- 1College of Animal Science and Technology, Gansu Agricultural University, Lanzhou, China
- 2The State Key Laboratory of Grassland Agro-Ecosystems, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
Excessive fat deposition in the tail of sheep will affect its feed efficiency, which will increase the feeding cost. The purpose of this study was to identify the single nucleotide polymorphisms (SNPs) of RAP1GAP and rBAT genes by PCR amplification and Sanger sequencing, the SNPs were genotyped by KASP genotyping assays to evaluate their association with tail fat deposition traits. The results showed that two intronic mutations of g.13561 G > A and g.1460 T > C were found in RAP1GAP and rBAT, respectively. There were three genotypes of GG, AG, AA and CC, CT and TT at these two loci, respectively. Association analysis showed that g.13561 G > A of RAP1GAP was associated with tail width, tail fat weight and relative tail fat weight (P < 0.05). The g.1460 T > C of rBAT was associated with tail width and tail fat weight (P < 0.05). Different combinations of genotypes also differed significantly with tail fat deposition traits. In the tail fat tissue, the expression levels of RAP1GAP gene was significantly higher in small-tailed sheep than in big-tailed sheep, and the expression levels of rBAT gene was significantly higher in big-tailed sheep than in small-tailed sheep. In the liver, the expression levels of RAP1GAP and rBAT gene was significantly higher at 6 months than at 0 and 3 months. In conclusion, RAP1GAP and rBAT polymorphisms can be used as a candidate molecular marker to reduce tail fat deposition in sheep.
Introduction
As a common domestic animal, sheep (Ovis aries) are important means of production and livelihood in farming areas, and their meat and wool products play a pivotal role in improving and enhancing local production and livelihood. According to historical records, the wild ancestors of sheep were thin-tailed. Later, the fat-tailed or fat-rumped sheep were bred by artificial selection (1). Tail fat of sheep provides enough energy to maintain normal needs in winter with a shortage of forage (2, 3). With the commercial farming of sheep, the excessive deposition of tail fat limits the commercial value of lamb. It also reduces the feed conversion rate and increases feeding costs. In actual production, it has been found that the mating conception rate of large-tailed sheep is lower than that of small-tailed sheep, because the fat tail hinders natural mating.
Ras-related regulatory protein 1 GTPase-activating protein (RAP1GAP) is a protein coding gene, an important regulator of small G protein Rap1 activation, and the first identified member of the family of GTPase-activating proteins (GAPs) (4, 5). It can inhibit extracellular signal-regulated kinase activity, cell proliferation, survival and migration of melanoma cells (6). Leptin is a crucial hormonal mediator to maintain normal weight. RAP1GAP is a specific negative regulator of Rap1. The activation of Rap1 will inhibit the cellular effect of leptin and promote fat production (7, 8). Studies have shown that Rap1 gene deletion in the forebrain can protect mice from neuroleptin resistance, obesity and glucose imbalance induced by HFD (9). The rBAT (related to b0, + amino acid transporter; SLC3A1) belong to the solute carrier family 3, have been identified (10). The rBAT encodes a type II membrane glycoprotein, which is a component of the renal amino acid transport protein that transports neutral and basic amino acids in the renal tubules and intestine. The rBAT protein is highly expressed in the apical membrane of epithelial cells of kidney and small intestine (11, 12), forming a heterodimer with b0, + AT (SLC7A9) and AGT1 (SLC7A13) (13, 14). Liver is the hub of energy metabolism in animals. It plays an important physiological role in regulating blood lipid level and cholesterol homeostasis. The liver stores glucose through gluconeogenesis and stores excess fat in adipose tissue. In the study of chickens (Pullus), it has been introduced that the liver mediates de novo lipogenesis (15). In pigs (Sus scrofa), liver and adipose tissue are also the main parts of adipogenesis (16, 17).
As a short fat-tailed breed native to China, Hu sheep is widely welcomed by farmers because of its high reproductive performance, fast growth, strong adaptability, early sexual maturity, annual estrus, large number of lambs and strong lactation ability (18, 19). The RAP1GAP and rBAT genes have been studied in human medicine, but it is not clear whether they are associated with tail fat deposition in Hu sheep. Therefore, we have two objectives in this study: (1) to explore the relationship between genetic variation of RAP1GAP and rBAT genes and tail fat deposition traits in sheep. (2) to detect the expression features of RAP1GAP and rBAT genes in Hu sheep at smallest and largest tail fat weight and at different developmental stages in liver. This study may provide an important candidate molecular markers and theoretical basis for the breeding of thin-tailed fat sheep.
Materials and methods
Statement of ethics
All experimental procedures in this study were approved by the Animal Care and Use Committee of Biological Research of Gansu Province, China. And the experimental protocol and sample collection were approved by the Ethics Committee of Gansu Agricultural University (permit number for conducting animal experiments: No. 2012–2-159).
Experimental animals and extraction of DNA
A total of 1,662 male Hu sheep were used in this study. All lambs purchased from five different companies (including Jinchang Zhongtian Sheep Industry Co. Ltd., Gansu Zhongsheng Huamei Sheep Industry Development Co. Ltd., Gansu Sanyangjinyuan Husbandry Co. Ltd., Shandong Runlin Sheep Industry Co. Ltd., and Wuwei Pukang Sheep Industry Co. Ltd). All lambs were weaned at 56 days of age and immunized before weaning. After weaning, they were transferred to Minqin Defu agricultural Co., Ltd. (Gansu Province, China) for separate feeding. The adaptation period was 14 days, the pre-test period was 10 days and the experimental period was 100 days. During the experiment, maintain the same feeding and environmental conditions. All lambs were raised in a single pen and were allowed to eat and drink freely. Before the end of the experiment, the experimental animals were fed with pellet feed purchased from Gansu Sanyang Jinyuan animal husbandry Co. Ltd. At 180 days, the body weight (BW) was measured with a calibrated electronic scale and tail length and width were measured by the same person using a clearly graduated leather ruler. Carcass and tail fat were weighed after slaughter and a small part of tail fat and other tissues were collected for use in subsequent experiments. We calculate the relative weight of tail fat according to the following formula: the relative weight of tail fat (body weight) = the weight of tail fat / BW, the relative weight of tail fat (Carcass) = the weight of tail fat / carcass weight. Venous blood samples (5 ml) were collected from 1,662 adult sheep and placed in heparin sodium vacuum tubes. Before DNA extraction, they were stored at −20°C. Blood DNA was extracted by EasyPure Blood Genomic DNA kit (TransGen Biotech, Beijing, China), and the DNA of 1,372 sheep was successfully extracted.
SNP identification and genotyping
The RAP1GAP and rBAT genes (GenBank Accession No. NC_040253.1 and NC_040254.1) was amplified by PCR. And PCR primers for the RAP1GAP and rBAT genes were designed according to the DNA base sequence (Table 1). The DNA of 10 Hu sheep was randomly selected as the template DNA for PCR amplification. The PCR reaction system included 17.5 μL of 2 × TSINGKE Master Mix (TSINGKE Biological Technology, Beijing, China), 1.12 μL of forward and reverse primers each, 1.4 μL of sheep template DNA and 14 μL of ddH2O. Amplification was performed according to the following procedure: 3 min at 94°C; followed by 30 s at 94°C, 30 s at 45–65°C, and 30 s at 72°C for 35 cycles; final 72°C for 5 min. SNPs in RAP1GAP and rBAT genes were identified by sequencing of PCR products. All individuals in the study were genotyped with the competitive allele-specific FRET-based PCR (KASPar) assays (LGC Genomics, Hoddesdon, UK). The primers used for genotyping are listed in Table 2.
Expression studies of RAP1GAP and rBAT genes in sheep tail fat and liver tissue
In order to explore the mRNA expression patterns of sheep RAP1GAP and rBAT genes in tail fat tissue. After slaughter, 12 lambs were selected from the same batch of sheep. According to the characteristics of tail fat deposition (tail width, tail fat weight and relative tail fat weight), they were divided into big-tail group (n = 6) and small-tail group (n = 6). Supplementary Table 1 shows individual information. Secondly, to explore the mRNA expression level of sheep RAP1GAP and rBAT genes in liver, the liver tissues of lambs at three different development stages (0, 3, and 6 months of age) were collected, three in each stage. Total RNA was extracted with transzol (TransGen Biotech, Beijing, China), followed by the detection of RNA concentration and purity. RNA extraction was performed by using a reverse transcriptase kit (Takara, Dalian, China), followed by reverse transcription to generate cDNA. The sample preparation and experimental program setup for Quantitative real-time PCR is based on our previous studies (20). Each sample was subjected to four technical repetitions. Data were analyzed using the 2−ΔΔCT method (21), and average relative expression levels of genes were calculated.
Statistical analysis
According to a previously published method (22), genetic indices such as genotype frequency and allele frequency were calculated based on genotyping results. The association analysis between genotypes and phenotype values based on a linear model in SPSS 25.0 software:
Where Yijkln is a vector of phenotypic observation, μ is a vector of mean, Gi represents the effect of the ith genotype, Fj stands for the effect of jth farm (j = 1, 2, 3, 4, 5, 6, 7), Sk represents the effect of season (k = winter, summer), Fl and Mn represent the family effect, and εijkln was a vector of random residuals. Correlation among genotypic and phenotypic was tested using Duncan test and Tukey test.
Results
SNP scanning of sheep RAP1GAP and rBAT genes
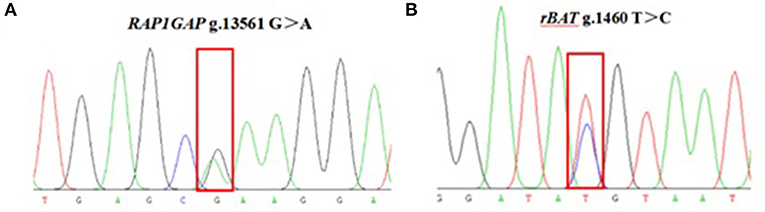
The 937 bp target fragment of RAP1GAP gene and 853 bp target fragment of rBAT gene were successfully amplified by previously designed primers (Figure 1). Two mutations were discovered within the RAP1GAP and rBAT genes after sequencing of PCR products (Figure 2). They are intronic located in intron 1 (g.13561 G > A) and intron 1 (g.1460 T > C), respectively.
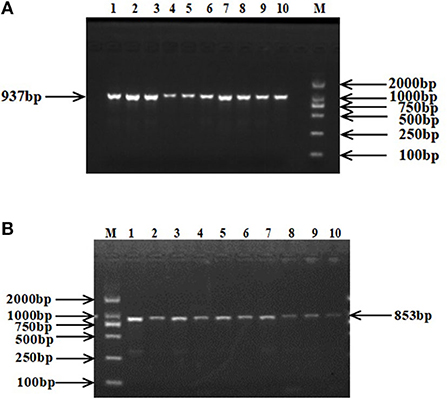
Figure 1. PCR amplification fragments of RAP1GAP (A) and rBAT (B) genes. M: DL2000 DNA Marker; 1–10: PCR products.
Genotyping and genotype and allele frequency analysis
Genotyping of SNPs in RAP1GAP and rBAT genes by KASPar assays using blood genomic DNA of Hu sheep as template. Three genotypes were detected in both genes: GG, AG, AA (RAP1GAP) and CC, CT, and TT (rBAT; Figure 3). Within the RAP1GAP g.13561 G > A locus, the genotype frequencies of GG, AG, and AA were 0.21, 0.51, and 0.28, respectively. Analysis of genotypic frequencies indicated that A was the dominant allele (0.54). In the rBAT g.1460 T > C locus, the genotype frequencies of CC, CT and TT were 0.31, 0.50, and 0.19, respectively. And the C allele was the dominant allele (0.56; Table 3). The PIC, Ne, Ho and He for the RAP1GAP gene were 0.38, 2, 0.50, and 0.50, and for the rBAT gene were 0.37, 1.96, 0.51, and 0.49, respectively.
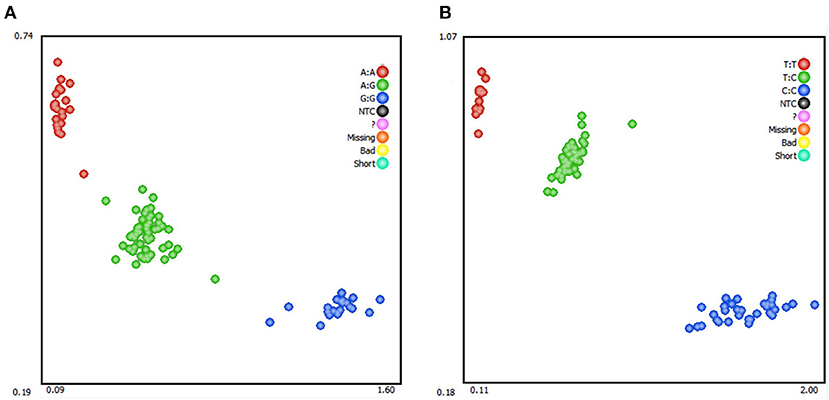
Figure 3. Kaspar-based single nucleotide polymorphism (SNP) genotyping of sheep RAP1GAP g.13561 G > A (A) and rBAT g.1460 T > C (B).
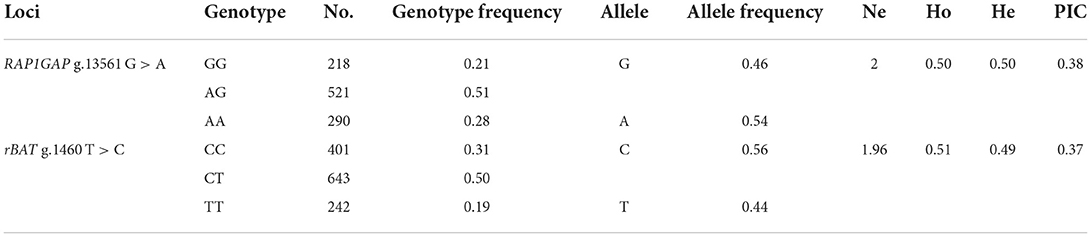
Table 3. Genotype frequency, allele frequency and genetic diversity at the RAP1GAP g.13561 G > A and rBAT g.1460 T > C loci.
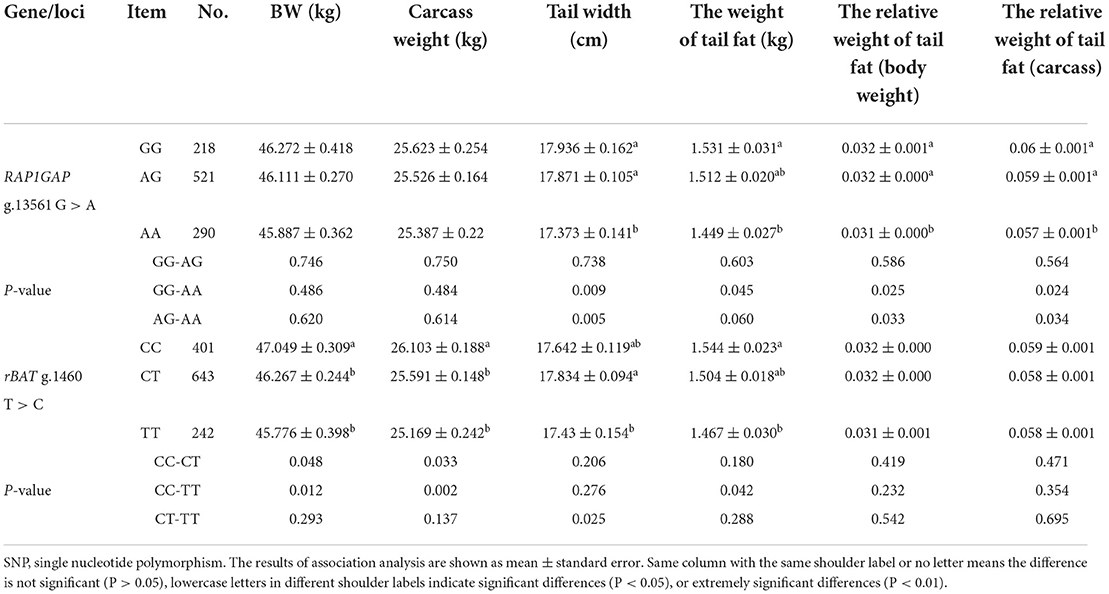
Association analysis of tail fat deposition traits with sheep RAP1GAP and rBAT genes
The phenotypic distribution of tail weight or relative tail weight in Supplementary Table 2. The association analysis between genotype and phenotype showed that SNPs at the RAP1GAP g.13561 G > A and rBAT g.1460 T > C locus were significantly associated with the traits of tail fat deposition (Table 4). Specifically, in the RAP1GAP gene, the tail width, relative tail fat weight (body weight), and relative tail fat weight (carcass) of individuals with AA genotype was significantly lower than with AG and GG genotypes, and the tail fat weight of individuals with AA genotype was significantly lower than with GG genotype (P < 0.05). In the rBAT gene, the tail width of individuals with TT genotype was significantly lower than with CT genotype, and the tail fat weight of individuals with TT genotype was significantly lower than with CC genotype (P < 0.05). However, the SNP at the RAP1GAP g.13561 G > A locus was no significantly associated with 180 day BW and carcass weight (P > 0.05). And the SNP at the rBAT g.1460 T > C locus was significantly associated with 180 days BW and carcass weight (P < 0.05). In a word, the AA and TT genotypes of RAP1GAP g.13561 G > A and rBAT g. 1460 T > C was the dominant genotypes associated with tail fat deposition in sheep, respectively.
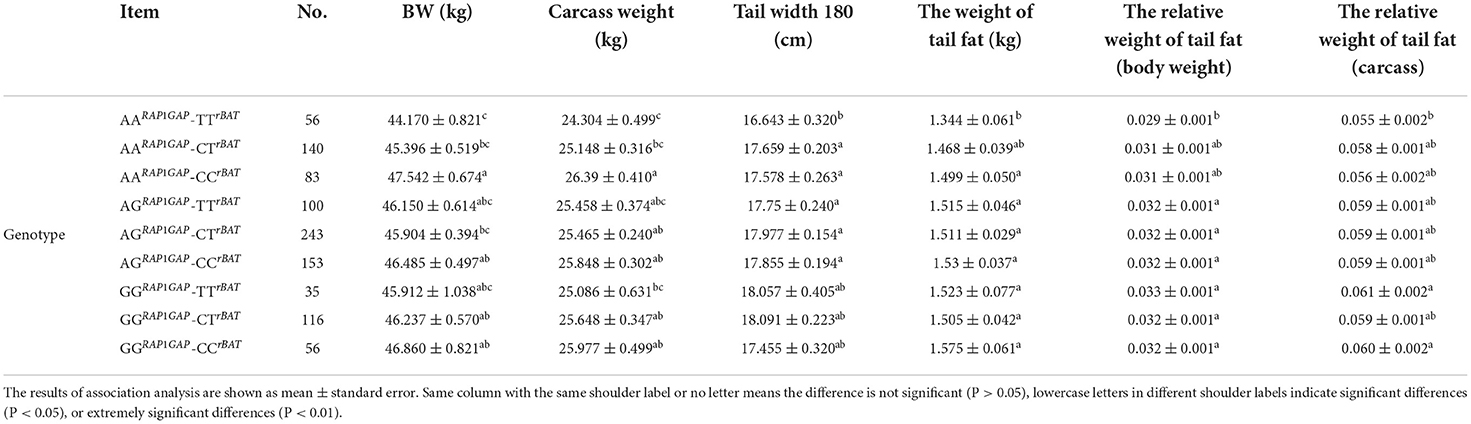
Analysis of RAP1GAP and rBAT genotypes combinations
As shown in Table 5, by combining the genotypes of the two SNPs loci (RAP1GAP g.13561 G > A and rBAT g.1460 T > C), we analyzed the effect of different combinations on tail fat deposition traits. The tail width of AARAP1GAP-TTrBAT genotype individuals was significantly lower than AARAP1GAP-CTrBAT, AARAP1GAP-CCrBAT, AGRAP1GAP-TTrBAT, AGRAP1GAP-CTrBAT, and AGRAP1GAP-CCrBAT genotypes individuals (P < 0.05). The weight of tail fat and the relative weight of tail fat (body weight) of individuals with AARAP1GAP-TTrBAT genotype was significantly lower compared with those of individuals with AGRAP1GAP-TTrBAT, AGRAP1GAP-CTrBAT, AGRAP1GAP-CCrBAT, GGRAP1GAP-TTrBAT, GGRAP1GAP-CTrBAT, and GGRAP1GAP-CCrBAT genotypes (P < 0.05). The relative weight of tail fat (carcass) of AARAP1GAP-TTrBAT genotype individuals was significantly lower than GGRAP1GAP-TTrBAT and GGRAP1GAP-CCrBAT genotypes individuals (P < 0.05). At the same time, combinations between genotypes have an effect on carcass weight and body weight (BW) at 180 days.
Group statistics
We chose big-tail sheep (n = 6) and small-tail sheep (n = 6) to study the expression of RAP1GAP and rBAT genes in tail fat, and calculated the relative weight of tail fat according to the statistics of previous phenotypic data. The tail width, tail fat weight and relative tail fat weight of grouped individuals are shown in Figures 4A–C. The results showed that there was significant difference between the big-tail group and the small-tail group (P < 0.05).
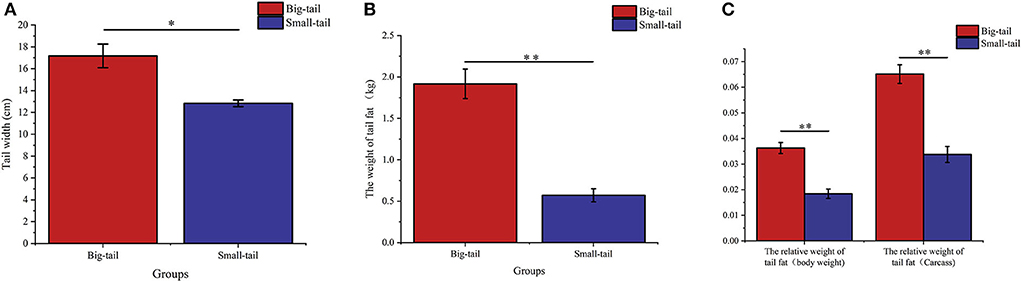
Figure 4. Phenotypic values of tail width (A), tail fat weight (B), and relative tail fat weight (C) of grouped individuals, * indicates a significant difference between the big-tail group and the small-tail group (P < 0.05), ** indicates a very significant difference between the big-tail group and the small-tail group (P < 0.01).
RAP1GAP and rBAT genes expression in sheep small-tail and big-tail groups
In the tail fat tissue, the results of qRT-PCR showed that the level of RAP1GAP gene expression was significantly higher in the small-tail sheep group than in the big-tail sheep group (P < 0.05). However, rBAT gene was significantly lower in the small-tail sheep group than in the big-tail sheep group (P < 0.05; Figure 5).
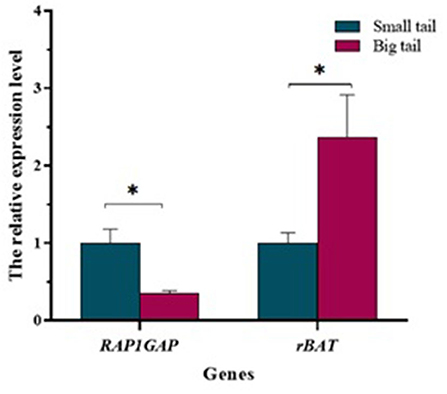
Figure 5. Relative expression of RAP1GAP and rBAT mRNA between the small-tail and big-tail groups. Asterisks indicate significant differences between the small- and big-tail groups (P < 0.05).
Expression of RAP1GAP and rBAT genes in liver at different developmental stages
To investigate RAP1GAP and rBAT gene expression in different developmental stages of the liver, we collected liver tissue samples from sheep at different stages of development (0, 3, and 6 months of age), three for each stage. The results show that in liver tissue, RAP1GAP and rBAT gene expression did not change significantly at 0–3 months of age, but increased dramatically at 6 months of age (P < 0.01; Figure 6).
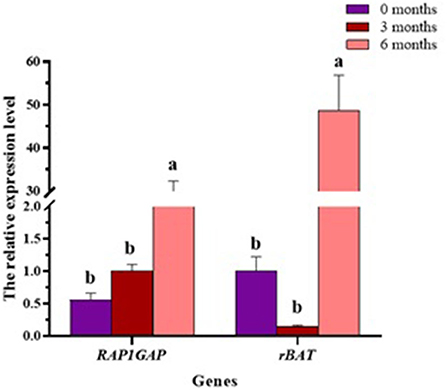
Figure 6. In the liver, the relative expression of RAP1GAP and rBAT at different developmental stages. The same or no letters indicate a non-significant difference (P > 0.05), different letters indicate a significant difference (P < 0.05).
Discussion
High fat diet is easy to lead to obesity and a series of diseases, which is one of the reasons for the change of people's diet structure. People prefer high protein and low-fat diet. In addition, in the intensive breeding of sheep, tail fat is no longer important in providing energy during the cold season, and the excessive deposition of tail fat increases the breeding cost and reduces the meat quality grade. Therefore, in order to maximize economic benefits and reduce feeding costs, commercial breeding urgently needs to cultivate sheep varieties with small tail shape. Nowadays, the mature application of marker assisted selection (MAS) may make this need a reality.
Adding the same amount of fat and lean tissue to the tail requires a greater energy cost. In addition, the price of fat tail per kilogram is lower than that of red meat. Therefore, sheep producing small fat tail have more economic value in actual production (23). Understanding the genetic mechanism of sheep tail size is very important for future sheep molecular breeding. Previous studies have found that overexpression of RAP1GAP has a negative regulatory effect on Rap1, which can enhance the cellular effect of leptin and reduce the production of obesity (6–8). rBAT mutation can cause damage to the transport of plasma membrane, and then affect the outflow of arginine in cells (24). Arginine supplementation increases lipolysis and inhibits lipogenesis in white adipose tissue (25). Therefore, it is very important to explore the potential effects of RAP1GAP and rBAT genes on important economic traits of sheep. Sanger sequencing showed that two novel polymorphic sites (g.13561 G > A and g.1460 T > C) were found in the intron region of RAP1GAP and rBAT genes in sheep. It is well-known that intronic mutations do not change amino acid sequences. However, relevant studies have shown that mutations located in introns may have similar effects to synonymous mutations (26–28), which can affect some important cellular processes, such as mRNA stability (29), protein folding and function (30), mRNA translation (31), mRNA folding (32), or splicing (33). The occurrence of intronic mutations can affect some important economic traits of animals, such as growth traits (20), tail fat deposition traits (34), feed efficiency (35), and litter size (36). To investigate whether intronic mutations affect phenotypes in sheep, this study assessed the relationship between different genotypes and tail fat deposition traits. The results revealed that the RAP1GAP gene with a SNP of g.13561 G > A and the rBAT gene with a SNP of g.1460 T > C were significantly associated with tail fat deposition traits (P < 0.05). We also found that the combined genotypes of RAP1GAP g.13561 G > A and rBAT g.1460 T > C also differed for tail fat deposition, which is consistent with previous reports (34).
At the same time, the expression of RAP1GAP and rBAT genes was detected in adipose tissue of big-tail group and small-tail group. The results showed that the expression of RAP1GAP gene was abundant in the small-tail group. This may be because the extensive expression of RAP1GAP gene in the small-tail group inhibits Rap1, enhances the cellular effect of leptin, and thus reduces fat deposition (9). It may also be that the extensive expression of RAP1GAP gene inhibits the proliferation and survival of tail adipocytes (6). The expression of rBAT gene is abundant in the big tail group, which may be because rBAT mutation leads to the weakening of plasma membrane transport capacity, affecting the outflow of arginine in cells, and the reduction of arginine promotes the production of fat (24, 25). In addition, the liver plays an important physiological role in fat production (37). In ruminants and humans, the endogenous fat released by fat decomposition increases the content of circulating non-esterified fatty acids into the liver, which is the main source of triglyceride accumulation in the liver (38). It was found that RAP1GAP and rBAT genes were widely expressed in the liver at the age of 6 months.
In conclusion, RAP1GAP and rBAT genes might play an important role in the development of adipocytes. Therefore, the polymorphisms of RAP1GAP and rBAT genes are of great significance for the breeding of smaller tail sheep, and can be used as candidate molecular markers. There are still some limitations in our research. We analyzed the association between polymorphisms of RAP1GAP and rBAT genes and tail fat deposition traits, which was verified in Hu sheep population. However, further studies are needed to verify its relationship with other breeds of big tailed sheep and its function at the cellular and protein levels.
Conclusion
In this study, a novel study was performed to explore the presence of polymorphisms on ovine RAP1GAP and rBAT genes, to investigate their association with tail fat deposition traits in Hu sheep, and to detect their expression features in tail fat and liver tissue. The results showed that two SNP g.13561 G > A and g.1460 T > C were identified in RAP1GAP and rBAT genes which were significantly associated with tail fat deposition traits. In addition, the expression level of RAP1GAP gene in the small-tail group was more abundant than that in the big-tail group, on the contrary, the expression of rBAT gene was more abundant in the big-tail group. During the development of liver, the expression levels of RAP1GAP and rBAT genes peaked at 6-month of age. Therefore, the two newly discovered polymorphic loci can be used as potential molecular markers for breeding smaller tailed sheep.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank Accession Nos. NC_040253.1 and 106 NC_040254.1.
Ethics statement
The animal study was reviewed and approved by Ethics Committee of Gansu Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XZh, WW, and ZM conceived and designed the study. YZhan, YZhao, JW, BZ, WL, CL, JC, DX, XZe, JL, RZ, YH, and PC collected samples. ZM, LZ, DZ, XL, and XY performed the experiments and analyzed the data. ZM wrote the paper. XZh revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31960653), the National Joint Research Program for Animal and Poultry Breeding (19210365), and the Gansu Provincial Key Research and Development Program (20YF3NA012).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.974513/full#supplementary-material
References
1. Moradi MH, Nejati-Javaremi A, Moradi-Shahrbabak M, Dodds KG, Mcewan JC. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genetics. (2012) 13:10. doi: 10.1186/1471-2156-13-10
2. Ermias E, Yami A, Rege J. Fat deposition in tropical sheep as adaptive attribute to periodic feed fluctuation. J Anim Breed Genet. (2002) 119:344. doi: 10.1046/j.1439-0388.2002.00344.x
3. Kashan N, Azar G, Afzalzadeh A, Salehi A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Ruminant Res. (2005) 60:267–71. doi: 10.1016/j.smallrumres.2005.01.001
4. Rubinfeld B, Munemitsu S, Clark R, Conroy L, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. (1991) 65:1033–42. doi: 10.1016/0092-8674(91)90555-D
5. Frische EW, Zwartkruis FJ. Rap1, a mercenary among the Ras-like GTPases. Dev Biol. (2010) 340:1–9. doi: 10.1016/j.ydbio.2009.12.043
6. Zheng H, Gao L, Feng Y, Yuan L, Zhao H, Cornelius LA. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. (2009) 69:449–57. doi: 10.1158/0008-5472.CAN-08-2399
7. Fukuda M, Williams KW, Gautron L, Elmquist JK. Induction of leptin resistance by activation of cAMP-Epac signaling. Cell Metab. (2011) 13:331–9. doi: 10.1016/j.cmet.2011.01.016
8. Sands WA, Woolson HD, Milne GR, Rutherford C, Palmer TM. Exchange protein activated by cyclic AMP (Epac)-mediated induction of suppressor of cytokine signaling 3 (SOCS-3) in vascular endothelial cells. Mol Cell Biol. (2006) 26:6333–46. doi: 10.1128/MCB.00207-06
9. Kaneko K, Xu P, Cordonier EL, Chen SS, Ng A, Xu Y, et al. Neuronal Rap1 regulates energy balance, glucose homeostasis, and leptin actions. Cell Rep. (2016) 16:3003–15. doi: 10.1016/j.celrep.2016.08.039
10. Fotiadis D, Kanai Y, Palacín M. The SLC3 and SLC7 families of amino acid transporters. Mol Aspects Med. (2013) 34:139–58. doi: 10.1016/j.mam.2012.10.007
11. Furriols M, Chillarón J, Mora C, Castelló A, Bertran J, Camps M, et al. rBAT, related to L-cysteine transport, is localized to the microvilli of proximal straight tubules, and its expression is regulated in kidney by development. J Biol Chem. (1993) 268:27060–8. doi: 10.1016/S0021-9258(19)74218-5
12. Pickel VM, Nirenberg MJ, Chan J, Mosckovitz R, Udenfriend S, Tate SS. Ultrastructural localization of a neutral and basic amino acid transporter in rat kidney and intestine. Proc Nat Acad Sci USA. (1993) 90:7779–83. doi: 10.1073/pnas.90.16.7779
13. Feliubadaló L, Mariona F, Jesús P. Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat Genet. (1999) 1999:12652. doi: 10.1038/12652
14. Nagamori S, Wiriyasermkul P, Guarch ME, Okuyama H, Kanai Y. Novel cystine transporter in renal proximal tubule identified as a missing partner of cystinuria-related plasma membrane protein rBAT/SLC3A1. Proc Natl Acad Sci USA. (2016) 113:1254–60. doi: 10.1073/pnas.1519959113
15. Richards MP, Poch SM, Coon CN, Rosebrough RW, Ashwell CM, Mcmurtry JP. Feed restriction significantly alters lipogenic gene expression in broiler breeder chickens. J Nutr. (2003) 133:707. doi: 10.1093/jn/133.3.707
16. O'Hea EK, Leveille GA. Significance of adipose tissue and liver as sites of fatty acid synthesis in the pig and the efficiency of utilization of various substrates for lipogenesis. J Nutr. (1969) 99:338–44. doi: 10.1093/jn/99.3.338
17. Mourot J, Kouba M, Peiniau P. Comparative study of in vitro lipogenesis in various adipose tissues in the growing domestic pig (Sus domesticus). Comparat Biochem Physiol B. (1995) 111:379–84. doi: 10.1016/0305-0491(95)00005-S
18. Diao Z, Yanghua QU, Liu C, Gao Y, Luo H. Effects of forage canola mixed silage on slaughter performance and meat quality in Hu sheep. China Anim Husbandry Vet Med. (2018) 45:1564–70. doi: 10.16431/j.cnki.1671-7236.2018.06.018
19. Feng X, Li F, Wang F, Zhang G, Pang J, Ren C, et al. Genome-wide differential expression profiling of mRNAs and lncRNAs associated with prolificacy in Hu sheep. Biosci Rep. (2018) 2018:BSR20171350. doi: 10.1042/BSR20171350
20. Zhao L, Li F, Yuan L, Zhang X, Zhang D, Li X, et al. Expression of ovine CTNNA3 and CAP2 genes and their association with growth traits. Gene. (2022) 807:145949. doi: 10.1016/j.gene.2021.145949
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
22. Zhao H, Wu X, Cai H, Pan C, Lei C, Chen H, et al. Genetic variants and effects on milk traits of the caprine paired-like homeodomain transcription factor 2 (PITX2) gene in dairy goats. Gene. (2013) 532:203–10. doi: 10.1016/j.gene.2013.09.062
23. Safdarian M, Zamiri MJ, Hashemi M, Noorolahi H. Relationships of fat-tail dimensions with fat-tail weight and carcass characteristics at different slaughter weights of Torki-Ghashghaii sheep. Meat Sci. (2008) 80:686–9. doi: 10.1016/j.meatsci.2008.03.007
24. Pineda M, Wagner CA, Bröer A, Stehberger PA, Bröer S. Cystinuria-specific rBAT(R365W) mutation reveals two translocation pathways in the amino acid transporter rBAT-b0,+AT. Biochem J. (2004) 377:665–74. doi: 10.1042/bj20030956
25. Jobgen W, Fu WJ, Gao H, Li P, Meininger CJ, Smith SB, et al. High fat feeding and dietary L-arginine supplementation differentially regulate gene expression in rat white adipose tissue. Amino Acids. (2009) 37:187–98. doi: 10.1007/s00726-009-0246-7
26. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. (2002) 3:285–98. doi: 10.1038/nrg775
27. Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, et al. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem. (2010) 285:28741–8. doi: 10.1074/jbc.M110.154575
28. Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. (2011) 12:683–91. doi: 10.1038/nrg3051
29. Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. (2003). doi: 10.1093/hmg/ddg055
30. Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Cakagno AM, Ambudkar SV, et al. A 'silent' polymorphism in the MDR1 gene changes substrate specificity. Science. (2007) 315:525–8. doi: 10.1126/science.1135308
31. Drummond D, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. (2008) 134:341–52. doi: 10.1016/j.cell.2008.05.042
32. Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. (2013) 342:475–9. doi: 10.1126/science.1241934
33. Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol Biol Evol. (2006) 23:301–9. doi: 10.1093/molbev/msj035
34. Lz A, Flb C, Teng LD, Ly B, Xz A, Dz A, et al. Ovine ELOVL5 and FASN genes polymorphisms and their correlations with sheep tail fat deposition. Gene. (2021) 807:145954. doi: 10.1016/j.gene.2021.145954
35. Zhang D, Zhang X, Li F, Yuan L, Zhang Y, Li X, et al. Polymorphisms in ovine ME1 and CA1 genes and their association with feed efficiency in Hu sheep. J Anim Breed Genet. (2021) 138:589–99. doi: 10.1111/jbg.12541
36. Wang F, Chu M, Pan L, Wang X, He X, Zhang R, et al. Polymorphism detection of GDF9 gene and its association with litter size in luzhong mutton sheep (Ovis aries). Animals. (2021) 11:20571. doi: 10.3390/ani11020571
37. Goceri E, Shah ZK, Layman R, Jiang X, Gurcan MN. Quantification of liver fat: a comprehensive review. Comput Biol Med. (2016) 71:174–89. doi: 10.1016/j.compbiomed.2016.02.013
Keywords: RAP1GAP, rBAT, single nucleotide polymorphisms, tail fat deposition, Hu sheep
Citation: Ma Z, Wang W, Zhang D, Zhang Y, Zhao Y, Li X, Zhao L, Lin C, Wang J, Zhou B, Cheng J, Xu D, Li W, Yang X, Huang Y, Cui P, Liu J, Zeng X, Zhai R and Zhang X (2022) Ovine RAP1GAP and rBAT gene polymorphisms and their association with tail fat deposition in Hu sheep. Front. Vet. Sci. 9:974513. doi: 10.3389/fvets.2022.974513
Received: 21 June 2022; Accepted: 08 August 2022;
Published: 25 August 2022.
Edited by:
Ran Di, Institute of Animal Sciences (CAAS), ChinaReviewed by:
Xiang Zhou, Huazhong Agricultural University, ChinaLyu Fenghua, China Agricultural University, China
Sangang He, Xinjiang Academy of Animal Science, China
Copyright © 2022 Ma, Wang, Zhang, Zhang, Zhao, Li, Zhao, Lin, Wang, Zhou, Cheng, Xu, Li, Yang, Huang, Cui, Liu, Zeng, Zhai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxue Zhang, emhhbmd4eEBnc2F1LmVkdS5jbg==
 Zongwu Ma
Zongwu Ma Weimin Wang
Weimin Wang Deyin Zhang2
Deyin Zhang2 Yuan Zhao
Yuan Zhao Jiangbo Cheng
Jiangbo Cheng Xiaoxue Zhang
Xiaoxue Zhang