
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci., 03 August 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1180621
 Ali Mazloum1*
Ali Mazloum1* Maxim Karagyaur2
Maxim Karagyaur2 Roman Chernyshev1
Roman Chernyshev1 Antoinette van Schalkwyk3,4
Antoinette van Schalkwyk3,4 Ma Jun5
Ma Jun5 Fu Qiang5
Fu Qiang5 Alexander Sprygin1*
Alexander Sprygin1*Gene editing tools have become an indispensable part of research into the fundamental aspects of cell biology. With a vast body of literature having been generated based on next generation sequencing technologies, keeping track of this ever-growing body of information remains challenging. This necessitates the translation of genomic data into tangible applications. In order to address this objective, the generated Next Generation Sequencing (NGS) data forms the basis for targeted genome editing strategies, employing known enzymes of various cellular machinery, in generating organisms with specifically selected phenotypes. This review focuses primarily on CRISPR/Cas9 technology in the context of its advantages over Zinc finger proteins (ZNF) and Transcription activator-like effector nucleases (TALEN) and meganucleases mutagenesis strategies, for use in agricultural and veterinary applications. This review will describe the application of CRISPR/Cas9 in creating modified organisms with custom-made properties, without the undesired non-targeted effects associated with virus vector vaccines and bioactive molecules produced in bacterial systems. Examples of the successful and unsuccessful applications of this technology to plants, animals and microorganisms are provided, as well as an in-depth look into possible future trends and applications in vaccine development, disease resistance and enhanced phenotypic traits will be discussed.
Targeted genome engineering enables the modification of specific genomic loci at predetermined sites to generate novel plants, animals, and microorganisms by bypassing the procedure of using random mutagenesis and long-term selection (1, 2). Understanding the functional significance of individual coding and regulatory genes creates unprecedented opportunities. These technologies make it possible to revolutionize agriculture and industry to solve research and practical problems in the areas of food and biological safety. The development of these technologies will enable us to further understand the function of individual genes, which can be exploited to improve nutrition and the taste of food, and increase the resistance of agricultural animals and plants to infection and parasites (3–5).
Among the main approaches to directly modify the genome, two technologies can be distinguished: ectopic gene insertion (cis- and trans-genesis) and editing the nucleotide sequence (6, 7). Each of these strategies is radically different from the previous approaches of induced mutagenesis and crossing that were once widely used. Their use required a clear understanding of the function of individual genes as well as their interactions and regulation. Thus, the use of direct genomic modification technologies is practically devoid of the element of randomness, and the resulting organisms, with a high degree of probability, acquire and transmit the desired properties and characteristics to their offspring. Each of these approaches has certain features, advantages and limitations, successes and failures, of which the most relevant will be discussed in this review.
Genetic engineering was first introduced into scientific practice in the 1970s, even before the advent of widespread modern nuclease-based gene editing platforms; however, a number of technical shortcomings limited its widespread use. The limiting factors of the methods initially used were relatively low efficiencies and specificity (depending on the specific object of research) as well as significant material and time costs.
At the early stages of genetic engineering, genome modification was based upon the natural mechanism of homologous recombination (very low efficiency–up to 0.001%), when a vector (usually a recombinant plasmid) containing a DNA construct corresponding to the sequence of the target gene with the necessary modifications exchanged nucleotide sequences with the intact target gene (8). The revolutionary work of Rouet et al. demonstrated the possibility of introducing double-strand breaks into the chromosomes of mouse cells of the transplanted COS1 cell line using an expression system of the rare-cut nuclease I-Sce I with amazing efficiency, up to 82% (9, 10).
The discovery of site-specific recombinases (SSR) of bacteriophage P1 and baker’s yeast Saccharomyces cerevisiae introduced the method of complete gene deletion into the practice of genome editing (11, 12). The two most common types of SSR are the Cre-loxP and Flp-FRT systems. Cre recombinase is an enzyme first discovered in bacteriophage P1 that excises a DNA fragment by homologous recombination between highly specific flanking sequences, known as Lox-P sites (13, 14). The Flp-FRT system operates in a similar manner; however, the Flp recombinase discovered in baker’s yeast recognizes FRT sequences (15, 16). During hybridization of cells expressing SSR and cells containing the gene of interest flanked by loxP or FRT sites, the target gene may be cleaved and inactivated (Figure 1) (17, 18). The main disadvantage of this method is the need for preliminary integration of loxP and FRT sites within the genome region to be deleted. Therefore, its main application was the creation of genomic modifications for basic animal research (19). The targets in this case, as a rule, were marker genes encoding green fluorescent protein (GFP), β-galactosidase, antibiotic resistance enzymes, and others (20).
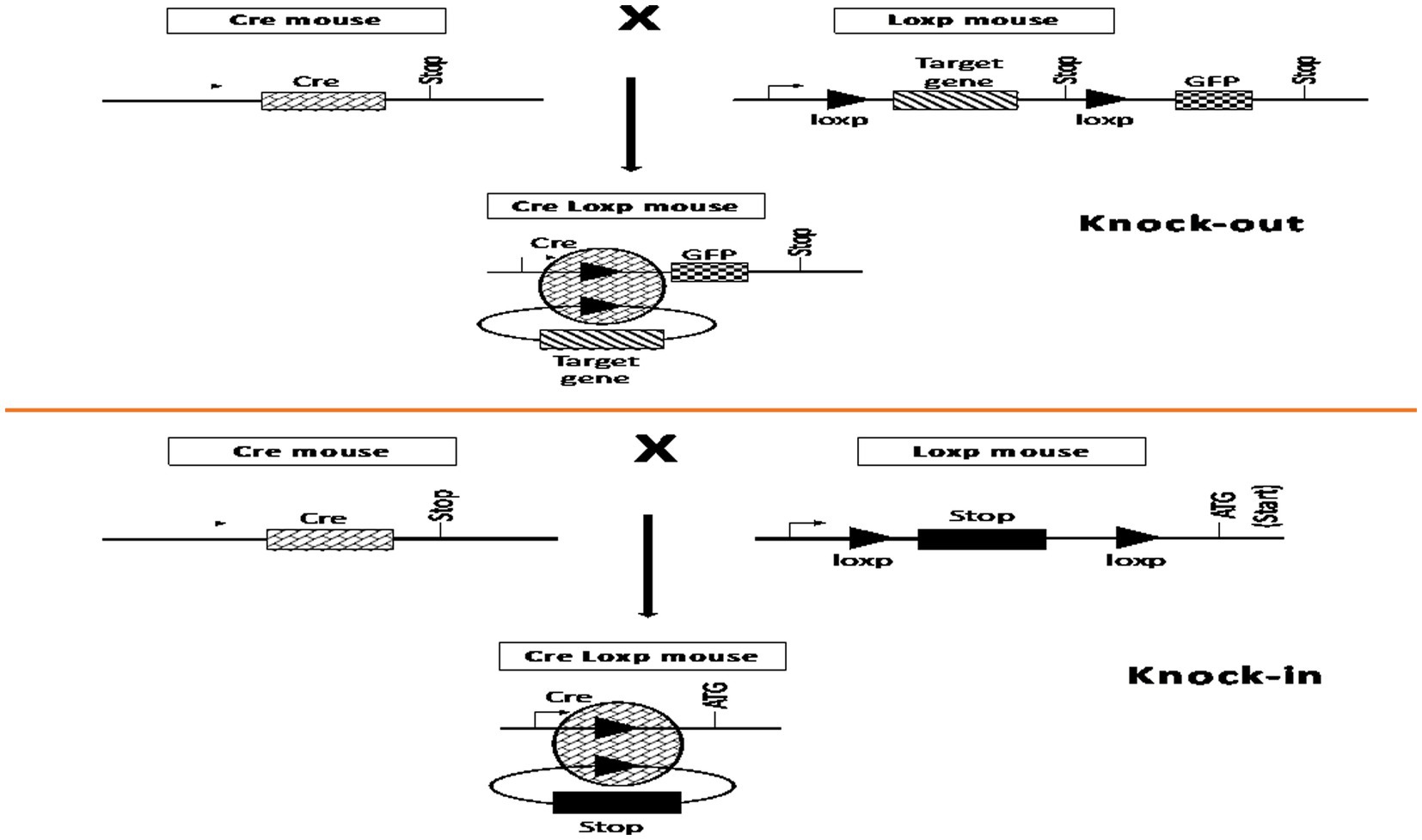
Figure 1. Mechanism of action of site-specific recombinases in mouse cells. This figure is based on previously published work (18).
Because it was found that the introduction of a double-stranded DNA break markedly increases the efficiency of genome editing, the search for endonucleases capable of generating such breaks within a desired region of the genome began. As a result, meganucleases were used for these purposes and a number of artificial nucleases were developed including zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). A feature of these nucleases is the recognition of the target DNA sequence due to amino acid residues, which required time-consuming optimization of the accuracy and efficiency of these systems. In 2012, the CRISPR/Cas9 bacterial “immunity” system was proposed for genome editing, which targets a region of the genome based on Watson-Crick interactions between the target region of the DNA and crRNA/gRNA, which is part of the CRISPR/Cas9 effector complex. The introduction of a double-stranded DNA break triggers the repair process, which includes non-homologous end connection (NHEJ) and homologous directed repair (HDR). NHEJ directly connects the ends of the cleaved DNA, whereas HDR uses a homologous sequence as a matrix to restore missing DNA sequences at the break point (21). A brief demonstration of the mechanism of action of these four nuclease systems is presented in Figure 2.
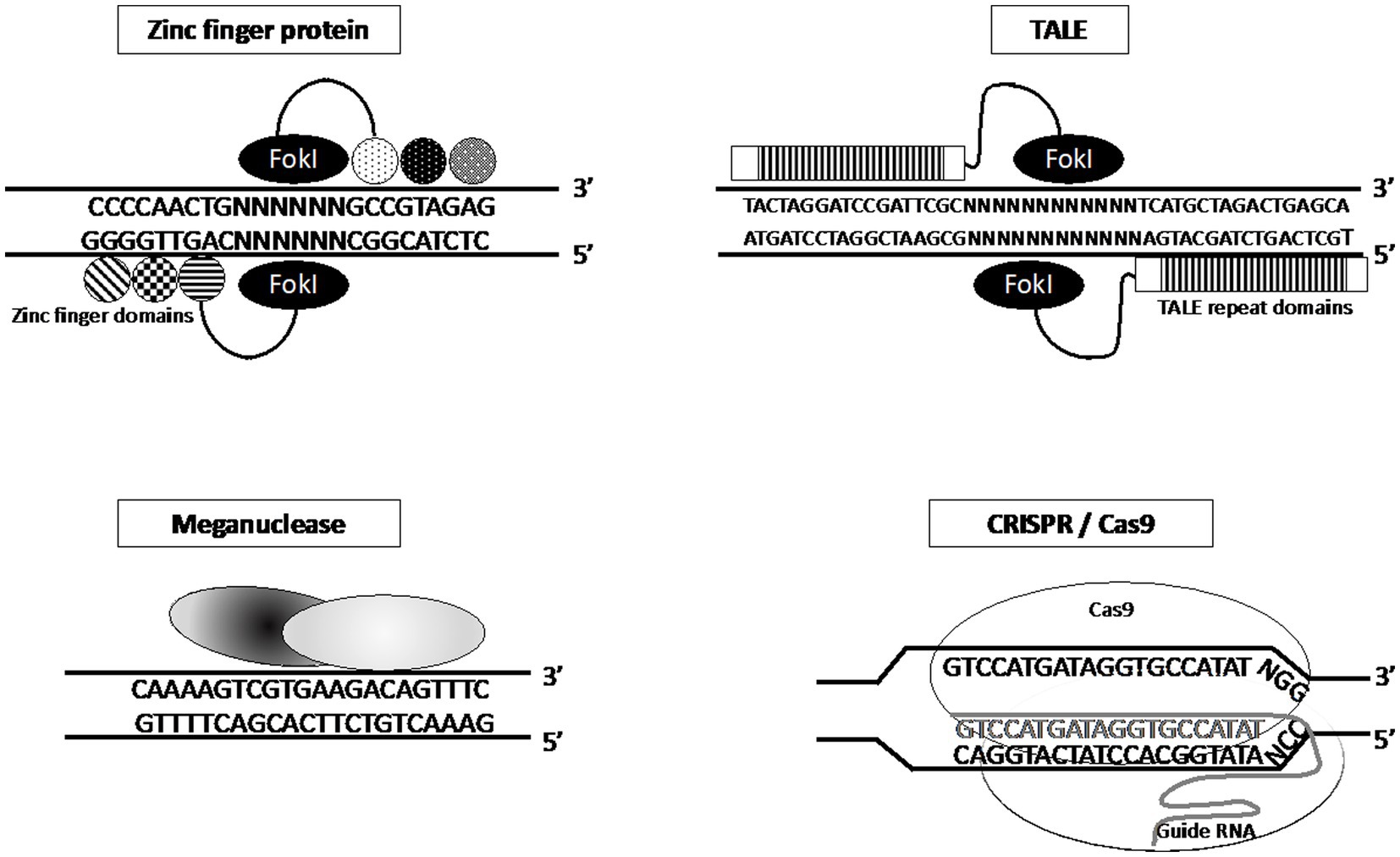
Figure 2. Mechanism of action of different genetically engineered nucleases for DNA restriction (22).
The first kind of nucleases that were used were Meganucleases, which are a highly specific enzymes characterized by an ability to recognize and cut longer DNA sequences from 14 to 40 base pairs (Figure 2) (22). There are five meganuclease families based on their structures: LAGLIDADG, His-Cys box, HNH, PD-(D/E)XK and GIY-YIG (23). The LAGLIDADG family of proteins exhibits conserved features, of which I-CreI, I-CeuI, I-AniI, I-SceI, I-PfuI, and I-DmoI, are the most widely used (24, 25). Because of their high specificity for the DNA target site, the number of candidate target genes for genome editing is relatively low, which requires the identification of new meganucleases for genetic modification of other genes. Currently, tens of thousands of such enzymes are available; however, identifying specific meganucleases for each purpose (modification) is a significant obstacle when conducting research (26).
The second type are Zinc-finger nucleases (ZFNs), a synthetic endonucleases constructed by fusing the DNA-binding domains of transcription factors with the nuclease domain of FokI restriction enzyme (27). Each DNA-binding domain can recognize 3 nucleotides and when combined into tandem structures of 3–6, the DNA-binding domains of individual ZFNs are able to recognize 9–18 pairs of nucleotides in the target DNA. ZFNs involve the use of a pair of editors designed in such a way that each pair recognizes DNA sites in the forward (5′-3′) and reverse (3′-5′) strands of the target gene flanking the area selected for editing. With this design, the nuclease domains of FokI dimerize and introduce a double-strand break in DNA (Figure 2) (28, 29). Despite a number of advantages, the use of ZFN is limited because of the high cost and complexity of designing and testing the editors, the limited choice of the target site, and the possibility of non-specific DNA cleavage.
Transcription activator-like effector nucleases (TALEN) are the third type of artificial programmable endonucleases, created by fusing transcription activator-like effector (TALE) DNA-binding domains to the DNA cleavage domain of FokI endonuclease (Figure 2). TALEs can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The TALE DNA-binding domain contains a repeated highly conserved 33–34 amino acid sequence with divergence at the 12th and 13th amino acids (30). These two positions, referred to as the Repeat Variable Di-residue (RVD), are highly variable and show a strong correlation with specific nucleotide recognition (31).
The main advantage of the TALEN design over ZFN is the greater number (>18 bp) of binding nucleotides. To date, there are approximately 20 constructed RVDs, 7 of which are used in 90% of the cases (32). Among other advantages of TALENs is the ability to edit a wider range of DNA sites compared with ZFNs, which are determined by the sequence of the DNA-binding domain. Nevertheless, TALENs have disadvantages manifested both in off-targeted binding effects and the difficulty in delivering enzymes to target cells because of their high molecular weight (33, 34).
The fourth type of nucleases are CRISPR/Cas, which revolutionized the science of genetic modification in the past decade, and will be subsequently discussed in more detail.
A recent and revolutionary genome editing tool is the CRISPR/Cas9 bacterial “immunity” system, which is based on the guiding ability of short RNAs and nuclease activity of the CRISPR-associated Cas9 enzyme. When combined, the complex (Cas9/gRNA) can cleave DNA at almost any desired site (Figure 2) (35, 36). In nature, CRISPR (clustered regularly interspaced short palindromic repeats) is an adaptive immunity mechanism for members of the Archaea and bacteria kingdoms. It consists of two key components; CRISPR array and CRISPR-associated nuclease. The CRISPR array stores genetic information pertaining to previous phage or plasmid infections, thus acting as a genetic “memory.” It consists of tandem nucleotide repeats (25–36 bp in length) separated by unique sequences of approximately the same length, also known as spacers. The spacers are acquired from the genome of parasitic elements during prokaryote infection and become the basis of prokaryote adaptive immunity, as they guide the CRISPR-associated nuclease (the second component of CRISPR/Cas9 system) to destroy the parasite genome during re-infection. A short RNA guiding CRISPR/Cas9 system that targets DNA is known as crRNA (CRISPR RNA). The genes encoding Cas proteins are often located in close proximity to the CRISPR template (Figure 3) (21, 37).
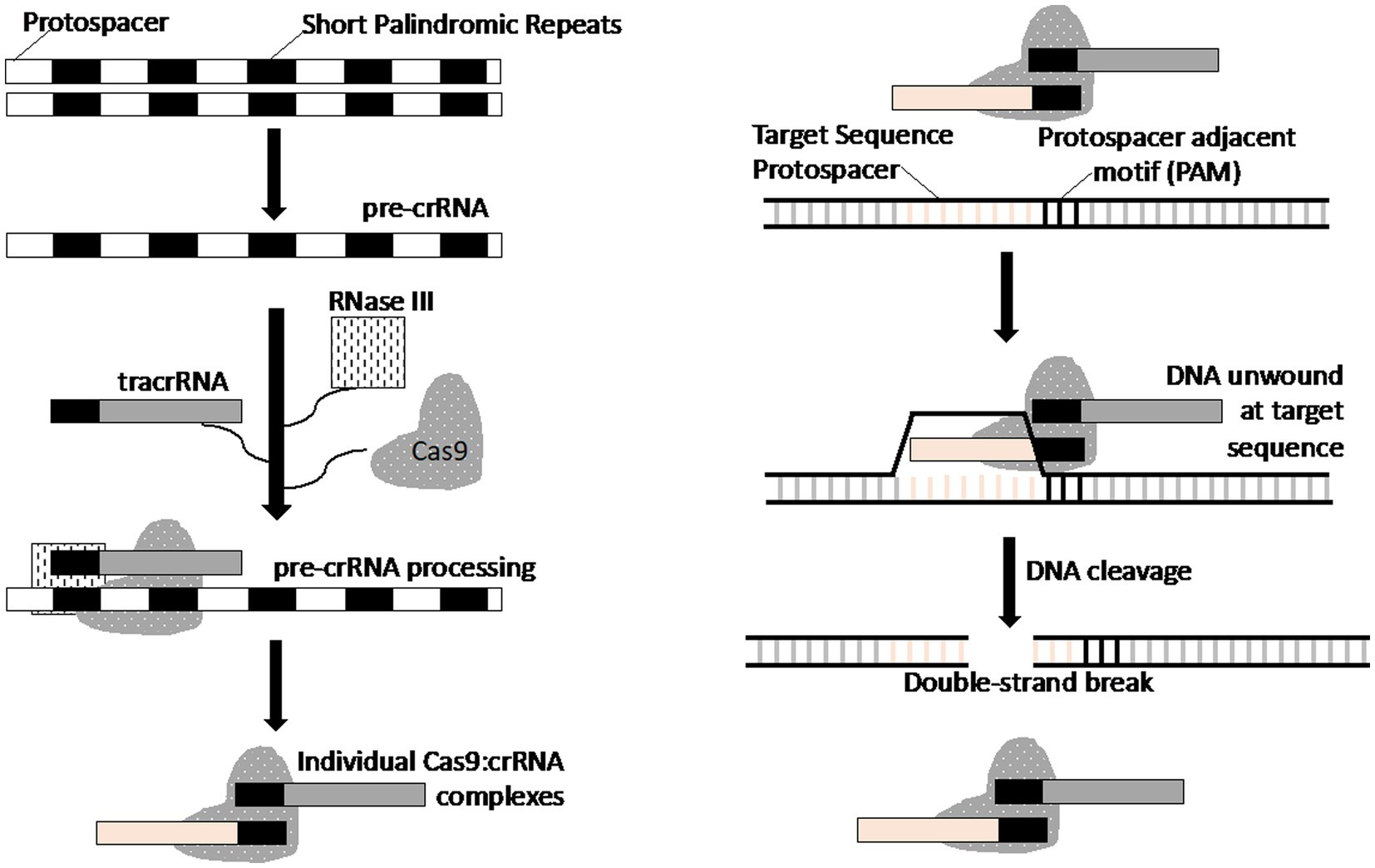
Figure 3. Schematic representation of the mechanism of the CRISPR-Cas system for double-stranded DNA cleavage. This figure is based on previously published work (21).
Three types (I–III) of CRISPR systems have been identified across a wide range of bacterial and archaeal hosts. Each system comprises a cluster of CRISPR-associated (Cas) genes, noncoding RNAs, and a distinctive array of repetitive elements (direct repeats). The Type II CRISPR system is well-characterized, consisting of the nuclease Cas9, the crRNA array that encodes the guide RNAs, and a required auxiliary trans-activating crRNA (tracrRNA) that facilitates the processing of the crRNA array into discrete crRNAs and mediates the crRNA interaction with the Cas9 apoenzyme. The synthetic crRNA analog obtained by fusion of crRNA and trans-crRNA is known as a single guide RNA (sgRNA or gRNA). Because of its convenience and easy customization, CRISPR/Cas9 technology has been used for genome editing of target cells. Since 2010, multiple modifications and improvements to this system have been implemented to make it easier to use and available to any researcher (38). This system enables one to control gene expression, make precise genome modifications, and even trigger chromosomal recombination.
As mentioned previously, CRISPR-Cas technology relies on the interaction between the crRNA/gRNA and target DNA (RNA–DNA interaction), which determines the exact position where Cas nucleases can perform double-strand DNA cleavage. Changing the sequence of the sgRNA changes the specificity of the CRISPR/Cas9 system. Because the synthesis of a new crRNA/gRNA is easy, CRISPR/Cas9 has a number of advantages compared with ZFNs and TALENs, such as the simplicity of design, versatility with respect to any target, and the ability to modify several genomic sites simultaneously (multiplex editing). The only significant disadvantage of CRISPR/Cas9 is the high molecular weight of the Cas9 nuclease, which can impede delivery of the genome editing components into cells. However, various easy-to-use delivery methods (e.g., lentiviral expression system, liposomes, electroporation, transfection by recombinant plasmids) have been developed to overcome this limitation (39, 40). A comparison of the advantages and disadvantages of the four methods for nuclease genome editing is presented in Table 1 (41). Based on the majority of the parameters evaluated in Table 1, CRISPR/Cas9 technology is the superior or most preferred technology.
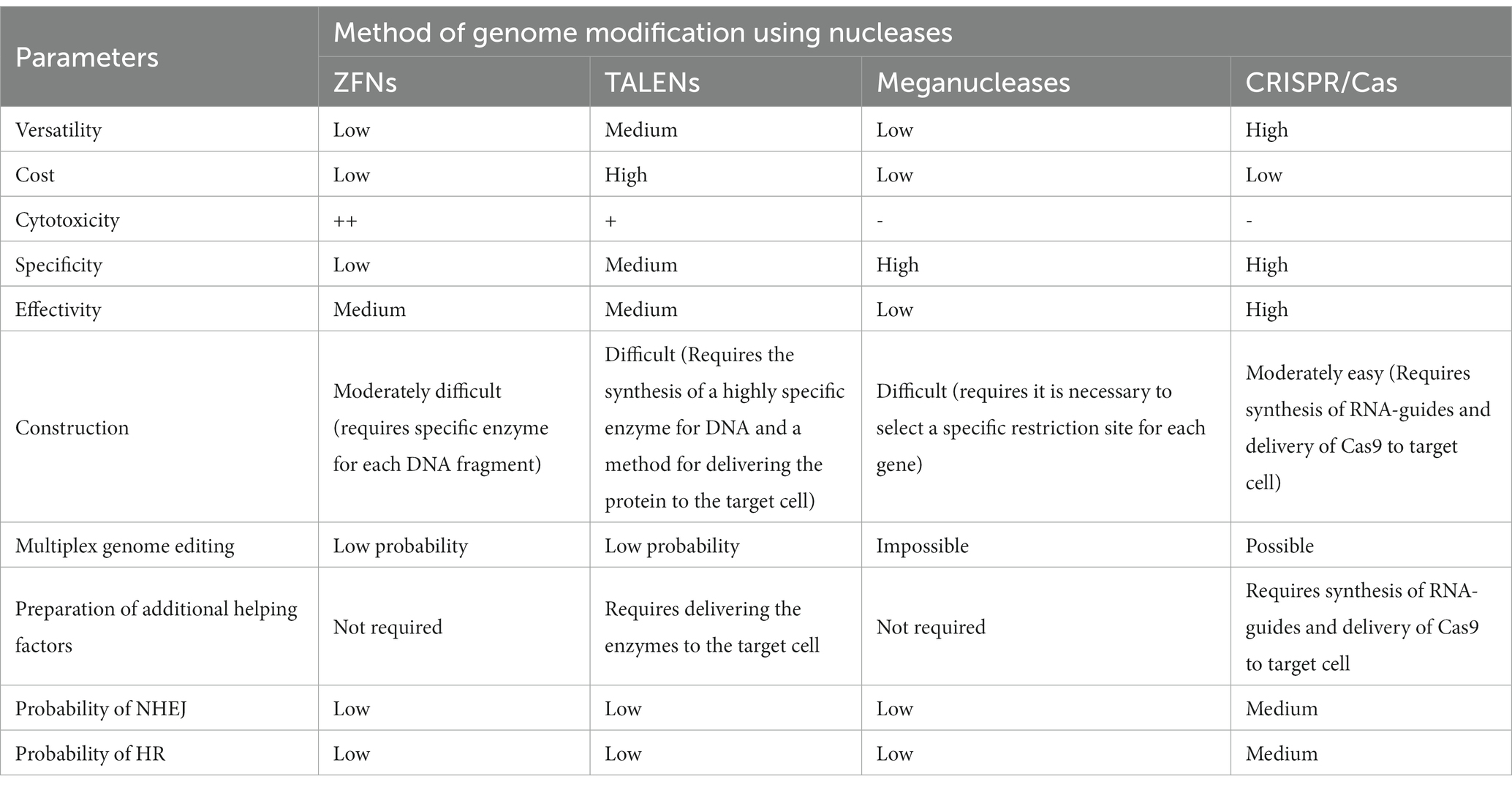
Table 1. Comparison of parameters for various methods of genetic editing using nucleases (41).
CRISPR -Cas9 technology facilitates knockin–knockout strategies to study and determine the influence of a particular gene on the virulence of a virus or its replication as well as the phenotypic traits of animals or prokaryotes. The results of such modifications are usually associated with one of two possible outcomes: knockout - deletion of any gene from the genome, or knockin–introduction of a new gene into the genome of a virus or animal. As a result, the implementation of a DIVA strategy (differentiation of infected from vaccinated animals), in which the gene is inactivated by integrating a marker gene (e.g., encoding a green or red fluorescent protein), facilitates the selection of the recombinant organism.
The major advantages of CRISPR/Cas 9 is the ease and versatility of design and cloning to generate guide RNA (gRNA) libraries for large-scale genetic screening. Genomic screenings enables the discovery of the function of new genes and to determine their role and mechanism of action within living cells. Before CRISPR/Cas9, genomic screenings were performed using two main approaches: “turning on” gene expression using a cDNA library and “turning off” genes by RNA interference (42). The efficacy of these approaches was rather restricted, whereas contemporary gRNA libraries enable one to edit not only the whole genome, but a subset of genes involved in a specific signal or metabolic pathway as well as ncRNAs. Moreover, no Cas9-related cytotoxicity has been reported and its activity depends solely on the complex of NHEJ proteins responsible for DNA repair following double-strand breaks. Currently, CRISPR/Cas 9-based screenings are used to analyze pathological processes, such as carcinogenesis, metastasis, and inflammation (21).
Various scientific groups around the world are conducting research on the development of prevention strategies for various animal diseases (e.g., African swine fever, classical swine fever, Marek’s disease, capripoxvirus infections, Aujeszky’s disease) in an effort to create a safe and effective biological product or vaccine that provides long-term protection against infection.
The increasing global population depends on the effective and enhanced productivity of livestock and poultry for food. One of the best strategies to increase the efficiency of food production is to select farm animals with the most desirable traits, either through extensive selective breeding campaign or by modifying the genetics of these animals (43). Microinjection of the structures required for CRISPR/Cas9 function, directly into zygotes results in a high probability of obtaining offspring with the desired recombinant traits. The CRISPR/Cas9 construct is capable of inactivating genes and creating site-specific mutations by cytoplasmic injection, pronuclear injection, or electroporation (44).
Because CRISPR/Cas9 has become an important tool for genetic engineering, it has been used to produce changes and correct undesirable defects in specific genes to improve the quality of livestock (e.g., cattle, pigs, horses). A large number of animals with edited genomes have been created. These include a pig with an altered myostatin gene, resulting in larger muscle mass, or a pig with resistance to the reproductive and respiratory syndrome virus, and transgenic cattle resistant to tuberculosis (45–47). Recently, scientists from China has created three different pig lines that are resistant to the classical swine fever virus (48–51). When studying the productivity of genetically modified livestock, significant improvements have been achieved with respect to meat yield, disease resistance, and other desirable qualities (52).
Virus replication depends on numerous factors in the target host cells. Due to the obligate host-dependent replication of viruses, it is more difficult to apply genome editing tools to the virus genome than compared with the editing of the eukaryote host genome. Success in generating novel viral mutants has been achieved with CRISPR/Cas9 in the absence of viral replication in the cell and, as a consequence, decontamination of cell cultures and sensitive organisms. Recently, various studies have successfully used CRISPR/Cas9 technology to obtain attenuated viral variants (viruses with both DNA- and RNA-containing genomes) as vaccine prototypes (53, 54).
The CRISPR/Cas9 system has been successfully applied to DNA-containing viruses, such as the smallpox vaccine virus (VACV) (55, 56), human polyomavirus 2 (JCPyV) (57), African swine fever virus (ASFV) (58–60), avian adenovirus 4 (FAdV-4) (61), Aujeszky’s disease virus (62, 63), and Marek disease virus (64) (Table 2). CRISPR/Cas9 technology guided by gRNA provides significantly greater efficiency for creating mutant VACVs without obvious side effects in recipient organisms compared to using a site-specific recombinase system. Additionally, it provides a marker-free system that can be used to efficiently construct VACV vectors carrying therapeutic genes in the TK or N1L regions (55, 56). A system was established to make changes to the non-coding region and the open reading frame (ORS) of the “late” gene in the genome of human polyomavirus 2, which subsequently inhibited viral replication and protein expression (57). The genomes of ASFVs were altered between positions 71 and 78 in the phosphoprotein p30 gene, encoded by the CP204L gene. This resulted in the mutated ASFVs not displaying the classical cytopathic effects when cultured in the infected wild boar lung cell line WSL (58). Additionally, the complete deletion of the gene encoding the immunogenic protein A238L (5EL) of the ASF virus genotype IX was the basis for implementing a differentiation between infected and vaccinated animal (DIVA) strategy for ASF (59). The CRISPR/Cas9 system was successfully applied to generate a unique nucleotide deletion (1966DEL) within the avirulent strain KR5 of the avian adenovirus 4, between ORS 42 and ORS 43 confirmed the absence of virulent function (61).
Currently, the CRISPR/Cas9 system is primarily used for editing genes of DNA-containing viruses, but technical obstacles related to its use for the mutagenesis of RNA-containing viruses remain. The recently discovered Cas9 nuclease from the Gram-negative bacterium Francisella novicida (FnCas9) is capable of targeting endogenous bacterial RNA. This FnCas9 can be directed by an engineered RNA-targeting guide RNA to target and inhibit a human +ssRNA virus, hepatitis C virus, within eukaryotic cells. This work revealed a versatile and portable RNA-targeting system that can effectively function in eukaryotic cells and be programmed as an antiviral defense (65). The recently developed CRISPR/Cas13a system effectively modified the NS3 region of the Dengue virus and the replication of the recombinant virus in mammalian cells (66). The loss of infectious activity of the Porcine reproductive and respiratory syndrome virus (PRRSV) resulted from the inactivation of the viral genes, ORC5 and ORC7, using the CRISPR/Cas13b system; thus, the cleavage of viral RNA was observed in virus-sensitive cells (67). These data demonstrate a new and effective technology for future gene editing of RNA-containing viruses.
Orthopoxviruses (OPXV) are suitable viral vectors for gene expression and therapy because they have a large transgenic capacity up to 25,000 base pairs (kbp) (68), a wide range of hosts (including humans) (69, 70), and they are capable of stimulating both long-term cellular and humoral immune responses against the viral vector (−s) antigen (s), despite the existing vector immunity (68, 71). They are also thermostable, easy to store, transport, and use, which ultimately ensures their high profitability in commercial production systems. The first OPXV successfully used as a vector was the smallpox vaccine virus (VACV), which was used as a vaccine to fight smallpox in humans. Because of the successful use of recombinant VACV-encoding hepatitis B antigens to stimulate the immune system against hepatitis B (72), replication-deficient orthopoxvirus-modified vaccinia virus Ankara (MVA) and NYVAC was adopted for constructing recombinant vectors and vaccines against various human and animal diseases (73, 74).
Recombinant OPXV can encode antigens for one or more infectious agents, including antigens and genes encoding specific immune-stimulating factors, such as cytokines or chemokines (75). Recombinant OPXV can be constructed by homologous recombination between transfected heterologous DNA and replicating viral DNA (76), in vitro ligation (77), or recombination of artificial bacterial chromosomes (78). Commonly used insertion sites include the thymidine kinase (TK) gene, the hemagglutinin (HA) gene, the intergenic region between the F12L and F13L genes, and sites of natural deletions in the OPXV genome (especially with respect to MVA) (76).
As a rule, these systems require multistage operations with low efficiency. The creation of candidate recombinant vaccines by homologous recombination is limited due to low recombination efficiency (<3%), the complexity of processes (for example, the creation of a plasmid with a transgene and selection of a recombinant virus), the instability of the transgene, the need for 200–500 bp flanking DNA sequence (which promotes recombination to non-target sites), and the absence of simultaneous multiple editing of several genes (76, 77, 79–82). Using CRISPR/Cas9, scientists have been able to circumvent all of these limitations to accelerate the process of obtaining recombinant viruses as vectors during the transfer of foreign genes in the prevention of various animal diseases.
To date, several recombinant vaccines and vectors based on OPXV have reached different stages of clinical trials. Various vaccines and vectors are based on VACV, MVA, NYVAC, raccoon poxvirus, lumpy skin disease virus (LSDV) and modified strains of OPXV Tian Tian smallpox vaccine (MVTT). They target malignant neoplasms (e.g., prostate, skin, colorectal cancer, breast and ovarian cancer) (83–88) and infectious diseases (e.g., AIDS, malaria, Ebola, tuberculosis, hepatitis, flu). In many parts of Europe, Canada, and the USA, the recombinant vaccine VACV–Raboral V-RG is widely used for the eradication of rabies virus in wild fox populations (76, 89). Several vector vaccines based on OPXV have been used to prevent various animal diseases (Table 3) (88).
Table 3. Current vaccines based on orthopoxviruses (88).
Recently, a genome-wide screen using CRISPR/Cas9 was conducted to identify host factors that may potentially participate in the replication of certain viruses, such as SARS-CoV-2 (90), HIV (91), arthritic alphaviruses (92), Coxsackie virus (93), Venezuelan equine encephalitis virus (VEEV) (94) and influenza virus (95). For the influenza virus, it was demonstrated that inactivation of specific host genes, such as DOCK5, Annexin-A1, IFIT2, IRF7 and ZDHHC22, can induce protection against cell death during infection (96–100). Because of the high throughput and precise editing features of the CRISPR/Cas9 screening system, one can quickly and accurately identify specific genes and proteins involved in the pathogenesis of viruses. The relative ease of use and reproducibility of the CRISPR/Cas9 system makes it a powerful tool for studying virus-host interactions and identifying new antiviral targets.
CRISPR-Cas9/Cas12a are well-studied nucleases optimized for the production of recombinant bacteria, which benefits the production of food, vitamins, and medicines. Bacteria may be used as biological “factories,” which can readily manufacture products using cheap raw materials.
A list of genetically modified bacteria using CRISPR/Cas technology are listed in Supplementary Table S1.
As described earlier, cis- and trans-genesis technologies enable the introduction of new genes into the genome to impart fundamentally new qualities on the host cell; for example, pest resistance, cold, or drought tolerance (101–103). Such foreign genes can also be introduced into the recipient genome using genome editing technologies (104). Genomic editing, for the most part, has a less radical effect on the physiology of the modified organism, because the changes are usually precise and minimal.
The genome and transcriptome of each organism may be considered a balanced system over the course of evolution where a significant change in the nucleotide sequence (more pronounced than spontaneous mutagenesis) could change the phenotype, or introduce foreign genes that may cause an imbalance to this system. Interference between existing and newly acquired molecules, signaling, and metabolic cascades can be unpredictable, which can reduce the value of productive Genetically Modified Organisms (GMOs) or render them potentially dangerous to the environment or for food consumption (105, 106).
A striking example of the unpredictable consequences of the genomic modification of a productive organism was the creation of GMO soybeans resistant to the herbicide glyphosate (Monsanto) (107). The resulting GMOs remained viable under the conditions of glyphosate treatment; however, it accumulated in their tissues, resulting in an unsafe food product, since glyphosate has genotoxic, immunotoxic, and potentially, carcinogenic effects (108, 109). Another example of an unpredictable outcome was a genetic modification for the development of a strain-producer of the amino acid L-tryptophan (“Showa Denko K.K.,” Japan), which is used as a dietary supplement (110, 111). An increase in the productivity of the strain unexpectedly led to the contamination of the final product with highly toxic dimerization products of tryptophan precursors, which the proposed purification methods did not remove. A study of the safety of genetically modified soy with reduced linoleic acid content (to improve the properties of soy oil and bring it closer to the quality of olive oil) in mice revealed that its long-term use reduced the likelihood of obesity and the development of insulin resistance; however, it stimulated the development of fatty hepatosis (112).
By themselves, the technologies for the genetic modification of productive animals are not dangerous, especially if we take into account the fact that there are natural mechanisms of horizontal transfer of genetic information even between evolutionarily distant organisms (113–115), which could potentially lead to the emergence of a natural “genetically modified organism.” However, because of the reasons listed above, GMOs or GMO products require thorough comprehensive study as neglect of this rule can, at times, have tragic consequences.
When assessing the safety of food obtained from GMOs, it is possible to use protocols similar to those used in the preclinical study of drugs by assessing acute, chronic, and reproductive toxicity as well as the mutagenic and carcinogenic potential (116). The safety of GMOs for the environment should be considered if there is a risk of “release” of GMOs into the environment. The possibility of transmission and distribution of genes in the wild population should be assessed as well as the “invasive potential” of GMOs to replace natural populations. In some cases, GMOs are designed to replace or modify existing species in nature. This approach is based on gene-drive technology and was proposed to combat the persistence of a parasite (e.g., pathogens of malaria, Dengue fever, Zika) in disease-carrying insects (117, 118).
It has become increasingly clear that genetic technologies and GMOs are the key to solving many problems associated with hunger, epizootics, depletion of natural resources, and environmental pollution (119–121). However, this requires the development of clear legal norms and the creation of an effective independent institute to monitor the use of genomic technologies and the study of the GMOs created.
Selective breeding for desirable phenotypic traits has been performed in agriculture since the first description of heredity by Gregor Mendel in 1865. This is a time-consuming practice and the desirable phenotype is not guaranteed, due to polygenic traits, complex gene regulation and epigenetics. The vast expansion of the human population following the industrial revolution necessitated large scale changes to agricultural practices in order to sustain and secure the increasing demand for food, medicine and vaccines. It was expected that science would not only contribute, but be at the forefront to develop and implement these changes.
Genome editing technologies have brought significant breakthroughs to routine laboratory work and have revolutionized research by facilitating the study of cells and organisms, on a molecular level. The function of an individual gene, protein or complex could be investigated, by deleting or inserting it into a new host. This in-depth study into the fundamental biology of organisms enabled the application of subsequent changes to the genetic composition of the organism, in order to obtain a highly desirable trait. These traits included the expression of novel proteins in order to transfer novel characteristics to an organism, resulting in a disease resistant or drought tolerant phenotype compared to the parental organism. None of the selective targeting and alteration of specific genes would be possible, if it is not based on a clear and fundamental understanding of the importance and function of the specific gene or protein complex.
The most widely used genome editing technologies include the CRISPR/Cas9 system, chimeric zinc-finger nucleases (ZFN) to create double-strand breaks, and transcription activator-like effector nucleases (TALEN). In recent years, CRISPR/Cas9 has become the preferred, most efficient and cost-effective method for use in editing the genomes of viruses, prokaryotes and eukaryotes. Relying on a single target molecule to guide RNA for DNA sequence recognition, the Cas9 enzyme may be steered toward a specific site with a low risk of off-target effects. This also enables multiplexing in contrast to the other existing gene-targeting technologies.
The applications of CRISPR/Cas9 have far exceeded the potential of only editing genomes, but have additionally been used in diagnostics and third generation sequencing technologies. The ability to timeously and efficiently identify and differentiate between pathogens causing disease, could result in the protection of the livestock by implementing control, prevention or mitigating strategies. The versatility of the implementations of CRISPR-based technologies add value throughout the livestock production chain, including animal production, diagnosis, protection and treatment. The future applications of these technologies would only be limited by the creativity of dedicated scientist.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the grant no. 075–15–2021-1054 from the Ministry of Education and Science of Russia to implement objectives of the Federal Scientific and Technical Program for the Development of genetic technologies during 2019–2027.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1180621/full#supplementary-material
1. Bawa, AS, and Anilakumar, KR. Genetically modified foods: safety, risks and public concerns-a review. J Food Sci Technol. (2013) 50:1035–46. doi: 10.1007/s13197-012-0899-1
2. Karalis, DT, Karalis, T, Karalis, S, and Kleisiari, AS. Genetically modified products, perspectives and challenges. Cureus. (2020) 12:e7306. doi: 10.7759/cureus.7306
3. Ibrahim, HMM, Ahmad, EM, Martínez-Medina, A, and Aly, MAM. Effective approaches to study the plant-root knot nematode interaction. Plant Physiol Biochem. (2019) 141:332–42. doi: 10.1016/j.plaphy.2019.06.009
4. Lovett, B, Bilgo, E, Diabate, A, and St Leger, R. A review of progress toward field application of transgenic mosquitocidal entomopathogenic fungi. Pest Manag Sci. (2019) 75:2316–24. doi: 10.1002/ps.5385
5. Bishop, SC. Genetic resistance to infections in sheep. Vet Microbiol. (2015) 181:2–7. doi: 10.1016/j.vetmic.2015.07.013
6. Spinoza, C, Schlechter, R, Herrera, D, Torres, E, Serrano, A, Medina, C, et al. Cisgenesis and intragenesis: new tools for improving crops. Biol Res. (2013) 46:323–31. doi: 10.4067/S0716-97602013000400003
7. Gupta, D, Bhattacharjee, O, Mandal, D, Sen, MK, Dey, D, Dasgupta, A, et al. CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci. (2019) 232:116636. doi: 10.1016/j.lfs.2019.116636
8. Smithies, O, Gregg, RG, Boggs, SS, Koralewski, MA, and Kucherlapati, RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. (1985) 317:230–4. doi: 10.1038/317230a0
9. Rouet, P, Smih, F, and Jasin, M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. (1994) 14:8096–106. doi: 10.1128/mcb.14.12.8096-8106.1994
10. Rouet, P, Smih, F, and Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. (1994) 91:6064–8. doi: 10.1073/pnas.91.13.6064
11. Iida, S, Meyer, J, Kennedy, KE, and Arber, W. A site-specific, conservative recombination system carried by bacteriophage P1. Mapping the recombinase gene cin and the cross-over sites cix for the inversion of the C segment. EMBO J. (1982) 1:1445–53. doi: 10.1002/j.1460-2075.1982.tb01336.x
12. Gimble, FS, and Thorner, J. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature. (1992) 357:301–6. doi: 10.1038/357301a0
13. Hoess, R, Abremski, K, and Sternberg, N. The nature of the interaction of the P1 recombinase Cre with the recombining site loxP. Cold Spring Harb Symp Quant Biol. (1984) 49:761–8. doi: 10.1101/SQB.1984.049.01.086
14. McLellan, MA, Rosenthal, NA, and Pinto, AR. Cre-loxP-mediated recombination: general principles and experimental considerations. Curr Protoc Mouse Biol. (2017) 7:1–12. doi: 10.1002/cpmo.22
15. Vetter, D, Andrews, BJ, Roberts-Beatty, L, and Sadowski, PD. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. (1983) 80:7284–8. doi: 10.1073/pnas.80.23.7284
16. Schweizer, HP. Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. J Mol Microbiol Biotechnol. (2003) 5:67–77. doi: 10.1159/000069976
17. Kim, H, Kim, M, Im, SK, and Fang, S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res. (2018) 34:147–59. doi: 10.5625/lar.2018.34.4.147
18. Miao, X. Recent advances in the development of new transgenic animal technology. Cell Mol Life Sci. (2013) 70:815–28. doi: 10.1007/s00018-012-1081-7
19. Zou, YR, Müller, W, Gu, H, and Rajewsky, K. Cre-loxP-mediated gene replacement: a mouse strain producing humanized antibodies. Curr Biol. (1994) 4:1099–103. doi: 10.1016/S0960-9822(00)00248-7
20. Lui, D, and Burgess, SM. Measurement of spatial proximity and accessibility of chromosomal loci in Saccharomyces cerevisiae using Cre/loxP site-specific recombination. Methods Mol Biol. (2009) 557:55–63. doi: 10.1007/978-1-59745-527-5_5
21. Martina, F, and Christos, E. The revolution of the CRISPR-CAS9 system. United Kingdom: CRISPR Biotech Engineering (2020).
22. Chevalier, BS, and Stoddard, BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. (2001) 29:3757–74. doi: 10.1093/nar/29.18.3757
23. Zhao, L, Bonocora, RP, Shub, DA, and Stoddard, BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. (2007) 26:2432–42. doi: 10.1038/sj.emboj.7601672
24. Stoddard, BL. Homing endonuclease structure and function. Q Rev Biophys. (2005) 38:49–95. doi: 10.1017/S0033583505004063
25. Pâques, F, and Duchateau, P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. (2007) 7:49–66. doi: 10.2174/156652307779940216
26. Xu, X, Hulshoff, MS, Tan, X, Zeisberg, M, and Zeisberg, EM. CRISPR/Cas derivatives as novel gene modulating tools: possibilities and in vivo applications. Int J Mol Sci. (2020) 21:3038. doi: 10.3390/ijms21093038
27. Carroll, D. Genome engineering with zinc-finger nucleases. Genetics. (2011) 188:773–82. doi: 10.1534/genetics.111.131433
28. Chuang, CK, and Lin, WM. Points of view on the tools for Genome/gene editing. Int J Mol Sci. (2021) 22:9872. doi: 10.3390/ijms22189872
29. Maeder, ML, and Gersbach, CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. (2016) 24:430–46. doi: 10.1038/mt.2016.10
30. Joung, JK, and Sander, JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. (2013) 14:49–55. doi: 10.1038/nrm3486
31. Wright, DA, Li, T, Yang, B, and Spalding, MH. TALEN-mediated genome editing: prospects and perspectives. Biochem J. (2014) 462:15–24. doi: 10.1042/BJ20140295
32. Mak, AN, Bradley, P, Cernadas, RA, Bogdanove, AJ, and Stoddard, BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. (2012) 335:716–9. doi: 10.1126/science.1216211
33. Hockemeyer, D, and Jaenisch, R. Gene targeting in human pluripotent cells. Cold Spring Harb Symp Quant Biol. (2010) 75:201–9. doi: 10.1101/sqb.2010.75.021
34. Mussolino, C, Morbitzer, R, Lütge, F, Dannemann, N, Lahaye, T, and Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. (2011) 39:9283–93. doi: 10.1093/nar/gkr597
35. Doudna, JA, and Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science. (2014) 346:1258096. doi: 10.1126/science.1258096
36. Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA, and Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. (2012) 337:816–21. doi: 10.1126/science.1225829
37. Ishino, Y, Krupovic, M, and Forterre, P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to Genome editing technology. J Bacteriol. (2018) 200:e00580–17. doi: 10.1128/JB.00580-17
38. Makarova, KS, Haft, DH, Barrangou, R, Brouns, SJ, Charpentier, E, Horvath, P, et al. Evolution and classification of the CRISPR-Cas systems. Microbiology. (2011) 9:467–77. doi: 10.1038/nrmicro2577
39. Nishimasu, H, Ran, FA, Hsu, PD, Konermann, S, Shehata, SI, Dohmae, N, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cells. (2014) 156:935–49. doi: 10.1016/j.cell.2014.02.001
40. Zhang, S, Shen, J, Li, D, and Cheng, Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics. (2021) 11:614–48. doi: 10.7150/thno.47007
41. Khan, SH. Genome-editing technologies: concept, pros, and cons of various Genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids. (2019) 16:326–34. doi: 10.1016/j.omtn.2019.02.027
42. Boutros, M, and Ahringer, J. The art and design of genetic screens: RNA interference. Nat Rev Genet. (2008) 9:554–66. doi: 10.1038/nrg2364
43. Van Eenennaam, AL. Application of genome editing in farm animals: cattle. Transgenic Res. (2019) 28:93–100. doi: 10.1007/s11248-019-00141-6
44. Navarro-Serna, S, Vilarino, M, Park, I, Gadea, J, and Ross, PJ. Livestock gene editing by one-step embryo manipulation. J Equine Vet Sci. (2020) 89:103025. doi: 10.1016/j.jevs.2020.103025
45. Wang, K, Ouyang, H, Xie, Z, Yao, C, Guo, N, Li, M, et al. Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci Rep. (2015) 5:1–11. doi: 10.1038/srep16623
46. Burkard, C, Lillico, SG, Reid, E, Jackson, B, Mileham, AJ, Ait-Ali, T, et al. Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. (2017) 13:e1006206. doi: 10.1371/journal.ppat.1006206
47. Gao, Y, Wu, H, Wang, Y, Liu, X, Chen, L, Li, Q, et al. Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol. (2017) 18:13. doi: 10.1186/s13059-016-1144-4
48. Zhao, Y, Wang, T, Yao, L, Liu, B, Teng, C, and Ouyang, H. Classical swine fever virus replicated poorly in cells from MxA transgenic pigs. BMC Vet Res. (2016) 12:169. doi: 10.1186/s12917-016-0794-5
49. Xie, Z, Jiao, H, Xiao, H, Jiang, Y, Liu, Z, Qi, C, et al. Generation of pRSAD2 gene knock-in pig via CRISPR/Cas9 technology. Antivir Res. (2020) 174:104696. doi: 10.1016/j.antiviral.2019.104696
50. Xie, Z, Pang, D, Yuan, H, Jiao, H, Lu, C, Wang, K, et al. Genetically modified pigs are protected from classical swine fever virus. PLoS Pathog. (2018) 14:e1007193. doi: 10.1371/journal.ppat.1007193
51. Lu, C, Pang, D, Li, M, Yuan, H, Yu, T, Huang, P, et al. CRISPR/Cas9-mediated hitchhike expression of functional shRNAs at the porcine miR-17-92 cluster. Cells. (2019) 8:113. doi: 10.3390/cells8020113
52. Tait-Burkard, C, Doeschl-Wilson, A, McGrew, MJ, Archibald, AL, Sang, HM, Houston, RD, et al. Livestock 2.0–genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. (2018) 19:204. doi: 10.1186/s13059-018-1583-1
53. Borca, MV, Berggren, KA, Ramirez-Medina, E, Vuono, EA, and Gladue, DP. CRISPR/Cas gene editing of a large DNA virus: African swine fever virus. Bio Protoc. (2018) 8:e2978. doi: 10.21769/BioProtoc.2978
54. Shen, Z, Wang, G, Yang, Y, Shi, J, Fang, L, Li, F, et al. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J Biol Chem. (2019) 294:13606–18. doi: 10.1074/jbc.RA119.009713
55. Yuan, M, Gao, X, Chard, LS, Ali, Z, Ahmed, J, Li, Y, et al. A marker-free system for highly efficient construction of vaccinia virus vectors using CRISPR Cas9. Mol Ther Methods Clin Dev. (2015) 2:15035. doi: 10.1038/mtm.2015.35
56. Di Gioia, C, Yuan, M, and Wang, Y. Vaccinia virus Genome editing using CRISPR. Methods Mol Biol. (2019) 2023:109–17. doi: 10.1007/978-1-4939-9593-6_6
57. Chou, Y-Y, Krupp, A, Kaynor, C, Gaudin, R, Ma, M, Cahir-McFarland, E, et al. Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Sci Rep. (2016) 6:36921. doi: 10.1038/srep36921
58. Hübner, A, Petersen, B, Keil, GM, Niemann, H, Mettenleiter, TC, and Fuchs, W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L). Sci Rep. (2018) 8:1449. doi: 10.1038/s41598-018-19626-1
59. Abkallo, HM, Svitek, N, Oduor, B, Awino, E, Henson, SP, Oyola, SO, et al. Rapid CRISPR/Cas9 editing of genotype IX African swine fever virus circulating in eastern and Central Africa. Front Genet. (2021) 12:733674. doi: 10.3389/fgene.2021.733674
60. Borca, MV, Holinka, LG, Berggren, KA, and Gladue, DP. CRISPR-Cas9, a tool to efficiently increase the development of recombinant African swine fever viruses. Sci Rep. (2018) 8:3154. doi: 10.1038/s41598-018-21575-8
61. Pan, Q, Wang, J, Gao, Y, Cui, H, Liu, C, Qi, X, et al. The natural large genomic deletion is unrelated to the increased virulence of the novel genotype fowl adenovirus 4 recently emerged in China. Viruses. (2018) 10:494. doi: 10.3390/v10090494
62. Liang, X, Sun, L, Yu, T, Pan, Y, Wang, D, Hu, X, et al. A CRISPR/Cas9 and Cre/lox system-based express vaccine development strategy against re-emerging pseudorabies virus. Sci Rep. (2016) 6:19176. doi: 10.1038/srep19176
63. Xu, A, Qin, C, Lang, Y, Wang, M, Lin, M, Li, C, et al. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol Lett. (2015) 37:1265–72. doi: 10.1007/s10529-015-1796-2
64. Hagag, IT, Wight, DJ, Bartsch, D, Sid, H, Jordan, I, Bertzbach, LD, et al. Abrogation of Marek’s disease virus replication using CRISPR/Cas9. Sci Rep. (2020) 10:10919. doi: 10.1038/s41598-020-67951-1
65. Price, AA, Sampson, TR, Ratner, HK, Grakoui, A, and Weiss, DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci U S A. (2015) 112:6164–9. doi: 10.1073/pnas.1422340112
66. Li, H, Wang, S, Dong, X, Li, Q, Li, M, Li, J, et al. CRISPR-Cas13a cleavage of dengue virus NS3 gene efficiently inhibits viral replication. Mol Ther Nucleic Acids. (2020) 19:1460–9. doi: 10.1016/j.omtn.2020.01.028
67. Cui, J, Techakriengkrai, N, Nedumpun, T, and Suradhat, S. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci Rep. (2020) 10:1–13. doi: 10.1038/s41598-020-66775-3
68. Smith, GL, and Moss, B. Infectious poxvirus vectors have capacity for at least 25,000 base pairs of foreign DNA. Gene. (1983) 25:21–8. doi: 10.1016/0378-1119(83)90163-4
69. Moss, B, Carroll, MW, Wyatt, LS, Bennink, JR, Hirsch, VM, Goldstein, S, et al. Host range restricted, non-replicating vaccinia virus vectors as vaccine candidates. Adv Exp Med Biol. (1996) 397:7–13. doi: 10.1007/978-1-4899-1382-1_2
70. Werden, SJ, Rahman, MM, and McFadden, G. Poxvirus host range genes. Adv Virus Res. (2008) 71:135–71. doi: 10.1016/S0065-3527(08)00003-1
71. Gudmundsdotter, L, Nilsson, C, Brave, A, Hejdeman, B, Earl, P, Moss, B, et al. Recombinant modified vaccinia Ankara (MVA) effectively boosts DNA-primed HIV-specific immune responses in humans despite pre-existing vaccinia immunity. Vaccine. (2009) 27:4468–74. doi: 10.1016/j.vaccine.2009.05.018
72. Smith, GL, Mackett, M, and Moss, B. Infectious vaccinia virus recombinants that express hepatitis B virus surface antigen. Nature. (1983) 302:490–5. doi: 10.1038/302490a0
73. Draper, SJ, and Heeney, JL. Viruses as vaccine vectors for infectious diseases and cancer. Nat Rev Microbiol. (2010) 8:62–73. doi: 10.1038/nrmicro2240
74. Ramirez, JC, Gherardi, MM, and Esteban, M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western reserve strain and advantages as a vaccine. J Virol. (2000) 74:923–33. doi: 10.1128/JVI.74.2.923-933.2000
75. Garcia-Arriaza, J, and Esteban, M. Enhancing poxvirus vectors vaccine immunogenicity. Hum Vaccin Immunother. (2014) 10:2235–44. doi: 10.4161/hv.28974
76. Yang, DK, Kim, HH, Lee, KW, and Song, JY. The present and future of rabies vaccine in animals. Clin Exp Vaccine Res. (2013) 2:19–25. doi: 10.7774/cevr.2013.2.1.19
77. Wyatt, LS, Earl, PL, and Moss, B. Generation of recombinant vaccinia viruses. Curr Protoc Mol Biol. (2017) 117:16.17.11–8. doi: 10.1002/cpmb.32
78. Merchlinsky, M, and Moss, B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: recombination-independent selectable cloning vectors. Virology. (1992) 190:522–6. doi: 10.1016/0042-6822(92)91246-Q
79. Domi, A, and Moss, B. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proc Natl Acad Sci U S A. (2002) 99:12415–20. doi: 10.1073/pnas.192420599
80. Paszkowski, P, Noyce, RS, and Evans, DH. Live-cell imaging of vaccinia virus recombination. PLoS Pathog. (2016) 12:e1005824. doi: 10.1371/journal.ppat.1005824
81. Ball, LA. High-frequency homologous recombination in vaccinia virus DNA. J Virol. (1987) 61:1788–95. doi: 10.1128/jvi.61.6.1788-1795.1987
82. Yuan, M, Wang, P, Chard, LS, Lemoine, NR, and Wang, Y. A simple and efficient approach to construct mutant vaccinia virus vectors. J Vis Exp. (2016) 116:54171. doi: 10.3791/54171
83. Wyatt, LS, Belyakov, IM, Earl, PL, Berzofsky, JA, and Moss, B. Enhanced cell surface expression, immunogenicity and genetic stability resulting from a spontaneous truncation of HIV env expressed by a recombinant MVA. Virology. (2008) 372:260–72. doi: 10.1016/j.virol.2007.10.033
84. McNeel, DG, Chen, Y-H, Gulley, JL, Dwyer, AJ, Madan, RA, Carducci, MA, et al. Randomized phase II trial of docetaxel with or without PSA-TRICOM vaccine in patients with castrate-resistant metastatic prostate cancer: a trial of the ECOG-ACRIN cancer research group (E1809). Hum Vaccin Immunother. (2015) 11:2469–74. doi: 10.1080/21645515.2015.1062190
85. Gulley, JL, Madan, RA, Tsang, KY, Jochems, C, Marté, JL, Farsaci, B, et al. Immune impact induced by PROSTVAC (PSA-TRICOM), a therapeutic vaccine for prostate cancer. Cancer Immunol Res. (2014) 2:133–41. doi: 10.1158/2326-6066.CIR-13-0108
86. Arlen, PM, Gulley, JL, Parker, C, Skarupa, L, Pazdur, M, Panicali, D, et al. A randomized phase II study of concurrent docetaxel plus vaccine versus vaccine alone in metastatic androgen-independent prostate cancer. Clin Cancer Res. (2006) 12:1260–9. doi: 10.1158/1078-0432.CCR-05-2059
87. Kaufman, HL, Deraffele, G, Mitcham, J, Moroziewicz, D, Cohen, SM, Hurst-Wicker, KS, et al. Targeting the local tumor microenvironment with vaccinia virus expressing B7.1 for the treatment of melanoma. J Clin Investig. (2005) 115:1903–12. doi: 10.1172/JCI24624
88. Okoli, AS, Asare, N, Gjøen, T, Klein, J, and Ytrehus, B. Knowledge Base for the assessment of environmental risks by the use of genetically modified virus-vectored vaccines for domesticated animals; scientific opinion of the panel on microbial ecology of the Norwegian scientific Committee for Food Safety (VKM). Oslo, Norway: Norwegian Scientific Committee for Food Safety (2016).
89. Fehlner-Gardiner, C, Rudd, R, Donovan, D, Slate, D, Kempf, L, and Badcock, J. Comparing ONRAB(R) and RABORAL V-RG(R) oral rabies vaccine field performance in raccoons and striped skunks. J Wildl Dis. (2012) 48:157–67. doi: 10.7589/0090-3558-48.1.157
90. Baggen, J, Persoons, L, Vanstreels, E, Jansen, S, Van Looveren, D, Boeckx, B, et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet. (2021) 53:435–44. doi: 10.1038/s41588-021-00805-2
91. Hultquist, JF, Hiatt, J, Schumann, K, McGregor, MJ, Roth, TL, Haas, P, et al. CRISPR–Cas9genome engineering of primary CD4+ T cells for the interrogation of HIV–host factor interactions. Nat Protoc. (2019) 14:1–27. doi: 10.1038/s41596-018-0069-7
92. Zhang, R, Kim, AS, Fox, JM, Nair, S, Basore, K, Klimstra, WB, et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature. (2018) 557:570–4. doi: 10.1038/s41586-018-0121-3
93. Shin, HJ, Ku, KB, Kim, S, Kim, HS, Kim, YS, Kim, BT, et al. A crucial role of ACBD3 required for coxsackievirus infection in animal model developed by AAV-mediated CRISPR Genome editing technique. Viruses. (2021) 13:237. doi: 10.3390/v13020237
94. Ma, H, Kim, AS, Kafai, NM, Earnest, JT, Shah, AP, Case, JB, et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nat Cell Biol. (2020) 588:308–14. doi: 10.1038/s41586-020-2915-3
95. Sharon, DM, Nesdoly, S, Yang, HJ, Gélinas, J-F, Xia, Y, Ansorge, S, et al. A pooled genome-wide screening strategy to identify and rank influenza host restriction factors in cell-based vaccine production platforms. Sci Rep. (2020) 10:1–15. doi: 10.1038/s41598-020-68934-y
96. Forst, CV, Zhou, B, Wang, M, Chou, T-W, Mason, G, Song, W-M, et al. Integrative gene network analysis identifies key signatures, intrinsic networks and host factors for influenza virus a infections. NPJ Syst Biol Appl. (2017) 3:35. doi: 10.1038/s41540-017-0036-x
97. Kim, TH, Kern, C, and Zhou, H. Knockout of IRF7 highlights its modulator function of host response against avian InfluenzaVirus and the involvement of MAPK and TOR Signaling pathways in chicken. Genes. (2020) 11:385. doi: 10.3390/genes11040385
98. Cui, J, Morgan, D, Cheng, DH, Foo, SL, Yap, GLR, Ampomah, PB, et al. RNA-sequencing-based transcriptomic analysis reveals a role for annexin-A1 in classical and in-fluenza a virus-induced autophagy. Cells. (2020) 9:1399. doi: 10.3390/cells9061399
99. Tran, V, Ledwith, MP, Thamamongood, T, Higgins, CA, Tripathi, S, Chang, MW, et al. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat Microbiol. (2020) 5:1490–503. doi: 10.1038/s41564-020-0778-x
100. Gadalla, MR, Morrison, E, Serebryakova, MV, Han, X, Wolff, T, Freund, C, et al. NS1-mediated upregulation of ZDHHC22 acyltransferase in influenza a virus infected cells. Cell Microbiol. (2021):e13322. doi: 10.1111/cmi.13322
101. Ricroch, AE, and Hénard-Damave, MC. Next biotech plants: new traits, crops, developers and technologies for addressing global challenges. Crit Rev Biotechnol. (2016) 36:675–90. doi: 10.3109/07388551.2015.1004521
102. Álvarez-Alfageme, F, Devos, Y, Camargo, AM, Arpaia, S, and Messéan, A. Managing resistance evolution to transgenic Bt maize in corn borers in Spain. Crit Rev Biotechnol. (2022) 42:201–19. doi: 10.1080/07388551.2021.1931018
103. Hu, W, Zheng, H, Li, Q, Wang, Y, Liu, X, Hu, X, et al. shRNA transgenic swine display resistance to infection with the foot-and-mouth disease virus. Sci Rep. (2021) 11:16377. doi: 10.1038/s41598-021-95853-3
104. Jaganathan, D, Ramasamy, K, Sellamuthu, G, Jayabalan, S, and Venkataraman, G. CRISPR for crop improvement: an update review. Front Plant Sci. (2018) 9:985. doi: 10.3389/fpls.2018.00985
105. De Santis, B, Stockhofe, N, Wal, JM, Weesendorp, E, Lallès, JP, van Dijk, J, et al. Case studies on genetically modified organisms (GMOs): potential risk scenarios and associated health indicators. Food Chem Toxicol. (2018) 117:36–65. doi: 10.1016/j.fct.2017.08.033
106. Pott, A, Otto, M, and Schulz, R. Impact of genetically modified organisms on aquatic environments: review of available data for the risk assessment. Sci Total Environ. (2018) 635:687–98. doi: 10.1016/j.scitotenv.2018.04.013
107. Bøhn, T, Cuhra, M, Traavik, T, Sanden, M, Fagan, J, and Primicerio, R. Compositional differences in soybeans on the market: glyphosate accumulates in roundup ready GM soybeans. Food Chem. (2014) 153:207–15. doi: 10.1016/j.foodchem.2013.12.054
108. Andreotti, G, Koutros, S, Hofmann, JN, Sandler, DP, Lubin, JH, Lynch, CF, et al. Glyphosate use and cancer incidence in the agricultural health study. J Natl Cancer Inst. (2018) 110:509–16. doi: 10.1093/jnci/djx233
109. Peillex, C, and Pelletier, M. The impact and toxicity of glyphosate and glyphosate-based herbicides on health and immunity. J Immunotoxicol. (2020) 17:163–74. doi: 10.1080/1547691X.2020.1804492
110. Roberts, L. L-tryptophan puzzle takes new twist. Science. (1990) 249:988. doi: 10.1126/science.2396102
111. Mann, LRB, Straton, B, and Christ, WE. The thalidomide of genetic engineering. Biosafety Information Centre. (2005).
112. Deol, P, Fahrmann, J, Yang, J, Evans, JR, Rizo, A, Grapov, D, et al. Omega-6 and omega-3 oxylipins are implicated in soybean oil-induced obesity in mice. Sci Rep. (2017) 7:12488. doi: 10.1038/s41598-017-12624-9
113. Guo, M, Ye, J, Gao, D, Xu, N, and Yang, J. Agrobacterium-mediated horizontal gene transfer: mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol Adv. (2019) 37:259–70. doi: 10.1016/j.biotechadv.2018.12.008
114. Huang, W, Tsai, L, Li, Y, Hua, N, Sun, C, and Wei, C. Widespread of horizontal gene transfer in the human genome. BMC Genomics. (2017) 18:274. doi: 10.1186/s12864-017-3649-y
115. Graham, LA, and Davies, PL. Horizontal gene transfer in vertebrates: a fishy Tale. Trends Genet. (2021) 37:501–3. doi: 10.1016/j.tig.2021.02.006
116. EFSA GMO Panel Working Group on Animal Feeding Trials. Safety and nutritional assessment of GM plants and derived food and feed: the role of animal feeding trials. Food Chem Toxicol. (2008) 46:70. doi: 10.1016/j.fct.2008.02.008
117. Hammond, A, Galizi, R, Kyrou, K, Simoni, A, Siniscalchi, C, Katsanos, D, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. (2016) 34:78–83. doi: 10.1038/nbt.3439
118. Nateghi Rostami, M. CRISPR/Cas9 gene drive technology to control transmission of vector-borne parasitic infections. Parasite Immunol. (2020) 42:e12762. doi: 10.1111/pim.12762
119. Okoli, AS, Blix, T, Myhr, AI, Xu, W, and Xu, X. Sustainable use of CRISPR/Cas in fish aquaculture: the biosafety perspective. Transgenic Res. (2022) 31:1–21. doi: 10.1007/s11248-021-00274-7
120. Carter, BE, Conn, CC, and Wiles, JR. Concern about hunger may increase receptivity to GMOs. Trends Plant Sci. (2016) 21:539–41. doi: 10.1016/j.tplants.2016.05.003
Keywords: genome editing, CRISPR-Cas, genetically modified organisms, vaccine, virus loxP mouse Cre mouse
Citation: Mazloum A, Karagyaur M, Chernyshev R, van Schalkwyk A, Jun M, Qiang F and Sprygin A (2023) Post-genomic era in agriculture and veterinary science: successful and proposed application of genetic targeting technologies. Front. Vet. Sci. 10:1180621. doi: 10.3389/fvets.2023.1180621
Received: 06 March 2023; Accepted: 20 July 2023;
Published: 03 August 2023.
Edited by:
Paul M. Coussens, Michigan State University, United StatesReviewed by:
Yingchun Liu, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2023 Mazloum, Karagyaur, Chernyshev, van Schalkwyk, Jun, Qiang and Sprygin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Mazloum, YWxpLm1hemxvdW02QGdtYWlsLmNvbQ==; Alexander Sprygin, c3ByaWdpbmF2QG1haWwucnU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.