- 1Key Laboratory of Clinical Diagnosis and Treatment Technology in Animal Diseases, Ministry of Agriculture, College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot, China
- 2Wushen Animal Disease Prevention and Control Center, Ordos, China
Haemonchus contortus is a highly pathogenic and economically important parasitic nematode that affects small ruminants worldwide. While omics studies hold great promise, there are fewer research tools available for analyzing subsequent gene function studies. RNA interference (RNAi) technology offers a solution to this problem, as it especially allows for the knockout or shutting off of the expression of specific genes. As a result, RNAi technology has been widely used to explore gene function and disease treatment research. In this study, we reviewed the latest advancements in RNAi research on Haemonchus contortus in China, with the aim of providing a reference for the identification of key genes involved in growth and development, anthelmintic resistance, diagnostic markers, and diagnostic drug targets for the treatment of Haemonchus contortus.
1. Introduction
Haemonchus contortus (H. contortus) is a type of parasitic nematode that belongs to the Haemonchus spp. (Trichostrongylidae) and is a major threat to the economically important gastrointestinal nematode of small ruminants. It primarily infects the abomasum, and when 2,000 or more worms are present there they can cause severe harm. These worms can suck up to 30 mL of blood per day, in addition to the blood loss caused by their departure. This can result in severe infection in livestock and cause many issues, such as anemia, weight loss, and even death (1). It is believed that H. contortus lays more eggs than other nematodes of the Trichostrongylidae family, with females laying 5,000–10,000 eggs every day. Third-stage larvae (L3) are more resistant and can generally survive for 3 months in pastures, and they can remain dormant for up to 1 year in adverse environments, leading to the contamination of pastures and the environment. Recent epidemiological studies showed that the positivity rate of H. contortus infection in sheep in Iran, India, Egypt, Brazil, and Ethiopia was 84.6, 44.7, 38.8, 76.4, and 68.75%, respectively (2–6). These developing countries have experienced significant losses in their dominant animal product industries due to H. contortus infections.
The prevention and treatment of H. contortus can be accomplished through the use of anthelmintics, with the selection and breeding of livestock breeds that are resistant to parasitic infection, biological control methods, and improved feed management (7, 8). Among these methods, anthelmintics are the most effective and economical means of control. Since the invention of highly effective and broad-spectrum anthelmintics, their prolonged use has led to serious resistance problems. Researchers have made extensive efforts to address these problems (9–11). With the release of the complete genome of H. contortus in 2013 (chromosome-level assembly was completed in 2019) (12) and the advancement of post-genomics in the last 15 years (13, 14), there are numerous molecular targets that can be exploited for control. Although these molecular targets have the potential to control H. contortus, they still require the evaluation of some techniques before they can be used as drug screening or vaccine candidates (15).
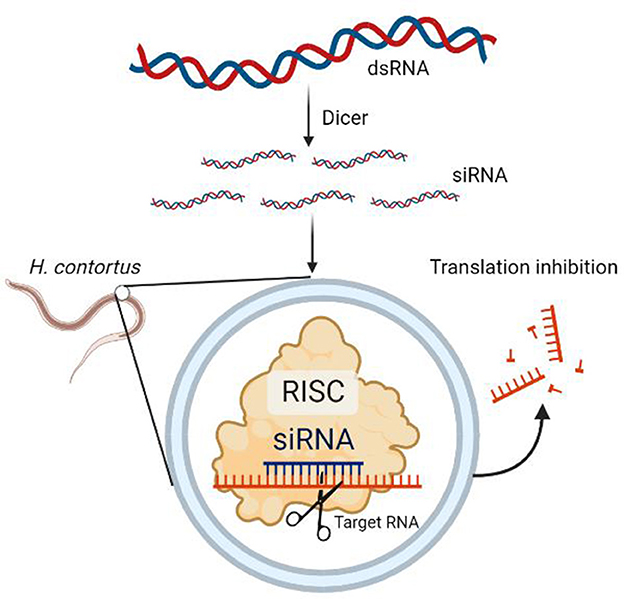
RNA interference (RNAi) is an important tool for evaluating the gene function. It is based on the principle that exogenous genes, such as viral genes, artificially transferred genes, transposons, and so on, are randomly integrated into the host cell genome and often produce some double-stranded RNAs (dsRNAs), which are transcribed using the host cell. Dicer, a nucleic acid endonuclease in the host cell, cleaves the dsRNA into multiple small fragments of RNA, commonly known as small interfering RNA (siRNA), with a specific length and structure. These siRNA fragments are unwound into a righteous strand and an antisense strand by intracellular RNA decapping enzymes and are then bound by antisense siRNA to some enzymes in vivo. The RNA-induced silencing complex (RISC) specifically binds to the homologous region of exogenous mRNA and functions as a nuclease, cleaving the mRNA at the binding site, which is the area where the antisense strand of the siRNA is complementary to it. The cleaved and fragmented mRNA is then degraded, leading to a degradation response in the host cell against these mRNAs (Figure 1).
In recent years, the availability of genomic and transcriptomic data for H. contortus has highlighted the need for functional genomics techniques. Since there are limited methods to directly study gene function in nematodes, RNAi and its related technologies have been developed over time to successfully identify key genes, provide a rational method to develop new control strategies, and apply them in relevant gene function studies (16). Comparative genomic analyses, especially using data from free-living Caenorhabditis elegans (C. elegans), can help predict gene function (17). In this study, we reviewed recent studies on the RNAi technique applied to H. contortus development genes and drug target screening and discussed the problems and limitations of the technique. It provides a theoretical basis for further in-depth research on the application of RNAi technology in parasitic nematodes.
2. Research progress of RNAi for Haemonchus contortus
2.1. Key genes for growth and development
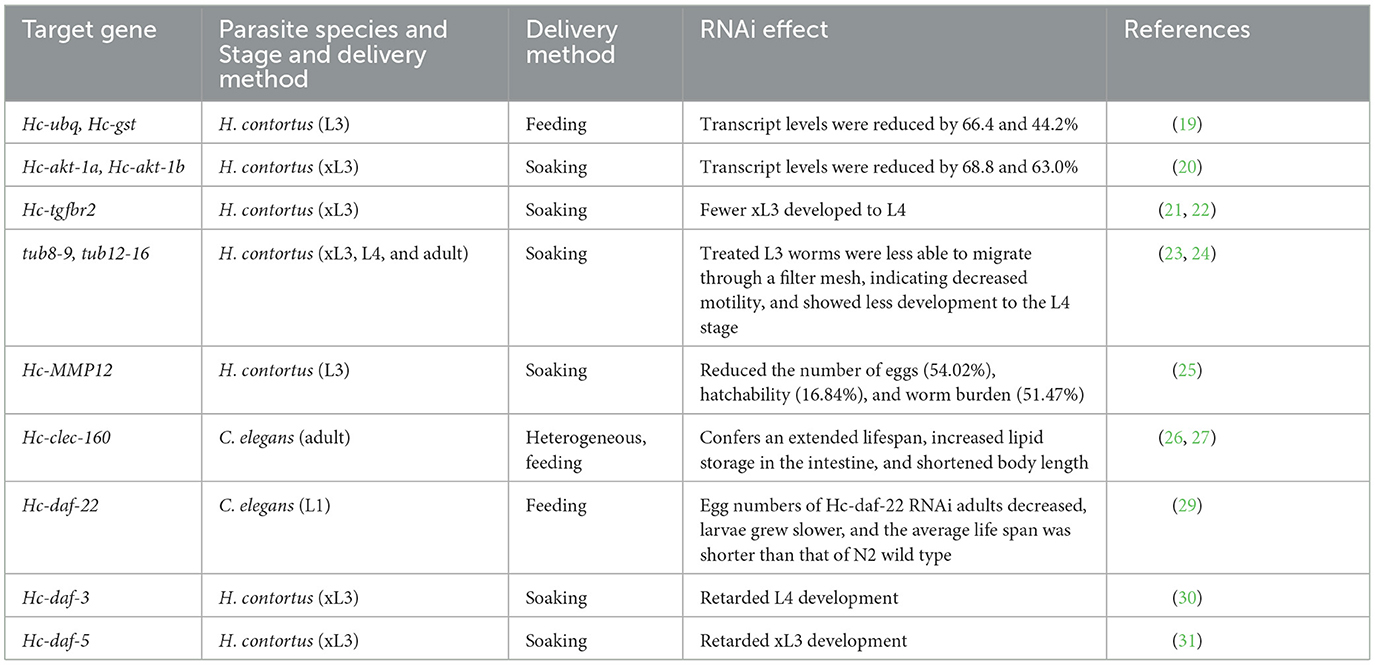
The exploration of the free-living and parasitic stages of H. contortus has never ceased. Free-living third-stage larvae (L3) of H. contortus can tolerate extreme weather conditions such as desiccation (18). To explore the molecular mechanism of desiccation survival, researchers screened two desiccation-related differentially expressed genes, Hc-ubq and Hc-gst, in L3 using RNA-seq (19). Quantitative RT-PCR results showed that expression was upregulated in L3 in desiccation environments, and silencing both Hc-ubq and Hc-gst reduced the survival rate of L3. Silencing of the homologs of C. elegans, Ce-ubq-2, and Ce-gst-7 also showed higher larval mortality, which suggests that ubq and gst may play an important role in nematode dehydration tolerance. It is reported that researchers significantly reduced the transcript abundance of two transcript isoforms of the serine/threonine-specific protein kinase, AKT, Hc-akt-1a, and Hc-akt-1b, by using the soaking RNAi method (20). The results showed that silencing these two genes significantly reduced in vitro larval stage development. In addition, research shows that silencing of the Hc-tgfbr2 genes significantly reduced the development of L3–L4 in vitro (21, 22). These genes play an important role in regulating development and the transition from free-living to parasitic stages (Figure 2).
The silencing of genes for motility at different stages of H. contortus was also reported by Andrew C Kotze, who found that in vitro dsRNA soaking of two β-microtubulin genes, tub8-9 and tub12-16, restricted motility in L3, but adults did not exhibit the phenomenon (23, 24). This may be because phenotypic disruption is caused by multiple factors, including the transient nature of gene repression and the variability of RNAi action at different developmental stages. The different stages of RNAi also simultaneously demonstrate that dsRNA uptake is non-specific and that L3 can silence specific genes without normal mouthparts. Effective silencing of the Dim-1 gene in H. contortus L3 resulted in reduced migration of L3 and slowed the larval development from L3 to early L4.
Matrix metalloprotease 12A (MMP-12) is known to have important roles during embryonic development, organ morphogenesis, and pathological processes in animals. Recent studies used three different siRNAs targeting the Hc-MMP-12 gene for silencing. Compared to the snRNA-treated control, siRNA-2 silencing of the Hc-MMP-12 gene resulted in a shorter H. contortus length (25). This was in addition to a 54.02% egg reduction, a 16.84% reduction in the hatching rate, and a 51.47% reduction in infection intensity in the treated group compared to the control. A new gene, Hc-clec-160, was screened by RT-PCR. To characterize the function of the Hc-clec-160 gene, earlier studies on the interference of clec-160 in C. elegans using heterogeneous RNAi resulted in shortened body length and reduced egg size, indicating that clec-160 plays an important role in the growth and reproduction of this helminth (26). Using the same approach, a new gene called the Hc-fau gene was also shown to play an important role in regulating the life cycle and reproduction in H. contortus (27).
The dauer of H. contortus is similar to that of C. elegans and hookworm in vivo (28). Growth and development cease at a precise point in the L3; metabolism levels are low, and seasonal variations are evident. Protection of the sheath and the dauer are the main survival strategies in the free-living and parasitic stages in the absence of the host or adverse conditions. The discovery of the steroid ligands of DAF opened a new avenue of small molecule physiology in C. elegans. These signaling pathways are present in parasitic nematodes and have been indicated to play a role in development regulation and the formation of L3 during the infectious process. Hc-daf is similar to ce-daf in affecting worm development and competence. This study demonstrated that daf-3, daf-5, daf-12, daf-16 and daf-22 inhibited the development of L3 into L4 after RNAi (29–34). If we can obtain some clues as to the genes or signal transduction mechanisms related to the dauer and prevent its stagnation in the host, we will solve the fundamental occurrence of spring H. contortus outbreaks (Table 1).
2.2. Screening of anthelmintic resistance genes and anthelmintic targets
The problem of anthelmintic resistance in H. contortus is a serious threat to parasitic control, and in some areas, anthelmintics such as macrolides and benzimidazoles have completely lost their deworming effect on gastrointestinal nematodes in livestock. The identification of alternative anthelmintic targets is an increasingly important task. Using RNAi for the functional identification of relevant genes and the search for new compounds that display antagonistic activity against these genes to enhance anthelmintic efficacy opens new prospects (35). An RNAi-based functional validation assay revealed HCON_00143950 as an IVM (ivermectin)-tolerance-associated gene in H. contortus. The possible role of this gene in IVM resistance could be the detoxification of xenobiotics in phase I of xenobiotic metabolism (36). In France, researchers performed an RNAi of the nhr-8 gene in an ivermectin-resistant strain of H. contortus (37). The nhr-8 silencing increased the susceptibility of the resistant strain to ivermectin and mediated the upregulation of genes associated with ivermectin detoxification. It is also reported that silencing the Cyt-P450 gene resulted in reduced resistance to ivermectin, which led to larval pharyngeal paralysis and thus starvation (36). Meanwhile, silencing the Cyt-P450 gene increased the sensitivity of H. contortus to ivermectin; these results also provide new approaches for further understanding ivermectin targets.
Earlier studies have demonstrated that normal movement can be restored in H. contortus by using RNAi on HcGlu Cl-α3 through the use of C. elegans as a model organism (38). Their results supported the claim that ivermectin has a paralyzing effect on parasitic nematodes by stimulating channels in the nervous motor system. Cholinergic agonists, such as levamisole and pyrantel, are widely used to treat nematode infections as anthelmintics. These drugs cause spastic paralysis in nematode parasites by activating acetylcholine receptors (AChRs) present in their body wall muscles. The critical role of acr-8 in vivo anthelmintic sensitivity is substantiated by the successful demonstration of RNAi gene silencing in Hc-acr-8, which reduced the sensitivity of H. contortus larvae to levamisole. Pyrantel sensitivity remained unchanged, thus providing new evidence for distinct modes of action of these important anthelmintics in parasitic species vs. C. elegans (39). As one of the largest protein families, they regulate nearly all processes within the cell and are considered important drug targets. Numerous studies have been conducted on inhibitors for protein kinases (protein kinases, PKs), leading to a wealth of compounds that target PKs, which have the potential to be lead anthelmintics. Some researchers used RNAi to identify 35 PK compounds against C. elegans and H. contortus. Of these, 18 compounds showed efficacy against C. elegans, and six other compounds also showed efficacy against at least one parasitic species (35). RNAi has great potential as a screening tool for identifying potential anthelmintic targets.
3. Analysis of the limitations of RNAi in the study on Haemonchus contortus
RNAi has been widely used in the functional identification genes of H. contortus. However, its problems are also evident and constantly arise in the following three respects.
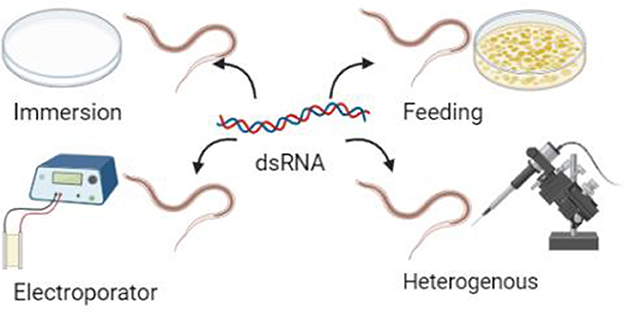
First, the sensitivity of H. contortus to RNAi varies with dsRNA delivery methods and target genes and is less reproducible. The results of many studies have shown that commonly used immersion, feeding, and injection methods can produce RNAi phenomena in H. contortus, but the immersion method is less efficient and more costly, and the storage time of larvae and batch effects have a significant impact on the results (40). The heterogeneous RNAi makes it difficult to accurately deliver dsRNA to specific sites and can easily lead to cellular damage. The feeding method is still the most widely used and is more likely to produce phenotypic effects and be easy to observe (41). When dsRNA or siRNA is delivered using electroporation, the phenotypic effects cannot be examined due to the high mortality of the treated larvae (42, 43). RNAi may inhibit gene expression under certain conditions. However, this only works on a limited number of genes, and in some cases, the effect is negligible, which may reflect differences in dsRNA uptake and transport to and from cells in H. contortus. The efficiency of gene interference in H. contortus has no particularly clear correlation with the gene itself or the delivery method; however, existing research found that gene interference may only occur in certain cell types. For example, the C. elegans rde-4 null mutants are amenable to RNAi with siRNA but not dsRNA (44). If parasitic nematodes lack rde-4 or a functional equivalent, RNAi should still be possible with siRNA delivery. They can still be delivered by siRNA; that is, siRNA is more effective than dsRNA. It is very likely that the ortholog of rde-4 or some undefined mechanism is still in operation. These examples can show that the efficiency of gene interference will become more controllable with further information and a more detailed search of genome sequencing projects. The insights gained from studies on other worms may also help optimize dsRNA delivery and determine which developmental stages may be best targeted.
Second, a single developmental stage for RNAi will make it difficult to assess phenotypes that may only be expressed at later developmental stages, as reliable culture systems have not been established. Further research is needed to develop reliable phenotyping systems. In vitro, reagents such as hypochlorite are widely used for sterilization and unsheathing of L3, which can affect larvae and subsequent stages of development even at very low concentrations (45). High concentrations of dsRNA delivery may be cytotoxic, so it may be necessary for us to determine the optimal concentration for each gene tested. It has also been reported that some of the phenotypes generated by RNAi can be replicated by chemical inhibitor target activity, providing proof of the principle that RNAi can replicate chemo-suppressive effects and confirming that RNAi screening is a potentially powerful and effective method for identifying novel drug targets. Based on RNAi, we need to further investigate the mechanism of these genes and improve the culture system of H. contortus to detect the long-term effects of RNAi; only then can RNAi become a reliable method for gene function detection.
In addition, there is also a large body of data indicating limitations in identifying potential H. contortus genes as a predictive model based on C. elegans data, with RNAi-mediated silencing being reliable for some genes but not others. H. contortus-specific genes are mainly located on autosomes, which have a higher recombination rate, and evolutionary mutations mainly occur in these regions, possibly being promoted by higher recombination. When these genes are silenced by RNAi, only a few genes can alter the phenotype, and in C. elegans, it is more difficult for us to observe the occurrence and alteration of these phenotypes. When conditions allow, we can further perform protein analyses to examine the decrease in mRNA and protein levels. Therefore, we need to be extra cautious and careful while using C. elegans as a model organism (46, 47). The RNAi technology will be more specific and targeted if these challenges are successfully overcome.
4. Discussion and perspective
The current database has a large amount of genomic and transcriptomic data, and many studies have identified certain important genes that can successfully eliminate H. contortus. We currently need to move forward to the next level; RNAi technology has significantly improved its efficacy and ease of use and can be used to develop a more reliable and functional genomics platform. This can also be combined with technologies such as CRISPR/Cas and will provide a more rational approach to gene function identification. In vivo experiments have currently demonstrated that site-specific targeting of genes can improve their success rate, making in vivo RNAi a feasible option for identifying essential gene functions during infection, which is particularly important for elucidating non-conserved genes and speculating on the genes that play a role in influencing host immune responses. However, greater efforts are still needed to improve our understanding of H. contortus biology and to identify new approaches for parasitic control and prevention. The conclusion that can be drawn from the outlined studies is that RNAi is possible in parasitic nematodes and, if effectively utilized, can be a valuable tool for screening and validating new gene and vaccine targets.
Author contributions
SH: conceptualization. BH: writing—original draft preparation and writing—review and editing. YH: visualization. YH, BB, and SH: supervision, project administration, and funding acquisition. All authors read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ordos Science and Technology Plan (2022EEDSKJZDZX026) and the Control Technology of Animal Diseases in Wushen Banner.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Flay KJ, Hill FI, Muguiro DH, A. Review: Haemonchus contortus infection in pasture-based sheep production systems, with a focus on the pathogenesis of anaemia and changes in haematological parameters. Animals. (2022) 12:1238. doi: 10.3390/ani12101238
2. Hosseinnezhad H, Sharifdini M, Ashrafi K, Atrkar Roushan Z, Mirjalali H, Rahmati B. Trichostrongyloid nematodes in ruminants of northern Iran: prevalence and molecular analysis. BMC Vet Res. (2021) 17:371. doi: 10.1186/s12917-021-03086-3
3. Bhat RA, Tak H, Bhat BA, Dar JA, Ahmad R. Gastrointestinal helminth parasites of wild ungulates in Hirpora Wildlife Sanctuary, Kashmir, India. J Parasit Dis. (2022) 46:804–10. doi: 10.1007/s12639-022-01493-3
4. Abbas I, Hildreth MB. Trichostrongyle infections in domestic ruminants from Egypt: a systematic review and meta-analysis. Vet Parasitol Reg Stud Reports. (2022) 34:100761. doi: 10.1016/j.vprsr.2022.100761
5. Moreira RT, Mota ALAA, Gonçalves VSP, Rocha GCD, Borges JRJ. Situation of and phenotypic markers of susceptibility to helminth infection among sheep on farms in the Brazilian cerrado biome. Rev Bras Parasitol Vet. (2021) 30:e021720. doi: 10.1590/s1984-296120201092
6. Habte A, Ibrahim N. Prevalence of Haemonchus contortus infection in sheep slaughtered at Jimma town municipal abattoir, Ethiopia. Trop Anim Health Prod. (2018) 50:1865–70. doi: 10.1007/s11250-018-1637-0
7. Naeem M, Iqbal Z, Roohi N. Ovine haemonchosis: a review. Trop Anim Health Prod. (2020) 53:19. doi: 10.1007/s11250-020-02439-8
8. Zajac AM, Garza J. Biology, Epidemiology, and control of gastrointestinal nematodes of small ruminants. Vet Clin North Am Food Anim Pract. (2020) 36:73–87. doi: 10.1016/j.cvfa.2019.12.005
9. Muchiut S, Fiel C, Lirón JP, Lloberas M, Ceriani C, Lorenzo R, et al. Population replacement of benzimidazole-resistant Haemonchus contortus with susceptible strains: evidence of changes in the resistance status. Parasitol Res. (2022) 121:2623–32. doi: 10.1007/s00436-022-07582-9
10. Estrada-Reyes ZM, Ogunade IM, Pech-Cervantes AA, Terrill TH. Copy number variant-based genome wide association study reveals immune-related genes associated with parasite resistance in a heritage sheep breed from the United States. Parasite Immunol. (2022) 44:e12943. doi: 10.1111/pim.12943
11. Hinney B, Wiedermann S, Kaiser W, Krücken J, Joachim A. Eprinomectin and moxidectin resistance of trichostrongyloids on a goat farm in Austria. Pathogens. (2022) 11:498. doi: 10.3390/pathogens11050498
12. Palevich N, Maclean PH, Baten A, Scott RW, Leathwick DM. The genome sequence of the anthelmintic-susceptible New Zealand Haemonchus contortus. Genome Biol Evol. (2019) 11:1965–70. doi: 10.1093/gbe/evz141
13. Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. (2013) 14:R88. doi: 10.1186/gb-2013-14-8-r88
14. Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. (2013) 14:R89. doi: 10.1186/gb-2013-14-8-r89
15. Geary TG. Haemonchus contortus: applications in drug discovery. Adv Parasitol. (2016) 93:429–63. doi: 10.1016/bs.apar.2016.02.013
16. Zawadzki JL, Presidente PJ, Meeusen EN, De Veer MJ, RNAi. in Haemonchus contortus: a potential method for target validation. Trends Parasitol. (2006) 22:495–9. doi: 10.1016/j.pt.2006.08.015
17. Britton C, Roberts B, Marks ND. Functional genomics tools for Haemonchus contortus and lessons from other helminths. Adv Parasitol. (2016) 93:599–623. doi: 10.1016/bs.apar.2016.02.017
18. Chylinski C, Lherminé E, Coquille M, Cabaret J. Desiccation tolerance of gastrointestinal nematode third-stage larvae: exploring the effects on survival and fitness. Parasitol Res. (2014) 113:2789–96. doi: 10.1007/s00436-014-3938-1
19. Yang Y, Ma Y, Chen X, Guo X, Yan B, Du A. Screening and analysis of Hc-ubq and Hc-gst related to desiccation survival of infective Haemonchus contortus larvae. Vet Parasitol. (2015) 210:179–85. doi: 10.1016/j.vetpar.2015.03.020
20. Di W, Gasser RB, He L, Li F, Liu X, Zhou C, et al. A serine/threonine-specific protein kinase of Haemonchus contortus with a role in the development. FASEB J. (2020) 34:2075–86. doi: 10.1096/fj.201900888RR
21. He L, Gasser RB, Korhonen PK Di W, Li F, Zhang H, et al. A TGF-β type I receptor-like molecule with a key functional role in Haemonchus contortus development. Int J Parasitol. (2018) 48:1023–33. doi: 10.1016/j.ijpara.2018.06.005
22. He L, Gasser RB, Li T, Di W, Li F, Zhang H, et al. A TGF-β type II receptor that associates with developmental transition in Haemonchus contortus in vitro. PLoS Negl Trop Dis. (2019) 13:e0007913. doi: 10.1371/journal.pntd.0007913
23. Kotze AC, Bagnall NH. RNA interference in Haemonchus contortus: suppression of beta-tubulin gene expression in L3, L4 and adult worms in vitro. Mol Biochem Parasitol. (2006) 145:101–10. doi: 10.1016/j.molbiopara.2005.09.012
24. Arenal A, Díaz A, Sprenger LK, Buzatti A, Fernandes MA, Dos Santos JN, et al. Gene silencing of Dim-1, a member of the disorganized muscle family, in Haemonchus contortus. Mol Biochem Parasitol. (2017) 211:71–4. doi: 10.1016/j.molbiopara.2016.10.004
25. Naqvi MA, Li H, Gao W, Naqvi SZ, Jamil T, Aimulajiang K, et al. Haemonchus contortus: siRNA mediated knockdown of matrix metalloproteinase 12A (MMP-12) results in reduction of infectivity. Parasit Vectors. (2020) 13:151. doi: 10.1186/s13071-020-04025-1
26. Yan B, Guo X, Zhou Q, Yang Y, Chen X, Sun W, et al. Hc-fau, a novel gene regulating diapause in the nematode parasite Haemonchus contortus. Int J Parasitol. (2014) 44:775–86. doi: 10.1016/j.ijpara.2014.05.011
27. Zhang L, Mou L, Chen X, Yang Y, Hu M, Li X, et al. Identification and preliminary characterization of Hc-clec-160, a novel C-type lectin domain-containing gene of the strongylid nematode Haemonchus contortus. Parasit Vectors. (2018) 11:430. doi: 10.1186/s13071-018-3005-3
28. Hotez P, Hawdon J, Schad GA. Hookworm larval infectivity, arrest and amphiparatenesis: the Caenorhabditis elegans Daf-c paradigm. Parasitol Today. (1993) 9:23–6. doi: 10.1016/0169-4758(93)90159-D
29. Huang Y, Zheng X, Zhang H, Ding H, Guo X, Yang Y, et al. Site-directed mutagenesis study revealed three important residues in Hc-DAF-22, a key enzyme regulating diapause of Haemonchus contortus. Front Microbiol. (2017) 8:2176. doi: 10.3389/fmicb.2017.02176
30. Di W, Liu L, Zhang T, Li F, He L, Wang C, et al. A DAF-3 co-Smad molecule functions in Haemonchus contortus development. Parasit Vectors. (2019) 12:609. doi: 10.1186/s13071-019-3855-3
31. Di W, Li F, He L, Wang C, Zhou C, Liu L, et al. A transcription factor DAF-5 functions in Haemonchus contortus development. Parasit Vectors. (2021) 14:529. doi: 10.1186/s13071-021-05036-2
32. Ma G, Wang T, Korhonen PK, Stroehlein AJ, Young ND, Gasser RB. Dauer signalling pathway model for Haemonchus contortus. Parasit Vectors. (2019) 12:187. doi: 10.1186/s13071-019-3419-6
33. Ma G, Wang T, Korhonen PK, Young ND, Nie S, Ang CS, et al. Dafachronic acid promotes larval development in Haemonchus contortus by modulating dauer signalling and lipid metabolism. PLoS Pathog. (2019) 15:e1007960. doi: 10.1371/journal.ppat.1007960
34. Lok JB, Kliewer SA, Mangelsdorf DJ. The ‘nuclear option' revisited: confirmation of Ss-daf-12 function and therapeutic potential in Strongyloides stercoralis and other parasitic nematode infections. Mol Biochem Parasitol. (2022) 250:111490. doi: 10.1016/j.molbiopara.2022.111490
35. Taylor CM, Martin J, Rao RU, Powell K, Abubucker S, Mitreva M. Using existing drugs as leads for broad spectrum anthelmintics targeting protein kinases. PLoS Pathog. (2013) 9:e1003149. doi: 10.1371/journal.ppat.1003149
36. Khan S, Nisar A, Yuan J, Luo X, Dou X, Liu F, et al. A whole genome re-sequencing based GWA analysis reveals candidate genes associated with ivermectin resistance in Haemonchus contortus. Genes. (2020) 11:367. doi: 10.3390/genes11040367
37. Ménez C, Alberich M, Courtot E, Guegnard F, Blanchard A, Aguilaniu H, et al. The transcription factor NHR-8: a new target to increase ivermectin efficacy in nematodes. PLoS Pathog. (2019) 15:e1007598. doi: 10.1371/journal.ppat.1007598
38. Cook A, Aptel N, Portillo V, Siney E, Sihota R, Holden-Dye L, et al. Caenorhabditis elegans ivermectin receptors regulate locomotor behaviour and are functional orthologues of Haemonchus contortus receptors. Mol Biochem Parasitol. (2006) 147:118–25. doi: 10.1016/j.molbiopara.2006.02.003
39. Blanchard A, Guégnard F, Charvet CL, Crisford A, Courtot E, Sauvé C, et al. Deciphering the molecular determinants of cholinergic anthelmintic sensitivity in nematodes: when novel functional validation approaches highlight major differences between the model Caenorhabditis elegans and parasitic species. PLoS Pathog. (2018) 14:e1006996. doi: 10.1371/journal.ppat.1006996
40. Samarasinghe B, Knox DP, Britton C. Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. Int J Parasitol. (2011) 41:51–9. doi: 10.1016/j.ijpara.2010.07.005
41. Zawadzki JL, Kotze AC, Fritz JA, Johnson NM, Hemsworth JE, Hines BM, et al. Silencing of essential genes by RNA interference in Haemonchus contortus. Parasitology. (2012) 139:613–29. doi: 10.1017/S0031182012000121
42. Issa Z, Grant WN, Stasiuk S, Shoemaker CB. Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis. Int J Parasitol. (2005) 35:935–40. doi: 10.1016/j.ijpara.2005.06.001
43. Geldhof P, Murray L, Couthier A, Gilleard JS, McLauchlan G, Knox DP, et al. Testing the efficacy of RNA interference in Haemonchus contortus. Int J Parasitol. (2006) 36:801–10. doi: 10.1016/j.ijpara.2005.12.004
44. Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. (2001) 7:1397–402.
45. Van WJA. Viability of nematode larvae after exsheathment with sodium hypochlorite. Parasitol Today. (1998) 14:474–5. doi: 10.1016/S0169-4758(98)01343-X
46. Geldhof P, Clark D, Molloy C, Knox DP. Assessment of Caenorhabditis elegans as a model in Haemonchus contortus vaccine research. Mol Biochem Parasitol. (2007) 152:220–3. doi: 10.1016/j.molbiopara.2006.12.006
Keywords: RNA interference, Haemonchus contortus, identification of gene function, limitation analysis, research progress
Citation: Hou B, Hai Y, Buyin B and Hasi S (2023) Research progress and limitation analysis of RNA interference in Haemonchus contortus in China. Front. Vet. Sci. 10:1079676. doi: 10.3389/fvets.2023.1079676
Received: 25 October 2022; Accepted: 01 February 2023;
Published: 24 February 2023.
Edited by:
Yadong Zheng, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Lan He, Huazhong Agricultural University, ChinaChunqun Wang, Huazhong Agricultural University, China
Copyright © 2023 Hou, Hai, Buyin and Hasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Surong Hasi,  c3Vyb25nJiN4MDAwNDA7aW1hdS5lZHUuY24=
c3Vyb25nJiN4MDAwNDA7aW1hdS5lZHUuY24=
 Bin Hou
Bin Hou Ying Hai2
Ying Hai2