
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 04 May 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1175701
 Zhaoyang Xu1,2†
Zhaoyang Xu1,2† Yifan Hu1†
Yifan Hu1† Junbo Li1,3
Junbo Li1,3 Ancheng Wang1
Ancheng Wang1 Xin Meng1,3
Xin Meng1,3 Lingchao Chen1
Lingchao Chen1 Jianchao Wei1
Jianchao Wei1 Wu Tong1
Wu Tong1 Ning Kong1
Ning Kong1 Lingxue Yu1
Lingxue Yu1 Hai Yu1
Hai Yu1 Tongling Shan1
Tongling Shan1 Guangzhi Tong1
Guangzhi Tong1 Guihua Wang2*
Guihua Wang2* Hao Zheng1,4*
Hao Zheng1,4*African swine fever is a highly lethal contagious disease of pigs for which there is no vaccine. Its causative agent African swine fever virus (ASFV) is a highly complex enveloped DNA virus encoding more than 150 open reading frames. The antigenicity of ASFV is still unclear at present. In this study, 35 proteins of ASFV were expressed by Escherichia coli, and ELISA was developed for the detection of antibodies against these proteins. p30, p54, and p22 were presented as the major antigens of ASFV, positively reacting with all five clinical ASFV-positive pig sera, and 10 pig sera experimentally infected by ASFV. Five proteins (pB475L, pC129R, pE199L, pE184L, and pK145R) reacted well with ASFV-positive sera. The p30 induced a rapid and strong antibody immune response during ASFV infection. These results will promote the development of subunit vaccines and serum diagnostic methods against ASFV.
African swine fever, caused by the African swine fever virus (ASFV), is a highly lethal contagious disease in domestic pigs and wild boars. It affects swine of all breeds and ages, giving rise to a variety of clinical signs and lesions, from acute hemorrhagic fever with high mortality to chronic infection with skin ulcers and joint swelling, and causes serious economic consequences in the pig industry (1). Therefore, it is included in the list of diseases obliged declaration to the World Organization for Animal Health (OIE).
African swine fever (ASF) was first described in 1921 in Kenya and soon became enzootic throughout sub-Saharan Africa. It first crossed continents to Portugal in 1957 and from there to other European and Latin American countries, from where the disease was successfully eradicated in the mid-1990s, except for Sardinia (2). A new transmission era started when ASF was introduced into Georgia from Southeast Africa in 2007. Subsequently, it spread progressively across the Eurasian continent (2). Currently, most countries in Central and Eastern Europe and East and Southeast Asia have reported ASF cases, and it has even appeared in several Western European countries (3, 4). ASF was first reported in China in August 2018 (5). Within 6 months, acute ASF with almost 100% mortality had spread throughout most provinces in China and caused devastation to thousands of Chinese pig farms (4, 6). After 2 years of major efforts, acute ASF has been controlled to a great extent. In early 2021, however, ASFV with low virulence was found on some Chinese pig farms (7). Then, low-virulent isolates became predominant in the ASFV epidemic in China. Currently, ASFV is classified as a Class I infectious pathogen, and ASF has become the number one swine disease in China (6).
African swine fever virus (ASFV) is the sole characterized member of Asfarviridae and has a linear double-stranded DNA genome of 170–194 kbp encoding 150–170 predicted open reading frames (ORFs) (3). The differences in viral genome length and gene number predominantly originate from the loss or gain of genes in multigene families at the genome termini. A total of 94 viral proteins were expressed in cell lines infected with ASFV (8). However, the expression profiles of ASFV proteins in different cell lines varied markedly. A total of 68 viral proteins have been identified in mature virions (9). Many viral proteins, especially those containing transmembrane domains, have an unknown function. Characterization of viral antigens is important for developing ASF vaccine and serodiagnostic methods against ASFV. Several viral proteins such as p30, p54, p72, and CD2v are the major antigens of ASFV, and subunit vaccines made from these proteins provide partial protection against virulent ASFV challenge (10). Pigs immunized with mixed DNA vaccines or recombinant vaccinia virus expressing these proteins are partially protected against virulent ASFV (11, 12). Immunization with a DNA expression library containing >4,000 plasmids cloned with ASFV genome fragments clearly improved the protection against ASFV (13). These results suggested that many protective antigens were unexplored.
In this study, 35 structural and highly expressed ASFV proteins were prepared by a prokaryotic expression system. To evaluate the antigenicity of these proteins in ASFV infection, the levels of specific antibodies against these proteins in ASFV-positive pig sera were detected by ELISA.
A total of 26 inactivated sera from pigs experimentally infected with the CD2v and UK gene-deleted ASFV vaccine candidate (ASFV-SY18-ΔCD2v/UK) were kindly provided by Dr. Sun at Harbin Veterinary Research Institute, Harbin, China. Among these, 10 sera from pigs infected with ASFV-SY18-ΔCD2v/UK at 49 days post-inoculation (dpi) were served as ASFV-positive sera (14). A total of 16 sera were collected from four pigs infected with ASFV-SY18-ΔCD2v/UK at 6, 10, 15, and 21 dpi. In total, five inactivated clinical sera were obtained from farm pigs that had recovered from ASFV infection and also served as ASFV-positive sera. A total of five sera as negative controls were collected from healthy pigs in 2017. The ASFV-positive or ASFV-negative sera were tested by the ID Vet ASFV p30 antibody detection kit and the Ingenasa ASFV p72 ELISA kit.
The genome of genotype II ASFV strain Pig/HLJ/2018 (MK333180) was used as a reference sequence. A total of 38 genes of ASFV including 32 major structural proteins and seven proteins highly expressed in cells were analyzed with Protean in the LaserGene (DNAStar, Madison, Wisconsin, USA) and the online software of ProtScale (URL), SignalP, and TMHMM. The signal peptide and transmembrane sequences were deleted from transmembrane proteins. When a protein was >500 amino acids in length, the major antigenic fragment with 200–300 amino acids was used for expression. After optimization with Escherichia coli codon, these sequences were synthesized by General Biol (Anhui, China) and cloned into the prokaryotic expression vector pCold I with the restriction enzymes NdeI/XbaI, respectively. All constructs were verified by DNA sequencing and transformed into competent E. coli strain BL21 (TaKaRa Biotechnology (Dalian), Dalian, China).
Transformed BL21 cells were grown overnight in Luria–Bertani (LB) liquid medium with ampicillin (100 μg/ml). The culture was diluted 1:100 in fresh LB medium and grown for 2.5 h. Expression of recombinant proteins was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16 h at 16°C, and cells were harvested by centrifugation at 5,000 rpm in an Eppendorf F-34-6-38 rotor for 10 min. Soluble proteins were purified using WorkBeads 40 Ni-NTA (Bio-Works, Upasala, Sweden). Insoluble proteins were purified after solubilization with urea. Cell pellets were resuspended in 10 ml 50 mM Tris/HCl, and lysozyme (1.25 mg) was added. The cells were lysed by five cycles of sonication and then centrifuged. The pellet was washed three times in 5 ml 50 mM Tris/HCl with 1% Triton X-100. Next, 8 M urea (2.5 ml) was added, and the mixture was sonicated until the pellet was dissolved. The suspension was centrifuged at 6,000 rpm in an Eppendorf F-34-6-38 rotor for 10 min. The denatured protein was purified with WorkBeads 40 Ni-NTA and then refolded by dialysis in PBS.
ELISA plates (Corning Inc., Corning, New York, USA) were coated with ASFV recombinant proteins (100 μl per well) diluted to the appropriate concentrations (1 μg/ml) in coating buffer (50 mM sodium carbonate/bicarbonate buffer, pH 9.6) and incubated overnight at 4°C. The wells were washed three times with PBS plus 0.05% Tween 20 and blocked with PBS plus 5% milk (200 μl per well) at 37°C for 1 h. After blocking, plates were washed five times as mentioned above and incubated for 1 h at 37°C with pig sera diluted 1:100 in PBS plus 5% milk (100 μl per well). The plates were washed again five times and incubated with horseradish-peroxidase-conjugated anti-pig IgG antibody (Sigma–Aldrich, Saint Louis, Missouri, USA) diluted 1:4,000 (100 μl per well) for 1 h at 37°C. Finally, the plates were washed again and developed with TMB in the dark at room temperature for 15 min. After stopping the reaction with 2M H2SO4 (50 μl per well), the A450 was read on a Bio-Rad microplate reader (Bio-Rad, Hercules, California, USA).
Data were analyzed using GraphPad Prism 6.0 software. All data are presented as the mean ± standard deviation (SD). A p-value of <0.05 was considered statistically significant.
The ASFV genome contains more than 150 ORFs, and the virion consists of nearly 70 proteins. To explore the antigenicity of ASFV in infected pigs, 38 viral genes (Table 1) were expressed by E. coli. Among these genes, O61R (p12 gene), D117L (p17 gene), and K78R (p10 gene) were fused together with linker sequences, and I73R was fused with B169L. The amino acid sequences of expressed ASFV proteins are presented in Supplementary material S1. After induction with IPTG, four proteins (p54, pA104R, p11.5, and pI73R-pB169L) were expressed in soluble form, and the other 31 proteins (p30, pH171R, pCP312R, pE199L, pC257L, pE120R, p72, p22, pI177L, pK196R, pE146L, pF317L, pC717R, pB475L, pB602L, pEP152R, pE248R, pE184L, pH240R, pM1249L, pB125R, pC129R, pK421R, pEP153R, pK205R, pK145R, p150, CD2v, p15, p34, and p12-p17-p10) were expressed in the form of inclusion bodies. As shown in Supplementary material S2, 35 proteins were purified with Ni-NTA and used to coat ELISA plates.
A total of 15 positive pig sera for anti-ASFV antibodies (10 from pigs experimentally infected with attenuated ASFV and five from clinical field samples) and five pig sera free of ASFV were detected with ELISA plates coated with ASFV proteins. As shown in Figure 1, 24 proteins reacted with ASFV sera in varying degrees. p30, p54, and p22 showed a positive reaction with all ASFV sera (the value of sample/negative ≥3), which suggested that they were the dominant antigens of ASFV. pA104R, pB475L, pE120R, pC129R, pE199L, pCP312R, pE184L, pB602L, pK205R, pH171R, and the fusion protein p12-p17-p10 showed a positive reaction with all five clinical ASFV sera and with only some of the experimentally infected ASFV sera. p34, p11.5, pB125R, pK145R, CD2v, pK421R, p15, pI177L, p72, and fusion protein pI73R-pB169L showed a positive reaction with some of the clinical and experimentally infected ASFV sera. The other 11 proteins showed a negative reaction with all ASFV sera (Supplementary material S3). These results indicated that some ASFV proteins such as p30, p54, and p22 have excellent antigenicity and stimulate strong immune responses. Some viral proteins, however, did not induce any antibody immune responses in ASFV infection.
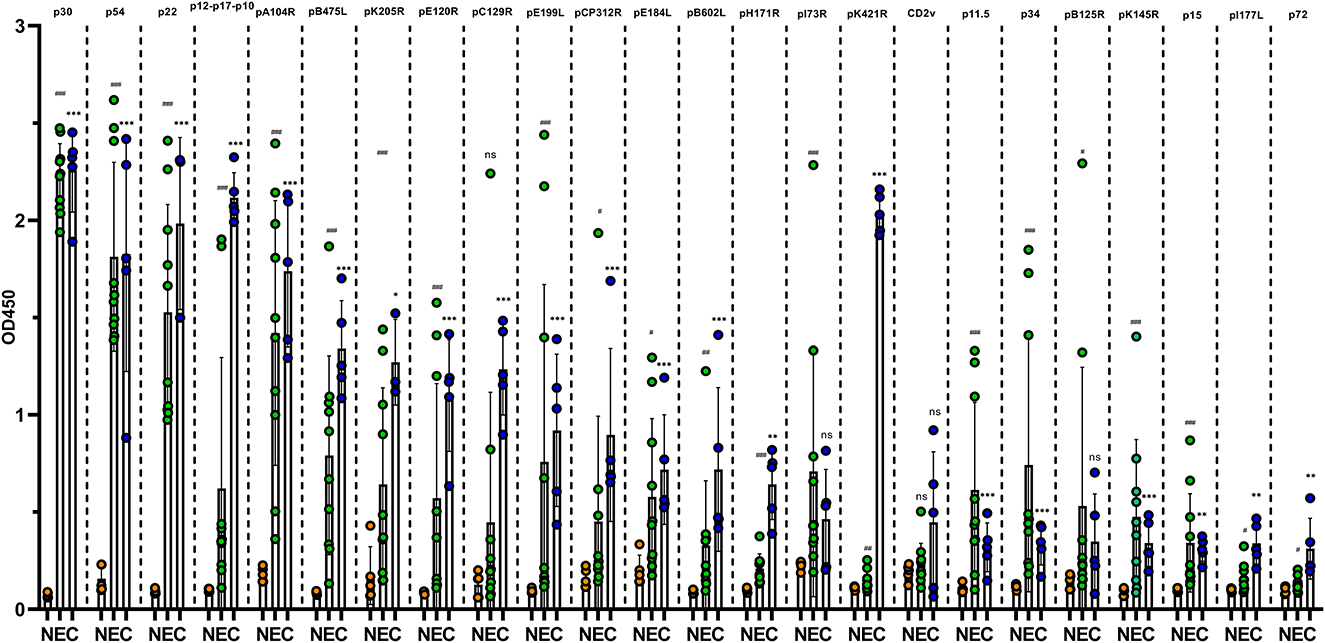
Figure 1. Reaction of ASFV recombinant proteins with antibodies in pig sera. ELISAs based on purified ASFV proteins were constructed and used to test five clinical ASFV-positive sera from farm pigs (C group, blue dot), 10 ASFV-positive pig sera experimentally infected with low-virulence ASFV (E group, green dot) and five ASFV-negative pig sera (N group, orange). The data of each group are presented as means and SD and were tested for significance using Student's t-test. * indicates a significant difference between the C group and N group (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). # indicates a significant difference between the E group and N group (#, P < 0.05; ##, P < 0.01; and ###, P < 0.001).
Some ASFV proteins were able to induce host antibody immunity. To further explore the properties of antibody generation during ASFV infection, pig sera were collected at 6, 10, 15, and 21 days after attenuated ASFV infection, and levels of antibodies against 15 ASFV proteins of high antigenicity were detected by ELISA. As shown in Figure 2, three of four pigs showed p30-antibody positive conversion (S/N ≥3) at 10 dpi. All four pig sera were p30-antibody positive, and S/N for p30 antibodies was >10 at 15 dpi. A total of three pigs became antibody positive for p22, p34, and p54 at 15 dpi and for pB475L and pK145R at 21 dpi, but the level of antibodies against these proteins remained negative in the rest of one pig. In total, one to two pigs developed antibodies against pA104R, pE184L, pC129R, pE199L, pE120R, and pCP312R and the fusion protein pI73R-pB169L. These results showed that attenuated ASFV infection induced a rapid and strong p30-antibody response.
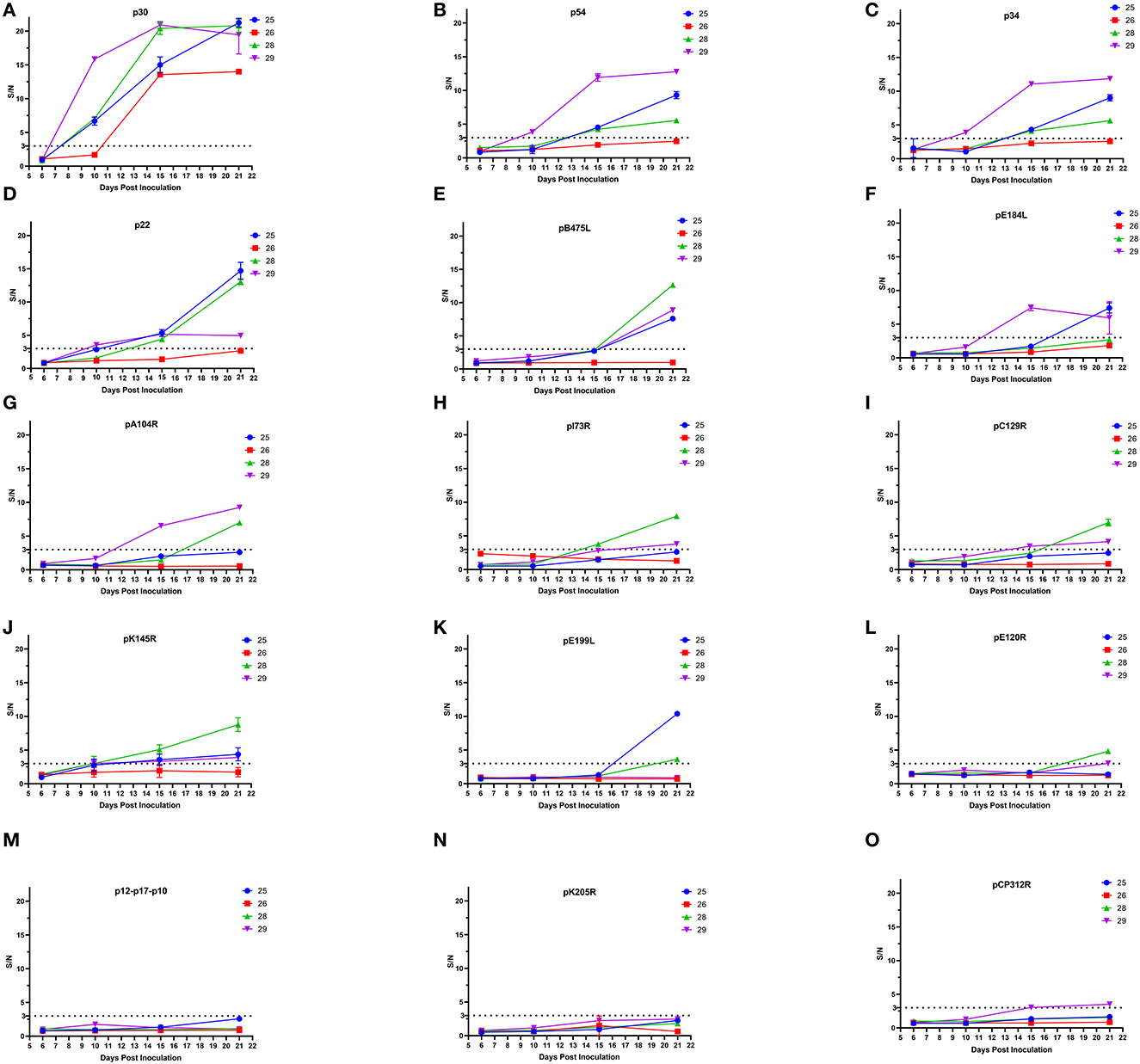
Figure 2. Development of ASFV-specific antibodies in pigs infected with low-virulence ASFV. ELISA methods based on 15 ASFV recombinant proteins including p30 (A), p54 (B), p34 (C), p22 (D), pB475L (E), pE184L (F), pA104R (G), pI73R-pB169L (H), pC129R (I), pK145R (J), pE199L (K), pE120R (L), p12-p17-p10 (M), pK205R (N), and pCP312R (O) were used to test the sera of four pigs (the number 25, 26, 28, and 29) at 6, 10, 15, and 21 dpi with low-virulence ASFV. A total of five ASFV-negative pig sera served as negative controls. If S/N≥3 (cutoff value), the sample was positive for antibodies against ASFV protein.
African swine fever virus (ASFV) is a highly complex enveloped DNA virus and encodes >150 ORFs (3). At present, it has been confirmed that ~100 proteins are expressed by ASFV, and 63 proteins form virion (8, 9). However, the roles of many structure proteins are uncharacterized. Clarifying the antigenicity of ASFV proteins is important to develop serological diagnostic methods and vaccines against ASFV. In this study, 33 genes of structural proteins with high abundance in virion and five genes of non-structure proteins with high expressing levels in host cells were expressed by E. coli to screen the dominant antigens of ASFV (8, 9). Among 33 structure proteins, many proteins are important components of the virion and play an important role in virus entry and morphogenesis, and the others are uncharacterized (Table 1). Indirect ELISAs based on these proteins were established to detect the antibody levels in the sera of pigs infected by ASFV. It was confirmed that ASFV infection induced strong antibody responses against p30, p54, and p22. pA104R, pB475L, p34, pE120R, pC129R, pE199L, pE184L, pCP312R, pK145R, and pB602L also induced distinct antibodies.
In this study, ASFV proteins were prepared using an E. coli expression system. To improve the expression levels of ASFV proteins in E. coli, the gene sequences were optimized. The signal peptides and transmembrane domains of proteins were removed. For several large ASFV proteins including pM1249L, pB602L, pCP717R, and p72, the predicted antigen dominant regions were expressed. Protein truncation may reduce detecting the special antibodies against natural proteins. Reis et al. showed strong antibody responses against pB602L (15). In our study, most sera from pigs infected with attenuated ASFV showed a weak reaction with the truncated pB602L protein. The genotype of ASFV may affect the generation of special antibodies. The sera from clinical samples in our study showed a stronger reaction with the truncated pB602L protein than the sera from the pigs infected with attenuated ASFV. The p72 protein is the major capsid protein and one of the most immunogenic ASFV proteins, which is an important target for test and vaccine development (16–19). In our study, however, the expressed p72 showed poor reactivity with the ASFV-positive sera, which may be because the p72 protein did not form the correct structure. It has been shown that correctly folded p72 needed the assistance of pB602L (20). Thus, the p72 protein which was expressed alone in E. coli may not be a suitable diagnostic antigen.
Among 35 ASFV proteins expressed in this study, p30, p54, and p22 were the dominant antigens during ASFV infection, which was consistent with previous studies (15, 17). To improve the sensitivity and specificity of p30 and p54-based ELISAs, most p30 and p54 were produced in insect cells using a baculovirus expression system (21). Although p30 protein was expressed in inclusion bodies in E. coli in our study, refolded p30 protein showed a strong reaction with all ASFV-positive sera. The p30 antibody developed early during attenuated ASFV infection and was the most sensitive target detected by ELISA. At 10 dpi, p30 antibodies became positive in most pigs. Even in one pig that remained negative for antibodies against other ASFV proteins, p30 antibodies became positive at 15 dpi. The indirect immunoperoxidase test is the best for ASF serological diagnosis due to its superior sensitivity. ASFV antibodies detected with immunoperoxidase became positive at 8–10 dpi in subacute or chronic infection, which is consistent with the seroconversion time of the p30 antibody in our study (22, 23). Therefore, the p30 antibody was an excellent target for ASF serodiagnosis. The p30 protein expressed in E. coli was also suitable as a diagnostic antigen for ELISA.
At present, there is no safe and effective vaccine available against ASFV (24). Several subunit vaccines including DNA- and peptide-based vaccines based on some major antigens, such as p30, p54, p72, CD2v, and/or pEP153R, provide partial protection against virulent ASFV challenge (25, 26). In addition to p54, p30, and hemagglutinin, Lacasta et al. suggested that some unidentified protective determinants within the ASFV genome could play an important role in preventing ASFV infection (13). Combining the results of ASFV proteins reacting with ASFV-positive sera and the development of ASFV antibodies in this study, pB475L, pC129R, pE199L, pE184L, and pK145R induced rapid antibody development in ASFV infection, besides p30, p54, and p22. pB475L is a non-structural protein, while pC129R, pE199L, pE184L, and pK145R are structural proteins. E199L can mediate virus entry but the other four proteins are uncharacterized (27). It is unclear whether these five proteins can induce protective immunity against ASFV.
In this study, p30, p54, and p22 were identified as the dominant antigens of ASFV. In particular, the development of the p30 antibody was rapid and strong during ASFV infection. Thus, it was an important target of serodiagnosis. pB475L, pC129R, pE199L, pE184L, and pK145R also induced antibody development in ASFV infection. These results promoted the development of subunit vaccines and serodiagnostic methods against ASFV.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
ZX and YH: methodology, investigation, and validation. JL, AW, and XM: formal analysis and validation. LC: investigation. JW, WT, LY, NK, and HY: methodology and resources. TS: review and editing. GT: project administration. GW: supervision and writing—review and editing. HZ: conceptualization, methodology, writing–original draft, supervision, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study was supported by the National Key Research and Development Program of China (Grant No. 2021YFD1801300), the Shanghai Science and Technology Innovation Action Plan (Grant No. 20392002500), and the Shanghai Municipal Natural Science Foundation (No. 20ZR1469500).
The authors are grateful to Dr. Yuan Sun at Harbin Veterinary Research Institute, CAAS, who kindly provided pig sera experimentally infected with attenuated ASFV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1175701/full#supplementary-material
1. Pikalo J, Zani L, Hühr J, Beer M, Blome S. Pathogenesis of African swine fever in domestic pigs and European wild boar-lessons learned from recent animal trials. Virus Res. (2019) 271:197614. doi: 10.1016/j.virusres.2019.04.001
2. Galindo I and Alonso C. African swine fever virus: A review. Viruses. (2017) 9:103–13. doi: 10.3390/v9050103
3. Dixon LK, Sun H, Roberts H. African swine fever. Antiviral Res. (2019) 165:34–41. doi: 10.1016/j.antiviral.2019.02.018
4. Gavier-Widen D, Stahl K, Dixon L. No hasty solutions for African swine fever. Science. (2020) 367:622–4. doi: 10.1126/science.aaz8590
5. Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, et al. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. (2018) 65:1482–4. doi: 10.1111/tbed.12989
6. Wu K, Liu J, Wang L, Fan S, Li Z, Li Y, et al. Current state of global African swine fever vaccine development under the prevalence and transmission of ASF in China. Vaccines. (2020) 8:531–58. doi: 10.3390/vaccines8030531
7. Sun E, Zhang Z, Wang Z, He X, Zhang X, Wang L, et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci China Life Sci. (2021) 64:752–65. doi: 10.1007/s11427-021-1904-4
8. Keßler C, Forth JH, Keil GM, Mettenleiter TC, Blome S, Karger A. The intracellular proteome of African swine fever virus. Sci Rep. (2018) 8:14714–23. doi: 10.1038/s41598-018-32985-z
9. Alejo A, Matamoros T, Guerra M, Andrés G. A proteomic atlas of the African swine fever virus particle. J Virol. (2018) 92:e01293–18. doi: 10.1128/JVI.01293-18
10. Sanchez EG, Perez-Nunez D, Revilla Y. Development of vaccines against African swine fever virus. Virus Res. (2019) 265:150–5. doi: 10.1016/j.virusres.2019.03.022
11. Argilaguet JM, Pérez-Martín E, Nofrarías M, Gallardo C, Accensi F, Lacasta A, et al. DNA vaccination partially protects against African swine fever virus lethal challenge in the absence of antibodies. PLoS ONE. (2012) 7:e40942. doi: 10.1371/journal.pone.0040942
12. Jancovich JK, Chapman D, Hansen DT, Robida MD, Loskutov A, Craciunescu F, et al. Immunization of pigs by DNA prime and recombinant Vaccinia virus boost to identify and rank African swine fever virus immunogenic and protective proteins. J Virol. (2018) 92:e02219–17. doi: 10.1128/JVI.02219-17
13. Lacasta A, Ballester M, Monteagudo PL, Rodríguez JM, Salas ML, Accensi F, et al. Expression library immunization can confer protection against lethal challenge with African swine fever virus. J Virol. (2014) 88:13322–32. doi: 10.1128/JVI.01893-14
14. Teklue T, Wang T, Luo Y, Hu R, Sun Y, Qiu H-J. Generation and evaluation of an African swine fever virus mutant with deletion of the CD2v and UK genes. Vaccines. (2020) 8:763–80. doi: 10.3390/vaccines8040763
15. Reis AL, Parkhouse RME, Penedos AR, Martins C, Leitão A. Systematic analysis of longitudinal serological responses of pigs infected experimentally with African swine fever virus. J Gen Virol. (2007) 88:2426–34. doi: 10.1099/vir.0.82857-0
16. Sanz A, García-Barreno B, Nogal ML, Viñuela E, Enjuanes L. Monoclonal antibodies specific for African swine fever virus proteins. J Virol. (1985) 54:199–206. doi: 10.1128/jvi.54.1.199-206.1985
17. Kollnberger SD, Gutierrez-Castañeda B, Foster-Cuevas M, Corteyn A, Parkhouse RME. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. J Gen Virol. (2002) 83:1331–42. doi: 10.1099/0022-1317-83-6-1331
18. Liu S, Luo Y, Wang Y, Li S, Zhao Z, Bi Y, et al. Cryo-EM structure of the African swine fever virus. Cell Host Microbe. (2019) 26:836–43. doi: 10.1016/j.chom.2019.11.004
19. Cubillos C, Gómez-Sebastian S, Moreno N, Nuñez MC, Mulumba-Mfumu LK, Quembo CJ, et al. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. (2013) 173:159–67. doi: 10.1016/j.virusres.2012.10.021
20. Liu Q, Ma B, Qian N, Zhang F, Tan X, Lei J, et al. Structure of the African swine fever virus major capsid protein p72. Cell Res. (2019) 29:953–5. doi: 10.1038/s41422-019-0232-x
21. Oviedo JM, Rodríguez F, Gómez-Puertas P, Brun A, Gómez N, Alonso C, et al. High level expression of the major antigenic African swine fever virus proteins p54 and p30 in baculovirus and their potential use as diagnostic reagents. J Virol Methods. (1997) 64:27–35. doi: 10.1016/S0166-0934(96)02140-4
22. Gallardo C, Fernandez-Pinero J, Arias M. African swine fever (ASF) diagnosis, an essential tool in the epidemiological investigation. Virus Res. (2019) 271:197676. doi: 10.1016/j.virusres.2019.197676
23. Gallardo C, Nieto R, Soler A, Pelayo V, Fernández-Pinero J, Markowska-Daniel I, et al. Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in Eastern European Union countries: How to improve surveillance and control programs. J Clin Microbiol. (2015) 53:2555–65. doi: 10.1128/JCM.00857-15
24. Urbano AC and Ferreira F. African swine fever control and prevention: an update on vaccine development. Emerg Microbes Infect. (2022) 11:2021–33. doi: 10.1080/22221751.2022.2108342
25. Sang H, Miller G, Lokhandwala S, Sangewar N, Waghela SD, Bishop RP, et al. Progress toward development of effective and safe African swine fever virus vaccines. Front Vet Sci. (2020) 7:84. doi: 10.3389/fvets.2020.00084
26. Revilla Y, Perez-Nunez D, Richt JA. African swine fever virus biology and vaccine approaches. Adv Virus Res. (2018) 100:41–74. doi: 10.1016/bs.aivir.2017.10.002
Keywords: African swine fever virus, dominant antigen, p30 protein, p54 protein, p22 protein
Citation: Xu Z, Hu Y, Li J, Wang A, Meng X, Chen L, Wei J, Tong W, Kong N, Yu L, Yu H, Shan T, Tong G, Wang G and Zheng H (2023) Screening and identification of the dominant antigens of the African swine fever virus. Front. Vet. Sci. 10:1175701. doi: 10.3389/fvets.2023.1175701
Received: 28 February 2023; Accepted: 27 March 2023;
Published: 04 May 2023.
Edited by:
Shixing Yang, Jiangsu University, ChinaReviewed by:
Fernando Costa Ferreira, University of Lisbon, PortugalCopyright © 2023 Xu, Hu, Li, Wang, Meng, Chen, Wei, Tong, Kong, Yu, Yu, Shan, Tong, Wang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guihua Wang, d2d1aWh1YTExMjZAMTYzLmNvbQ==; Hao Zheng, aGFvemhlbmdAc2h2cmkuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.