- Division of Microbiology, Brewing, and Biotechnology, School of Biosciences, Sutton Bonington Campus, University of Nottingham, Loughborough, Leicestershire, United Kingdom
Introduction: Rotavirus A is a major cause of acute dehydrating diarrhea in neonatal pigs resulting in significant mortality, morbidity, reduced performance and economic loss. Commercially available prebiotic galacto-oligosaccharides are similar to those of mammalian milk and stimulate the development of the microbiota and immune system in neonates. Little is known about the effects of supplementing sows' diets with galacto-oligosaccharides during gestation. This study aimed to determine if dietary galacto-oligosaccharide supplementation during gestation could improve immunity, reduce rotavirus infection and modulate the microbiota in sows and neonates in a commercial farm setting with confirmed natural endemic rotavirus challenge.
Methods: In a randomized controlled trial, control sows received lactation diet with no galacto-oligosaccharide supplementation and test sows received lactation diet with 30 g/day galacto-oligosaccharide top-dressed into feed daily, seven days before farrowing. Colostrum was collected from sows 24 hours post-partum and tested for rotavirus specific antibodies. Fecal samples were collected from sows and piglets three days post-partum, tested for rotavirus A by qPCR and the microbiome composition assessed by 16s rRNA gene sequencing.
Results: Supplementation with galacto-oligosaccharides during gestation significantly increased rotavirus-specific IgG and IgA in sow colostrum and reduced the number of rotavirus positive piglet fecal samples. Abundance of potential pathogens Treponema and Clostridiales were higher in fecal samples from non-galacto-oligosaccharide fed sows, their piglets and rotavirus positive samples.
Discussion: This study demonstrates that galacto-oligosaccharide supplementation during gestation significantly increases rotavirus specific IgG and IgA in sow colostrum thereby reducing neonatal rotavirus infection and suppresses potential pathogenic bacteria in nursing sows and neonatal piglets.
Introduction
Rotaviruses are classified into at least ten serogroups (1, 2) with A, B, and C affecting humans (3), whilst groups A to H have been found in pigs (2). The most common groups are A, B and C, with Rotavirus A (RVA) representing the most prevalent group causing acute dehydrating diarrhea in public and veterinary health settings (2). RVA fecal-oral infection results in destruction of small intestinal enterocytes, the development of malabsorptive diarrhea (4) and promotes gut dysbiosis through alteration of the microbiota (5).
The effects on pigs are significant mortality and morbidity in neonates, reduced performance in surviving growers and significant economic loss (1, 2, 6). RV is endemic in UK pig farms. A range of RVA genotypes has been identified in UK pigs: six G types (VP7); G2, G3, G4, G5, G9, and G11 and six P types (VP4); P6, P7, P8, P13, P23, and P32 (7). Furthermore, the common human genotype P8 can infect pigs highlighting the need for surveillance of porcine rotavirus genotypes to safeguard human and porcine health (7).
Previous livestock vaccination strategies have focussed on the induction of active (immune cell mediated) and passive (antibody mediated) immunity by oral administration of attenuated RV vaccines (8). However, these have lacked efficacy, in contrast to engineered virus-like particles (VLP) designed as vaccines to boost antibodies in bovine and porcine mammary secretions which have shown promise when administered with attenuated vaccines (9). The wide variety of RV genotypes in pigs complicates effective vaccine production. This is further complicated by attenuated replicating porcine RVA vaccines which may contribute to the diversity of porcine RVs, through re-assortment of vaccine strains with wild type strains and the emergence of novel genetic variants that can evade herd immunity (2, 7). Whilst vaccination remains popular in the farming community, a more pragmatic view may be to focus on cleaning and disinfection with efficacious detergents that not only limit the spread and infectivity of RV but also other microbial pathogens (10, 11). Nevertheless, endemic porcine RV infection still needs alternative strategies to boost lactogenic immunity in sows, thus providing RV antibodies to the neonate with colostrum and milk (2).
Galacto-oligosaccharides (GOS) are a major constituent of mammalian milk (12, 13) primarily stimulating the development of the microbiota in neonates and conferring a variety of health benefits including innate and adaptive immune development (14, 15). Milk oligosaccharides are typically composed of three to ten monosaccharide units, including glucose (Glc), galactose (Gal) and N-acetyl-glucosamine (GlcNAc) as well as fucose and sialic acids. The core moiety present at the reducing end of milk oligosaccharides is either lactose (Gal(β1–4)Glc) or N-acetyl-lactosamine (Gal(β1–4)GlcNAc) (16). Most animal milk oligosaccharides are sialylated, containing N-acetylneuraminic acid (Neu5Ac) and/or N-glycolylneuraminic acid (Neu5Gc) (17). Compared with other domestic animals, porcine milk contains the highest percentage of neutral oligosaccharides (20%), the most abundant variety of mono-sialylated and di-sialylated large oligosaccharides and are the closest to human milk oligosaccharide composition (13). In addition, porcine milk oligosaccharides (PMOs) decrease in abundance by ~43% during the first week of lactation with the relative concentration of acidic PMOs decreasing and neutral PMOs increasing (18), indicating a change in functionality during lactation.
In pigs there is evidence that GOS is readily fermented in the gastrointestinal tract (GIT) increasing short-chain fatty acid (SCFA) concentrations and increasing beneficial probiotic bacteria numbers (19, 20). Furthermore, GOS may reduce adhesion of pathogens to cells, (21) inhibit pathogen colonization (21), improve gut architecture (20) and reduce expression of pro-inflammatory cytokines (22). Specific effects of GOS on RVs have been demonstrated. For example, GOS/fructo-oligosaccharide mixtures reduce RV induced diarrhea and modulate dysbiosis in suckling rats (5, 23). Human milk oligosaccharides (HMOs) inhibit RV infectivity in vitro (24, 25), in acutely infected piglets (24) and reduce the duration of RV-induced diarrhea in piglets whilst modulating the colonic microbiota in vivo. (26). Also, RV specific antibodies from Human breast milk neutralize RV infectivity in vitro (27). However, most studies have focussed on feeding neonatal to pre-weaning piglets GOS, whilst few have considered supplementing the diets of gestational sows to determine effects on the neonate. It has been reported that the combination of GOS and casein glycomacropeptides (CGMP) fed to gestational sows modulated the neonatal microbiota colonization, promoted gut development and growth performance of piglets, thus demonstrating that manipulation of the maternal gestational immune/microbiome axis has positive effects on offspring, but without RVA challenge (28). The aims of this study were to determine if GOS supplementation in gestational sows conferred immunity, reduced infectivity and modulated the microbiome in neonatal piglets in a commercial pig farm where RV challenge is naturally endemic and as confirmed by previous veterinary reports.
Materials and methods
Experimental design
Animals
A randomized controlled trial was performed on a commercial farrow-to-finish pig farm in Yorkshire UK, between October and December 2018. The trial was approved by the farm veterinary consultant and by the University of Nottingham ethics committee on 12-9-18, approval reference number 190. Landrace x Large white sows crossed with a Piétran boar were paired with respect to parity. Gestating sows of similar weight were moved to 3.0 × 1.8 m farrowing pens with a 0.8 × 2.2 m farrowing crate, seven days before farrow. Pens had a slatted floor and were heated with industry standard heat lamps. Temperature was kept at range 18–20°C for sows and 23–24°C for piglets with light periods from 8:00 am to 17:00 pm. Relative humidity was 50 to 70% for farrowing units and 24 to 30% for weaning units. Metal chain toys with plastic balls were provided as environmental enrichment. Sows received a wheat-based lactation diet (Gold Lactator, Noble Foods, Stokesley, UK) containing 18.4% protein, 5.6% ash, 4.6% oil, 4.1% fiber, 1.13% lysine, 0.9% calcium, 0.34% methionine and 0.49% phosphorous. New-born pigs received a 1 ml intramuscular iron injection (Gleptosil, Alstoe Ltd, York, UK) 24 h after birth. Sows and gilts were vaccinated with a combined Rotavirus OSU 6 strain and E. coli strains 0101:K99 vaccine two weeks prior to farrowing as per manufacturer's instructions and as according to standard farm practice (Rokovac Neo, Bioveta, Czech Republic). Piglets and sows did not receive any creep feed supplementation or prophylactic antibiotic treatment during the trials. Sows were individually housed and randomized in a homogenous pattern to either basal control diet or supplementation with GOS powder (DP2+ GOS, Nutrabiotic, Saputo Dairy UK, Weybridge UK). Sows received the lactation diet with no GOS supplementation (non-GOS sows) or received the lactation diet with 30 g/day GOS top-dressed into feed daily, seven days before farrowing (GOS sows). Piglets born to non-GOS sows were referred to as non-GOS piglets and those born to GOS sows were referred to as GOS piglets. Trial size was determined using a power calculation accessed at: https://clincalc.com/stats/samplesize.aspx on 02-08-18, where α = 0.05, β = 0.2, and power = 0.8, giving thirty-six replicates per control and treatment groups with a total of seventy-two pens, with one sow per pen. Trials were repeated six times, from week one to week six, in order to obtain the desired number of replicates. Models were fixed effect, whereby sows from the production cohort were randomly allocated to farrowing pens pre-assigned for non-GOS or GOS feed (independent variables). All animals were kept in identical environmental conditions, housed in identical pens and in the same building. Pens were cleaned and disinfected prior to trial replicates from the end of week one to week six onwards throughout the entire standard farm production methods. Once born, neonatal piglets were cross fostered within treatment groups, as per commercial farm standard practice, to equilibrate litter size and for welfare reasons. All trial personnel, including investigators were blinded to treatment allocation. All animals were monitored daily by trained farm personnel for any signs of scour, disease, lameness and/or distress. No animals were euthanized, or invasive samples taken during studies.
Sample collection
Trained farm personnel collected samples for biosecurity reasons. Colostrum from sows was collected within 24 hours post parturition by massaging the two teats closest to the head of sows and immediately frozen at −20°C, in a freezer, in 30 ml sterile plastic universal tubes (Thermo Scientific, Loughborough, UK). Approximately 2–3 g of freshly voided fecal samples were collected from sows and piglets per pen, in sterile nuclease free 2 ml micro tubes (Sarstedt, Leicester, UK) three days post partition and immediately frozen at−20°C. Fecal samples from piglets were pooled from each pen, whilst those of sows were kept separately. Frozen samples were delivered by refrigerated courier service to the University of Nottingham for storage at −80°C and further laboratory analyses.
ELISA for RVA IgG and IgA in colostrum
Samples were defrosted and 1 ml aliquots centrifuged at 13,000 g for 15 min to separate the fat from the colostrum. Aqueous phase colostrum was pipetted from underneath the fat layer and into sterile 2 ml micro tubes for subsequent analysis. The Ingezim rotavirus porcine ELISA kit (Immunologia Y Genetica Aplicada S.A. Madrid, Spain) was used to determine specific anti-RVA IgG and anti-RVA IgA activity in the colostrum samples according to manufacturer's instructions. For the detection of anti-RVA IgA antibodies, ELISA was performed as with IgG, but the secondary antibody was substituted with peroxidise-labeled goat anti-porcine IgA (Thermo Fisher Scientific, Bonn, Germany) at a dilution of 1/10,000 as according to Kreuzer et al. (29). The positive control serum supplied with the kit, was assayed on each occasion and the mean value from these measurements used to obtain a normalized absorbance ratio to reduce assay-to-assay variation (30). Total non-specific IgG and IgA in colostrum were assayed using IgG and IgA Pig ELISA Kits obtained from (Abcam plc, Cambridge, UK).
DNA and RNA extraction
Bacterial DNA was extracted from 200 mg sow and piglet fecal samples using the QIAamp PowerFecal QIAcube HT Kit and QIAcube HT robot according to manufacturer's instructions (Qiagen, Hilden, Germany). Viral RNA was extracted from sow and piglet feces by mixing 100 mg with 900 μl isotonic 0.9% NaCl (Merck, Gillingham, UK), prepared in diethyl pyrocarbonate (DEPC) treated nuclease free water (Fisher Scientific UK Ltd, Loughborough UK), vortexed and centrifuged at 16,000 g for 5 min. All glassware was treated with 0.1% v/v DEPC (Merck, Gillingham, UK), to remove RNase enzymes and autoclaved at 121°C at 15 psi to eliminate residual DEPC. 200 μl of the clear supernatant was used for viral nucleic acid extraction using the QIAamp 96 Virus QIAcube HT Kit and QIAcube HT robot according to manufacturer's instructions (Qiagen, Hilden, Germany). DNA was digested in samples by including an optional DNase digestion step in the QIAcube HT protocol using the Qiagen RNase Free DNase Set (Qiagen, Hilden, Germany) to prevent the possibility of interference with RNA assays in downstream applications. Bespoke software for loading onto the QIAcube HT robot was provided by Qiagen for this step. During viral RNA extraction 4 μl per sample of a Techne qPCR Rotavirus A kit internal extraction control RNA was spiked into the lysis buffer as a positive control for the extraction process (Cole-Parmer, Stone, Staffordshire UK).
Detection of RVA RNA in RNA samples
The Techne qPCR Rotavirus A kit was used to detect the presence of RVA in samples with an amplification protocol using OneStep 2x Reverse Transcription-qPCR MasterMix according to manufacturer's instructions (Cole-Parmer, Stone, Staffordshire UK). RVA specific primer probe mix was used to detect the presence of RVA non-structural protein 5 (NSP5) genomes. Standard curves were prepared with RVA positive control template with copy numbers from 2 × 105 per μl to 2 per ul. Real-time quantitative PCR data were collected using the Roche LightCycler 480 (Hoffman La Roche, Basel, Switzerland). The amplification protocol was reverse transcription for 10 min at 42°C, enzyme activation for 2 min at 95°C, then 50 cycles of denaturation for 10 s at 95°C and fluorogenic data collection for 60 s at 60°C followed by one cycle of cooling. The detection format was dual color hydrolysis/Universal Probe Library (UPL), with dynamic integration time mode and a filter combination of duplexing TaqMan probes, FAM and VIC. Amplification curves were initially analyzed using the LightCycler 480 Software release 1.5.0.39. as obtained from https://pim-eservices.roche.com/eLD/web/gb/en/products/3.8.1.4.4.8 accessed 20-02-20.
PCR amplification of 16S rRNA gene sequences
Using the extracted DNA as a template, the V4 region of the bacterial 16S rRNA genes were PCR amplified using primers 515f (5' GTGCCAGCMGCCGCGGTAA 3') and 806r (5' GGACTACHVGGGTWTCTAAT 3') (31). The full preparation and sequencing of 16S rRNA gene sequencing libraries were conducted according to the MiSeq Wet Lab SOP accessed at https://github.com/SchlossLab/MiSeq_WetLab_SOP/blob/master/MiSeq_WetLab_SOP on the 19-02-20. Amplicons were sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using 2 × 250 bp cycles (32). Sequence data were deposited in the NCBI database within Bioproject PRJNA884280.
Microbiota diversity analysis
The 16S rRNA sequence analyses were performed using Mothur v. 1.43, (33) open source software and accessed at: (https://github.com/mothur/mothur/releases accessed 12-03-20). Analysis was performed according to the MiSeq SOP accessed at: (https://mothur.org/wiki/miseq_sop/ accessed 12-03-20). The 16S rRNA gene sequences were aligned against a reference alignment based on the SILVA rRNA database for use in Mothur available at: (https://mothur.org/wiki/silva_reference_files accessed 12-03-20) (34) and clustered into OTUs using the “opticlust” clustering algorithm (35). The consensus taxonomy of the OTUs was generated using the “classify.otu” command in Mothur with reference data from the Ribosomal Database Project (version 14) (36, 37) adapted for use in Mothur available at: (https://mothur.org/wiki/rdp_reference_files accessed 12-03-20).
Statistical analyses
Analyses were performed in R version 4.1.1 using R Studio (2021.09.0) (38) unless otherwise stated. Shapiro Wilk tests (39) were used to determine normality for the results of ELISA, log10 copy numbers for RVA positive fecal samples and microbiota α-diversity metrics. For ELISA and log10 copy numbers of RVA positive samples, significant differences between groups were tested using Mann–Whitney U-tests. Significant differences in the number of RVA infected piglet fecal samples were tested using the Binomial test. Coverage and α-diversity expressed as Inverse Simpson diversity (40), Chao (41) Richness, Shannon (42) Index, and ACE Estimator (43), were calculated using the “summary.single” command in Mothur (33). Significant differences were tested for using Kruskal–Wallis rank sum tests. Estimates of β-diversity were calculated in Mothur as Yue and Clayton (44) Dissimilarity (θYC), Bray and Curtis (45) Dissimilarity and Jaccard (46) Similarity. Analysis of molecular variance executed in Mothur (AMOVA) was used to test for differences in β-diversity between samples (47, 48). Linear discriminant analysis effect size (LEfSe) was used to examine differential OTU abundances at genus level in Mothur (49). Where appropriate, multiple comparisons (AMOVA, Kruskal–Wallis rank sum tests) were adjusted for false discovery rates (FDR) by the Benjamini and Hochberg procedure (50) (P = 0.05, FDR = 25%).
Results
RVA specific and total antibody titres in sow colostrum
RVA specific and total antibody levels in sow colostrum are shown in Figure 1. Median RVA specific antibody levels in sow colostrum were, IgG non-GOS sows 0.179, IgG GOS sows 0.285, IgA non-GOS sows 1.771, IgA GOS sows 2.182 (normalized absorbance ratios). Median total antibody levels in sow colostrum were, IgG non-GOS sows 38.06, IgG GOS sows 33.25, IgA non-GOS sows 12.83, IgA GOS sows 11.55 (mg/ml colostrum). Shapiro-Wilk normality tests indicated colostrum concentrations of RVA specific and non-specific antibodies were not normally distributed (P < 0.05 in each case). Colostrum RVA specific IgG and IgA concentrations expressed as ELISA normalized absorbance ratio were significantly higher in GOS fed sows compared with non-GOS sows (P = 0.03 and P = 0.049 respectively, Mann–Whitney U-tests). However, total IgG and IgA colostrum contents were not significantly different between GOS fed sows compared with non-GOS sows (P = 0.587 and P = 0.886 respectively, Mann–Whitney U-tests) (Figure 1).
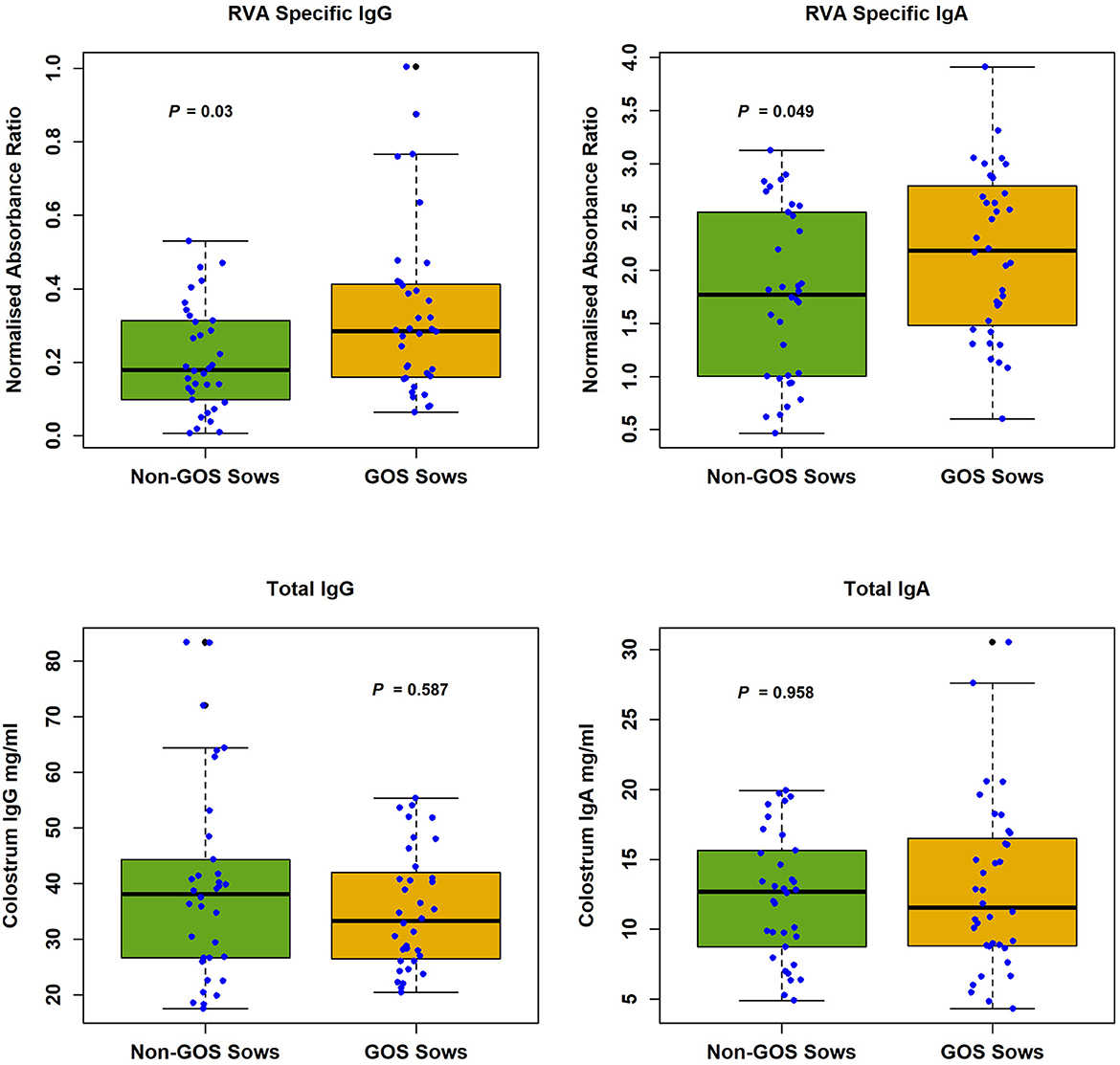
Figure 1. RVA specific IgG and IgA and total IgG and IgA in colostrum from non-GOS and GOS fed sows.
qPCR identification of RVA infected fecal samples
Internal extraction control RNA spiked into lysis buffer during viral RNA extraction was positive for all samples indicating successful RNA extraction and qPCR amplification using the LightCycler 480 VIC channel. Log10 copy numbers per g of fecal material for RVA positive samples from non-GOS piglets and GOS piglets were non-normally distributed (P = 5.7 × 10−4 and P = 0.024, respectively using Shapiro–Wilk tests). Median log10 copy numbers per g of fecal material were 16.25 for non-GOS piglets and 17.12 for GOS piglets. There was no significant difference in the RVA log10 copy number between non-GOS piglets or GOS piglets (P = 0.7007, Mann–Whitney U-tests). Out of thirty-four non-GOS piglet fecal samples, twelve (35%) tested negative and twenty-two (65%) positive for RVA. Out of thirty-six GOS piglet fecal samples, twenty (55%) tested negative and sixteen (45%) positive for RVA. There was a significant difference in the number of piglet fecal samples testing RVA positive between groups, P = 0.0085, Binomial test. Out of seventy-one sow fecal samples analyzed seven proved RVA positive, four non-GOS sow fecal samples (8.15–14.23 log10 copy number per g) and three GOS sow fecal samples (7.72–11.75 log10 copy number per g).
Fecal microbiota diversity and composition
In total 3,333,385 high quality 16S rRNA, V4 sequences were obtained from 141 sow and piglet fecal samples. Of these, 2,189,090 were recovered from seventy-one sow fecal samples and 1,144,295 from seventy piglet fecal samples. By treatment groups, 1,021,516 sequences were recovered from thirty-five non-GOS fed sows, 1,167,574 from 36 GOS fed sows, 449,463 from thirty-four piglets born to non-GOS fed sows and 694,832 from thirty-six piglets born to GOS fed sows. Sequences were subsampled to 11,210 per sample with a Good's coverage (51) of 97.8 to 99.9%. Metrics for α-diversity were not normally distributed (Shapiro–Wilk tests). There were no significant differences in α-diversity metrics between non-GOS fed sows and GOS fed sows, or piglets born to non-GOS fed sows and piglets born to GOS fed sows, P > 0.05 in each case (Kruskal–Wallis rank sum tests). α-diversity for all four metrics were significantly higher in non-GOS sows as opposed to non-GOS piglets and GOS sows as opposed to GOS piglets P < 0.005 in each case (Figure 2). Calculated β-diversity θYC, Bray and Curtis (45) and Jaccard (46) distances between non-GOS fed sows and GOS fed sows were not significantly different, as determined by AMOVA (48), P = 0.707, P = 0.581, and P = 0.285, respectively. θYC, Bray-Curtis and Jaccard distances were not significantly different between non-GOS piglets and GOS piglets, P =0.11, P = 0.102, and P = 0.075. There was a highly significant difference between sows and piglets for all three β-diversity metrics, P < 0.001 in each case (Figure 3).
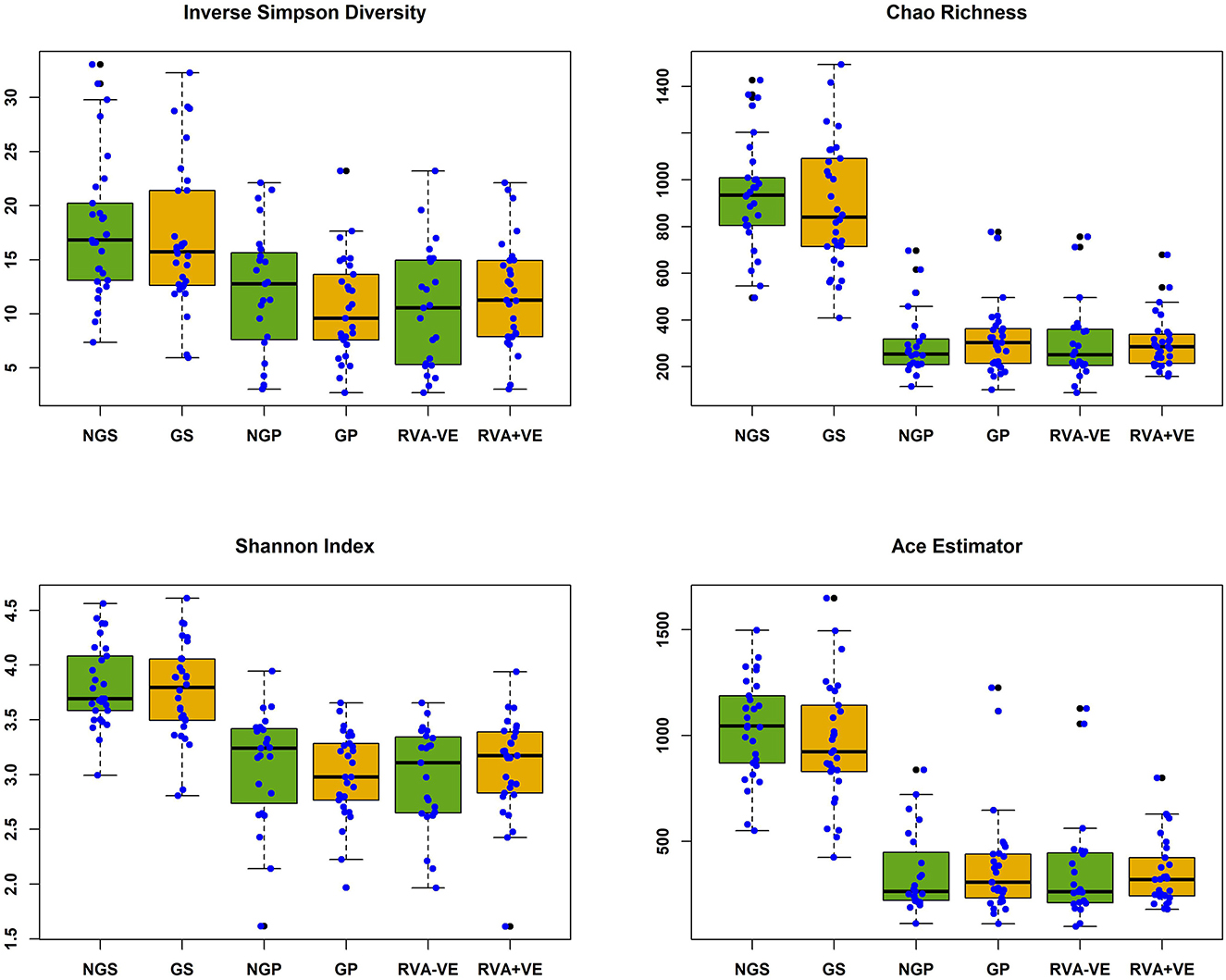
Figure 2. Alpha diversity of fecal samples collected from sows and piglets during suckling. NGS, non-GOS fed sow; GS, GOS fed sow; NGP, non-GOS piglet; GP, GOS piglet; RVA-VE, RVA negative piglet; RVA+VE, RVA positive piglet. Significant difference between NGS and NGP, GS, and GP in all cases P < 0.005.
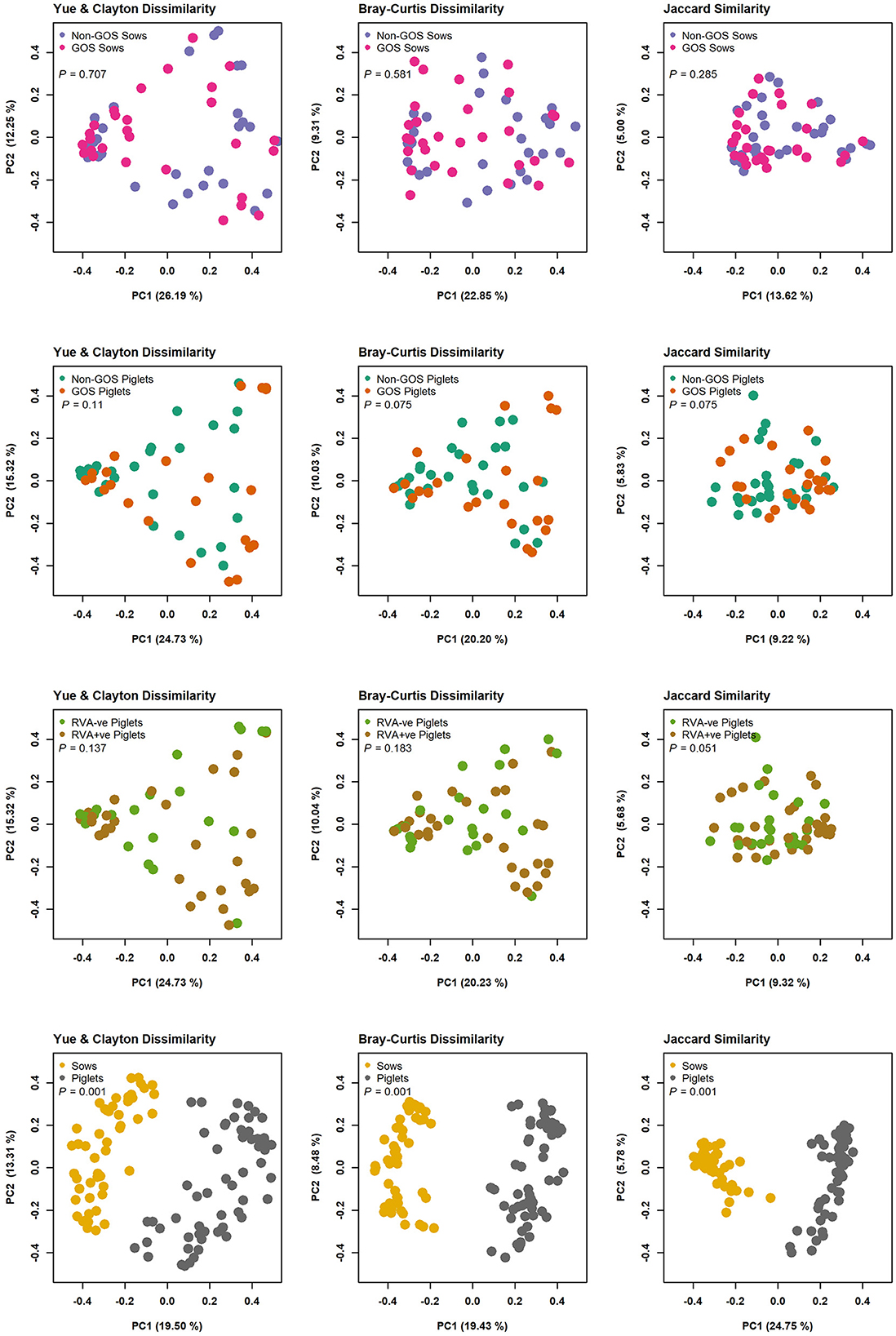
Figure 3. β-diversity measures for fecal samples from non-GOS sows vs. GOS fed sows, non-GOS piglets vs. GOS piglets, RVA negative vs. RVA positive piglets and sows vs. piglets.
Figure 4 shows relative abundance of bacterial taxa at phylum and genus level for fecal samples from non-GOS sows, GOS sows, non-GOS piglets and GOS piglets. For sow fecal samples, sequences were clustered into 5629 OTUs and classified into 19 unique phyla, 43 classes, 80 orders, 171 families and 397 genera. In total, the top ten taxa allocated to OTUs at phylum level were Firmicutes (60.09%), Proteobacteria (17.23%), Bacteroidetes (9.10%), Actinobacteria (5.71%), Spirochaetes (4.98%), Planctomycetes (1.37%), Bacteria unclassified (1.18%), Synergistetes (0.11%), Verrucomicrobia (0.06%) and Fusobacteria (0.03%). The top ten taxa allocated to OTUs at genus level were, Clostridium sensu stricto (18.63%), Acinetobacter (6.89%), Enterobacteriaceae unclassified (6.43%), Terrisporobacter (5.19%), Lactobacillus (4.85%), Romboutsia (3.07%), Planococcaceae unclassified (3.02%), Turicibacter (2.02%), Streptococcus (1.99%) and Bacteroides (0.95%). For piglet fecal samples, sequences were clustered into 2273 OTUs and classified into 19 unique phyla, 40 classes, 73 orders, 154 families and 349 genera. The top ten taxa allocated to OTUs at phylum level were Firmicutes (46.66%), Bacteroidetes (25.03%), Proteobacteria (15.21%), Fusobacteria (10.01%), Actinobacteria (2.76%), Verrucomicrobia (0.14%), Bacteria unclassified (0.09%), Synergistetes (0.04%), Spirochaetes (0.02%) and Planctomycetes (0.002%). The top ten taxa allocated to OTUs at genus level were, Bacteroides (20.47%), Clostridium Senso Stricto (13.17%), Enterobacteriaceae unclassified (12.37%), Lactobacillus (8.85%), Streptococcus (2.59%), Terrisporobacter (0.29%), Romboutsia (0.21%), Planococcaceae unclassified (12%), Acinetobacter (0.11%) and Turicibacter (0.05%). LEfSe identified significant differences in the abundance of differential OTUs annotated to taxa at genus level between treatment groups (Figure 5). In total non-GOS sows had eight OTUs occurring at significantly higher relative abundance compared with GOS sows, five of these being Treponema and one each to Phascolarctobacterium, Megasphaera, and Clostridiales unclassified. Non-GOS piglets had seven OTUs occurring at significantly higher relative abundance compared with GOS piglets, two of these being Ruminococcaceae unclassified and one each to Lactobacillus, Phascolarctobacterium, Aerococcus, Actinobacillus, and Clostridiales unclassified. GOS piglets had three OTUs occurring at a differentially higher abundance than non-GOS piglets, these being Peptoniphilus, Lachnospiriaceae unclassified, and Collinsella.
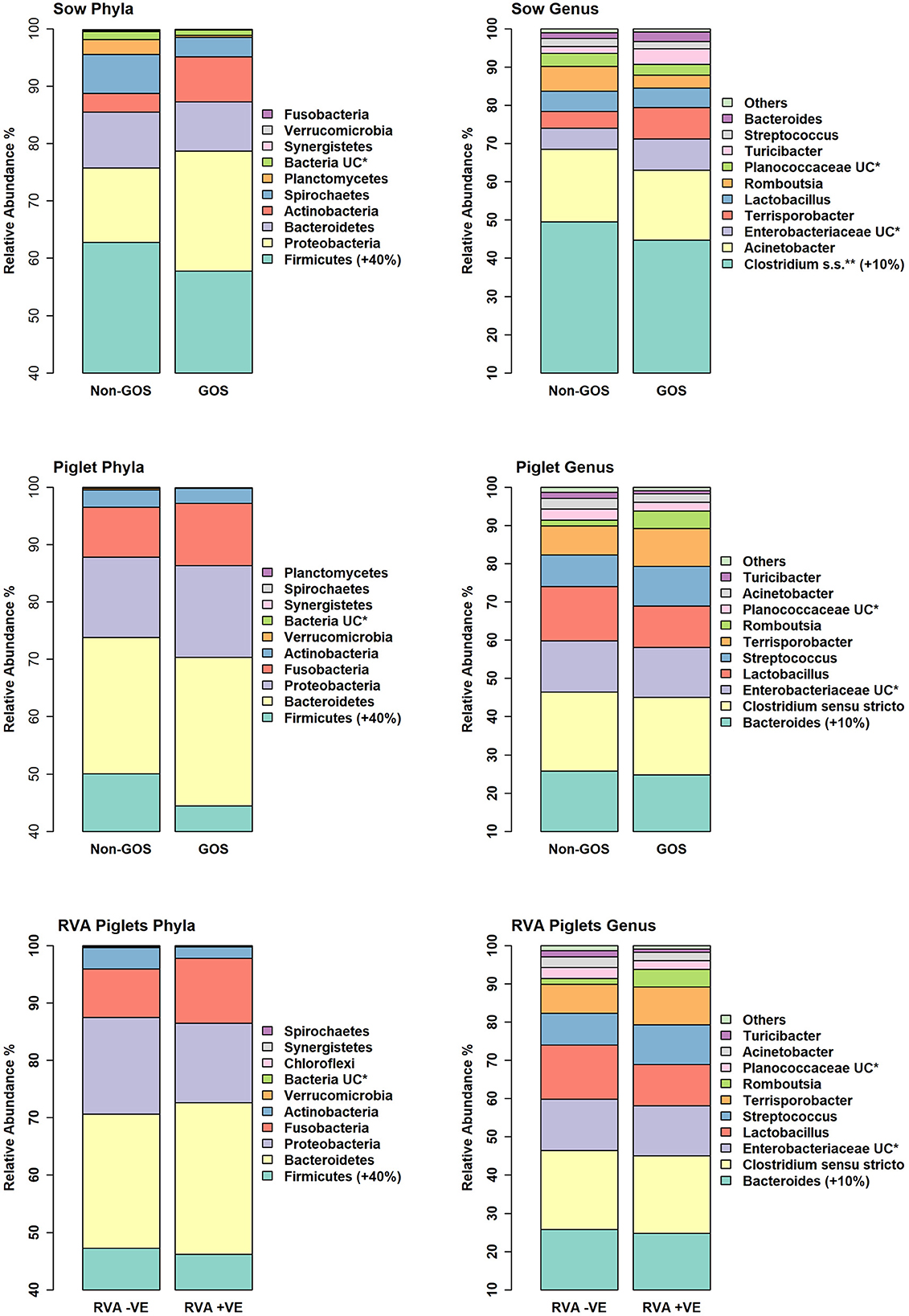
Figure 4. Relative abundance of bacterial taxa at phylum and genus level for fecal samples from non-GOS sows, GOS sows, non-GOS piglets, GOS piglets, RVA negative piglets, and RVA positive piglets.
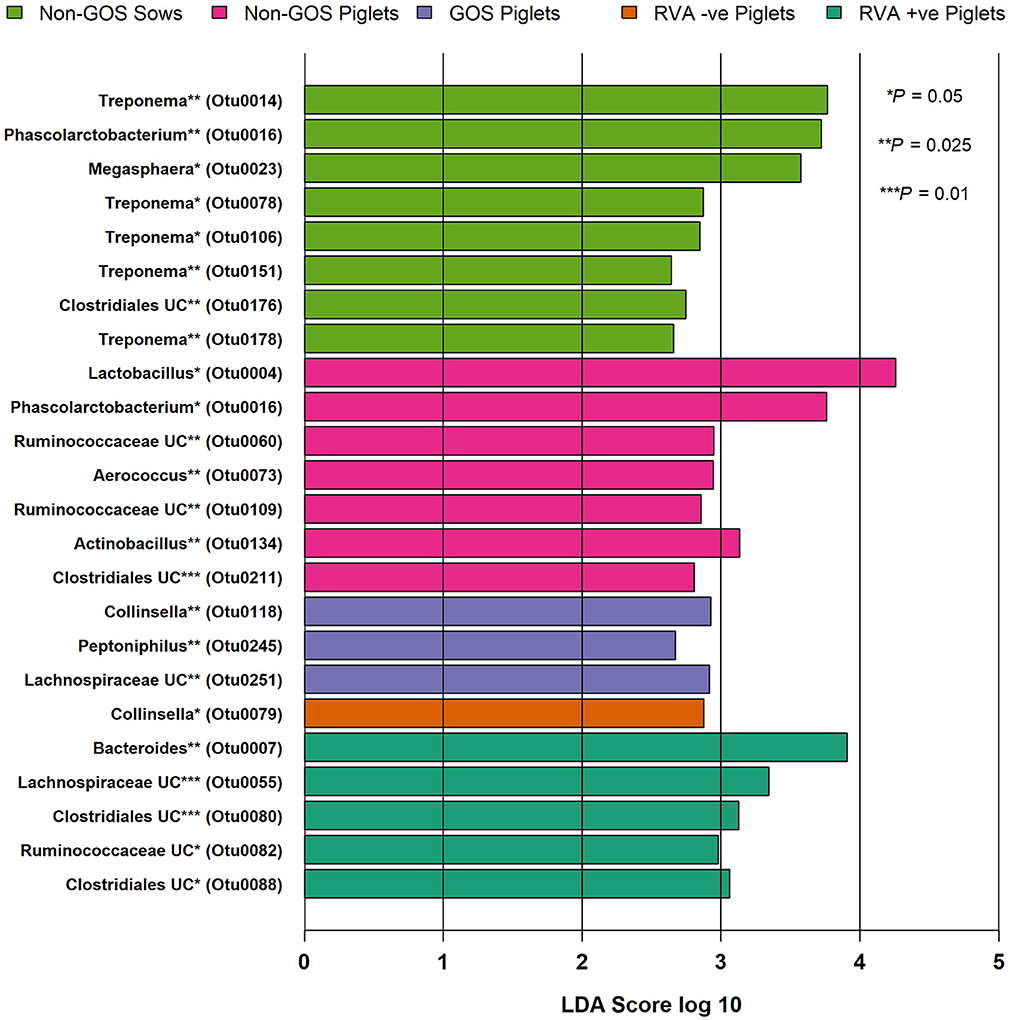
Figure 5. Significant differences in differential abundance of taxa at genus level for fecal samples from non-GOS sows, non-GOS piglets, GOS piglets, RVA negative piglets, and RVA positive piglets (LEfSe). No attributable differential abundance features for GOS sows, therefore not shown. UC = unclassified.
Fecal microbiota diversity and composition in non-infected and RVA infected piglets
In separate analyses by Mothur, 1,144,334 high quality 16S rRNA, V4 sequences were obtained from seventy piglet fecal samples. Of these, 531,797 were recovered from thirty-two RVA negative samples and 612,537 from thirty-eight RVA positive samples. Sequences were subsampled to 8078 per sample with a Good's coverage of 97.8 to 99.9%. Metrics for α-diversity were not normally distributed (Shapiro–Wilk tests). There were no significant differences in α diversity (Kruskal–Wallis Rank sum tests) or β-diversity (AMOVA) (48). Sequences were clustered into 2188 OTUs and classified into 19 unique phyla, 40 classes, 74 orders, 157 families and 348 genera. Figure 4 shows relative abundance of bacterial taxa at phylum and genus level for RVA negative and RVA positive fecal samples. In total, relative abundance of the top ten OTUs annotated to taxa at phylum level were Firmicutes (46.68%), Bacteroidetes (25.03%), Proteobacteria (15.21%), Fusobacteria (10.01%), Actinobacteria (2.76%), Verrucomicrobia (0.14%), Bacteria unclassified (0.07%), Chloroflexi (0.04%), Synergistetes (0.03%), and Spirochaetes (0.02%). The top ten OTUs annotated to taxa at genus level were Bacteroides (20.47%), Clostridium sensu stricto (13.17%), Enterobacteriacea unclassified (12.37%), Fusobacterium (9.42%), Lactobacillus (8.85%), Prevotella (3.14%), Streptococcus (2.59%), Peptostreptococcus (2.43%), Enterococcus (1.20%), and Phascolarctobacterium (1.11%). LEfSe identified significant differences in the abundance of differential OTUs annotated to taxa at genus level between RVA negative piglets and RVA positive piglets (Figure 5). RVA negative piglets expressed an increased differential abundance of Collinsella in contrast with RVA positive piglets. RVA positive piglets expressed a significant differential abundance in five OTUs, two being ascribed to Clostridiales unclassified and three others being Bacteroides, Lachnospiraceae unclassified, and Ruminococcacae unclassified.
Discussion
The objectives of this study were to determine if GOS supplementation in gestational sows conferred immunity, reduced infectivity and modulated the microbiome in neonatal piglets in a commercial pig farm where RVA challenge was endemic. Whilst PMOs are expressed naturally in sow colostrum (13), supplementation with GOS top-fed at 30 g per day was associated with significantly increased RVA specific IgG and IgA in sow colostrum (P = 0.03 and P = 0.049 respectively), but not the expression of total IgG and IgA (Figure 1). The maternal gut microbiome breast axis and the importance of entero-mammary pathways in programming the mammary gland to face the nutritional, microbiological, immunological, and neuroendocrine requirements of the growing infant have been well described in humans (52). However, humans possess a hemochorial placenta whereas pigs have an epitheliochorial placenta (53), one which, in contrast, is a relatively impenetrable barrier to maternal immunoglobulins during gestation, particularly IgG. Thus, piglets are born “agammaglobulinemic” and survival depends on early acquisition of maternal immunity through colostrum (54) before gut closure within 24 to 48 h post-partum and reduced intestinal enterocyte ability to sequester immunoglobulins from protein rich colostrum (55). Moreover, colostrum intake is the main determinant of piglet survival through energy provision and immune protection with long-term effects on growth and immunity (56). Few animal studies have investigated how pre- and/or probiotics fed to epitheliochorial pregnant mammals interact with the immune composition of mammary secretions. In dogs, pregnant bitches fed a mixture of fructo-oligosaccharides, mannan-oligosaccharides, E. faecium and L. acidophilus expressed significantly more IgG, IgM and IgA in colostrum (57). Possible mechanisms are the modulation of immunoglobulin secretion by the maternal microbiome. In murine models, gut microbiome induced maternal IgG is transferred to the neonatal intestine through milk via neonatal Fc receptors and directly inhibits pathogen colonization (58). For IgA, the gut microbiota induces Peyer's-patch dependent secretion of maternal IgA into milk. Antigen sampling by M cells in Peyer's-patches are the major source of migratory IgA plasma cells in mammary glands that produce maternal IgA found in milk (59). Similar mechanisms are found in sows with IgA secreted by mammary gland recruited plasma cells exhibiting specificity for antigens in the maternal digestive tract. This “entero-mammary” link is due to the migration of lymphocytes originating in gut associated lymphoid tissue via the bloodstream to the mammary gland (54). Other mechanisms may include viral triggering of goblet cell associated pathways, which present antigens to the immune system and serve as mechanisms of tolerance or translocation outside the gut (60).
In this study 65% of non-GOS piglet fecal samples tested positive for RVA as opposed to 45% for GOS-fed piglet fecal samples representing a significant reduction in infectivity of RVA in the maternally GOS fed group (P = 0.008). This reduction in infectivity can be explained by the significantly higher levels of RVA specific IgG and IgA expressed in the GOS fed sows colostrum as possibly modulated by entero-mammary pathways. Nevertheless, there may be other factors affected and/or modulated by GOS feeding such as the many unique proteins, cytokines, exosomes and leucocytes found in sow colostrum (61), which may require further investigation. Previous work has shown that human milk oligosaccharide supplementation can protect pigs against RV infection, as evidenced by shorter diarrhea duration, inhibiting RV binding and/or replication, enhancing mucosal T helper cell and T helper cell 2 cytokine responses and modulating microbiota composition (24). However, this is with direct feeding of GOS to piglets in contrast to the present study where colostrum and then milk were the only source of nutrition for piglets during the study period. In this respect, this study may be one of the first to demonstrate a significant increase in RVA colostrum viral specific immunoglobulins expressed following prebiotic gestational feeding with GOS to sows and concomitant reduction in infectivity in neonates in a commercial farm setting. Out of seventy-one sow fecal samples only seven (9.9%) were RVA positive with no significant difference between non-GOS and GOS fed sows. RVA prevalence rates in pigs varies from 3.3 to 67.3% (2) and prevalence in this study may have been low. Sows are usually immune to RVA, but the virus has been detected in the feces of sows as early as 5 days before farrowing and up to 2 weeks thereafter. Moreover, sows immune to RVA can shed the virus as a result of transient re-infection, or as asymptomatic carriers and at a time when piglets are susceptible to infection (62). Nevertheless, piglets may acquire RVA from their immediate environment given the prevalence of the virus and its stability in feces over time and at ambient temperatures (63). This demonstrates the circulation of RVA from adult sows to piglets and to the environment with resultant re-infection from environmental sources contaminated with RVA positive fecal matter. Animal and environmental RVA reservoirs indicate the need for efficacious detergents that limit the spread and infectivity of RVA and other microbial pathogens as previously described (10, 11) and in this respect GOS supplementation of gestational sows as an adjunct to these practices to reduce the RVA burden in neonates may be useful.
There were no significant differences in α or β-diversity metrics between non-GOS fed sows and GOS fed sows, or piglets born to non-GOS fed sows and piglets born to GOS fed sows. However, highly significant differences in α and β-diversity were seen between non-GOS fed sows and their piglets and GOS fed sows and their piglets (Figures 2, 3) demonstrating major differences in richness, evenness, community membership and structure. Notably, 2.5 times the number of OTUs were recovered from sow fecal samples as opposed to piglet fecal samples. The suckling pig microbiota is particularly different from that of sows and shows a lower bacterial diversity (64). This is not unexpected since piglets have high a high protein and PMO diet compared with the fiber rich diet of sows that support different microbial communities. Moreover, microbial gut diversity increases with age and with longitudinal changes in structure at different growth stages (65). However, it should be considered that both the environment and the sow influence the development of the piglet microbiome. In early lactation, the piglets' GIT microbiota composition is similar to the bacteria found on pen floors, in sow's milk and the nipple surface with the fecal microbiota of piglets becoming more similar to the sow as lactation progresses (66).
Predominant phyla in sows and piglets irrespective of GOS supplementation to sows were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria in keeping with other studies (65, 67, 68) (Figure 4). However, piglets had a higher relative abundance of taxa at phylum level belonging to Fusobacteria (10%) compared with sows (0.3%), which are associated with diarrhea and may be indicative of infection with enteric viruses such as porcine epidemic diarrhea virus which is known to affect the balance of beneficial gut bacteria as opposed to potential bacterial pathogens (69). Irrespective of GOS supplementation, Clostridium sensu stricto, Acinetobacter, Enterobacteriaceae unclassified, Terrisporobacter, and Lactobacillus dominated taxa at genus level in sow fecal samples as did Bacteroides, Clostridium Senso Stricto, Enterobacteriaceae unclassified, Lactobacillus, and Streptococcus in piglet fecal samples (Figure 4). These results were consistent with those from sow and piglet fecal microbiota taken from commercial pig farms (64) and as analyzed by similar methods. However, analyses of differential abundance of taxa at genus level by LEfSe revealed a significant increase in five OTUs belonging to the genus Treponema in non-GOS fed sows, but not GOS fed sows (Figure 5). Treponema spp are a cause of ear necrosis and shoulder ulcers in pigs leading to animal welfare problems and economic losses for producers (70). LEfSe also indicated a significant and increased differential abundance of Clostridiales in both non-GOS sows and non-GOS piglets (Figure 5). Whilst the majority of these organisms are commensal, some have potential to cause severe and sometimes lethal enteric infections in pigs (71). These results may indicate a direct effect of GOS in the sow GIT, thus indicating the capacity for GOS to inhibit pathogen colonization (20, 21). Reduction of Clostridial spp in GOS piglets may be explained by piglets inheriting fewer organisms from GOS fed sows with low abundance. Alternatively, sampling and translocation of maternal gut bacteria into colostrum and presentation of antigens to T helper cells by migratory dendritic cells may explain the reduction in Clostridiales in piglets (72). In non-GOS fed sows the occurrence of OTUs attributed to Treponema and Clostridia may indicate that sows harbor potentially pathogenic organisms that may cause pathologies in down-stream production and therefore, GOS supplementation to sows may suppress potential bacterial pathogens in the GIT microbiome, that otherwise may be transmitted allochthonously.
There were no significant differences in microbiota diversity and composition of RVA negative and RVA positive fecal samples taken from piglets when analyzed separately from sow fecal samples (Figure 3). Predominant phyla were Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria in keeping with other studies (65, 67, 68) (Figure 4). Abundance of Fusobacterium at genus level was higher than that of Lactobacillus, which is indicative of viral enteric infection (69). The only OTU occurring at significantly differential levels in RVA negative fecal samples from piglets was Collinsella (Figure 5). This bacterium is a member of the Coriobacteriaceae and has been strongly and positively correlated with intestinal and circulating rotavirus specific IFN-γ producing CD8+ T helper cell responses, which are known to correlate with protection against rotavirus diarrhea (73). Moreover, Collinsella produces ursodeoxycholate which reportedly inhibits binding of SARS-CoV-2 to angiotensin-converting enzyme, suppresses pro-inflammatory cytokines such as TNF-α, IL-1β, IL-2, IL-4, IL-6, and is protective against COVID-19 infection reducing mortality rates (74, 75). Collinsella also occurred at significantly differential levels in GOS piglets as opposed to non-GOS piglets although any true link between GOS feeding to gestational sows and occurrence of Collinsella in piglets requires further research. In RVA positive piglets two OTUs attributed to Clostridiales occurred at significantly differential levels possibly indicating how enteric viruses can favor potential pathogens as opposed to beneficial community members (69, 76) (Figure 5). Indeed, RVA infection favors shifts in ileal microbiome structure with a significant increase in mucin digesting Bacteroides as verified by this study in RVA positive piglets (76).
Conclusions
This study is one of the first to demonstrate that GOS supplementation to sows during gestation significantly increases RVA specific IgG and IgA in colostrum, which confers immunity to neonates and reduces infectivity presumably through the effect of GOS on entero-mammary pathways. The implications for commercial pig farming are that gestational fed GOS could be used as a useful adjunct to other anti-virals and/or cleaning with efficacious detergents that can reduce infectivity in neonates by 20%, which would represent a significant economic gain for commercial herds. Whilst there was no demonstrable effect on microbial diversity of GOS in sows and their offspring, it should be considered that only fecal samples were collected in this study and may not be a true proxy of intestinal contents, which may be different in community membership and structure. In this respect, more research is required. However, non-GOS sows compared with GOS fed sows had a significant and increased differential abundance of potentially pathogenic organisms Treponema and Clostridiales suggesting GOS modulates the maternal microbiome by suppressing these organisms. The occurrence of Collinsella at significantly differential levels in GOS and RVA negative piglet fecal samples as opposed to the occurrence of Clostridiales and Bacteroides in non-GOS and RVA positive samples suggests modulation of the piglet microbiome through gestational feeding with GOS. Nevertheless, any true link between gestational GOS feeding to sows and occurrence of viral suppressing Collinsella in piglets, or indeed any other member of the microbiota requires further research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA884280.
Ethics statement
The animal study was reviewed and approved by the Farm Veterinary Consultant and by the University of Nottingham Ethics Committee.
Author contributions
AL, IC, and KM: conceptualization. AL, LL, and PC: methodology. AL: bioinformatics, formal analyses, and writing–original draft. AL, LL, PC, IC, and KM: validation. AL and KM: investigation. IC and KM: writing–review and editing. IC: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Saputo Dairy UK (formerly Dairy Crest), 5 The Heights, Brooklands, Weybridge, Surrey, KT13 0NY under funding code RG35FQ.
Acknowledgments
We would like to wholeheartedly thank Mr. Brian Bainbridge and his staff of Worsall Manor Farms Limited, Worsall Manor, Worsall, Yarm-On-Tees, TS15 9PJ for administrating GOS into commercial farm animal diets and sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Estes M, Kapikian A. “Rotaviruses”. In:Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, , editors. Fields Virology. Philadelphia, PA: Kluwer Health/Lippincott, Williams and Wilkins (2007). p. 1917–74.
2. Vlasova AN, Amimo JO, Saif LJ. Porcine rotaviruses: Epidemiology, immune responses and control strategies. Viruses. (2017) 9:E48. doi: 10.3390/v9030048
3. Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR. et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus classification working group (RCWG). Arch Virol. (2011) 156:1397–413. doi: 10.1007/s00705-011-1006-z
4. Estes MK, Kang G, Zeng CQ, Crawford SE, Ciarlet M. Pathogenesis of rotavirus gastroenteritis. Novartis Found Symp. (2001) 238:82–96. doi: 10.1002/0470846534.ch6
5. Azagra-Boronat I, Massot-Cladera M, Knipping K. van't Land B, Stahl Bernd, Garssen J, et al. Supplementation with 2'-FL and sc GOS/IcFOS ameliorates Rotavirus-induced diarrhea in suckling rats. Front Cell Infect Microbiol. (2018) 8:372. doi: 10.3389/fcimb.2018.00372
6. Svensmark B, Jorsal SE, Nielsen K, Willeberg P. Epidemiological studies of piglet diarrhoea in intensively managed Danish sow herds. I Pre-weaning diarrhoea Acta Vet Scand. (1989) 30:43–53. doi: 10.1186/BF03548067
7. Chandler-Bostock R, Hancox LR, Nawaz S, Watts O, Iturriza-Gomara M, Mellits KH. Genetic diversity of porcine group A rotavirus strains in the UK. Vet Microbiol. (2014) 173:27–37. doi: 10.1016/j.vetmic.2014.06.030
8. Saif LJ, Fernandez FM. Group A rotavirus veterinary vaccines. J Infect Dis. (1996) 174:S98–S106. doi: 10.1093/infdis/174.Supplement_1.S98
9. Azevedo M. Vlasova, AN, Saif LJ. Human rotavirus virus-like particle vaccines evaluated in a neonatal gnotobiotic pig model of human rotavirus disease. Expert Rev Vaccines. (2013) 12:169–81. doi: 10.1586/erv.13.3
10. Chandler-Bostock R, Mellits KH. Efficacy of disinfectants against porcine rotavirus in the presence and absence of organic matter. Lett Appl Microbiol. (2015) 61:538–43. doi: 10.1111/lam.12502
11. Hancox LR, Le Bon M, Dodd CE, Mellits KH. Inclusion of detergent in a cleaning regime and effect on microbial load in livestock housing. Vet Rec. (2013) 173:167. doi: 10.1136/vr.101392
12. Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. (2006) 54:7471–80. doi: 10.1021/jf0615810
13. Albrecht S, Lane JA, Mariño K, Al Busadah KA, Carrington SD, Hickey RM, et al. A comparative study of free oligosaccharides in the milk of domestic animals. Br J Nutr. (2014) 111:1313–28. doi: 10.1017/S0007114513003772
14. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. (2012) 22:1147–62. doi: 10.1093/glycob/cws074
15. Donovan SM1, Comstock SS. Human milk oligosaccharides influence neonatal mucosal and systemic immunity. Ann Nutr Metab. (2016) 69:42–51. doi: 10.1159/000452818
16. Urashima T, Saito T, Nakamura T, Messer M. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj J. (2001) 18:357–71. doi: 10.1023/A:1014881913541
17. Urashima T, Taufik E, Fukuda K, Asakuma S. Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci Biotechnol Biochem. (2013) 77:455–66. doi: 10.1271/bbb.120810
18. Difilippo E, Bettonvil M, Willems RH, Braber S, Fink-Gremmels J, Jeurink PV, et al. Oligosaccharides in urine, blood, and feces of piglets fed milk replacer containing galacto-oligosaccharides. J Agric Food Chem. (2015) 63:10862–72. doi: 10.1021/acs.jafc.5b04449
19. Difilippo E, Pan F, Logtenberg M, Willems RH, Braber S, Fink-Gremmels J, et al. (2016). In vitro fermentation of porcine milk oligosaccharides and galacto-oligosaccharides using piglet fecal inoculum. J Agric Food Chem. (2016) 64:2127–33. doi: 10.1021/acs.jafc.5b05384
20. Lee A, Mansbridge SC, Liang L, Connerton IF, Mellits KH. Galacto-oligosaccharides increase the abundance of beneficial probiotic bacteria and improve gut architecture and goblet cell expression in poorly performing piglets, but not performance. Animals. (2023) 13:230. doi: 10.3390/ani13020230
21. Yan YL, Hu Y, Simpson DJ, Gänzle MG. Enzymatic synthesis and purification of galactosylated chitosan oligosaccharides reducing adhesion of enterotoxigenic Escherichia coli K88. J Agric Food Chem. (2017) 65:5142–50. doi: 10.1021/acs.jafc.7b01741
22. Bouwhuis MA, McDonnell MJ, Sweeney T, Mukhopadhya A, O'Shea CJ, O'Doherty JV. Seaweed extracts and galacto-oligosaccharides improve intestinal health in pigs following Salmonella Typhimurium challenge. Animal. (2017) 11:1488–96. doi: 10.1017/S1751731117000118
23. Azagra-Boronat I, Massot-Cladera M, Knipping K, Van Land B, Tims S, Stahl B, et al. Oligosaccharides modulate Rotavirus-associated dysbiosis and TLR gene expression in neonatal rats. Cells. (2019) 8:876. doi: 10.3390/cells8080876
24. Hester SN, Chen X, Li M, Monaco MH, Comstock SS, Kuhlenschmidt TB, et al. Human milk oligosaccharides inhibit rotavirus infectivity in vitro and in acutely infected piglets. Br J Nutr. (2013) 110:1233–42. doi: 10.1017/S0007114513000391
25. Laucirica DR, Triantis V, Schoemaker R, Estes MK, Ramani S. Milk oligosaccharides inhibit human rotavirus infectivity in MA104 cells. J Nutr. (2017) 147:1709–14. doi: 10.3945/jn.116.246090
26. Li M, Monaco MH, Wang M, Comstock SS, Kuhlenschmmidt TB, Fahey GC, et al. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic bacteria. ISME J. (2014) 8:1609–20. doi: 10.1038/ismej.2014.10
27. Asensi MT, Martínez-Costa C, Buesa J. Anti-rotavirus antibodies in Human milk: Quantification and neutralizing activity. J Pediatr Gastroenterol Nutr. (2006) 42:560–7. doi: 10.1097/01.mpg.0000221892.59371.b3
28. Wu Y, Zhang X, Tao S, Pi Y, Han D, Ye H. Maternal supplementation with combined galactooligosaccharides and casein glycomacropeptides modulated microbial colonization and intestinal development of neonatal piglets. J Funct Foods. (2020) 74:104170. doi: 10.1016/j.jff.2020.104170
29. Kreuzer S, Machnowska P, Aßmus, Sieber M, Pieper R, Schmidt MFG, et al. Feeding of the probiotic bacterium Enterococcus faecium NCIMB 10415 differentially affects shedding of enteric viruses in pigs. Vet Res. (2012) 43:58. doi: 10.1186/1297-9716-43-58
30. Ramanakumar AV, Thomann P, Candeias JM, Ferreira S, Villa LL, Franco EL. Use of the normalized absorbance ratio as an internal standardization approach to minimize measurement error in enzyme-linked immunosorbent assays for diagnosis of human papillomavirus infection. J Clin Microbiol. (2010) 48:791–6. doi: 10.1128/JCM.00844-09
31. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. (2011) 108:4516–22. doi: 10.1073/pnas.1000080107
32. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. (2013) 79:5112–20. doi: 10.1128/AEM.01043-13
33. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
34. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. (2007) 35:7188–96. doi: 10.1093/nar/gkm864
35. Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere. (2017) 2:17. doi: 10.1128/mSphereDirect.00073-17
36. Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. (2014) 42:D633–42. doi: 10.1093/nar/gkt1244
37. Wang Q, Garrity GM, Tiedje JM, Cole J. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
38. Development Core Team R,. R: A Language Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. (2013). Available online at: http://www.R-project.org (accessed December 15, 2017).
39. Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples). Biometrika. (1965) 52:591–611. doi: 10.1093/biomet/52.3-4.591
41. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. (1984) 11:265–70.
42. Shannon CE. A mathematical theory of communication. Bell System Tech J. (1948) 27:379–423. doi: 10.1002/j.1538-7305.1948.tb00917.x
43. Chao A, Lee SM. Estimating the number of classes via sample coverage. J Am Stat Assoc. (1992) 87:210–17. doi: 10.1080/01621459.1992.10475194
44. Yue JC, Clayton MK, A. similarity measure based on species proportions. Commun Stat Theor Methods. (2005) 34:2123–31. doi: 10.1080/STA-200066418
45. Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. (1957) 27:326–49. doi: 10.2307/1942268
46. Jaccard P. Etude comparative de la distribution florale dans une portion des Alpe's et du Jura. Bullet Soci Vaudoise Sci Nat. (1901) 37:547–79. doi: 10.5169/seals-266450
47. Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. (1992) 131:479–91. doi: 10.1093/genetics/131.2.479
48. Anderson MJ, A. new method for non-parametric multivariate analysis of variance. Austral Ecol. (2001) 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x
49. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
50. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
51. Good IJ, Toulmin GH. The number of new species, and the increase in population coverage, when a sample is increased. Biometrika. (1956) 43:45–63. doi: 10.1093/biomet/43.1-2.45
52. Rodríguez JM, Fernández L, Verhasselt V. The gut-breast axis: Programming health for life. Nutrients. (2021) 13:606. doi: 10.3390/nu13020606
53. Furukawa S, Kuroda Y, Sugiyama A. A comparison of the histological structure of the placenta in experimental animals. J Toxicol Paothol. (2014) 27:11–18. doi: 10.1293/tox.2013-0060
54. Salmon, H. Mammary gland immunology and neonate protection in pigs. In:Clegg RA, , editor. Advances in Experimental Medicine and Biology. Biology of the Mammary Gland: Homing of lymphocytes into the MG. Boston, MA: Springer (2002). p. 279–86. doi: 10.1007/0-306-46832-8_32
55. Weström BR, Ohlsson BG, Svendsen J, Tagesson C, Karlsson BW. Intestinal transmission of macromolecules (BSA and FITC-Dextran) in the neonatal pig: enhancing effect of colostrum, proteins and proteinase inhibitors. Biol Neonate. (1985) 47:359–66. doi: 10.1159/000242140
56. Devillers N, Le Dividich J, Prunier A. Influence of colostrum intake on piglet survival and immunity. Animal. (2011) 5:1605–12. doi: 10.1017/S175173111100067X
57. Alonge S, Aiudi GG, Lacalandra GM, Leoci MM. Pre-and Probiotics to increase the immune power of colostrum in dogs. Front Vet Sci. (2020) 7:570414. doi: 10.3389/fvets.2020.570414
58. Sanidad KZ, Amir M, Ananthanarayanan, Singaraju A, Shiland NB, Hong HS, et al. Maternal gut microbiome–induced IgG regulates neonatal gut microbiome and immunity. Sci Immunol. (2022) 7:eabh3816 doi: 10.1126/sciimmunol.abh3816
59. Usami K, Niimi K, Matsuo A, Suyama Y, Sakai Y, Sato S, et al. The gut microbiota induces Peyer's-patch-dependent secretion of maternal IgA into milk. Cell Rep. (2021) 36:109655. doi: 10.1016/j.celrep.2021.109655
60. Cortez V, Schultz-Cherry S. The role of goblet cells in viral pathogenesis. FEBS J. (2021) 288:7060–72. doi: 10.1111/febs.15731
61. Inoue R, Tsukahara T. Composition and physiological functions of the porcine colostrum. Anim Sci J. (2021) 92:e13618 doi: 10.1111/asj.13618
62. Benfield DA, Stotz I, Moore R, McAdaragh JP. Shedding of rotavirus in feces of sows before and after farrowing. J Clin Microbiol. (1982) 16:186–90. doi: 10.1128/jcm.16.1.186-190.1982
63. Woode GN, Bohl EH. Rotavirus. In:Leman AD, Glock RD, Mengeling WL, Penny RHE, Scholl E, and Straw B, , editors. Diseases of Swine. Ames, IA: Iowa State University Press (1981). p. 310–22.
64. Lührmann A, Ovadenko K, Hellmich J, Sudendey C, Belik V, Zentek J. et al. Characterization of the fecal microbiota of sows and their offspring from German commercial pig farms. PLoS ONE. (2021) 16:e0256112. doi: 10.1371/journal.pone.0256112
65. Wang X, Tsai T, Deng F, Wei X, Chai J, Knapp J. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome. (2019) 7:109. doi: 10.1186/s40168-019-0721-7
66. Cheng C, Wei H, Yu H, Xu C, Jiang S, Peng J. Co-occurrence of early gut colonisation in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe. (2018) 49:30–40. doi: 10.1016/j.anaerobe.2017.12.002
67. Zhang L, Mu C, He X, Su Y, Mao S, Zhang J, et al. Effects of dietary fibre source on microbiota composition in the large intestine of suckling piglets. FEMS Microbiol Lett. (2016) 363:fnw138. doi: 10.1093/femsle/fnw138
68. Lee A, Le Bon M, Connerton IF, Mellits KH. Common colonic community indicators of the suckling pig microbiota where diversity and abundance correlate with performance. FEMS Microbiol Ecol. (2022) 98:48. doi: 10.1093/femsec/fiac048
69. Tan Z, Dong W, Ding Y, Ding X, Zhang Q, Jiang L. Porcine epidemic diarrhea altered colonic microbiota communities in suckling piglets. Genes. (2020) 11:44. doi: 10.3390/genes11010044
70. Svartström O, Karlsson F, Fellstr?m C, Pringle M. Characterization of Treponema spp. isolates from pigs with ear necrosis and shoulder ulcers. Vet Microbiol. (2013) 166:617–23. doi: 10.1016/j.vetmic.2013.07.005
71. Songer JG, Uzal FA. Clostridial enteric infections in pigs. J Vet Diagn Invest. (2005) 17:528–36. doi: 10.1177/104063870501700602
72. Aliberti J. Immunity and tolerance induced by intestinal mucosal dendritic cells. Mediat Inflamm. (2016) 2016:3104727. doi: 10.1155/2016/3104727
73. Twitchell EL, Tin C, Wen K, Zhang H, Becker-Dreps S, Azcarate-Peril MA. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut Pathog. (2016) 8:51. doi: 10.1186/s13099-016-0136-y
74. Ko WK, Lee SH, Kim SJ, Jo MJ, Kumar H, Han IB, et al. Anti-inflammatory effects of ursodeoxycholic acid by lipopolysaccharide-stimulated inflammatory responses in RAW 264.7 macrophages. PLoS ONE. (2017) 12:e0180673. doi: 10.1371/journal.pone.0180673
75. Hirayama M, Nishiwaki H, Hamaguchi T, Ito M, Ueyama J, Maeda T, et al. Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PloS ONE. (2021) 16:e0260451. doi: 10.1371/journal.pone.0260451
Keywords: rotavirus, microbiota, pigs, galacto-oligosaccharides, antibodies, colostrum
Citation: Lee A, Liang L, Connerton PL, Connerton IF and Mellits KH (2023) Galacto-oligosaccharides fed during gestation increase Rotavirus A specific antibodies in sow colostrum, modulate the microbiome, and reduce infectivity in neonatal piglets in a commercial farm setting. Front. Vet. Sci. 10:1118302. doi: 10.3389/fvets.2023.1118302
Received: 07 December 2022; Accepted: 17 January 2023;
Published: 07 February 2023.
Edited by:
Juan D. Latorre, University of Arkansas, United StatesReviewed by:
Adil Sabr Al-Ogaili, Middle Technical University, IraqFernanda Rosa, Texas Tech University, United States
Copyright © 2023 Lee, Liang, Connerton, Connerton and Mellits. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam Lee,  YWRhbS5sZWVAbm90dGluZ2hhbS5hYy51aw==
YWRhbS5sZWVAbm90dGluZ2hhbS5hYy51aw==
 Adam Lee
Adam Lee Lu Liang
Lu Liang Kenneth H. Mellits
Kenneth H. Mellits