
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 08 November 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1177302
This article is part of the Research Topic Pathogenic and Symbiotic Bacteria in Ruminants: Antimicrobial Resistance and Microbial Homeostasis View all 10 articles
Intramammary infections (IMI) in animals reared for milk production can result in large economic losses and distress to the animals. Staphylococcus aureus is an important causative agent of IMI in dairy cows, but its prevalence in water buffaloes has not been determined. Therefore, the current study was conducted to investigate the prevalence of subclinical mastitis in water buffaloes and the antimicrobial susceptibility, virulence genes and biofilm formation abilities of Staphylococcus aureus isolates recovered from water buffaloes in Guangdong, China. Staphylococcus aureus strains were isolated from milk samples of water buffaloes with subclinical mastitis, and twofold microdilution, PCR and crystal violet staining methods were used to determine antimicrobial susceptibility, distributions of virulence and antimicrobial resistance genes and biofilm formation ability, respectively. Our results indicated that 29.44% of water buffaloes were diagnosed with subclinical mastitis, and the most prevalent pathogens were Escherichia coli (96.17%), coagulase-negative staphylococci (CoNS) (67.60%) and S. aureus (28.57%). Most S. aureus isolates showed resistance to bacitracin, doxycycline, penicillin, florfenicol, and tetracycline but were susceptible to ciprofloxacin, ceftizoxime, cefoquinoxime, and ofloxacin. Moreover, 63.72% of S. aureus isolates were positive for tetM, and the prevalence of msrB, blaZ, mecA, fexA, and tetK ranged from 21.24 to 6.19%. All S. aureus isolates harbored clfB and icaA genes, and the virulence genes hla (93.8%), hld (91.15%), clfA (90.27%), fnbA (86.73%), and hlb (83.19%), and tsst, icaD, sec, see, fnbB, and sea showed a varied prevalence ranging from 3.5 to 65.49%. All S. aureus isolates possessed the ability to form biofilms, and 30.09% of isolates showed strong biofilm formation abilities, while 19.47% of isolates were weak biofilm producers. Our results indicated that subclinical mastitis is prevalent in water buffaloes in Guangdong, China, and S. aureus is prevalent in samples from water buffaloes with subclinical mastitis. Most S. aureus isolates were susceptible to cephalosporins and fluoroquinolones; thus, ceftizoxime and cefoquinoxime can be used to treat subclinical mastitis in water buffaloes.
Water buffalo (Bubalus bubalis) is of importance in the milk industry and contributes to approximately 15% of milk production (1). Many parts of the world have traditionally produced buffalo milk, including Asia, Egypt, and Europe. China produces approximately 5% of global buffalo milk, and Guangdong, Guangxi and Hunan are its primary producers (2). Italy is the main producer of buffalo milk in Europe because of the popularity of buffalo mozzarella cheese (3), which retails for twice the price of bovine milk cheese (4).
Mastitis is one of the most prevalent diseases among dairy animals, causing economic losses to milk producers due to a decrease in milk quality and production, an increase in veterinary and labor costs and an increased rate of culling (5). Bacteria are the primary causative agents of mastitis, although physical trauma and mechanical injury also contribute (1). The primary bacterial causes of mastitis in both dairy cows and water buffaloes are Staphylococcus aureus, Streptococcus agalactiae, Streptococcus dysgalactiae, and Escherichia coli. Staphylococcus aureus is one of the primary pathogens causing mastitis in dairy cows at a level of approximately 40% in China (6).
Among antimicrobial agents, antibiotics are normally used to treat infections caused by bacteria, including intramammary infections (IMI) in domestic animals. In dairy animals, mastitis and reproductive diseases often require prolonged use of antimicrobial agents (7). Unfortunately, the widespread use of antimicrobial agents has resulted in high levels of antimicrobial resistance, leading to clinical treatment failures. Therefore, monitoring systems for antimicrobial resistance of bacterial pathogens can provide essential information for the rational use of antimicrobial agents when treating infections (8). Knowledge regarding the prevalence of mastitis and the knowledge of pathogens causing mastitis is critical in preventing the occurrence of mastitis and can provide effective measures for control and appropriate therapeutic protocols (9).
There are two groups of virulence genes in S. aureus, including surface-localized structural components serving as virulence factors and secreted virulence factors involved in evading host defenses and colonizing mammary glands (10). Surface-localized structural components include membrane-bound factors (fibrinogen binding protein, collagen binding protein and elastin binding protein), cell wall-bound factors (lipoteichoic acid, peptidoglycan, protein A and protease) and cell surface-associated factors (capsule). Secretory factors include toxins (staphylococcal enterotoxins, leucocidin, toxic shock syndrome toxin, and hemolysins) and enzymes (staphylokinase, coagulase, lipase, DNase, and hyaluronidase). Moreover, biofilm formation also contributes to adhesion and invasion into mammary epithelial cells and thus provides an escape from the host immune system. Enterotoxins often lead to food poisoning and include staphylococcal enterotoxins A to F and G to Q (11). Enterotoxins G to Q are more prevalent in isolates from dairy cows with mastitis than in isolates from cows without mastitis; this has implicated these virulence factors in the occurrence of mastitis (12). However, the clear mechanisms of virulence in mastitis of dairy cows need further study.
In China, the number of water buffalo farms is increasing, and thus, mastitis is occurring more frequently in these animals. Mastitis can be divided into two forms: clinical and subclinical. In clinical mastitis, clots and flakes can be observed in milk, and the quarters become swollen with severe conditions leading to the formation of lacerations, necrosis and cord formation of the teat. While no clinical signs or symptoms can be seen in subclinical mastitis, there is a reduction in milk production and deterioration of milk quality (13). Subclinical infection is 15–40 times more prevalent than clinical infection and can rapidly spread on a farm (14).
To reduce the prevalence of subclinical mastitis in water buffaloes, it is essential to have knowledge of prevalence data and understand the antimicrobial susceptibility profiles of subclinical mastitis caused by S. aureus. Therefore, this study was conducted to investigate the prevalence of subclinical mastitis in water buffaloes in Guangdong, China, and to determine the antimicrobial susceptibility of S. aureus isolates and their antimicrobial resistance and virulence determinants.
Ausvet epidemiological calculators1 were used to calculate the sample sizes according to the method from Charan and Biswas at a 95% confidence level (15) as follows:
where equals 2.58 with a 1% type error (p < 0.01). The expected proportion of water buffaloes in Guangdong Province is 0.05, and the expected precision (d) is 0.01.
A total of 3,900 samples from 975 water buffaloes were included in this study from the following regions: Qingyuan (n = 884), Guangzhou (n = 1,076), Jiangmen (n = 992) and Zhaoqing (n = 948). The average sample number per farm was 342.9 (range 174 to 584).
The milking of animals on all farms was performed twice a day, and the sample collection procedure consisted of fore-stripping (3–5 squirts of milk) followed by teat disinfection with 0.25% iodine solution and drying with a clean towel. The milking clusters were then attached and removed automatically when finished, followed by postmilking teat disinfection with 0.5% iodine. Duplicate milk samples for each quarter were aseptically collected according to standard protocols of the National Mastitis Council (16). Briefly, milk samples (3 mL) were collected from all quarters of each animal after the first 3 streams of milk were discarded and placed in an ice box and transferred to the laboratory within 6 h. Presumptive evidence of subclinical mastitis (17) was determined using a commercial California Mastitis Test kit (CMT) (ImmuCell, Portland, ME, United States) following the recommendations of the manufacturer. Briefly, 2 mL of milk sample was mixed with an equal amount of CMT solution in the paddle and stirred for 30 s. Thickening indicated elevated somatic cell counts (SCC), and these samples were then used for bacterial isolation and identification.
Identification of S. aureus from milk samples was carried out as previously described (18). Briefly, a 0.1 mL milk sample was inoculated into 3 mL tryptic soy broth (TSB; AoBox, Beijing, China) containing 10% NaCl and incubated at 37°C for 24 h. A loopful of enrichment broth culture was streaked onto a Baird-Parker agar plate (AoBox) and incubated at 37°C for 24 h. The suspected S. aureus colonies had a black and shiny appearance with a thin white border surrounded by a light area. The suspected colonies were streaked onto chromogenic S. aureus agar plates (CHROMagar, Paris, France). At least 3 positive colonies per sample were then confirmed as coagulase-positive S. aureus using commercial API STAPH test strips (bioMérieux, Marcy-l’Ѐtoile, France). Staphylococcus aureus isolates were stored at −80°C in cryogenic vials (Biologix, Shandong, China) containing 1 mL TSB and 30% glycerin. Coagulase-negative staphylococci were isolated as previously described (19). In brief, milk samples were cultured on blood agar plates (AoBox) at 37°C for 48 h. Typical colonies were selected and identified using classical biochemical methods, including Gram staining and oxidase, catalase and DNase tests, and the ability to coagulate rabbit plasma using commercial kits (Sigma Chemical, Shanghai, China). If the suspected strains failed biochemical identification, molecular identification using PCR amplification and sequencing of the sodA gene was performed as previously described (19).
Escherichia coli isolation utilized a 0.1 mL milk sample inoculated in 3 mL Mueller-Hinton broth (MHB; Aobox), which was incubated at 37°C for 24 h. Samples were then streaked onto MacConkey Agar plates, and plates were kept at 37°C for 24 h. Presumptive identification of E. coli were pink colonies that were then subjected to matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) using a Microflex LT instrument (Bruker Daltonics, Bremen, Germany).
Streptococcus agalactiae and S. dysgalactiae were identified as previously described (20) using EN medium, and positive isolates were then transferred to Columbia Blood Agar Base Medium containing 5% sheep blood (Hope Bio-Technology, Qingdao, China) and incubated at 37°C for 24 h. Colonies with typical Streptococcus morphologies were then subjected to catalase and 6.5% NaCl tests. Isolates were further identified as S. agalactiae and S. dysgalactiae according to reactions using the sodium hippurate test, esculin hydrate, and CAMP tests.
Antimicrobial resistance phenotypes of S. aureus isolates were determined using the microdilution method in MH broth according to the Clinical Laboratory and Standards Institute guidelines (CLSI) (21). A loopful of each S. aureus isolate preserved in glycerinated TSB was streaked on a Baird-Parker agar plate and incubated at 37°C for 24 h. The colonies were inoculated in MH broth, and the cultures were diluted in sterile normal saline and adjusted to a turbidity of 0.5 McFarland standard (105–106 colony-forming units (CFU)/mL). The suspension was then swabbed on Muller-Hinton agar plates and incubated at 37°C for 24 h as previously described. Staphylococcus aureus ATCC 25923 was used as the reference strain. Each experiment was repeated at least three times. Breakpoints for different antimicrobial agents were based on CLSI guidelines (21).
Antimicrobial agents included ceftiofur (CTF) (0.02–16 μg/mL), cefoquinoxime (CFQ, 0.03–32 μg/mL), ceftizoxime (CFT, 0.03–32 μg/mL), cefoxitin (CFX, 0.03–32 μg/mL), florfenicol (FLO, 0.125–128 μg/mL), ciprofloxacin (CIP, 0.03–32 μg/mL), enrofloxacin (ENO, 0.03–32 μg/mL), ofloxaxin (OFX, 0.06–64 μg/mL), erythromycin (ERY, 0.125–128 μg/mL), azithromycin (ATM, 0.125–128 μg/mL), gentamycin (GEN, 0.125–128 μg/mL), penicillin (PEN, 0.03–32 μg/mL), ampicillin (AMP, 0.125–128 μg/mL), tetracycline (TET, 0.125–128 μg/mL), doxycycline (DXC, 0.03–32 μg/mL) and bacitracin (BTC, 4–1024 μg/mL).
Strains used for testing were taken from frozen stocks, cultures were streaked onto TSA plates containing 5% sterile defibrinated sheep blood, and the plates were incubated at 37°C for 24 h. Single colonies were inoculated into 3 mL TSB and cultured with shaking at 37°C for 24 h. The cultures were centrifuged at 3000 × g, and cell pellets were suspended in phosphate buffered saline (PBS, pH 7.4; Solarbio, Beijing, China) containing 20 mg/mL lysostaphin (Meilunbio, Dalian, China). The mixture was kept at 37°C for 30 min, genomic DNA was extracted using a TIANamp bacterial DNA extraction kit (TianGen, Beijing, China), and DNA quality was evaluated by UV spectroscopy with a NanoDrop-2000 instrument (Thermo Fisher, Shanghai, China). The extracted DNA was diluted to 50 mg/L in sterile deionized water for PCR assays (see below).
Antimicrobial resistance genes (ARGs) were detected using multiplex PCR as previously described (22). Briefly, PCRs included gene-specific primers for the following ARG groups: penicillin (blaZ), macrolide (msrA and msrB), erythromycin (ermA and ermC), streptogramin acetyltransferase genes (vatA, vatB, and vatC), aminoglycoside (aacA-D), tetracycline (tetK and tetM), lincosamide (linA), methicillin (mecA), florfenicol (fexA), oxazolidine ketone (cfr and optrA) and vancomycin (vgaA and vgaC).
Virulence genes (hla, hlb, hld, sea, seb, sec, sed, see, tst, and lukF), biofilm-related genes (bap, icaA, and icaD) and adhesion-related genes (fnbA, fnbB, clfA, and clfB) were detected using PCR as previously described (23). The primers were provided by Sangon Biotech (Shanghai, China), and water rather than DNA template was added as a contamination control. DNA from isolates that harbored virulence genes or ARGs was used as a positive control. These were included in all PCRs. Gene amplifications were performed using a commercial PCR instrument (Bio-Rad, Hercules, CA, United States) as previously described (24). Briefly, the PCR mixture contained DNA (1 μL), 0.2 μL of each primer, Prime STAR Max DNA polymerase (12.5 μL), and ddH2O (11.1 μL). The PCR cycling conditions consisted of an initial denaturation at 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, annealing at appropriate temperatures for 30 s (Supplementary Table S1) and extension at 72°C for 1 min.
Biofilm formation was determined in 96-well microtiter plate assays using minimal medium M9 (Sigma Chemical) as previously described (25). Briefly, the overnight cultures in TSB were diluted 1:100, and 200 μL was transferred into each well of the microtiter plate that was incubated at 37°C for 72 h. Each well was washed with 200 μL PBS after the supernatant was discarded and fixed with 200 μL methanol for 20 min and washed again with PBS 3 × and then stained with 0.4% crystal violet (Meilunbio, Dalian, China) for 15 min. The biofilms were then dissolved in 200 μL 33% (w/v) acetic acid for 30 min. The biofilm formation was measured at 590 nm optical density (OD590 nm) in a Bio-Rad plate reader (Shanghai, China). The strong biofilm-forming strain Salmonella enterica Typhimurium ATCC 14028 was used as the positive control, and sterile TSB was used as the negative control for the biofilm formation assay (26). All assays were performed in triplicate. The OD590 nm value of 0.6 was applied as the cutoff point to distinguish between biofilm producers and nonbiofilm producers (10). Biofilm formation was classified as strong +++ (OD590 nm > 1.8), moderate ++ (1.8 > OD590 nm > 1.2), weak + (1.2 > OD590 nm > 0.6), and negative − (OD590 nm < 0.6).
Pearson correlation analysis was applied to differences in antimicrobial resistance rates in correlation to antimicrobial resistance genes harbored by S. aureus isolates. T tests were used to analyze the significance of biofilm formation between S. aureus isolates. All analyses were carried out using Prism 8 (GraphPad, Boston, MA, United States).
Our screening of 975 water buffaloes indicated that 287 (29.44%) were considered to have subclinical mastitis according to the CMT tests. Strongly positive (+++) results were observed in 53.31% (153/287) of the cases, while mild and moderate intensity results occurred in 26.13% (75/287) and 20.56% (59/287) of the cases, respectively. Escherichia coli (276/287) was the most common bacteria isolated from these positive samples, followed by coagulase-negative staphylococci (CNS) (194/287). Staphylococcus aureus (113/287), S. agalactiae (82/287) and S. dysgalactiae (41/287).
Resistance to bacitracin, doxycycline, penicillin, florfenicol and tetracycline was observed in 90.27, 84.07, 84.96, 81.42 and 82.3% of the examined S. aureus isolates, respectively (Table 1). A lower prevalence of resistance was noted for ciprofloxacin (7.08%), ceftizoxime (15.04%), cefoquinoxime (18.58%) and ofloxacin (16.81%). Among S. aureus isolates, only 12 (10.62%) were resistant to cefoxitin, and these were classified as phenotypic methicillin-resistant S. aureus (MRSA).
The ARGs possessed by S. aureus isolates included tetM, ermC, vatC, and aacA-D in 63.72% (72/113), 37.17% (42/113), 32.74% (37/113), and 27.43% (31/113), respectively. Moreover, the prevalence of msrB, blaZ, mecA, fexA, and tetK was 21.24, 19.47, 16.81, 15.04, and 6.19%, respectively (Figure 1). Interestingly, ARGs for macrolide resistance msrA, erythromycin resistance ermA, streptogramin resistance vatA and vatB, oxazolidinone resistance cfr and optrA and vancomycin resistance vgaA and vgaC were not detected. These results indicated a lack of a correlation between resistance phenotypes and ARG distributions among the S. aureus isolates.
The virulence-associated genes we detected in this study were distributed with varying frequencies among S. aureus isolates (n = 113) (Figure 2). In particular, clfB and icaA were present in all S. aureus isolates, and nearly all harbored hla (93.8%), hld (91.15%), clfA (90.27%), fnbA (86.73%) and hlb (83.19%). In contrast, a lower prevalence was found for tsst (27.43%), icaD (19.47%), sec (15.93%), see (9.73%), fnbB (65.49%) and sea (3.54%). The virulence genes seb, sed, bap, and lukF were not detected in any isolates.
All our milk samples produced isolates able to form biofilms. The rates of strong, moderate and weak biofilm producers were 30.09, 50.44, and 19.47%, respectively. In particular, most Qingyuan isolates (70.83%, 17/24) were strong biofilm producers, while 20.83% displayed moderate phenotypes. In contrast, only 9.76% (4/41) of the Guangzhou isolates were strong biofilm producers, and 65.85% (27/41) were moderate producers. Biofilm formation in S. aureus isolates from Guangzhou, Qingyuan, Jiangmen and Zhaoqing was 0.69 ± 0.24, 1.16 ± 0.35, 1.05 ± 0.25, and 1.03 ± 0.28, respectively. The biofilm formation of isolates from Guangzhou was significantly lower (p < 0.01) than that of isolates from other areas (Figure 3; Table 2).
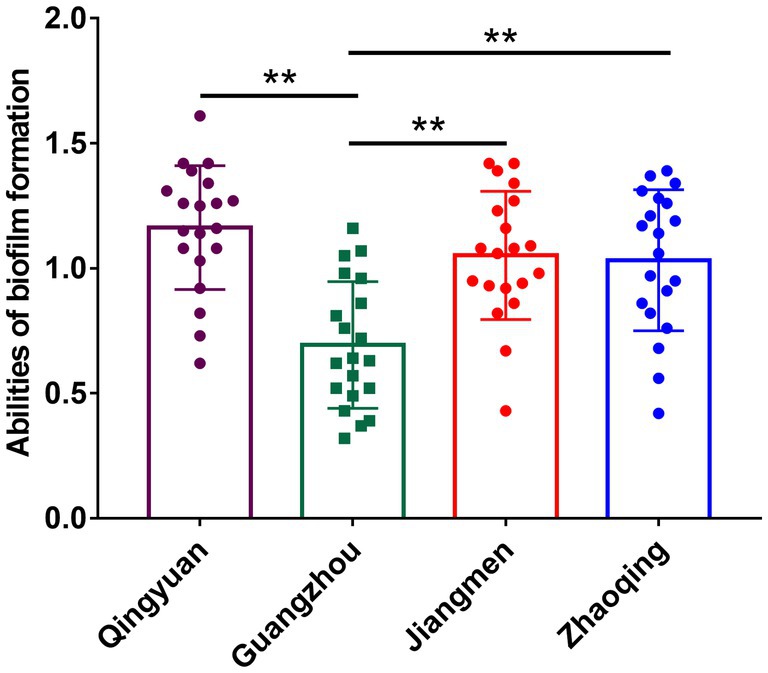
Figure 3. Biofilm formation of S. aureus isolates in different areas of Guangdong. Note: There were no significant difference between S. aureus isolates from Qingyuan, Jiangmen, and Zhaoqing. But S. aureus isolates from Guangzhou were significantly lower (p < 0.01) in comparison with isolates from Qingyuan, Jiangmen, and Zhaoqing. ** means p < 0.01.
Mastitis is a disease that is globally prevalent in dairy animals (1), and water buffaloes are generally less susceptible to this infection in comparison with dairy cows because of strong muscles at the opening of the teat canal (27). In the current study, 29.44% of water buffaloes were diagnosed with subclinical mastitis, which was consistent with a previous report for these animals (28). Previous studies have indicated that the prevalence of subclinical mastitis in water buffaloes ranges from 6.0 to 87% (27). It seems that factors such as animal age, stage of lactation, management style and farm environment may have contributed to these variations (13). To our knowledge, few studies have been carried out to investigate the bacteriology of subclinical mastitis in water buffaloes. In this study, we found that CoNS, E. coli and S. aureus were the dominant bacterial pathogens and that S. agalactiae and S. dysgalactiae were present to a lesser degree. Escherichia coli infections often lead to severe systemic clinical symptoms, and this was the most prevalent pathogen in our study. Similarly, E. coli was the most prevalent pathogen in subclinical mastitis infections in a Nepal water buffalo study (28), while Streptococcus (39.2%) was the most prevalent pathogen in mastitis dairy cows, and only 8.4% were present as E. coli (29). Therefore, it seems that other factors (see above) may influence the occurrence of mastitis. Moreover, S. aureus (61%) was the dominant pathogen in cattle from Jammu and Kashmir with mastitis, and E. coli (13%), CoNS (13.04%), Streptococcus uberis (4.35%) and S. dysgalactiae (8.69%) were also isolated from samples (30). Similar results were reported by other researchers (27); the reasons for this might be topographical and management conditions and the difference between dairy cows and water buffaloes.
Antimicrobial susceptibility can provide important information in choosing antimicrobial agents when treating infections. In this study, S. aureus isolates showed high resistance to penicillin, florfenicol, erythromycin, tetracycline, doxycycline and bacitracin. Similar results were observed in S. aureus isolates from dairy cows with mastitis in northern China (31). However, other researchers reported lower rates of antimicrobial resistance in S. aureus isolates in Pakistan (13). Cephalosporins are important antimicrobial agents, and S. aureus isolates resistant to ceftiofur have been reported (23). Similarly, antimicrobial resistance to cephalosporins, including cefoquinoxime, ceftizoxime, and ceftiofur, was detected in our study. β-lactams, fluoroquinolones, and aminoglycosides are commonly used to treat dairy mastitis (31), and this most likely contributed to the high levels of resistance we found to these agents. Saini and his colleagues also found that the herd level of antimicrobial agents used when treating mastitis in bovines was positively correlated with antimicrobial resistance among isolates from mastitic animals (32). Unfortunately, the use of antimicrobial agents in these farms was not documented in our study, so we cannot directly correlate the use of antimicrobial agents and antimicrobial resistance. Moreover, we detected antimicrobial resistance genes among isolates, although we found no significant correlation between phenotype and genotype. For example, only 22.92% (22/96) of isolates resistant to penicillin carried the blaZ gene, similar to previous findings (33, 34). These inconsistencies indicated that the presence of a particular ARG was not an indicator of phenotypic resistance, and this can be influenced by numerous genetic and environmental conditions (35).
MRSA is a global health concern since it is not only resistant to β-lactams but also nonsusceptible to other commonly used antimicrobial agents (36). In the Philippines and Pakistan, the MRSA prevalence was 25.81 and 19.6%, respectively, in water buffaloes with mastitis (13, 37), while a much lower rate (2.2%) of MRSA was detected in water buffaloes with mastitis in Iran (38). Several factors, such as age, feeding status, body conditions, and hand or machine hygiene on the farm, may contribute to this phenomenon. Several technologies, such as nanoparticles and antibiotics combined with plant extracts or microparticles, are widely used in food, veterinary and animal science. For example, a report indicated that antibiotics coupled with zinc oxide nanoparticles can significantly increase the zone of inhibition; similarly, amoxicillin showed the highest increase in inhibitory effects against MRSA when combined with Calotropis procera extract (39). These technologies are believed to be promising methods for treating infections caused by MRSA.
Biofilms can increase the resistance of S. aureus to antimicrobial agents and are responsible for persistent infections (40). Biofilms are composed of multiple layers of bacteria, which prevents the permeability of antimicrobial agents and thus increases tolerance. In our study, we investigated the biofilm formation ability of S. aureus isolates grouped by area. Interestingly, S. aureus isolates from Guangzhou showed significantly lower levels of biofilm formation in comparison with isolates from other areas (p < 0.01). However, the antimicrobial resistance of S. aureus isolates did not differ by region (data not shown). Similarly, a previous study indicated that the biofilm formation ability of ST7 and ST188 strains was much higher than that of other lineages even though their phenotypic antimicrobial resistance was comparable with that of other lineages (41). These data indicated that gene mutations, horizontal gene transfer and modifications of antibiotic molecules are the primary modes of antimicrobial resistance in S. aureus isolates from water buffaloes and that biofilm formation plays only a secondary role (42).
Virulence genes contribute to the pathogenesis of S. aureus infections. Adhesion is the first step for S. aureus to invade host cells and immune responses (43) and involves clfA, clfB, fnbA, and fnbB. In our study, all isolates carried clfB, and most isolates carried clfA, fnbA, and fnbB. These results were similar to previous reports where the clfB gene was detected in all isolates from bovine mastitis samples, and fnbA and clfB were comparable with the levels we found (44, 45). In contrast, much lower detection levels were reported for fnbB in S. aureus isolates from Algeria and Australia (43, 46).
Hemolysins are also involved in invasion and the host immune response (44, 46). In our study, over 80% of our total isolates carried hla, hlb, and hld, consistent with previous reports (41, 44). Toxic shock syndrome toxin, a superantigen encoded by the tsst gene, can lead to toxic shock syndrome in humans (47). and the tsst prevalence in S. aureus isolates ranged from 2.1 to 40.0% (10, 44) and was 25% in our study. It is therefore important to monitor the epidemiology of such super antigenic toxin genes to protect public health from this threat.
In conclusion, subclinical mastitis was prevalent among water buffaloes in Guangdong, China, and S. aureus was identified as a significant pathogen associated with subclinical mastitis of water buffaloes. The majority of S. aureus isolates exhibited resistance against bacitracin, doxycycline, penicillin, florfenicol, and tetracycline while maintaining susceptibility to other antimicrobial agents, including ciprofloxacin, ceftizoxime, cefoquinoxime, and ofloxacin. Furthermore, the S. aureus isolates harbored various virulence genes, such as hla, hld, clfA, fnbA, and hlb. Notably, all S. aureus isolates showed the ability to form biofilms, with nearly one-third of the isolates possessing strong biofilm formation abilities. Given these findings, antibiotics should be cautiously used when treating subclinical mastitis in water buffaloes within this region. Additionally, the impact of biofilm formation on the transmission of antibiotic resistance must be investigated in further studies.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal studies were approved by the Ethical Committee of Foshan University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
DZ, NZ, and HY: conceptualization and writing – review and editing. XL and XS: methodology and data curation. XF and QL: analysis. DZ: writing original draft preparation and funding acquisition. HY: supervision and project administration. All authors contributed to the article and approved the submitted version.
This research was supported by National Natural Science Foundation of China (31772795) and Innovative Team Project of Guangdong Provincial Department of Education (2022KCXTD028).
We would like to acknowledge the technical support from veterinarians and owners of water buffalo farms during sample collection and providing essential information.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1177302/full#supplementary-material
1. Ali, T, Kamran,, Raziq, A, Wazir, I, Ullah, R, Shah, P, et al. Prevalence of mastitis pathogens and antimicrobial susceptibility of isolates from cattle and buffaloes in northwest of Pakistan. Front Vet Sci. (2021) 8:746755. doi: 10.3389/fvets.2021.746755
2. Tong, F, Wang, T, Gao, N, Liu, Z, Cui, K, Duan, Y, et al. The microbiome of the buffalo digestive tract. Nat Commun. (2022) 13:823. doi: 10.1038/s41467-022-28402-9
3. Locatelli, C, Piepers, S, De Vliegher, S, Barberio, A, Supré, K, Scaccabarozzi, L, et al. Effect on quarter milk somatic cell count and antimicrobial susceptibility of Streptococcus rostri causing intramammary infection in dairy water buffaloes. J Dairy Sci. (2013) 96:3799–805. doi: 10.3168/jds.2012-6275
4. Pisanu, S, Cacciotto, C, Pagnozzi, D, Puggioni, GMG, Uzzau, S, Ciaramella, P, et al. Proteomic changes in the milk of water buffaloes (Bubalus bubalis) with subclinical mastitis due to intramammary infection by Staphylococcus aureus and by non-aureus staphylococci. Sci Rep. (2019) 9:15850. doi: 10.1038/s41598-019-52063-2
5. Lamey, AE, Ammar, AM, Zaki, ER, Khairy, N, Moshref, BS, and Refai, MK. Virulence factors of Escherichia coli isolated from recurrent cases of clinical and subclinical mastitis in buffaloes. Int J Microbiol Res. (2013) 4:86–94.
6. Ren, Q, Liao, GH, Wu, ZH, Lv, JF, and Chen, W. Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J Dairy Sci. (2020) 103:3368–80. doi: 10.3168/jds.2019-17420
7. Nobrega, DB, De Buck, J, Naqvi, SA, Liu, G, Naushad, S, Saini, V, et al. Comparison of treatment records and inventory of empty drug containers to quantify antimicrobial usage in dairy herds. J Dairy Sci. (2017) 100:9736–45. doi: 10.3168/jds.2017-13116
8. WHO. World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva, Switzerland: WHO (2014) Available at: http://www.who.int/drugresistance/documents/surveillancereport/en/.
9. Gao, J, Barkema, HW, Zhang, L, Liu, G, Deng, Z, Cai, L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. (2017) 100:4797–806. doi: 10.3168/jds.2016-12334
10. Wang, W, Lin, X, Jiang, T, Peng, Z, Xu, J, Yi, L, et al. Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front Microbiol. (2018) 9:1123. doi: 10.3389/fmicb.2018.01123
11. Vaughn, JM, Abdi, RD, Gillespie, BE, and Dego, OK. Genetic diversity and virulence characteristics of Staphylococcus aureus isolates from cases of bovine mastitis. Microb Pathog. (2020) 144:104171. doi: 10.1016/j.micpath.2020.104171
12. Piccinini, R, Borromeo, V, and Zecconi, A. Relationship between S. aureus gene pattern and dairy herd mastitis prevalence. Vet Microbiol. (2010) 145:100–5. doi: 10.1016/j.vetmic.2010.03.005
13. Javed, S, McClure, J, Syed, MA, Obasuyi, O, Ali, S, Tabassum, S, et al. Epidemiology and molecular characterization of Staphylococcus aureus causing bovine mastitis in water buffaloes from the Hazara division of Khyber Pakhtunkhwa, Pakistan. PLoS One. (2022) 17:e0268152. doi: 10.1371/journal.pone.0268152
14. Zhang, K, Sparling, J, Chow, BL, Elsayed, S, Hussain, Z, Church, DL, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol. (2004) 42:4947–55. doi: 10.1128/JCM.42.11.4947-4955.2004
15. Charan, J, and Biswas, T. How to calculate sample size for different study designs in medical research? Indian J Psychol Med. (2013) 35:121–6. doi: 10.4103/0253-7176.116232
16. Adkins, PRF, and Middleton, JR. Laboratory handbook on bovine mastitis. Madison, WI: National Mastitis Council (2017).
17. Schalm, OW, and Noorlander, DO. Experiments and observations leading to development of the California mastitis test. J Am Vet Med Assoc. (1957) 130:199–204.
18. Dai, J, Wu, S, Huang, J, Wu, Q, Zhang, F, Zhang, J, et al. Prevalence and characterization of Staphylococcus aureus isolated from pasteurized milk in China. Front Microbiol. (2019) 10:641. doi: 10.3389/fmicb.2019.00641
19. Yang, F, Shi, W, Meng, N, Zhao, Y, Ding, X, and Li, Q. Antimicrobial resistance and virulence profiles of staphylococci isolated from clinical bovine mastitis. Front Microbiol. (2023) 14:1190790. doi: 10.3389/fmicb.2023.1190790
20. Tian, XY, Zheng, N, Han, RW, Ho, H, Wang, J, Wang, YT, et al. Antimicrobial resistance and virulence genes of streptococcus isolated from dairy cows with mastitis in China. Microb Pathog. (2019) 131:33–9. doi: 10.1016/j.micpath.2019.03.035
21. CLSI. Performance standards for antimicrobial susceptibility testing; CLSI document M100-S27. Wayne, PA: Clinical Laboratory Standards Institute (CLSI) (2017).
22. Dan, M, Yehui, W, Qingling, M, Jun, Q, Xingxing, Z, Shuai, M, et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, Northwest China. J Glob Antimicrob Resist. (2019) 16:98–104. doi: 10.1016/j.jgar.2018.08.024
23. Liu, K, Tao, L, Li, J, Fang, L, Cui, L, Li, J, et al. Characterization of Staphylococcus aureus isolates from cases of clinical bovine mastitis on large-scale Chinese dairy farms. Front Vet Sci. (2020) 7:580129. doi: 10.3389/fvets.2020.580129
24. Pimentel de Araujo, F, Pirolo, M, Monaco, M, Del Grosso, M, Ambretti, S, Lombardo, D, et al. Virulence determinants in Staphylococcus aureus clones causing osteomyelitis in Italy. Front Microbiol. (2022) 13:846167. doi: 10.3389/fmicb.2022.846167
25. Müsken, M, Di Fiore, S, Römling, U, and Häussler, S. A 96-well-plate-based optical method for the quantitative and quanlitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc. (2010) 5:1460–9. doi: 10.1038/nprot.2010.110
26. Yan, Q, Wang, J, Gangiredla, J, Cao, Y, Martins, M, Gopinath, GR, et al. Comparative genotypic and phenotypic analysis of Cronobacter species cultured from four powdered infant formula production facilities: indication of pathoadaptation along the food chain. Appl Environ Microbiol. (2015) 81:4388–402. doi: 10.1128/AEM.00359-15
27. Nayan, V, Singh, K, Iquebal, MA, Jaiswal, S, Bhardwaj, A, Singh, C, et al. Genome-wide DNA methylation and its effect on gene expression during subclinical mastitis in water buffalo. Front Genet. (2022) 13:828292. doi: 10.3389/fgene.2022.828292
28. Bhandari, S, Subedi, D, Tiwari, BB, Shrestha, P, Shah, S, and AI-Mustapha, AI. Prevalence and risk factors for multidrug-resistant Escherichia coli isolated from subclinical mastitis in the western Chiwan region of Nepal. J Dairy Sci. (2021) 104:12765–72. doi: 10.3168/jds.2020-19480
29. Dyson, R, Charman, N, Hodge, A, Rowe, SM, and Taylor, LF. A survey of mastitis pathogens including antimicrobial susceptibility in southeastern Australian dairy herds. J Dairy Sci. (2022) 105:1504–18. doi: 10.3168/jds.2021-20955
30. Singha, S, Ericsson, CD, Chowdhury, S, Nath, SC, Paul, OB, Hoque, MA, et al. Occurrence and aetiology of subclinical mastitis in water buffalo in Bangladesh. J Dairy Res. (2021) 88:314–20. doi: 10.1017/S0022029921000698
31. Liu, H, Li, S, Meng, L, Dong, L, Zhao, S, Lan, X, et al. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. (2017) 100:8796–803. doi: 10.3168/jds.2017-13370
32. Saini, V, McClure, JT, Scholl, T, DeVries, TJ, and Barkema, HW. Herd-level association between antimicrobial use and antimicrobial resistance in bovine mastitis Staphylococcus aureus isolates on Canadian dairy farms. J Dairy Sci. (2012) 95:1921–9. doi: 10.3168/jds.2011-5065
33. Jamali, H, Radmehr, R, and Ismail, S. Prevalence and antibiotic resistance of Staphylococcus aureus isolated from bovine clinical mastitis. J Dairy Sci. (2014) 97:2226–30. doi: 10.3168/jds.2013-7509
34. Qu, Y, Zhao, H, Nobrega, DB, Cobo, ER, Han, B, Zhao, Z, et al. Molecular epidemiology and distribution of antimicrobial resistance genes of staphylococcus species isolated from Chinese dairy cows with clinical mastitis. J Dairy Sci. (2019) 102:1571–83. doi: 10.3168/jds.2018-15136
35. Bissong, MEA, and Ateba, CN. Genotypic and phenotypic evaluation of biofilm production and antimicrobial resistance in Staphylococcus aureus isolated from milk, north West Province, South Africa. Antibiotics. (2020) 9:156. doi: 10.3390/antibiotics9040156
36. Shoaib, M, Aqib, AI, Muzammil, I, Majeed, N, Bhutta, ZA, Kulyar, MF, et al. MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front Microbiol. (2023) 13:1067284. doi: 10.3389/fmicb.2022.1067284
37. Badua, AT, Boonyayatra, S, Awaiwanont, N, Gaban, PBV, and Mingala, CN. Methicillin-resistant Staphylococcus aureus (MRSA) associated with mastitis among water buffaloes in the Philippines. Heliyon. (2020) 6:e05663. doi: 10.1016/j.heliyon.2020.e05663
38. Panahi, M, and Saei, HD. Genetic diversity and methicillin resistance of Staphylococcus aureus originating from buffaloes with mastitis in Iran. Comp Immunol Microbiol Infect Dis. (2019) 62:19–24. doi: 10.1016/j.cimic.201811.014
39. Aqib, AI, Saqib, M, Khan, SR, Ahmad, T, Shah, SAR, Naseer, MA, et al. Non-steroidal anti-inflammatory drug, plant extracts, and characterized microparticles to modulate antimicrobial resistance of epidemic mec a positive S. aureus of dairy origin. Appl Nanosci. (2021) 11:553–63. doi: 10.1007/s13204-020-01628-z
40. Cucarella, C, Tormo, MA, Ubeda, C, Trotonda, MP, Monzon, M, Peris, C, et al. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. (2014) 72:2177–85. doi: 10.1128/IAI.72.4.2177-2185.2004
41. Li, T, Lu, H, Wang, X, Gao, Q, Dai, Y, Shang, J, et al. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol. (2017) 7:127. doi: 10.3389/fcimb.2017.00127
42. Munita, JM, and Arias, C. Mechanisms of antibiotic resistance. Microbiol Spectr. (2016) 4:2. doi: 10.1128/microbiolspec.VMBF-0016-2015
43. Zaatout, N, Ayachi, A, Kecha, M, and Kadlec, K. Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J Appl Microbiol. (2019) 127:1305–14. doi: 10.1111/jam.14402
44. Wang, D, Zhang, L, Zhou, X, He, Y, Yong, C, Shen, M, et al. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J Dairy Sci. (2016) 99:9560–9. doi: 10.3168/jds.2016-11625
45. Pereyra, EA, Picech, F, Renna, MS, Baravalle, C, Andreotti, CS, Russi, R, et al. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet Microbiol. (2016) 183:69–77. doi: 10.1016/j.vetmic.2015.12.002
46. Gogoi-Tiwari, J, Waryah, CB, Eto, KY, Tau, M, Wells, K, Costantino, P, et al. Relative distribution of virulence-associated factors among Australian bovine Staphylococcus aureus isolates: potential relevance to development of an effective bovine mastitis vaccine. Virulence. (2015) 6:419–23. doi: 10.1080/21505594.2015.1043508
Keywords: Staphylococcus aureus , antimicrobial resistance, virulence genes, biofilm formation, subclinical mastitis in water buffaloes
Citation: Zhang D, Lu X, Feng X, Shang X, Liu Q, Zhang N and Yang H (2023) Molecular characteristics of Staphylococcus aureus strains isolated from subclinical mastitis of water buffaloes in Guangdong Province, China. Front. Vet. Sci. 10:1177302. doi: 10.3389/fvets.2023.1177302
Received: 01 March 2023; Accepted: 13 October 2023;
Published: 08 November 2023.
Edited by:
Heba Sayed El-Mahallawy, Suez Canal University, EgyptReviewed by:
Waqas Ahmad, University of Veterinary and Animal Sciences, PakistanCopyright © 2023 Zhang, Lu, Feng, Shang, Liu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Zhang, emhhbmduYW5AZm9zdS5lZHUuY24=; Hong Yang, eWhvbmcwMDdAZm9zdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.