- 1Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 2Department of Agriculture, Ministry of Environment, Water, and Agriculture, Riyadh, Saudi Arabia
- 3King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia
- 4Laboratory of Infectious Diseases, Joint Faculty of Veterinary Medicine, Kagoshima University, Kagoshima, Japan
- 5Department of Zoology, Abdul Wali Khan University Mardan, Mardan, Pakistan
Ticks are important ectoparasites that transmit various pathogens causing morbidity and mortality in humans and animals. Saudi Arabia faces several challenges that can contribute to the emergence and spread of antimicrobial resistance (AMR) bacteria. These challenges require collaborative efforts to successfully achieve significant control of AMR in the country. The present study aims to isolate bacteria from camels' tick Hyalomma dromedarii in Al-Jouf province to identify and determine these isolates' antimicrobial susceptibilities. Forty-nine ticks were collected from dromedary camels and morphologically classified as H. dromedarii. Ticks were then homogenized and plated individually, which resulted in the isolation of 55 bacteria. The results showed that the bacterial isolates belong to 20 different species. About 71% (n = 39) of the total isolates were identified as Gram-positive bacteria comprised of 11 different species, while 29% (n = 16) of the total isolates were Gram-negative bacteria comprised of 9 different species. The most prevalent isolate within the total samples was Staphylococcus lentus (22.45%, 11/49), followed by Staphylococcus pseudintermedius (18.37%, 9/49) and Sphingomonas paucimobilis (16.33% 8/49). The antimicrobial susceptibility profile of Gram-positive bacteria showed that 100% (n = 31) were resistant to benzylpenicillin; 90.3% (n = 28) were resistant to oxacillin; 58.1% (n = 18) were resistant to clindamycin; 48.4% (n = 15) were resistant to vancomycin. In addition, 32.3% (n = 10) were resistant to trimethoprim/sulfamethoxazole and rifampicin; 25.8% (n = 8) were resistant to erythromycin; 16.1% (n = 5) were resistant to teicoplanin; 6.5% (n = 2) were resistant to tetracycline. All Gram-positive bacteria were 100% susceptible to linezolid, gentamicin, tobramycin, levofloxacin, moxifloxacin, tigecycline, and nitrofurantoin. In antimicrobial susceptibility tests for the Gram-negative bacteria, 57.14% (n = 8) of the identified bacteria were resistant to ampicillin, whereas 50% (n = 7) were resistant to cefoxitin and ceftazidime. About 28.57% (n = 4) of the Gram-negative bacteria were resistant to ceftriaxone, trimethoprim/sulfamethoxazole. In addition, 21.43% (n = 3) were resistant to amoxicillin/clavulanic acid and cephalothin; 14.29% (n = 2) were resistant to cefepime and nitrofurantoin; 7.14% (n = 1) were resistant to piperacillin/tazobactam and tigecycline. However, all Gram-negative bacteria were susceptible to other examined antimicrobials. This is the first study that investigates the role of the hard tick as a potential reservoir for AMR pathogens within our region.
1 Introduction
Ticks are recognized as one of the main arthropods' vectors of disease agents to both humans and animals (1–3). Ticks can transmit a wide spectrum of pathogenic and non-pathogenic microorganisms such as bacteria, protozoa and viruses (4–6). Different bacteria have been detected in hard ticks at different developmental stages (7–10). Some microorganisms are life-threatening to animals (11), whereas others have risks to human health (12). The major losses caused by ticks are due to their ability to transmit diseases to livestock, which are of great economic importance worldwide. In addition, blood sucking by ticks causes anemia and reduction in weight among livestock, while their bites also reduce the quality of hides (13). These factors result in a substantial reduction in milk and meat production, increasing the morbidity and mortality among livestock.
Antimicrobial resistance (AMR) bacteria are microorganisms, mainly bacteria, that show resistance to one or more classes of antimicrobial agents (14). Multi-drugs resistance (MDR) are microorganism resistance to at least one antimicrobial in 3 or more different classes (15). The emergence and spread of AMR pathogens, which have acquired novel resistance mechanisms, is currently one of the most important threats to public and animal health. AMR is associated with a remarkable burden of increased morbidity, mortality, healthcare costs, and antibiotic use (15–17). AMR bacterial infections kill approximately 700,000 individuals globally each year, which is expected to rise to 10 million by 2050 (18). The AMR bacteria have been found in humans, animals, foods, plants, and the environment (water, soil, and air). They can spread from person to person or between humans and animals (19).
The present study aims to isolate and identify bacterial species from the camel's tick H. dromedarii in Al-Jouf province in Saudi Arabia. In addition, we aim to determine if the isolated bacterial species are resistant to clinical useful antimicrobial agents. To the best of our knowledge, this is the first report that investigates the hard tick's role as a potential reservoir for AMR bacteria in our region.
2 Materials and methods
2.1 Selection of the study area
The current study used hard ticks collected from dromedary camels throughout Al-Jouf province (29.8874° N and 39.3206° E) in the northern part of Saudi Arabia (Figure 1). Al-Jouf region is one of the most fertile regions in Saudi Arabia as a soil and water treasure. In addition, it has a suitable atmosphere which lies between the climate of the Arabian Peninsula and the climate of the Mediterranean Sea. These conditions have made the region suitable for cultivating various crops. Therefore, the agricultural sector occupies most of the economic activities in the Al-Jouf region. With the availability of agricultural resources, the region has developed the livestock sector. Approximately 7,398 camels were recorded in the Al-Jouf region in 2005 (20). These factors made the area suitable for our study due to the availability of a large number of animals and the frequent human-animal interaction.
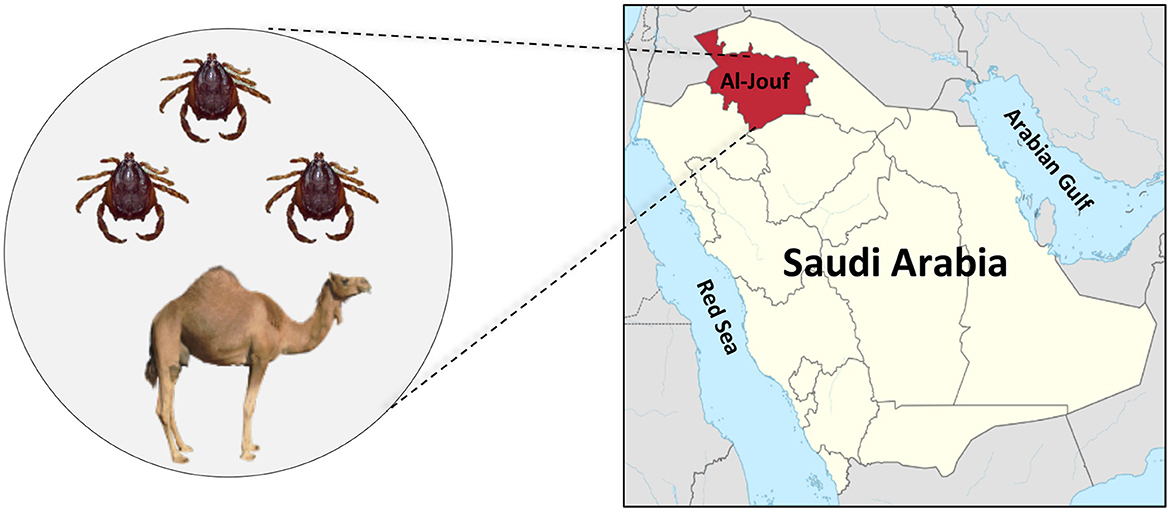
Figure 1. The geographic location of Al-Jouf province within Saudi Arabia where ticks (H. dromedarii) were collected from camels.
2.2 The collection and identification of the ticks
In total, 49 ticks were collected in August 2022 from 13 female dromedary camels (Camelus dromedaries) from Al-Jouf camel market (Supplementary Table 1). The collected ticks were stored in a jar containing 70% ethanol and transported to the Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, for further analysis. Collected specimens were subjected to taxonomic identification based on the external morphology to the species level, with developing stages and gender recorded using standard morphological keys by means of the stereomicroscope (Leica EZ4HD) (21).
2.3 Bacterial isolation from ticks
Ticks were washed with 70% ethanol and rinsed 3 times with phosphate-buffered saline (PBS) (PanReac, AppliChem). Tick's exoskeletons were removed using sterile forceps and blades, and the internal organs were cut into small pieces and then transferred into tubes. Each tick was homogenized individually with PBS using an electric homogenizer. Each homogenate was inoculated into 3 ml of nutrient broth media (VWR Chemicals, USA) in a 15 ml tube and incubated for 24 h at 37°C with shaking (250 rpm) (10, 22). The growing cultures were plated into different media, including blood agar base (VWR Chemicals, USA) and MacConkey agar (OXOID, UK) to allow the growth of a large spectrum of bacteria (23). After 24 h of incubation at 37°C, different colonies (1–2 colonies from each plate) were selected based on morphology (color, structure, shape, size). The bacterial isolates were stored with glycerol at −80°C for further analysis.
2.4 Identification of the bacterial isolates
Bacterial isolates were subjected to Gram staining to differentiate the Gram-positive and Gram-negative bacteria (23). After that, bacteria were identified by the Vitek 2 compact system (bioMérieux Inc. USA) (24). The identification was conducted using GP ID REF21342 card (for Gram-positive) and GN ID REF21341 card (for Gram-negative). All procedures were followed according to the manufacturer's instructions.
2.5 Determination of antimicrobail agents susceptibility of the bacterial isolates
Antimicrobial susceptibility test was conducted for 45 identified isolates using Vitek 2 compact system (bioMérieux Inc. USA) (23). Antimicrobial susceptibility tests were run on cards that contain dilutions of antimicrobials to detect the breakpoint minimum inhibitory concentration (MIC) against the bacteria. AST-P580 card (for Staphylococcus spp., Enterococcus spp., and Streptococcus agalactiae), and AST-N291 card (for Gram-negative bacilli) cards (bioMérieux Inc. USA) were used to determine antimicrobial agent's susceptibility. The used antimicrobial agents' classes include: penicillins (ampicillin, amoxicillin/clavulanic acid, piperacillin/tazobactam, benzylpenicillin, oxacillin); aminoglycosides (gentamicin, tobramycin, amikacin); cephalosporin (cephalothin, cefoxitin, ceftazidime, ceftriaxone, cefepime); carbapenem (imipenem, meropenem); fluoroquinolone (ciprofloxacin, levofloxacin, moxifloxacin), tetracyclines (tetracycline, tigecycline); glycopeptide (teicoplanin, vancomycin); macrolides (erythromycin); lincomycin (clindamycin); oxazolidinone (linezolid); rifamycin (rifampicin); nitrofuran (nitrofurantoin); and Sulfonamides (trimethoprim/sulfamethoxazole). According to their known or expected primary activity, these agents were tested against either Gram-positive or Gram-negative species. The following quality control strains were included in all tests: E. coli ATCC 25922 and 35218, Staphylococcus aureus ATCC 29213, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, Haemophilus influenzae ATCC 49247 and 49766, and Streptococcus pneumoniae ATCC 49619. The MIC cutoff values distinguishing sensitive, moderate, and resistant bacteria to antimicrobial agents were programmed into the system per the National Committee for Clinical Laboratory Standards (NCCLS), USA guidelines. The results were interpreted using Vitek 2 compact software version 07.01.
3 Results
3.1 Identification of the isolated bacteria
The collected 49 ticks from 13 female dromedary camels were classified as Hyalomma dromedarii and they were either nymphs (79.6%, n = 39) or males (20.4%, n = 10). From the collected ticks, a total of 55 bacterial species were isolated and subjected to Gram staining, followed by identification by Vitek 2 compact system. The results showed that the bacterial isolates belong to 20 different bacterial species. About 71% (n = 39) of the total isolates were identified as Gram-positive bacteria comprised of 11 different species: Staphylococcus lentus (n = 11), Staphylococcus pseudintermedius (n = 9), Aerococcus viridans (n = 4), Staphylococcus aureus (n = 3), Staphylococcus vitulinus (n = 3), Staphylococcus sciuri (n = 2), Staphylococcus haemolyticus (n = 2), Enterococcus casseliflavus (n = 2), Staphylococcus hominis (n = 1), Staphylococcus epidermidis (n = 1), and Streptococcus equi ssp zooepidemicus (n = 1). The Gram-negative bacteria were represented by 29% (n = 16) of the total isolates comprised of 9 different species: Sphingomonas paucimobilis (n = 8), Klebsiella pneumoniae ssp ozaenae (n = 1), Klebsiella pneumoniae ssp pneumoniae (n = 1), Pseudomonas aeruginosa (n = 1), Pseudomonas putida (n = 1), Pseudomonas fluorescens (n = 1), Stenotrophomonas maltophilia (n = 1), Rhizobium radiobacter (n = 1) and Cronobacter sakazakii group (n = 1). The most prevalent isolated bacteria was S. lentus (20%, n = 11), followed by S. pseudintermedius (16.4%, n = 9) and S. paucimobilis (14.5%, n = 8). Interestingly, each collected tick was found positive for one bacterial specie, however, 4 out of the 49 ticks were found to have 2 or 3 bacterial species (Supplementary Table 1).
3.2 Determination of the antimicrobial agents susceptibility of the isolated bacteria
Fourty-five bacterial isolates were tested for antimicrobial susceptibility using Vitek 2 compact system (among them 31 isolates are Gram-positive and 14 isolates are Gram-negative). Our results showed that Gram-positive and Gram-negative bacteria exhibited resistance to several antimicrobial agents (Table 1). The antimicrobial susceptibility profile of Gram-positive bacteria showed that 100% (n = 31) were resistant to benzylpenicillin; 90.3% (n = 28) were resistant to oxacillin; 58.1% (n = 18) were resistant to clindamycin; 48.4% (n = 15) were resistant to vancomycin. In addition, 32.3% (n = 10) were resistant to trimethoprim/sulfamethoxazole and rifampicin; 25.8% (n = 8) were resistant to erythromycin; 16.1% (n = 5) were resistant to teicoplanin; 6.5% (n = 2) were resistant to tetracycline (Table 1). All Gram-positive bacteria were 100% susceptible to linezolid, gentamicin, tobramycin, levofloxacin, moxifloxacin, tigecycline, and nitrofurantoin (Table 1).
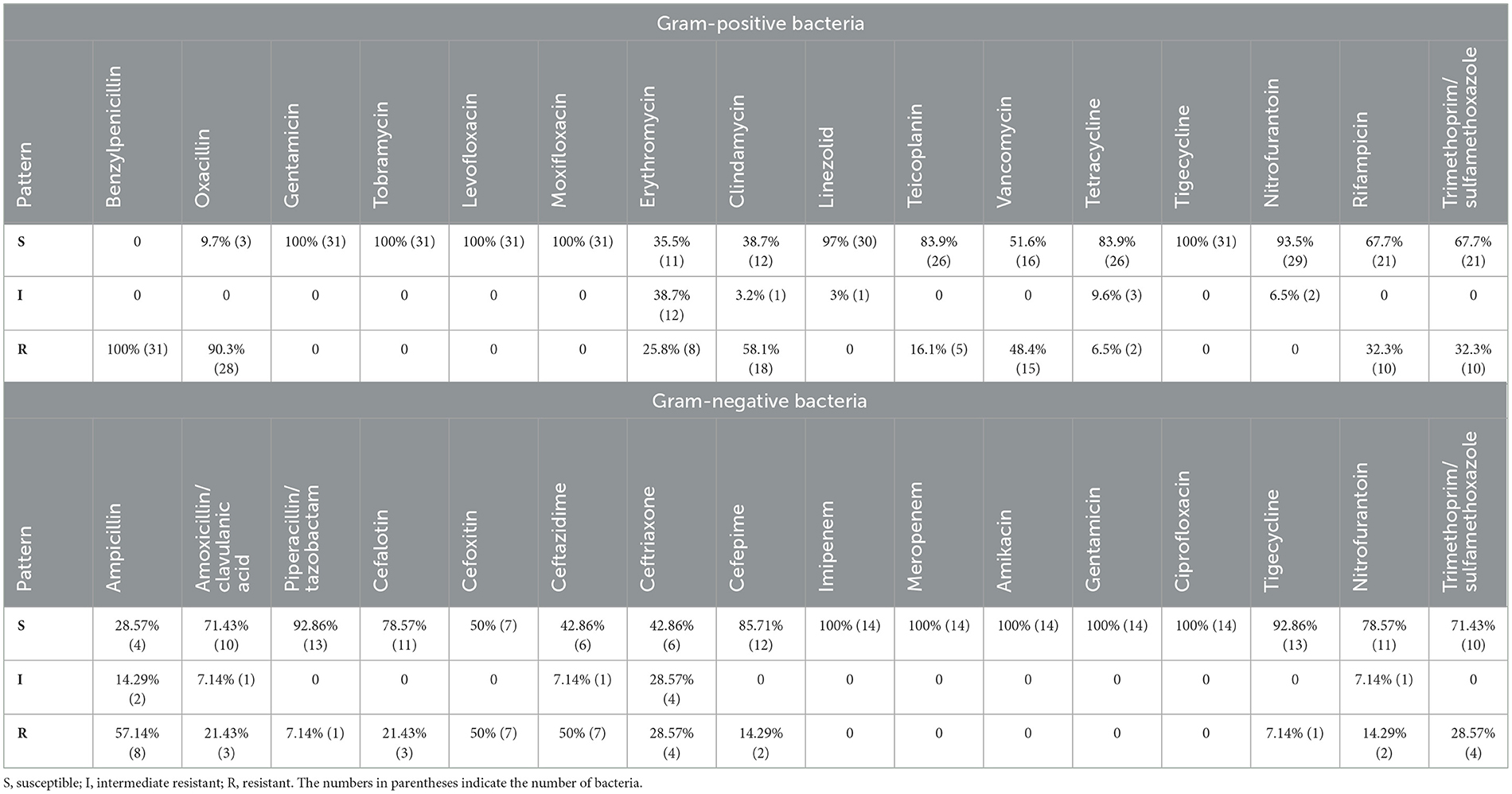
Table 1. Antimicrobials susceptibility of Gram-positive and Gram-negative bacteria isolated from H. dromedarii.
For the Gram-negative bacterial antimicrobial agents susceptibility testing, 57.14% (n = 8) of the identified Gram-negative bacteria were resistant to ampicillin, whereas 50% (n = 7) were resistant to cefoxitin and ceftazidime. About 28.57% (n = 4) of the Gram-negative bacteria were resistant to ceftriaxone, trimethoprim/sulfamethoxazole. In addition, 21.43% (n = 3) were resistant to amoxicillin/clavulanic acid and cephalothin; 14.29% (n = 2) were resistant to cefepime and nitrofurantoin; 7.14% (n = 1) were resistant to piperacillin/tazobactam and tigecycline (Table 1). However, all Gram-negative bacteria were susceptible to other antimicrobials including imipenem, meropenem, amikacin, gentamicin, and ciprofloxacin (Table 1).
All Gram-positive bacteria tested for antimicrobial agents susceptibility showed resistance to one or more classes of antimicrobials (Figure 2). Among S. lentus isolates, all isolates (n = 11/11) showed resistance to benzylpenicillin and clindamycin, 10 isolates (n = 10/11) showed resistance to oxacillin, 5 isolates (n = 5/11) showed resistance to rifampicin, 4 isolates (n = 4/11) showed resistance to erythromycin, 3 isolates (n = 3/11) showed resistance to vancomycin, only one isolate of S. lentus showed resistance to teicoplanin (Figure 2). All S. pseudintermedius isolates (n = 9/9) showed resistance to benzylpenicillin and oxacillin, 7 isolates (n = 7/9) showed resistance to trimethoprim/sulfamethoxazole, 4 isolates (n = 4/9) showed resistance to vancomycin, 2 isolates (n = 2/9) showed resistance to rifampicin and clindamycin, only one isolate (n = 1/9) showed resistance to erythromycin (Figure 2). For the S. aurues, all isolates (n = 3/3) showed resistance to benzylpenicillin, two isolates (n = 2/3) showed resistance to oxacillin, teicoplanin and vancomycin, and only one isolate of S. aurues (n = 1/3) showed resistance to clindamycin, tetracycline and trimethoprim/sulfamethoxazole (Figure 2). For S. sciuri, two isolates (n = 2/2) showed resistance to benzylpenicillin, oxacillin, clindamycin, teicoplanin, vancomycin and rifampicin, one isolate (n = 1/2) showed resistance to erythromycin and tetracycline. E. casseliflavus isolates (n = 2/2) showed resistance to benzylpenicillin, oxacillin, and vancomycin. One isolate of E. casseliflavus (n = 1/2) showed resistance to erythromycin, rifampicin, and trimethoprim/sulfamethoxazole (Figure 2). For S. haemolyticus, two isolates (n = 2/2) showed resistance to benzylpenicillin and oxacillin, one isolate (n = 1/2) showed resistance to clindamycin. S. hominis isolate showed resistance to benzylpenicillin, oxacillin, erythromycin, clindamycin, and vancomycin. S. epidermidis showed resistance to benzylpenicillin.
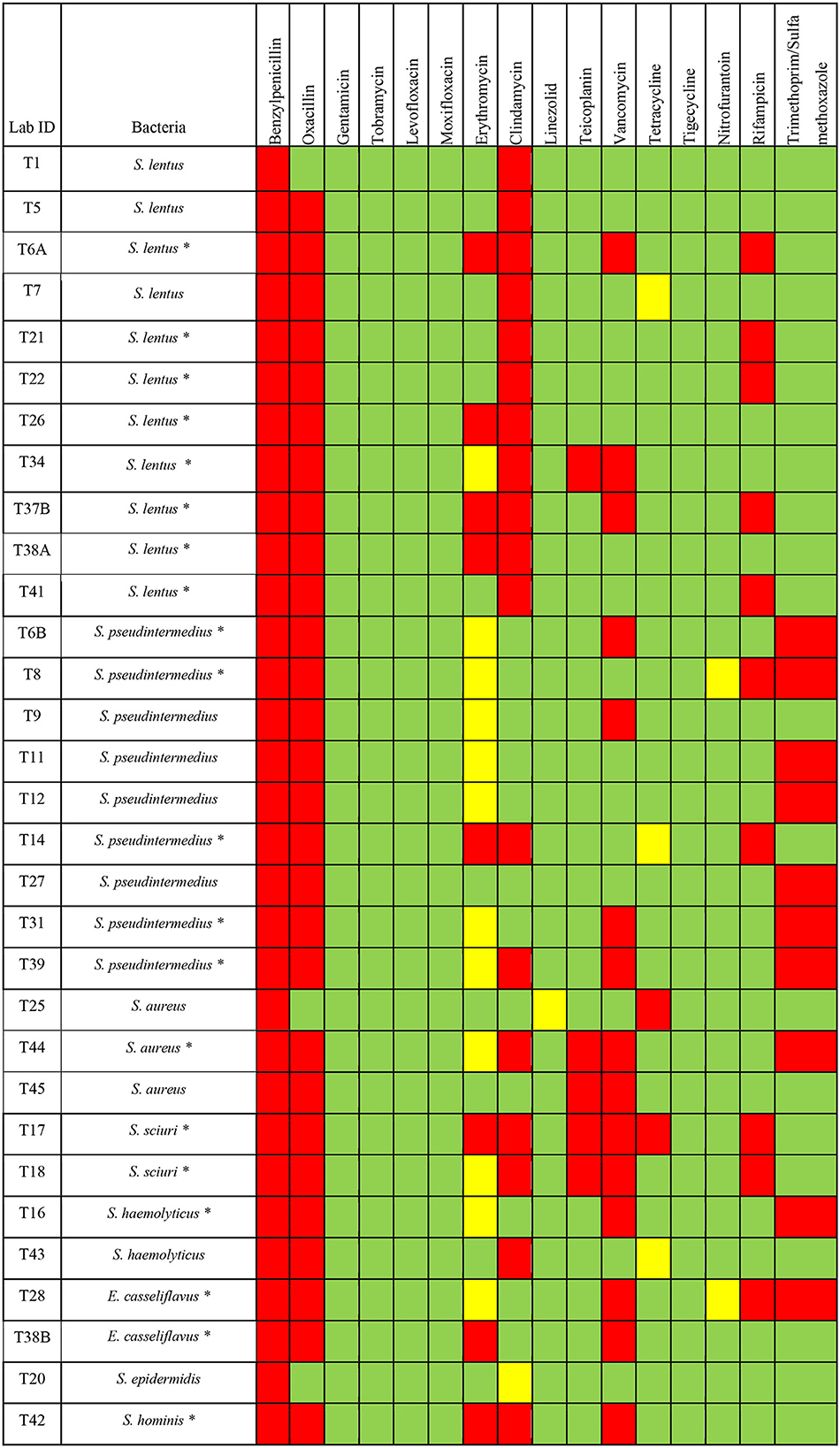
Figure 2. Antimicrobials susceptibility of different Gram-positive bacteria isolated from H. dromedarii. Green, susceptible; Yellow, intermediate resistant; Red, resistant. The star symbol next to the bacteria name denotes the MDR bacteria.
Among the Gram-negative bacterial species tested for susceptibility, 11 out of 14 isolates showed resistance to one or more classes of antimicrobials. For S. paucimobilis, 5 isolates (n = 5/8) showed resistance to ceftazidime, 4 isolates (n = 4/8) showed resistance to ampicillin, 3 isolates (n = 3/8) showed resistance to cefoxitin. Only one isolate of S. paucimobilis (n = 1/8) showed resistance to amoxicillin/clavulanic acid, ceftriaxone, and trimethoprim/sulfamethoxazole (Figure 3). C. sakazakii group showed resistance to cefoxitin and cefalotin. K. pneumoniae ssp ozaenae showed resistance to ampicillin, cephalothin, cefoxitin, ceftazidime, ceftriaxone, cefepime. K. pneumoniae ssp pneumonia showed resistance to ampicillin, cephalothin, ceftazidime, ceftriaxone, cefepime, and trimethoprim/sulfamethoxazole. P. putida showed resistance to ampicillin, amoxicillin/clavulanic acid, cefoxitin, nitrofurantoin, and trimethoprim/sulfamethoxazole. P. aeruginosa showed resistance to ampicillin, amoxicillin/clavulanic acid, piperacillin/tazobactam, cefoxitin, ceftriaxone, tigecycline, nitrofurantoin and trimethoprim/sulfamethoxazole (Figure 3).
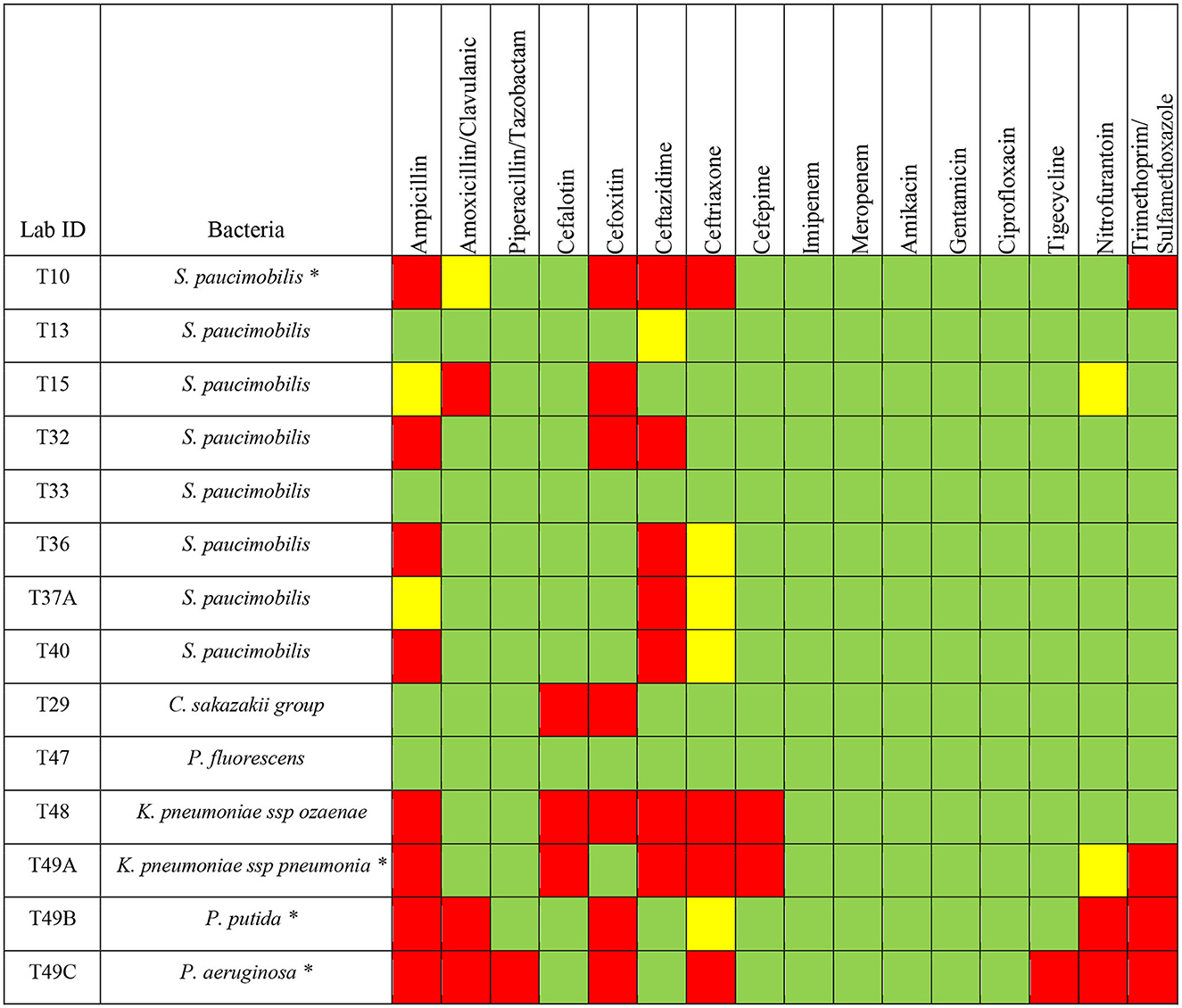
Figure 3. Antimicrobials susceptibility of different Gram-negative bacteria isolated from H. dromedarii. Green, susceptible; Yellow, intermediate resistant; Red, resistant. The star symbol next to the bacteria name denotes the MDR bacteria.
For MDR bacteria, we looked for any bacterial isolate that is resistant to three or more of the antimicrobial agent classes. Among the Gram-positive bacteria, we found that 64.5% (n = 20) are considered as MDR bacteria with some isolates resistant to even more than 4 classes of antimicrobial agents (Figure 2). The percentage of MDR bacteria among the Gram-positive bacterial species was as the following: S. lentus (72.7%, n = 8/11), S. pseudintermedius (55.5%, n = 5/9), S. sciuri (100%, n = 2/2), E. casseliflavus (100%, n = 2/2), S. hominis (100%, n = 1/1), S. aurues (33.3%, n = 1/3), S. haemolyticus (50%, n = 1/2) (Figure 2).
Among the Gram-negative bacteria, we found that 28.5% (n = 4) are MDR bacteria with some isolates resistant to even more than 4 classes of antimicrobial agents (Figure 3). One isolate of S. paucimobilis (n = 1/8), K. pneumoniae ssp pneumoniae, P. aeruginosa, and P. putida, showed resistance to 3 or more different classes of antimicrobials (Figure 3).
4 Discussion
The increasing prevalence of AMR bacteria is a global concern affecting both animal and human health. The role of ticks in disseminating AMR bacteria is not completely understood. To address this issue, different bacteria were isolated from H. dromedarii ticks from Al-Jouf province and their antimicrobial susceptibilities to different clinically utilized antimicrobials were determined. We concluded that isolated bacteria from ticks showed significant resistance to different antimicrobial agents, including benzylpenicillin; oxacillin, clindamycin, vancomycin, trimethoprim/sulfamethoxazole, rifampicin, erythromycin, teicoplanin, tetracycline, ampicillin, cefoxitin, ceftazidime, ceftriaxone, trimethoprim/sulfamethoxazole, amoxicillin/clavulanic acid, cephalothin, cefepime and nitrofurantoin, piperacillin/tazobactam, and tigecycline. Collectively, these data indicate the possibility of the existence of different AMR bacteria within the ticks that needs further research impetus. Furthermore, almost all isolated bacterial species can infect and spread between animals and humans, causing economic and health-related problems (25–31). To our knowledge, this is the first report in the region about ticks and their association with AMR bacteria.
During our study, several genera of Gram-positive and Gram-negative bacteria were identified which included species of the genera of Staphylococcus, Enterococcus, Streptococcus, Klebsiella Pseudomonas, and Stenotrophomonas which is similar to the findings from other studies (32–34). Some of these bacteria of the genera of Enterococcus, Streptococcus, Pseudomonas, and Klebsiella are potential pathogens to both humans and animals. Ticks may play a role as reservoir hosts for pathogenic bacteria leading to public and veterinary health risks. Other tick species may be screened for AMR bacteria to minimize any zoonotic consequences.
About 42 bacterial samples isolated from the ticks showed resistance to one or more antimicrobial agents from 9 different classes including pipracillins (ampicillin, amoxicillin/clavulanic acid, piperacillin/tazobactam, benzylpenicillin, oxacillin); cephalosporin (cephalothin, cefoxitin, ceftazidime, ceftriaxone, cefepime); tetracyclines (tetracycline, tigecycline); glycopeptide (teicoplanin, vancomycin); macrolides (erythromycin); lincomycin (clindamycin); rifamycin (rifampicin); nitrofuran (nitrofurantoin); and Sulfonamides (trimethoprim/sulfamethoxazole). Interestingly, among the 42 AMR bacteria, 24 bacterial isolates were resistant to 3 or more antimicrobials, considering MDR bacteria. This needs further research to investigate antimicrobial-resistant genes (ARGs) against the tested antimicrobial agents. The observed resistant phenotypes are concerning and supporting the active role of ticks as carriers of AMR bacteria. Scientific research has already established a direct correlation between antimicrobial use and the degree of resistance (35, 36). However, further studies must examine the correlation between microbiota, ARGs, and antimicrobial use in ticks. Monitoring efforts must be further emphasized where antimicrobials are widely utilized and tick-borne diseases are endemic. Even our results showed that ticks might act as a vector that transmits different AMR bacteria. Further studies are essential to answer questions related to the role of ticks in the spread and transmission of AMR bacteria among different hosts, including humans, animals, and the environment.
Tick microbiota could be influenced by the environment and blood meals (37, 38). The environment contains many species of bacterial genera, for example: Staphylococcus and Pseudomonas. We found these bacterial genera in ticks, which might suggest that these bacteria were acquired by the ticks from the surrounding environment. Previous studies have also observed the presence of these bacterial genera in ticks. However, there are still debates whether these bacteria are just environmental contaminants, or they are belonging to the tick microbiota (39–41).
The tick-host interaction facilitates the transmission of AMR bacteria between the ticks and their hosts (42). During our study, different AMR bacteria were identified and some of them could be potential pathogens. On the other hand, we identified different species of the Staphylococcus including S. aureus, which could be part of the tick's microbiota. However, there is a possibility that Staphylococcus spp. has been acquired from other hosts or environments since they are resistant to different classes of antimicrobial agents. The presence of different bacteria within the ticks combined with different resistance patterns could indicate the active transmission of these AMR bacteria between the ticks and their hosts. Further studies should be encouraged to determine the role of ticks in the transmission of AMR bacteria to animals as well as humans.
5 Conclusion
Our study highlighted the risk of camels' ticks as a reservoir for AMR bacteria. There is a significant risk of transmitting AMR bacteria among camels, humans, and other animals that are meditated through ticks leading to a public health concern. This study will lay a foundation for future research on AMR pathogens transmitted by ticks and to increase the awareness of tick-transmitted pathogens that threaten the public and animal health.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization: MMA, AAlo, and AAli. Methodology: ATA, MMA NA, AOA, and ED. Validation: MMA, AAlo, and WA. Analysis: ATA, MMA, ED, MA, TT, and AAli. Resources: MMA. Writing—original draft preparation: ATA, MMA, and AAli. Writing—review and editing: ATA, MMA, AAlo, AAli, and AOA. All authors have read and agreed to the published version of the manuscript.
Funding
The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1227908/full#supplementary-material
References
1. Perveen N, Muzaffar SB, Al-Deeb MA. Ticks and tick-borne diseases of livestock in the Middle East and North Africa: a review. Insects. (2021) 12:83. doi: 10.3390/insects12010083
2. Goldstein SA, Weiss SR. Origins and pathogenesis of Middle East respiratory syndrome-associated coronavirus: recent advances. F1000Res. (2017) 6:1628. doi: 10.12688/f1000research.11827.1
3. Springer A, Glass A, Topp AK, Strube C. Zoonotic tick-borne pathogens in temperate and cold regions of europe—a review on the prevalence in domestic animals. Front Vet Sci. (2020) 7:604910. doi: 10.3389/fvets.2020.604910
4. Colwell DD, Colwell DD, Dantas-Torres F, Otranto D. Vector-borne parasitic zoonoses: emerging scenarios and new perspectives. Vet Parasitol. (2011) 182:14–21. doi: 10.1016/j.vetpar.2011.07.012
5. Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. (2004) 129:S3–14. doi: 10.1017/S0031182004005967
6. Orkun Ö, Karaer Z, Çakmak A, Nalbantoglu S. Identification of tick-borne pathogens in ticks feeding on humans in Turkey. PLoS Negl Trop Dis. (2014) 8:e3067. doi: 10.1371/journal.pntd.0003067
7. Adejinmi J, Ayinmode A. Transmission of bacteria isolates through all the developmental stages of dog ticks (bacteriological evidence). J Animal Veter Adv. (2008) 7:959–962.
8. Andreotti R, Pérez de León AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplusthrough tag-encoded pyrosequencing. BMC Microbiol. (2011) 11:6. doi: 10.1186/1471-2180-11-6
9. El Kammah KM, Oyoun LMI, Abdel-Shafy S. Detection of microorganisms in the saliva and midgut smears of different tick species (Acari: Ixodoidea) in Egypt. J Egypt Soc Parasitol. (2007) 37:533–9.
10. Jalil WI, Zenad MM. Isolation of aerobic bacteria from ticks infested sheep in Iraq. Asian Pac J Trop Biomed. (2016) 6:67–70. doi: 10.1016/j.apjtb.2015.09.020
11. Heyman P, Cochez C, Hofhuis A, van der Giessen J, Sprong H, Porter SR, et al. A clear and present danger: tick-borne diseases in Europe expert. Rev Anti Infect Ther. (2010) 8:33–50. doi: 10.1586/eri.09.118
12. CDC. Diseases Transmitted by Ticks. (2022). Available online at: https://www.cdc.gov/ticks/diseases/index.html (accessed September 11, 2023).
13. Gerem B, Angesom E, Desta H. Review on the impact of ticks on livestock health and productivity. J Biol Agric Healthc. (2016) 6:1–7.
14. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. (2015) 109:309–18. doi: 10.1179/2047773215Y.0000000030
15. van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. (2016) 30:377–90. doi: 10.1016/j.idc.2016.02.004
16. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. (2022) 399:629–655. doi: 10.1016/S0140-6736(21)02724-0
17. Serra-Burriel M, Keys M, Campillo-Artero C, Agodi A, Barchitta M, Gikas A, et al. Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: systematic review and meta-analysis. PLoS ONE. (2020) 15:e0227139. doi: 10.1371/journal.pone.0227139
18. O'Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review on Antimicrobial Resistance. London: Wellcome Trust. (2016).
19. WHO. Antimicrobial resistance. (2022). Available online at: https://www.who.int/news-room/questions-and-answers/item/antimicrobial-resistance (accessed July 27, 2017).
20. Al-Jawf Province. (2023). Available online at: https://en.wikipedia.org/wiki/Al-Jawf_Province (accessed October 15, 2023).
21. Estrada-Peña A, Bouattour AJLC, Walker A. Ticks of Domestic Animals in the Mediterranean Region. A Guide to Identification of Species. (2004).
22. Al myahii MH, Khaleel H, Ali Nasir HH. Isolation and identification of staphylococcus aureus from ticks on the cattle in Basra city. Bas J Vet Res. (2019) 18:244–251. doi: 10.33762/BVETR.2019.162855
24. biomerieux Inc. VITEK® 2: Healthcare. (2022). Available online at: https://www.biomerieux-usa.com/vitek-2 (accessed December 4, 2023).
25. CDC. Pseudomonas aeruginosa in Healthcare Settings. (2019). Available online at: https://wwwcdcgov/hai/organisms/pseudomonashtml (accessed November 13, 2019).
26. Alkhatib B, Veytsman E, Klumpp L, Hayes E. Sphingomonas paucimobilis septic shock in an immunocompetent patient. Cureus. (2022) 14:e26720. doi: 10.7759/cureus.26720
27. Hay CY, Sherris DA. Staphylococcus lentus sinusitis: a new sinonasal pathogen. Ear Nose Throat J. (2020) 99:NP62–3. doi: 10.1177/0145561319848990
28. Bhooshan S, Negi V, Khatri PK. Staphylococcus pseudintermedius: an undocumented, emerging pathogen in humans. GMS Hyg Infect Control. (2020) 15:Doc32. doi: 10.3205/dgkh000367
29. Roberts S, Chambers S. Diagnosis and management of Staphylococcus aureus infections of the skin and soft tissue. Intern Med J. (2005) 35:S97–105. doi: 10.1111/j.1444-0903.2005.00983.x
30. Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, DC: ASM press (2002). doi: 10.1128/9781555817923
31. Narnaware SD, Dahiya SS, Ranjan R, Tuteja FC. Fatal neonatal infection with Klebsiella pneumoniae in dromedary camels: pathology and molecular identification of isolates from four cases. Trop Anim Health Prod. (2020) 52:3923–9. doi: 10.1007/s11250-020-02421-4
32. Nana W, Jinmiao L, Yi D, Shibo L. Profiles of microbial community and antibiotic resistome in wild tick species. mSystems. (2022) 7:e00037–22. doi: 10.1128/msystems.00037-22
33. Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, Tomsho LP, et al. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS ONE. (2011) 6:e25604–e25604. doi: 10.1371/journal.pone.0025604
34. Zhuang L, Wang CY, Tong YG, Tang F, Yang H, Liu W, et al. Discovery of Rickettsia species in Dermacentor niveus Neumann ticks by investigating the diversity of bacterial communities. Ticks Tick Borne Dis. (2014) 5:564–8. doi: 10.1016/j.ttbdis.2014.04.004
35. Goossens H, Ferech M, Vander Stichele R, Elseviers M, Project Group ESAC. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. (2005) 365:579–87. doi: 10.1016/S0140-6736(05)17907-0
36. Wang B, Yan J, Li G, Zhang J, Zhang L, Li Z, et al. Risk of penicillin fermentation dreg: Increase of antibiotic resistance genes after soil discharge. Environ Pollut. (2020) 259:113956. doi: 10.1016/j.envpol.2020.113956
37. Thapa S, Zhang Y, Allen MS. Effects of temperature on bacterial microbiome composition in Ixodes scapularis ticks. Microbiologyopen. (2019) 8:e00719. doi: 10.1002/mbo3.719
38. Swei A, Kwan JY. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. (2017) 11:813–6. doi: 10.1038/ismej.2016.152
39. Li SS, Zhang XY, Zhou XJ, Chen KL, Masoudi A, Liu JZ, et al. Bacterial microbiota analysis demonstrates that ticks can acquire bacteria from habitat and host blood meal. Exp Appl Acarol. (2022) 87:81–95. doi: 10.1007/s10493-022-00714-x
40. Narasimhan S, Swei A, Abouneameh S, Pal U, Pedra JHF, Fikrig E. Grappling with the tick microbiome. Trends Parasitol. (2021) 37:722–33. doi: 10.1016/j.pt.2021.04.004
41. Díaz-Sánchez S, Hernández-Jarguín A, Torina A, de Mera IGF, Blanda V, Caracappa S, et al. Characterization of the bacterial microbiota in wild-caught Ixodes ventalloi. Ticks Tick Borne Dis. (2019) 10:336–43. doi: 10.1016/j.ttbdis.2018.11.014
Keywords: ticks, Hyalomma dromedarii, tick-borne bacteria, antimicrobial resistance, antimicrobial agents
Citation: Aljasham AT, Damra EM, Alkahtani NS, Alouffi A, Al Salem WS, Alshabanah AO, Alotaibi M, Tanaka T, Ali A and Almutairi MM (2023) Isolation, identification and antimicrobial susceptibility of the bacteria isolated from Hyalomma dromedarii infesting camels in Al-Jouf province, Saudi Arabia. Front. Vet. Sci. 10:1227908. doi: 10.3389/fvets.2023.1227908
Received: 24 May 2023; Accepted: 20 November 2023;
Published: 14 December 2023.
Edited by:
Kursat Altay, Cumhuriyet University, TürkiyeCopyright © 2023 Aljasham, Damra, Alkahtani, Alouffi, Al Salem, Alshabanah, Alotaibi, Tanaka, Ali and Almutairi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mashal M. Almutairi, bW1hbG11dGFpcmlAa3N1LmVkdS5zYQ==
 Alanoud T. Aljasham
Alanoud T. Aljasham Eman M. Damra2
Eman M. Damra2 Nora S. Alkahtani
Nora S. Alkahtani Moureq Alotaibi
Moureq Alotaibi Tetsuya Tanaka
Tetsuya Tanaka Abid Ali
Abid Ali Mashal M. Almutairi
Mashal M. Almutairi