
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 20 July 2023
Sec. Veterinary Epidemiology and Economics
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1074127
 Ahmed Elfadadny1,2
Ahmed Elfadadny1,2 Jumpei Uchiyama3
Jumpei Uchiyama3 Kazuyoshi Goto3
Kazuyoshi Goto3 Ichiro Imanishi4
Ichiro Imanishi4 Rokaia F. Ragab1
Rokaia F. Ragab1 Wedad M. Nageeb5
Wedad M. Nageeb5 Keita Iyori6
Keita Iyori6 Yoichi Toyoda6
Yoichi Toyoda6 Toshihiro Tsukui7
Toshihiro Tsukui7 Kaori Ide1,8
Kaori Ide1,8 Keiko Kawamoto9
Keiko Kawamoto9 Koji Nishifuji1,8*
Koji Nishifuji1,8*The strong bond between dogs and their owners creates a close association that could result in the transfer of antibiotic-resistant bacteria from canines to humans, potentially leading to the spread of antimicrobial resistance genes. Pseudomonas aeruginosa, a common causative agent of persistent ear infections in dogs, is often resistant to multiple antibiotics. Assessing the antimicrobial resistance profile and genotype of P. aeruginosa is crucial for the appropriate use of veterinary pharmaceuticals. However, in recent years, few studies have been conducted on this bacterium in Japan. We determined the antimicrobial resistance profile and genotype of P. aeruginosa isolated from the ear canal of dogs in Japan in 2020. Analysis of antimicrobial resistance using disk diffusion tests indicated a high frequency of resistance to most antimicrobial agents. Particularly, 29 isolates from the ear canals of the 29 affected dogs (100%) were resistant to cefovecin, cefpodoxime, and florfenicol; however, they were susceptible to cefepime and piperacillin/tazobactam. Only 3.4, 10.3, and 10.3% of the isolates were resistant to ceftazidime, tobramycin, and gentamicin, respectively. Furthermore, upon analyzing the population structure using multilocus sequence typing, a considerably large clonal complex was not observed in the tested isolates. Three isolates, namely ST3881, ST1646, and ST532, were clonally related to the clinically isolated sequence types in Japan (such as ST1831, ST1413, ST1812, and ST1849), which is indicative of dog-to-human transmission. Considering the variation in antibiotic resistance compared to that reported by previous studies and the potential risk of dog-to-human transmission, we believe that the survey for antimicrobial resistance profile and population structure should be continued regularly. However, the prevalence of multidrug-resistant P. aeruginosa in dogs in Japan is not a crisis.
Canine otitis is one of the most commonly reported issues under the dermatology section of veterinary hospitals (1). It encompasses two common types: otitis externa, which refers to inflammation of the outer ear canal and pinna, and otitis media, which involves inflammation of the middle ear (2, 3). In dogs, otitis externa occurs more commonly than otitis media. However, in certain cases, otitis externa can progress and penetrate the tympanic membrane, thereby spreading the infection to the middle ear and leading to otitis media (2, 4). The major causes of canine otitis are the prolonged exposure of the canine to an etiological agent within its environment and/or the failure of the antimicrobial treatment in eliciting a response from the microbes, when the disease is complicated with bacterial strains (4). Repeated episodes of inflammation can cause severe histopathological changes in the ear canal. These changes include epithelial and glandular hyperplasia, calcification, and increased cerumen production along the external ear canal, which can lead to further failure of otitis treatment and potentially result in end-stage ear infection (5).
Canine otitis also has a detrimental effect on the owner’s quality of life (6). The prevalence of otitis cases that are diagnosed in clinical veterinary practice ranges from 8.7 to 20% for otitis externa (7–9) and 50 to 80% for otitis media, as a complication to external otitis (10). In a one-year-prevalence study conducted in the United Kingdom, the prevalence of otitis was notably higher in certain canine species, such as Basset Hound (28.81%), Chinese Shar Pie (17.76%), Labradoodle (17.71%), Beagles (14.72%) Golden Retrievers (14.11%), and Cockapoo (12.97%) (11). In Japan, the incidence of otitis in dogs admitted to the animal hospital in Osaka prefecture was primarily confined to miniature poodles and cocker spaniel dogs (12). The incidence of otitis in a specific species has been attributed to the anatomical confirmation of the ear pinnae and ear canal (13). In a previous study (11), the incidence odds of otitis were 1.76 and 1.84 times in the breeds with pendulous and V-shaped drop pinnae, respectively, compared to that in breeds with the erect-shaped ear. These results suggested that the retention of heat and moisture, as well as accumulation of foreign materials in pendulous and V-shaped drop pinnae may provide a favorable environment for bacterial colonization (14, 15). Canine otitis is driven by secondary bacterial pathogens in 98% of the cases in North America (16, 17), especially coagulase-positive Staphylococcus species, Pseudomonas, Proteus, Escherichia coli, Klebsiella, and Malassezia species, and mixed infections are common (10, 18). The major organisms isolated from dogs suffering from otitis are Staphylococcus spp., especially S. pseudintermedius, which account for 10–70% of cases (19). Pseudomonas aeruginosa is the second major otitis pathogen, which accounts for 20–60% of cases and can cause intractable otitis (4, 19–21). The canine otitis caused by P. aeruginosa is challenging to cure because of its biofilm formation and intrinsic and acquired drug resistance (22).
Previous studies have demonstrated that the extensive utilization of antimicrobial agents in pets, especially in dogs and cats, may pose a potential threat to public health because of the likelihood of such pets serving as a means for the spread of antimicrobial-resistant bacteria and elements (23). A study demonstrated that the resistance of P. aeruginosa to antimicrobials includes not only penicillin, aminoglycosides, and fluoroquinolones groups (24), but also recently formulated drugs, such as ceftolozone-tazobactam and ceftazidime-avibactam (25, 26). Such antimicrobial resistance leads to challenges in the treatment of otitis caused by P. aeruginosa in dogs in the clinical setting (27). Thus, studies are required to monitor the efficacy of drugs used in dogs infected by P. aeruginosa to maintain the potential of a critical therapeutic agent and develop an effective management plan for the long run (28, 29). Several molecular typing techniques have been used to type P. aeruginosa strains, such as pulsed-field gel electrophoresis (30), polymerase chain reaction (PCR) fingerprinting (31), and multilocus sequence typing (MLST) (32). Among these techniques, MLST was developed in 2004 for P. aeruginosa, and this method is based on the sequencing of the allelic difference of seven housekeeping genes, which collectively provide the sequence types (STs) to characterize isolates. The selection of the seven loci allows scientists to track variability among strains such that one can be different in one or more loci and still be included in the global clonal history of the species with the highest possible accuracy (32, 33).
The One Health Perspective approach links the safety and health of humans with that of their surroundings (34), and this approach has recently raised the consideration of a survey for antimicrobial drug–resistant bacteria in small animals (35). The close contact between pets and humans offers excellent opportunities for the interspecies transmission of resistant bacteria in either direction, such as through licking, petting, and physical injuries. In contrast to other diseases, the horizontal transmission of antimicrobial resistance genes could occur either in the human host and/or environment. For example, P. aeruginosa bacterium of dogs and humans can meet outside their hosts and allow the transfer of resistance genes (23). In comparison to food-borne zoonoses, pet-associated zoonoses remain a neglected area globally. A few epidemiological studies on the antimicrobial resistance profile of P. aeruginosa in companion animals have been conducted in Japan (36, 37). Despite the importance of conducting a continuous survey on this topic, which should involve a large number of antibiotics, there has been a scarcity of studies in recent years. Our study investigated the antimicrobial resistance profile and genotype structure of P. aeruginosa isolated from ear canals of dogs in Japan during the year 2020, to determine their genetic relationship to other P. aeruginosa clones circulating in Japan.
The samples were collected using sterile swabs from the ear canals of 29 dogs affected by unilateral and/or bilateral otitis, which were diagnosed from January to December 2020 by local veterinarians in Japan. The swabs were transported to VDT Co. Ltd. after being placed in the Stuart transport medium (Copan Diagnostics Inc., United States). Furthermore, the swabs were immediately smeared onto cetrimide agar (Wako, Fujifilm, Japan), a selective media for P. aeruginosa species identification, and incubated overnight at 35–37°C. The isolated bacteria were identified using a Microflex system (Bruker Daltonics, Bremen, Germany). The protein profile of each bacterium was analyzed using the matrix-assisted laser desorption ionization (MALDI) Biotyper v3.1.
The signalment information of dogs, including breed, age, and sex is listed in Table 1. The Biosafety Management Committee of the Tokyo University of Agriculture and Technology reviewed and approved all experiments using P. aeruginosa strains (#R2-10).
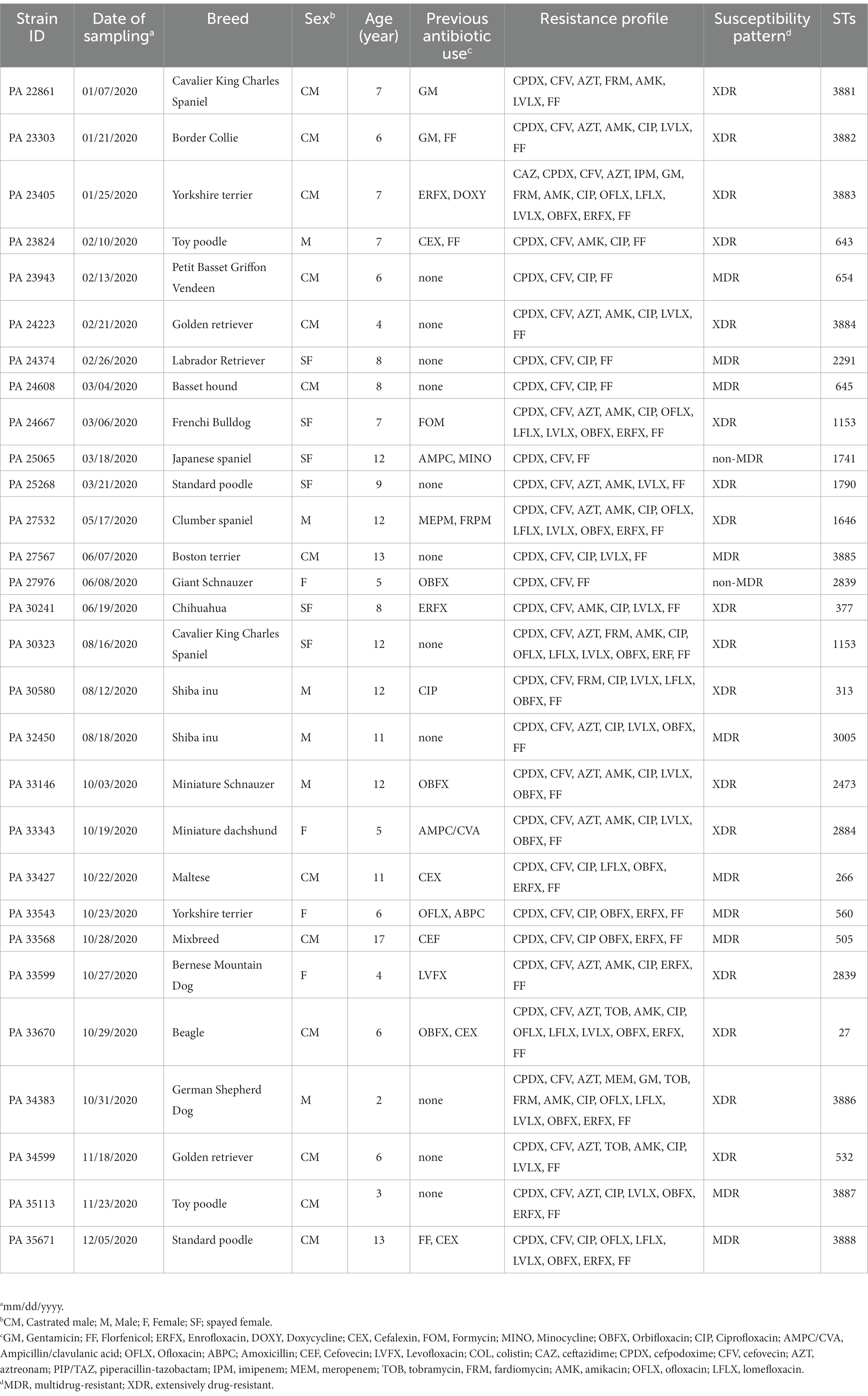
Table 1. Signalment information of 29 Pseudomonas aeruginosa isolates obtained from the ear canals of dogs in Japan.
The antibiotic susceptibility test was performed using the disk diffusion and minimum inhibitory concentration (MIC) methods, based on the Clinical and Laboratory Standards Institute guidelines (CLSI) for Vet-08 and M-100-30thED (38). Briefly, the test was carried out using BD sensei-Disk of the designated antibiotic drug (agar diffusion test; Kirby–Bauer disk diffusion method) on bacteria-inoculated Müller–Hinton Agar (Becton Dickinson GmbH, Heidelberg, Germany). We analyzed the antimicrobial resistance patterns of P. aeruginosa isolates using a panel of 20 antibiotics, which could be categorized into six types based on their mode of action, including fluoroquinolones, aminoglycoside, cephalosporins, carbapenems, piperacillin-tazobactam, and polymyxin. The following antibiotics were used: colistin (COL), ceftazidime (CAZ), cefpodoxime (CPDX), cefovecin (CFV), cefepime (CFPM), aztreonam (AZT), piperacillin-tazobactam (PIP/TAZ), imipenem (IPM), meropenem (MEM), gentamicin (GM), tobramycin (TOB), fradiomycin (FRM), amikacin (AMK), ciprofloxacin (CIP), ofloxacin (OFLX), lomefloxacin (LFLX), levofloxacin (LVLX), orbifloxacin (OBFX), enrofloxacin (ERFX), and florfenicol (FF). The isolates were classified into three categories, namely susceptible, intermediate, and resistant, based on the breakpoints described by CSLI for Vet-08 and M-100-30th ED. The MIC of IPM, MEM, and COL were also determined against P. aeruginosa isolates using the broth microdilution technique, according to the relevant CLSI guidelines. Based on criteria provided by previous classification (39), the classification of the P. aeruginosa isolate as either multidrug-resistant (MDR), extensively drug-resistant (XDR), pan-drug resistant (PDR), or non-MDR (non-multidrug resistant) was based on its susceptibility to various antimicrobial agents. Specifically, an isolate was deemed MDR if it showed resistance to at least one agent in three or more antimicrobial categories. Conversely, an isolate was classified as XDR if it was susceptible to at least one agent in two or fewer categories. An isolate was labeled as PDR if it was resistant to all available antimicrobials. Finally, an isolate was considered non-MDR if it did not meet the criteria for MDR classification (39).
The bacterial DNA was extracted using the Kaneka Easy DNA Extraction Kit (Version 2, Kaneka, Tokyo, Japan). The extracted DNA was purified using a Wizard Genomic DNA Purification kit (Promega, Tokyo, Japan). The sequences for the seven housekeeping genes acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE of P. aeruginosa were amplified via PCR using appropriate primer sets (32). The PCR product was sequenced with a Big Dye Terminator 3.1 Cycle Sequencing Kit using an ABI 3100 genetic analyzer (Applied Biosystems).
The allelic number and ST were determined using the MLST database.1 If no matches were found in the database, STs were newly registered to the database (Supplementary Table S1). For phylogenetic analysis, the concatenated sequences of the seven housekeeping genes of all STs for P. aeruginosa (including the STs of the current study) were downloaded from the MLST website.2 The sequences were aligned using Multiple Sequence Comparison by Log-Expectation, and the phylogenetic tree was generated by the neighbor-joining method, using MEGA software version 10.2.4. The tree was visualized by the online iTOL software version 6.1.1.3
All data in the current study were statistically analyzed using a computer software. For detection of age prevalence in the affected dogs, data were checked by the Mann–Whitney U test (GraphPad Software, Inc., San Diego, California). In all analyzes, p < 0.05 was considered statistically significant. To avoid any discrepancy in the results of the antimicrobial resistance profile of each isolate, all disk diffusion and MIC tests were performed in triplicate. The genotype structure of P. aeruginosa isolates was analyzed by the goeBURST algorithmic method using PHYLOViZ v2.0 software (40).4 The clonal relatedness among the STs of the 29 clinical isolates was checked based on their allelic number. STs were categorized into the same group if they had mutually identical alleles at six of the seven loci with one other member of the group. Conversely, the STs that did not share identical alleles in at least two out of seven loci were considered singleton STs. Using the iTOL software version 6.1.1 (see footnote 3), a heatmap with hierarchical clustering was created to show the overall distribution of antibiotic resistance phenotypes in the isolates.
Information regarding the affected dogs with otitis is described in Table 1. All 29 dogs included in the study were found to have 29 P. aeruginosa isolates (100%) in their ear canals. In this cohort, 65.5% (19/29) of dogs with otitis were male either intact (n = 6) or castrated (n = 13). No breed predisposition was observed for otitis. However, the incidence of the disease was significantly higher (Mann–Whitney U test, p < 0.0001) in senior dogs (above 7 years), who had reached the last quarter of their life expectancy as indicated in a previous study (41), with a mean ± standard deviation (SD) of 11.28 ± 2.37 than in mature adult ones with a mean (±SD) of 5.57 ± 1.39 years old. In addition, 37.9% (11/29) of the dogs with otitis had been previously treated with fluoroquinolones.
The detailed number and percentage of frequency of susceptibility, intermediate susceptibility, and resistance toward each antimicrobial agent are expressed in Table 2. All the isolates: n = 29/29 (100%) expressed resistance phenotype toward CPDX, CFV, and FF. Additionally, the isolates of P. aeruginosa showed variable frequencies of resistance to the group of fluoroquinolones antibiotics CIP: n = 25/29 (86.2%), ERFX: n = 13/29 (44.8%), LVLX: n = 19/29 (65.5%), OFLX: n = 7/29 (24.1%), LFLX: n = 9/29 (31%), and OBFX: n = 16/29 (55.2%). In addition, P. aeruginosa isolates harbored resistance ranging from 10.3 to 55.1% toward the aminoglycoside group of antibiotics, such as GM: n = 3/29 (10.3%), TOB: n = 3/29 (10.3%), AMK: n = 16/29 (55.2%), and FRM: n = 5/29 (17.2%). In contrast, the isolates showed the lowest frequency of resistance toward cephalosporins CFPM: n = 0/29 (0.0%) and CAZ: n = 1/29 (3.4%) and PIP/TAZ: n = 0/29 (0.0%) (Figure 1). The frequency of resistance toward carbapenems antibiotics was 3.4% for IPM and MEM. All isolates showed intermediate susceptibility to COL (Table 3). Additionally, in terms of susceptibility to antimicrobials, 10/29 (34.5%) and 17/29 (58.6%) of P. aeruginosa isolates were classified as MDR and XDR, respectively, while 2/29 were non-MDR isolates. The antibiotic pattern and resistance profile for each isolate are shown in Table 1.
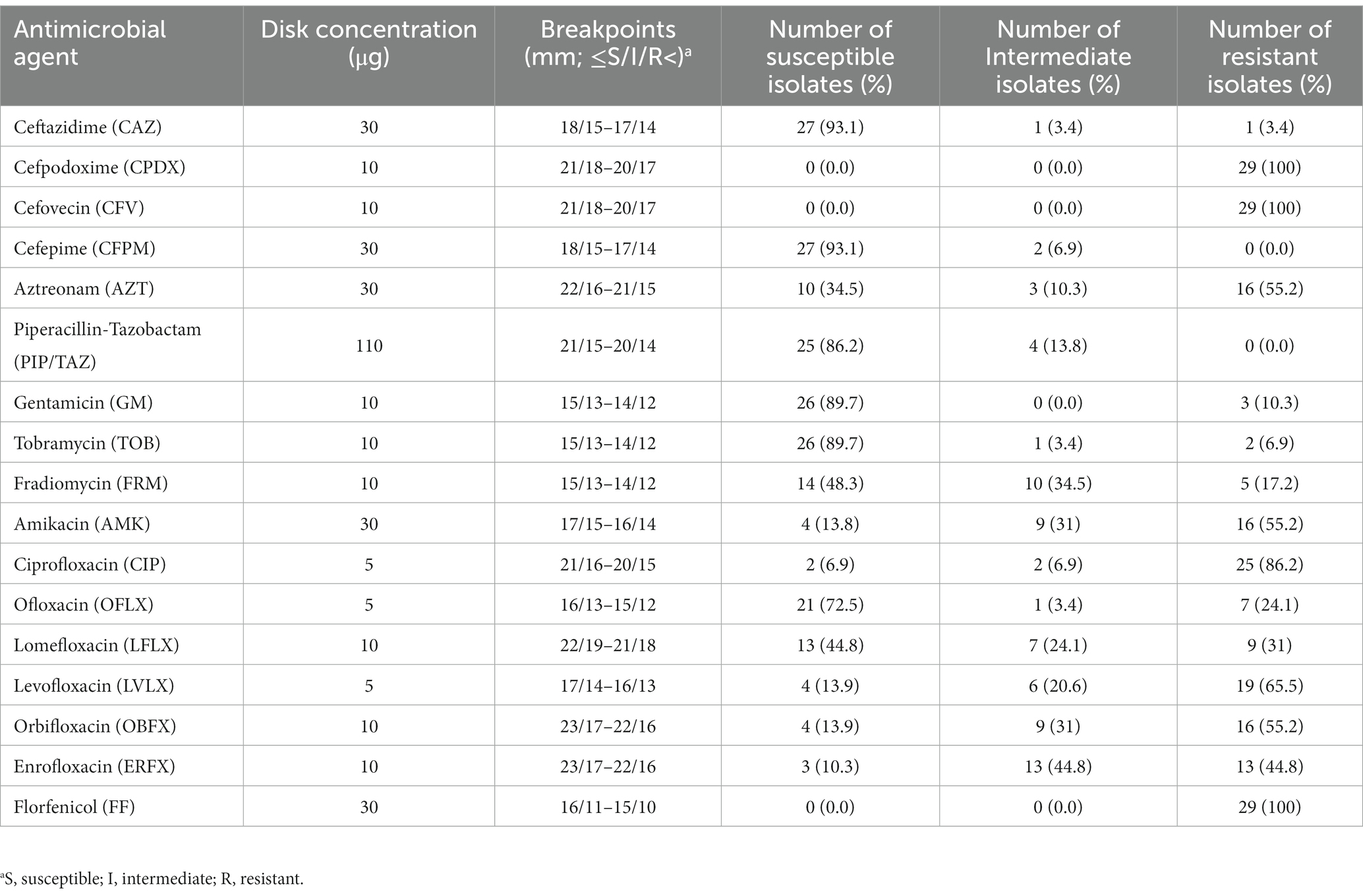
Table 2. Antimicrobial susceptibility percentage (susceptible, intermediate, and resistant) of the 29 P. aeruginosa isolates obtained from the ear canals of dogs.
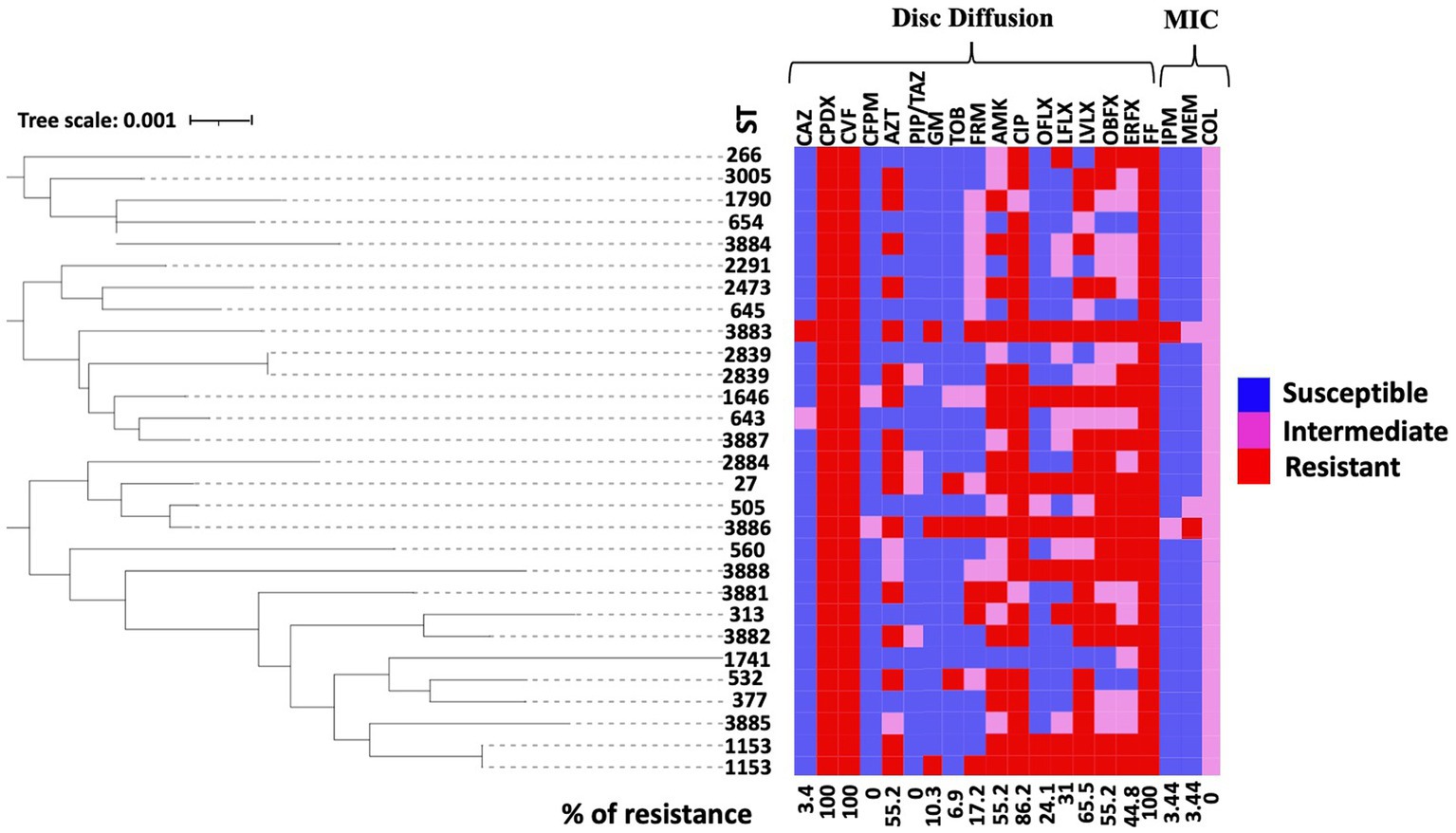
Figure 1. Antimicrobial susceptibility pattern of the 29 isolates isolated from the ear canals of dogs. A rectangular neighbor-joining tree was constructed based on the MLST data of all isolates. A heat map representing the antimicrobial susceptibility profile for each strain was annotated to the phylogenetic tree. Antimicrobial tests for colistin (COL), ceftazidime (CAZ), cefpodoxime (CPDX), cefovecin (CFV), cefepime (CFPM), aztreonam (AZT), piperacillin-tazobactam (PIP/TAZ), imipenem (IPM), meropenem (MEM), gentamicin (GM), tobramycin (TOB), fardiomycin (FRM), amikacin (AMK), ciprofloxacin (CIP), ofloxacin (OFLX), lomefloxacin (LFLX), levofloxacin (LVLX), orbifloxacin (OBFX), enrofloxacin (EREX), and florfenicol (FF) were performed using the disk diffusion method and MIC test based on CSLI. The figure shows the resistance status of antimicrobials (red, resistant; pink, intermediate; blue, sensitive).
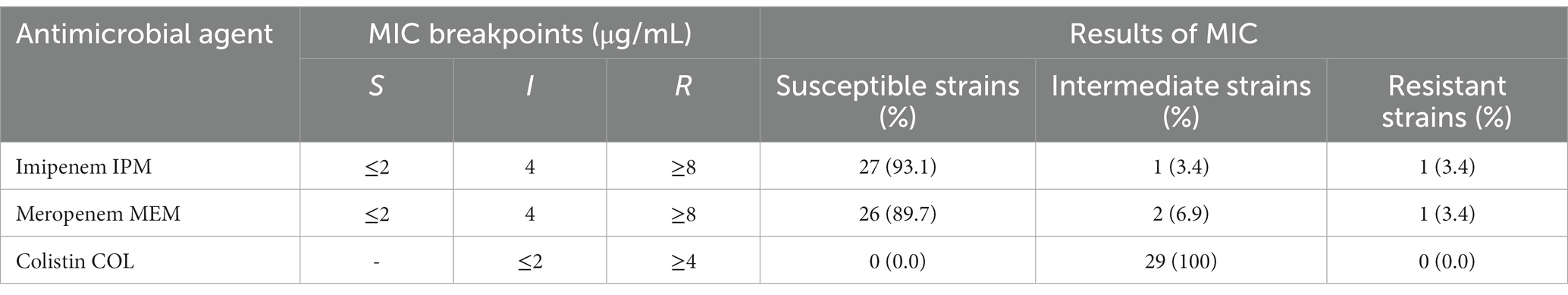
Table 3. Minimum inhibitory concentration (MIC) for imipenem, meropenem, and colistin for the 29 P. aeruginosa isolates obtained from the ear canals of dogs.
Using the MLST analysis, 27 STs were identified among the 29 isolates from the ear canals of dogs affected with otitis. Two STs were assigned to four isolates: ST 1153 was assigned to PA 24667 and PA 30323 isolates, while ST2839 was assigned to PA 27976 and PA 33599 isolates. The same STs mean identical sequences in all housekeeping genes. Other STs were assigned for other isolates. We then analyzed the CCs using these STs. The present STs did not exhibit a CC formation among the isolated strains except for two groups clonally related by a single locus variant: ST313 with ST3882 and ST505 with ST3886. The remaining 25 STs were classified as singletons (Figure 2A), indicating the high genetic diversity between the isolates. The genetic relatedness of the strains isolated in this study with the STs prevalent in Japan was examined. A relationship was observed between two STs, ST3881 and ST1646, which are clonally related by a single locus variant to the previously identified ST1831 and ST1413 from Japan, respectively, while ST532 is the group founder of two Japanese STs, ST1812 and ST1849 (Figure 2A). Also, three previously identified human high-risk clones were identified in the current study: ST27, ST532, and ST654.
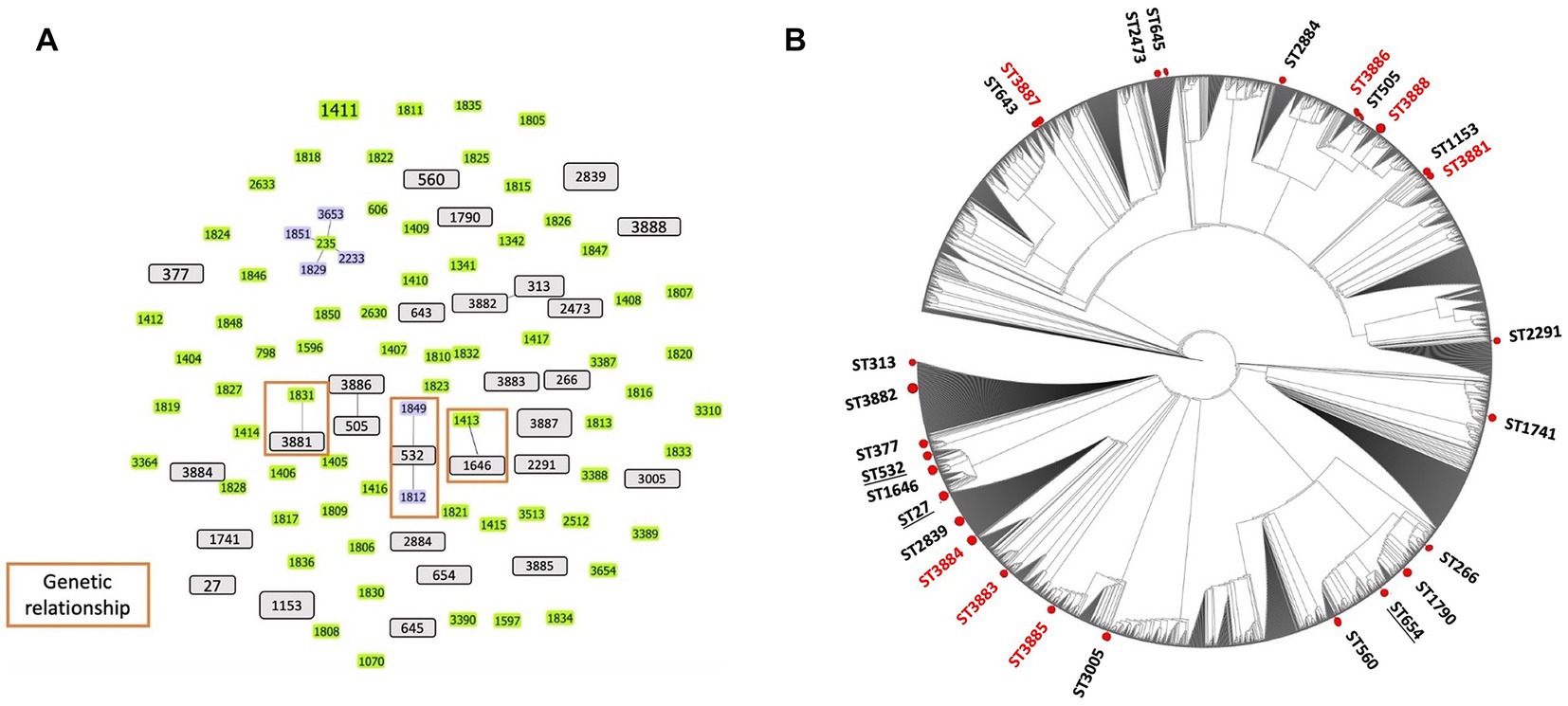
Figure 2. Population structure of Pseudomonas aeruginosa strains: (A) Genetic relatedness between all known sequence types (STs) and the canine P. aeruginosa STs identified in Japan. Three clonal complexes were displayed via single locus variant analysis using goeBURST software (orange rectangular). Gray bars represent all STs identified in this study from dogs. Green and blue (founder ST) bars indicate the STs identified in Japan based on the pubMLST database. (B) The phylogenetic tree represents the distribution of the STs identified in this study from dogs among all known STs from the pubMLST database. Red circles indicate the positions of STs in this study (red STs are new STs identified). No cluster formation was observed among the STs of P. aeruginosa isolated from dogs. Underlined STs represent the high-risk clones of P. aeruginosa identified from dog isolates in this study.
The present study conducted a phylogenetic analysis using concatenated sequences of MLST alleles, which involved both the STs of this study and the entire MLST database as of the date of analysis. The circular tree obtained from this analysis indicates that the STs examined in our study display diversity and are dispersed among all known STs for the P. aeruginosa organism, without forming any distinct clusters. The results of this analysis are illustrated in Figure 2B. Furthermore, the heat map for antimicrobial susceptibility patterns was annotated to the rectangular phylogenetic tree of the STs to examine the dissemination of resistant phenotypes on the tree. The antibiotic resistance phenotypes (i.e., susceptible, intermediate, and resistant) were distributed in all clusters. The highly resistant strains did not vary meaningfully among clusters (Figure 1).
It has been reported in various countries that opportunistic infection causing agents, such as P. aeruginosa and other companion-animal-borne organisms, are developing antibiotic resistance (42, 43). The spread of bacteria resistant to antibiotics from animals to people is a major public health concern, especially when those bacteria are resistant to antimicrobials used in human medicine (23). The One Health approach emphasizes the necessity of P. aeruginosa antimicrobial resistance surveillance, particularly in human-animal-environment interactions. However, studies on continuous monitoring of antimicrobial patterns and the genetic characterization of P. aeruginosa isolates from dogs are limited in Japan. From the review of the frequency of resistance to the 20 antibiotics demonstrated in our study, we concluded that the therapeutic lines for these isolates are challenging. The high variation in resistance frequency toward commonly used veterinary antibiotics such as fluoroquinolones (ranging from 24.1 to 86.2%), makes choosing a suitable antimicrobial difficult, and this could be attributed to the discrepancy between different methods used for checking susceptibility (the disk diffusion test in our study). Nonetheless, when otitis associated with a P. aeruginosa infection is diagnosed, an antibiotic sensitivity test remains the reliable method for identifying and guiding the optimal treatment (44). Compared to the scientific investigations on P. aeruginosa isolated from dogs, the antibiotic resistance profile was slightly higher in the isolates of the present study, with the appearance of a high frequency of MDR and XDR isolates (36, 37). This increase in the number of isolated resistant strains in this study may be attributed to the well-recognized resistant and sometimes multi-resistant nature of the bacterium against the antibiotics commonly used in the veterinary field. Hence, the choice of antimicrobials to treat the disease in clinical settings is limited (45). The overproduction of chromosomally encoded AmpC genes, the outer membrane porin OprD, and the multidrug efflux system are the predominant therapeutic challenges of this organism (46). The phylogenetic tree showed that the antimicrobial resistance phenotypes are distributed among all strains without a significant cluster formation, indicating a weak resistance-related genetic association between the current isolates. Notably, two strains possess the same STs (ST2839 and ST1153) and are shown to have variable resistant rates to antibiotics. However, they possess a common susceptibility to CAZ, CFPM, GM, and TOB, suggesting the possible treatment of strains sharing identical genetic material.
Interestingly, the Infectious Diseases Control Law in Japan did not describe a carbapenem-resistant isolate of P. aeruginosa and MDR isolates originating from companion animals. However, the current study detected that 34.5 and 58.6% of P. aeruginosa isolates were classified as MDR and XDR, emphasizing the need for regular resistance investigation. The rates of GM and CAZ resistance in our study was 10.3, and 3.4%, respectively, and are similar to that reported previously against P. aeruginosa in dogs, which is 19.4 and 1.4%, respectively (37, 47, 48). The CAZ, GM, PIP/TAZ, CFPM, and TOB retained bactericidal activity against the isolates from the ear canals of dogs.
Companion animals represent an insufficiently researched reservoir of P. aeruginosa strains that exhibit antimicrobial resistance. In instances of MDR-P. aeruginosa infections in humans, polymyxin (colistin) and carbapenems (imipenem and meropenem) are recognized as the ultimate therapeutic options (49). In this study, we investigated the prevalence of resistant strains of P. aeruginosa, isolated from dogs in Japan, toward COL, IPM, and MEM using the MIC test, which is reliable and accurate for these antimicrobials. Our investigation revealed that all P. aeruginosa isolates exhibited intermediate susceptibility to colistin (COL). This finding emphasizes the need for continuous monitoring of isolated strains from companion animals in Japan. At the same time, the resistance toward carbapenems is also crucial. The carbapenems resistant isolates of P. aeruginosa have been detected by the MIC test and found to be 3.4% for IPM and MEM. The epidemiological investigation on the increase in the frequency of carbapenems resistance was seen to vary from 0.0% in 2003–2010 (37), 0.5% in 2014–2015 (36), 6.67% in 2017–2018 (50) to 3.4% in 2020 (the current study). In Japan, the carbapenem-resistant P. aeruginosa isolates from companion animals were found to fluctuate over time (50), indicating that a continuous survey of carbapenem-resistant P. aeruginosa from the pets’ environment is necessary. Although the carbapenems resistance status in Japan is not a crisis as reported by the Japanese Veterinary Antimicrobial Resistance Monitoring (JVARM) system, recent scientific studies (36, 37, 50) reported an increase in the carbapenem-resistant isolates from dogs and cats. However, the use of antimicrobials in Japan seems to be prudent, and this increase might be due to the prescriptions of such medicines by local drug suppliers, personnel importation of drugs, and the use of human antimicrobials in the treatment of companion animals, which are not monitored under the JVARM and are excluded from calculations (51). These findings indicate that the veterinary niches, especially the dog-human interface, represent a potential reservoir of resistant P. aeruginosa for animals and humans. The use of COL in human medicine was relatively low owing to its neurotoxicity and nephrotoxicity (52). However, in veterinary and human clinical practice, COL has been reintroduced to treat pathogens resistant to other available antimicrobials. The recommended treatment of otitis in dogs includes recognizing and managing the predisposing factors, ear cleaning, topical treatment, and systemic injection of antimicrobials (if necessary). Polymyxins, including COL, has been introduced in some countries as authorized active ingredients for topical ear application in canine otitis treatment (43) and for intravenous treatment of food-producing animals and equine septicemia (53, 54). Restriction on the use of COL, IPM, and MEM in the veterinary section is required for saving these antibiotics as the last lines of defense against MDR-P. aeruginosa in humans, especially cystic fibrosis (49, 55).
The inappropriate use of antibiotics may induce an annual increase in antibiotic resistance, particularly for antibiotics considered crucial therapeutic agents for treating P. aeruginosa infections in humans. The high level of resistance toward fluoroquinolones (i.e., CIP and ERFX) among isolates that originated from dogs is alarming. The ERFX antibiotic is a potential therapeutic option for the treatment of many human diseases (56). The Nippon AMR One Health Report5 stated in 2021 that fluoroquinolones such as ERFX and OBFX have been used in a persistent amount (between 0.81 and 0.91 ton every year from 2013 to 2018) for treatment of dogs and cats. The amount of CIP used, which is an active metabolite of ERFX, cannot be measured because its prescription has been restricted for veterinarians in Japan. Thus, attention should be paid to monitoring the jumping of resistance to ERFX and CIP. Collectively, the isolated P. aeruginosa strains from dogs in the current study exhibited a high frequency of antibiotic resistance, yet CFPM, GM, TOB, PIP/TAZ, and CAZ possess the ability to kill the P. aeruginosa isolated from the ear canal of affected dogs. A widely utilized approach to preserve susceptibility to these therapeutic agents for clinical purposes is combining phage therapy with alternative antimicrobial methods, such as phage-antibiotic synergy. This method is particularly effective against drug-resistant bacteria (57).
Notably, animals, particularly companion animals have been recognized as potential reservoirs of multidrug-resistant bacteria and resistance genes that can be transmitted to humans (58–60). The genetic analysis of P. aeruginosa isolates from dogs provides important information about the geographical distribution of these isolates in the veterinary community related to humans (59). In the present study, three previously identified human high-risk clones (ST532, ST27, and ST654) were identified from the isolates of infected dogs in this study, which represent their ability to colonize in different niches including the animal environment, and is indicative of a possible zoonotic transmission to humans (61). We chose the MLST technique for our study to easily distinguish the strains of P. aeruginosa and compare STs recovered from dogs with that circulating in the same geographical area, i.e., Japan. Our study supports the nonclonal epidemic structure of P. aeruginosa strains (62–64), whereas the genetic relatedness between some clones isolated from dogs (i.e., ST3881, ST1646, and ST532) and the Japanese STs isolated from wounds and urinary tract infections in humans (i.e., ST1831, ST1413, ST1812, and ST1849) support the theory of horizontal transmission of P. aeruginosa from dog to human. Previous studies have reported MDR-P. aeruginosa isolates from an infected dog, its owner, and the domestic environment, which suggests the occurrence of zoo anthroponotic transmission (65, 66). In addition, the phylogenetic analysis of the resulting data facilitates a clear demonstration of the relative intraspecies evolutionary position of dogs-inhabiting strains indicated by the non-clustered distribution of dog STs among the whole MLST database of the P. aeruginosa. The findings of the current study (i.e., the genetic association between dog STs and those circulating in humans in Japan and the dog STs that matched the STs in the MLST database isolated from different clinical samples) promoted the concept that P. aeruginosa isolates are merely environmental strains that transmit to humans after becoming environmentally adapted. Here, we can support the hypothesis of the zoonotic transmission from either the dog itself or the environment of an infected dog to his immunocompromised owner, especially in the presence of pet-human life sharing. The current findings hypothetically support the previous reports of P. aeruginosa transmission, either directly or indirectly through contamination of environmental objects (67–69). Since pet ownership, especially dogs and cats, become progressively popular in Japan, awareness and knowledge about related public health concerns should be raised. While we have made efforts to ensure the reliability of each experimental result presented in this study, it is important to acknowledge certain limitations that may affect the interpretation of the empirical findings. These limitations include the small sample size, which needs to be addressed in future studies to further support and strengthen the conclusions drawn.
Continuously updating the antimicrobial resistance profiles is necessary to establish the appropriate antibiotic drugs for treating P. aeruginosa infections in dogs and monitoring the emergence of new clones of P. aeruginosa from the pets’ environment. In addition, our study showed that CFPM, PIP/TAZ, CAZ, TOB, and GM could be reliable therapeutic options for treating pseudomonal canine otitis.
The original contributions presented in the study are included in the article/Supplementary materials, further inquiries can be directed to the corresponding author.
AE, JU, and KN conceived and designed the study, and substantially revised the paper. AE, JU, KG, KIY, and YT carried out the experiments, data analysis, and acquisition. AE, II, RR, WN, TT, and KK drafted the original manuscript. JU, KID and KN supervised the study. All authors read and approved the final manuscript.
AE has received financial support from Damanhour University and the Mission Sector, Ministry of Higher Education, Egypt to accomplish the current study (program 2018/2019).
TT was employed by Zenoaq Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1074127/full#supplementary-material
1. ^https://pubmlst.org/bigsdb?db=pubmlst_paeruginosa_seqdef
2. ^https://pubmlst.org/paeruginosa/; accessed on 5 February 2022
1. Hill, PB, Lo, A, Eden, CAN, Huntley, S, Morey, V, Ramsey, S, et al. Survey of the prevalence, diagnosis and treatment of dermatological conditions in small animals in general practice. Vet Rec. (2006) 158:533–9. doi: 10.1136/vr.158.16.533
2. Souza, CP, Foss, KD, Mascarenhas, MB, and Clegg, JL. Otitis media with effusion in two Boston terrier dogs. Vet Med Sci. (2023) 9:1069–73. doi: 10.1002/vms3.1092
5. Huang, H-P, Little, CJL, and McNeil, PE. Histological changes in the external ear canal of dogs with otitis externa. Vet Dermatol. (2009) 20:422–8. doi: 10.1111/j.1365-3164.2009.00853.x
6. Noli, C, Colombo, S, Cornegliani, L, Ghibaudo, G, Persico, P, Vercelli, A, et al. Quality of life of dogs with skin disease and of their owners. Part 2: administration of a questionnaire in various skin diseases and correlation to efficacy of therapy. Vet Dermatol. (2011) 22:344–51. doi: 10.1111/j.1365-3164.2011.00956.x
7. Angus, JC. Otic cytology in health and disease. Vet Clin North Am Small Anim Pract. (2004) 34:411–24. doi: 10.1016/j.cvsm.2003.10.005
8. Topală, R, Burtan, I, Fântânaru, M, Ciobanu, S, and Burtan, LC. Epidemiological studies of otitis externa at carnivores. Luc Ştiinłifice Med Vet. (2007) 40:247–51.
9. August, JR. Otitis externa. A disease of multifactorial etiology. Vet Clin North Am Small Anim Pract. (1988) 18:731–42. doi: 10.1016/s0195-5616(88)50076-1
10. Cole, LK, Kwochka, KW, Kowalski, JJ, and Hillier, A. Microbial flora and antimicrobial susceptibility patterns of isolated pathogens from the horizontal ear canal and middle ear in dogs with otitis media. J Am Vet Med Assoc. (1998) 212:534–8.
11. O’Neill, DG, Volk, AV, Soares, T, Church, DB, Brodbelt, DC, and Pegram, C. Frequency and predisposing factors for canine otitis externa in the UK–a primary veterinary care epidemiological view. Canine Med Genet. (2021) 8:7. doi: 10.1186/s40575-021-00106-1
12. Baba, E, and Fukata, T. Incidence of otitis externa in dogs and cats in Japan. Vet Rec. (1981) 108:393–5. doi: 10.1136/vr.108.18.393
13. Griffin, C. Classifying cases of otitis externa the PPSP system. Proceedings of ESVD Workshop on Otitis St Helens. (2010).
14. O’Neill, DG, Lee, YH, Brodbelt, DC, Church, DB, Pegram, C, and Halfacree, Z. Reporting the epidemiology of aural haematoma in dogs and proposing a novel aetiopathogenetic pathway. Sci Rep. (2021) 11:21670. doi: 10.1038/s41598-021-00352-0
15. Harvey, R. A review of recent developments in veterinary otology. Vet. Sci. (2022) 9:161. doi: 10.3390/vetsci9040161
16. Schaefer, P, and Baugh, RF. Acute otitis externa: an update. Am Fam Physician. (2012) 86:1055–61.
17. Rosenfeld, RM, Brown, L, Cannon, CR, Dolor, RJ, Ganiats, TG, Hannley, M, et al. Clinical practice guideline: Acute otitis externa. Otolaryngol Head Neck Surg. (2006) 134:S4–S23. doi: 10.1016/j.otohns.2006.02.014
18. Schick, AE, Angus, JC, and Coyner, KS. Variability of laboratory identification and antibiotic susceptibility reporting of Pseudomonas spp. isolates from dogs with chronic otitis externa. Vet Dermatol. (2007) 18:120–6. doi: 10.1111/j.1365-3164.2007.00581.x
19. Rosenfeld, RM, Schwartz, SR, Cannon, CR, Roland, PS, Simon, GR, Kumar, KA, et al. Clinical practice guideline: acute otitis externa. Otolaryngol Head Neck Surg. (2014) 150:S1–S24. doi: 10.1177/0194599813517083
20. De Martino, L, Nocera, FP, Mallardo, K, Nizza, S, Masturzo, E, Fiorito, F, et al. An update on microbiological causes of canine otitis externa in Campania region, Italy. Asian Pac J Trop Biomed. (2016) 6:384–9. doi: 10.1016/j.apjtb.2015.11.012
21. Nocera, FP, Ambrosio, M, Fiorito, F, Cortese, L, and De Martino, L. On gram-positive- and gram-negative-Bacteria-associated canine and feline skin infections: a 4-year retrospective study of the university veterinary microbiology diagnostic Laboratory of Naples, Italy. Animals. (2021) 11:1603. doi: 10.3390/ani11061603
22. Von Silva-Tarouca, MSE, Wolf, G, and Mueller, RS. Determination of minimum inhibitory concentrations for silver sulfadiazine and other topical antimicrobial agents against strains of Pseudomonas aeruginosa isolated from canine otitis externa. Vet Dermatol. (2019) 30:145–e42. doi: 10.1111/vde.12718
23. Guardabassi, L, Schwarz, S, and Lloyd, DH. Pet animals as reservoirs of antimicrobial-resistant bacteria: review. J Antimicrob Chemother. (2004) 54:321–32. doi: 10.1093/jac/dkh332
24. Rubin, J, Walker, RD, Blickenstaff, K, Bodeis-Jones, S, and Zhao, S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet Microbiol. (2008) 131:164–72. doi: 10.1016/j.vetmic.2008.02.018
25. MacVane, SH, Pandey, R, Steed, LL, Kreiswirth, BN, and Chen, L. Emergence of Ceftolozane-Tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother. (2022) 61:e01183. doi: 10.1128/AAC.01183-17
26. Yiwei, Z, Jie, C, Han, S, Zhongju, C, Qi-wen, Y, Jin, Z, et al. Emergence of ceftazidime- and avibactam-resistant Klebsiella pneumoniae Carbapenemase-producing Pseudomonas aeruginosa in China. mSystems. (2022) 6:e0078721. doi: 10.1128/mSystems.00787-21
27. Nuttall, T, and Cole, LK. Evidence-based veterinary dermatology: a systematic review of interventions for treatment of Pseudomonas otitis in dogs. Vet Dermatol. (2007) 18:69–77. doi: 10.1111/j.1365-3164.2007.00575.x
28. Moradali, MF, Ghods, S, and Rehm, BHA. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. (2017) 7:39. doi: 10.3389/fcimb.2017.00039
29. Brock, MT, Fedderly, GC, Borlee, GI, Russell, MM, Filipowska, LK, Hyatt, DR, et al. Pseudomonas aeruginosa variants obtained from veterinary clinical samples reveal a role for cyclic di-GMP in biofilm formation and colony morphology. Microbiology. (2017) 163:1613–25. doi: 10.1099/mic.0.000541
30. Hu, H, and Manos, J. Pulsed-field gel electrophoresis of Pseudomonas aeruginosa. Methods Mol Biol. (2015) 1301:157–70. doi: 10.1007/978-1-4939-2599-5_14
31. Syrmis, MW, O’Carroll, MR, Sloots, TP, Coulter, C, Wainwright, CE, Bell, SC, et al. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J Med Microbiol. (2004) 53:1089–96. doi: 10.1099/jmm.0.45611-0
32. Curran, B, Jonas, D, Grundmann, H, Pitt, T, and Dowson, CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. (2004) 42:5644–9. doi: 10.1128/JCM.42.12.5644-5649.2004
33. Vernez, I, Hauser, P, Bernasconi, MV, and Blanc, DS. Population genetic analysis of Pseudomonas aeruginosa using multilocus sequence typing. FEMS Immunol Med Microbiol. (2005) 43:29–35. doi: 10.1016/j.femsim.2004.06.024
34. Aslam, B, Khurshid, M, Arshad, MI, Muzammil, S, Rasool, M, Yasmeen, N, et al. Antibiotic resistance: one health one world outlook. Front Cell Infect Microbiol. (2021) 11:771510. doi: 10.3389/fcimb.2021.771510
35. Marco-Fuertes, A, Marin, C, Lorenzo-Rebenaque, L, Vega, S, and Montoro-Dasi, L. Antimicrobial resistance in companion animals: a new challenge for the one health approach in the European Union. Vet Sci. (2022) 9:208. doi: 10.3390/vetsci9050208
36. Yukawa, S, Tsuyuki, Y, Sato, T, Fukuda, A, Usui, M, and Tamura, Y. Antimicrobial resistance of Pseudomonas aeruginosa isolated from dogs and cats in primary veterinary hospitals in Japan. Jpn J Infect Dis. (2017) 70:461. doi: 10.7883/yoken.JJID.2016.536
37. Harada, K, Arima, S, Niina, A, Kataoka, Y, and Takahashi, T. Characterization of Pseudomonas aeruginosa isolates from dogs and cats in Japan: current status of antimicrobial resistance and prevailing resistance mechanisms. Microbiol Immunol. (2012) 56:123–7. doi: 10.1111/j.1348-0421.2011.00416.x
38. Clinical and Laboratory Standards Institute. Institute CaLS. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals CLSI supplement VET08. Pennsylvania: Clinical and Laboratory Standards Institute (2018).
39. Magiorakos, AP, Srinivasan, A, Carey, RB, Carmeli, Y, Falagas, ME, Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
40. Francisco, AP, Bugalho, M, Ramirez, M, and Carriço, JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. (2009) 10:1–15. doi: 10.1186/1471-2105-10-152
41. Harvey, ND. How old is my dog? Identification of rational age groupings in pet dogs based upon normative age-linked processes. Front Vet Sci. (2021) 8:643085. doi: 10.3389/fvets.2021.643085
42. Marshall, BM, Ochieng, DJ, and Levy, SB. Commensals: underappreciated reservoir of antibiotic resistance. Microbe. (2009) 4:231–8. doi: 10.1128/microbe.4.231.1
43. Scott, A, Pottenger, S, Timofte, D, Moore, M, Wright, L, Kukavica-Ibrulj, I, et al. Reservoirs of resistance: polymyxin resistance in veterinary-associated companion animal isolates of Pseudomonas aeruginosa. Vet Rec. (2019) 185:206. doi: 10.1136/vr.105075
44. Zamankhan Malayeri, H, Jamshidi, S, and Zahraei, ST. Identification and antimicrobial susceptibility patterns of bacteria causing otitis externa in dogs. Vet Res Commun. (2010) 34:435–44. doi: 10.1007/s11259-010-9417-y
45. Nielsen, SS, Bicout, DJ, Calistri, P, Canali, E, Drewe, JA, Garin-Bastuji, B, et al. Assessment of listing and categorisation of animal diseases within the framework of the animal health law (regulation (EU) no 2016/429): antimicrobial-resistant Pseudomonas aeruginosa in dogs and cats. EFSA J. (2022) 20:e07310. doi: 10.2903/j.efsa.2022.7080
46. Lister, PD, Wolter, DJ, and Hanson, ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. (2009) 22:582–610. doi: 10.1128/CMR.00040-09
47. Bourély, C, Cazeau, G, Jarrige, N, Leblond, A, Madec, JY, Haenni, M, et al. Antimicrobial resistance patterns of bacteria isolated from dogs with otitis. Epidemiol Infect. (2019) 147:e121. doi: 10.1017/S0950268818003278
48. Bugden, DL. Identification and antibiotic susceptibility of bacterial isolates from dogs with otitis externa in Australia. Aust Vet J. (2013) 91:43–6. doi: 10.1111/avj.12007
49. Jeannot, K, Hagart, K, Dortet, L, Kostrzewa, M, Filloux, A, Plesiat, P, et al. Detection of Colistin resistance in Pseudomonas aeruginosa using the MALDIxin test on the routine MALDI Biotyper Sirius mass spectrometer. Front Microbiol. (2021) 12:725383. doi: 10.3389/fmicb.2021.725383
50. Hayashi, W, Izumi, K, Yoshida, S, Takizawa, S, Sakaguchi, K, Iyori, K, et al. Antimicrobial resistance and type III secretion system Virulotypes of Pseudomonas aeruginosa isolates from dogs and cats in primary veterinary hospitals in Japan: identification of the international high-risk clone sequence type 235. Microbiol Spectr. (2022) 9:e0040821. doi: 10.1128/Spectrum.00408-21
51. Makita, K, Sugahara, N, Nakamura, K, Matsuoka, T, Sakai, M, and Tamura, Y. Current status of antimicrobial drug use in Japanese companion animal clinics and the factors associated with their use. Front Vet Sci. (2021) 8:705648. doi: 10.3389/fvets.2021.705648
52. Justo, JA, and Bosso, JA. Adverse reactions associated with systemic polymyxin therapy. Pharmacotherapy. (2015) 35:28–33. doi: 10.1002/phar.1493
53. Werners, AH. Treatment of endotoxaemia and septicaemia in the equine patient. J Vet Pharmacol Ther. (2017) 40:1–15. doi: 10.1111/jvp.12329
54. Catry, B, Cavaleri, M, Baptiste, K, Grave, K, Grein, K, Holm, A, et al. Use of colistin-containing products within the European Union and European economic area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. (2015) 46:297–306. doi: 10.1016/j.ijantimicag.2015.06.005
55. Langendonk, RF, Neill, DR, and Fothergill, JL. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa: implications for current resistance-breaking therapies. Front Cell Infect Microbiol. (2021) 11:665759. doi: 10.3389/fcimb.2021.665759
56. Baggio, D, and Ananda-Rajah, MR. Fluoroquinolone antibiotics and adverse events. Aust Prescr. (2021) 44:161–4. doi: 10.18773/austprescr.2021.035
57. Uchiyama, J, Shigehisa, R, Nasukawa, T, Mizukami, K, Takemura-Uchiyama, I, Ujihara, T, et al. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Arch Virol. (2018) 163:1941–8. doi: 10.1007/s00705-018-3811-0
58. Tartor, YH, Gharieb, RMA, Abd El-Aziz, NK, El Damaty, HM, Enany, S, Khalifa, E, et al. Virulence determinants and plasmid-mediated Colistin resistance mcr genes in gram-negative Bacteria isolated from bovine Milk. Front Cell Infect Microbiol. (2021) 11:761417. doi: 10.3389/fcimb.2021.761417
59. Argudín, MA, Deplano, A, Meghraoui, A, Dodémont, M, Heinrichs, A, Denis, O, et al. Bacteria from animals as a Pool of antimicrobial resistance genes. Antibiotics. (2017) 6:12. doi: 10.3390/antibiotics6020012
60. Hamame, A, Davoust, B, Cherak, Z, Rolain, J-M, and Diene, SM. Mobile Colistin resistance (mcr) genes in cats and dogs and their zoonotic transmission risks. Pathogens. (2022) 11:698. doi: 10.3390/pathogens11060698
61. del Barrio-Tofiño, E, López-Causapé, C, and Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. (2020) 56:106196. doi: 10.1016/j.ijantimicag.2020.106196
62. Maatallah, M, Cheriaa, J, Backhrouf, A, Iversen, A, Grundmann, H, Do, T, et al. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One. (2011) 6:–e25617. doi: 10.1371/journal.pone.0025617
63. Nageeb, W, Amin, DH, Mohammedsaleh, ZM, and Makharita, RR. Novel molecular markers linked to Pseudomonas aeruginosa epidemic high-risk clones. Antibiotics. (2021) 10:35. doi: 10.3390/antibiotics10010035
64. Haenni, M, Hocquet, D, Ponsin, C, Cholley, P, Guyeux, C, Madec, JY, et al. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet Res. (2015) 11:9–5. doi: 10.1186/s12917-015-0324-x
65. Wang, Y, Wang, X, Schwarz, S, Zhang, R, Lei, L, Liu, X, et al. IMP-45-producing multidrug-resistant Pseudomonas aeruginosa of canine origin. J Antimicrob Chemother. (2014) 69:2579–81. doi: 10.1093/jac/dku133
66. Fernandes, MR, Sellera, FP, Moura, Q, Carvalho, MPN, Rosato, PN, Cerdeira, L, et al. Zooanthroponotic transmission of drug-resistant Pseudomonas aeruginosa. Brazil Emerg Infect Dis. (2018) 24:1160–2. doi: 10.3201/eid2406.180335
67. Muscarella, LF. Contribution of tap water and environmental surfaces to nosocomial transmission of antibiotic-resistant Pseudomonas aeruginosa. Infect Control Hosp Epidemiol. (2004) 25:342–5. doi: 10.1086/502402
68. Suarez, C, Peña, C, Arch, O, Dominguez, MA, Tubau, F, Juan, C, et al. A large sustained endemic outbreak of multiresistant Pseudomonas aeruginosa: a new epidemiological scenario for nosocomial acquisition. BMC Infect Dis. (2011) 11:272. doi: 10.1186/1471-2334-11-272
Keywords: otitis, dog, antimicrobial resistance, multilocus sequence typing, Pseudomonas aeruginosa
Citation: Elfadadny A, Uchiyama J, Goto K, Imanishi I, Ragab RF, Nageeb WM, Iyori K, Toyoda Y, Tsukui T, Ide K, Kawamoto K and Nishifuji K (2023) Antimicrobial resistance and genotyping of Pseudomonas aeruginosa isolated from the ear canals of dogs in Japan. Front. Vet. Sci. 10:1074127. doi: 10.3389/fvets.2023.1074127
Received: 19 October 2022; Accepted: 22 May 2023;
Published: 20 July 2023.
Edited by:
Mihai Mares, Iasi University of Life Sciences, RomaniaReviewed by:
Yasmine Hasanine Tartor, Zagazig University, EgyptCopyright © 2023 Elfadadny, Uchiyama, Goto, Imanishi, Ragab, Nageeb, Iyori, Toyoda, Tsukui, Ide, Kawamoto and Nishifuji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koji Nishifuji, a29qaW1haWxAY2MudHVhdC5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.