
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 15 May 2023
Sec. Parasitology
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1167070
This article is part of the Research Topic Dog Filariosis: The Threat Walks Not Only in The Blood Stream View all 6 articles
 Chandler C. Roe1,2*
Chandler C. Roe1,2* Olivia Holiday3
Olivia Holiday3 Kelly Upshaw-Bia3
Kelly Upshaw-Bia3 Gaven Benally3
Gaven Benally3 Charles H. D. Williamson1
Charles H. D. Williamson1 Jennifer Urbanz4
Jennifer Urbanz4 Guilherme G. Verocai5
Guilherme G. Verocai5 Chase L. Ridenour1,2,6
Chase L. Ridenour1,2,6 Roxanne Nottingham1
Roxanne Nottingham1 Morgan A. Ford7
Morgan A. Ford7 Derek P. Lake1
Derek P. Lake1 Theodore A. Kennedy7
Theodore A. Kennedy7 Crystal M. Hepp1,2,6
Crystal M. Hepp1,2,6 Jason W. Sahl1
Jason W. Sahl1Onchocerca lupi (Rodonaja, 1967) is an understudied, vector-borne, filarioid nematode that causes ocular onchocercosis in dogs, cats, coyotes, wolves, and is also capable of infecting humans. Onchocercosis in dogs has been reported with increasing incidence worldwide. However, despite the growing number of reports describing canine O. lupi cases as well as zoonotic infections globally, the disease prevalence in endemic areas and vector species of this parasite remains largely unknown. Here, our study aimed to identify the occurrence of O. lupi infected dogs in northern Arizona, New Mexico, and Utah, United States and identify the vector of this nematode. A total of 532 skin samples from randomly selected companion animals with known geographic locations within the Navajo Reservation were collected and molecularly surveyed by PCR for the presence of O. lupi DNA (September 2019–June 2022) using previously published nematode primers (COI) and DNA sequencing. O. lupi DNA was detected in 50 (9.4%) sampled animals throughout the reservation. Using positive animal samples to target geographic locations, pointed hematophagous insect trapping was performed to identify potential O. lupi vectors. Out of 1,922 insects screened, 38 individual insects and 19 insect pools tested positive for the presence of O. lupi, all of which belong to the Diptera family. This increased surveillance of definitive host and biological vector/intermediate host is the first large scale prevalence study of O. lupi in companion animals in an endemic area of the United States, and identified an overall prevalence of 9.4% in companion animals as well as multiple likely biological vector and putative vector species in the southwestern United States. Furthermore, the identification of these putative vectors in close proximity to human populations coupled with multiple, local zoonotic cases highlight the One Health importance of O. lupi.
Onchocerca lupi (Nematoda: Onchocercidae) is an emerging zoonotic filarial nematode that was first described in a gray wolf from Caucasia in the Republic of Georgia in 1967 (1). Within the last decade O. lupi has shown to be endemic to regions of the United States (US), Europe, Northern Africa, and the Middle East (2–13). The most important definitive hosts of O. lupi are domestic dogs (Canis lupus familiaris); however, this parasite has also been reported to infect humans (1, 10, 14–16) and wild carnivores like coyotes (Canis latrans) and more rarely, wolves (Canis lupus), and cats (Felis catus). While the increase in incidence of this parasite has renewed interest world-wide, low occurrence of disease within human populations coupled with O. lupi classified as a non-reportable veterinary disease in the United States and the current lack of a commercial diagnostic severely impedes our understanding of disease prevalence and geographic distribution. Due to the implications for both animal and human populations, the purpose of this study was to determine the occurrence of O. lupi in companion animal populations on the Navajo Reservation, located in the southwestern United States, as it has overlapping reports of O. lupi in humans, dogs, and coyotes (15, 17–19) and determine potential vector species.
While O. lupi is considered a vector-borne parasite that is putatively transmitted by dipteran hematophagous insects, as are other congeneric species (20), demonstration of active transmission by a suspected vector species remains absent for this parasite. There is limited knowledge on the life cycle of O. lupi; the current dogma is grounded in knowledge from more heavily studied Onchocerca species (e.g., Onchocerca volvulus, Onchocerca ochengi) which suggest the blackfly Simulium spp. (Diptera, Simuliidae) as the putative intermediate host of O. lupi (21). Recent research has identified Simulium tribulatum as a putative intermediate host for this parasite along the San Gabriel River watershed in southern California (United States), but vector competency has yet to be explored (22). An additional study has identified O. lupi DNA sequences acquired from head and bodies of Simulium griseum, but has not yet been published in peer-reviewed literature. Subsequent black fly sampling in areas of California failed to detect O. lupi DNA but identified DNA of other Onchocerca genetic lineages likely associated with wild ungulates (23, 24).
Blackflies, such as Simulium spp., are the most widespread of the small biting Diptera (25). The genus Simulium can thrive in a wide range of climates from tropical to temperate conditions, but the presence of a running water habitat is a necessary component of their life cycle (25). However, reports of autochthonous O. lupi in canines from regions lacking the necessary water habitats required for blackfly breeding suggests the possibility of an O. lupi vector outside the Simuliidae family. The principal method for control of vector-borne diseases, such as lymphatic filariasis and malaria, is through vector control (e.g., targeted deployment of insecticides) and requires extensive knowledge of the vector species life cycle (26). There is an urgent need to extend the insect sampling spectrum beyond urban waterways to explain increasing endemic disease in anhydrous regions such as Navajo Reservation (43,452km2) in the southwestern United States; this is essential for implementing effective vector control strategies to mitigate O. lupi transmission.
The dogma around O. lupi considers canid species as definitive hosts and an unknown arthropod species as intermediate host. Sexually mature nematode develop in the sclera of the eye within the definitive host where females release microfilariae into canid skin, predominantly in the head, ears, interscapular, and lumbar regions (21). While the putative simuliid intermediate host takes a blood meal, unsheathed microfilariae are ingested and migrate to the midgut followed by the thoracic muscles and develop from L1 (non-infectious) to L3 (infectious) larvae. Once infectious, the L3 migrates to the mouthparts of the intermediate host, ready to infect a vertebrate host during its next blood meal. It is important to consider that, to date, L3 larvae have not been identified in the head of an intermediate host for O. lupi, which would serve as unequivocal biological proof for vector suitability. However, current research from closely related Onchocerca species suggests L3 larvae are the only developmental stage present in the head of intermediate hosts (27, 28). Current PCR-based methods for O. lupi identification alone cannot differentiate viable from nonviable or immature (L1 and L2) from infective (L3) larvae within wild-caught insects and are therefore insufficient when screening individuals or pools of whole insects as potential intermediate hosts. Therefore, the identification of O. lupi DNA within the heads of suspected vectors suggests active transmission as only infectious L3 larva DNA should be present in the head of the arthropod, serving as a molecular confirmation of vector suitability.
Current knowledge regarding the patterns of epidemiology and vector-parasite interactions of O. lupi is lacking, which is unfortunate as reported incidence in dogs as well as human cases seem to be rising (2–4, 13, 17). O. lupi is considered endemic in canids within the southwestern United States, which includes California, Arizona, New Mexico, and Texas (3, 4, 15, 22). Worldwide, there have been 18 reported human cases of zoonotic O. lupi, five of which were in the southwestern United States (9, 10, 13, 17, 19) within the past 12 years (21). These clinical reports coupled with the presence of positive O. lupi coyotes in northern Arizona and southern New Mexico (15) indicate a need for increased surveillance of hosts and vectors in regions with overlapping O. lupi infections in human, dog, and wildlife populations. Furthermore, identifying where O. lupi occurs in the environment is crucial for determining the risk of spread to non-endemic regions as well as determining risk for human populations. In the present study, we conducted surveillance of O. lupi in canine populations on the Navajo Reservation, located in the southwestern United States (Arizona, New Mexico, and Utah), and used these data to target geographic locations for collection and molecular screening of hematophagous insects to identify potential vectors of the parasite in this region.
This study was approved by IACUC of Northern Arizona University (Approved protocol 19-016). Written informed consent was obtained from owners for animal participation. Geographic coordinates for positive dog and insect trap site locations were excluded from this manuscript for privacy purposes.
The Navajo Reservation is the largest federally recognized sovereign region retained by the Navajo Nation within the southwestern United States and is located largely on the Colorado Plateau. The Reservation covers 43,452km2 spanning the states of Arizona, New Mexico, Colorado, and Utah. The Navajo Reservation varies in altitude (940 to 3,153 m) and encompasses several distinct landscapes with varying climates (29). Regardless of the distinct terrains, water is sparse throughout most of the Reservation (30). Three distinct topographies, each with a different climate and vegetation, makeup the Navajo Reservation: the cold, subhumid climate of the mountains, the intermediate steppe climate, and the warm, arid desert climate. Ponderosa pine-covered high plateaus account for 8% of the region with a reported cold and subhumid climate with lows and highs between −15.6°C and 26.7°C and an annual rainfall of 40.64–68.58 cm (31). With a described climate as cold and dry, the mesas and high plains account for 37% of the region with temperatures ranging between −12.22°C and 31.11°C and annual rainfall between 30.48–40.64 cm and reported vegetation is listed as grasses, sagebrush, and pinyon-juniper (29, 32). Lastly, the warm, arid desert landscape accounts for 55% of Navajo land with temperatures ranging between −11.67°C to 43.33°C and annual rainfall between 17.78–27.96 cm (31). Desert vegetation such as grasses and browse plants are limited (29). A 2010 census reported the total population on the Navajo Reservation as 173,637 people (33) whom are widely dispersed across the reservation in part because of the scarce availability of water (34). This study conducted companion animal and insect sampling within all three climates on the Navajo Reservation.
From September 2019 through July 2022, we collected 532 skin tissue samples from companion animals (including both domestic and stray dogs and cats) throughout the Navajo Reservation (Arizona, New Mexico, and Utah) to gain thorough surveillance data. Samples were obtained primarily through spay and neuter clinics as well as from any animal undergoing a surgical procedure with owner consent at three veterinary clinics and a mobile unit. Selection criteria included all animals with a minimum estimated age of 6 months. Upon sample collection, animals in this study did not display clinical disease symptoms typical of canine ocular onchocercosis such as nodule formation. However, animals were selected for this study regardless of their health status. All animal handling and sample collection was in accordance with IACUC regulations. Skin samples were obtained using a disposable 0.2 cm skin punch from the inter ocular frontal area of the head, which was previously identified as one of the predilection areas for presence of O. lupi microfilariae on the canine host (35), and immediately fixed and stored in 80% ethanol.
Upon receipt in the laboratory, skin samples were stored at 4°C until processing. DNA was extracted from 532 skin samples using the Qiagen Dneasy Blood and Tissue kit with a preliminary overnight incubation at 56°C following manufacturer’s recommendations within the tissue lysis protocol (Qiagen). The conventional PCR (cPCR) screening for O. lupi from skin samples was based on the partial sequence of the cytochrome c oxidase gene (COI; COIF: 5′-TGATTGGTGGTTTTGGTAA-3′, COIR: 5′-CATAAGTACGAGTATCAATATC-3′) as described previously (36). The cPCR amplification was carried out with a final 25 μL reaction volume consisting of 1X KAPA 2G Master Mix, 8 μL of genomic DNA, and 100 nM forward and reverse COI primers. The following thermocycler conditions were used: 95°C × 3 min, 35 cycles of (95°C × 15 s, 60°C × 30s, 72°C × 1.5 min), followed by 72°C × 1 min. The presence of PCR product was determined by visualization using gel electrophoresis with a 2.0% TAE agarose gel and sequenced directly with capillary electrophoresis using a BigDye Terminator v3.1 Cycle Sequencing Kit on the 3,130 Genetic Analyzer platform (Applied Biosystems). Fifty-two Forward and reverse sequences were assembled using SeqMan v17 (DNAStar) and were queried with BLASTN (37) against the NCBI Nucleotide database (nt) to confirm their identity as O. lupi.
Insects were collected between May and August 2021 from 15 individual sites spanning Arizona and New Mexico on the Navajo Reservation using BG-Sentinel mosquito traps (Biogents) baited with both CO2 and the BG-Lure chemical attractant (Biogents). Funding dictated a single insect trapping season. Twelve of the 15 trap sites were targeted collection points; COI cPCR positive dogs identified from phase 1 of this study informed these locations. Three additional non-informed trap sites were included based on accessibility, permissions, and close proximity to dry creek beds. Due to remote trapping locations, traps generally collected insects continuously for 7 days with nonstop CO2 emittance. At the end of 7 days, insects were collected and flash frozen for transport to the laboratory at Northern Arizona University. Traps were moved between the 15 locations throughout the insect trapping season depending on accessibility and permissions. Generally, each trap site collected insects during a single week only, however, when technical difficulties (CO2 emittance issues) arose, traps were left at the same site an additional week. Insects were stored at −20°C until further processing. A Leica S8 AP0 dissecting microscope with an attached Canon EOS Rebel T3 camera was used to process all insects which included morphological examination, counting, photographing, and sorting. The small/min size and great abundance of biting midges at most locations informed insect pooling from the same trap in sets of 50 biting midges. Biting midges were pooled into pools between 2 and 50 insects based on the following criteria: the same species, the same site, the same collection week. Seventy biting midge pools were initially processed as whole insects, however, once positive pools were identified, the remaining 15 pools had DNA extracted from the heads and bodies separately.
Total DNA was extracted from heads and bodies of Dipteran insects using the Dneasy Blood and Tissue Kit (Qiagen) with the addition of 5 min bead beating at 1,500 Hz followed by an overnight proteinase-K digestion at 56°C following their tissue lysis protocol. Extracted DNA was screened for parasite presence using previously published mitochondrial COI gene primers as described above. All samples were evaluated by gel electrophoresis and samples displaying a band were sequenced directly using Sanger sequencing and sequence data was assembled, queried, and deposited using the same methods listed above.
To molecularly determine Diptera species identification where parasite presence was detected, partial sequence using Diptera universal primers (38) for the COI gene (LCO1490F: 5′-GGTCAACAAATCATAAAGATATTGG-3′ and HC02198R: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) and were amplified as with the same parameters of the parasite screening, but with modified cPCR conditions: 95°C × 3 min, 35 cycles of (95°C × 15 s, 55°C × 30 s, 72°C × 1.5 min), followed by 72°C × 1 min. Results were visualized on a 2.0% TAE gel and subsequently sequenced, assembled, and examined using the same methods as listed above. The target amplicon size was 710 bp. Sequences were queried against the NCBI nt database using BLASTN. To further confirm and identify insect species, a database of cytochrome oxidase subunit I sequences downloaded from NCBI was generated with the RESCRIPt QIIME 2 plugin version 2021.11.0 + 3.g8aa880e (39) using the taxonomy ids 7147 (Diptera), 6231 (Nematoda), and 2 (Bacteria). Sequences were removed from the database if they contained 5 or more degenerate bases, 10 or more homopolymers, were <500 or >1,600 nt in length, or did not have a top BLASTX hit to a COI sequence in the Uniprot Reviewed (Swiss-Prot) database (40) downloaded 18 May 2022. Taxonomy was assigned to sequences using this database and the q2-feature-classifier with the classify-sklearn naïve Bayes approach (41) within QIIME 2 version qiime2-2022.2 (42). Diptera COI DNA sequences from positive O. lupi insects were deposited to the GenBank sequence database under BioProject PRJNA890670. Sequencing data was deposited in GenBank under the BioProject PRJNA890670.
Of the 532 animals screened in this study, the initial partial COI cPCR results identified 52 skin samples as positive for the presence filarioid nematode DNA. As the COI cPCR primers used in this study are not species specific for O. lupi, these 52 samples of interest were directly sequenced using Sanger sequencing technology to confirm species identity. BLAST results showed 50 of the 52 COI sequences had the closest sequence similarity with 100% identity over a minimum of 411 bases with Onchocerca lupi (accession number YP_010142643.1). All 50 O. lupi sequences were identical. The overall occurrence of O. lupi in companion animals on the Navajo Reservation was 9.4%. Two of the 52 samples had BLAST results with the closest sequence similarity (99.42%–100%) to the canine heartworm, Dirofilaria immitis (accession number MT027229.1). Each O. lupi positive animal was considered as a local hotspot for insect trapping.
Insect trapping resulted in more than 5,000 insects collected from May 2021 through August 2021. Of these, 1,922 insects (38.44%, 70 pools ranging from 2 to 50 whole insects along with 15 pools and 205 individual insects with heads and bodies processed separately) were extracted and screened for parasite presence. The remaining insects were identified morphologically either as non-biting insects (e.g., fruit flies, etc.) or redundant pools of biting midges from trap sites where O. lupi positive insects were already identified. A total of 38 insects (29 bodies and 9 heads) and 19 pools were positive for O. lupi from 12 of the 15 trap sites (Table 1). These insects were then molecularly identified using Diptera COI gene primers. Unknown Ceratopogonidae sp. (biting midges) were the most abundant insect trapped and had the highest rates of presence of O. lupi DNA; 15 pools across four trap sites were O. lupi positive (Table 2). Initially, the size and profusion of biting midge pools imposed pool processing as whole insects; however, once O. lupi positive biting midge pools were identified, remaining pools were processed with heads and bodies separated. O. lupi DNA was identified in both the head and body in biting midge pools. Three pools of biting midges Culicoides sonorensis were identified as O. lupi-positive at 3 separate trap sites as well as 1 pool of Culicoides variipennis at a single trap site. O. lupi DNA was also found in the heads of Culicoides sonorensis but was detected at lower concentrations than in the pools of the unknown Ceratopogonidae. A total of 19 stable flies (11 bodies and 8 heads), Stomoxys calcitrans, were positive for O. lupi DNA at 3 different trap sites. Single insect positives from individual sites included eye gnats of the genus Hippelates sp., Tachinidae sp., Anthomyiidae sp., Coenosia attenuata, Lucilia sericata, Oscinelinae sp., Delia platura, Ravinia errabunda, and seven single fly, single site undetermined species (Table 2). Two Ceratopogonidae sp. biting midges from a single site were positive for an undescribed species of Onchocerca. The highest partial COI identity for this parasite was to Onchocerca lienalis (94.4%, accession number KX853325; Supplementary Figure 1). Additionally, DNA of the horse stomach nematode Habronema muscae was identified in a single house fly, Musca domestica, at a single site.
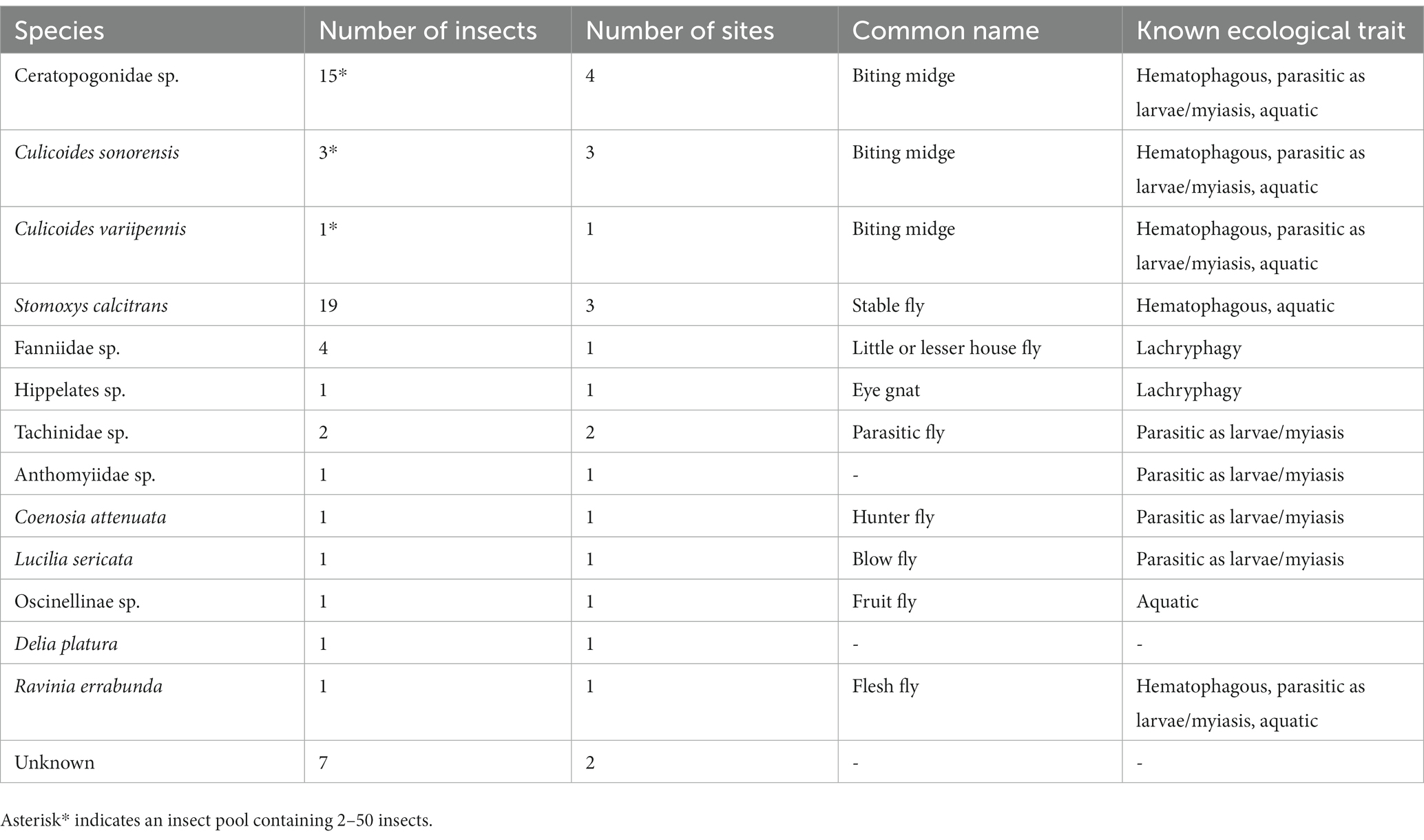
Table 2. Insects positive for the presence of Onchocerca lupi along with common names and known ecological traits.
This study conducted molecular screening of companion animals to identify the occurrence of the filarial nematode O. lupi and field surveillance of hematophagous insects to demonstrate possible intermediate hosts of O. lupi. This study identified 9.4% occurrence of O. lupi in asymptomatic companion animals on the Navajo Reservation in Arizona, New Mexico, and Utah in the southwestern United States from 2019 to 2022. This is the first large-scale study on the prevalence of O. lupi in dogs and cats in the US, as most published reports focus on clinical cases, diagnostics, and new geographic records (3, 4, 43–45). Only a few epidemiological studies have been conducted for determining the prevalence of O. lupi in canine populations, none of these in North America. Previous studies in Greece, Portugal, and Spain reported positivity rates in dogs ranging from 4.8–8% (43, 44). Interestingly, studies investigating prevalence rates of other Onchocerca species demonstrated a correlation between statistically higher prevalence rates and host age (46). Many of our samples were collected from spay and neuter clinics focusing on young animals; because of the heavy sampling of these younger animals, it is possible we are under-estimating the prevalence rate of O. lupi, particularly because we lack detailed knowledge of various biological parameters of its life cycle, including pre-patent period and patency. Furthermore, it is known that various Onchocerca species have an uneven distribution of microfilariae in the dermis of their hosts (47), which may have generated some false negative results in this study as our O. lupi detection relied solely upon microfilariae presence within skin snips from a single anatomic location of companion animal hosts. It is important to consider that the animals that were positive for O. lupi appeared to be healthy and did not display symptoms of disease at the time of sample collection. This could be due to their young age combined with Onchocerca’s long development period into gravid adults. It is also feasible to consider canines with varying levels of tolerance of O. lupi infections. The absence of ocular and skin symptoms in conjunction with cPCR positive tests brings into question the pathogenic role of O. lupi within its definitive hosts.
All O. lupi positive insects belonged to the Order Diptera; we identified 13 different species (Table 1). Among the 38 positive flies, seven could not be identified to species-level morphologically or molecularly. It is not surprising that this study identified unknown Diptera species on the Navajo Reservation, as to date, no comprehensive seasonal entomological survey has been conducted; however, there have been over 150,000 species described within the Order Diptera (48). It is estimated that 5 million morphologically distinct dipteran species exist but have not yet been described (49). Furthermore, the Sanger sequencing data for four of these unknown flies shows mixed calls across the COI gene indicating mixed samples or contamination. Given that these unknown flies were identified as singletons at single locations suggests it’s unlikely any of these unknown non-biting flies are true vectors of O. lupi. Additionally, it is unlikely that the eight O. lupi positive singleton non-biting flies (Tachinidae sp., Anthomyiidae sp., Coenosia attenuata, Lucilia sericata, Oscinelinae sp., Ravinia errabunda, Delia platura, Hippelates sp.) found at single sites are the vector species responsible for disease transmission on the Navajo Reservation. We hypothesize these flies (Tachinidae sp., Coenosia attenuate, Oscinelinae sp.) are either predators of smaller insects and acquired O. lupi DNA by feeding on insects that took blood meals from an infected host or may have only fed on serosanguinolent secretions of an infected definitive host (Lucilia sericata, Ravinia errabunda, Anthomyiidae sp., Delia platura, Hippelates sp.). The four Fanniidae sp. found at a single site have potential as O. lupi vectors. Three species of Fanniidae, Fannia canicularis, Fannia benjamini, and Fannia thelaziae, have been implicated as the vector of the eye worm Thelazia californiensis by ingesting L1 larvae while feeding on lacrimal secretions. These larvae mature into L3 infectious larvae within the fly and are transmitted to a new host while feeding on lacrimal secretions (50). However, we hypothesize it is unlikely these serve as competent O. lupi vectors as they were present only at a single trap site. Seven cryptic fly species found at two different trap sites were unable to be morphologically or molecularly identified. Morphological identification determined these flies were seven different species and unlikely true vectors of O. lupi given their infrequency and non-biting appearance.
In this study, we identified the stable fly, S. calcitrans (Diptera: Muscidae) containing O. lupi DNA, but its role as a suitable vector remains unclear. Stable flies are known to feed on dogs, particularly dog’s ears causing bleeding, lesions/crusts. This would likely attract other non-blood feeding dipteran for an opportunistic meal of organic matter/nutrients. While stable flies are a disease vector (Habronema microstoma; horse stomach worm), they are largely considered deficient biological vectors of disease in comparison to other blood feeding flies (51). To date, pathogen/parasite development or reproduction has not been demonstrated within the stable fly other than the horse stomach worm. However, despite being inefficient biological vectors, as a mechanical vector, stable flies are still culpable for pathogen/parasite transmission. In this study, we identified 19 stable flies, including 11 bodies and 8 heads, as positive for O. lupi DNA. Positive stable flies originated from 3 different locations on the Navajo Reservation. We hypothesize the O. lupi positive stable flies likely fed on an infected mammal and while they are not biological vectors, it is still plausible they are mechanical vectors. Even so, we expect both human populations and companion animals are at low risk of O. lupi transmission from stable flies.
Multiple Ceratopogonidae sp. (biting midges) were identified as the most abundant O. lupi positive insect in our study (O. lupi was present in the heads and bodies) and likely an intermediate host in Northern Arizona and New Mexico. The Ceratopogonidae (Diptera) family, which includes the genus Culicoides, commonly called biting midges or “no-see-ums,” are considered hematophagous pests and vector several infectious agents including viruses, protozoans, and filarioid nematodes worldwide (52–54). Biting midges are most notable as vectors of several arbovirus livestock diseases such as bluetongue virus (BTV), African horse sickness virus (AHSV), epizootic hemorrhagic disease virus (EHDV), and Schmallenberg virus (SBV), all of which are of considerable veterinary and economic relevance (55). Importantly, biting midges have been implicated as vectors for several filarial nematodes including Mansonella ozzardi (Nematoda: Onchocercidae) with humans as the primary host (56), as well as seven reported species of Onchocerca infecting a range of hosts including mammals from the biological families Bovidae, Cervidae, and Equidae (20, 54). The Ceratopogonidae biting midges, which we identify here as potential vectors for O. lupi, are widespread throughout the United States (57). When considering the potential of vector-borne disease dissemination into non-endemic regions, it is imperative to examine the potential range expansion of the vector species. In terms of range expansion, there are two aspects of vector dispersal: (1) wing-propelled flight of females to find nearby blood-meals (upwards of 5 km) (58), and (2) wind-borne dispersal from wind streams with potential to spread these vector-borne diseases into non-endemic regions upwards of 200 km away (59). Research has demonstrated recent range expansion of Culicoides spp. in the southeastern United States (60), and when coupled with wind expansion potential, highlights the immediate risk for vector establishment, and dissemination of O. lupi into non-endemic regions.
We hypothesize that O. lupi has adapted to two families of hematophagous Diptera as competent vectors depending on the variable geographic area—Simuliidae and Ceratopogonidae. The Navajo Reservation is bordered by the San Juan River in the north, the Little Colorado River (flow in much of this river is ephemeral) to the south, and the Colorado River to the west (61). Despite these major waterways surrounding the Navajo Reservation, the Reservation itself is vast, with large regions lacking moving water habitats necessary for blackfly development. Transmission of the filarial nematode M. ozzardi, first described in 1897, further supports our hypothesis of transmission of O. lupi by two families of dipteran vectors (62). Mansonella ozzardi is a human parasite transmitted by biting midges (mostly members of the genus Culicoides) and blackflies (genus Simulium) depending on the endemic location. For example, blackflies of the Simulium genus were identified as M. ozzardi vectors in Central and South America while Culicoides spp. were implicated in transmission in Mexico and several Caribbean islands (62). Additionally, an important factor that further supports our hypothesis is that biting midges are superficial blood feeders, as are black flies, which allows for involvement in transmitting skin-dwelling filarioid nematodes as opposed to solenophagy Diptera that feed directly from blood capillaries of vertebrate hosts (e.g., mosquitoes).
Additionally, a pool consisting of two biting midges from a single site (site 9) showed the presence of parasite DNA in the initial COI cPCR. The COI DNA sequence belongs within the Onchocerca genus, however, the species has yet to be described (Supplementary Figure 1). Blast results showed the closest match to this unknown sample was Onchocerca lienalis with 94.56% identity. Little is known regarding unknown Onchocerca species in wildlife and arthropods (23), however, a more comprehensive examination of unknown Onchocerca species within North America is needed to catalog the diversity of both Onchocerca and the arthropods vectoring them.
Current O. lupi surveillance tools targeting the COI mitochondrial gene are limited to detection and cannot provide insight into population structure, at least within the United States. Two recent studies published the first mitochondrial genome as well the first draft genome for O. lupi (18, 63). These studies should facilitate the development of high-resolution surveillance tools for O. lupi to understand the genetic diversity and relationships among isolates globally. Further research regarding vector competency of Ceratopogonidae and Culicoides species is necessary to confirm their role as intermediate hosts for O. lupi. Subsequent knowledge of the vector life cycle is paramount to break the cycle of O. lupi transmission on the Navajo Reservation. Mitigation efforts to control the vector species will likely decrease and prevent O. lupi disease in companion animals thereby reducing the risk of human infection.
Our increased surveillance of definitive hosts represents the first large scale study of O. lupi in companion animals from an endemic area of the US. We identified a prevalence rate of 9.4% and, from these data points, conducted targeted insect trapping to define putative vectors in the southwestern United States. We found strong evidence that biting midge species may be putative vectors in Northern Arizona and New Mexico. We hypothesize that O. lupi has adapted to two families of hematophagous Diptera as competent intermediate hosts depending on the geographic area, Simuliidae and Ceratopogonidae. Further research regarding vector competency of Ceratopogonidae and Culicoides species is necessary to confirm their role as intermediate hosts for O. lupi.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by IACUC of Northern Arizona University (19-016). Written informed consent was obtained from the owners for the participation of their animals in this study.
CRo, OH, KU-B, GB, GV, RN, CH, and JS contributed to conception and study design. CRo and CRi created and organized metadata database. CRo and CW performed sequence analyses. MF, JU, TK, DL, OH, KU-B, and GV contributed to sample collection. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the Pacific Southwest Center of Excellence in Vector-Borne Diseases under grant no. (1004601).
Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1167070/full#supplementary-material
1. Rodonaja, TE. A new species of nematode, Onchocerca lupi n Sp., from Canis lupus cubanensis. Soobshchenyia Akad Nuak Gruz SSR. (1967);715–719.
2. Wallitsch, K, Jaffey, J, Ferguson, S, Verocai, G, Sobotyk, C, van Eerde, E, et al. Extensive aberrant migration of Onchocerca lupi in a dog. Top Companion Anim Med. (2022) 49:100666. doi: 10.1016/j.tcam.2022.100666
3. Verocai, GG, Sobotyk, C, Lamison, A, Borst, MM, and Edwards, E. Autochthonous, zoonotic Onchocerca lupi in a South Texas dog, United States. Parasit Vectors. (2021) 14:203. doi: 10.1186/s13071-021-04707-4
4. McLean, NJ, Newkirk, K, and Adema, CM. Canine ocular onchocerciasis: a retrospective review of the diagnosis, treatment, and outcome of 16 cases in New Mexico (2011-2015). Vet Ophthalmol. (2017) 20:349–56. doi: 10.1111/vop.12433
5. Verocai, GG, Conboy, G, Lejeune, M, Marron, F, Hanna, P, MacDonald, E, et al. Onchocerca lupi nematodes in dogs exported from the United States into Canada. Emerg Infect Dis. (2016) 22:1477–9. doi: 10.3201/eid2208.151918
6. Faísca, P, Morales-Hojas, R, Alves, M, Gomes, J, Botelho, M, Melo, M, et al. A case of canine ocular onchocercosis in Portugal. Vet Ophthalmol. (2010) 13:117–21. doi: 10.1111/j.1463-5224.2010.00763.x
7. Széll, Z, Erdélyi, I, Sréter, T, Albert, M, and Varga, I. Canine ocular onchocercosis in Hungary. Vet Parasitol. (2001) 97:243–9. doi: 10.1016/S0304-4017(01)00397-1
8. Komnenou, A, Egyed, Z, Sréter, T, and Eberhard, ML. Canine onchocercosis in Greece: report of further 20 cases and molecular characterization of the parasite and its Wolbachia endosymbiont. Vet Parasitol. (2003) 118:151–5. doi: 10.1016/j.vetpar.2003.09.007
9. Otranto, D, Dantas-Torres, F, Gürlü, VP, Sakru, N, Yakar, K, Bain, O, et al. First evidence of human zoonotic infection by Onchocerca lupi (Spirurida, Onchocercidae). Am J Trop Med Hyg. (2011) 84:55–8. doi: 10.4269/ajtmh.2011.10-0465
10. Eberhard, ML, Ostovar, GA, Chundu, K, Hobohm, D, Feiz-Erfan, I, Mathison, BA, et al. Zoonotic Onchocerca lupi infection in a 22-month-old child in Arizona: first report in the United States and a review of the literature. Am J Trop Med Hyg. (2013) 88:601–5. doi: 10.4269/ajtmh.12-0733
11. Radwan, AM, Ahmed, NE, Elakabawy, LM, Ramadan, MY, and Elmadawy, RS. Prevalence and pathogenesis of some filarial nematodes infecting donkeys in Egypt. Vet World. (2016) 9:888–92. doi: 10.14202/vetworld.2016.888-892
12. Mowlavi, G, Farzbod, F, Kheirkhah, A, Mobedi, I, Bowman, DD, and Naddaf, SR. Human ocular onchocerciasis caused by Onchocerca lupi (Spirurida, Onchocercidae) in Iran. J Helminthol. (2014) 88:250–5. doi: 10.1017/S0022149X13000060
13. Otranto, D, Dantas-Torres, F, Cebeci, Z, Yeniad, B, Buyukbabani, N, Boral, OB, et al. Human ocular filariasis: further evidence on the zoonotic role of Onchocerca lupi. Parasit Vectors. (2012) 5:1–7. doi: 10.1186/1756-3305-5-1
14. Labelle, AL, Daniels, JB, Dix, M, and Labelle, P. Onchocerca lupi causing ocular disease in two cats. Vet Ophthalmol. (2011) 14:105–10. doi: 10.1111/j.1463-5224.2011.00911.x
15. Roe, C, Yaglom, H, Howard, A, Urbanz, J, Verocai, G, Andrews, L, et al. Coyotes as a reservoir for the emerging zoonotic parasite, Onchocerca lupi, in the southwestern United States. Emerg Infect Dis Rev. (2019) 26:2989–93. doi: 10.3201/eid2612.190136
16. Maia, C, Annoscia, G, Latrofa, MS, Pereira, A, Giannelli, A, Pedroso, L, et al. Onchocerca lupi Nematode in Cat. Portugal Emerg Infect Dis. (2015) 21:2252–4. doi: 10.3201/eid2112.150061
17. Otranto, D, Sakru, N, Testini, G, Gürlü, VP, Yakar, K, Lia, RP, et al. Case report: first evidence of human zoonotic infection by Onchocerca lupi (Spirurida, Onchocercidae). Am J Trop Med Hyg. (2011) 84:55–8. doi: 10.4269/ajtmh.2011.10-0465
18. Roe, CC, Urbanz, J, Andrews, L, Verocai, GG, Engelthaler, DM, Hepp, CM, et al. Mitochondrial DNA part B complete mitochondrial genome of Onchocerca lupi (Nematoda, Onchocercidae). (2021). Available at: https://www.tandfonline.com/action/journalInformation?journalCode=tmdn20 (Accessed 21 April 2022).
19. Wu, DB, Ko, B, Lopez Hernandez, G, Botros, J, Spader, H, Sapp, S, et al. Case report Neuroinvasive Onchocerca lupi infection in a ten-year-old girl. Infect Dis. (2022) 2022:9773058. doi: 10.1155/2022/9773058
20. Lefoulon, E, Giannelli, A, Makepeace, BL, Mutafchiev, Y, Townson, S, Uni, S, et al. Whence river blindness? The domestication of mammals and host-parasite co-evolution in the nematode genus Onchocerca. Int J Parasitol. (2017) 47:457–70. doi: 10.1016/j.ijpara.2016.12.009
21. Rojas, A, Morales-Calvo, F, Salant, H, Otranto, D, and Baneth, G. Zoonotic ocular Onchocercosis by Onchocerca lupi. Yale J Biol Med. (2021) 94:331–41.
22. Hassan, HK, Bolcen, S, Kubofcik, J, Nutman, TB, Eberhard, ML, Middleton, K, et al. Isolation of onchocerca lupi in dogs and black flies, California, USA. Emerg Infect Dis. (2015) 21:789–96. doi: 10.3201/eid2105.142011
23. Kulpa, M, Nelson, KJ, Morales, AM, Ryan, BM, Koschik, ML, Scott, JJ, et al. Presence of a cryptic Onchocerca species in black flies of northern California, USA. Parasit Vectors. (2021) 14:478. doi: 10.1186/s13071-021-04990-1
24. Verocai, GG, Nelson, KJ, Callahan, RT, Wekesa, JW, Hassan, HK, and Hoberg, EP. A cryptic species of Onchocerca (Nematoda: Onchocercidae) in blackflies (Simulium spp.) from southern California, USA. Parasit Vectors. (2018) 11:11. doi: 10.1186/s13071-018-3133-9
26. Wilson Id, AL, Courtenay, O, Kelly-Hope, LA, Scott Id, TW, Takken, W, Torr, SJ, et al. The importance of vector control for the control and elimination of vector-borne diseases. PLoS Negl Trop Dis. (2020) 14:e0007831. doi: 10.1371/journal.pntd.0007831.t002
27. Alhassan, A, Makepeace, BL, Lacourse, EJ, Osei-Atweneboana, MY, and CKS, C. A simple isothermal DNA amplification method to screen Black flies for Onchocerca volvulus infection. PLoS One. (2014) 9:e0108927. doi: 10.1371/journal.pone.0108927
28. Pilotte, N, Unnasch, TR, and Williams, SA. The current status of molecular Xenomonitoring for lymphatic Filariasis and onchocerciasis. Trends Parasitol. (2017) 33:788–98. doi: 10.1016/j.pt.2017.06.008
29. Kluckhohn, C, and Leighton, DC. The Navaho. Revised ed. Cambridge, MA: Harvard University Press (1992). 374 p.
30. The Navajo Nation’s Shifting Sands of climate change | climate central. (2023). Available at: https://www.climatecentral.org/news/navajo-nation-climate-change-17326 (Accessed 1 February 2023).
31. Navajo Nation. Available at: http://navajopeople.org/blog/navajo-nation/ (2023). (Accessed 1 February 2023).
32. Geography of Diné Bikéyah. Available at: http://xroads.virginia.edu/~MA97/dinetah/geo.html (2023). (Accessed 2 February 2023).
33. Navajo Nation Wind. Available at: https://navajoprofile.wind.enavajo.org/ (2023). (Accessed 1 February 2023).
34. IHS. Navajo Nation. Navajo area. Available at: https://www.ihs.gov/navajo/navajonation/ (2023). (Accessed 2 February 2023).
35. Otranto, D, Dantas-Torres, F, Giannelli, A, Abramo, F, Ignjatović Ćupina, A, Petrić, D, et al. Cutaneous distribution and circadian rhythm of Onchocerca lupi microfilariae in dogs. PLoS Negl Trop Dis. (2013) 7:e2585. doi: 10.1371/journal.pntd.002585
36. Casiraghi, M, Anderson, TJC, Bandi, C, Bazzocchi, C, and Genchi, C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. (2001) 122:93–103. doi: 10.1017/S0031182000007149
37. Morgulis, A, Coulouris, G, Raytselis, Y, Madden, TL, Agarwala, R, and Schäffer, AA. Database indexing for production MegaBLAST searches. Bioinformatics. (2008) 24:1757–64. doi: 10.1093/bioinformatics/btn322
38. Black, M, Hoeh, W, Lutz, R, and Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. (1994) 3:294–9.
39. Robeson, MS, O’Rourke, DR, Kaehler, BD, Ziemski, M, Dillon, MR, Foster, JT, et al. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput Biol. (2021) 17:e1009581. doi: 10.1371/journal.pcbi.1009581
40. UniProt consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. (2021) 49:D480–9. doi: 10.1093/nar/gkaa1100
41. Bokulich, NA, Kaehler, BD, Rideout, JR, Dillon, M, Bolyen, E, Knight, R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. (2018) 6:1–17. doi: 10.1186/s40168-018-0470-z
42. Bolyen, E, Ram Rideout, J, Dillon, MR, Bokulich, NA, Abnet, CC, Al-Ghalith, GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:850–2. doi: 10.1038/s41587-019-0190-3
43. Miró, G, Montoya, A, Checa, R, Gálvez, R, Mínguez, JJ, Marino, V, et al. First detection of Onchocerca lupi infection in dogs in southern Spain. Parasit Vectors. (2016) 9:290. doi: 10.1186/s13071-016-1587-1
44. Otranto, D, Dantas-Torres, F, Giannelli, A, Latrofa, MS, Papadopoulos, E, Cardoso, L, et al. Zoonotic Onchocerca lupi infection in dogs, Greece and Portugal, 2011-2012. Emerg Infect Dis. (2013) 19:2000–3. doi: 10.3201/eid1912.130264
45. Unterköfler, MS, Huck, A, Silbermayr, K, and Fuehrer, HP. Autochthonous Onchocerca lupi infection of a domestic dog in Austria. Parasit Vectors. (2023) 16:46. doi: 10.1186/s13071-023-05681-9
46. Traversa, D, Mackenzie, CD, Cork, SC, Papini, RA, Lubas, G, and Sgorbini, M. Incidental detection of Onchocerca microfilariae in donkeys (Equus asinus) in Italy: report of four cases. Front Vet Sci. (2020) 7:569916. doi: 10.3389/fvets.2020.569916
47. Mellor, PS. Studies on Onchocerca cervicalis Railliet and Henry 1910: I. Onchocerca cervicalis in British horses. J Helminthol. (1973) 47:97–110. doi: 10.1017/S0022149X00023774
48. Yeates, DK, and Wiegmann, BM. The evolutionary biology of flies In: DK Yeates and BM Wiegmann, editors.United States: Columbia University Press. System Entomology, vol. 31 (2006). 566–7.
49. Brown, BV, Borkent, A, Adler, PH, de Amorim, DS, Barber, K, Bickel, D, et al. Comprehensive inventory of true flies (Diptera) at a tropical site. Commun Biol. (2018) 1:1–8. doi: 10.1038/s42003-018-0022-x
50. Sobotyk, C, Foster, T, Callahan, RT, McLean, NJ, and Verocai, GG. Zoonotic Thelazia californiensis in dogs from New Mexico, USA, and a review of north American cases in animals and humans. Vet Parasitol Reg Stud Rep. (2021) 24:100553. doi: 10.1016/j.vprsr.2021.100553
51. Taylor, DB. Stable Fly (Stomoxys calcitrans, Muscidae). Encycl Infect Immun. (2022):903–13. doi: 10.1016/B978-0-12-818731-9.00008-2
52. Mellor, PS, Boorman, J, and Baylis, M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. (2003) 45:307–40. doi: 10.1146/annurev.ento451307
53. Carpenter, S, Groschup, MH, Garros, C, Felippe-Bauer, ML, and Purse, BV. Culicoides biting midges, arboviruses and public health in Europe. Antivir Res. (2013) 100:102–13. doi: 10.1016/j.antiviral.2013.07.020
54. Marquardt, WC, Kondratieff, BC, and Marquardt, WH. Biology of disease vectors. Burlington: Elsevier Science & Technology (2004).
55. Brand, SPC, and Keeling, MJ. The impact of temperature changes on vector-borne disease transmission: Culicoides midges and bluetongue virus. J R Soc Interface. (2017) 14:20160481. doi: 10.1098/rsif.2016.0481
56. Shelley, AJ, and Coscarón, S. Simuliid blackflies (Diptera: Simuliidae) and ceratopogonid midges (Diptera: Ceratopogonidae) as vectors of Mansonella ozzardi (Nematoda: Onchocercidae) in northern Argentina. Mem Inst Oswaldo Cruz. (2001) 96:451–8. doi: 10.1590/S0074-02762001000400003
57. Holbrook, FR, Tabachnick, WJ, Schmidtmann, ET, Mckinnon, CN, Bobian, RJ, and Grogan, WL. Sympatry in the Culicoides variipennis Complex (Diptera: Ceratopogonidae): a taxonomic reassessment. J Med Entomol. (2000) 37:65–76. doi: 10.1603/0022-2585-37.1.65
58. Kasi, Z, Schreiberov, A, Kim Akov, A, and Sov, KC. Blood meal analysis: host-feeding patterns of biting midges (Diptera, Ceratopogonidae, Culicoides Latreille) in Slovakia. Parasite. (2021) 28:58. doi: 10.1051/parasite/2021058
59. Marquardt, WC, and Kondratieff, BC. Biology of disease vectors. WC Marquardt and BC Kondratieff (C Boris), editors. Burlington, MA: Elsevier Academic Press; (2005).
60. Vigil, SL, Ruder, MG, Shaw, D, Wlodkowski, J, Garrett, K, Walter, M, et al. Apparent range expansion of Culicoides (Hoffmania) insignis (Diptera: Ceratopogonidae) in the southeastern United States. J Med Entomol. (2018) 55:1043–6. doi: 10.1093/jme/tjy036
61. Nation, N. History, Navajo Nation Available at: http://www.navajo-nsn.gov/history.htm
62. Lima, NF, Veggiani Aybar, CA, Dantur Juri, MJ, and Ferreira, MU. Mansonella ozzardi: a neglected New World filarial nematode. Pathog Glob Health. (2016) 110:97–107. doi: 10.1080/20477724.2016.1190544
Keywords: Onchocerca lupi, prevalence, nematodes, Ceratopogonidae, biting midges
Citation: Roe CC, Holiday O, Upshaw-Bia K, Benally G, Williamson CHD, Urbanz J, Verocai GG, Ridenour CL, Nottingham R, Ford MA, Lake DP, Kennedy TA, Hepp CM and Sahl JW (2023) Biting midges (Diptera: Ceratopogonidae) as putative vectors of zoonotic Onchocerca lupi (Nematoda: Onchocercidae) in northern Arizona and New Mexico, southwestern United States. Front. Vet. Sci. 10:1167070. doi: 10.3389/fvets.2023.1167070
Received: 15 February 2023; Accepted: 28 April 2023;
Published: 15 May 2023.
Edited by:
Ettore Napoli, University of Messina, ItalyReviewed by:
Alicia Rojas, University of Costa Rica, Costa RicaCopyright © 2023 Roe, Holiday, Upshaw-Bia, Benally, Williamson, Urbanz, Verocai, Ridenour, Nottingham, Ford, Lake, Kennedy, Hepp and Sahl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chandler C. Roe, Y2hhbmRsZXIucm9lQG5hdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.