
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 15 September 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.952319
 Abdallah A. Hassanin1*
Abdallah A. Hassanin1* Ali Osman2
Ali Osman2 Osama Osman Atallah3
Osama Osman Atallah3 Mohamed T. El-Saadony4*
Mohamed T. El-Saadony4* Sameh A. Abdelnour5
Sameh A. Abdelnour5 Heba S. A. Taha1
Heba S. A. Taha1 Mohamed F. Awad6
Mohamed F. Awad6 Hany Elkashef7
Hany Elkashef7 Ahmed Ezzat Ahmed8,9
Ahmed Ezzat Ahmed8,9 Ibrahim Abd El-Rahim10,11
Ibrahim Abd El-Rahim10,11 Abdullah Mohamed12
Abdullah Mohamed12 Ahmed S. Eldomiaty1
Ahmed S. Eldomiaty1Caseins determine the physicochemical, physiological, and biological characteristics of milk. Four caseins—alpha-S-1, alpha-S-2, beta, and kappa—were analyzed phylogenetically and in silico and characterized regarding chemical, antimicrobial, and antioxidant features in five dairy animals: Arabian camels, sheep, goats, cattle, and water buffalos. The sequence of full-length amino acids of the four caseins for the five species was retracted from the NCBI GenBank database. Multiple sequence alignment is used to examine further the candidate sequences for phylogenetic analysis using Clustal X and NJ-Plot tools. The results revealed that sheep and goats possess strong similarities (98.06%) because of their common ancestor. The same was observed with cattle and water buffalos (96.25%). The Arabian camel was located in a single subclade due to low similarity in casein residues and compositions with other dairy animals. Protein modeling showed that alpha-S1- and alpha-S2-caseins possess the highest number of phosphoserine residues. The in silico computed chemical properties showed that β-casein recorded highest hydrophobicity index and lowest basic amino acid content, while α-S2-casein showed the opposite. The computed biological parameters revealed that α-S2-casein presented the highest bactericidal stretches. Only Arabian camel β-casein and k-casein showed one bactericidal stretches. The analysis also revealed that β-casein, particularly in Arabian camels, possesses the highest antioxidant activity index. These results support the importance of the bioinformatics resources to determine milk casein micelles' chemical and biological activities.
Casein in milk is widely consumed in the human diet worldwide, particularly in developing countries. Because of its widespread availability and enormous production quantities, cattle milk is the most consumed milk worldwide as a valued source of human nutrition. Non-bovine milk is essential to people in developing countries (1, 2). For example, buffalo milk in Asian countries, sheep milk in Europe and the Middle East, camel milk in Africa and some Asian countries, and goat milk in Africa and southern Asia (1, 3).
The majority of studies on milk casein have been conducted on cow milk as the most produced and consumed milk around the world (4); however, very few studies have been conducted on the comparison of milk from different domestic dairy animal species (5) and non-dairy animals, where these properties are explicit and may provide insight (6). The main nutritional benefit of milk is its protein content (of which around 80% is casein) (7). The casein proteins including four classes, namely, alpha S-1, alpha S-2, beta, and kappa, are assembled into a structure called casein micelles (8). Milk casein of different animal species possesses variations in amino acid sequences. The average size of casein micelles can also vary significantly from one species to another (9, 10). Animal species have variations in casein ratios (11) and micelle sizes (12). Caseins are almost similar in molecular weight in different domestic dairy species (13).
Comparative genomic analysis can provide new insights into the functionality of casein genes with respect to the caseins. Comparative genomic analysis is a rapidly emerging field in computational biology whereby two or more genomes are compared to obtain a global view of genomes and their deduced proteomes and assign previously unknown functions to genes with respect to their proteins (14).
Computational chemical properties can also pave the way to predicting the properties of deduced proteins. The calculated propensity index, which is deduced from the associated half-maximal inhibitory concentration (IC50) value for each amino acid alteration, serves as a useful benchmark for evaluating protein sequence determinants. Because low IC50 values imply greater antimicrobial activity, amino acids having a lower bactericidal propensity value (PV) are more likely to be used in antimicrobial peptides. Residues with positive charges (Arginine, Lysine, and Histidine) and some hydrophobic residues (Tryptophane, Tyrosine, and Valine) are unfavorable and present a low propensity index, whereas negatively charged residues are unflavored and show a high propensity index. Antimicrobial proteins would require positively charged residues to drive them to the negatively charged bacterial cell wall and cytoplasmic membrane, where they exert their antimicrobial effect (15). To form pores or other destabilizing structures that lead to membrane depolarization or local disruption and, finally, bacterial cell death, hydrophobic residues would need to interact with lipophilic regions of lipid bilayers (16). Interestingly, tryptophane (W) has the lowest PV value among the hydrophobic residues, whereas leucine (L) has the highest value, and isoleucine (I) and valine (V) are favored over L. Moreover, W residues are known to play a role in peptide antibacterial activity (17).
Several studies have shown that milk casein has hydrophilic, hydrophobic, antimicrobial, antioxidant, and anticancer properties (18–21). These properties offer great potential use of casein micelles as food additives in food industries instead of the use of synthetic additives, which cause some side effects: allergic, intoxications, cancer, and other degenerative diseases. The presence of phosphate groups near the peptide chain results in polar, acidic domains that are good for sequestering divalent metals, such as calcium, zinc, copper, manganese, and iron. An anionic triplet embedded in the bioactive peptide (SerP–SerP–SerP–Glu–Glu) is a distinguishing property of every functional CPP generated from whole and individual casein micelles (22, 23).
In silico analysis for caseins enables the accurate prediction of the computed chemical properties and antimicrobial, and antioxidant activities. Thus, this study was conducted in silico to determine the phylogenetic relationship, three-dimensional structure, chemical, and biological features of caseins in five domestic dairy animals: Arabian camels, sheep, goats, cattle, and water buffalos.
The caseins—alpha-S1-, alpha-S2-, beta-, and kappa-casein—were comprehensively in silico and characterized in five milk-producing animals, namely Arabian camels, sheep, goats, cattle, and water buffalos, to determine their genetics, chemical, and biological features.
Amino acid sequences for casein subunits—alpha-S1, alpha-S2, β-, and k-casein—were retracted from the NCBI (https://www.ncbi.nlm.nih.gov/genbank/) in FASTA format. The accession number of each protein is presented in Table 1. Protein sequences were manually curated and then aligned using the CLUSTAL-Wtool provided by MEGA X software (24). Phylogeny was performed using the same software. The trees were calculated using the rooted Neighbor-Joining (NJ) method (25) on distance matrices employing NEIGHBOR from the Clustal-X package. The Default P-distance method was used for distance analysis. Rattus norvegicus (rat) was used as the out-group. To generate a consensus tree, these trees were evaluated using the Clustalx software. The NJ Plot software application was used to plot a rooted tree. Because of the enormous number of sequences in the alignment, the bootstrapping of the “alignment dataset” was limited to 1,000 times. Sequences with more than 90% bootstrap support value were confirmed and categorized.
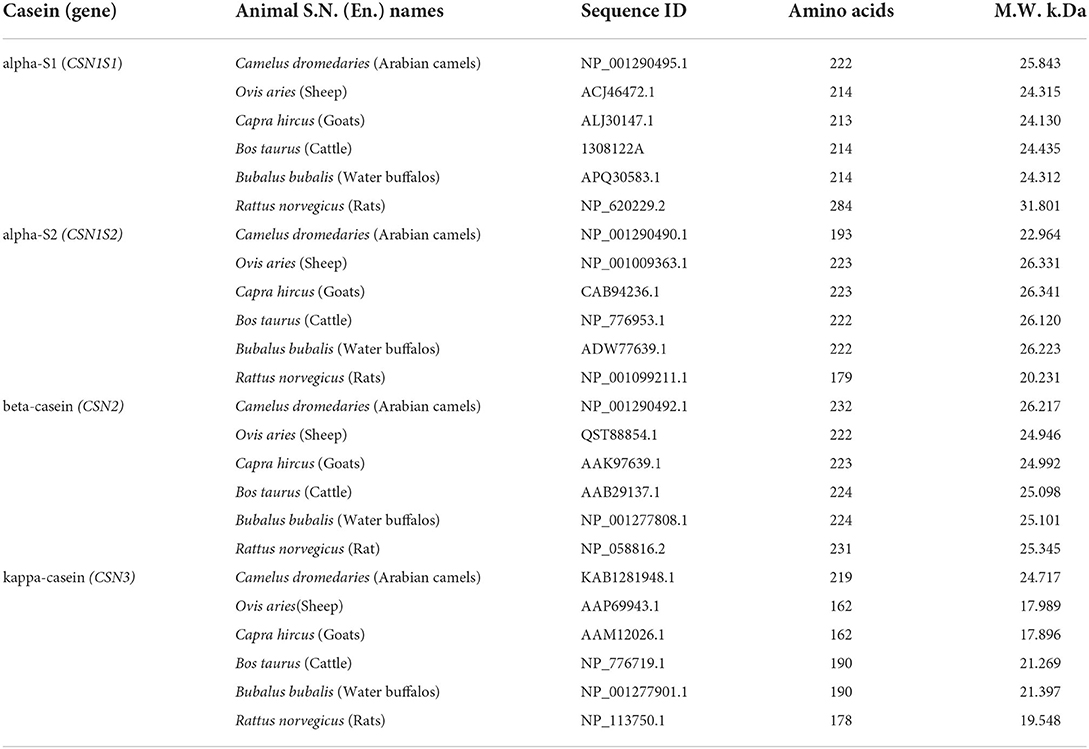
Table 1. Animal S.N. (En.) names, sequence ID, amino acids, and molecular weight of caseins in five dairy animals.
For further analysis, combined alpha-S1-, alpha-S2-, beta-, and kappa-casein amino acid sequences in sheep, goats, cattle, Arabian camels, and water buffalos, respectively, were subjected to multiple sequence alignment using the Clustal Omegadatabase [CLUSTAL O (1.2.4)] (https://www.ebi.ac.uk/Tools/ msa/clustalo/), assisted by some manual adjustments to indicate the regions of similarity, identifying probably functional, structural, and evolutionary relationships between the sequences. A phylogenetic tree was rooted in a taxonomically distant organism (Rattus norvegicus).
Genomic organization of chromosome six illustrating structure of casein-encoding genes CSN1S1, CSN1S2, CSN2, CSN3. The domains and phosphorylation sites associated with each protein subunit are presented from the NCBI databases. Protein 3D structures were predicted using the SWISS-MODEL homology modeling (swissmodel.expasy.org) method under the default parameters (26).
Three-dimensional representations of casein subunits were deduced from Kumosinski and Brown (27) for alpha-s1-casein and k-casein—Kumosinski, Brown (28) for beta-casein and Farrell Jr, Malin (29) for alpha-S2-casein. Protein structures exhibiting the highest homology were selected and developed as a template. Sequence homology for each sequence exceeded 70% and was thus considered highly reliable. Schematic illustrations were produced with the software package UCSF Chimera, candidate version 1.11.2.
The chemical properties of caseins were in silico, computed by the ProtParam tool (https://web.expasy.org/protparam/), a tool of the ExPASy database that allows the computation of various physical and chemical features for a certain protein stored in Swiss-Prot or TrEMBL or for a user-entered protein sequence. The computed parameters include the molecular weight, instability index, hydrophobicity index, basic amino acids (%), negatively charged residues, and positively charged residues (30).
The publicly available AMPA tool (http://tcoffee.crg.cat/apps/ampa/do) was used to predict antimicrobial peptides. AMPA: an automated web server for the prediction of antimicrobial protein regions (31, 32). Whole-protein sequences were run in the AMPA using the default parameter values, i.e., a propensity threshold of 0.225 and a window size of 7 amino acids.
The antioxidant activity index was calculated based on the casein content of hydrophobic amino acids, particularly tryptophan, methionine, isoleucine, leucine, and proline.
Four casein sequences, alpha-S1, alpha-S2, beta, and kappa-casein, in five milk production animal species (Arabian camels, sheep, goats, cattle, and water buffalos) were individually (Figure 1) and in combination (Figure 2) analyzed by multiple sequence alignment for phylogeny study using the Clustal-X package and the rooted Neighbor-Joining (NJ) method (Supplementary Figure 1). This analysis was conducted to determine the phylogenetic relationship among sheep, goats, cattle, Arabian camels, and water buffalos through the alignment of caseins. The tree was rooted in a taxonomically distant organism (Rattus norvegicus). The phylogeny analysis revealed that the Arabian camel, which belongs to the Camelidae family, possesses low similarity with other domestic animals that belong to the Bovidae family, ranging between 60.55 with sheep to 62.22 with water buffalos (Table 2). Thus, the Arabian camel was sub-grouped individually (Figures 1, 2). The highest similarity (98.06) (Table 2) appeared between sheep and goats, which belong to the Bovidae family and are sub-grouped together by phylogeny analysis (Figures 1, 2). The phylogeny analysis revealed that cattle and water buffalo that belong to the Bovinae family possess a high similarity value (96.25) (Table 2), which included them in one subgroup (Figures 1, 2).
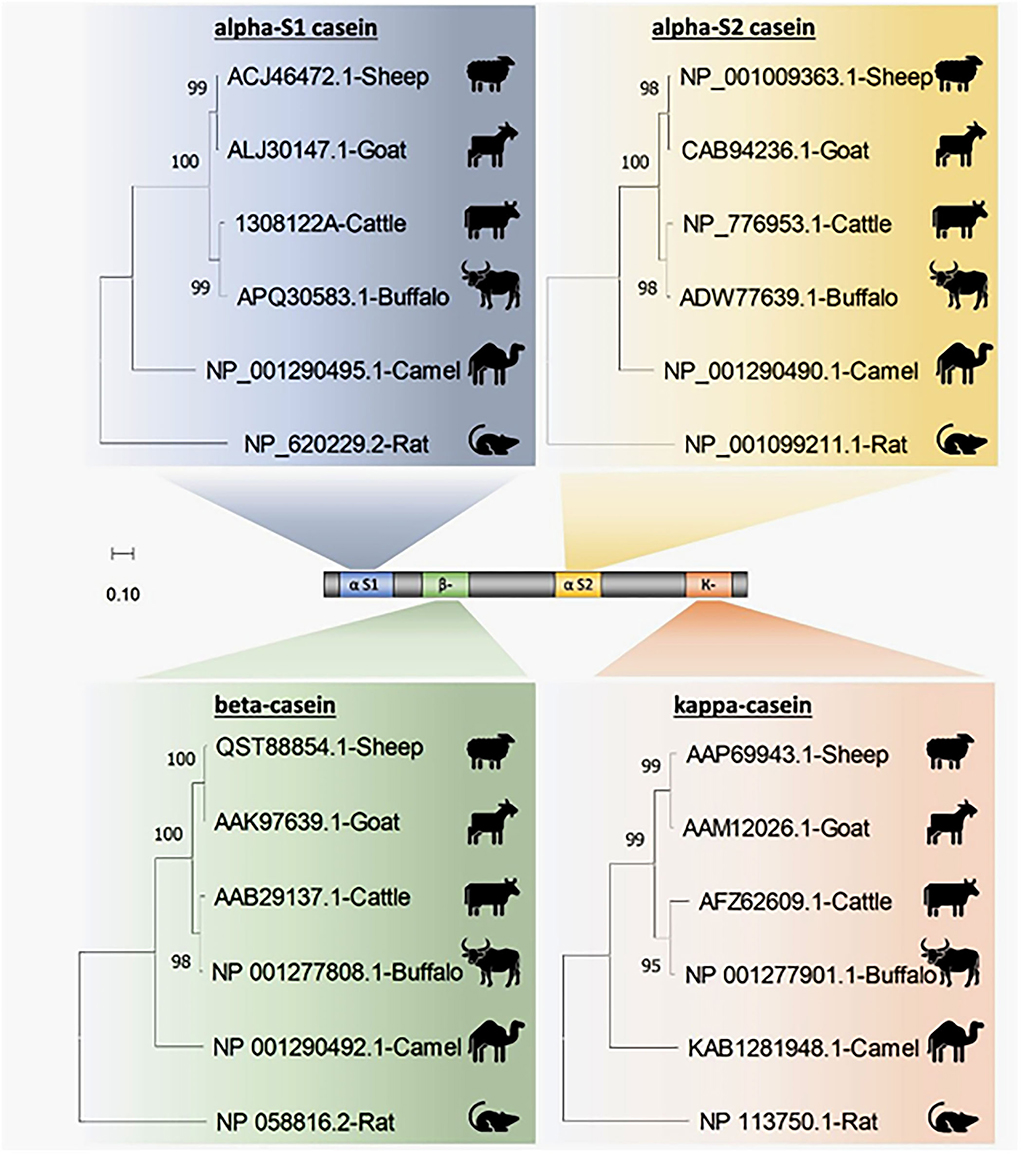
Figure 1. Phylogenetic analysis of casein subunits from different domestic animals. A neighbor-joining tree representing the phylogenetic relationship of sheep, goats, cattle, Arabian camels, and water buffalos was derived from the alignment of alpha-S1-, alpha-S2-, beta-, and kappa-casein amino acid sequences. The tree was rooted in Rattus norvegicus. Bootstrap support values are indicated on the nodes. Bootstrap values were calculated from 1,000 replications.
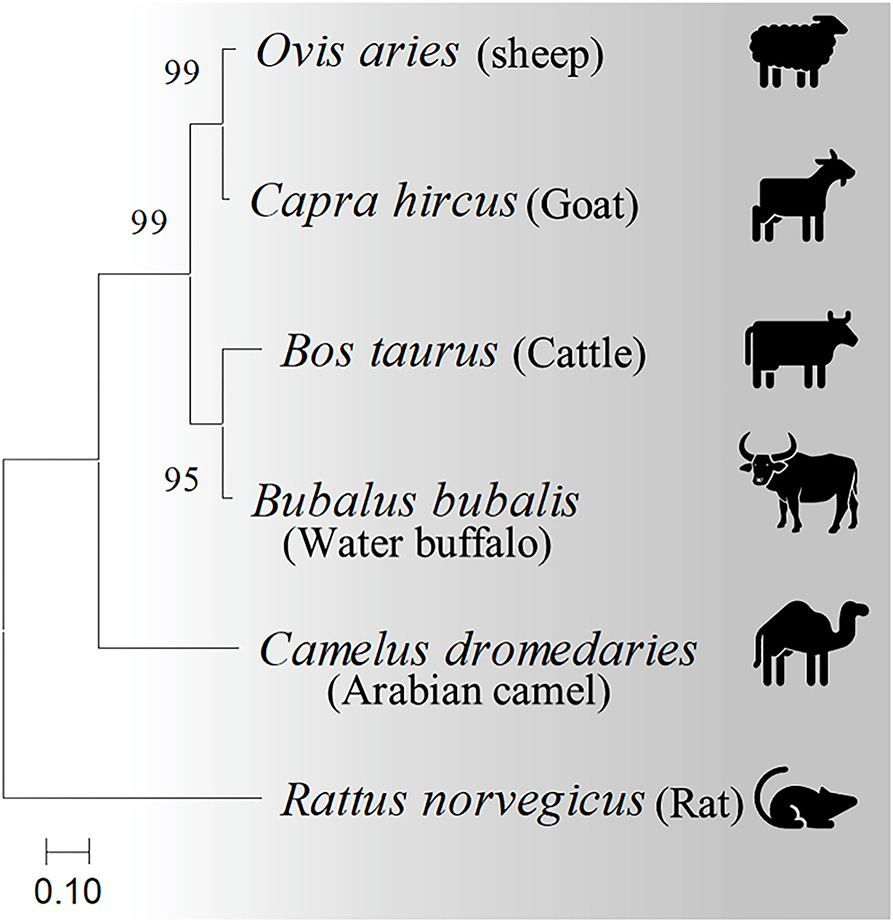
Figure 2. Phylogenetic relationships among casein homologs from different domestic animals. A neighbor-joining tree inferred from combined alpha-S1-, alpha-S2-, beta-, and kappa-casein amino acid sequences shows the phylogenetic positions of five other animals. The phylogenetic tree is derived from the alignment of four concatenated casein from sheep, goats, cattle, Arabian camels, and water buffalos. The tree was rooted in Rattus norvegicus. Bootstrap support values are indicated on the nodes. Bootstrap values were calculated from 1,000 replications.
Protein modeling is considered a routine approach to provide structural models of proteins when no experimental structures are available. Protein modeling uses template protein structures to predict the conformations of other proteins with similar amino acids because small changes in the protein sequence usually result in small changes in the 3D structure. The 3D structure of caseins of cattle was used as a model for the rest of the domestic animals because it is the most popular and the most studied (Figure 3).
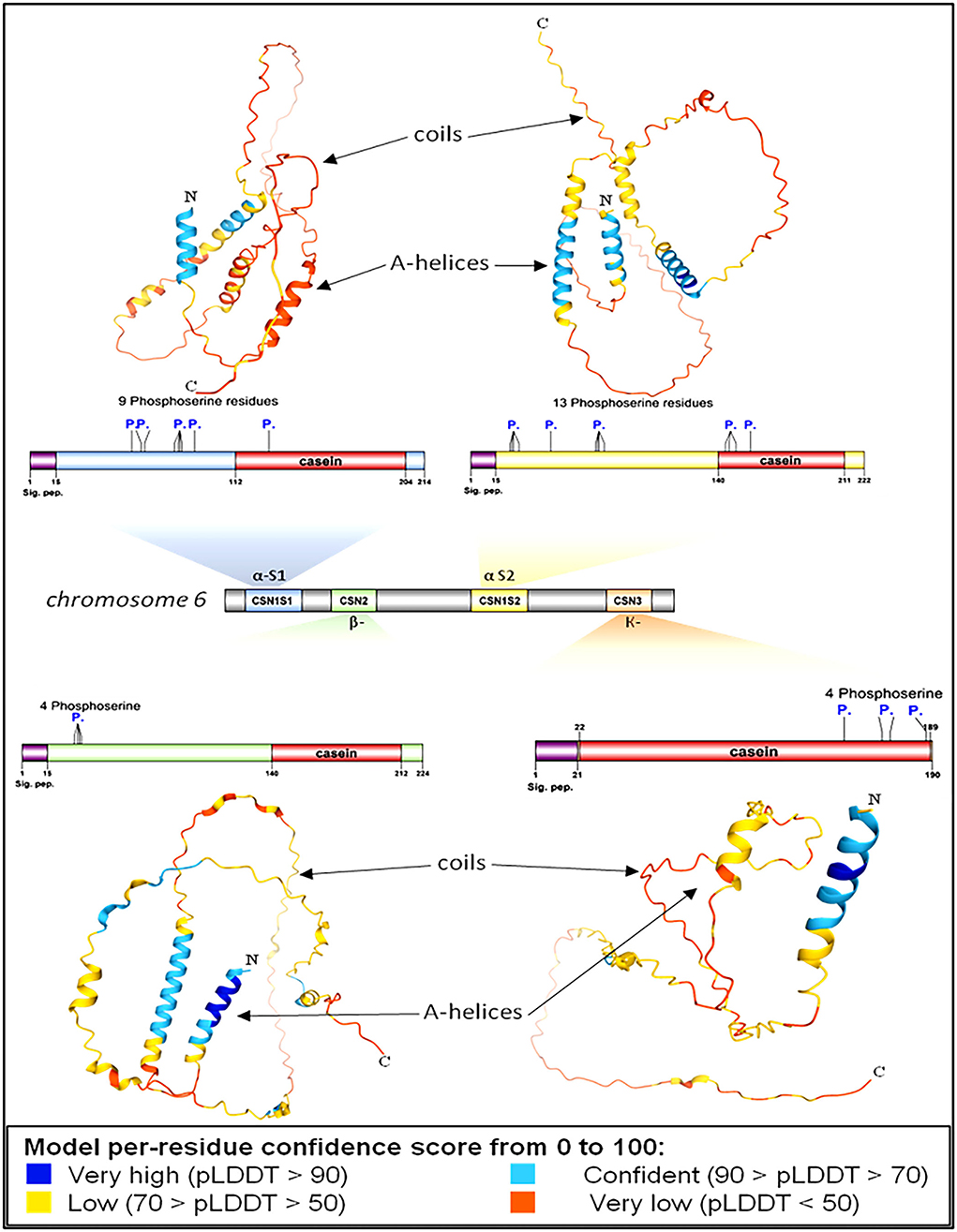
Figure 3. Genomic and structural characterization of milk caseins from Bos taurus (cattle). Casein subunit organization within chromosome number 6 (middle panel). The characteristics and genetic features of each casein subunit are detailed. Signal peptide domains (sig. pep.) and phosphorylation sites (P.) are indicated on each protein diagram. Ribbon diagrams illustrating the high-resolution structures of alpha-S1-, alpha-S2-, beta-, and kappa-casein. Proteins' N- and C-termini are indicated on protein models.
Protein modeling analysis revealed that the C-terminal ends had fewer secondary structures than the N-terminal of all caseins (Figure 3). In contrast to the C-terminal, the N-terminal exhibits a high degree of hydrophobicity that is not hydrolyzed in milk due to the presence of hydrophobic amino acid residues leucine and tryptophan in alpha S1; casein, proline, and tryptophan in alpha S2; casein, proline, and isoleucine in beta-casein; and alanine and valine in kappa casein. The analysis showed the presence of various numbers of phosphoserine residues in all caseins 9, 13, 4, and 4 in each alpha-S1-, alpha-S2-, beta-, and kappa-casein, respectively.
Casein micelles (αs1-, αs2-, β-, and κ-casein) are present in all types of milk as self-assembled particles (33). The chemical structure of the casein micelles in milk production animals has not yet been sufficiently studied, except in cattle milk. The current in silico study comprehensively investigated the chemical characteristics of caseins as computed protein parameters; amino acid composition (Figure 4), protein size, molecular weight, hydrophobicity index (the percentage of hydrophobic amino acids), basic amino acid residues, negatively charged residues (Asp + Glu) and positively charged residues (Arg + Lys) in five milk production animals: Arabian camels, sheep, goats, cattle, and water buffalos (Figures 5, 6).
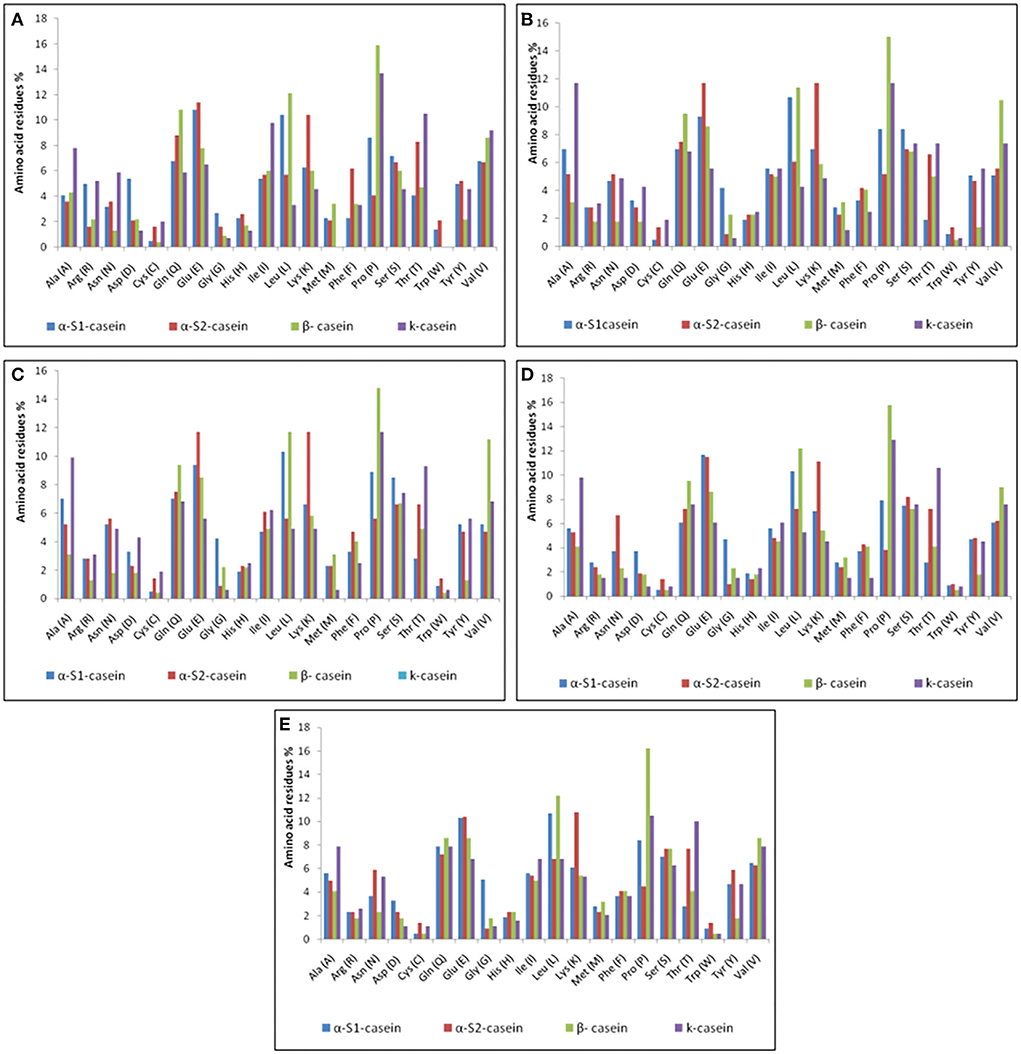
Figure 4. Amino acid compositions of caseins. (A) Arabian camel caseins. (B) Sheep caseins. (C) Goat caseins. (D) Cattle caseins. (E) Water buffalo caseins.
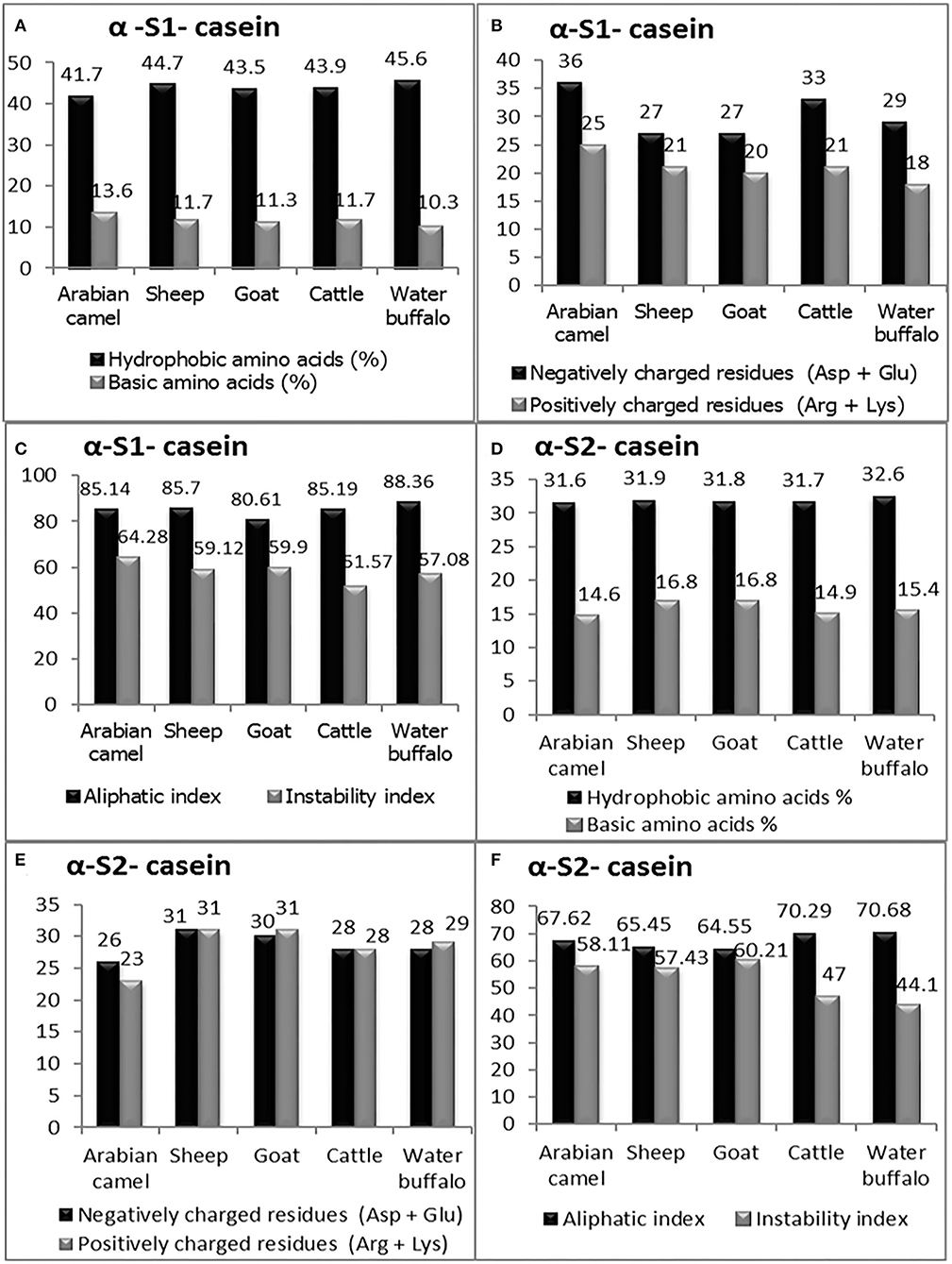
Figure 5. The computed parameters of α-S1-casein; (A) hydrophobic amino acids (%), basic amino acids (%). (B) Negatively charged residues (Asp + Glu), positively charged residues (Arg + Lys). (C) Aliphatic and instability indexes. The computed parameters of α-S2-casein (D) hydrophobic amino acids (%), and basic amino acids (%). (E) Negatively charged residues (Asp + Glu), positively charged residues (Arg + Lys). (F) Aliphatic and instability indexes.
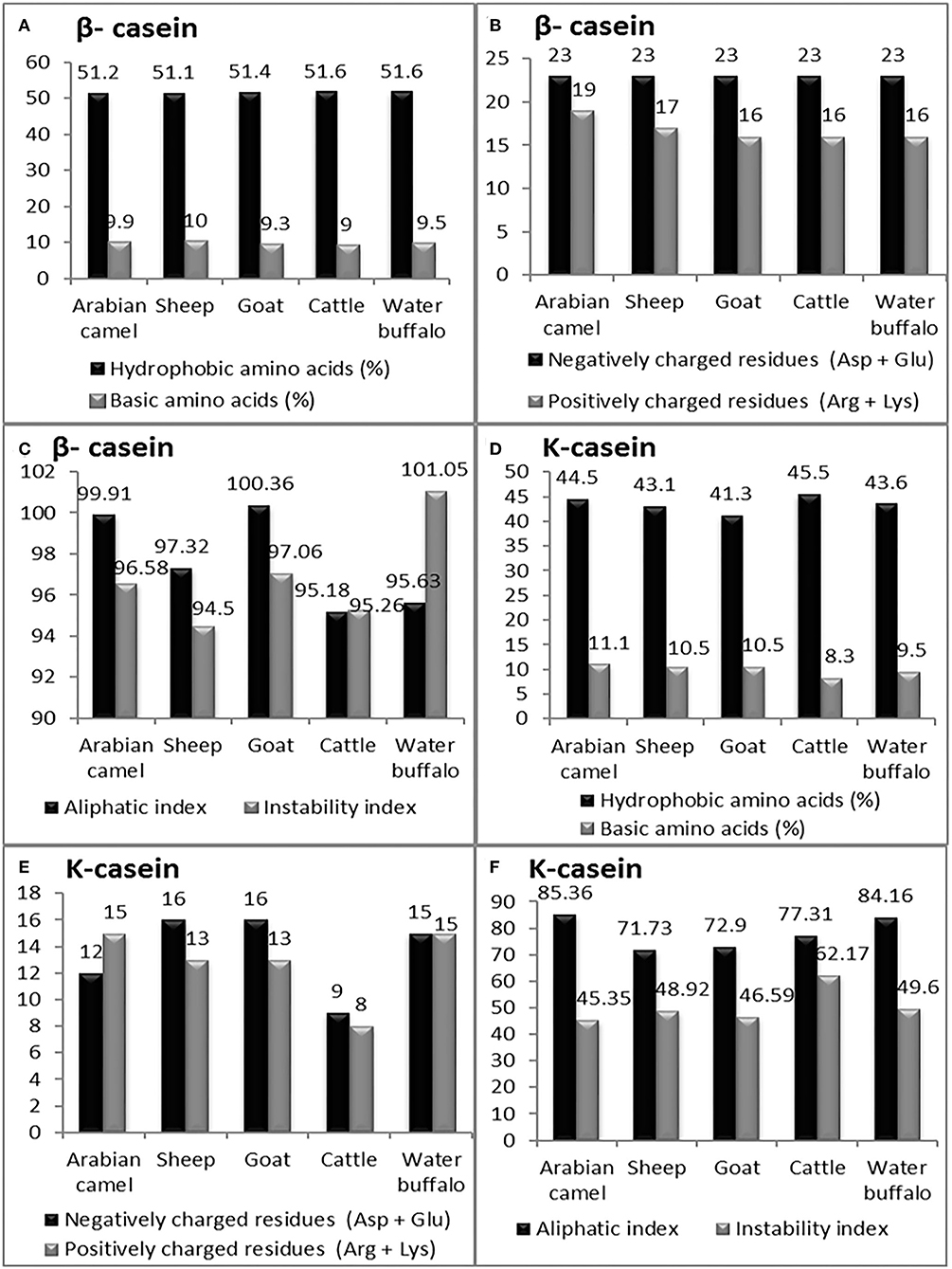
Figure 6. The computed parameters of β-casein; (A) Hydrophobic amino acids (%), Basic amino acids (%). (B) Negatively charged residues (Asp + Glu), positively charged residues (Arg + Lys). (C) Aliphatic and instability indexes. The computed parameters of K-casein. (D) Hydrophobic amino acids (%), Basic amino acids (%). (E) Negatively charged residues (Asp + Glu), positively charged residues (Arg + Lys). (F) Aliphatic and instability index.
The primary structure and percentage of each amino acid residue of alpha-S1-casein and alpha-S2-casein are presented in Table 3. The computed protein parameters indicated that the alpha-S1-casein of Arabian camels has an intermediate hydrophobicity index correlated with the presence of hydrophobic side chains in glycine, alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, and tryptophan, which ranged between 41.7% in Arabian camels and 45.6% in water buffalos, as well as the intermediate content of basic amino acid residues, which ranged between 10.3 and 13.6% in water buffalos and Arabian camels, respectively (Figure 5A). The analysis revealed that the Arabian camels and water buffalos showed the highest and lowest content of negatively charged residues (Asp + Glu) and positively charged residues (Arg + Lys) (Figure 5B). The alpha-S1-casein in Arabian camels showed the highest instability index (64.28), while the alpha-S1-casein in cattle was the lowest (51.57). On the other hand, the highest aliphatic side chains (aliphatic index), which are due to the presence of alanine, valine, isoleucine, and leucine residues, was 88.36 in water buffalos, while the lowest was 80.61 in goats—alpha-S1-casein (Figure 5C).
The computed protein parameters indicated that the alpha–S2-casein of Arabian camels had a low hydrophobicity index, ranging between 31.60 in Arabian camels to 32.60 in water buffalos; the content of basic amino acid residues was the highest among the four caseins, ranging between 14.60 in Arabian camels to 16.80 in both sheep and goats (Figure 5D). The analysis revealed that sheep and Arabian camels showed the highest and lowest content of negatively charged residues and positively charged residues (Figure 5E). The alpha–S2-casein in goats showed the highest instability index (60.21), while the alpha–S2-casein in water buffalos presented the lowest (44.10); the highest aliphatic index was 70.68 in water buffalos; however, the lowest was 64.55 in goats—alpha–S2-casein (Figure 5F).
The presence of hydrophobic domains in β-casein is featured; β-casein is the most hydrophobic casein and contains more prolyl residues than any other casein micelles. Hence, a molecular structure dominated by hydrophobic interactions of its surface than the α-S1, α-S2and k-casein would be expected. These characteristics appear to be manifested in the physicochemical properties of this protein. The hydrophobicity index of β-casein ranged between 51.10 in sheep and 51.60 in both cattle and water buffalos; unlike the hydrophobicity index, β-casein has the lowest basic amino acid content among the four caseins, ranging between nine in cattle to ten in sheep (Figure 6A). The analysis revealed that β-casein has the same content of negatively charged residues in all examined animals, while the positively charged residues ranged from 16 in goats, cattle, and water buffalos to 19 in Arabian camels (Figure 6B). The β-casein in goats exhibited the lowest instability index (94.50), whereas buffalos presented the highest (101.05). The highest aliphatic index was 100.36 in goats, while the lowest was 95.18 in cattle β-casein (Figure 6C).
The computed analysis revealed that K-casein is less hydrophobic than β-casein and has a lower frequency of prolyl residues. The analysis showed that the hydrophobicity index of K-casein ranged from 41.30 in goats to 45.50 in cattle, and β-casein has the lowest basic amino acid content (9.5) in water buffalos to 11.10 in Arabian camels (Figure 6D). The analysis revealed that k-casein has a low content of negatively and positively charged residues in all examined animals. The negatively charged residues ranged from 9 in cattle to 16 in sheep and goats, while the positively charged residues ranged from 8 in cattle to 15 in Arabian camels (Figure 6E). The k-casein in Arabian camels showed the lowest instability index (45.35), while cattle presented the highest (62.17). The highest aliphatic index was 84.16 in water buffalos, while the lowest was 71.73 in sheep β-casein (Figure 6F).
In silico, antimicrobial index and bactericidal stretches were calculated for caseins αs1-, αs2-, β-, and κ-casein in the five animals understudy to discover the antimicrobial patterns of milk casein. The analysis showed that all caseins have hydrophobicity indexes (Table 4), which enable milk casein to hydrophobically interact with pathogens with the consideration that showed the highest hydrophobicity index. The analysis also revealed that α-S2-casein and β-casein showed the highest and the lowest content of basic amino acids, respectively (Table 4), while α-S1-casein and k-casein showed intermediate content of basic amino acids, which means that all caseins can have bacteriostatic interactions with microorganisms. The more interesting result was that α-S2-casein showed the highest number of bactericidal stretches (three in Arabian camels, one in each sheep and goats, three in cattle, and two in water buffalos) compared to only one bactericidal stretch in Arabian camel β-casein and Arabian camel k-casein (Table 4). These results gave preference to Arabian camel casein due to containing 3, 1, and 1 bactericidal stretches in each of α-S2-casein, β-casein, and k-casein, respectively.
The presence of phosphorylated amino acids dramatically affects the antimicrobial activity of the caseins. The analysis of cattle caseins as the most popular and most studied model among animals showed the presence of phosphoserine residues in all casein micelles. The α-S2-casein contained the largest number of phosphoserine residues (13), followed by α-S1-casein (9), while each β-casein and k-caseins contained four phosphoserine residues (Figure 7).
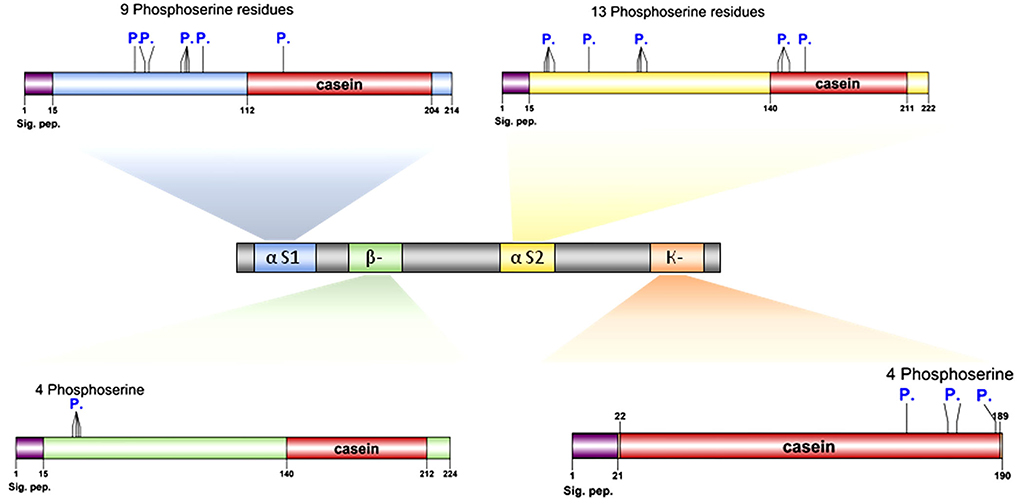
Figure 7. Proteomic and structural characterization of milk caseins from Bos taurus (cattle). Signal peptide domains (sig. pep.) and phosphorylation sites (P.) are indicated on each protein diagram.
The antioxidant activity index is attributed to the interesting antioxidant properties of the protein due to its content of hydrophobic amino acids, particularly tryptophan, methionine, isoleucine, leucine, and proline. The computed analysis revealed the hydrophobic amino-acid content of caseins αs1-, αs2-, β-, and κ-casein. The analysis showed that β-casein possessed the highest antioxidant activity index, ranging between 34.5 in goats and 37.4 in Arabian camels (Table 4). Both alpha-S1-casein and k-casein possessed an intermediate antioxidant activity index, whereas the alpha -S1-casein ranged between 26.2 in goats to 27.5 in both sheep and water buffalos (Table 4), while the K-casein ranged between 22.8 in sheep to 26.2 in water buffalos. The alpha-S2-casein possessed the lowest antioxidant activity index, ranging between 17.6 in Arabian camels and 19.6 in goats (Table 4).
Phylogenetic comparative analysis is crucial in determining phylogenetic relationships between two or more genomes to discover the differences and similarities among organisms (34–41).
The phylogeny analysis divided the five domestic animals according to combined and caseins into three subgroups, which were completely consistent with the genetic background of the species. The observed data assumed that the five domestic animals had evolved following a close evolutionary model. Interestingly, the result of our analysis divided the domestic animals based on caseins into three subgroups; one consisted of only one species (Arabian camels), the second contained both cattle and water buffalos, and the third included sheep and goats. This result could be explained by the zoological taxa of these animals. The Arabian camel belongs to the Camelidae family; the Bovidae family comprises cows, buffalos, sheep, and goats, and the Bovinae family consists of both cows and buffalos.
The positions of the Arabian camel in all phylogenetic tree topologies are the same. In the same context, the rest of all species topologies are mostly the same, whether in the tree constructed based on the combined caseins or in the trees constructed based on the individual caseins. The genetic distances and common ancestors, either between sheep and goats or cattle and water buffalos in all trees, are quite similar.
The 3D modeling of casein molecules identifies regions with different levels of organization within each casein type that affect the biological functions of the protein. When purified caseins were utilized as substrates, the proteinases were efficiently hydrolyzed at the sites with the least secondary structure. The C-terminal ends of all casein sequences are highly unstructured compared to their N-terminal ends, which are well-structured. The phosphorylated sites of the casein were poorly hydrolyzed, which is in agreement with previous studies (42–44).
Divergence in casein genes and the presence or absence of one or more genes encoding caseins explain the different biological and chemical functions of the casein micelles (45). The genes encoding α-caseins are absent in most mammalian species; for example, human milk lacks αs2-casein, and it may be possible that its role in casein micelle formation could be shifted to αs1-casein (14). In contrast, genes encoding β-and κ-caseins are widely distributed among mammals.
Amino acid composition and the type of protein residue sequence determine the structure, hydrophobicity, biological functions, and nutritional value of milk casein (46). Milk casein displays antimicrobial activity by preventing pathogen adhesion and invasion by either directly interacting with the pathogen or modifying the host environment, resulting in microorganism growth inhibition (47, 48).
The antimicrobial activity of caseins against pathogens is specific due to their affinity for polarized bacterial membranes rather than the depolarized membranes of eukaryotic cells (49). There is mounting evidence suggesting that the antibacterial properties of milk hydrolysates are linked to the production of peptides with α-helical structures. The antibacterial action is greatly affected by the presence of phosphorylated residues of certain amino acids or chemical changes in C- and N- termini (49, 50). Antimicrobial peptides can also kill bacteria by aggregating in the cytoplasmic membrane, altering their membrane permeability and triggering cell death (51, 52).
Peptides and proteins' antibacterial activity could be related to their net charge or hydrophobic characteristics. Because the majority of antibacterial peptides are positively charged, they electrostatically bond to the negatively charged components of the bacterial cell wall, potentially causing cell wall disintegration (53, 54).
Our in silico analysis indicated the antimicrobial characteristics of caseins based on the hydrophobicity, basic amino acid content, negatively charged residues (Asp + Glu), and positively charged residues (Arg + Lys) were consistent with the results of several studies (19, 55–63). Because α-S2 casein is not found in human milk, hydrolysates of bovine α-S2 casein have shown promise as a novel microbiota-modifying agent in the human gastrointestinal tract (56). Strömqvist and Falk (57) found that Helicobacter pylori, an early-life pathogen, is prevented from adhering to human stomach mucosa by the small subunit of k-casein.
Our in silico analysis gave the lion's share to the casein of the Arabian camel, which is consistent with the in vitro results of several studies (19, 64, 65), indicating the value of bioinformatics tools besides the recent genome editing approaches (66, 67) to accurate prediction and determination of antimicrobial features of casein micelles.
The antioxidant activity index presented in Table 4 revealed that all caseins may possess antioxidant activity levels. The β-casein exhibited greater antioxidant activity, particularly Arabian camel β- casein. These results were in agreement with the research by Salami, and Moosavi-Movahedi [68], who reported that camel β-casein presented higher antioxidant activity when compared to the camel's other caseins. These researchers attributed camel casein's intriguing antioxidant characteristics to the protein's highest hydrophobicity index and its main sequence, which play a key role in free radical scavenging. This is due to the greater antioxidant amino-acid content, such as Tyr, Met, Ile, Leu, and Pro of camel casein, compared to other caseins. Proteins' antioxidant capabilities are influenced not only by their amino-acid residue composition but also by their location and accessibility (68).
This study is a step toward a better understanding of the great importance and utility of bioinformatics tools in the accurate prediction, comprehensive characterization, and in silico determination of the biological activities of milk proteins. This study determined the relationship between five domestic dairy animals based on phylogenetic analysis of caseins. The results enhanced our knowledge of casein micelles, their chemical features, and potential biological functions. Therefore, it is likely that the presence of such features in the milk structure could have additional beneficial health effects, such as reduction of oxidative stress in the stomach and antimicrobial agents against harmful pathogens.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AH, MA, and AE: conceptualization and investigation. AH and OA: methodology. AH, AA, and OA: software. AH, AO, and SA: validation. AH, HE, and AO: formal analysis. ME-S, IA, HE, and OA: resources. OA, AM, AO, and AH: data curation. AH, HE, and SA: writing original draft preparation. AH and AE: writing, review, and editing. AH and MA: visualization and supervision. AA, MA, and IA: funding acquisition. All authors have read and agreed to the published version of the manuscript.
The authors are grateful to the deanship of scientific research at King Khalid University, Abha, Saudi Arabia, for supporting this work under the grant number (R.G.P2/117/43), and Taif University for funding the current work under Taif University Researchers Supporting Project number (TURSP-−2020/111), Taif University, Taif, Saudi Arabia. Also, the authors extend their appreciation to the Deanship of Scientific Research at Umm Al-Qura University for supporting this work through Grant Code 22UQU4320609DSR04.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.952319/full#supplementary-material
NCBI, National Center for Biotechnology Information; NJ, Neighbor-Joining; CSN1S1, Casein Alpha S1; CSN1S2, Casein Alpha S2; CSN2, Casein Beta; CSN3, Casein Kappa; IC50, inhibitory concentration; PV, propensity value; 3D, Three-dimensional; C-terminal, carboxyl-terminus or carboxy-terminus; N-terminal, amino-terminus or NH2-terminus.
1. Gonzalez-Gonzalez C, Tuohy K, Jauregi P. Production of angiotensin-I-converting enzyme (ACE) inhibitory activity in milk fermented with probiotic strains: effects of calcium, pH and peptides on the ACE-inhibitory activity. Int Dairy J. (2011) 219:615–22. doi: 10.1016/j.idairyj.2011.04.001
2. Park YW, Haenlein GF, Wendorff W. Handbook of Milk of Non-Bovine Mammals. John Wiley & Sons (2006). doi: 10.1002/9780470999738
3. Verduci E, D'Elios S, Cerrato L, Comberiati P, Calvani M, Palazzo S, et al. Cow's milk substitutes for children: nutritional aspects of milk from different mammalian species, special formula and plant-based beverages. Nutrients. (2019) 118:1739. doi: 10.3390/nu11081739
4. Roy D, Ye A, Moughan PJ, Singh H. Composition, structure, and digestive dynamics of milk from different species—A review. Front Nutr. (2020) 7:577759. doi: 10.3389/fnut.2020.577759
5. Wang X, Yu Z, Zhao X, Han R, Huang D, Yang Y, et al. Comparative proteomic characterization of bovine milk containing β-casein variants A1A1 and A2A2, and their heterozygote A1A2. J Sci Food Agric. (2021) 1012:718–25. doi: 10.1002/jsfa.10684
6. Madende M, Osthoff G, Patterton HG, Patterton HE, Martin P, Opperman DJ. Characterization of casein and alpha lactalbumin of African elephant (Loxodonta africana) milk. Int J Dairy Sci. (2015) 9812:8308–18. doi: 10.3168/jds.2014-9195
7. Xiong X, Ho MT, Bhandari B, Bansal N. Foaming properties of milk protein dispersions at different protein content and casein to whey protein ratios. Int Dairy J. (2020) 109:104758. doi: 10.1016/j.idairyj.2020.104758
8. Sadiq U, Gill H, Chandrapala J. Casein Micelles as an emerging delivery system for bioactive food components. Foods. (2021) 108:1965. doi: 10.3390/foods10081965
9. Holt C. Casein and casein micelle structures, functions and diversity in 20 species. Int Dairy J. (2016) 60:2–13. doi: 10.1016/j.idairyj.2016.01.004
10. Jandal JM. Comparative aspects of goat and sheep milk. Small Rumin Res. (1996) 222:177–85. doi: 10.1016/S0921-4488(96)00880-2
11. Mestawet T, Girma A, Ådnøy T, Devold T, Narvhus J, Vegarud G. New insights in goat breeds of Ethiopia: high content of αs1-CN and its association with coagulation properties, whey syneresis and micelle size. Small Rumin. (2014) 119:146–155. doi: 10.1016/j.smallrumres.2014.02.011
12. Bijl E, de Vries R, van Valenberg H, Huppertz T, Van Hooijdonk T. Factors influencing casein micelle size in milk of individual cows: genetic variants and glycosylation of κ-casein. Int Dairy J. (2014) 341:135–41. doi: 10.1016/j.idairyj.2013.08.001
13. Głab TK, Boratyński J. Potential of casein as a carrier for biologically active agents. Top Curr Chem. (2017) 375:71. doi: 10.1007/s41061-017-0158-z
14. Madende M, Osthoff G. Comparative genomics of casein genes. J Dairy Res. (2019) 863:323–30. doi: 10.1017/S0022029919000414
15. Matsuzaki K, Nakamura A, Murase O, Sugishita K-i, Fujii N, Miyajima K. Modulation of magainin 2– lipid bilayer interactions by peptide charge. Biochemistry. (1997) 368:2104–11. doi: 10.1021/bi961870p
16. Sitaram N, Nagaraj R. Host-defense antimicrobial peptides: importance of structure for activity. Curr Pharm Des. (2002) 89:727–42. doi: 10.2174/1381612023395358
17. Chan DI, Prenner EJ, Vogel HJ. Tryptophan-and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta. (2006) 17589:1184–202. doi: 10.1016/j.bbamem.2006.04.006
18. Guntero VA, Gutierrez L, Kneeteman MN, Ferretti CA. In silico study of the interaction between casein with tocopherols: preliminary evaluation of lipophilic substrate inclusion on proteic matrix. Chem Proc. (2020) 31:49. doi: 10.3390/ecsoc-24-08345
19. Lajnaf R, Gharsallah H, Attia H, Ayadi M. Comparative study on antioxidant, antimicrobial, emulsifying and physico-chemical properties of purified bovine and camel β-casein. LWT. (2021) 140:110842. doi: 10.1016/j.lwt.2020.110842
20. Wang Z, Qiao X, Hao S, Ji R. Demonstration of hepatoprotective action of camel milk through improving antioxidant activity and regulating gene expression in mice. J Camel Pract Res. (2017) 242:169–74. doi: 10.5958/2277-8934.2017.00026.1
21. Abdel-Hamid M, Romeih E, Saporito P, Osman A, Mateiu RV, Mojsoska B, et al. Camel milk whey hydrolysate inhibits growth and biofilm formation of Pseudomonas aeruginosa PAO1 and methicillin-resistant Staphylococcus aureus. Food Control. (2020) 111:107056. doi: 10.1016/j.foodcont.2019.107056
22. Kitts D. Antioxidant properties of casein-phosphopeptides. Trends Food Sci Technol. (2005) 1612:549–54. doi: 10.1016/j.tifs.2005.08.009
23. Lang Y, Li B, Gong E, Shu C, Si X, Gao N, et al. Effects of α-casein and β-casein on the stability, antioxidant activity and bioaccessibility of blueberry anthocyanins with an in vitro simulated digestion. Food Chem. (2021) 334:127526. doi: 10.1016/j.foodchem.2020.127526
24. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 356:1547–9. doi: 10.1093/molbev/msy096
25. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 44:406–25.
26. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. (2018) 46W1:W296–303. doi: 10.1093/nar/gky427
27. Kumosinski T, Brown E, Farrell H Jr. Three-dimensional molecular modeling of bovine caseins: αs1-casein. Int J Dairy Sci. (1991) 749:2889–95. doi: 10.3168/jds.S0022-0302(91)78470-1
28. Kumosinski T, Brown E, Farrell H Jr. Three-dimensional molecular modeling of bovine caseins: an energy-minimized β-casein structure. Int J Dairy Sci. (1993) 764:931–45. doi: 10.3168/jds.S0022-0302(93)77420-2
29. Farrell H Jr, Malin E, Brown E, Mora-Gutierrez A. Review of the chemistry of αS2-casein and the generation of a homologous molecular model to explain its properties. J Dairy Sci. (2009) 924:1338–53. doi: 10.3168/jds.2008-1711
30. Walker JM. The Proteomics Protocols Handbook. Hatfield: Springer (2005). doi: 10.1385/1592598900
31. Torrent M, Di Tommaso P, Pulido D, Nogués MV, Notredame C, Boix E, et al. AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics. (2011) 281:130–1. doi: 10.1093/bioinformatics/btr604
32. Torrent M, Nogués VM, Boix E. A theoretical approach to spot active regions in antimicrobial proteins. BMC Bioinformatics. (2009) 101:373. doi: 10.1186/1471-2105-10-373
33. De Kruif CG, Huppertz T, Urban VS, Petukhov AV. Casein micelles and their internal structure. Adv Colloid Interface Sci. (2012) 171:36–52. doi: 10.1016/j.cis.2012.01.002
34. Hassanin AA, Saad AM, Bardisi EA, Salama A, Sitohy MZ. Transfer of anthocyanin accumulating Delila and Rosea1 genes from the transgenic tomato micro-tom cultivar to moneymaker cultivar by conventional breeding. J Agric Food Chem. (2020) 6839:10741–9. doi: 10.1021/acs.jafc.0c03307
35. Raza SHA, Hassanin AA, Dhshan AIM, Abdelnour SA, Khan R, Mei C, et al. In silico genomic and proteomic analyses of three heat shock proteins (HSP70, HSP90-α, and HSP90-β) in even-toed ungulates. Electron J Biotechnol. (2021). 53:61–70. doi: 10.1016/j.ejbt.2021.07.002
36. Fathy DM, Eldomiaty AS, El-Fattah HIA, Mahgou E-SI, Hassanin AA. Morphological, biochemical and molecular characterization of rhizobia of faba bean plants grown in North Nile Delta Egypt. Pak J Biol Sci. (2021) 24:672–9. doi: 10.3923/pjbs.2021.672.679
37. Fang EF, Hassanien AA, Wong JH, Bah CS, Soliman SS, Ng TB. Isolation of a new trypsin inhibitor from the Faba bean (Vicia faba cv. Giza 843) with potential medicinal applications. Protein Pept Lett. (2011) 181:64–72. doi: 10.2174/092986611794328726
38. Fang EF, Hassanien AAE, Wong JH, Bah CSF, Soliman SS, Ng TB. Purification and modes of antifungal action by Vicia faba cv. Egypt trypsin inhibitor. J Agric Food Chem. (2010) 5819:10729–35. doi: 10.1021/jf102277k
39. Farrag ME, Soliman SS, Amer IM, Heakel RM, Hassanin AA. Analysis of genetic diversity among a population of canola genotypes as reveled by ISSR-PCR and their associations to seed yield and oil content. Ann Agric Sci Moshtohor. (2019) 572:425–34. doi: 10.21608/assjm.2019.44917
40. Hassanin AA, Haidar Abbas Raza S, Ahmed Ujjan J, Aysh Alrashidi A, Sitohy BM, Al-Surhanee AA, et al. Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: benefits of getting vaccinated and common questions. Saudi J Biol Sci. (2021) doi: 10.1016/j.sjbs.2021.12.020
41. Zhang L, Hong Y, Liao Y, Tian K, Sun H, Liu X, et al. Dietary Lasia spinosa Thw. improves growth performance in broilers. Front Nutr. (2022) 8:775223. doi: 10.3389/fnut.2021.775223
42. Deutsch S-M, Molle D, Gagnaire V, Piot M, Atlan D, Lortal S. Hydrolysis of sequenced β-casein peptides provides new insight into peptidase activity from thermophilic lactic acid bacteria and highlights intrinsic resistance of phosphopeptides. Appl Environ Microbiol. (2000) 6612:5360–7. doi: 10.1128/AEM.66.12.5360-5367.2000
43. Perpetuo EA, Juliano L, Lebrun I. Biochemical and pharmacological aspects of two bradykinin-potentiating peptides obtained from tryptic hydrolysis of casein. J Protein Chem. (2003) 227:601–6. doi: 10.1023/B:JOPC.0000008724.98339.ff
44. Souza Júnior EC, Santos MPF, Sampaio VS, Ferrão SPB, Fontan RCI, Bonomo RCF, et al. Hydrolysis of casein from different sources by immobilized trypsin on biochar: effect of immobilization method. J Chromatogr B. (2020) 1146:122124. doi: 10.1016/j.jchromb.2020.122124
45. Müller H, Naumann F, Freytag J-C. Data Quality in Genome Databases. Humboldt-Universität zu Berlin; Mathematisch-Naturwissenschaftliche Fakultät II (2003). doi: 10.18452/9205
46. Mauer L. Protein | Heat Treatment for Food Proteins, in Encyclopedia of Food Sciences Nutrition (Second Edition). Oxford: Academic Press (2003). p. 4868–72.
47. Gauthier SF, Pouliot Y, Saint-Sauveur D. Immunomodulatory peptides obtained by the enzymatic hydrolysis of whey proteins. Int Dairy J. (2006) 1611:1315–23. doi: 10.1016/j.idairyj.2006.06.014
48. Hernández-Ledesma B, García-Nebot MJ, Fernández-Tomé S, Amigo L, Recio I. Dairy protein hydrolysates: peptides for health benefits. Int Dairy J. (2014) 382:82–100. doi: 10.1016/j.idairyj.2013.11.004
49. Benkerroum N. Antimicrobial peptides generated from milk proteins: a survey and prospects for application in the food industry. A review. Int J Dairy Technol. (2010). 633:320–38. doi: 10.1111/j.1471-0307.2010.00584.x
50. Dashper SG, Liu SW, Reynolds EC. Antimicrobial peptides and their potential as oral therapeutic agents. Int J Pept Res Ther. (2007) 134:505–16. doi: 10.1007/s10989-007-9094-z
51. Zambrowicz A, Timmer M, Polanowski A, Lubec G, Trziszka T. Manufacturing of peptides exhibiting biological activity. Amino acids. (2013) 442:315–320. doi: 10.1007/s00726-012-1379-7
52. Hartmann R, Meisel H. Food-derived peptides with biological activity: from research to food applications. Curr Opin Biotechnol. (2007) 182:163–9. doi: 10.1016/j.copbio.2007.01.013
53. Osman A, Goda HA, Abdel-Hamid M, Badran SM, Otte J. Antibacterial peptides generated by Alcalase hydrolysis of goat whey. LWT. (2016) 65:480–6. doi: 10.1016/j.lwt.2015.08.043
54. Abdel-Hamid M, Goda HA, De Gobba C, Jenssen H, Osman A. Antibacterial activity of papain hydrolysed camel whey and its fractions. Int Dairy J. (2016) 61:91–8. doi: 10.1016/j.idairyj.2016.04.004
55. Nagpal R, Behare P, Rana R, Kumar A, Kumar M, Arora S, et al. Bioactive peptides erived from milk proteins and their health beneficial potentials: an update. Food Funct. (2011) 21:18–27. doi: 10.1039/C0FO00016G
56. Lahov E, Regelson W. Antibacterial and immunostimulating casein-derived substances from milk: casecidin, isracidin peptides. Food Chem Toxicol. (1996) 341:131–45. doi: 10.1016/0278-6915(95)00097-6
57. Strömqvist M, Falk P, Bergström S, Hansson L, Lönnerdal B, Normark S, et al. Human milk kappa-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. (1995) 213:288–96. doi: 10.1097/00005176-199510000-00006
58. Pellegrini A, Thomas U, Bramaz N, Hunziker P, von Fellenberg R. Isolation and identification of three bactericidal domains in the bovine α-lactalbumin molecule. Biochim Biophys Acta. (1999) 14263:439–48. doi: 10.1016/S0304-4165(98)00165-2
59. Malkoski M, Dashper SG, O'Brien-Simpson NM, Talbo GH, Macris M, Cross KJ, et al. Kappacin, a novel antibacterial peptide from bovine milk. Antimicrob Agents Chemother. (2001) 458:2309–15. doi: 10.1128/AAC.45.8.2309-2315.2001
60. Hayes M, Ross R, Fitzgerald G, Hill C, Stanton C. Casein-derived antimicrobial peptides generated by Lactobacillus acidophilus DPC6026. Appl Environ Microbiol. (2006) 723:2260–4. doi: 10.1128/AEM.72.3.2260-2264.2006
61. McCann K, Shiell B, Michalski W, Lee A, Wan J, Roginski H, et al. Isolation and characterisation of a novel antibacterial peptide from bovine αS1-casein. Int Dairy J. (2006) 164:316–23. doi: 10.1016/j.idairyj.2005.05.005
62. Nebbia S, Lamberti C, Lo Bianco G, Cirrincione S, Laroute V, Cocaign-Bousquet M, et al. Antimicrobial potential of food lactic acid bacteria: bioactive peptide decrypting from caseins and bacteriocin production. Microorganisms. (2021) 91:65. doi: 10.3390/microorganisms9010065
63. Ouertani A, Chaabouni I, Mosbah A, Long J, Barakat M, Mansuelle P, et al. Two new secreted proteases generate a casein-derived antimicrobial peptide in Bacillus cereus food born isolate leading to bacterial competition in milk. Front Microbiol. (2018) 9:1148. doi: 10.3389/fmicb.2018.01148
64. Abd EL, Rahim A. Antioxidant and antimicrobial activities of enzymatic hydrolysates of camel's milk whey protein and casein. J Food Dairy Sci. (2020) 112:45–50. doi: 10.21608/jfds.2020.78877
65. Almi-Sebbane D, Adt I, Degraeve P, Jardin J, Bettler E, Terreux R, et al. Casesidin-like anti-bacterial peptides in peptic hydrolysate of camel milk β-casein. Int Dairy J. (2018) 86:49–56. doi: 10.1016/j.idairyj.2018.06.016
66. Raza SHA, Hassanin AA, Pant SD, Bing S, Sitohy MZ, Abdelnour SA, et al. Potentials, prospects and applications of genome editing technologies in livestock production. Saudi J Biol Sci. (2021) 29:1928–35. doi: 10.1016/j.sjbs.2021.11.037
67. Abdelnour SA, Xie L, Hassanin AA, Zuo E, Lu Y. The potential of CRISPR/Cas9 gene editing as a treatment strategy for inherited diseases. Front Cell Dev Biol. (2021) 9:699597. doi: 10.3389/fcell.2021.699597
Keywords: casein protein, phylogenetic analysis, bioinformatics, antimicrobial, antioxidant
Citation: Hassanin AA, Osman A, Atallah OO, El-Saadony MT, Abdelnour SA, Taha HSA, Awad MF, Elkashef H, Ahmed AE, Abd El-Rahim I, Mohamed A and Eldomiaty AS (2022) Phylogenetic comparative analysis: Chemical and biological features of caseins (alpha-S-1, alpha-S-2, beta- and kappa-) in domestic dairy animals. Front. Vet. Sci. 9:952319. doi: 10.3389/fvets.2022.952319
Received: 25 May 2022; Accepted: 11 August 2022;
Published: 15 September 2022.
Edited by:
Jean-Christophe Bambou, Institut National de Recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), FranceReviewed by:
Amr Rashad, Alexandria University, EgyptCopyright © 2022 Hassanin, Osman, Atallah, El-Saadony, Abdelnour, Taha, Awad, Elkashef, Ahmed, Abd El-Rahim, Mohamed and Eldomiaty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdallah A. Hassanin, YXNhZmFuQHp1LmVkdS5lZw==; Mohamed T. El-Saadony, bV90bGF0ZWxzYWRvbnlAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.