
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 03 April 2023
Sec. Livestock Genomics
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1160384
 Fei Shui1,2†
Fei Shui1,2† Guiru Qiu1†
Guiru Qiu1† Shenqiang Pan1
Shenqiang Pan1 Xin Wang1,2
Xin Wang1,2 Fumin Jia1,2
Fumin Jia1,2 Tingting Jiang1
Tingting Jiang1 Yongsheng Li3
Yongsheng Li3 Zhaoyu Geng1,2
Zhaoyu Geng1,2 Sihua Jin1,2*
Sihua Jin1,2*Quantitative polymerase chain reaction (qPCR) is an important method to detect gene expression at the molecular level. The selection of appropriate housekeeping genes is the key to accurately calculating the expression level of target genes and conducting gene function studies. In this study, the expression of eight candidate reference genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-actin (β-actin), 18S ribosomal RNA (18S rRNA), hydroxymethylbilane synthase (HMBS), hypoxanthine phosphoribosyltransferase 1 (HPRT1), TATA box binding protein (TBP), ribosomal protein L13 (RPL13), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAZ), in the duodenal epithelial tissue of 42-day-old meat-type ducks were detected using qPCR. Furthermore, their expression stability was analyzed using the geNorm, NormFinder, and BestKeeper programs. The results indicated that HMBS and YWHAZ were the most stably expressed genes. All three programs indicated that the expression of 18S rRNA was the least stable, making it unsuitable for the study of gene expression in meat-type duck tissues. This study provides stable reference genes for gene expression analysis and contributes to further studies on the gene function of meat-type ducks.
Reference genes, often referred to as housekeeping genes, are typically used as a reference to normalize mRNA levels between different samples since the expression levels of genes may vary across tissues or cells in some cases (1, 2). Quantitative polymerase chain reaction (qPCR) is an important tool in molecular biology research. It has the advantages of high specificity, high sensitivity, a high degree of automation, and accurate quantification (3). Nowadays, qPCR has become a common method to study the genetic mechanisms of some growth traits that are beneficial to human beings and livestock. Reference genes have an important influence on the process of qPCR because the standardization of these genes controls the accuracy of qPCR (4). In scientific research, the best internal reference gene should have a relatively stable expression level in different stages of development and different tissues of organisms. However, many experiments have shown that no single gene can reach a constant level of expression under such different conditions (5–7).
A total of eight commonly used reference genes were selected, based on existing studies. Godornes et al. (8) reported that the hypoxanthine phosphoribosyltransferase 1 (HPRT1) gene is a stable reference gene in rabbits. Another study on chickens found that the most suitable reference gene in the liver is ribosomal protein L13 (RPL13); in the jejunum it is glyceraldehyde-3-phosphate dehydrogenase (GAPDH), while hydroxymethylbilane synthase (HMBS) is compatible in all tissues (9). In pigs, HPRT1 and HMBS are reference genes for skeletal muscles and affect their postnatal growth (10). In goats, beta-actin (β-actin) was the most stable in the stomach, small intestine, and ovary; 18S ribosomal RNA (18S rRNA) in the heart and spleen; HMBS in the uterus and lungs; TATA box binding protein (TBP) in the liver; HPRT1 in the kidney; and GAPDH in the muscles (11). While all the above genes can be employed as stable reference genes for rabbits, pigs, and other model animals, there is a lack of stable reference genes for conducting gene expression studies in meat-type ducks. Filling this gap in research has been made more pertinent by the growth of the meat-type duck industry in China, becoming an important livestock and poultry industry in the country's rural economic development.
This study, therefore, considered meat-type duck as the research subject and, based on qPCR technology and comprehensive use of the geNorm, NormFinder, and BestKeeper programs, assessed the stability of multiple candidate reference genes, aiming to identify a list of effective reference genes for quantitative gene expression analysis in meat-type ducks, which will be beneficial to further gene function studies.
All the study procedures that included animals were strictly carried out under the regulations and guidelines established by the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004). All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of Anhui Agricultural University (SYXK 2016-007). The animals in the study had ad libitum access to water and feed (Table 1) and were humanely sacrificed.
A total of sixteen 42-day-old male meat-type ducks were provided by Huangshan Qiangying Duck Breeding Co. Ltd. (Anhui, China). At 21 days, all ducks were reared in the individual cages (55 × 50 × 40 cm) for feeding until 42 days of age. All ducks were exposed to continuous illumination (24 L:0 D) for the first 72 h after hatching, followed by a 20 L:4 D lighting regime until the end of the experiment. All ducks were kept in the same house and fed the same basal diet at room temperature. After slaughtering, duodenal tissues of the meat-type ducks were collected and immediately placed in a 2.0 mL centrifuge tube with RNALater at 4°C overnight and stored at −80°C for further use.
The total RNA from duodenal epithelial tissues was extracted using the total RNA Kit (Omega Bio-Tek, Doraville, GA, USA) following the manufacturer's instructions. The RNA concentration and purity of samples were determined by the NanoDrop spectrophotometer (Thermo Fisher Scientific, New York, NY, USA) and 1.0% agarose gel electrophoresis. SuperMix (Yeasen, Shanghai, China) was used to reverse the transcription of the isolated total RNA into cDNA; then, qPCR was performed. All experimental procedures were performed as per the manufacturer's protocol and conducted on a sterilized bench in a clean room.
According to the duck gene sequence published in GenBank, Table 2 shows that the primers of eight candidate reference genes were designed using Primer Premier 5.0 software and UCSC In-Silico PCR were used to test the specificity and sensitivity (http://genome.ucsc.edu/). The cDNA obtained in the previous step served as a template; the reference genes GAPDH, β-actin, 18S rRNA, HMBS, HPRT1, TBP, RPL13, and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAZ) were used for conventional PCR amplification; and the primer specificity and amplification efficiency were detected by gel electrophoresis and qPCR. PCR was implemented in a 20.0 μL reaction containing 8.2 μL ddH2O, 10.0 μL SYBR Green Master Mix (Yeasen, Shanghai, China), 1.0 μL template DNA, and 0.4 μL of each primer. The reaction conditions were 95°C for 5 min; 95°C for 10 s, 60°C for 20 s, 72°C for 20 s, 40 cycles; 95°C for 15 s, 60°C for 1 min, 95°C for 15 s. The sample copy number of logarithmic scales was taken as the abscissa and the Ct value as the ordinate, and the testing purpose gene amplification curve, melting curve, and standard curve were obtained. The qualified primers could then be used for subsequent experiments.
Based on the mRNA expression of reference genes, three types of stability evaluation software, geNorm, NormFinder, and BestKeeper, were used for statistical analysis to select the stable reference genes. The test data obtained from qPCR of the reference genes were exported to Microsoft Excel 2019 (Microsoft, Redmond, WA, USA). All three software packages were used as per the manufacturer's instructions.
geNorm is a program designed by Vandesompele et al. (4) for selecting the most stable reference genes and determining the optimal reference genes in qPCR. geNorm achieves the rank of internal reference genes by calculating the average expression stability. Each dataset was transformed to relative quantities using the 2−ΔCt method (ΔCt = the corresponding Ct value – minimum Ct value). NormFinder (https://moma.dk/normfinder-software) is a program designed by Andersen et al. (12) to screen stable reference genes. The computational mechanism of this program is the same as that of the geNorm program. NormFinder also uses the 2−ΔCt method, derives relative quantities from raw Ct values, and then calculates and ranks each dataset. BestKeeper (https://www.gene-quantification.de/bestkeeper.html) is a program written by Pfaffl et al. (13) for the expression analysis of housekeeping and target genes. BestKeeper can directly calculate the stability using the Ct value of gene expression. The Ct value of each sample gene was entered into an Excel spreadsheet, and then BestKeeper internal stability analysis software was imported for calculation. Through the input of Ct, the program calculated the correlation coefficient (r), standard deviation (SD), and coefficient of variation (CV) for each gene to generate the pairing and then compared the size of each value to determine the reference gene with good stability. The larger the R, the smaller the SD and CV, and the better the stability of the reference gene. SD > 1 indicated that the expression of a housekeeping gene was not stable. Finally, by averaging the rank of the eight reference genes obtained using the three software, the stability of the genes was comprehensively evaluated, and the most suitable housekeeping genes were derived.
Total RNA samples were analyzed using 1.0% agarose gel electrophoresis (Supplementary Figure 1). Two bands (representing 28 S and 18 S) were detected; the 28 S band was brighter than the 18 S band. The OD260/OD280 ratios of the samples' RNA were all 1.9–2.1, which showed that the isolated total RNA was of sufficient purity and without degradation.
The specificity was assessed by visualization of specific amplified product sizes by electrophoresis in agarose gel (1.5%). As shown in Figure 1, the results of the amplification revealed that the fragment lengths of the products of GAPDH, β-actin, 18S rRNA, HMBS, HPRT1, TBP, RPL13, and YWHAZ were consistent with their theoretical lengths. This indicated that the cDNA generated by reverse transcription had good integrity and amplification product specificity and could be used in the follow-up experiment.
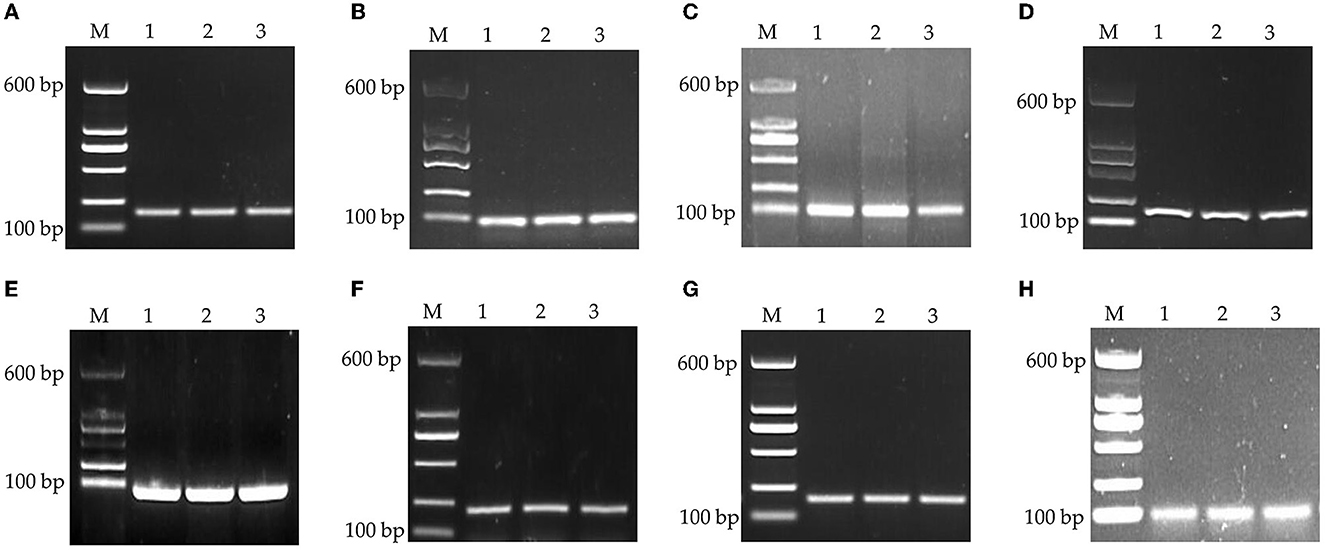
Figure 1. PCR products of housekeeping genes by agarose gel electrophoresis. (A) GAPDH, (B) β-actin, (C) 18S rRNA, (D) HMBS, (E) HPRT1, (F) TBP, (G) RPL13, and (H) YWHAZ. M: DNA marker.
geNorm analyzed the expression stability of the eight candidate housekeeping genes in the duodenum of meat-type ducks. As per the principle that the smaller the average stability value (M), the more stable the gene expression, the ranking of the eight housekeeping genes is represented in Table 3. Using the transformed data, geNorm generated a graph considering the stability value in which low stability values characterized genes with the most stable expression. The stability rank of each housekeeping gene was ranked as follows: HMBS = YWHAZ > β-actin > RPL13 > GAPDH > TBP > HPRT1 > 18S rRNA. The results showed that the most stable internal genes were HMBS and YWHAZ, while the least stable was 18S rRNA.
NormFinder calculated the stable values of the eight reference genes in the duodenal epithelial cells of meat-type ducks. The smaller the stability value (S) of a gene, the more stable it is. The stability of each reference gene was ranked as follows (Table 4): HMBS > YWHAZ > β-actin > RPL13 > GAPDH > TBP > HPRT1 > 18S rRNA. The results revealed that the most stable internal gene was HMBS, while the least stable was 18S rRNA.
The determination of reference gene stability by BestKeeper program analysis was done by directly inputting the Ct value of each gene into an Excel table and importing it into the BestKeeper program. The stability rank of each reference gene by BestKeeper was as follows (Table 5): HMBS = YWHAZ > β-actin > RPL13 > HPRT1 > GAPDH > TBP > 18S rRNA. The results indicated that HMBS and YWHAZ were the most stably expressed genes; 18S rRNA was the least stably expressed.
The comprehensive evaluation results of the eight housekeeping genes selected by geNorm, NormFinder, and BestKeeper are shown in Table 6 and Figure 2. From geNorm, HMBS and YWHAZ were determined to be the most stable genes in duodenal epithelial cells, followed by β-actin, RPL13, GAPDH, TBP, HPRT1, and 18S rRNA. From NormFinder, HMBS was identified to be the most stably expressed reference gene, followed by YWHAZ, β-actin, RPL13, GAPDH, TBP, HPRT1, and 18S rRNA. BestKeeper revealed that HMBS and YWHAZ were the most stable genes, followed by β-actin, RPL13, HPRT1, GAPDH, TBP, and 18S rRNA. The comprehensive analysis results showed that HMBS and YWHAZ are relatively the most stable reference genes in the duodenal epithelial cells of meat-type ducks, followed by β-actin, while 18S rRNA, HPRT1, and TBP are the least stable. Therefore, HMBS and YWHAZ can be selected as the best reference genes in the duodenal epithelial cells of meat-type ducks.
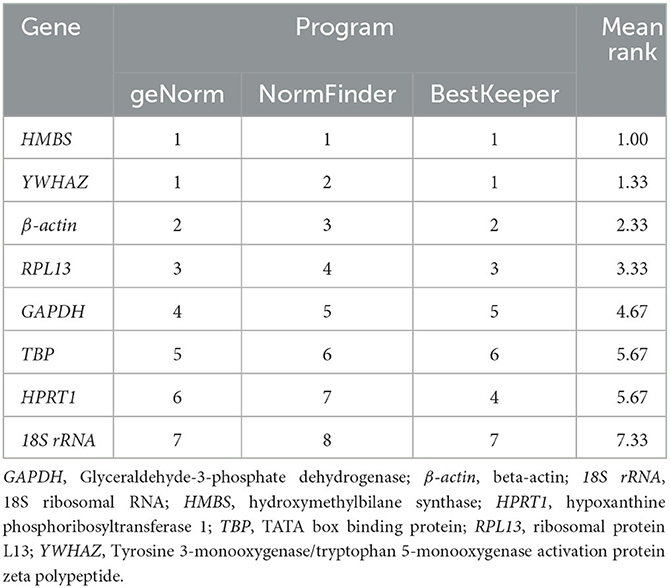
Table 6. Integrated table of reference gene expression stability values by the three different statistical methods.
In animal breeding research, gene expression is important for understanding biological processes. The qPCR technique has become widely used to detect gene expression functions (14). Many factors in qPCR, including the selection of housekeeping genes, may impact the results. To make the analysis of the expression of a particular gene more accurate, there must be a reliable way to calculate the expression. Therefore, a suitable internal reference gene is needed to reliably quantify gene transcripts. However, research pertaining to the suitable confirmation of the stability of expression levels when these housekeeping genes are used is scarce.
Housekeeping gene expression vary in different tissues and under different experimental conditions (15). For example, as the most used housekeeping gene, GAPDH was observed to be the most available internal reference gene for expression investigations in human reticulocytes (16–18). However, a study on virus-infected salmon reached the opposite conclusion (19, 20). We also observed that GAPDH was not the best internal reference gene in the duodenum of meat-type ducks. Therefore, different housekeeping genes should be identified for study in different animals or experiments.
Previous studies have shown that HMBS is the most stable housekeeping gene in human hepatocellular carcinoma tissue, blood samples (21), and brain tissue (22). Moreover, studies have found that HMBS has good stability in broilers (9, 23), layers (24), and quails (25). The above research results are consistent with the results of the experiments in this study, suggesting that HMBS may be the most suitable gene as an internal reference gene in birds. Another study mentioned that HMBS is highly specific to avian species (26). Based on this, HMBS can help reduce the possibility of false negative tests caused by insufficient sampling or storage degradation.
Moreover, Dai et al. (27) found that YWHAZ ranked high in the search for reference genes of fetal mice, which is suitable to be used in the study together with other reference genes. YWHAZ is considered the most stable reference gene in sheep leukocytes (28) and hypothalamic-pituitary-gonadal axis tissues of sows (29), which can serve for accurate and repeatable qPCR data analysis. Previous studies have shown that YWHAZ is a relatively suitable reference gene in multiple avian species, allowing for accurate normalization and quantification of gene expression levels in a variety of avian species (30–34).
In our study, we applied qPCR to detect the expression of eight candidate reference genes, GAPDH, β-actin, 18S rRNA, HMBS, HPRT1, TBP, RPL13, and YWHAZ, in the duodenal epithelial tissue of 42-day-old meat-type ducks and used the geNorm, NormFinder, and BestKeeper programs to investigate the expression stability. These software are commonly used to identify reference genes stably expressed (32, 35–37). The stability of each gene, as analyzed by different software programs, was inconsistent owing to different algorithms. geNorm prefers to compute the stability value of gene expression and estimate the number of housekeeping genes, whereas NormFinder focuses on the CV of a gene across all samples, and BestKeeper directly analyzes Ct values. Although some differences were observed in the final results owing to the distinct algorithms of the three programs, the ranking of the eight genes studied was the same. Considering the evaluation results of the three programs, HMBS and YWHAZ can be considered relatively stable internal reference genes in duodenal epithelial cells, and 18S rRNA, HPRT1, and TBP relatively unstable. The results of this study yield a reference basis for the applicable normalization of housekeeping genes under diverse experimental conditions.
In summary, our results show that HMBS and YWHAZ are the most stable reference genes in meat-type ducks, while 18S rRNA is the least stable and is not suitable for studying gene expression in meat-type duck tissues. Our study highlights the value of reference genes in the use of qPCR and lays the foundation for the application of quantitative gene expression analysis to elucidate gene function studies in modern poultry breeding programs.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
All the study procedures that included animals were strictly carried out under the regulations and guidelines established by the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004). All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of Anhui Agricultural University (SYXK 2016-007). The animals in the study had ad libitum access to water and feed (Table 1) and were humanely sacrificed.
SJ and ZG conceived this study. FS, GQ, SP, XW, YL, TJ, and FJ collected the samples and performed the experiments. FS, SP, and SJ analyzed the data. FS, GQ, and SJ wrote the manuscript. SJ, FS, GQ, and ZG participated in the planning of experiments and revising the manuscript. All authors have read and approved the final version of the manuscript.
This study was funded by Quality Project of Department of Education of Anhui Province (2021jxjy026), Program for Young Outstanding Scientists of University (gxyqZD2022017), Natural Science Foundation from Department of Anhui Provincial Education (2022AH050928), University-Industry Collaborative Education Program of Ministry of Education (221000488095409), the University Synergy Innovation Program of Anhui Province (GXXT-2021-052), and the Science and Technology Major Project of Anhui Province (201903a06020018).
We would like to thank Jiangfa Wang from Huangshan Qiangying Duck Breeding Co. Ltd., China for raising the ducks and for their assistance in the collection of samples.
YL was employed by Huangshan Qiangying Duck Breeding Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1160384/full#supplementary-material
Supplementary Figure S1. The agarose electrophoresis of total RNA.
1. Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. (2004) 313:856–62. doi: 10.1016/j.bbrc.2003.11.177
2. Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, et al. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS One. (2012) 7:1–10. doi: 10.1371/journal.pone.0033278
3. Gachon C, Mingam A, Charrier B. Real-time PCR: what relevance to plant studies? J Exp Bot. (2004) 55:1445–54. doi: 10.1093/jxb/erh181
4. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:34. doi: 10.1186/gb-2002-3-7-research0034
5. Haberhausen G, Pinsl J, Kuhn CC, Markert-Hahn C. Comparative study of different standardization concepts in quantitative competitive reverse transcription-PCR assays. J Clin Microbiol. (1998) 36:628–33. doi: 10.1128/JCM.36.3.628-633.1998
6. Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. (1999) 75:291–5. doi: 10.1016/S0168-1656(99)00163-7
7. Lovdal T, Lillo C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem. (2009) 387:238–42. doi: 10.1016/j.ab.2009.01.024
8. Godornes C, Leader BT, Molini BJ, Centurion-Lara A, Lukehart SA. Quantitation of rabbit cytokine mRNA by real-time RT-PCR. Cytokine. (2007) 38:1–7. doi: 10.1016/j.cyto.2007.04.002
9. Zhang J, Gao YY, Huang YQ, Fan Q, Lu XT, Wang CK. Selection of housekeeping genes for quantitative gene expression analysis in yellow-feathered broilers. Ital J Anim Sci. (2018) 17:540–6. doi: 10.1080/1828051X.2017.1365633
10. Ishida A, Ashihara A, Nakashima K, Katsumata M. Expression of cationic amino acid transporters in pig skeletal muscles during postnatal development. Amino Acids. (2017) 49:1805–14. doi: 10.1007/s00726-017-2478-2
11. Zhang Y, Zhang XD, Liu X, Li YS, Ding JP, Zhang XR, et al. Reference gene screening for analyzing gene expression across goat tissue. Asian-Australas J Anim Sci. (2013) 26:1665–71. doi: 10.5713/ajas.2013.13199
12. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. (2004) 64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496
13. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. (2004) 26:509–15. doi: 10.1023/B:BILE.0000019559.84305.47
14. Shakeel M, Rodriguez A, Tahir UB, Jin F. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol Lett. (2018) 40:227–36. doi: 10.1007/s10529-017-2465-4
15. Walker CG, Meier S, Mitchell MD, Roche JR, Littlejohn M. Evaluation of real-time PCR endogenous control genes for analysis of gene expression in bovine endometrium. BMC Mol Biol. (2009) 10:100. doi: 10.1186/1471-2199-10-100
16. Buldak L, Labuzek K, Buldak RJ, Kozlowski M, Machnik G, Liber S, et al. Metformin affects macrophages' phenotype and improves the activity of glutathione peroxidase, superoxide dismutase, catalase and decreases malondialdehyde concentration in a partially AMPK-independent manner in LPS-stimulated human monocytes/macrophages. Pharmacol Rep. (2014) 66:418–29. doi: 10.1016/j.pharep.2013.11.008
17. Bronkhorst AJ, Aucamp J, Wentzel JF, Pretorius PJ. Reference gene selection for in vitro cell-free DNA analysis and gene expression profiling. Clin Biochem. (2016) 49:606–8. doi: 10.1016/j.clinbiochem.2016.01.022
18. Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. (2006) 7:33. doi: 10.1186/1471-2199-7-33
19. Jorgensen SM, Kleveland EJ, Grimholt U, Gjoen T. Validation of reference genes for real-time polymerase chain reaction studies in Atlantic salmon. Mar Biotechnol. (2006) 8:398–408. doi: 10.1007/s10126-005-5164-4
20. Kang Y, Wu Z, Cai D, Lu B. Evaluation of reference genes for gene expression studies in mouse and N2a cell ischemic stroke models using quantitative real-time PCR. BMC Neurosci. (2018) 19:3. doi: 10.1186/s12868-018-0403-6
21. Ahn HR, Baek GO, Yoon MG, Son JA, You D, Yoon JH, et al. HMBS is the most suitable reference gene for RT-qPCR in human HCC tissues and blood samples. Oncol Lett. (2021) 22:791. doi: 10.3892/ol.2021.13052
22. Coulson DT, Brockbank S, Quinn JG, Murphy S, Ravid R, Irvine GB, et al. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol Biol. (2008) 9:46. doi: 10.1186/1471-2199-9-46
23. Na W, Wang Y, Gong P, Zhang X, Zhang K, Zhang H, et al. Screening of reference genes for RT-qPCR in chicken adipose tissue and adipocytes. Front Physiol. (2021) 12:676864. doi: 10.3389/fphys.2021.676864
24. Rodriguez-Hernandez R, Oviedo-Rondon EO, Rondon-Barragan IS. Identification of reliable reference genes for expression studies in the magnum of laying hens housed in cage and cage-free systems. Vet Med Sci. (2021) 7:1890–8. doi: 10.1002/vms3.507
25. de Sousa FCB, do Nascimento CS, Macario MDS, Araujo RDS, Barbosa LT, Bayao GFV, et al. Selection of reference genes for quantitative real-time PCR normalization in European quail tissues. Mol Biol Rep. (2021) 48:67–76. doi: 10.1007/s11033-020-06134-7
26. Wang Y, Zhang J, Patrick K, Li M, Gong J, Xu B, et al. Hydroxymethylbilane synthase (HMBS) gene-based endogenous internal control for avian species. AMB Express. (2020) 10:181. doi: 10.1186/s13568-020-01112-5
27. Dai Y, Kou H, Guo X, Gong Z, Liu H, Liu Y, et al. Identification and validation of reference genes for RT-qPCR analysis in fetal rat pancreas. Reprod Toxicol. (2021) 105:211–20. doi: 10.1016/j.reprotox.2021.09.009
28. Mahakapuge TA, Scheerlinck JP, Rojas CA, Every AL, Hagen J. Assessment of reference genes for reliable analysis of gene transcription by RT-qPCR in ovine leukocytes. Vet Immunol Immunopathol. (2016) 171:1–6. doi: 10.1016/j.vetimm.2015.10.010
29. Kim HD, Jo CH, Choe YH, Lee HJ, Jang M, Bae SG, et al. PPIA, HPRT1, and YWHAZ are suitable reference genes for quantitative PCR assay of the hypothalamic-pituitary-gonadal axis in sows. Anim Biosci. (2022) 35:1850–9. doi: 10.5713/ab.22.0083
30. Boo SY, Tan SW, Alitheen NB, Ho CL, Omar AR, Yeap SK. Identification of reference genes in chicken intraepithelial lymphocyte natural killer cells infected with very-virulent infectious bursal disease virus. Sci Rep. (2020) 10:8561. doi: 10.1038/s41598-020-65474-3
31. Hassanpour H, Bahadoran S, Farhadfar F, Chamali ZF, Nazari H, Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in lung and heart of pulmonary hypertensive chickens. Poult Sci. (2018) 97:4048–56. doi: 10.3382/ps/pey258
32. Hassanpour H, Aghajani Z, Bahadoran S, Farhadi N, Nazari H, Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in ovary and uterus of laying hens under heat stress. Stress. (2019) 22:387–94. doi: 10.1080/10253890.2019.1574294
33. Olias P, Adam I, Meyer A, Scharff C, Gruber AD. Reference genes for quantitative gene expression studies in multiple avian species. PLoS ONE. (2014) 9:e99678. doi: 10.1371/journal.pone.0099678
34. Bages S, Estany J, Tor M, Pena RN. Investigating reference genes for quantitative real-time PCR analysis across four chicken tissues. Gene. (2015) 561:82–7. doi: 10.1016/j.gene.2015.02.016
35. Fragoulis A, Biller K, Fragoulis S, Lex D, Uhlig S, Reiss LK. Reference gene selection for gene expression analyses in mouse models of acute lung injury. Int J Mol Sci. (2021) 22:7853. doi: 10.3390/ijms22157853
36. Wang GH, Wang SH, Zhang WZ, Liang CC, Cheng G, Wang XY, et al. Analysis of stability of reference genes for qPCR in bovine preadipocytes during proliferation and differentiation in vitro. Gene. (2022) 830:146502. doi: 10.1016/j.gene.2022.146502
Keywords: duodenal epithelial, gene expression, meat-type ducks, reference genes, qPCR
Citation: Shui F, Qiu G, Pan S, Wang X, Jia F, Jiang T, Li Y, Geng Z and Jin S (2023) Identification of stable reference genes for quantitative gene expression analysis in the duodenum of meat-type ducks. Front. Vet. Sci. 10:1160384. doi: 10.3389/fvets.2023.1160384
Received: 07 February 2023; Accepted: 20 March 2023;
Published: 03 April 2023.
Edited by:
Yanyan Sun, Institute of Animal Sciences (CAAS), ChinaReviewed by:
Hao Bai, Yangzhou University, ChinaCopyright © 2023 Shui, Qiu, Pan, Wang, Jia, Jiang, Li, Geng and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sihua Jin, anNoMzIzNUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.