
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 16 September 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.984703
This article is part of the Research TopicApplication of Genomics in Livestock Populations under Selection or ConservationView all 20 articles
 Haonan Yuan1
Haonan Yuan1 Xuanbo Liu1
Xuanbo Liu1 Zhengwen Wang1
Zhengwen Wang1 Yue Ren2
Yue Ren2 Yongqing Li3
Yongqing Li3 Caixia Gao4
Caixia Gao4 Ting Jiao1,5
Ting Jiao1,5 Yuan Cai1
Yuan Cai1 Yanan Yang1*
Yanan Yang1* Shengguo Zhao1*
Shengguo Zhao1*Alternative splicing (AS) allows the generation of multiple transcript variants from a single gene and affects biological processes by generating protein diversity in organisms. In total, 41,642 AS events corresponding to 9,924 genes were identified, and SE is the most abundant alternatively spliced type. The analysis of functional categories demonstrates that alternatively spliced differentially expressed genes (DEGs) were enriched in the MAPK signaling pathway and hypoxia-inducible factor 1 (HIF-1) signaling pathway. Proteoglycans in cancer between the normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups, such as SLC2A1, HK1, HK2, ENO3, and PFKFB3, have the potential to rapidly proliferate alveolar type II epithelial (ATII) cells by increasing the intracellular levels of glucose and quickly divert to anabolic pathways by glycolysis intermediates under hypoxia. ACADL, EHHADH, and CPT1A undergo one or two AS types with different frequencies in ATII cells between TN and TL groups (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups), and a constant supply of lipids might be obtained either from the circulation or de novo synthesis for better growth of ATII cells under hypoxia condition. MCM7 and MCM3 undergo different AS types between LN and LL groups (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups), which may bind to the amino-terminal PER-SIM-ARNT domain and the carboxyl terminus of HIF-1α to maintain their stability. Overall, AS and expression levels of candidate mRNAs between Tibetan pigs and Landrace pigs revealed by RNA-seq suggest their potential involvement in the ATII cells grown under hypoxia conditions.
Tibetan pigs adapt well to hypoxic environments compared to other pigs, as the native breeds live in the Qinghai-Tibet Plateau (1). Studies have shown that Tibetan pigs have evolved typical characteristics to adapt to high-altitude hypoxia, especially with developed lungs, denser pulmonary arterioles, and larger alveoli (2, 3). Hypoxia could induce epithelial injury, influence alveolar homeostasis, and cause a series of pulmonary diseases, such as pulmonary hypertension (4, 5), chronic obstructive pulmonary disease (6, 7), and pulmonary fibrosis (8). Alveolar type I epithelial (ATI) and alveolar type II epithelial (ATII) cells have covered the alveolar surface. ATII can transform into ATI and is responsible for the lungs' repair, recycling, and production (9, 10). ATII could undergo cell death and replace by myofibroblasts in hypoxia-induced IPF, which prevents the repairing and renewal of the alveolar wall (11). The injury of regeneration and transdifferentiation in alveolar epithelial cells are vital points that lead to the disease under hypoxia-induced, which may result in breaks in epithelial basement membranes of alveoli (9). Activation of endoplasmic reticulum stress (12), a different expression of ROS (13), and hemoglobin (14) could involve in the oxygen-sensing pathway in alveolar epithelial cells. Alternative splicing (AS) is one of the essential mechanisms in post-transcriptional regulation and could be regulated by many biotic and abiotic stress factors, especially tightly associated with hypoxic adaptation of cells (15). For example, splicing targets of alternative first exon usage, exon skipping, and intron retention could potentially contribute to cancer cell hypoxic adaptation by promoting cancer cell proliferation, transcriptional regulation, and migration (16–18). Large-scale alterations in alternative splicing of ribosomal protein mRNAs were influenced by hypoxia (19). Promotes expression of the angiogenesis inhibitory alternatively spliced hypoxia-inducible factor-3α (HIF-3α) IPAS isoform, and HIF-1α splicing during angiogenesis could be regulated by hypoxia (18, 20). Recent studies have identified alternative splicing events that exist in lung (21–23), heat (24), and ovary (25). Until now, the analysis of alternative splicing in ATII was rarely reported. Here, we carried out a comparative study of AS in ATII during normoxic (21% O2) and hypoxic (21% O2) to explore the patterns and conservation of AS between Tibetan pigs and Landrace pigs. Our results supported further development of hypoxia-associated splicing events in ATII, representing one of the steps forward in the hypoxic adaptation of Tibetan pigs.
Alveolar type II epithelial primary cells from newborn male Tibetan pigs and Landrace pigs were isolated and cultured as described previously (26) with minor modifications. ATII cells were collected at 48 h, which were cultured under normoxic conditions (21% O2, 5% CO2, and 79% N2) between Tibetan pigs (TN, n = 3) and Landrace pigs (LN, n = 3), and under hypoxic conditions (2% O2, 5% CO2, and 98% N2) between Tibetan pigs (TL, n = 3) and Landrace pigs (LL, n = 3), respectively.
Total RNA was extracted from ATII cells using a TRIzol reagent kit (Invitrogen, Carlsbad, CA, USA), treated, removed, and precipitated using DNase I (NEB, Beijing, China) phenol-chloroform, and ethanol. Total RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and checked using Rnase-free agarose gel electrophoresis.
The mRNAs and non-coding RNAs (ncRNAs) were obtained by removing ribosome RNAs (rRNAs) from total RNA, fragmented into short fragments using fragmentation buffer and reverse transcribed into complementary DNA (cDNA) with random primers, and synthesized to second-strand cDNA. Next, the cDNA fragments were ligated to Illumina sequencing adapters by purifying with a QiaQuick PCR extraction kit (Qiagen, Venlo, The Netherlands), and the second-strand cDNA was digested. The twelve cDNA libraries were generated, purified, and sequenced using Illumina HiSeq™ 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Clean, high-quality reads were obtained and filtered from raw reads using fastp (27) (version 0.18.0) and removing the rRNA mapped reads to the rRNA database. The RefSeq (Sus scrofa 11.1) databases were mapped using HISAT2 (28). Transcripts reconstruction was carried out with software Stringtie and HISAT2. HTSeq counted the number of reads aligned to each gene and exon. A fragment per kilobase of transcript per million mapped reads (FPKM) value was calculated to quantify its expression abundance. We carried out differentially expressed genes (DEGs) using a threshold of |log2(fold_change)| ≥2 and a false discovery rate (FDR) adjusted p-value <5%.
Paired-end raw data were first evaluated using FastQC v0.11.8 (29), and quality control using the FASTX toolkit to trim bases in 5' sequences and trimmomatic to trim adaptor sequences and low-quality reads (30, 31). High-quality reads were aligned to the reference genome sequence (Sus scrofa 11.1) and merged using TopHat2 v2.1.1 (32) and Cufflinks v2.2.1 (33). Differential AS events were identified and analyzed using rMATS (version 4.0.1, http://rnaseq-mats.sourceforge.net/index.html) and AS variations of each transcription region by using StringTie software among four groups. The FDR <0.05 in the comparison was used to identify different AS events. The classification of AS was as follows: alternative 5′ splice sites (A5SSs), alternative 3' splice sites (A3SSs), retained introns (Ris), skipped exons (Ses), and mutually exclusive exons (MXEs) were the main categories of selective splicing.
We analyzed alternatively spliced DEGs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) in the online tool database for annotation, visualization, and integrated discovery (DAVID, version 6.7, https://david.ncifcrf.gov/). GO was used to determine and explore the functions of the alternatively spliced DEGs as molecular function, biological process, and cellular component. KEGG analyzed alternatively spliced DEGs to reveal their roles, regulation processes, and enrichment in different biological pathways. The p-values <0.05 were considered significantly different enriched biological pathways. The co-expression regulatory network of alternatively spliced DEGs is generated using the PCC, and the diagram only shows the top 250. The potential regulatory network was constructed by Cytoscape (34).
The four groups randomly selected three alternatively spliced DEGs for Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) verification. Total RNA was extracted from ATII cells to synthesize cDNA using a FastQuant cDNA first-strand synthesis kit (TianGen, China). The cDNA was subjected to qRT-PCR analysis. Transcript-specific primers (Supplementary Table S1 in Supplementary material 1) were designed based on the unique regions of selected alternatively spliced DEGs using Primer 5.0 software, β-actin was used as reference genes, and expression levels were calculated using the 2−ΔΔCt method. PCR conditions were performed as follows: 95°C for 30 s, forty cycles at 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s.
The average 11,245,202,725 bp clean data were obtained from 11,516,010,250 bp raw reads after filtering out low-quality data among twelve libraries (Supplementary Table S2 in Supplementary material 1). The 41,642 AS events corresponding to 9,924 genes were identified using genomic information and transcript data from the RNA-seq dataset (Figure 1, Supplementary material 2). Hypoxia-induced generally increased the number of AS events. Therefore, a total of five alternative splicing forms were obtained through data mapping analysis, such as A5SS, MXE, A3SS, SE, and RI, which revealed Ses as the most abundant event type (73.01%), followed by MXE (10.70%), A3SS (7.95%), and A5SS (5.02%); mutually RI occurred in only 3.32% of AS events among the four groups (Figure 2). Furthermore, 1,444, 2,192, 2,522, 954, and 9,238 alternatively spliced genes undergo A5SS, A3SS, MXE, RI, and SE events, respectively. The results demonstrated that almost all DEGs underwent at least one AS event. The frequencies of AS events were similar among different groups. The highest frequency (64) of AS events was in the TNC gene (ncbi_397460) in the TN group (Supplementary material 3), and the highest frequency of AS events was in the OBSCN gene among the four groups.
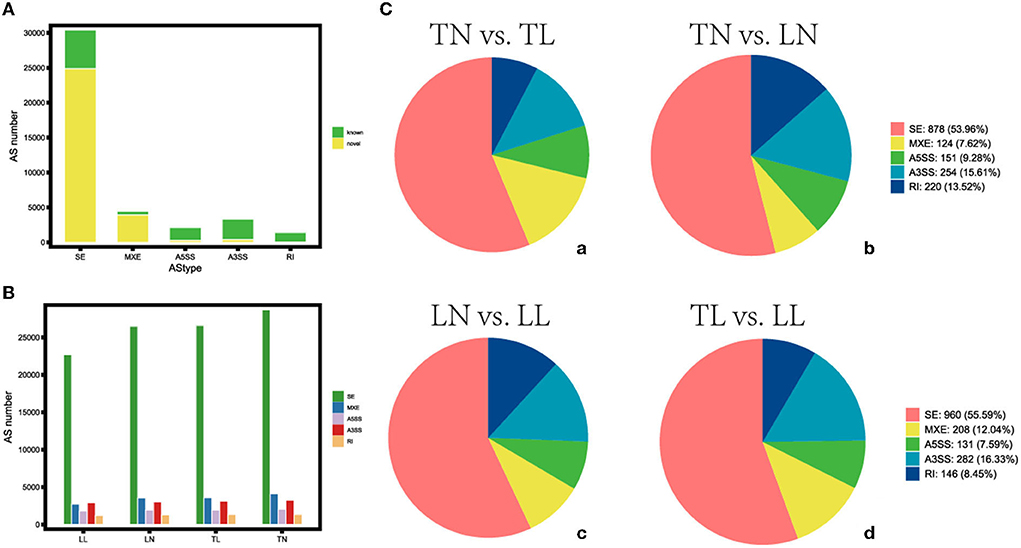
Figure 1. Distribution of the different types of alternative splicing (AS) events. (A) Distribution of the known and novel AS. (B) The number of AS events among four groups. (C) Different types of AS events (a) between TN and TL, (b) between TN and LN, (c) between LN and LL, and (d) between TL and LL groups.
The analysis found that most of the DEGs underwent AS events. Approximately, 33,985 AS events of the total expressed genes and 1,763 significant AS events were screened between TN and TL groups (Supplementary Table S3 in Supplementary material 1). We further selected the 1,470 intersection genes between normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups for significant hypoxia-related genes to identify their AS events, which revealed that 75.00% of them undergo diverse AS (Supplementary material 4). The AS of 901 intersection genes associated with hypoxia, such as EPAS1, NREP, and VPS13B, were only mediated by one or two events. Another 189 genes, such as CCDC14, NKTR, and ATRX, exhibited complex AS. For example, AS in NFAT5, ECM1, ZBTB20, KMT2E, ZMYM1, and PLAGL1 were classified as five basic types between normoxic and hypoxic groups. We found that 233 differential splicing events of 4,514 AS circumstances were present in 1,470 differentially expressed intersection genes between the TN and TL groups (Figure 3, Supplementary Table S4 in Supplementary material 1).
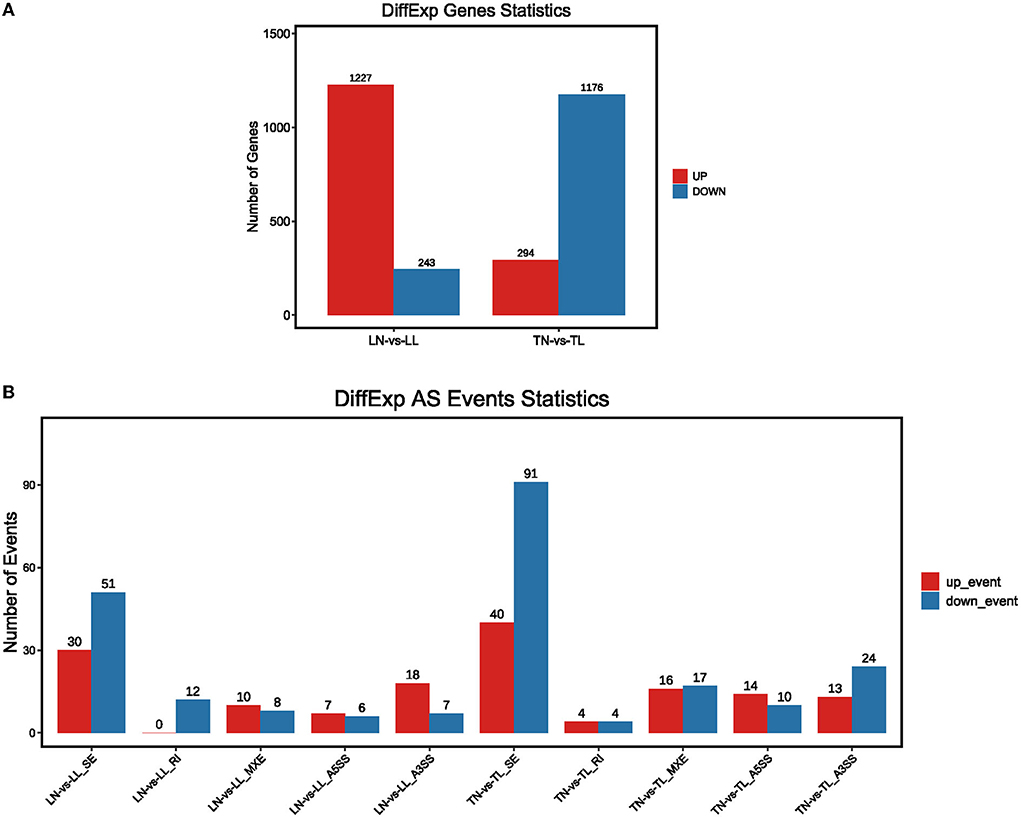
Figure 3. The presence of alternate spliced DEGs in the cross region of normoxic and hypoxic groups. (A) DEGs with alternate splicing were distributed in Tibetan pigs and landrace pigs in normoxia and hypoxia groups; (B) Distribution of DEGs with alternate splicing in five splicing events of Tibetan pigs and Landrace pigs in normoxia and hypoxia groups, respectively.
The enrichment analyses of alternatively spliced DEGs were performed by GO analysis to investigate the biological function of AS events between normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups. The results showed that 817 biological processes, 163 molecular functions, and 114 cellular components were significantly enriched (p < 0.05) (Figures 4A,B). For AS genes of DEGs, biological processes were enriched considerably, such as regulation of nucleobase-containing compound metabolic process (GO: 0019219), nucleic acid metabolic process (GO: 0090304), and nucleobase-containing compound metabolic process (GO: 0006139). Several genes were significantly enriched in the nucleus (GO: 0005634), intracellular part (GO: 0044424), and intracellular (GO: 0005622) cellular component. Binding (GO: 0005488), heterocyclic compound binding (GO: 1901363), and organic cyclic compound binding (GO: 0097159) of molecular functions were most significantly enriched. In a comparison of TN and TL (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups), pyruvate metabolic process (GO: 0006090), binding (GO: 0005488), and intracellular part (GO: 0044424) of biological processes, molecular functions, and cellular components were most significantly enriched (Figures 4C,D). In a comparison of LN and LL (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups), cellular metabolic process (GO: 0044237), catalytic activity (GO: 0003824), and intracellular part (GO: 0044424) of biological processes, molecular functions, and cellular components were most significantly enriched (Figures 4E,F).
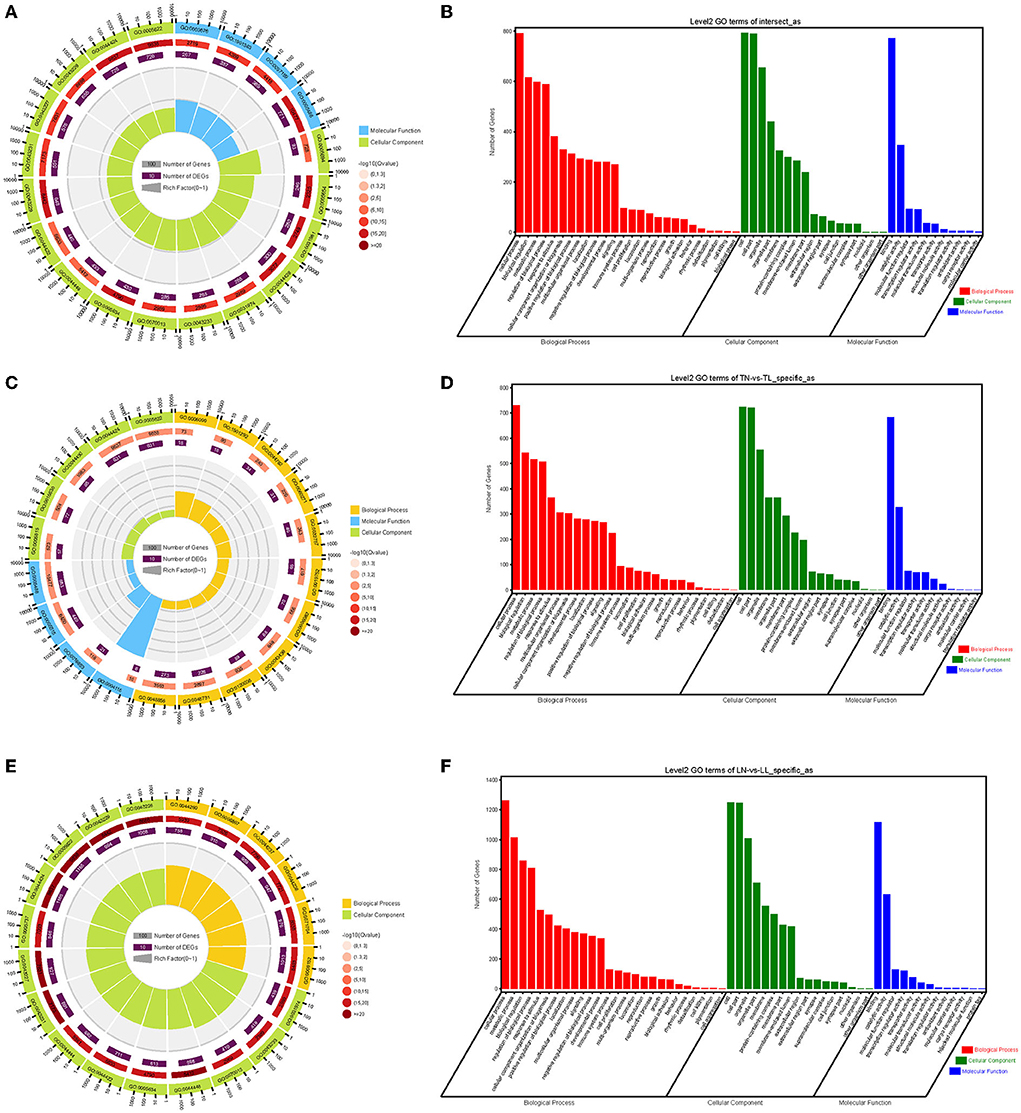
Figure 4. GO functional annotation (A,B) between normoxic (TN and LN) and hypoxic (TL and LL) groups, (C,D) between TN and TL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups), and (E,F) between LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) groups.
As the AS of mRNAs is directly related to functional characteristics, the function of alternatively spliced DEGs was analyzed by KEGG enrichment. A total of 279 pathways were enriched with 89 pathways significantly enriched (p < 0.05), of them MAPK signaling pathway (ko04010), HIF-1 signaling pathway (ko04066), and proteoglycans in cancer (ko05205) were most significantly enriched between normoxic (21% O2, TN vs. LN) and hypoxic (2% O2, TL vs. LL) groups (Figures 5A,B). When TL was compared with TN (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups) groups, alternatively spliced DEGs were found to be significantly enriched in carbon metabolism (ko01200), glycolysis/gluconeogenesis (ko00010), and fatty acid metabolism (ko01212) pathways (Figures 5C,D). Cell cycle (ko04110), metabolic pathways (ko01100), and RNA transport (ko03013) were most significantly enriched by abundant genes between LN and LL groups (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups) (Figures 5E,F).

Figure 5. KEGG enrichment pathways of alternatively spliced DEGs (A,B) between normoxic (TN and LN) and hypoxic (TL and LL) groups, (C,D) between TN and TL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups), and (E,F) between LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) groups. The ordinate is the pathway, and the abscissa is the enrichment factor. Darker colors indicate smaller q-values.
Three hypoxia-related co-expression networks of alternatively spliced DEGs were constructed. The top 250 relationship pair network diagrams are listed, such as comparison groups of normoxia and hypoxia, TN and TL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups), LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) (Figure 6, Supplementary Figures S1, S2). The intersection of comparisons between normoxic (TN, LN) and hypoxic (TL, LL) represented the main differences of ATII cells at different oxygen concentrations gradient. ROCK2 (ncbi_397445), KIF5B (ncbi_595132), and ZFP91 (ncbi_100525558) were selected as the most affected mRNAs, and there were strong correlations with several RNAs undergoing AS events between normoxic (TN, LN) and hypoxia (TL, LL) groups. Interestingly, VCAN (ncbi_397328), HSD3B1 (ncbi_445539), and FAM13C (ncbi_100525364) were most significantly correlated with a large number of alternatively spliced DEGs between TN and TL groups (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups). Meanwhile, ITGAV (ncbi_397285), ADAM9 (ncbi_397344), and MYOF (ncbi_100154898) were most significantly correlated with a large number of alternatively spliced DEGs between LN and LL groups (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups).
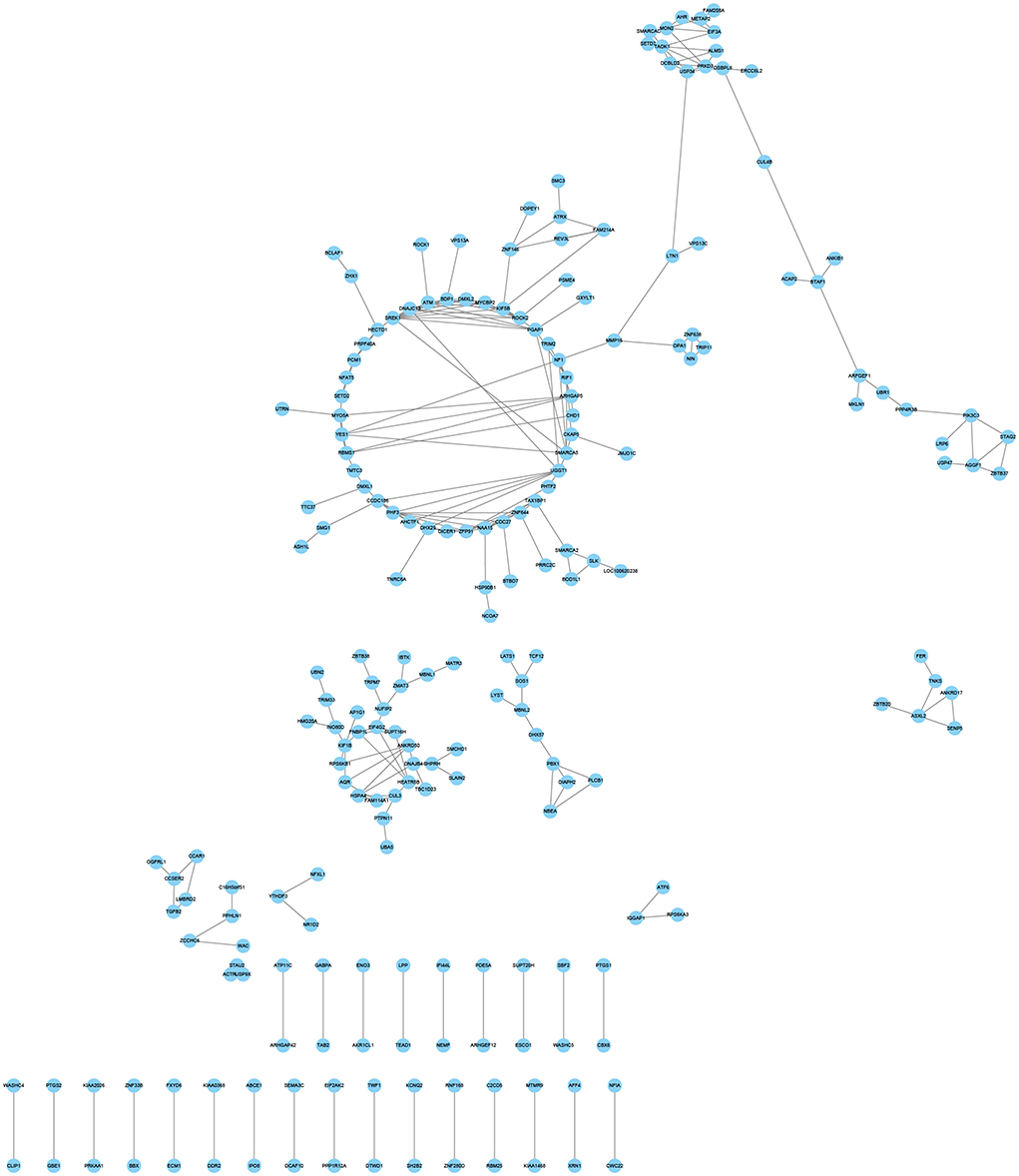
Figure 6. Coexpression network analyses of alternatively spliced DEGs between normoxic (TN and LN) and hypoxic (TL and LL) groups.
Three alternatively spliced DEGs were randomly selected to further test the accuracy of RNA-seq data using qRT-PCR. HP1BP3, NECTIN2, and DDX11 were predicted and identified as having two transcripts, and the type of alternative splicing is SE. The expression levels of the transcript with inclusion and skipping are higher than that of skipping transcript among four groups (Supplementary Figure S3), indicating that the alternative splicing prediction based on RNA-seq data was reliable.
The identification, characterization, and post-transcriptional regulation of alternatively spliced DEGs were widely studied by attracting the interest of researchers (15, 35, 36), such as Xiang pig gilts, bovine, and human. Animals have a more complicated and more extensive intron than plants (37). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis (37–39), and their most common AS events were exon skipping and intron retention, respectively (40, 41). A5SS, MXE, A3SS, SE, and RI were components of five essential AS forms in our study, and this distribution pattern is also similar to that of other animals reported previously (15, 35, 42, 43), indicated that animals might possess similar alternative splicing forms. Alternative splicing events were numerous occur during organ development, tissue maturation, and cell differentiation, suggesting that alternative splicing supports proper development (15). The phenotype may be influenced by modification of gene transcription or translation induced by a hypoxia condition (44). SE may be the primary source of proteomic and transcriptomic and plays a significant role in hypoxia response by regulating genes and determining phenotype as the most abundant event types (73.01%) in ATII cells among four groups (45, 46).
According to the KEGG enrichment, a total of 1,088 alternatively spliced DEmRNAs were enriched in 279 pathways between normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups, of which 35, 17, and 25 genes were enriched in MAPK signaling pathway, HIF-1 signaling pathway, and proteoglycans in cancer. MAPK may arise and upregulate the transcription of anti-apoptotic genes under exposure to hypoxia and play critical roles in opposing the inflammatory response and regulating cell proliferation, differentiation, and apoptosis, which may be a novel strategy for the treatment of chronic obstructive pulmonary fibrosis (6, 47–50). Insulin-like growth factors (IGF1 and IGF2) enriched in 30 pathways and underwent one type of AS event between normoxic and hypoxic groups and might act as cross-talk between MAPK pathways and HIF signaling pathway (51), which may reduce ATII cell apoptosis under hypoxic conditions (52). The increase of HIF-1 transcriptional activity under a hypoxia environment is due to a decrease of cellular NAD+, which downregulates Sirt1 to enhance HIF-1α acetylation (53). As expected, we also discovered that several glycolysis-related genes (such as SLC2A1, HK1, HK2, ENO3, and PFKFB3) undergo one or two AS event types. The frequency of HK2 was higher in normoxia than that of hypoxia groups, and the frequency of SE events in ENO3 was lower in LN groups than any others. The frequency of SE events in HK1 was lowest in LL groups, enriched in the HIF-1 signaling pathway, and may promote anaerobic metabolism by elevating interstitial pressure and alleviating cell damage through glucose metabolism under hypoxia conditions (54, 55). The energy and metabolic intermediates produced through cells rely on glycolysis by hypoxia availability. HK1 and HK2, responsible for the initial steps of glycolysis, convert glucose to glucose-6-phosphate (G-6-P) through phosphorylation, initiating glycolysis and producing pyruvate and lactic acid as energy sources (56, 57). PFKFB enzymes catalyze the synthesis of fructose-2,6-bisphosphate (F-2,6-P2) as one of the numerous glycolytic regulators. PFKFB3 plays a dominant role in vascular cells, leukocytes, and many transformed cells and catalyzes the conversion of fructose-6-phosphate to fructose-1,6-biphosphate as the number of the four isoforms of PFKFBs (58, 59). PFKFB3 undergoes SE events and has a lower frequency in LN groups than in any other groups, may control the steady-state concentration of F-2,6-P2, and glycolysis also mediated the generation of growth factors and proinflammatory cytokines in ATII cells under hypoxia condition (59, 60). Thus, glycolysis intermediates can be increased in the intracellular levels of glucose and quickly diverted to anabolic pathways under hypoxia as substrates for lipid and protein biosynthesis and DNA replication to rapidly proliferate ATII cells (61, 62).
ROCK2, KIF5B, and Zinc finger protein 91 (ZFP91) regulated several mRNAs under hypoxia conditions stimulation as essential hypoxia-inducible genes between normoxia and hypoxia groups. Under hypoxia conditions, pulmonary arterial endothelial cells' proliferation and cell cycle via activation of the ROCK2 signaling pathway (63). Cell migration of macrophages and bladder cancer cells may inhibit ROCK2 expression (64, 65). ZFP91 could upregulate the expression of HIF-1α via binds to its promoter region and is involved in various biological processes (66, 67). In summary, the present study shows that the A3SS and SE AS events of ZFP91 and higher frequency of SE events in ROCK2 and under normoxia (LN and TN) groups may influence proliferation, apoptosis, and epithelial–mesenchymal transition of ATII cells (63–67).
Although the alternatively spliced DEGs in the same oxygen concentration of Tibetan pigs and Landrace pigs should have similar alternative splicing, 18, 12, and 10 alternatively spliced DEGs were most significantly enriched in carbon metabolism, glycolysis/gluconeogenesis, and fatty acid metabolism among the TN and TL groups (excluding alternatively spliced DEGs shared between normoxic and hypoxic groups). LDHA undergoes SE and MXE events and is significantly enriched in the glycolysis/gluconeogenesis pathway of ATII cells in Tibetan pigs under normoxia and hypoxia, a net charge of −6, and preferentially converts pyruvate to lactate, and occupies plasma membrane and mitochondrial with LDHB isoforms (68). Previous research reveals that CD36 and intracellular lipid expression and content were augmented in hypoxic hepatocytes. The membrane-bound sterol regulatory element-binding protein (SREBP) transcription factors could respond to lipid availability and regulate lipid uptake and synthesis as central regulators of lipid homeostasis (69). Fatty acids were identified as a physiological modulator of HIF and have similar functions to oxygen, defining a mechanism for lipoprotein regulation (70). Several alternatively spliced DEGs, such as ACADL, EHHADH, and CPT1A, undergo one or two AS types of ATII cells, and different types and frequencies of AS event may be a constant supply of lipids were obtained either from the circulation or de novo synthesis for ATII cells growth better under hypoxia condition (71, 72).
In contrast to the results obtained from the comparison of ATII cells of Landrace pigs under normoxic and hypoxic conditions, the pathways related to the alternatively spliced DEGs identified from the comparison between LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) were associated with cell cycle, metabolic pathways, RNA transport, and apoptosis. Available evidence suggests hypoxia compensates for cell cycle arrest with decreased S-phase entry in mature ECs and progenitor differentiation during angiogenesis (73). The p53 is a cell cycle regulator and apoptosis in the white shrimp in response to hypoxia (74), and the miR-493-STMN-1 pathway could promote hypoxia-induced epithelial cell cycle arrest in G2/M phase (75). CDK2 (cyclin-dependent kinase 2) undergo AS event between LN and LL groups, which could be activated by either CCNE (cyclin E) or CCNA (cyclin A) at the G1/S phase transition or S phase, and mediates degradation of HIF-1α at the G1/S change (76). MCM7 and MCM3 undergo AS events between LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) groups, bind to the amino-terminal PER-SIM-ARNT (PAS) domain, and the carboxyl terminus of HIF-1α to maintain their stability (77).
In this study, we disclosed features of AS events in ATII cells through RNA-seq data. The results indicated that different types of AS and regulatory networks might partially contribute to the significant variance in ATII cells of Tibetan pigs and Landrace pigs under different oxygen concentrations. ACADL, EHHADH, and CPT1A may be a constant supply of lipids were obtained either from the circulation or de novo synthesis for ATII cells of Tibetan pigs growth better under hypoxia conditions. Therefore, this study provided a better understanding of the effects of different AS of candidate functional genes on ATII cells' response to hypoxia.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by Livestock Care Committee of Gansu Agricultural University.
SZ and YY were the overall project leader who provided financial support and experimental conception. HY was involved in data analyses, statistical analyses, language revisions, journal selection, and manuscript submissions and revisions. XL and ZW contributed to the experimental design and implementation. CG contributed to the supervision and assistance of students in managing animals and collecting and analyzing samples. YL and YR were responsible for the trial implementation, supervision of students collecting and analyzing samples, and manuscript preparation. YC and TJ contributed to supervision of sample collection and analysis and manuscript editing. All authors contributed to the article and approved the submitted version.
The study was supported by the National Natural Science Foundation of China (32060730).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.984703/full#supplementary-material
Supplementary Material 1
Supplementary Table S1. Primers used to detect alternatively spliced differentially expressed genens (DEGs) in ATII cells of pigs by qRT-PCR.
Supplementary Table S2. Overview of the reads and quality filtering of mRNA libraries.
Supplementary Table S3. The number of genes underwent alternative splicing (AS) events.
Supplementary Table S4. AS events of Tibetan pigs and Landrace pigs were present in DEGs between normoxic and hypoxic groups.
Supplementary Material 2
2.1. The A3SS events corresponding to genes were identified.
2.2. The A5SS events corresponding to genes were identified.
2.3. The MXE events corresponding to genes were identified.
2.4. The RI events corresponding to genes were identified.
2.5. The SE events corresponding to genes were identified.
Supplementary Material 3
3.1. The frequency of AS events in the TN group.
3.2. The frequency of AS events in the TL group.
3.3. The frequency of AS events in the LN group.
3.4. The frequency of AS events in the LL group.
Supplementary Material 4
4.1. Differentially expressed genes between normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups.
4.2. Alternatively spliced DEGs between normoxic (21% O2, TN and LN) and hypoxic (2% O2, TL and LL) groups.
Supplementary Material 5
Supplementary Figure S1. Coexpression network analyses of alternatively spliced differentially expressed genes (DEGs) between TN and TL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) groups.
Supplementary Figure S2. Coexpression network analyses of alternatively spliced DEGs between LN and LL (excluding alternatively spliced DEGs shared between normoxia and hypoxia groups) groups.
Supplementary Figure S3. (A) Expression patterns of three alternatively spliced DEGs. (B) Expression patterns of eight randomly selected DEGs. Histogram represents the change in transcript level according to the FPKM value of RNA-seq (left y-axis), and broken line indicates that relative expression level defense by RT-PCR (right y-axis).
DEGs, differentially expressed genes; TN, ATII cells of Tibetan pigs were cultured under 21% O2; TL, ATII cells of Tibetan pigs were cultured under 2% O2; LN, ATII cells of Landrace pigs were cultured under 21% O2; LL, ATII cells of Landrace pigs were cultured under 2% O2.
1. Ma YF, Han XM, Huang CP, Zhong L, Adeola AC, Irwin DM, et al. Population genomics analysis revealed origin and high-altitude adaptation of tibetan pigs. Sci Rep. (2019) 9:11463. doi: 10.1038/s41598-019-47711-6
2. Yang Y, Gao C, Yang T, Sha Y, Cai Y, Wang X, et al. Characteristics of Tibetan pig lung tissue in response to a hypoxic environment on the Qinghai-Tibet Plateau. Arch Anim Breed. (2021) 64:283–92. doi: 10.5194/aab-64-283-2021
3. Yang Y, Yuan H, Yang T, Li Y, Gao C, Jiao T, et al. The expression regulatory network in the lung tissue of tibetan pigs provides insight into hypoxia-sensitive pathways in high-altitude hypoxia. Front Genet. (2021) 12:691592. doi: 10.3389/fgene.2021.691592
4. Sydykov A, Mamazhakypov A, Maripov A, Kosanovic D, Weissmann N, Ghofrani HA, et al. Pulmonary Hypertension In Acute And Chronic High Altitude Maladaptation Disorders. Int J Environ Res Public Health. (2021) 18:1692. doi: 10.3390/ijerph18041692
5. Nathan SD, Barbera JA, Gaine SP, Harari S, Martinez FJ, Olschewski H, et al. Pulmonary hypertension in chronic lung disease and hypoxia. Eur Respir J. (2019) 53:1801914. doi: 10.1183/13993003.01914-2018
6. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. (2017) 389:1931–40. doi: 10.1016/S0140-6736(17)31222-9
7. Labaki WW, Rosenberg SR. Chronic obstructive pulmonary disease. Ann Intern Med. (2020) 173:ITC17–32. doi: 10.7326/AITC202008040
8. Tanguy J, Goirand F, Bouchard A, Frenay J, Moreau M, Mothes C, et al. [18F]FMISO PET/CT imaging of hypoxia as a non-invasive biomarker of disease progression and therapy efficacy in a preclinical model of pulmonary fibrosis: comparison with the [18F]FDG PET/CT approach. Eur J Nucl Med Mol Imaging. (2021) 48:3058–74. doi: 10.1007/s00259-021-05209-2
9. Guillot L, Nathan N, Tabary O, Thouvenin G, Le Rouzic P, Corvol H, et al. Alveolar epithelial cells: master regulators of lung homeostasis. Int J Biochem Cell Biol. (2013) 45:2568–73. doi: 10.1016/j.biocel.2013.08.009
10. Aspal M, Zemans RL. Mechanisms of ATII-to-ATI cell differentiation during lung regeneration. Int J Mol Sci. (2020) 21:3188. doi: 10.3390/ijms21093188
11. Alvarez-Palomo B, Sanchez-Lopez LI, Moodley Y, Edel MJ, Serrano-Mollar A. Induced pluripotent stem cell-derived lung alveolar epithelial type II cells reduce damage in bleomycin-induced lung fibrosis. Stem Cell Res Ther. (2020) 11:213. doi: 10.1186/s13287-020-01726-3
12. Delbrel E, Uzunhan Y, Soumare A, Gille T, Marchant D, Planès C, et al. Stress is involved in epithelial-to-mesenchymal transition of alveolar epithelial cells exposed to a hypoxic microenvironment. Int J Mol Sci. (2019) 20:1299. doi: 10.3390/ijms20061299
13. Sherman MA, Suresh MV, Dolgachev VA, McCandless LK, Xue X, Ziru L, et al. Molecular characterization of hypoxic alveolar epithelial cells after lung contusion indicates an important role for HIF-1α. Ann Surg. (2018) 267:382–91. doi: 10.1097/SLA.0000000000002070
14. Grek CL, Newton DA, Spyropoulos DD, Baatz JE. Hypoxia up-regulates expression of hemoglobin in alveolar epithelial cells. Am J Respir Cell Mol Biol. (2011) 44:439–47. doi: 10.1165/rcmb.2009-0307OC
15. Ule J, Blencowe BJ. alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. (2019) 76:329–45. doi: 10.1016/j.molcel.2019.09.017
16. Han J, Li J, Ho JC, Chia GS, Kato H, Jha S, et al. Hypoxia is a key driver of alternative splicing in human breast cancer cells. Sci Rep. (2017) 7:4108. doi: 10.1038/s41598-017-04333-0
17. Liu Z, Sun L, Cai Y, Shen S, Zhang T, Wang N, et al. Hypoxia-induced suppression of alternative splicing of MBD2 promotes breast cancer metastasis via activation of FZD1. Cancer Res. (2021) 81:1265–78. doi: 10.1158/0008-5472.CAN-20-2876
18. Farina AR, Cappabianca L, Sebastiano M, Zelli V, Guadagni S, Mackay AR, et al. Hypoxia-induced alternative splicing: the 11th Hallmark of Cancer. J Exp Clin Cancer Res. (2020) 39:110. doi: 10.1186/s13046-020-01616-9
19. Brumwell A, Fell L, Obress L, Uniacke J. Hypoxia influences polysome distribution of human ribosomal protein S12 and alternative splicing of ribosomal protein mRNAs. RNA. (2020) 26:361–71. doi: 10.1261/rna.070318.119
20. Heikkilä M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol Life Sci. (2011) 68:3885–901. doi: 10.1007/s00018-011-0679-5
21. Wu DD, Yang CP, Wang MS, Dong KZ, Yan DW, Hao ZQ, et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl Sci Rev. (2020) 7:952–63. doi: 10.1093/nsr/nwz213
22. Yan JQ, Liu M, Ma YL, Le KD, Dong B, Development LiGH, et al. of alternative splicing signature in lung squamous cell carcinoma. Med Oncol. (2021) 38:49. doi: 10.1007/s12032-021-01490-1
23. Xu Z, Wei J, Qin F, Sun Y, Xiang W, Yuan L, et al. Hypoxia-associated alternative splicing signature in lung adenocarcinoma. Epigenomics. (2021) 13:47–63. doi: 10.2217/epi-2020-0399
24. Weeland CJ, van den Hoogenhof MM, Beqqali A, Creemers EE. Insights into alternative splicing of sarcomeric genes in the heart. J Mol Cell Cardiol. (2015) 81:107–13. doi: 10.1016/j.yjmcc.2015.02.008
25. Tang LT, Ran XQ, Mao N, Zhang FP, Niu X, Ruan YQ, et al. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology. (2018) 119:60–8. doi: 10.1016/j.theriogenology.2018.06.022
26. Wang X, Zhang L, Sun B. Neonatal type II alveolar epithelial cell transplant facilitates lung reparation in piglets with acute lung injury and extracorporeal life support. Pediatr Crit Care Med. (2016) 17:e182–92. doi: 10.1097/PCC.0000000000000667
27. Singh S, Gupta M, Pandher S, Kaur G, Goel N, Rathore P. Using de novo transcriptome assembly and analysis to study RNAi in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae). Sci Rep. (2019) 9:13710. doi: 10.1038/s41598-019-49997-y
28. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
29. Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data, Babraham Bioinformatic. Cambridge, United Kingdom: Babraham Institute (2010).
30. Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. (2010) 38:e131. doi: 10.1093/nar/gkq224
31. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30. (2014) 2114–20. doi: 10.1093/bioinformatics/btu170
32. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. (2013) 14:R36. doi: 10.1186/gb-2013-14-4-r36
33. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks [published correction appears in Nat Protoc. 2014 Oct;9:2513]. Nat Protoc. (2012) 7:562–78. doi: 10.1038/nprot.2012.016
34. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. (2015) 43:D447–52. doi: 10.1093/nar/gku1003
35. Bhadra M, Howell P, Dutta S, Heintz C, Mair WB. Alternative splicing in aging and longevity. Hum Genet. (2020) 139:357–69. doi: 10.1007/s00439-019-02094-6
36. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E, et al. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist Updat. (2020) 53:100728. doi: 10.1016/j.drup.2020.100728
37. Iwata H, Gotoh O. Comparative analysis of information contents relevant to recognition of introns in many species. BMC Genomics. (2011) 12:45. doi: 10.1186/1471-2164-12-45
38. Song H, Wang L, Chen D, Li F. The function of pre-mRNA alternative splicing in mammal spermatogenesis. Int J Biol Sci. (2020) 16:38–48. doi: 10.7150/ijbs.34422
39. Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. (2012) 22:1184–95. doi: 10.1101/gr.134106.111
40. Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. (2003) 34:177–80. doi: 10.1038/ng1159
41. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. (2008) 456:470–6. doi: 10.1038/nature07509
42. Wang X, Du L, Wei H, Zhang A, Yang K, Zhou H, et al. Identification of two Stat3 variants lacking a transactivation domain in grass carp: new insights into alternative splicing in the modification of teleost Stat3 signaling. Fish Shellfish Immunol. (2018) 77:13–21. doi: 10.1016/j.fsi.2018.03.022
43. Fang X, Xia L, Yu H, He W, Bai Z, Qin L, et al. Comparative genome-wide alternative splicing analysis of longissimus dorsi muscles between Japanese Black (Wagyu) and Chinese Red Steppes Cattle. Front Vet Sci. (2021) 8:634577. doi: 10.3389/fvets.2021.634577
44. Singh RS. Darwin's legacy II: why biology is not physics, or why it has taken a century to see the dependence of genes on the environment. Genome. (2015) 58:55–62. doi: 10.1139/gen-2015-0012
45. Cossins AR, Crawford DL. Fish as models for environmental genomics. Nat Rev Genet. (2005) 6:324–33. doi: 10.1038/nrg1590
46. Xia JH, Li HL, Li BJ, Gu XH, Lin HR. Acute hypoxia stress induced abundant differential expression genes and alternative splicing events in heart of tilapia. Gene. (2018) 639:52–61. doi: 10.1016/j.gene.2017.10.002
47. Singh M, Yadav S, Kumar M, Saxena S, Saraswat D, Bansal A, et al. The MAPK-activator protein-1 signaling regulates changes in lung tissue of rat exposed to hypobaric hypoxia. J Cell Physiol. (2018) 233:6851–65. doi: 10.1002/jcp.26556
48. Shologu N, Scully M, Laffey JG, O'Toole D. Human mesenchymal stem cell secretome from bone marrow or adipose-derived tissue sources for treatment of hypoxia-induced pulmonary epithelial injury. Int J Mol Sci. (2018) 19:2996. doi: 10.3390/ijms19102996
49. Estaras M, Gonzalez-Portillo MR, Fernandez-Bermejo M, Mateos JM, Vara D, Blanco-Fernandez G, et al. Melatonin induces apoptosis and modulates cyclin expression and MAPK Phosphorylation in pancreatic stellate cells subjected to hypoxia. Int J Mol Sci. (2021) 22:5555. doi: 10.3390/ijms22115555
50. Dong Q, Jie Y, Ma J, Li C, Xin T, Yang D, et al. Renal tubular cell death and inflammation response are regulated by the MAPK-ERK-CREB signaling pathway under hypoxia-reoxygenation injury. J Recept Signal Transduct Res. (2019) 39:383–91. doi: 10.1080/10799893.2019.1698050
51. Zeng Y, Zhang L, Hu Z. Cerebral insulin, insulin signaling pathway, and brain angiogenesis. Neurol Sci. (2016) 37:9–16. doi: 10.1007/s10072-015-2386-8
52. Fan J, Shi S, Qiu Y, Zheng Z, Yu L. MicroRNA-486-5p down-regulation protects cardiomyocytes against hypoxia-induced cell injury by targeting IGF-1. Int J Clin Exp Pathol. (2019) 12:2544–51.
53. Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. (2010) 40:294–309. doi: 10.1016/j.molcel.2010.09.022
54. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci. (2019) 20:238. doi: 10.3390/ijms20020238
55. Li X, Wang M, Li S, Chen Y, Wang M, Wu Z, et al. HIF-1-induced mitochondrial ribosome protein L52: a mechanism for breast cancer cellular adaptation and metastatic initiation in response to hypoxia. Theranostics. (2021) 11:7337–59. doi: 10.7150/thno.57804
56. Xu S, Catapang A, Doh HM, Bayley NA, Lee JT, Braas D, et al. Hexokinase 2 is targetable for HK1 negative, HK2 positive tumors from a wide variety of tissues of origin. J Nucl Med. (2018) 60:212–7. doi: 10.2967/jnumed.118.212365
57. Xu S, Zhou T, Doh HM, Trinh KR, Catapang A, Lee JT, et al. An HK2 antisense oligonucleotide induces synthetic lethality in HK1-HK2+ multiple myeloma. Cancer Res. (2019) 79:2748–60. doi: 10.1158/0008-5472.CAN-18-2799
58. Okar DA, Wu C, Lange AJ. Regulation of the regulatory enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Adv Enzyme Regul. (2004) 44:123–54. doi: 10.1016/j.advenzreg.2003.11.006
59. Chesney J. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and tumor cell glycolysis. Curr Opin Clin Nutr Metab Care. (2006) 9:535–9. doi: 10.1097/01.mco.0000241661.15514.fb
60. Cao Y, Zhang X, Wang L, Yang Q, Ma Q, Xu J, et al. PFKFB3-mediated endothelial glycolysis promotes pulmonary hypertension. Proc Natl Acad Sci USA. (2019) 116:13394–403. doi: 10.1073/pnas.1821401116
61. Lottes RG, Newton DA, Spyropoulos DD, Baatz JE. Alveolar type II cells maintain bioenergetic homeostasis in hypoxia through metabolic and molecular adaptation. Am J Physiol Lung Cell Mol Physiol. (2014) 306:L947–55. doi: 10.1152/ajplung.00298.2013
62. Arthur SA, Blaydes JP, Houghton FD. Glycolysis regulates human embryonic stem cell self-renewal under hypoxia through HIF-2α and the glycolytic sensors CTBPs. Stem Cell Rep. (2019) 12:728–42. doi: 10.1016/j.stemcr.2019.02.005
63. Qiao F, Zou Z, Liu C, Zhu X, Wang X, Yang C, et al. ROCK2 mediates the proliferation of pulmonary arterial endothelial cells induced by hypoxia in the development of pulmonary arterial hypertension. Exp Ther Med. (2016) 11:2567–72. doi: 10.3892/etm.2016.3214
64. Luo J, Lou Z, Zheng J. Targeted regulation by ROCK2 on bladder carcinoma via Wnt signaling under hypoxia. Cancer Biomark. (2019) 24:109–16. doi: 10.3233/CBM-181949
65. Chen H, Du J, Zhang S, Tong H, Zhang M. Ghrelin suppresses migration of macrophages via inhibition of ROCK2 under chronic intermittent hypoxia. J Int Med Res. (2020) 48:300060520926065. doi: 10.1177/0300060520926065
66. Ma J, Mi C, Wang KS, Lee JJ, Jin X. Zinc finger protein 91 (ZFP91) activates HIF-1α via NF-κB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget. (2016) 7:36551–62. doi: 10.18632/oncotarget.9070
67. Hanafi M, Chen X, Neamati N. Discovery of a napabucasin PROTAC as an effective degrader of the E3 ligase ZFP91. J Med Chem. (2021) 64:1626–48. doi: 10.1021/acs.jmedchem.0c01897
68. Hussien R, Brooks GA. Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol Genomics. (2011) 43:255–64. doi: 10.1152/physiolgenomics.00177.2010
69. Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol. (2011) 3:a004754. doi: 10.1101/cshperspect.a004754
70. Shao W, Hwang J, Liu C, Mukhopadhyay D, Zhao S, Shen MC, et al. Serum lipoprotein-derived fatty acids regulate hypoxia-inducible factor. J Biol Chem. (2020) 295:18284–300. doi: 10.1074/jbc.RA120.015238
71. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. (2007) 7:763–77. doi: 10.1038/nrc2222
72. Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. (2013) 6:1353–63. doi: 10.1242/dmm.011338
73. Acosta-Iborra B, Tiana M, Maeso-Alonso L, Hernández-Sierra R, Herranz G, Santamaria A, et al. Hypoxia compensates cell cycle arrest with progenitor differentiation during angiogenesis. FASEB J. (2020) 34:6654–74. doi: 10.1096/fj.201903082R
74. Nuñez-Hernandez DM, Felix-Portillo M, Peregrino-Uriarte AB, Yepiz-Plascencia G. Cell cycle regulation and apoptosis mediated by p53 in response to hypoxia in hepatopancreas of the white shrimp Litopenaeus vannamei. Chemosphere. (2018) 190:253–9. doi: 10.1016/j.chemosphere.2017.09.131
75. Liu T, Liu L, Liu M, Du R, Dang Y, Bai M, et al. MicroRNA-493 targets STMN-1 and promotes hypoxia-induced epithelial cell cycle arrest in G2/M and renal fibrosis. FASEB J. (2019) 33:1565–77. doi: 10.1096/fj.201701355RR
76. Hubbi ME, Semenza GL. An essential role for chaperone-mediated autophagy in cell cycle progression. Autophagy. (2015) 11:850–1. doi: 10.1080/15548627.2015.1037063
Keywords: alternative splicing, hypoxia, ATII cells, swine, MAPK signaling pathway, glycolysis/gluconeogenesis
Citation: Yuan H, Liu X, Wang Z, Ren Y, Li Y, Gao C, Jiao T, Cai Y, Yang Y and Zhao S (2022) Alternative splicing signature of alveolar type II epithelial cells of Tibetan pigs under hypoxia-induced. Front. Vet. Sci. 9:984703. doi: 10.3389/fvets.2022.984703
Received: 02 July 2022; Accepted: 18 August 2022;
Published: 16 September 2022.
Edited by:
Anupama Mukherjee, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Liu Jianbin, Lanzhou Institute of Animal Science and Veterinary Medicine (CAAS), ChinaCopyright © 2022 Yuan, Liu, Wang, Ren, Li, Gao, Jiao, Cai, Yang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanan Yang, eWFuZ3lhbmFuNDA0QDE2My5jb20=; Shengguo Zhao, emhhb3NnQGdzYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.