
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 25 May 2023
Sec. Veterinary Experimental and Diagnostic Pathology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1183360
Classical swine fever virus (CSFV), African swine fever virus (ASFV), and Erysipelothrix rhusiopathiae (E. rhusiopathiae) remain endemic in many parts of China. Co-infections make distinguishing their clinical symptoms and pathological changes difficult. This study developed a multiplex real-time quantitative reverse transcription polymerase chain reaction (multiplex qRT-PCR) that can simultaneously detect CSFV, ASFV, and E. rhusiopathiae. Three sets of primers and probes were designed to target the CSFV 5΄ untranslated region, ASFV p72 gene, and E. rhusiopathiae 16sRNA gene. Multiplex qRT-PCR for simultaneous differential detection of these three pathogens was developed after optimizing reaction parameters such as annealing temperature, primer and probe concentrations, amplification cycles, etc. The multiplex qRT–PCR could detect CSFV, ASFV, and E. rhusiopathiae simultaneously but could not amplify other porcine pathogens. The assay’s limit of detection (LOD) was 2.89 × 102 copies/μL for CSFV, ASFV, and E. rhusiopathiae. All correlation coefficients (R2) at higher than 0.99, and the amplification efficiency was 98, 90, and 84%, respectively. All correlation coefficients (R2) were higher than 0.99, and the efficacy of amplification was 84%. In a repeatability test utilizing standard recombinant plasmids, the intra- and inter-assay coefficients of variation (CVs) were less than 2.27 and 3.79 percent, respectively. Lastly, 150 clinical samples were used to evaluate the assay’s applicability in the field. The positive rates of CSFV, ASFV, and E. rhusiopathiae were 1.33%, 0, and 3.33%, respectively. And no co-infection among the three pathogens was found. The concordance rate between the multiplex qRT-PCR and single-plex commercial PCR kits reached 100%. This study’s multiplex qRT-PCR could provide a rapid, sensitive, and specific method for the simultaneous and differential detection of CSFV, ASFV, and E. rhusiopathiae.
Classical swine fever virus (CSFV), a single-stranded, positive-sense RNA virus in the Pestivirus genus of the Flaviviridae family (1), is the causative agent of classical swine fever (CSF), which frequently manifests as fever, organ hemorrhage, and immunosuppression (2). The African swine fever virus (ASFV), an enveloped double-stranded DNA virus, is currently the sole member of the Asfivirus genus within the Asfarviridae family (3). This virus can cause African swine fever (ASF), typified by high fever, pulmonary edema, severe hemorrhage, and extensive necrosis of lymphoid tissue, with close to 100% higher morbidity and mortality (4). Erysipelothrix rhusiopathiae (E. rhusiopathiae), a nonsporulating, gram-positive, rod-shaped bacterium, pertains to the Erysipelothrix genus. E. rhusiopathiae can cause swine erysipelas, displaying the primary clinical manifestation with fever, lameness, diamond-shaped lesions on the skin, and sudden mortality in growing and adult swine (5, 6). CSFV, ASFV, and E. rhusiopathiae are still prevalent in many countries, resulting in significant economic losses for the global swine industry. Similar clinical signs and pathological alterations make it difficult to distinguish between CSF, ASF, and swine erysipelas in the field. To diagnose these diseases accurately and quickly, developing a specific, sensitive, and rapid test method to detect these pathogens simultaneously and discriminately is essential.
Multiplex real-time quantitative polymerase chain reaction (multiplex qRT-PCR) offers excellent performance in clinical testing, which is based on constant measurements of the change of fluorescent signals during the amplification reaction (7). It provides greater detection capacity, faster speed, and lower labor costs (8). Most multiplex qRT-PCR assays are based on target-specific TaqMan probes, which have higher specificity than conventional RT-PCR, and it is widely used for pathogen diagnosis and monitoring (9). Currently, several qRT-PCR and qPCR assays have been established for the detection of CSFV (10, 11), ASFV (12, 13), E. rhusiopathiae (14, 15), and CSFV+ASFV (16, 17). However, a multiplex qRT–PCR assay that can simultaneously and differentially detect CSFV, ASFV, and E. rhusiopathiae has not been reported. Hence, developing a rapid and specific method for differentiating between these three pathogens is essential. This study aimed to establish multiplex qRT-PCR method-based TaqMan probes for simultaneously and differentially detecting CSFV, ASFV, and E. rhusiopathiae.
Eight ASFV-positive nucleic acid samples were provided by the Guangdong Animal Disease Prevention and Control Center. Type O foot-and-mouth disease virus (FMDV-O) nucleic acids, type A foot-and-mouth disease virus (FMDV-A) nucleic acids, CSFV nucleic acids, E. rhusiopathiae, porcine epidemic diarrhea virus (PEDV), porcine delta coronavirus (PDCoV), transmissible gastroenteritis virus (TGEV), emerging swine acute diarrhea syndrome coronavirus (SADS-CoV), porcine reproductive and respiratory syndrome virus (PRRSV), porcine rotavirus (PRV), porcine circovirus 2 (PCV2), swine Escherichia coli, and swine Pasteurella multocida were preserved by the Institute of Animal Health of the Guangdong Academy of Agricultural Sciences. A total of 150 clinical samples were obtained from pig farms in the southern Chinese province of Guangdong between 2021 and 2022, including 107 blood samples, 35 lymph node samples, and 8 kidney samples.
The study involved the design of specific primers and corresponding TaqMan probes targeted toward the 5′ untranslated region (UTR) of CSFV, the p72 gene of ASFV, and the 16sRNA gene of E. rhusiopathiae. Then the primers and probes were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). Table 1 lists detailed data relating to the primers and probes.
Major swine pathogens and clinical samples nucleic acids were extracted using GeneRotex96 automatic nucleic acid extractor with Tianlong virus DNA / RNA extraction kit and bacterial genomic DNA extraction kit (Xi’an TianLong Science and Technology Co., Ltd., Xi’an, China) according to the manufacturer’s instructions and stored at −80°C until use.
Recombinant plasmid pUC57-CSFV-ASFV-ER contained the 267 bp gene sequence, including three target amplified fragments was synthesized by Tsingke Biotechnology Co., Ltd. DNA concentration was determined by Thermo Scientific NanoDrop Lite (Wilmington, DE, United States). And the copy number was calculated according to the following equation:
where c = concentration of pUC57-CSFV-ASFV-ER (g/μL); n = number of base pairs in a single pUC57-CSFV-ASFV-ER.
Optimizing multiplex qRT-PCR conditions, encompassing annealing temperature, primer and probe concentrations, amplification cycles, and other relevant parameters, was conducted using Analytik Jena qTOWER3/G (Jena, Germany). The optimal reaction conditions for the multiplex qRT-PCR were determined using the following 20 μL basic systems:10 μL of One Step PrimeScript III RT-qPCR (2×) (Takara, Dalian, China), 2.0 μL of standard plasmid (containing 2.89 × 104 copies/μL), and three pairs of primers and probes with varying final concentrations, along with distilled water. The amplification parameters were as follows: reverse transcription at 52°C for 5 min, predenaturation at 95°C for 10 s, 40 cycles of 95°C for 5 s, varying annealing temperature for 30 s, and scanning for the fluorescence signal at the final stage in each process. The following reaction conditions parameters were obtained through an arrangement and combination test: the annealing temperature was from 53.1 to 59.9°C; the concentrations of the probe were 50, 75, 100, and 125 nM, respectively; the concentrations of the primers were100, 150, 200, 250 nM, respectively; amplification cycles were 35, 40, 45, respectively. Optimizing the final concentrations of the primers, probes, and amplification conditions were conducted to achieve the highest fluorescence intensity and the lowest threshold cycle (CT) by utilizing the standards plasmid of 2.89 × 104 copies/μL as templates. The specificity assay, sensitivity assay, reproducibility assay, and detection of clinical samples in the following study were conducted under the optimized reaction conditions.
The specificity of the established multiplex qRT-PCR was estimated by utilizing standard DNAs or RNAs of prominent swine pathogens, such as CSFV, ASFV, E. rhusiopathiae, PEDV, PDCoV, TGEV, SADS-CoV, FMDV-O, FMDV-A, PRRSV, PRV, PCV2, swine Escherichia coli, and swine Pasteurella multocida as templates for amplification. Nuclease-free water was utilized as a negative control, while standard plasmids were employed as a positive control.
To determine the assay’s sensitivity, the pUC57-CSFV-ASFV-ER was serially diluted 10-fold 10 times with concentrations ranging from 2.89 × 10−1 to 2.89 × 108 copies/μL. The detection limit was verified by amplifying diluted standard plasmids using the optimized qRT-PCR system. The optimized RT-qPCR assay ran the pUC57-CSFV-ASFV-ER dilutions of 2.89 × 102 to 2.89 × 107 copies/μL and established three standard curves calculated and plotted automatically by qPCRsoft 4.0.
The repeatability assay included intra- and inter-assay measurements. The DNA templates used in the repeatability test were diluted to 2.89 × 103, 2.89 × 104, and 2.89 × 105 copies/μL of pUC57-CSFV-ASFV-ER. Intra-assay was conducted by performing one repeat of the assay on each of the three dilutions, with nine replicates for each dilution. Inter-assays were conducted by performing the assay three times on each of the three dilutions on three separate occasions.
A total of 150 clinical samples were extracted and tested with the optimized multiplex real-time PCR assay. Positive (RT-qPCR system with pUC57-CSFV-ASFV-ER plasmid as template) and negative (RT-qPCR system with nuclease-free water as template) controls were included in each run.
Two CSFV-positive and five E. rhusiopathiae-positive nucleic acids from the clinical samples, along with eight ASFV-positive nucleic acids, were confirmed through Sanger sequencing. These nucleic acids, including 20 negative clinical samples, were subsequently amplified using both the established multiplex qRT-PCR assay and the single-plex commercial PCR kits. The ASFV commercial PCR kit was produced by Beijing SCENK biotechnology development Co., Ltd. (Beijing, China), while the CSFV and E. rhusiopathiae commercial PCR kits were produced by Aodong Inspection & Testing Co., Ltd. (Shenzhen, China). The diagnostic sensitivity and specificity of the assay were evaluated using commercial PCR kits as the standard.
The multiplex qRT-PCR was performed using probes concentrations ranging from 50 to 125 nM and primers with concentrations ranging from 100 to 250 nM. The fluorescence intensity and Ct values of all possible combinations were compared. After optimization, the final concentrations for probes and primers were determined to be 125 and 250 nM, respectively (Table 2; Figure 1). The developed multiplex qRT-PCR with a total volume of 20 μL, containing 10 μL 2× One-Step PrimerScript III RT-PCR (TaKaRa), 0.50 μL forward primer (10 μM), 0.50 μL reverse primer (10 μM), 0.25 μL probe (10 μM), 4.25 μL RNase-free ddH2O, and 2.0 μL template. The amplification process was conducted with the following parameters: reverse transcription at 52°C for 5 min, predenaturation at 95°C for 10 s, followed by 45 cycles of denaturation at 95°C for 5 s, and annealing and extension at 59°C for 30 s. Fluorescent signals were collected at the end of each cycle.
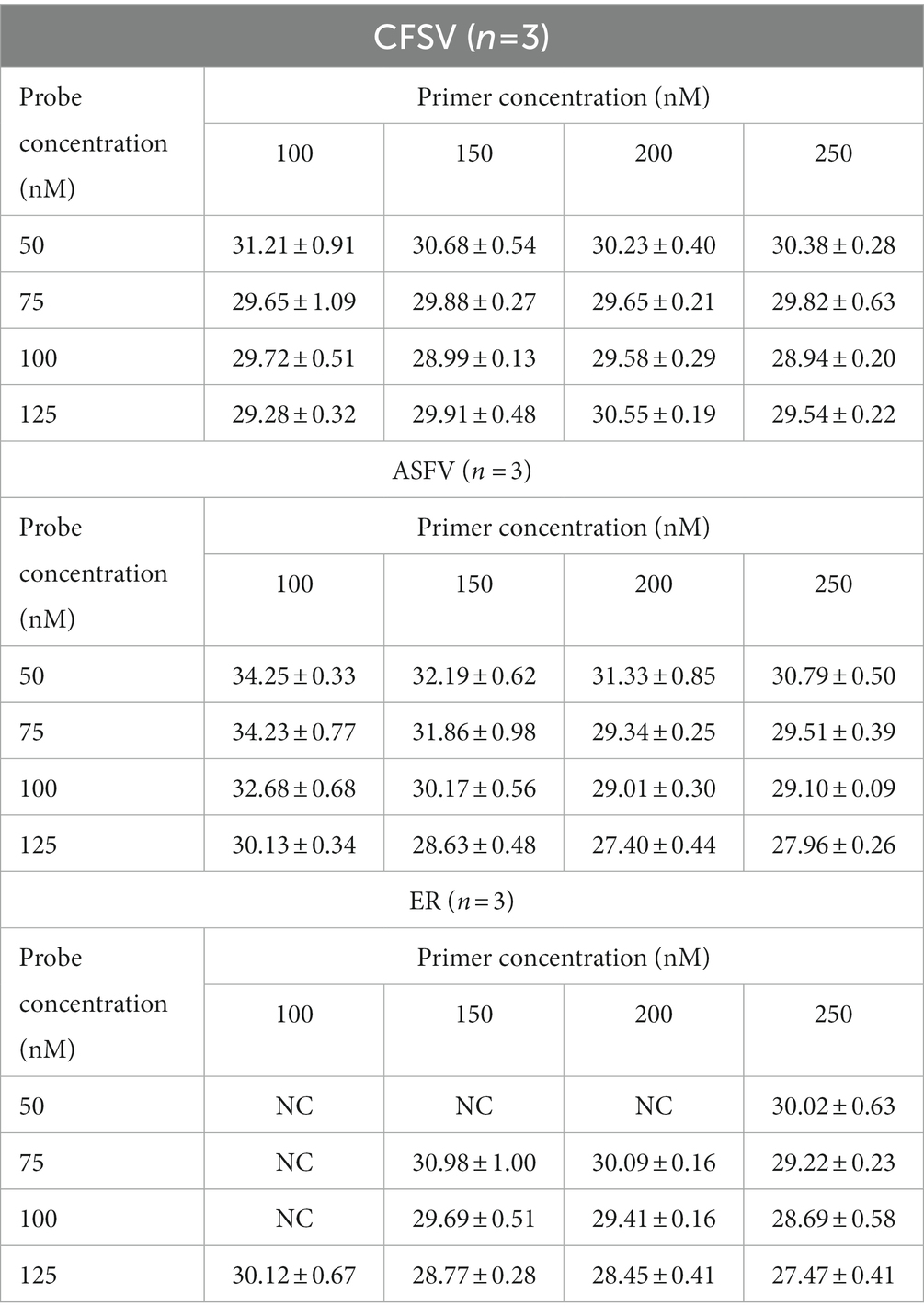
Table 2. This multiplex real-time PCR assay detected mean Ct values of CSFV, ASFV, and ER with different probe and primer concentrations.
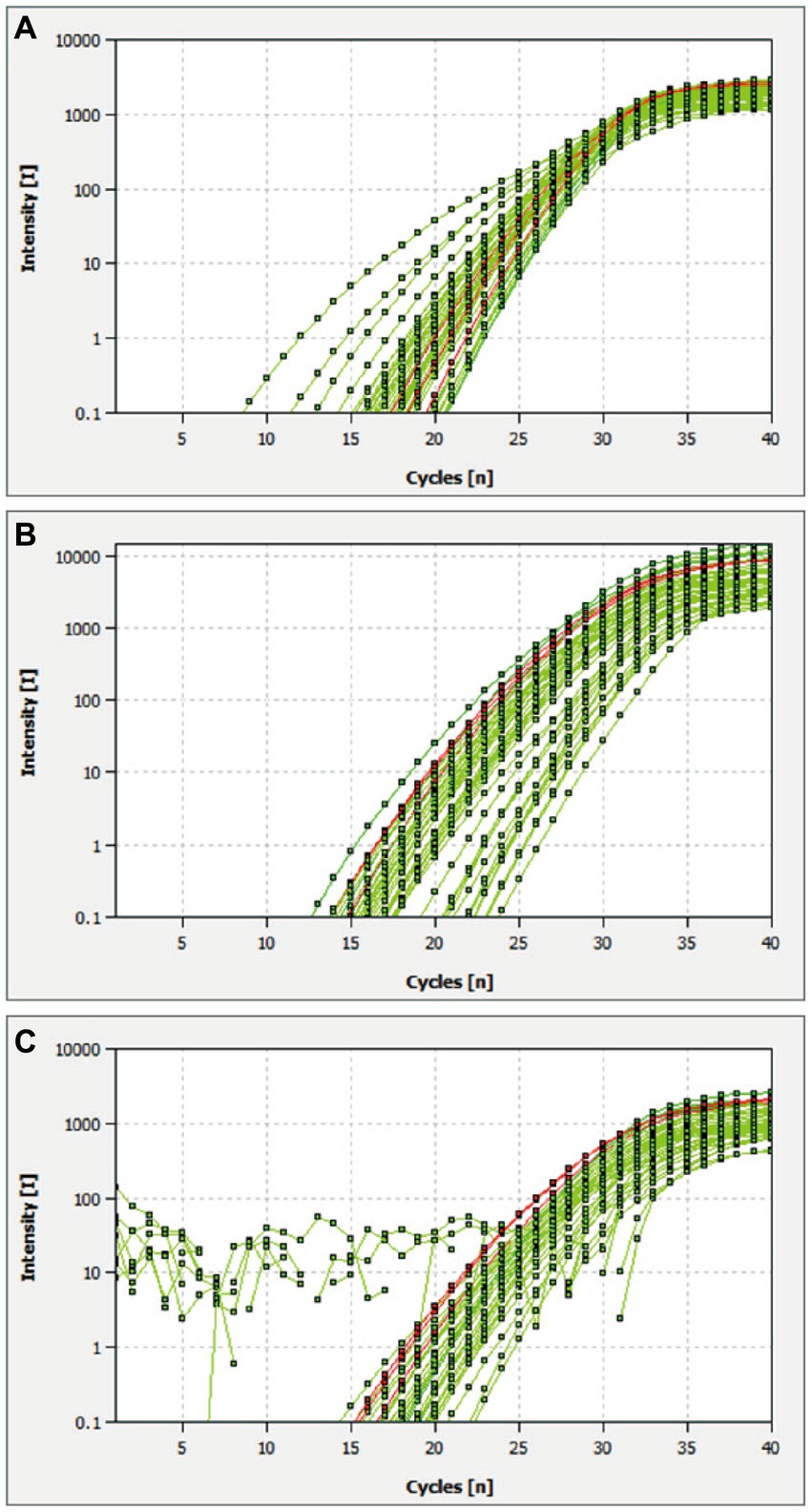
Figure 1. Optimal reaction conditions of the multiplex qRT-PCR. (A–C) Amplification curves of CSFV, ASFV, and E. rhusiopathiae detected by multiplex real-time PCR with different probe and primer concentrations. Plasmid standards with a concentration of 2.89 × 104 copies/μL were chosen as reaction templates. The red lines are the amplification curves of three fluorescence of the most suitable reaction tube.
The multiplex qRT-PCR demonstrated high sensitivity, as evidenced by the valid positive result obtained at the lowest copy number of 2.89 × 102 copies/μL (Figure 2). Multiplex qRT-PCR standard curves were constructed by 10-fold serial dilution of plasmids ranging from 2.89 × 107 copies/μL to 2.89 × 10−1 copies/μL as templates. The correlation coefficient (R2) and amplification efficiency (Eff%) of the equation were as follows: CSFV (R2 = 0.999, Eff% = 98); ASFV (R2 = 0.999, Eff% = 90); E. rhusiopathiae (R2 = 0.996, Eff% = 84), indicating a strong linear correlation between the initial template and the Ct value (Figure 2).
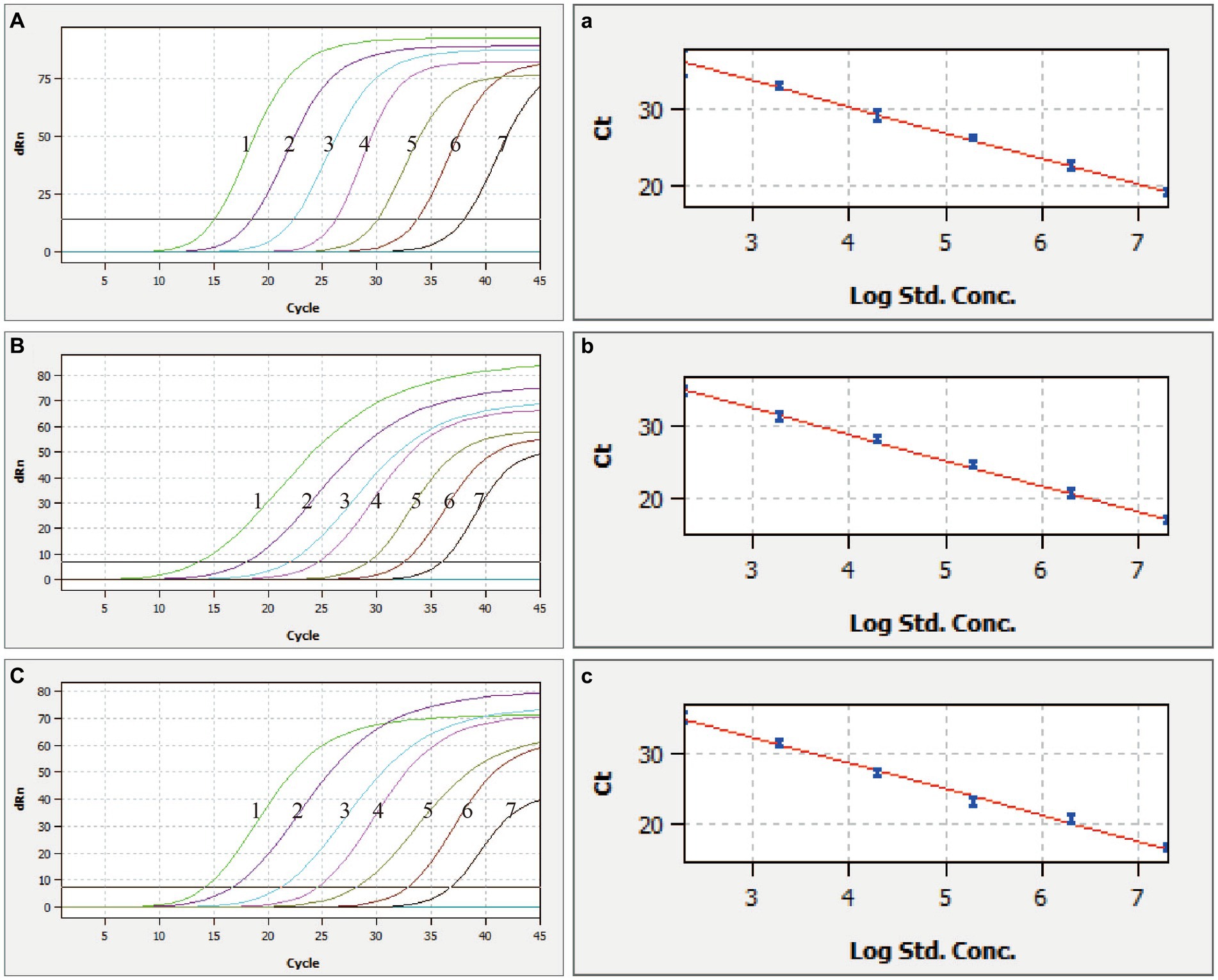
Figure 2. Sensitivity and standard curves of the multiplex qRT-PCR. (A–C) Fluorescence curve of CSFV, ASFV, and E. rhusiopathiae, (a–c) the standard curve of CSFV, ASFV, and E. rhusiopathiae, 1–7: 2.89 × 108 copies/μL – 2.89 × 102 copies/μL of the standard plasmids.
The DNA or RNA from various porcine pathogens were used as templates for the multiplex qRT-PCR. The amplification curves were observed only for CSFV, ASFV, and E. rhusiopathiae. Other viruses or bacterium, including PEDV, PDCoV, TGEV, SADS-CoV, FMDV-O, FMDV-A, PRRSV, PRV, PCV2, swine Escherichia coli, swine Pasteurella multocida, and nuclease-free water exhibited no fluorescent signals or amplification curves, indicating the assay’s high specificity (Figure 3).
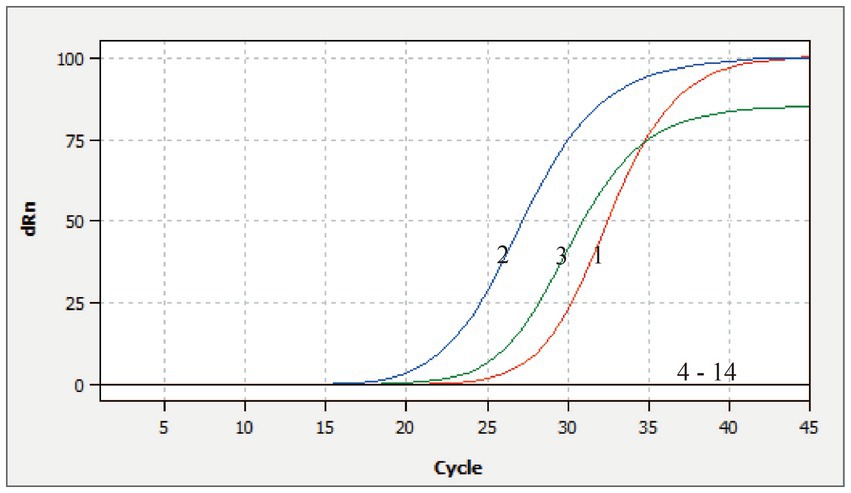
Figure 3. Specificity analysis of this multiplex qRT-PCR. 1–3: Positive templates of CSFV, ASFV, and E. rhusiopathiae. 4–14: PEDV, PDCoV, TGEV, SADS-CoV, FMDV-O, FMDV-A, PRRSV, PRV, PCV2, swine Escherichia coli, swine Pasteurella, and nuclease-free water control.
To assess the repeatability of the multiplex qRT-PCR, three different concentrations of standard plasmids (2.89 × 103, 2.89 × 104, and 2.89 × 105 copies/μL) were utilized as templates for both intra- and inter-assay comparisons. The results reveal that the coefficients of variation (CVs) for both intra- and inter-assay of the Ct values were less than 5% (Table 3), suggesting the assay’s excellent repeatability.
A total of 150 clinical samples were gathered from Guangdong Province, Southern China, during the period spanning from February 2021 to December 2022. These samples were subsequently analyzed using the established multiplex qRT-PCR method to verify its suitability. As a result, the positive rates of CSFV, ASFV, and E. rhusiopathiae were 1.33% (2/150), 0 (0/75), and 3.33% (5/150), respectively (Supplementary Table 1). And no co-infection among the three pathogens was found.
Upon comparing the results obtained from the multiplex qRT-PCR and single-plex commercial PCR, the analysis revealed a 100% concordance rate for both positive and negative samples (Table 4). Additionally, the diagnostic sensitivity and specificity were both 100%, indicating the high clinical value of the assay.
CSFV, ASFV, and E. rhusiopathiae are significant pathogens severely damaging the global swine industry. In certain pig herds, there may be cases of co-infection between CSFV and ASFV, resulting in clinical manifestations and pathological changes that are difficult to differentiate in the field. Similar observations also exist between CSFV and E. rhusiopathiae (18, 19). The clinical symptoms and pathological changes exhibited by these pathogens can be similar, posing a challenge in accurately identifying the causative agent just through clinical indicators. Hence, it is essential to differentially detect these three pathogens through laboratory procedures to diagnose these diseases accurately. Multiplex qPCR is one of the best options among the different methods for diagnosing because it can rapidly, precisely, sensitively, and accurately identify several pathogenic nucleic acids in a single reaction. It is possible to achieve simultaneous rapid detection of multiple pathogens, which is significant for rapid diagnosis, prevention, and epidemiological study of mixed infectious diseases. It is expected to be more useful in some sudden outbreak areas (20). In this study, we used three sets of specific primers and corresponding probes to develop a multiplex TaqMan probe-based qRT-PCR system that can detect CSFV, ASFV, and E. rhusiopathiae simultaneously and differentially. With a detection limit of 289 copies per reaction, The assay demonstrated the ability to precisely detect CSFV, ASFV, and E. rhusiopathiae, while exhibiting no cross-reactivity with other porcine pathogens. Moreover, the intra- and inter-assay CVs were all less than 3.79%. Thus demonstrating a high level of specificity, sensitivity, and repeatability. Furthermore, the overall concordance rate between the multiplex qRT-PCR and single-plex commercial PCR kits was 100%. Lastly, the positive rates of CSFV, ASFV, and E. rhusiopathiae in 150 clinical samples were 1.33%, 0, and 3.33%, respectively, indicating that CSFV and E. rhusiopathiae were still sporadically prevalent in pig herds in southern China. Even without co-infection among the three pathogens found, significant efforts are necessary to prevent and control these pathogens.
In summary, using the genomic sequences of CSFV, ASFV, and E. rhusiopathiae, specific primers and probes were designed. After optimizing the reaction conditions, a TaqMan-probe-based multiplex qRT-PCR capable of simultaneous and differential detection of CSFV, ASFV, and E. rhusiopathiae was developed with high specificity, sensitivity, repeatability, and clinical value.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
LZ, X-HW, and QZ designed the primers and probes, optimized the reaction conditions, and wrote the manuscript. C-LJ, X-RZ, and S-JL tested the diagnostic method. D-HL and QZ designed the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the following grants: the Project of President Funding of Guangdong Academy of Agricultural Sciences (202024), Guangzhou Basic and Applied Basic Research Foundation (202201011182), the Key-Area Research and Development Program of Guangdong Province (grant number 2019B020211005), and the grants from the Department of Agriculture and Rural Affairs of Guangdong Province (2022KJ119 and 2023KJ119).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1183360/full#supplementary-material
1. Brown, VR, and Bevins, SN. A review of classical swine fever virus and routes of introduction into the United States and the potential for virus establishment. Front Vet Sci. (2018) 5:31. doi: 10.3389/fvets.2018.00031
2. Li, H, Li, K, Bi, Z, Gu, J, Song, D, Lei, D, et al. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay for the detection of porcine pegivirus. J Virol Methods. (2019) 270:59–65. doi: 10.1016/j.jviromet.2019.04.019
3. Fan, X, Li, L, Zhao, Y, Liu, Y, Liu, C, Wang, Q, et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front Microbiol. (2020) 11:1696. doi: 10.3389/fmicb.2020.01696
4. Chen, Y, Shi, K, Liu, H, Yin, Y, Zhao, J, Long, F, et al. Development of a multiplex qRT-PCR assay for detection of African swine fever virus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus. J Vet Sci. (2021) 22:e87. doi: 10.4142/jvs.2021.22.e87
5. Wu, C, Lv, C, Zhao, Y, Zhu, W, Liu, L, Wang, T, et al. Characterization of Erysipelothrix rhusiopathiae isolates from diseased pigs in 15 Chinese provinces from 2012 to 2018. Microorganisms. (2021) 9:2615. doi: 10.3390/microorganisms9122615
6. Cabrera-García, AI, Müller, F, Rödler, FS, Traub, F, and Heilmann, RM. Infective endocarditis due to Erysipelothrix rhusiopathiae in a dog - a case report. BMC Vet Res. (2020) 16:328. doi: 10.1186/s12917-020-02546-6
7. Zhu, H, Zhang, H, Xu, Y, Laššáková, S, Korabečná, M, and Neužil, P. PCR past, present and future. Biotechniques. (2020) 69:317–25. doi: 10.2144/btn-2020-0057
8. Pan, Z, Lu, J, Wang, N, He, WT, Zhang, L, Zhao, W, et al. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence. (2020) 11:707–18. doi: 10.1080/21505594.2020.1771980
9. Heid, CA, Stevens, J, Livak, KJ, and Williams, PM. Real time quantitative PCR. Genome Res. (1996) 6:986–94. doi: 10.1101/gr.6.10.986
10. Tu, F, Yang, X, Xu, S, Chen, D, Zhou, L, Ge, X, et al. Development of a fluorescent probe-based real-time reverse transcription recombinase-aided amplification assay for the rapid detection of classical swine fever virus. Transbound Emerg Dis. (2021) 68:2017–27. doi: 10.1111/tbed.13849
11. McGoldrick, A, Lowings, JP, Ibata, G, Sands, JJ, Belak, S, and Paton, DJ. A novel approach to the detection of classical swine fever virus by RT-PCR with a fluorogenic probe (TaqMan). J Virol Methods. (1998) 72:125–35. doi: 10.1016/s0166-0934(97)00208-5
12. Gao, Q, Feng, Y, Yang, Y, Luo, Y, Gong, T, Wang, H, et al. Establishment of a dual real-time PCR assay for the identification of African swine fever virus genotypes I and II in China. Front Vet Sci. (2022) 9:882824. doi: 10.3389/fvets.2022.882824
13. Trinh, TBN, Truong, T, Nguyen, VT, Vu, XD, Dao, LA, Nguyen, TL, et al. Development of a novel real-time PCR assay targeting P54 gene for rapid detection of African swine fever virus (ASFV) strains circulating in Vietnam. Vet Med Sci. (2021) 7:2268–72. doi: 10.1002/vms3.605
14. To HKoyama, T, Nagai, S, Tuchiya, K, and Nunoya, T. Development of quantitative real-time polymerase chain reaction for detection of and discrimination between Erysipelothrix rhusiopathiae and other Erysipelothrix species. J Vet Diagn Investig. (2009) 21:701–6. doi: 10.1177/104063870902100518,
15. Pal, N, Bender, JS, and Opriessnig, T. Rapid detection and differentiation of Erysipelothrix spp. by a novel multiplex real-time PCR assay. J Appl Microbiol. (2010) 108:1083–93. doi: 10.1111/j.1365-2672.2009.04560.x
16. Haines, FJ, Hofmann, MA, King, DP, Drew, TW, and Crooke, HR. Development and validation of a multiplex, real-time RT PCR assay for the simultaneous detection of classical and African swine fever viruses. PLoS One. (2013) 8:e71019. doi: 10.1371/journal.pone.0071019
17. Nishi, T, Okadera, K, Fukai, K, Yoshizaki, M, Nakasuji, A, Yoneyama, S, et al. Establishment of a direct PCR assay for simultaneous differential diagnosis of African swine fever and classical swine fever using crude tissue samples. Viruses. (2022) 14:498. doi: 10.3390/v14030498
18. Cabezón, O, Muñoz-González, S, Colom-Cadena, A, Pérez-Simó, M, Rosell, R, Lavín, S, et al. African swine fever virus infection in classical swine fever subclinically infected wild boars. BMC Vet Res. (2017) 13:227. doi: 10.1186/s12917-017-1150-0
19. Liu, H, Shi, K, Sun, W, Zhao, J, Yin, Y, Si, H, et al. Development a multiplex RT-PCR assay for simultaneous detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. J Virol Methods. (2021) 287:114006. doi: 10.1016/j.jviromet.2020.114006
Keywords: classical swine fever virus (CSFV), African swine fever virus (ASFV), Erysipelothrix rhusiopathiae , multiplex qRT-PCR, detection
Citation: Zhao L, Wen X-H, Jia C-L, Zhou X-R, Luo S-J, Lv D-H and Zhai Q (2023) Development of a multiplex qRT-PCR assay for detection of classical swine fever virus, African swine fever virus, and Erysipelothrix rhusiopathiae. Front. Vet. Sci. 10:1183360. doi: 10.3389/fvets.2023.1183360
Received: 10 March 2023; Accepted: 03 May 2023;
Published: 25 May 2023.
Edited by:
Fernando Costa Ferreira, University of Lisbon, PortugalReviewed by:
Denis V. Kolbasov, Federal Research Center of Virology and Microbiology, RussiaCopyright © 2023 Zhao, Wen, Jia, Zhou, Luo, Lv and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dian-Hong Lv, Z2RyZWRAMTYzLmNvbQ==; Qi Zhai, emhhaXhpYW9xaUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.