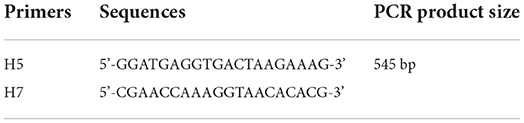- Laboratory of Animal Epidemic Diseases, College of Animal Science, Yangtze University, Jingzhou, China
A strain of avian leukosis virus (ALV) belonging to a new envelope subgroup J (ALV-J) emerged in 1988 as a new subgroup of ALV and spread rapidly throughout the world. Due to the infection and spread of ALV-J, the global poultry industry experienced a significant loss. Although the disease had been prevented and controlled effectively by culling domestic chickens in the infected zone, a few field cases of ALV-J infection were reported in China in recent years. This study was conducted to characterize the genome and analyze the lesions and histopathology of the ALV-J strain named HB2020, which was isolated from layer chickens in Hubei Province, China. The full-length proviral genome sequence analysis of ALV-J HB2020 revealed that it was a recombinant strain of ev-1 and HPRS-103 in the gag gene in comparison to ALV-J prototype HPRS-103. In the 3′-untranslated region (3'UTR) of the nucleotide sequence, there were found 205-base pairs (bp) deletion, of which 175 were detected in the redundant transmembrane (rTM) region. Besides, the surface glycoprotein gene gp85 had five mutations in a conservative site, whereas the transmembrane protein gene gp37 was relatively conserved. The animal experiments conducted later on this strain have shown that HB2020 can cause various neoplastic lesions in chickens, including enlarged livers with hemangiomas and spleens with white nodules. Additionally, as the exposure time increased, the number of tumor cells that resembled myelocytes in the blood smears of infected chickens gradually increased. These results indicated that HB2020 on recombination with ALV subgroup E (ALV-E) and ALV-J could induce severe hemangiomas and myelocytomas. This inference might provide a molecular basis for further research about the pathogenicity of ALV and emphasize the need for control and prevention of avian leukosis.
Introduction
Avian leukosis virus (ALV) is an retrovirus that causes a variety of neoplastic diseases in poultry and has spread worldwide since its discovery in the early twentieth century (1). ALV infection in chickens could not only induce tumors and mortality but also cause a variety of diseases as a result of immunosuppression, which could affect their immunological response to vaccination and production performance (2). Furthermore, ALV has the potential to propagate vertically, causing injury to the progeny chickens and affecting the quality of commercial chickens.
The outbreak of avian leukosis subgroup J (ALV-J) among broiler breeders in 1988 was a major setback for the global chicken industry. The first evidence that ALV-J affects both layers and broilers appeared from the 2002 discovery of ALV-J in commercial layers in China (3). This discovery was significant as it laid the foundation for ALV-related research while also serving as a caution to the poultry industry, particularly layer farms.
ALVs are highly diverse RNA viruses with multiple subtypes that cause cancer in poultry (4). According to the properties of the viral envelope proteins, ALVs are divided into 10 subgroups (A-J), and the most recently identified subgroup is K (5).
ALVs isolated from chickens are classified into seven subgroups, such as ALV-A, ALV-B, ALV-C, ALV-D, ALV-E, ALV-J, and ALV-K (6). The genome of ALV is about 7.6–7.8 kb in length and possesses the classic slow transformation retrovirus genome structure 5'-R-U5-gag-pol-env-U3-R-3'. A variety of proteins are encoded by the gag gene, among which the major gag proteins include capsid protein (CA, p27), matrix protein (MA, p19), p10, nucleocapsid protein (NC, p12), and protease (PR, p15) (7). A previous study has revealed that a mutation in the pol gene could lead to increased reverse transcriptase activity, replication capacity, and increased vertical transmission capability of ALV-K; furthermore, this mutation is genetically stable, indicating the importance of the pol gene in ALV transmission (8).
The ALV env gene encodes envelope glycosylation protein, which is linked together with the surface glycoprotein subunit (SU) encoded by the gp85 gene, and the transmembrane glycoprotein subunit (TM) encoded by the gp37 gene into a rod-shaped dimer that is attached to the envelope surface, namely virus glycoprotein (9). Gp85 is the ligand of the virus utilized to invade host cells, which also serves as the foundation for subgroup categorization (10). The significance of long terminal repeat (LTR) and gp85 in the etiology of ALV-J has been confirmed by previous studies (11).
As a non-coding regulatory platform, the mRNA 3′-untranslated region (3′UTR) may influence mRNA stability and translation (12). The 3'UTR is critical to viral RNA replication as viral RNA molecules are typically replicated from the 3'-end to the 5'-end of the genome (13). Within the 3′UTR, ALV unique region 3 (U3) is unconservative (14). The typical transcriptional regulatory elements, namely CAAT, CArG, PRE, TATA, and Y boxes present in U3 were well conserved previously but began to alter in the most recent strain. The 11 bp deletion in U3 of ALV-J was suggested to be connected to the incidence of hemangioma in 2011 (15).
As ALV is retrovirus, it undergoes regular recombination, resulting in the emergence of new strains and subgroups. ALV-J is the most common novel subtype obtained from recombination, and this subtype was also suspected of being a recombinant virus (16). Until now, many recombinant strains have been discovered to be made up of various subpopulations, and it has been demonstrated that ALV infection ability can be altered following recombination (17). As a result, attention should be paid more to ALV recombination, and more in-depth studies should be conducted for disease prevention and control.
In this study, an ALV strain named HB2020 was isolated from infected layer chickens obtained from Jingzhou, Hubei province, which was identified as a J subtype subsequently using polymerase chain reaction (PCR). To analyze the sequence homology and divergence of HB2020, the whole genome was sequenced and compared with other ALV strains. As a result, the strain HB2020 was determined to be a hybrid of ALV-E and ALV-J. Following that, the embryos of 11-day-old chickens were intravascularly intravenously inoculated with the HB2020 strain, and their clinical symptoms were meticulously documented to study its pathogenicity. To understand the pathogenicity and variation of the ALV-J, these results are expected to provide a basis for further investigation. A theoretical framework for recent study on the pathogenicity and variation of the ALV-J strain circulated in China recently might be supplied by the foregoing experimental results.
Methods
Clinical examination
In April 2020, a large number of layer chickens belonging to a commercial indigenous chicken farm in Hubei province of China displayed thinness and decreased egg production rate, and some chickens passed away before they attained 60 days of age. The chicken flocks' morbidity and mortality were 14.9 and 14.6%, respectively. From the farm mentioned previously, five 20-weeks-old layer chickens with substantial clinical symptoms in egg production were submitted to our laboratory for further diagnosis.
Virus isolation
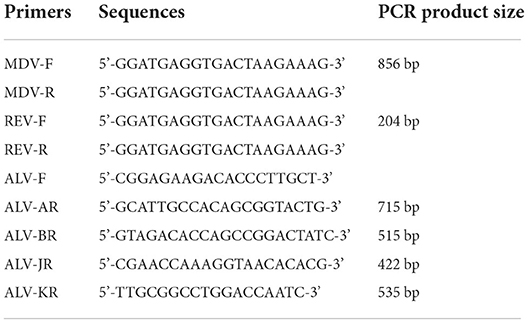
Virus isolation was performed using DF-1 chicken fibroblast cell line without endogenous ALV gene to exclude endogenous ALV interference. The DF-1 cells were planted in a six-well plate containing Dulbecco's Modified Eagle Medium (DMEM, Invitrogen®) with 10% fetal bovine serum (FBS, Gemini®). At 70% confluence, the DF-1 cells were co-cultured with filtered liver homogenates obtained from the positive sample and incubated for 2 h at 37°C. The cells were then continuously grown in DMEM containing 1% FBS in an incubator with 5% CO2 at 37°C. The supernatant from infected cells was collected after three blind passes to identify ALV using an ALV group-specific antigen (p27) enzyme-linked immunosorbent assay (ELISA, IDEXX®). Positive cells were collected for proviral DNA extraction employing the Genomic DNA Extraction Kit (GK0222, Generay Biotech. Co. Ltd., Shanghai, China) based on the manufacturer's instructions, subsequently eluted with 50 μL of DNase-free water, and kept at −80°C. The DNA extracted from P27-positive cells was identified using PCR to exclude Marek's disease virus (MDV) (18) and reticuloendotheliosis virus (REV) (19) (Table 1). The MDV and REV cases used as positive controls in PCR were archived in our laboratory (20, 21). Primer pairs A/B/J/K-F/R (Table 1) were used to detect ALV and define its subgroup (22, 23). The median tissue culture infectious dose (TCID50) in these cells' supernatant was calculated by the Reed-Muench method.
Phylogenetic and recombination analysis
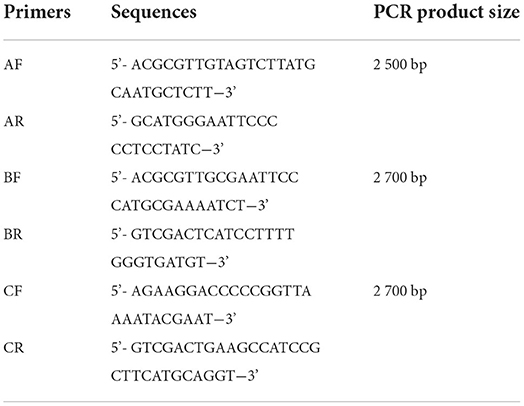
The entire proviral genome of HB2020 was amplified by a segmented PCR experiment (Table 2), using the template DNA extracted from the ELISA-positive cells as previously described. The whole proviral genomic DNA of HB2020 was sequenced and spliced using DNASTAR software. Sequence alignment with other ALV strains was performed using the DNASTAR software and the NCBI BLAST tool. Major genome-wide genes, such as gag, pol, gp37, gp85, and LTR, were compared with other ALV strains (Table 3). In molecular evolutionary genetics analysis version 6.0 (MEGA 6), the neighbor-joining approach was used to perform phylogenetic analysis with 1,000 bootstrap repetitions. Recombination detection program version 5 (RDP5) and SimPlot software (V3.5.1) were used to examine the possible recombination events for the entire nucleotide sequence of HB2020; the Kimura two-parameter model was used to map them, and 100 bootstrap replicates were employed to evaluate the reliability of the recombination event. The parental threshold cut-off value was set above 70%.
Analysis of the pathogenicity
Sixty embryonated specific pathogen-free (SPF) eggs (Specific Pathogen Free Avian Supplies) were split into two groups (an infection group and a negative control group) randomly, each comprising 30 embryonated eggs. At the age of 12 embryonic days, eggs in the infection group were intravenously inoculated with ALV-J of 104 TCID50 (24). The rest of the cohort was subjected to DMEM inoculation in the same manner. The chicks that attained the age of 3 days were vaccinated against MDV. Thereupon, PCR amplification of ALV-J was performed with H5/H7 primer to detect if the chicks were infected with ALV-J (Table 4). To generate blood smears, blood was withdrawn from each chick at the age of 4, 10, and 16 weeks throughout the chicken cultivation phase; a drop of fresh blood was taken and smeared evenly on a clean slide and stained with Wright-Giemsa (BASO BA-4017, Zhuhai, China) for examination. All chicks were subjected to bloodletting, and a thorough necropsy was performed on them at the age of 21 weeks.
Results
Virus isolation and identification
An ALV-J strain, named HB2020 (GenBank accession number: ON840093), was isolated and identified from the affected layer chickens using DF-1 cell co-culture and a PCR assay. Seven days after inoculation, the supernatants of infected DF-1 cells yielded a positive result using the ALV group-specific antigen (p27) ELISA (IDEXX), indicating the presence of ALV in the samples. PCR with the primers for MDV and REV exhibited negative results (Figure 1). The multi-PCR for A/B/J and A/J/K generated a 422 bp fragment (Figure 2), indicating that HB2020 is ALV-J. The Reed-Muench method determined that each 0.1 mL of DF-1 cellular supernatant had 104 TCID50.

Figure 1. (A) The PCR result with primer MDV-F/R. M:maker, 1: negative control, 2: positive control, 3:HB2020. (B) The PCR result with primer REV-F/R. M:maker, 1: negative control, 2: positive control, 3:HB2020.
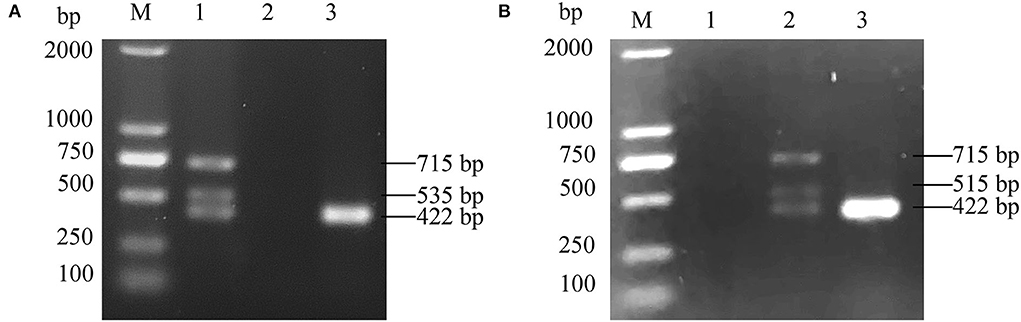
Figure 2. (A) The PCR result of A/J/K Multi-PCR. M:maker, 1: positive control, 2: negative control, 3:HB2020. (B) The PCR result of A/B/J Multi-PCR. M:maker, 1: negative control, 2: positive control, 3:HB2020.
Molecular characterization of the genome
The entire genome of HB2020 was 7471 bp long, assembled by the SeqMan program of DNASTAR software. In the heat map, HB2020 clustered in the same region as the other ALV-J (Figure 3). Phylogenetic analysis of the gag genes revealed two distinct branches: gag regions of HB2020 clustered with ev-1, and the ALV prototype HPRS-103 clustered with Rous sarcoma virus (strain Prague C) (RSV-PrC) and Schmidt-Ruppin D strain of RSV (SR-RSV-D(H) (Figure 4B). The genes and functional regions (LTR, pol, gp85, and gp37) of HB2020 and HPRS-103 were found in the same branch in their respective phylogenetic trees (Figure 4). The analysis of recombination events revealed that the major recombination sites of HB2020 and ev-1 were located in gag regions (Figure 5).
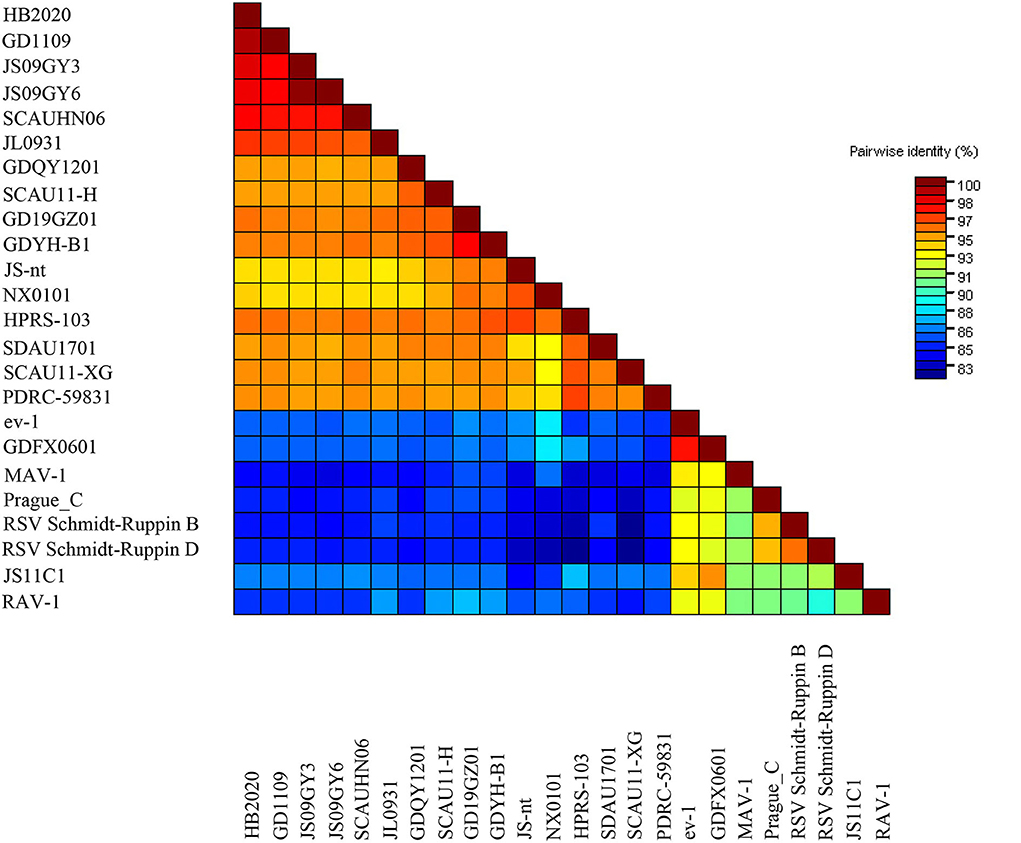
Figure 3. The pairwise identities plot of the entire gene for the HB2020 isolates reported in this study (red colored), aligned by ClustalW and displayed by the Sequence Demarcation Tool (SDT) software.
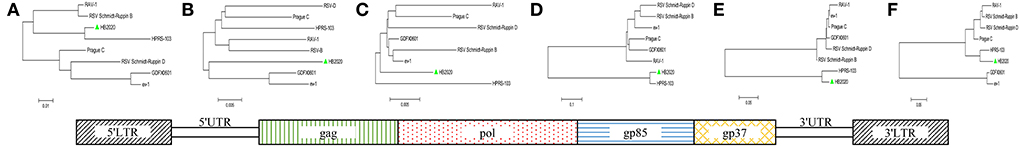
Figure 4. ALV isolate HB2020 and other ALVs were compared in terms of segmental sequences. The genome sequences were divided into functional areas, and each segmental sequence underwent sequence alignment. The genome structure of the isolate HB2020 is shown in the bottom box. Phylogenetic tree: (A) 5'LTR. (B) gag. (C) pol. (D) gp85. (E) gp37. (F) 3'LTR. The Neighbor-Joining method with 1,000 bootstrap replicates was used to infer the evolutionary relationships of each fragment (MEGA 6).
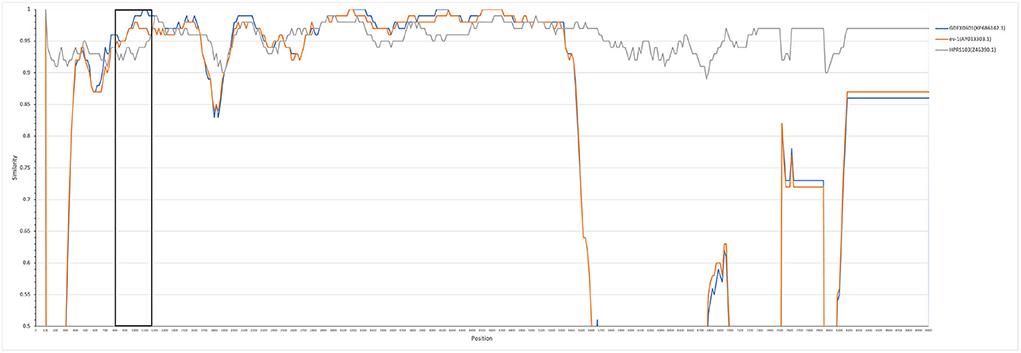
Figure 5. HB2020 recombination analysis SimPlot software (V3.5.1) was used to analyze recombination events, which were then mapped using the Kimura (2-parameter) model. The reliability of the recombination event was assessed using a 100-bootstrap, and a parental threshold of > 70% was chosen as the cut-off value.
Sequence analysis of the gp85 gene
Mutations were more likely to occur in the hypervariable regions 1 (hr1), 2 (hr2), and variable region 3 (vr3) of HB2020, with 32 mutations found in the gp85-encoded surface glycoprotein (SU) (Figure 6). Five (N58, K70, A71, K108, and N112) of the nine (N58, D60, K70, A71, K108, N112, N113, N121, and R272) frequent mutations in ALV-J were discovered in SU of HB2020. In addition, mutations in three amino acids (E109, R112, and N118) were found in the hr1 region of ALV-J that causes myeloid leukosis (Figure 6). The mutation of amino acid Q235 in the vr3 of HB2020 had not been reported in viruses that induce both myeloid and hemangiomatous avian leukosis cited in this study (JL093-1, GD1109, JS09GY3, JS09GY6, PDRC-5983, SCAU11-H, SCAU11-XG) (Figure 6).
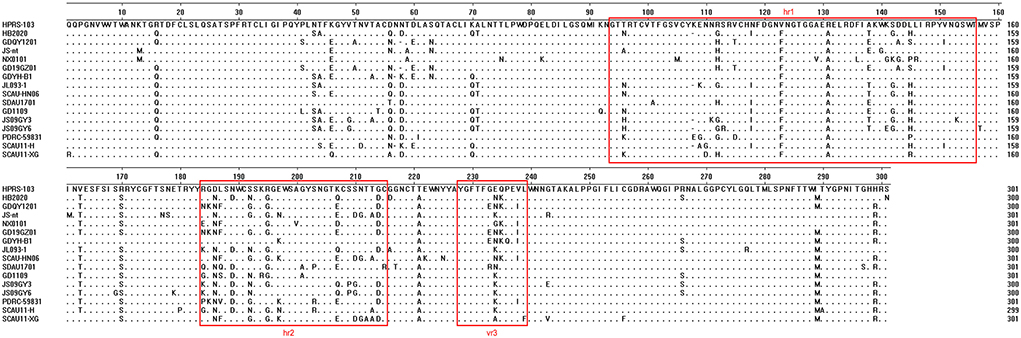
Figure 6. Comparison of the amino acid sequences of gp85 from ALV-J isolates and the HB2020 strain. The inferred gp85 was compared to other ALV strains. The colored boxes represent the gp85 hr1, hr2, and vr3 regions.
Sequence analysis of the 3'UTR gene
The 3'UTR of HB2020 was 542 bp long, containing the rTM region, direct repeat 1 (DR1), and the E element. There was a 175 bp deletion in rTM of HB2020, which was consistent with ALV-J (JL093–1) isolated from a hemangioma case and numerous ALV-J strains (GD1109, JS09GY3, and JS09GY6) that potentially induced both hemangiomas and myeloma (Figure 7). The DR1 was exceedingly conservative in these referenced ALV-J, and the complete initial E element of HB2020 was 174 bp long (Figure 7). The SoftBerry NSITE online service analysis system was employed to analyze the transcriptional regulatory elements; the result revealed that in the U3 region of the HB2020 genome, the transcriptional regulatory elements, including C/EBP, E2BP, CArG box, and Y box, were substantially conserved in comparison to HPRS-103 strain (Figure 8). In contrast to the HPRS-103 strain, the AIB REP1 transcription factor of HB2020 possessed two mutant bases (Figure 8).
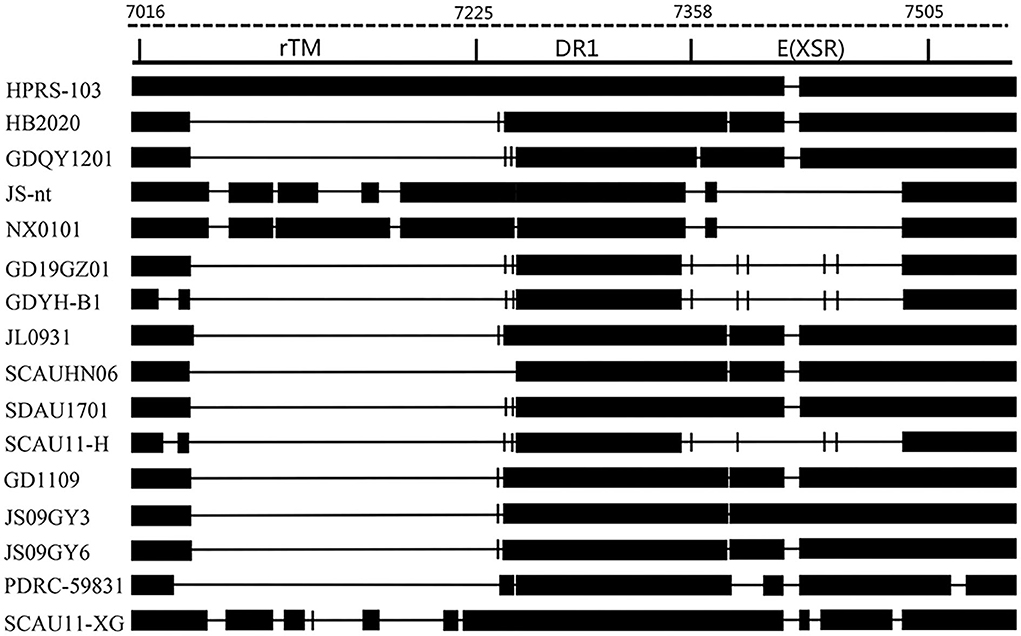
Figure 7. Comparison of the nucleotide and important regulatory elements of the HB2020 and other ALVs' 3'UTR regions.
Gross and histological lesions
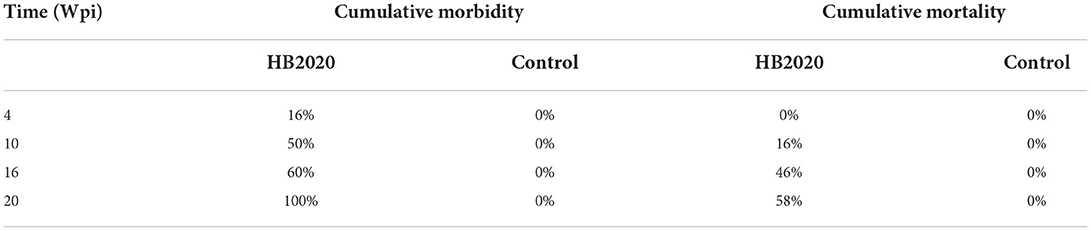
After incubation, 24 infected chicks survived, in comparison to 26 survivors in the uninfected control group. Blood cells' DNA was extracted from two chickens of the negative control group chosen randomly and tested using PCR with the H5/H7 primer. The result revealed that there were no positive findings, indicating that the chicken embryos had not been infected with ALV-J. At 4 weeks post infections (wpi), all of the chickens in the infected group were positive when PCR was performed with the H5/H7 primer, and four chickens in the infected group exhibited clinical symptoms, such as loss of appetite, listlessness, and diarrhea. At 10 wpi, the symptoms worsened (Table 5). The first instance of the clinical tumor was detected at 16 wpi, and nine further cases were detected at 20 wpi.
The dead affected chickens were immediately examined by necropsies. Most layer chickens in the infected group exhibited typical necropsy lesions of avian leukosis. Numerous hemangiomas in their dark red liver were spotted (Figure 9A), and the spleen was grossly enlarged with numerous white necrotic lesions of varying sizes (Figure 9C). The kidneys were swollen and covered with white nodules (Figure 9E). Under the microscope, vascular endothelial cells were hyperplastic and clustered in the liver (Figure 9B); splenic vascular endothelial cells were hyperplastic and clustered, with partial lymphocyte nuclei being absent (Figure 9D), and the kidney revealed a large number of myeloid tumor cells (Figure 9F).

Figure 9. Anatomical and histological lesions. (A) Hemangioma on the liver surface. (B) Vascular endothelial hyperplasia, HE, 40×. (C) Lesions on the surface of the spleen. (D) Proliferation of capillary vessels, HE, 400×. (E) Kidney enlarged with multifocal lesions. (F) Kidney cells without nucleolus, HE, 400×.
Blood analysis
Blood smears revealed the presence of a few myeloid tumor cells in the blood of four-week-old chickens of the infected group (Figures 10A,B). Myeloid tumor cells were large and spherical, with a round or oval nucleus on one side of the cell and a substantial number of acidophilic granules in the cytoplasm (Figures 10A,B). An increase in myeloid tumor cells was observed in the blood smears of 10-week-old chickens (Figures 10C,D). A large number of myeloid tumor cells were observed microscopically in the blood smears of 16-week-old chickens (Figures 10E,F).
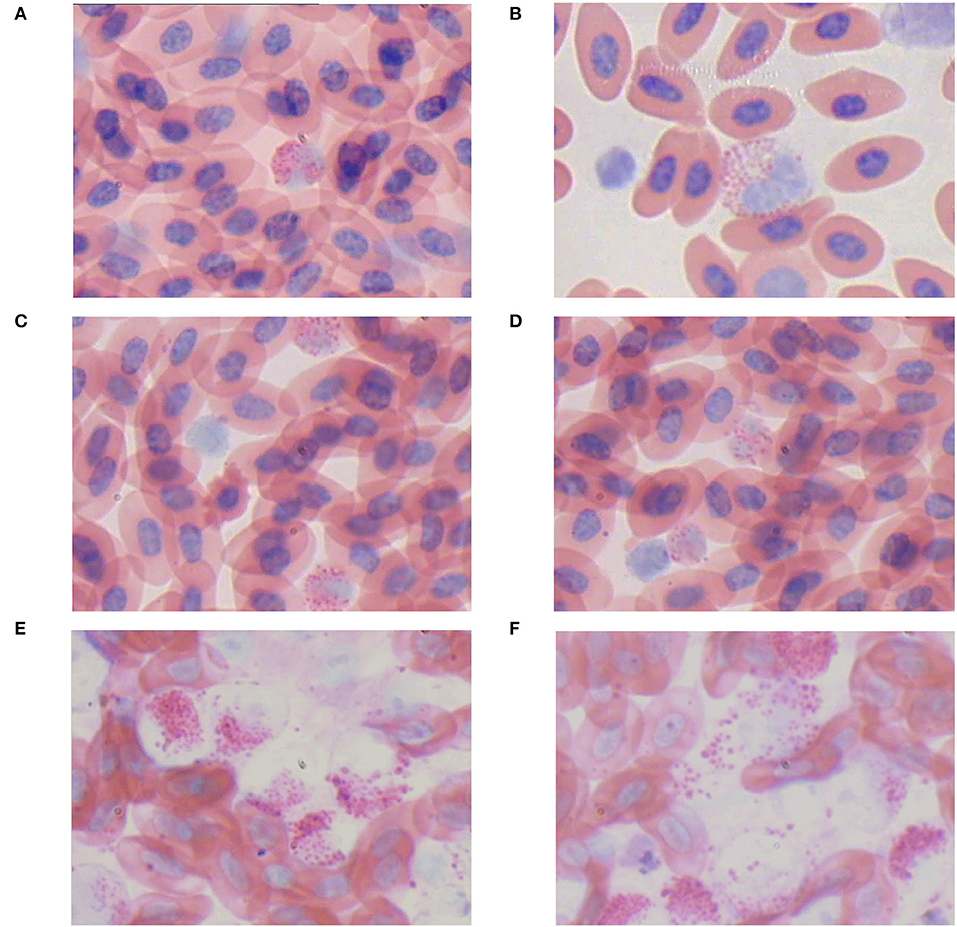
Figure 10. Blood swears (ME, 400×). (A,B) An myelocyte-like tumor cell. (C,D) The number of myelocyte-like tumor cell was increased. (E,F) The lysis of myelocyte-like tumor cells.
Discussion
In 1999, it was first reported that some broiler flocks from China had been infected with ALV-J. Subsequently, ALV-J spread rapidly in Chinese chicken flocks, causing huge losses to the Chinese chicken industry. This, along with the continued expansion of the virus' host range and the emergence of new strains with novel symptoms and lesions induced, sparked people's widespread concern (25). Therefore, an Eradication Program for avian leukosis was implemented in China about 10 years ago, which effectively prevented and controlled the disease. However, since 2017, there has been another ALV-J infection outbreak in several provinces of China, leading to an increase in serious tumor diseases and a sharp increase in mortality (26). Therefore, in this study, a strain of ALV-J was isolated from layer chickens, and some typical ALV cases were successfully replicated in white-feathered chickens. We discovered that liver with hemangiomas, kidney and spleen enlargement with white nodules to be common symptoms in the reproduced cases. Some chickens had numerous hemangiomas and bony nodules. The symptoms of the cases reproduced in this study were more severe than the clinical symptoms of HPRS-103 (27), suggesting that HB2020 is more virulent. The clinical symptoms as well as the various tumor types induced by HB2020 indicated that HB2020 might possess different molecular characteristics.
In 2009, approximately 70% of avian leukosis cases in China were infected with ALV-J (28). ALV-J is another subtype with a high mutation rate; the env gene of ALV-J shares 80–85% homology with the nucleotide sequences of its subgroups but only 40% homology with the nucleotide sequences of other subgroups (29). The HB2020's gag gene was closely related to ev-1 and GDFX0601 based on the level of sequence similarities, but the remaining genes and regions were on the same branches as HPRS-103 in their respective trees, therefore we speculated that the virus isolated, HB2020, was a recombinant strain belonging to subgroup J that had an ALV-E-like gag. The same recombination in gag regions was also detected in ALV-J strain LH20180301, isolated from a commercial farm in Shandong province of China (30). This mutation emerged very recently, and it may represent a new direction of variation in ALV-J isolated from China.
The envelope glycosylation protein (ENV) is composed of gp85-encoded surface glycoprotein and gp37-encoded transmembrane glycoprotein, and it is closely related to the virus' antigenicity, tumorigenic type, and host range (31). We confirmed that the HB2020 strain isolated in this study had some variation in ENV compared to all other ALV-J strains. The results revealed that the mutations of HB2020 were primarily concentrated in the vr3, hr1, and hr2 regions of the SU in env gene. Furthermore, we found that the genotypes of ALV-J gp85 that cause various pathological symptoms follow a pattern. Three specific amino acid mutations (E109, R112, and N118) were found in the SU of ALV-J that cause hemangiomas. However, a unique amino acid mutation Q235 could only be observed in the SU of the ALV-J strains that cause myeloid leukosis. This phenomenon indicated that these gene mutations might be associated with infectivity and pathogenicity; however, the mechanism of this phenomenon remains unclear. So far, several studies have shown that the sequences spanning amino acids 38 to 131 and 159 to 283 of ALV-J SU could influence the binding efficiency of chicken Na+/H+ Exchanger 1 (chNHE1), which was important to receptor binding and viral entry for N6, N11 sites and C3, C9 cysteines of ALV-J gp85 (32). The mechanism of the phenomenon observed in this study requires further investigation.
The 3' UTR of ALV is frequently mutated and deleted (33). Since 2012, a few ALVs with a deletion of 205 bp in the 3' UTR have been discovered in China, indicating that these viruses are more oncogenic (34, 35). The HB2020 also had 205 bp deletions in the 3'UTR, and animal testing revealed that it was highly pathogenic, and could cause systemic tumors, which was consistent with previous findings. It had an affinity for the LTR's U3 region and had been implicated in the propagation of the avian retroviruses (36). Many important protein binding motifs have been identified in the U3 region, including COAAT/enhancer, CArG box, and Y box elements. C/EBP, E2BP, CArG box, AIB REP1, and Y box could be found in the U3 region of HB2020. It is note worthy that AIB REP1 binding sites have been found in all myeloid leukosis (ML) ALV-Js, including HB2020. However, the AIB REP1 in the U3 region of HB2020 had two mutant bases. The AIB REP1 gene has been linked to the vascular invasion of hepatocellular carcinoma in humans (37). In this study, a lot of hemangiomas were observed in the chicken's liver with the most obvious pathological symptoms; perhaps this phenomenon was related to the AIB REP1 mutation and the role of AIB REP1 in ALV-induced tumorigenesis requires further investigation.
So far, ALV is still spreading all over the world. The molecular biological analysis and tumorigenicity experiment of HB2020 might help to monitor the mutation tendency and study the prevalence of ALV-J, and guide preventive actions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics statement
The animal study was reviewed and approved by the Academic Committee of the College of Animal Science, Yangtze University.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the National Nature Science Foundation of China (31972646), State Key Laboratory of Veterinary Biotechnology Foundation, China (SKLVBF201708), and Scientific Research Project of Education Department of Hubei Province (B2019028).
Acknowledgments
The authors thank the managers of the chicken farm for providing test samples. We gratefully acknowledge the authors and laboratories who have generated and submitted sequences of ALVs to the GenBank.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Venugopal K, Howes K, Barron GS, Payne LN. Recombinant Env-Gp85 of Hprs-103 (Subgroup J) avian leukosis virus: antigenic characteristics and usefulness as a diagnostic reagent. Avian Dis. (1997) 41:283–8. doi: 10.2307/1592179
2. Li H, Wang P, Lin L, Shi M, Gu Z, Huang T, et al. The emergence of the infection of subgroup j avian leucosis virus escalated the tumour incidence in commercial yellow chickens in Southern China in recent years. Transbound Emerg Dis. (2019) 66:312–6. doi: 10.1111/tbed.13023
3. Xu B, Dong W, He Z, Feng X, Yu C, Zhang L, et al. Rapid diagnosis of J subgroup avian leukemia of hailan brown hens by indirect fluorescent antibody method. Chin J Vet Med. (2002) (09):7–9.
4. Payne LN, Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. (2012) 41:11–9. doi: 10.1080/03079457.2011.646237
5. Chesters PM, Howes K, Petherbridge L, Evans S, Payne LN, Venugopal K. The viral envelope is a major determinant for the induction of lymphoid and myeloid tumours by avian leukosis virus subgroups a and j, respectively. J Gen Virol. (2002) 83:2553–61. doi: 10.1099/0022-1317-83-10-2553
6. Su Q, Cui Z, Zhang Z, Cui Z, Chang S, Zhao P. Whole-genome analysis of an emerging recombinant avian leukosis virus in yellow chickens, South China. Transbound Emerg Dis. (2020). doi: 10.1111/tbed.13574
8. Su Q, Li Y, Cui Z, Chang S, Zhao P. The emerging novel avian leukosis virus with mutations in the pol gene shows competitive replication advantages both in vivo and in vitro. Emerg Microbes Infect. (2018) 7:117. doi: 10.1038/s41426-018-0111-4
9. Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res Vet Sci. (1999) 67:113–9. doi: 10.1053/rvsc.1998.0283
10. Chen X, Wang H, Fang X, Gao K, Fang C, Gu Y, et al. Identification of a novel epitope specific for gp85 protein of avian leukosis virus subgroup K. Vet Immunol Immunopathol. (2020) 230:110143. doi: 10.1016/j.vetimm.2020.110143
11. Li T, Yao X, Li C, Zhang J, Xie Q, Wang W, et al. Gp37 regulates the pathogenesis of avian leukosis virus subgroup J via its C terminus. J Virol. (2020) 94. doi: 10.1128/JVI.02180-19
12. Navarro E, Mallen A, Hueso M. Dynamic variations of 3'utr length reprogram the MRNA regulatory landscape. BIOMEDICINES. (2021) 9. doi: 10.3390/biomedicines9111560
13. Rasekhian M, Roohvand F, Habtemariam S, Marzbany M, Kazemimanesh M. The role of 3'utr of Rna viruses on mrna stability and translation enhancement. Mini Rev Med Chem. (2021) 21:2389–98. doi: 10.2174/1389557521666210217092305
14. Zavala G, Cheng S, Jackwood MW. Molecular epidemiology of avian leukosis virus subgroup j and evolutionary history of its 3' untranslated region. Avian Dis. (2007) 51:942–53. doi: 10.1637/0005-2086(2007)51942:MEOALV2.0.CO;2
15. Shi M, Tian M, Liu C, Zhao Y, Lin Y, Zou N, et al. Sequence analysis for the complete proviral genome of subgroup j avian leukosis virus associated with hemangioma: a special 11 Bp deletion was observed in U3 region of 3'utr. Virol J. (2011) 8:158. doi: 10.1186/1743-422X-8-158
16. Li X, Yu Y, Ma M, Chang F, Muhammad F, Yu M, et al. Molecular characteristic and pathogenicity analysis of a novel multiple recombinant Alv-K strain. Vet Microbiol. (2021) 260:109184. doi: 10.1016/j.vetmic.2021.109184
17. Li J, Liu L, Niu X, Li J, Kang Z, Han C, et al. Research note: a novel recombinant subgroup E isolate of the avian leukosis virus with a subgroup B-like Gp85 region in China. Poult Sci. (2021) 100:101137. doi: 10.1016/j.psj.2021.101137
18. Gopal S, Manoharan P, Kathaperumal K, Chidambaram B, Divya KC. Differential detection of avian oncogenic viruses in poultry layer farms and turkeys by use of multiplex Pcr. J Clin Microbiol. (2012) 50:2668–73. doi: 10.1128/JCM.00457-12
19. Davidson I, Borovskaya A, Perl S, Malkinson M. Use of the polymerase chain reaction for the diagnosis of natural infection of chickens and turkeys with marek's disease virus and reticuloendotheliosis virus. Avian Pathol. (1995) 24:69–94. doi: 10.1080/03079459508419050
20. Meng X, Liang X, Cai Y, Gu Y. Diagnosis of chicken clinical tumor cases caused by mixed infection of Mdv and Rev. China Poultry. (2016) 38:71−3. doi: 10.16372/j.issn.1004-6364.2016.21.017
21. Zhu L, Wang H, Chen X, Gao K, Fang C, Liang X, et al. Prokaryotic expression,antibody preparation and identification of rev envelope protein. Acta Agriculturae Universitatis Jiangxiensis. (2021) 43:1159–66.
22. Gao Q, Yun B, Wang Q, Jiang L, Zhu H, Gao Y, et al. Development and application of a multiplex Pcr method for rapid differential detection of subgroup a, B, and J avian leukosis viruses. J Clin Microbiol. (2014) 52:37–44. doi: 10.1128/JCM.02200-13
23. Liang X. Biological Characteristics of the Isolates of Avian Leukosis Virus Subgroup a K Associated with Myeloid Avian Leukosis [doctor]. Jingzhou: Yangtze University. (2018). Available online at: www.cnki.net
24. Li Y. Epidemiological Investigation of Alv in Chinese Indigenous Breeds and the Research on the Biological Characteristics of Alv-K [doctor]: Shandong Agricultural University (2017).
25. Cui Z, Du Y, Zhang Z, Silva RF. Comparison of Chinese Field strains of avian leukosis subgroup J viruses with prototype strain Hprs-103 and United States Strains. Avian Dis. (2003) 47:1321–30. doi: 10.1637/6085
26. Li Y, Fu J, Cui S, Meng F, Cui Z, Fan J, et al. Gp85 genetic diversity of avian leukosis virus subgroup J among different individual chickens from a native flock. Poult Sci. (2017) 96:1100–7. doi: 10.3382/ps/pew407
27. Payne LN, Gillespie AM, Howes K. Induction of myeloid leukosis and other tumours with the Hprs-103 Strain of Alv. Vet Rec. (1991) 129:447–8. doi: 10.1136/vr.129.20.447
28. Gao Y, Shao H, Luo Q, Pan W, Qin L, Sun F, et al. Molecular epidemiology of a via n leukosis virus isolates from some regions of China in 2009. Chin J Prev Vet. (2010) 32:32–5+43.
29. Wang P, Li M, Li H, Bi Y, Lin L, Shi M, et al. Alv-J-contaminated commercial live vaccines induced pathogenicity in three-yellow chickens: one of the transmission routes of Alv-J to commercial chickens. Poult Sci. (2021) 100:101153. doi: 10.1016/j.psj.2021.101153
30. Cui N, Cui X, Huang Q, Yang S, Su S, Xu C, et al. Isolation and identification of subgroup J avian leukosis virus inducing multiple systemic tumors in parental meat-type chickens. Front Vet Sci. (2020) 7:614854. doi: 10.3389/fvets.2020.614854
31. Ye J, Fan Z, Shang J, Tian X, Yang J, Chen H, et al. Alv-J Gp37 molecular analysis reveals novel virus-adapted sites and three tyrosine-based env species. PLoS ONE. (2015) 10:e0122887. doi: 10.1371/journal.pone.0122887
32. Zhang Y, Yu M, Xing L, Liu P, Chen Y, Chang F, et al. The bipartite sequence motif in the N and C termini of Gp85 of subgroup J avian leukosis virus plays a crucial role in receptor binding and viral entry. J Virol. (2020) 94. doi: 10.1128/JVI.01232-20
33. Han C, Hao R, Liu L, Zeng X. Molecular characterization of 3'utrs of J subgroup avian leukosis virus in passerine birds in China. Arch Virol. (2015) 160:845–9. doi: 10.1007/s00705-014-2321-y
34. Ma M, Yu M, Chang F, Xing L, Bao Y, Wang S, et al. Molecular characterization of Avian leukosis virus subgroup J in Chinese local chickens between 2013 and 2018. Poult Sci. (2020) 99:5286–96. doi: 10.1016/j.psj.2020.08.004
35. Gao Y, Yun B, Qin L, Pan W, Qu Y, Liu Z, et al. Molecular epidemiology of avian leukosis virus subgroup J in layer flocks in China. J Clin Microbiol. (2012) 50:953–60. doi: 10.1128/JCM.06179-11
36. Tsichlis PN, Donehower L, Hager G, Zeller N, Malavarca R, Astrin S, et al. Sequence comparison in the crossover region of an oncogenic avian retrovirus recombinant and its nononcogenic parent: genetic regions that control growth rate and oncogenic potential. Mol Cell Biol. (1982) 2:1331–8. doi: 10.1128/mcb.2.11.1331-1338.1982
Keywords: avian leukosis virus subgroup J, recombination, hemangioma, myelocytoma, layer chicken
Citation: Wu L, Li Y, Chen X, Yang Y, Fang C, Gu Y, Liu J, Liang X and Yang Y (2022) Isolation and characterization of avian leukosis virus subgroup J associated with hemangioma and myelocytoma in layer chickens in China. Front. Vet. Sci. 9:970818. doi: 10.3389/fvets.2022.970818
Received: 24 June 2022; Accepted: 31 August 2022;
Published: 23 September 2022.
Edited by:
Elisa Rampacci, University of Perugia, ItalyCopyright © 2022 Wu, Li, Chen, Yang, Fang, Gu, Liu, Liang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiongyan Liang, MTgyMjk3NDY3QHFxLmNvbQ==; Yuying Yang, eWFuZ3l5Y25AeWFuZ3R6ZXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Lan Wu
Lan Wu Youjun Li
Youjun Li Xueyang Chen
Xueyang Chen Yuxin Yang
Yuxin Yang Chun Fang
Chun Fang Xiongyan Liang
Xiongyan Liang Yuying Yang
Yuying Yang