulcata infesting sheep showed 99.7% maximum identity with Hepatozoon colubri. Similarly, the Hepatozoon sp. detected in R. haemaphysaloides infesting goats shared 99.49% maximum identity with Hepatozoon ayorgbor, and the Hepatozoon sp. detected in R. sanguineus infesting dogs exhibited 99.7% identity with Hepatozoon canis. Having an overall infection rate (9.3%; 16/172), the highest infection rate was recorded for each H. canis, and H. colubri (3.5%; 6/172), followed by H. ayorgbor (2.3%; 4/172). In the phylogenetic tree, H. colubri clustered with corresponding species from Iran, H. ayorgbor clustered with the same species from Croatia, Ghana, and Portugal, and H. canis clustered with the conspecifics from Iran, Israel, Romania, and Zambia. Regarding the potential spillover of Hepatozoon spp. from wildlife through ticks, free ranging animals was at higher risk compared to confined animals (RR = 3.05), animals consuming food from wildlife habitats were at higher risk compared to those consuming domestic food (RR = 3.06), and animals residing in farm buildings located in wildlife habitats were at higher risk compared to those residing in farm buildings located in villages (RR = 3.28). In addition to the first report on H. canis in R. sanguineus in Pakistan, this is the earliest data showing H. ayorgbor in R. haemaphysaloides and H. colubri in Ha. sulcata and Hy. anatolicum. These preliminary findings suggest a potential spillover of Hepatozoon spp. from wild to domestic animals via ticks under certain risk factors.
Hepatozoon is a diverse genus of apicomplexan, primarily comprised of haemoparasites transmitted by arthropods, especially by ticks to all classes of terrestrial vertebrates, including wild and domestic animals (1). Hepatozoon is comprised of more than 300 apicomplexan haemoparasites, with ~50 species in mammals and 130 species in snakes (1–3). This genus was assigned to Haemogregarinidae prior to allocating to the family Hepatozoidae (4). Due to encompassing some distinct lineages, certain studies have proposed that Hepatozoon spp. may not belong to a single genus (5, 6). Hepatozoon spp. are globally distributed depending on multiple factors, including the abundance of vertebrate and arthropod hosts (1). With a heteroxenous life cycle, Hepatozoon spp. exhibit sporogonic development and oocyst generation occurs in arthropods, while merogony and gametogony in their vertebrate hosts (1, 7, 8).
As intermediate hosts, Hepatozoon spp. infect all classes of terrestrial vertebrates, including wild and domestic animals. With low host specificity, depending on the species, some Hepatozoon spp. are more prevalent in a particular group of hosts (9). For instance, Hepatozoon ayorgbor largely infects wild snakes and wild rodents, whereas Hepatozoon colubri mainly infects wild snakes, while Hepatozoon canis primarily infects domestic and wild canids. However, Hepatozoon spp. can cross-over from their specific wild hosts to unnatural wild and domestic hosts through different agents such as ticks (10–13). In the case of spillover, Hepatozoon spp. are considered more pathogenic in unnatural hosts compared to their specific hosts (14–16).
Hematophagous arthropods get infected with Hepatozoon spp. by feeding on vertebrate hosts and subsequently act as the definitive host and vectors (17). For instance, mosquitoes serve as principal vectors for H. ayorgbor (1, 18), while Rhipicephalus sanguineus is the main vector for H. canis (1, 19, 20). The principal vectors of many Hepatozoon spp., such as H. colubri, are not well-established yet. Unlike other tick-borne pathogens which are mainly transmitted during tick bite, Hepatozoon spp. are transmitted when vertebrate hosts ingest infected ticks (21, 22). Other transmission routes include via predation of prey, such as H. ayorgbor, transmission to snakes through feeding on rodents (18), and vertical transmission such as H. canis in R. sanguineus (23, 24).
The morphometrics of the peripheral blood gamonts seen in the hosts' blood smears were previously the most widely employed techniques for the identification of Hepatozoon spp. However, the limited number of apparent structures and few morphological features make it difficult and unreliable to differentiate Hepatozoon spp. (25). A sensitive method for detecting Hepatozoon spp. is molecular analysis based on the amplification and sequencing of 18S rRNA fragments (26, 27). Furthermore, ticks have been largely neglected as vectors of Hepatozoon spp. and little data is available on Hepatozoon spp. of wildlife origin in ticks infesting domestic animals. Therefore, the purpose of this study was to detect Hepatozoon spp. in ticks infesting domestic animals and to provide insight into their possible spillover.
The Advanced Studies and Research Board of Abdul Wali Khan University, Mardan (AWKUM) granted ethical permission (Dir/A&R/AWKUM/2018/1410) for the conduction of this study. Additionally, the owners of the animals also gave their oral permission to collect ticks from their animals.
This study was conducted in four districts of Khyber Pakhtunkhwa, including Bajaur (34.7865° N, 71.5249° E), Malakand (34.5030° N, 71.9046° E), Mohmand (34.5356° N, 71.2874° E) in the north-western and Lakki Marwat (32.6135° N, 70.9012° E) in the south-eastern. Global Positioning System was used for finding the actual coordinates of collection points in the studied area and subsequently used in designing the map using ArcGIS v 10.3.1 (ESRI, Redlands, CA, USA; Figure 1).
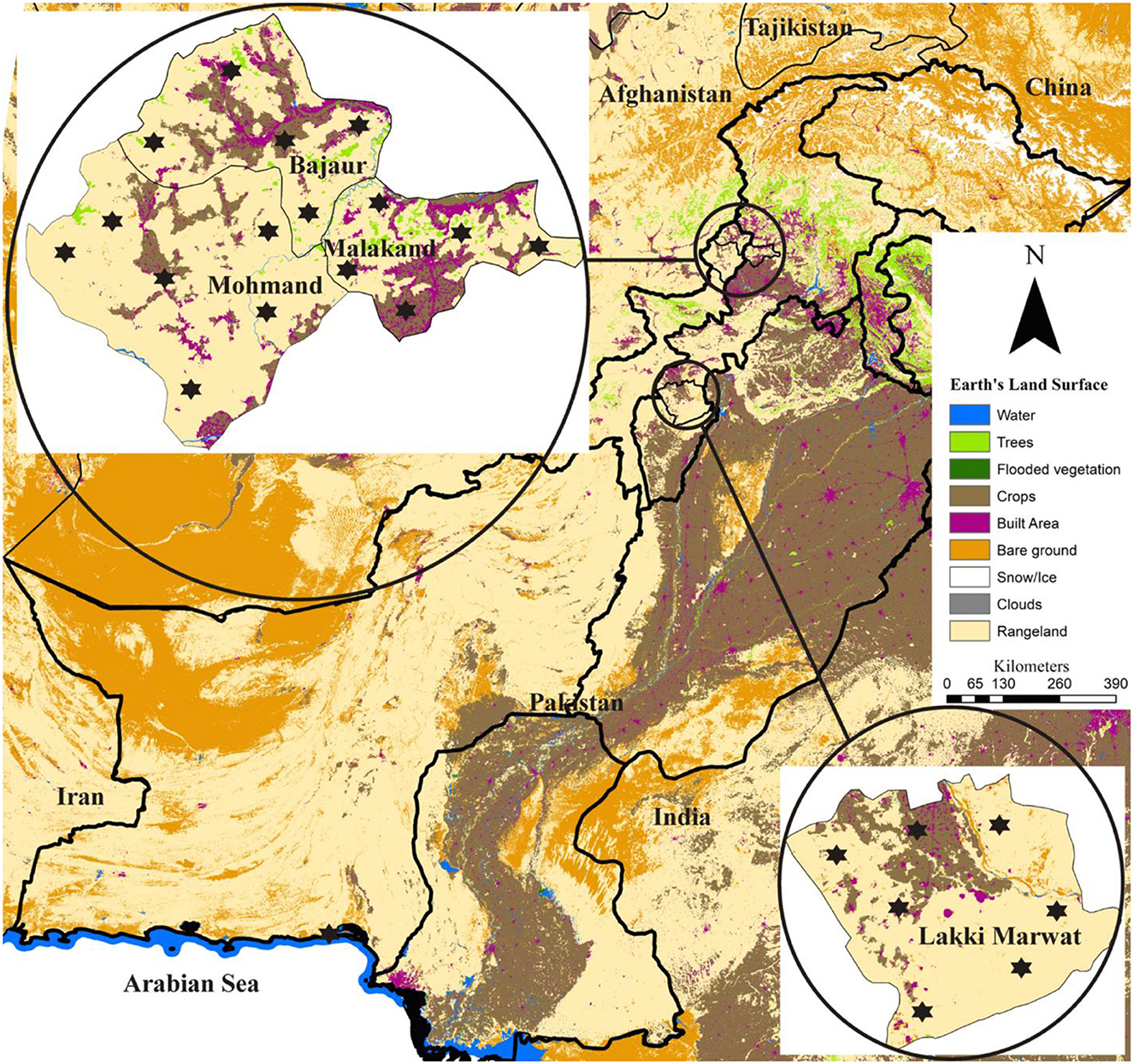
Figure 1. Locations (marked by an asterisk) where domestic hosts were examined for ticks are shown on the land-use and land-cover based map.
Ticks were randomly collected using forceps from different asymptomatic domestic animals, including cows, livestock guardian dogs, goats, and sheep from March 2022 to September 2022. The appropriate information was recorded during tick collection, including the host type, collection date, and collection site, as well as general observations about the field. Furthermore, the owners of domestic animals were given a questionnaire to complete in order to gather possible risk factors for Hepatozoon spp. spillover. The questionnaire consisted of closed-ended questions about host-related details such as gender, age, living status, farming type, food supply, and farm building location, nature, and its altitude. It also contained open-ended questions regarding wildlife and anthropogenic activities such as land use and land cover changes, and human-wildlife conflict. Ticks were stored in a mixture of 95% ethanol, 4% distilled water, and 1% glycerol in properly labeled tubes after cleaning with distilled water followed by 70% ethanol. Using a stereomicroscope (Stemi 508, Zeiss, Germany), tick specimens were morphologically identified based on various morphological features, and validated up to the species level using the standard taxonomic identification keys (28–32).
A sum of 172 tick specimens (one adult female, one male, and two nymphs per species per district) were selected and subjected to DNA extraction individually. Prior to DNA extraction using the phenol-chloroform method with minor modifications (33), the homogenization of ticks was individually carried out using a hygienic scissor coupled with a sterile pestle in a 1.5 ml Eppendorf tube. The extracted DNA pellet was hydrated with 20–30 μl (depending on the tick size) of “nuclease-free” water and subjected to NanoDrop spectrophotometer (Nano-Q, Optizen, South Korea) to measure its quality and quantity.
For the amplification of 18S rRNA fragments of Hepatozoon spp., the extracted genomic DNA samples were subjected to conventional PCR (BIOER, China) using specific primers: HEP2 144-169-F (5′-GGTAATTCTAGAGCTAATACATGAGC-3′) and HEP2 743-718-R (5′-ACAATAA AGTA AAAAACAYTTCAAAG-3′), and PCR experimental conditions were set as previously described (34). Each PCR reaction of 25 μL volume contained the following components: 8.5 μL of PCR water “nuclease free,” 1 μL of each primer with a concentration of 10 pmol/μL, 2 μL of extracted DNA (50–100 ng/μL), and 12.5 μL DreamTaq green MasterMix (2X). All PCR procedures employed PCR water as a negative control and H. canis DNA as a positive control. The obtained amplicons were run on a 2% agarose gel followed by observing via Gel Documentation (BioDoc-It™ Imaging Systems, Upland, CA, USA) and purified using the GeneClean II Kit (Qbiogene, Illkirch, France).
The purified amplicons were submitted for DNA sequencing (Macrogen, Inc., Seoul, South Korea) by Sanger sequencing method using an ABI 373XL system. Low-quality nucleotide sequences and contaminated reads were eliminated during trimming and assembling of raw sequences through SeqMan version 5.0 (DNASTAR; DNASTAR, Inc., Madison, WI, USA). With the aim to determine the closest identities with other sequences previously deposited in the GenBank, the cleaned sequences were subjected to BLAST (Basic Local Alignment Search Tool) at NCBI (National Center for Biotechnology Information). For the subsequent phylogenetic tree using Molecular Evolutionary Genetics Analysis (MEGA-X) (35), the homologous sequences found in the BLAST results were downloaded in FASTA format. These sequences along with a suitable outgroup were aligned in BioEdit alignment editor v 7.0.5 (36) using ClustalW Multiple alignment (37). The alignment was used for constructing a phylogenetic tree with 1,000 bootstrap replicates under Maximum Likelihood method.
The questionnaire data along with PCR data were collected and assembled in spreadsheets using Excel (Microsoft V, 2016). Using GraphPad Prism version 5.0 (GraphPad Software Inc., San Diego, CA, USA), the assembled data was subjected to chi-square test (χ2), selecting significance at P < 0.05, and relative risk (RR), with the 95% confidence interval.
Altogether, 537 ticks were morphologically identified, which were belonging to three distinct genera of hard ticks, including Haemaphysalis, Hyalomma, and Rhipicephalus. Herein, majority of the ticks were Haemaphysalis cornupunctata (69, 12.8%), followed by Haemaphysalis kashmirensis (62, 11.5%), Rhipicephalus microplus (58, 10.8%), Haemaphysalis montgomeryi (51, 9.5%), R. sanguineus (49, 9.1%), each Haemaphysalis bispinosa and Haemaphysalis sulcata (43, 8.0%), each Hyalomma anatolicum and Rhipicephalus turanicus (37, 6.9%), Rhipicephalus haemaphysaloides (33, 6.1%) Hyalomma scupense (30, 5.6%), and Hyalomma isaaci (25, 4.7%). The highest prevalence of tick infestation was recorded on dogs (68.4%: 13/19), followed by cows (64.7%: 22/34), goats (58%: 29/50), and sheep (56.3%: 27/48), resulting in overall prevalence (60.9%; 92/151). The details about each tick species, including life stage, host association, and collection site, are provided in Table 1.
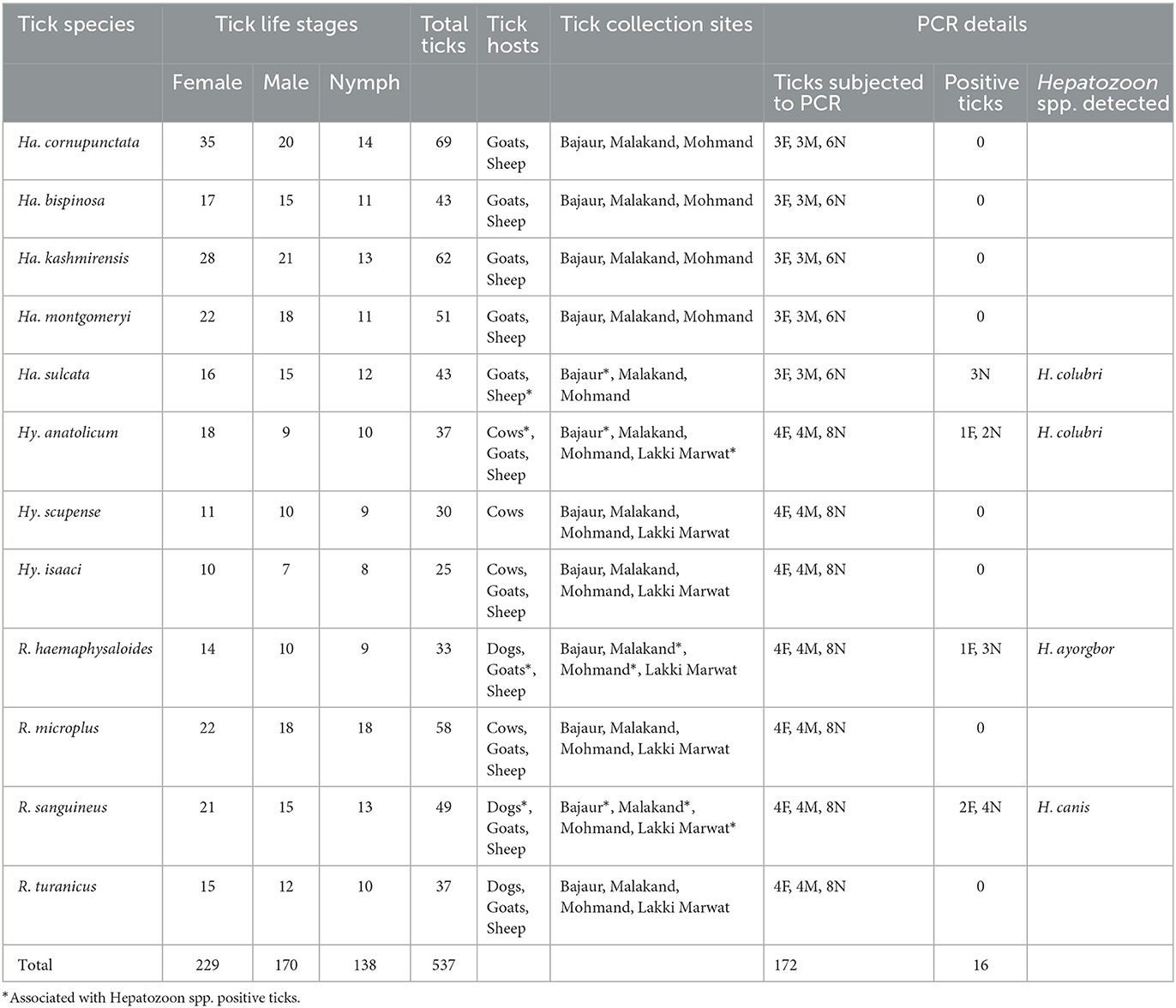
Table 1. Data about tick species, associated Hepatozoon spp., corresponding vertebrate hosts, locality, and molecular analysis.
According to BLAST analysis, Hepatozoon sp. detected in Hy. anatolicum from cows and Ha. sulcata from sheep shared a maximum identity of 99.66% with Hepatozoon colubri, whereas Hepatozoon sp. detected in R. haemaphysaloides from goats revealed a maximum identity of 99.49% with Hepatozoon ayorgbor, and Hepatozoon sp. detected in R. sanguineus from dogs showed a maximum identity of 99.66% with Hepatozoon canis. With a total infection rate (9.3%, 16/172), the highest rate was recorded for each H. canis, and H. colubri (3.5%, 6/172), followed by H. ayorgbor (2.3%, 4/172). Table 1 provides additional information regarding Hepatozoon spp., including the associated vertebrate hosts and the corresponding locality.
The obtained sequences for 18S rRNA were submitted to GenBank under the following accession numbers; OR241141 (H. ayorgbor), OR241161 (H. colubri), and OR241147 (H. canis).
A sum of 64 sequences (four sequences, two being forward and two reverse per detection) were obtained from all amplified fragments. All sequences of Hepatozoon sp. amplified from the genomic DNA of Hy. anatolicum and Ha. sulcata were identical to each other, resulting in a consensus sequence of 590 bp. Similarly, all sequences of Hepatozoon sp. amplified from the genomic DNA of R. haemaphysaloides were identical, while all sequences amplified from the genomic DNA of R. sanguineus were identical, yielding a consensus sequence of 588 and 596 bp, respectively. The Ha. sulcata and Hy. anatolicum based sequences showed the highest identity of 99.66% with H. colubri (MN723844), while the R. haemaphysaloides based sequence displayed the highest identity of 99.49% with H. ayorgbor (EF157822.1), as well as with two undetermined Hepatozoon spp. (MT919387 and MT919388), and R. sanguineus based sequence revealed maximum identity of 99.66% with H. canis (KX712126 and KX880505). The details regarding the infection rate and the association of Hepatozoon spp. with tick species in each district are shown in Table 1.
Based on 18S rRNA, a phylogenetic tree was obtained encompassing the three Hepatozoon spp. detected in the present survey. Hepatozoon ayorgbor clustered with the same species previously detected in Apodemus flavicollis, Python regius, and an unknown tick species from Croatia, Ghana, and Portugal, respectively. Hepatozoon colubri clustered with the corresponding species found in Zamenis lineatus from Iran, and H. canis clustered with conspecifics detected in Canis lupus, Canis aureus, and Canis familiaris from Iran, Romania, and Israel and Zambia, respectively (Figure 2).
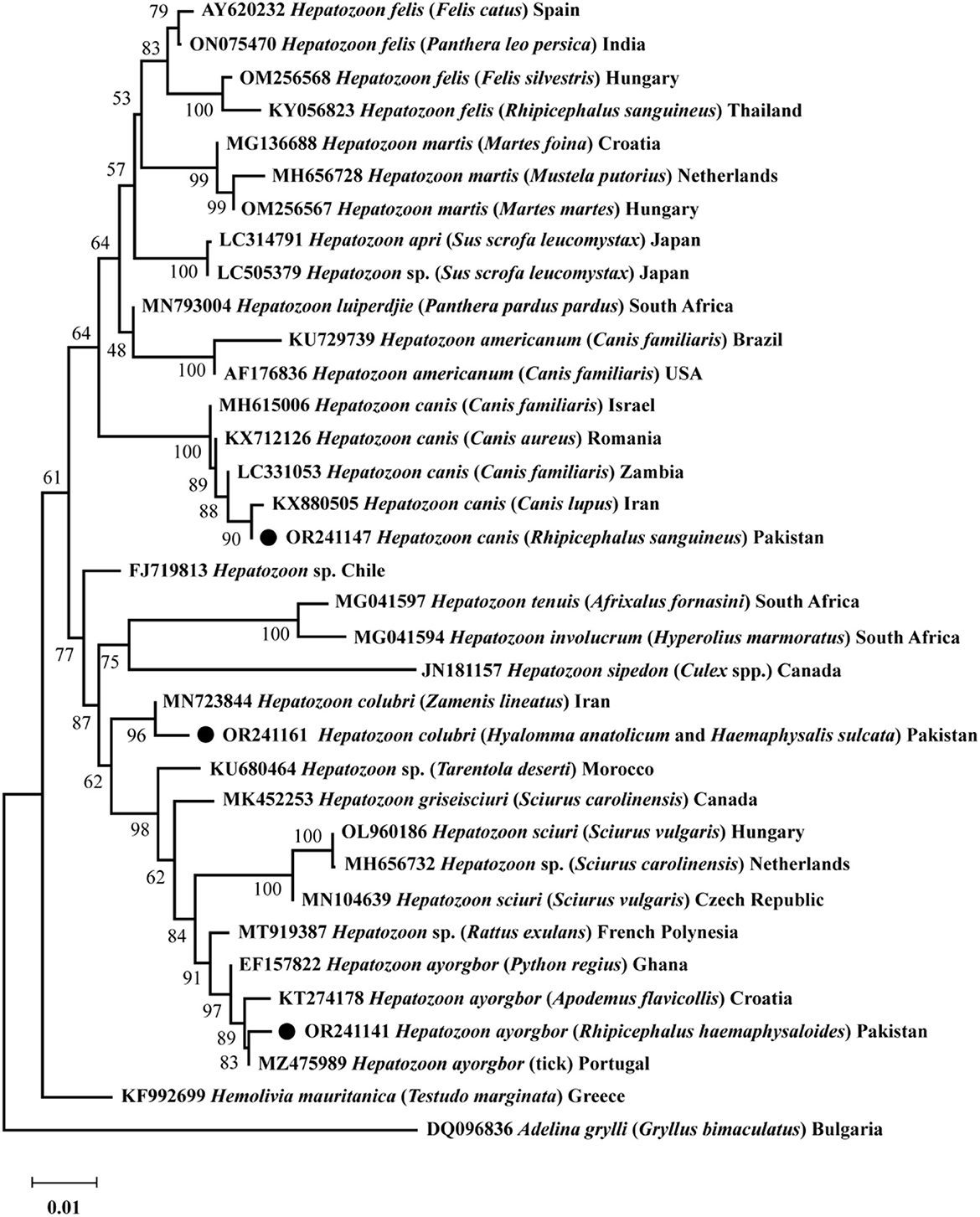
Figure 2. The phylogenetic tree was constructed for Hepatozoon spp. based on 18S rRNA sequences. The Neighbor-Joining method was used with 1,000 replicates for bootstrap analysis. The tree was rooted and subrooted using the 18S rRNA sequence of Adelina grylli and Hemolivia mauritanica, respectively. The GenBank accession numbers, species names, sources, and geographic locations were assigned to all sequences. The Hepatozoon spp. detected in this study are marked with a black circle.
Different factors were noted which could lead to the spillover of Hepatozoon spp. from wild to domestic hosts via ticks. For instance, locals were found handling, poaching, and hunting of wild animals using dogs. Moreover, dogs were found freely wandering between the habitats of wild and domestic animals, and occasionally feeding on dead wild canids. The overlapped area between domestic and wild animals was found to be increasing as a result of expansion of human settlements and deforestation. Deforestation, which is associated with climate change, was observed to create space for agriculture land and pastures for animal grazing. Under these circumstances, alongside an increase in the population of small generalist hosts like rodents, there was a noticeable decline in overall biodiversity. The invasion of the territory of domestic animals by wild animals were noted in the studied area. Furthermore, the habitat, food sources, and climate were found suitable for both wild and domestic animals, as well as ticks. Using a questionnaire, several other risk factors were gathered (Table 2). Herein, with statistical significance (P < 0.05), free ranging animals were at greater risk compared to those confined in shelters (RR = 3.05), animals getting food from wildlife habitat were at greater risk compared to those getting domestic food (RR = 3.06), and animals living in farm buildings located in wildlife habitats were at greater risk compared to those living in farm buildings located in villages (RR = 3.28).
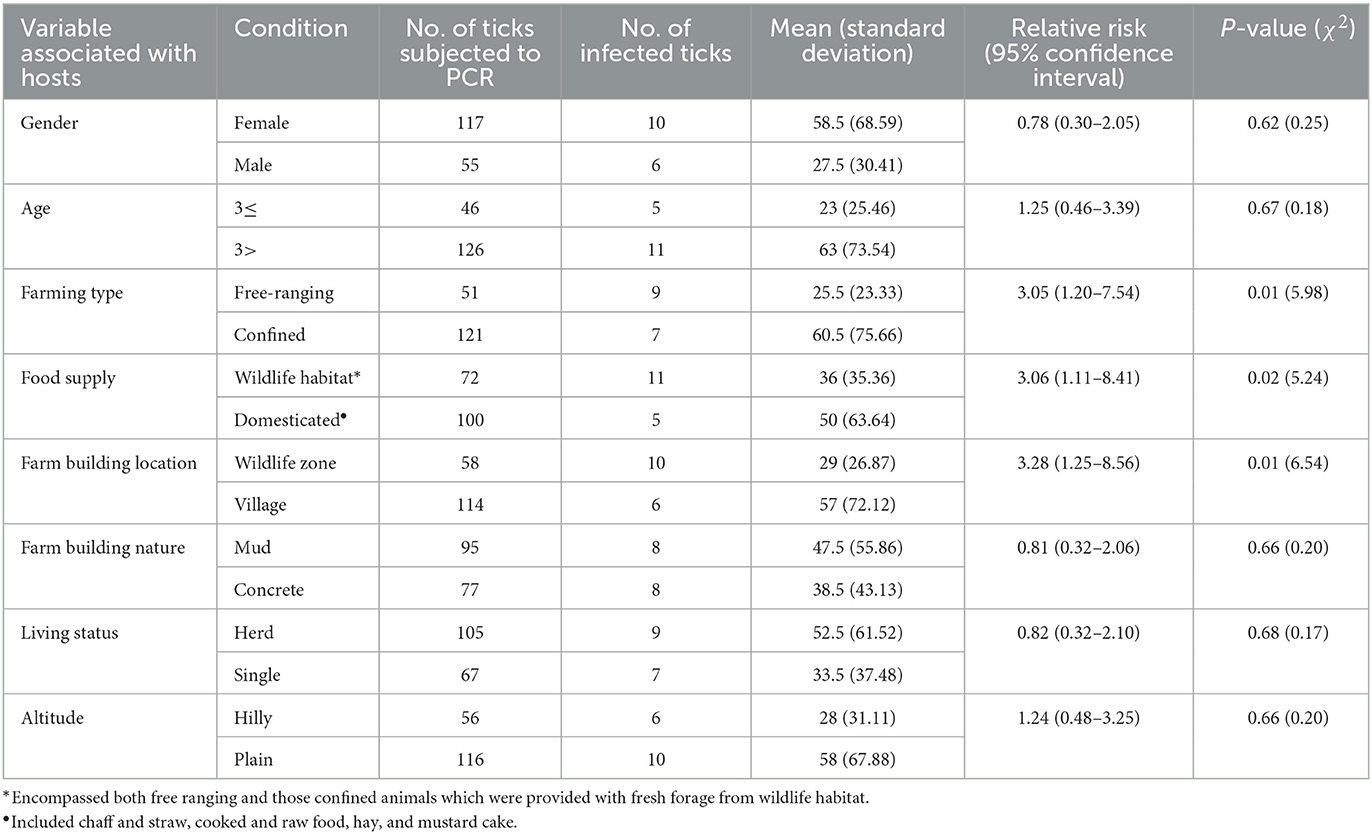
Table 2. Details on coupling questionnaire-based risk factors with molecular survey for potential spillover of Hepatozoon spp.
Despite potential agents of spillover of Hepatozoon spp. from wildlife to domestic animals, ticks have been widely overlooked as vectors of Hepatozoon spp., and there is a scarcity of knowledge regarding Hepatozoon spp. of wildlife origin in ticks infesting domestic animals. However, it is crucial to address this knowledge as these parasites pose a potential zoonotic threat, which, in turn could negatively impact food security within ecosystem (38). In addition to highlighting the possible spillover of Hepatozoon spp., to the best of our knowledge, it is the first report on the occurrence of H. canis in R. sanguineus in Pakistan, H. ayorgbor in R. haemaphysaloides and H. colubri in Ha. sulcata and Hy. anatolicum.
Having a large number of small ruminants, the northern part of Pakistan is considered endemic to Haemaphysalis ticks compared to Hyalomma and Rhipicephalus ticks (39–48). Haemaphysalis ticks being the most abundant in this study could be attributed to two main factors. Firstly, a larger portion of the study focused on the north-western KP, and secondly, the inclusion of its two common domestic hosts, goats and sheep. This study revealed that dogs exhibited the highest prevalence of tick infestation, highlighting their status as one of the most overlooked hosts regarding tick control practices. Examined dogs were in close interaction with other domestic animals, including cows, goats, and sheep for their role in protecting animals from predators. Furthermore, knowing the scarcity of DNA based information of Hepatozoon and the effectiveness of 18S rRNA in their accurate detection and differentiation (26, 27, 49), the collected ticks in the current endeavor were subjected to PCR targeting 18S rRNA of Hepatozoon. On comparison of the obtained sequences, like (26, 50), the differences were considerable to distinguish them as three distinct Hepatozoon spp. By clustering of query sequences of the Hepatozoon spp. with their corresponding homologous sequences, the phylogenetic analysis confirmed the evolutionary relationship suggested by previous reports (26, 27, 49, 51).
Looking at the details of the vertebrate hosts of the corresponding three Hepatozoon spp.; H. colubri in snakes (Zamenis lineatus and Zamenis longissimus), H. ayorgbor in snakes (Python regius) and the closely related Hepatozoon spp. in rats and gerbils (Rattus exulans, Rattus rattus, and Rhombomys opimus), and H. canis in jackals (Canis aureus) and dogs (Canis lupus familiaris) (20, 52–57), their wildlife origin is prominent. With H. ayorgbor detection for the first time in R. haemaphysaloides in the present study, previous studies have molecularly detected H. ayorgbor-like sequences in ticks, including Amblyomma rotundatum infesting snakes (Chironius multiventris, Corallus hortulanus, Oxyrhopus melanogenys, and Philodryas viridissima) in Brazil, and Amblyomma fimbriatum infesting monitor lizard (Varanus panoptes) and snake (Liasis fuscus) in Australia (58, 59), Ixodes tasmani infesting Tasmanian devils (Sarcophilus harrisii) in Australia and Ha. sulcata (host-seeking) in Turkey (60, 61). Some sequences in GenBank such as MZ475989 belonging to H. ayorgbor have been attributed to unknown tick species. Besides the detection of H. colubri for the first time in Hy. anatolicum and Ha. sulcata in the current study, previously H. colubri-like species has been detected in Amblyomma nitidum infesting reptiles in Japan (62). Unlike previous studies on the detection of H. ayorgbor-like sequences and H. colubri-like sequences in ticks infesting wild reptiles, herein, H. ayorgbor and H. colubri were detected in ticks infesting domestic animals. Such preliminary findings require further investigation because the presence of pathogen DNA in a tick species does not guarantee its role as a biological vector. Like previous studies that described R. sanguineus as the main vector of H. canis in many regions of the world (1, 19, 20, 63), H. canis was detected in R. sanguineus infesting domestic dogs. Furthermore, the absence of infection in male ticks compared to females and nymphs could indicate that males may not play a role as reservoirs of these parasites due to the fact that they may not feed on wild animals and they die soon after mating with females. Based on the documented evidence of tick infestation on wildlife in the region (44, 64–66), it could be anticipated that the ticks of the current study have acquired the corresponding three Hepatozoon spp. from wildlife somewhere during their lifecycle.
A pathogen spillover could be influenced by several factors associated with the pathogen itself, its different types of hosts and climate changes (67, 68), and human behavior. Hepatozoon spp. spillover could be determined by their abilities such as infecting a wide host range (1, 69), viability in the environment, adaptability and vector-borne transmission. An increase in the population of generalist small hosts, such as rodents, which are capable of hosting a broad spectrum of pathogens, may contribute to pathogen spillover by different routes, including ticks (68, 70). Additionally, decline in the diversity and abundance of wildlife, and their close proximity to public and domestic animals, may also amplify the spillover events of tick-borne pathogens (68, 71). The decline in biodiversity could be associated with rapid transformations in land use and land cover, as well as their conflict with human due to fears, attacks on crops and livestock, and financial benefits. The factors associated with domestic animals, including grazing in wildlife habitat (free-ranging), feeding on fresh forage from wildlife habitat, and their staying in farm buildings located in the wildlife habitat were noted as considerable risk factors associated with potential spillover. Hepatozoon spp. spillover may be facilitated by domestic dogs moving back and forth between wildlife habitats and human settlement (68), which was noted in the current study. Pakistan is the seventh most vulnerable country to climate change (72), which may influence vector dynamics, including ticks, potentially leading to cross-species transmission of Hepatozoon spp. Like previous studies, which provided evidences for the spillover of Hepatozoon species, including Hepatozoon americanum, H. canis, and Hepatozoon silvestris (10–12, 73–76), the current study proposes the spillover of Hepatozoon spp. from wild to domestic hosts via ticks. Hepatozoon spp. may have pathogenicity in unnatural hosts compared to their natural hosts (14–16), potentially posing a significant threat to domestic animals.
Besides the scarcity of knowledge on ticks as vectors of Hepatozoon spp., there is little information on Hepatozoon spp. of wildlife origin in ticks infesting domestic animals. Besides detecting H. canis in R. sanguineus, this is the earliest report on the detection of H. ayorgbor in R. haemaphysaloides and H. colubri in Ha. sulcata and Hy. anatolicum. By detecting Hepatozoon spp. of wildlife origin in ticks infesting domestic animals, this study proposed a possible spillover of Hepatozoon spp. from wild to domestic animals through ticks. The current study could assist in understanding the epidemiological surveillance and adopting preventive measures against tick-borne Hepatozoon spp. by minimizing the exposure of domestic animals to key risk factors. Further studies are needed in order to uncover ticks' role in the life cycle of Hepatozoon spp.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal studies were approved by the owners of the animals also gave their oral permission to collect ticks from their animals. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
AAli: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Writing—original draft, Writing—review and editing. HT: Formal analysis, Investigation, Methodology, Visualization, Writing—original draft, Writing—review and editing. MK: Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft, Writing—review and editing. MA: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Writing—original draft, Writing—review and editing. AAlo: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Writing—original draft, Writing—review and editing. HA: Data curation, Software, Writing—original draft, Writing—review and editing. TT: Data curation, Formal analysis, Methodology, Project administration, Software, Writing—review and editing. K-HT: Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Software, Visualization, Writing—original draft, Writing—review and editing.
The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia. This research was also partially funded by the National Science and Technology Council, grant number: 112-2327-B-002-008.
The authors acknowledge the financial support provided by the Higher Education Commission (HEC), Pakistan and Pakistan Science Foundation (PSF). This research was supported by King Saud University, Riyadh, Saudi Arabia (project number RSP2023R494).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Smith TG. The genus Hepatozoon (apicomplexa: adeleina). J Parasitol. (1996) 1996:565–85. doi: 10.2307/3283781
2. Jacobson ER. Infectious Diseases and Pathology of Reptiles: Color Atlas and Text. Boca Raton, FL: CRC Press (2007).
3. Vincent-Johnson N, Baneth G, Allen KE. Hepatozoonosis. In:Greene's Infectious Diseases of the Dog and Cat. No., Ed. 5. Philadelphia, PA: WB Saunders (2021). p. 1230–47.
4. Barta JR. Molecular approaches for inferring evolutionary relationships among protistan parasites. Vet Parasitol. (2001) 101:175–86. doi: 10.1016/S0304-4017(01)00564-7
5. Kvičerová J, Hypša V, Dvoráková N, Mikulíček P, Jandzik D, Gardner MG, et al. Hemolivia and Hepatozoon: haemogregarines with tangled evolutionary relationships. Protist. (2014) 165:688–700. doi: 10.1016/j.protis.2014.06.001
6. Karadjian G, Chavatte JM, Landau I. Systematic revision of the adeleid haemogregarines, with creation of Bartazoon ng, reassignment of Hepatozoon argantis Garnham, 1954 to Hemolivia, and molecular data on Hemolivia stellata. Parasite. (2015) 22:2015031. doi: 10.1051/parasite/2015031
7. Hamšíková Z, Silaghi C, Rudolf I, Venclíková K, Mahríková L, Slovák M, et al. Molecular detection and phylogenetic analysis of Hepatozoon spp. in questing Ixodes ricinus ticks and rodents from Slovakia and Czech Republic. Parasitol Res. (2016) 115:3897–904. doi: 10.1007/s00436-016-5156-5
8. Aktas M, Özübek S. Transstadial transmission of Hepatozoon canis by Rhipicephalus sanguineus (Acari: Ixodidae) in field conditions. J Med Entomol. (2017) 54:1044–8. doi: 10.1093/jme/tjx050
9. Duszynski DW, Kvicerova J, Seville RS. The Biology and Identification of the Coccidia (Apicomplexa) of Carnivores of the World. Cambridge, MA: Academic Press (2018).
10. Jarquín-Díaz VH, Barbachano-Guerrero A, Maldonado-Rodríguez R, Vásquez-Aguilar AA, Aguilar-Faisal JL. First molecular evidence of Hepatozoon canis in domestic dogs and ticks in fragmented rainforest areas in Mexico. Vet Parasitol. (2016) 6:4–8. doi: 10.1016/j.vprsr.2016.11.001
11. Da Silva MRL, Fornazari F, de Castro Demoner L, Teixeira CR, Langoni H, O'Dwyer LH. Didelphis albiventris naturally infected with Hepatozoon canis in southeastern Brazil. Ticks Tick Borne Dis. (2017) 8:878–81. doi: 10.1016/j.ttbdis.2017.07.005
12. Giannelli A, Latrofa MS, Nachum-Biala Y, HodŽić A, Greco G, Attanasi A, et al. Three different Hepatozoon species in domestic cats from southern Italy. Ticks Tick Borne Dis. (2017) 8:721–4. doi: 10.1016/j.ttbdis.2017.05.005
13. Squarre D, Nakamura Y, Hayashida K, Kawai N, Chambaro H, Namangala B, et al. Investigation of the piroplasm diversity circulating in wildlife and cattle of the greater Kafue ecosystem, Zambia. Parasit Vectors. (2020) 13:1–11. doi: 10.1186/s13071-020-04475-7
14. Pessôa SB, Biasi PD, Puorto G. Transferência do Hepatozoon tupinambis, parasita do lagarto Tupinambis teguixin, para a serpente cascavel (Crotalus durissus terrificus), por intermédio do mosquito Culex fatigans. Memórias do Instituto Oswaldo Cruz. (1974) 72:295–9. doi: 10.1590/S0074-02761974000200013
15. Simonato G, Franco V, Salvatore G, Manzocchi S, Dotto G, Morelli S, et al. First autochthonous clinical case of Hepatozoon silvestris in a domestic cat in Italy with unusual presentation. Parasit Vectors. (2022) 15:1–7. doi: 10.1186/s13071-022-05534-x
16. Wozniak EJ, Telford Jr SR. The fate of Hepatozoon species naturally infecting Florida black racers and watersnakes in potential mosquito and soft tick vectors, and histological evidence of pathogenicity in unnatural host species. Int J Parasitol. (1991) 21:511–6. doi: 10.1016/0020-7519(91)90055-C
17. Baneth GAD, Samish M, Alekseev E, Aroch I, Shkap V. Transmission of H. canis to dogs by naturally-fed or percutaneously-injected R sanguineus ticks. J Parasitol. (2001) 87:606–11. doi: 10.1645/0022-3395(2001)087[0606:TOHCTD]2.0.CO;2
18. Sloboda M, Kamler M, Bulantová J, Votypka J, Modry D. Rodents as intermediate hosts of Hepatozoon ayorgbor (Apicomplexa: Adeleina: Hepatozoidae) from the African ball python. Python regius? Folia Parasitologica. (2008) 55:13–6. doi: 10.14411/fp.2008.003
19. Baneth G. Perspectives on canine and feline hepatozoonosis. Vet Parasitol. (2011) 181:3–11. doi: 10.1016/j.vetpar.2011.04.015
20. Geurden T, Becskei C, Six RH, Maeder S, Latrofa MS, Otranto D, et al. Detection of tick-borne pathogens in ticks from dogs and cats in different European countries. Ticks Tick Borne Dis. (2018) 9:1431–6. doi: 10.1016/j.ttbdis.2018.06.013
21. Ali A, Zeb I, Alouffi A, Zahid H, Almutairi MM, Ayed Alshammari F, et al. Host immune responses to salivary components-a critical facet of tick-host interactions. Front Cell Infect Microbiol. (2022) 12:171. doi: 10.3389/fcimb.2022.809052
22. Ebani VV, Nardoni S, Mancianti F. Arthropod-borne pathogens in wild canids. Vet Sci. (2023) 10:165. doi: 10.3390/vetsci10020165
23. Murata T, Inoue M, Tateyama S, Taura Y, Nakama S. Vertical transmission of H. canis in dogs. J Vet Med Sci. (1993) 55:867–8. doi: 10.1292/jvms.55.867
24. Schäfer I, Müller E, Nijhof AM, Aupperle-Lellbach H, Loesenbeck G, Cramer S, et al. First evidence of vertical H. canis transmission in dogs in Europe. Parasit Vectors. (2022) 15:296. doi: 10.1186/s13071-022-05392-7
25. Smith TG, Kim B, Desser SS. Phylogenetic relationships among Hepatozoon species from snakes, frogs and mosquitoes of Ontario, Canada, determined by ITS-1 nucleotide sequences and life-cycle, morphological and developmental characteristics. Int J Parasitol. (1999) 29:293–304. doi: 10.1016/S0020-7519(98)00198-2
26. Inokuma H, Okuda M, Ohno K, Shimoda K, Onishi T. Analysis of the 18S rRNA gene sequence of a Hepatozoon detected in two Japanese dogs. Vet Parasitol. (2002) 106:265–71. doi: 10.1016/S0304-4017(02)00065-1
27. Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe: part I. Epizootiological aspects. Vet Parasitol. (2003) 113:189–201. doi: 10.1016/S0304-4017(03)00078-5
28. Hoogstraal H, Kaiser MN. Observations on Egyptian Hyalomma ticks (Ixodoidea, Ixodidae). 5 Biological notes and differences in identity of H. anatolicum and its subspecies anatolicum Koch and excavatum Koch among Russian and other workers identity of H. lusitanicum Koch. Ann Entomol Soc Am. (1959) 52:243–61. doi: 10.1093/aesa/52.3.243
29. Nosek J, Sixl W. Central-European ticks (Ixodoidea). Mitt Abt Zool Landesmus Joanneum. (1972) 1:480.
30. Apanaskevich DA. Differentiation of closely related species Hyalomma anatolicum and H. excavatum (Acari: Ixodidae) based on a study of all life cycle stages, throughout entire geographical range. Parazitologiia. (2003) 37:259–80.
31. Walker JB, Keirans JE, Horak IG. The Genus Rhipicephalus (Acari, Ixodidae): A Guide to the Brown Ticks of the World. Cambridge: Cambridge University Press (2000).
32. Kamran K, Ali A, Villagra C, Siddiqui S, Alouffi AS, Iqbal A. A cross-sectional study of hard ticks (acari: ixodidae) on horse farms to assess the risk factors associated with tick-borne diseases. Zoonoses Public Health. (2021) 68:247–62. doi: 10.1111/zph.12809
33. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual (No. Ed. 2). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (1989).
34. Almeida AP, Marcili A, Leite RC, Nieri-Bastos FA, Domingues LN, Martins JR, et al. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: Argasidae). Ticks Tick Borne Dis. (2012) 3:203–6. doi: 10.1016/j.ttbdis.2012.02.003
35. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547. doi: 10.1093/molbev/msy096
36. Hall T, Biosciences I, Carlsbad CJGBB. BioEdit: an important software for molecular biology. GERF Bull Biosci. (2011) 2:60–1.
37. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
38. Fitzpatrick JL. Global food security: the impact of veterinary parasites and parasitologists. Vet Parasitol. (2013) 195:233–48. doi: 10.1016/j.vetpar.2013.04.005
39. Karim S, Budachetri K, Mukherjee N, Williams J, Kausar A, Hassan MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl Trop Dis. (2017) 11:e0005681. doi: 10.1371/journal.pntd.0005681
40. Ali A, Zahid H, Zeb I, Tufail M, Khan S, Haroon M, et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit Vectors. (2021) 14:1–12. doi: 10.1186/s13071-021-04836-w
41. Ahmad I, Ullah S, Alouffi A, Almutairi MM, Khan M, Numan M, et al. Description of male, redescription of female, host record, and phylogenetic position of Haemaphysalis danieli. Pathogens. (2022) 11:1495. doi: 10.3390/pathogens11121495
42. Alam S, Khan M, Alouffi A, Almutairi MM, Ullah S, Numan M, et al. Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms. (2022) 10:1663. doi: 10.3390/microorganisms10081663
43. Khan SM, Khan M, Alouffi A, Almutairi MM, Numan M, Ullah S, et al. Phylogenetic position of Haemaphysalis kashmirensis and Haemaphysalis cornupunctata, with notes on Rickettsia spp. Genes. (2023) 14:360. doi: 10.3390/genes14020360
44. Ali A, Numan M, Ullah S, Khan M, Kamran K. Genetic characterization of Haemaphysalis (Rhipistoma) indica and Haemaphysalis (Segalia) montgomeryi ticks (Ixodoidea: Ixodidae). Ticks Tick Borne Dis. (2023) 14:102105. doi: 10.1016/j.ttbdis.2022.102105
45. Ali A, Obaid MK, Almutairi M, Alouffi A, Numan M, Ullah S, et al. Molecular detection of Coxiella spp. in ticks (ixodidae and argasidae) infesting domestic and wild animals: with notes on the epidemiology of tick-borne Coxiella burnetii in Asia. Front Microbiol. (2023) 14:1229950. doi: 10.3389/fmicb.2023.1229950
46. Ullah S, Alouffi A, Almutairi MM, Islam N, Rehman G, Ul Islam Z, et al. First report of Rickettsia conorii in Hyalomma kumari ticks. Animals. (2023) 13:1488. doi: 10.3390/ani13091488
47. Ali A, Khan MA, Zahid H, Yaseen PM, Qayash Khan M, Nawab J, et al. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front Physiol. (2019) 10:793. doi: 10.3389/fphys.2019.00793
48. Khan Z, Shehla S, Alouffi A, Kashif Obaid M, Zeb Khan A, Almutairi MM, et al. Molecular survey and genetic characterization of Anaplasma marginale in ticks collected from livestock hosts in Pakistan. Animals. (2022) 12:1708. doi: 10.3390/ani12131708
49. Li Y, Wang C, Allen KE, Little SE, Ahluwalia SK, Gao D, et al. Diagnosis of canine Hepatozoon spp. infection by quantitative PCR. Vet Parasitol. (2008) 157:50–8. doi: 10.1016/j.vetpar.2008.06.027
50. Baneth G, Barta JR, Shkap V, Martin DS, Macintire DK, Vincent-Johnson N. Genetic and antigenic evidence supports the separation of H. canis and Hepatozoon americanum at the species level. J Clin Microbiol. (2000) 38:1298–301. doi: 10.1128/JCM.38.3.1298-1301.2000
51. Hornok S, Boldogh SA, Takács N, Kontschán J, Szekeres S, Sós E, et al. Molecular epidemiological study on ticks and tick-borne protozoan parasites (Apicomplexa: Cytauxzoon and Hepatozoon spp.) from wild cats (Felis silvestris). Mustelidae and red squirrels (Sciurus vulgaris) in central Europe, Hungary. Parasit Vectors. (2022) 15:1–11. doi: 10.1186/s13071-022-05271-1
52. Sloboda M, Kamler M, Bulantová J, Votýpka J, Modrý D. A new species of Hepatozoon (Apicomplexa: Adeleorina) from Python regius (Serpentes: Pythonidae) and its experimental transmission by a mosquito vector. J Parasitol. (2007) 93:1189–98. doi: 10.1645/GE-1200R.1
53. Aktas M, Özübek S, Altay K, Balkaya I, Utuk AE, Kirbas A, et al. A molecular and parasitological survey of Hepatozoon canis in domestic dogs in Turkey. Vet Parasitol. (2015) 209:264–7. doi: 10.1016/j.vetpar.2015.02.015
54. Mitková B, Hrazdilová K, D'Amico G, Duscher GG, Suchentrunk F, Forejtek P, et al. Eurasian golden jackal as host of canine vector-borne protists. Parasit Vectors. (2017) 10:1–11. doi: 10.1186/s13071-017-2110-z
55. Ji N, Chen X, Liu G, Zhao S, Tan W, Liu G, et al. Theileria, Hepatozoon and Taenia infection in great gerbils (Rhombomys opimus) in northwestern China. Int J Parasitol. (2021) 15:79–86. doi: 10.1016/j.ijppaw.2021.04.002
56. Hrazdilová K, Cervená B, Blanvillain C, Foronda P, Modrý D. Quest for the type species of the genus Hepatozoon–phylogenetic position of hemogregarines of rats and consequences for taxonomy. System Biodivers. (2021) 19:622–31. doi: 10.1080/14772000.2021.1903616
57. Zechmeisterová K, Javanbakht H, Kvičerová J, Široký P. Against growing synonymy: identification pitfalls of Hepatozoon and Schellackia demonstrated on North Iranian reptiles. Eur J Protistol. (2021) 79:125780. doi: 10.1016/j.ejop.2021.125780
58. Vilcins IME, Old JM, Deane E. Detection of a Hepatozoon and spotted fever group Rickettsia species in the common marsupial tick (Ixodes tasmani) collected from wild Tasmanian devils (Sarcophilus harrisii), Tasmania. Vet Parasitol. (2009) 162:23–31. doi: 10.1016/j.vetpar.2009.02.015
59. Mendoza-Roldan JA, Ribeiro SR, Castilho-Onofrio V, Marcili A, Simonato BB, Latrofa MS, et al. Molecular detection of vector-borne agents in ectoparasites and reptiles from Brazil. Ticks Tick Borne Dis. (2021) 12:101585. doi: 10.1016/j.ttbdis.2020.101585
60. Vilcins IME, Ujvari B, Old JM, Deane E. Molecular and morphological description of a Hepatozoon species in reptiles and their ticks in the Northern Territory, Australia. J Parasitol. (2009) 95:434–42. doi: 10.1645/GE-1725.1
61. Orkun Ö, Çakmak A, Nalbantoglu S, Karaer Z. Turkey tick news: a molecular investigation into the presence of tick-borne pathogens in host-seeking ticks in Anatolia; Initial evidence of putative vectors and pathogens, and footsteps of a secretly rising vector tick, Haemaphysalis parva. Ticks Tick Borne Dis. (2020) 11:101373. doi: 10.1016/j.ttbdis.2020.101373
62. Qiu Y, Kidera N, Thu MJ, Hayashi M, Fujishima K, Tamura H. First molecular detection of Hemolivia and Hepatozoon parasites in reptile-associated ticks on Iriomote Island, Japan. Parasitol Res. (2021) 120:4067–72. doi: 10.1007/s00436-021-07345-y
63. Aktas M, Özübek S, Ipek DNS. Molecular investigations of Hepatozoon species in dogs and developmental stages of Rhipicephalus sanguineus. Parasitol Res. (2013) 112:2381–5. doi: 10.1007/s00436-013-3403-6
64. Khan M, Islam N, Khan A, Islam ZU, Muñoz-Leal S, Labruna MB, et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia sp. Ticks Tick Borne Dis. (2022) 13:102047. doi: 10.1016/j.ttbdis.2022.102047
65. Hoogstraal H, McCarthy VC. Hosts and distribution of Haemaphysalis kashmirensis with descriptions of immature stages and definition of the subgenus Herpetobia Canestrini (resurrected). J Parasitolo. (1965) 1965:674–9. doi: 10.2307/3276257
66. Ali A, Shehla S, Zahid H, Ullah F, Zeb I, Ahmed H, et al. Molecular survey and spatial distribution of Rickettsia spp. in ticks infesting free-ranging wild animals in Pakistan (2017–2021). Pathogens. (2022) 11:162. doi: 10.3390/pathogens11020162
67. Estrada-Peña A, Ostfeld RS, Peterson AT, Poulin R, de la Fuente J. Effects of environmental change on zoonotic disease risk: an ecological primer. Trends Parasitol. (2014) 30:205–14. doi: 10.1016/j.pt.2014.02.003
68. Ellwanger JH, Chies JAB. Zoonotic spillover: understanding basic aspects for better prevention. Genet Mol Biol. (2021) 44:355. doi: 10.1590/1678-4685-gmb-2020-0355
69. Laakkonen J, Sukura A, Oksanen A, Henttonen H, Soveri T. Haemogregarines of the genus Hepatozoon (Apicomplexa: Adeleina) in rodents from northern Europe. Folia Parasitol. (2001) 48:263–7. doi: 10.14411/fp.2001.043
70. Bordes F, Blasdell K, Morand S. Transmission ecology of rodent-borne diseases: new frontiers. Integr Zool. (2015) 10:424–35. doi: 10.1111/1749-4877.12149
71. Dantas-Torres F. Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int J Parasitol. (2015) 4:452–61. doi: 10.1016/j.ijppaw.2015.07.001
73. Baneth G, Mathew JS, Shkap V, Macintire DK, Barta JR, Ewing SA. Canine hepatozoonosis: two disease syndromes caused by separate Hepatozoon spp. Trends Parasitol. (2003) 19:27–31. doi: 10.1016/S1471-4922(02)00016-8
74. Tomanović S, Radulović Ž, Cakić S, Mihaljica D, Sukara R, Penezić A, et al. Tick species (acari: Ixodidae) of red foxes (Vulpes vulpes) in Serbia. In: 2nd International Symposium on Hunting “modern Aspects of Sustainable Management of Game”. Novi Sad: University of Novi Sad, Faculty of Agriculture (2013). p. 229–35.
75. Kegler K, Nufer U, Alic A, Posthaus H, Olias P, Basso W. Fatal infection with emerging apicomplexan parasite Hepatozoon silvestris in a domestic cat. Parasit Vectors. (2018) 11:1–5. doi: 10.1186/s13071-018-2992-4
Keywords: ticks, Hepatozoon ayorgbor, Hepatozoon colubri, Hepatozoon canis, spillover
Citation: Tila H, Khan M, Almutairi MM, Alouffi A, Ahmed H, Tanaka T, Tsai K-H and Ali A (2023) First report on detection of Hepatozoon ayorgbor in Rhipicephalus haemaphysaloides and Hepatozoon colubri in Haemaphysalis sulcata and Hyalomma anatolicum: risks of spillover of Hepatozoon spp. from wildlife to domestic animals. Front. Vet. Sci. 10:1255482. doi: 10.3389/fvets.2023.1255482
Received: 08 July 2023; Accepted: 28 August 2023;
Published: 18 September 2023.
Edited by:
Münir Aktaş, Firat University, TürkiyeReviewed by:
Sezayi Özübek, Firat University, TürkiyeCopyright © 2023 Tila, Khan, Almutairi, Alouffi, Ahmed, Tanaka, Tsai and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abid Ali, dW9wX2FsaUB5YWhvby5jb20=; Kun-Hsien Tsai, a3VuaHRzYWlAbnR1LmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.