
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 15 August 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.951554
 Zimin Xie1,2†
Zimin Xie1,2† Junqin Zhang1†
Junqin Zhang1† Minhua Sun1
Minhua Sun1 Qinghang Zeng1
Qinghang Zeng1 Yunzhen Huang1
Yunzhen Huang1 Jiawen Dong1
Jiawen Dong1 Linlin Li1
Linlin Li1 Shujian Huang2*
Shujian Huang2* Ming Liao1,3,4*
Ming Liao1,3,4*Since 2015, fowl adenovirus (FAdV) has been frequently reported worldwide, causing serious economic losses to the poultry industry. In this study, a FAdV-2, namely GX01, was isolated from liver samples of chickens with hepatitis and hydropericardium in Guangxi Province, China. The complete genome sequence of GX01 was determined about 43,663 base pairs (bp) with 53% G+C content. To our knowledge, this is the first FAdV-2 complete genome in China. There was a deleting fragment in ORF25 gene. Phylogenetic analysis based on the hexon loop-1 gene showed that GX01 is most closely related to FAdV-2 strain 685. Pathogenicity experiment of GX01 in 3-day-old and 10-day-old specific-pathogen-free chickens showed that although no mortality was observed within 21 days post infection (dpi), strain GX01 significantly inhibited weight gain of infected chickens. Moreover, FAdV-2 was still detectable in the anal swabs of infected chickens at 21 dpi. Necropsy analysis showed that the main lesions were observed in liver, heart, and spleen. Of note, hepatitis and hydropericardium were observed in the infected chickens. In addition, massive necrosis of lymphocyte was observed in spleen of infected 3-days-old chickens. We concluded that FAdV-2 strain GX01 is capable of causing hepatitis and hydropericardium, which will make serious impact on the growth of chickens. Our research lays a foundation to investigate the molecular epidemiology and etiology of FAdV.
Fowl adenoviruses (FAdVs), a non-enveloped, linear double-stranded DNA virus with a genome of 43–45 kbp in size, are common and harmful pathogens in chickens (1). FAdVs belongs to the family Aviadenovirus. According to the basis of phylogeny, genome organization and the lack of significant cross-neutralization (2, 3), it has currently been divided into five species (FAdV-A to FAdV-E) and 12 serotypes (FAdV-1 to FAdV-8a and FAdV-8b to FAdV-11) (4). FAdV-1 belongs to the species FAdV-A; FAdV-5 belongs to the species FAdV-B; FAdV-4 and FAdV-10 belong to the species FAdV-C; FAdV-2, 3, 9, and 11 belong to the species FAdV-D; FAdV-6, 7, 8a, and 8b belong to the species FAdV-E (1, 2, 5).
FAdVs are transmitted both vertically and horizontally, causing several diseases in chickens (6). These diseases include inclusion body hepatitis (IBH), hepatitis hydropericardium syndrome (HHS) and gizzard erosion (GE) (7–9). IBH is caused by all 12 serotypes (9–11). HHS is primarily related to FAdV-4 (12). And GE is mainly induced by FAdV-1, 8a, and 8b (13, 14). Since 2015, the epidemic trend of FAdVs has been gradually increasing worldwide, posing great threats to the poultry industry (6, 15). FAdV-2 has been detected in many countries, such as China (16, 17), South Africa (18, 19), Japan (2, 20), and Poland (21, 22). It mainly infected 3–5 weeks broiler, resulting in severe IBH with mortality rates ranging from 0–17% (16, 20, 23). Up till now, only one complete genome sequence of defined FAdV-2 is available in GenBank. Moreover, the pathogenicity of FAdV-2 is still unclear, especially the impact on weight and viral shedding (2, 24, 25).
In our study, we successfully isolated a FAdV-2 strain GX01 from commercial broiler with hepatitis and hydropericardium. The complete genome sequence of GX01 was determined and characterized. The pathogenicity of GX01 was evaluated in 3-day-old and 10-days-old Specific Pathogen Free (SPF) chickens. To our knowledge, this work reports the first FAdV-2 complete genome sequence in China. The pathogenicity experiment showed that FAdV-2 strain GX01 is capable of causing hepatitis and hydropericardium. Moreover, GX01 significantly inhibited weight gain and caused viral shedding for a long period of time in the infected chickens. We concluded that the isolated GX01 may have serious impact on the production of the poultry industry, especially the broiler industry.
In July 2020, the liver samples were collected from 20-days-old commercial broiler with hepatitis and hydropericardium in Guangxi Province, China. Total RNA and DNA were extracted using Tianlong Nucleic Acid Extraction & Purification Kit T180H (Tianlong Technology Co., Ltd., China). The extracted DNA was subjected to polymerase chain reaction (PCR) amplification to detect FAdV (26). Other pathogens include avian influenza virus (AIV), Newcastle disease virus (NDV), infectious bursal disease virus (IBDV), infectious laryngotracheitisand virus (ILTV), chicken infectious anemia virus (CIAV), avian leukosis virus (ALV), Marek's disease virus (MDV), egg drop syndrome virus (EDSV) and avian reovirus (ARV) were detected by PCR or reverse transcription-polymerase chain reaction (RT-PCR) assays. The positive PCR product was sequenced. And the obtained nucleotide sequence was blasted in NCBI. The primers used for detecting pathogens here were shown in Supplementary Table 1. And the primers were synthesized by Sangon Biotech (Guangzhou, China). The liver sample was homogenized in phosphate buffered saline (PBS) to obtain a 10% tissue suspension. After three freeze-thaw cycles, the suspensions were centrifuged at 12,000 × g at 4°C for 10 min. And then the suspensions were filtered through a 0.22-μm pore-size sterile filter (Millipore, Bedford, MA, United States). The filtered solution was inoculated into leghorn male hepatocellular (LMH) cells and propagated for three passages (27, 28). The infected LMH cells and the mock LMH cells were sent to Guangzhou Sevier Biotechnology Co., Ltd., China for transmission electron microscopy (TEM) analysis.
The 50% tissue culture infective dose (TCID50) assay was performed as previously described (29). Viral cytopathic effect (CPE) was observed for approximately 5 days. The virus titer was calculated as TCID50 according to the Reed-Muench method (30). The virus growth kinetics were determined as previously described with a few modifications (31). Briefly, the supernatant and infected LMH cells were harvested at 0, 12, 24, 36, 48, 60, 72, 84, and 96 h post infection (hpi), respectively. After three freeze-thaw cycles, the virus titers of each time point were determined by TCID50 assay.
Based on published sequences of FAdVs, a set of primers were designed to determine the complete genome sequence of GX01 (Supplementary Table 2). And the primers were synthesized by Sangon Biotech (Guangzhou, China). The fragments of GX01 were amplified by PCR assay. The amplified fragments were determined by sequencing (Shanghai Sangon Biotechnology Co., Ltd., China). Finally, all sequences were assembled using the SeqMan of the DNASTAR software package (version 7.1, Madison, WI, United States). The open reading frames (ORFs) of complete genome sequence were annotated using the Snapgene software (version 2.3.2, United States).
Based on the sequences of GX01 complete genome and hexon loop-1 gene and amino acid sequences of hexon, fiber, and DNA polymerase gene, the homology analysis of GX01 was carried out with other FAdV-D strains reference sequences using the MegAlign of the DNASTAR package. Moreover, Phylogenetic trees were constructed based on the sequences of DNA polymerase amino acid and hexon loop-1 gene, respectively, by the maximum likelihood method in MEGA 7.0 software (32). Bootstrap values were determined based on the original data from 1,000 replicates.
Twenty 3-days-old and 20 10-days-old SPF leghorn chickens (Guangdong Dahuanong Animal Health Products Co., Ltd., China) were raised in separated negative-pressure isolators and randomly divided into two groups, respectively. Chickens in the challenge groups were inoculated intravenously with dose of 0.2 ml (1.0 × 107 TCID50/ml) virus. The control groups were inoculated intravenously with an equal volume of DMEM/F12 basic medium (Gibco, Australia). All chickens were monitored daily for clinical signs. The anal swabs of all chickens were collected from 0 to 21 dpi. The weight of all chickens was recorded weekly. At 21 dpi, all chickens were humanely euthanized. The liver, heart, spleen, and kidney were collected. And the weight of spleen was recorded. The spleen indexes were calculated by the spleen (milligram, mg) / body weight (gram, g). For histopathological examinations, samples were fixed in 4% paraformaldehyde. The remaining samples were stored at−80°C. The animal experiment protocol used in this study was approved by and performed under the guidance of the Committee on the Ethics of Animal Experiments of Institute of Animal Health, Guangdong Academy of Agricultural Sciences Experimental Animal Welfare Ethics Committee on 8 November, 2021 (Approve ID: SPF2021027). All efforts were made to minimize animal suffering.
Total DNA of anal swabs and tissue samples were extracted. Quantitative real-time PCR analysis was carried out using SYBR Green master mix (Roche, United states). Absolute quantitative real-time PCR was conducted as described previously with small modifications (33). Briefly, 200 microliter (μl) was taken from anal swab to obtain DNA in 60 μl volume. 200 mg was taken from tissue sample to obtain DNA in 60 μl volume. And then 1 μl was taken to conduct real-time PCR. Primers were designed according to the conserved 52K gene of FAdVs (Supplementary Table 3) (33). A plasmid containing the 52K gene of FAdV-2 strain was used to construct standard curve in each reaction.
In July 2020, chickens on a chicken farm in Guangxi Province were suffering from severe hepatitis and mild hydropericardium. The liver samples were tested positive for FAdV but tested negative for AIV, NDV, IBDV, ILTV, CIAV, ALV, MDV, EDSV, and ARV. After inoculating the filtered tissue supernatant in LMH cells, the infected LMH cells were shedding, round-shaped and strongly refracted at 48 dpi to 72 dpi (Figure 1A). According to the growth curve of strain GX01, the virus titer increased dramatically from 12 hpi to 24 hpi, and then it reached a plateau at 24 hpi and peaked at 72 hpi, approximately 1.0 × 107 TCID50/ml (Figure 1B). Moreover, the virus particles were observed by TEM. It displayed a spherical shape and latticed distribution (Figure 2). These results indicated that the virus was successfully isolated. The isolated virus was designated as GX01.
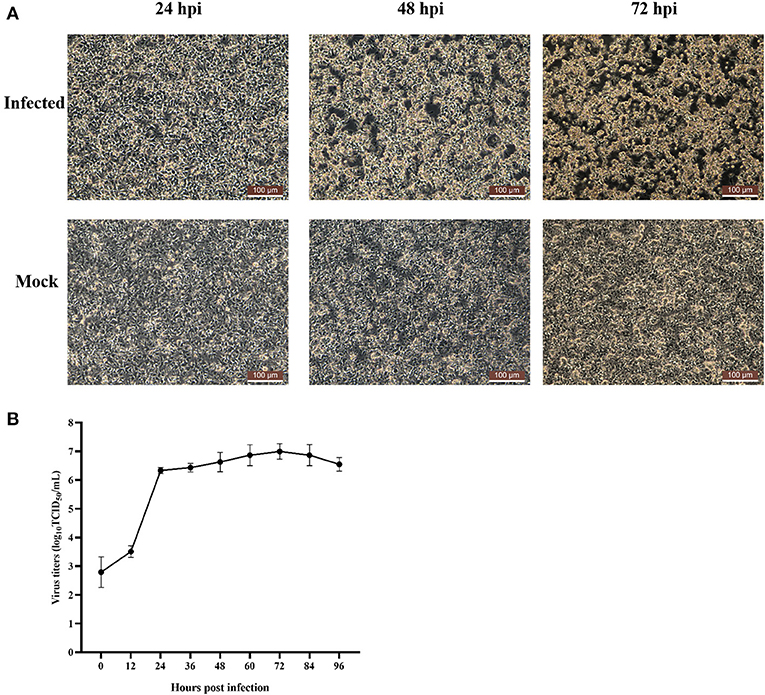
Figure 1. Cytopathic effect of FAdV-2 strain GX01 infected LMH cells and one-step growth curve. (A) Cytopathic effect of FAdV-2 strain GX01 infected LMH cells and non-infected LMH cells (Mock) at 24, 48, and 72 hpi. Scale bar = 100 μm. (B) One-step growth curve of FAdV-2 strain GX01 in LMH cells.
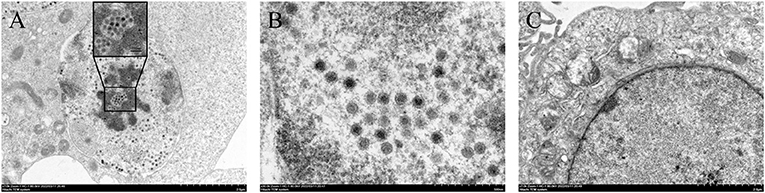
Figure 2. Transmission electron micrographs of LMH cells. (A) and (B) are transmission electron micrographs of viral particles; (C) showing transmission electron micrographs of normal LMH cells.
To obtain the complete genome sequence of strain GX01, all the sequences were assembled together. Finally, we obtained a 43,663 bp viral genome sequence of strain GX01, with 53% G + C content. And the complete genome sequence contained 36 ORFs (Figure 3A). The complete genome sequence is deposited in GenBank under the accession number ON014843. In addition, there was a deleting fragment in ORF25 gene (Figure 3B). The two coding sequences (CDS) of GX01 ORF25 gene are contiguous. The ORF25 gene of other FAdV-2 strains has a non-coding sequence in the middle of the CDS at both ends.
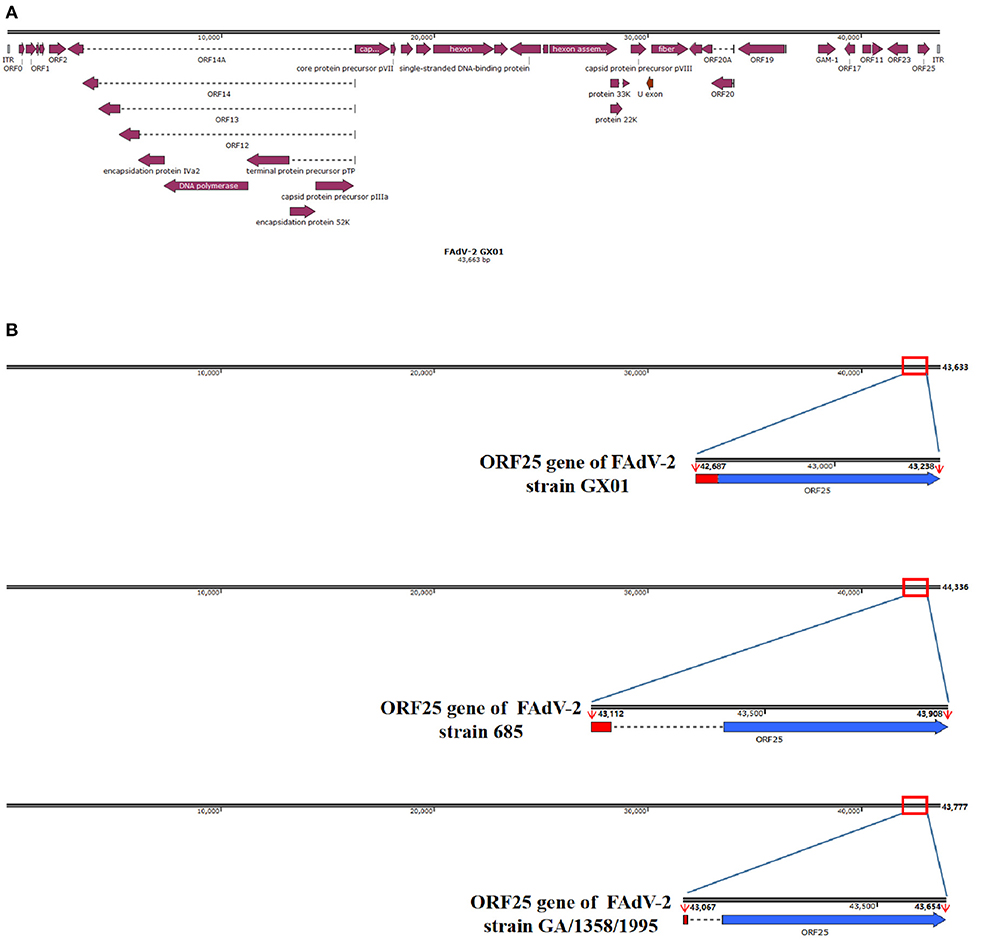
Figure 3. Schematic representation of FAdV-2 strain GX01 and ORF25 gene. (A) The complete genome schematic representation of FAdV-2 stain GX01; (B) Comparison of ORF25 gene of FAdV-2 strain GX01 with ORF25 gene of other FAdV-2 strains (red region: the first coding sequence of ORF25; blue region: the second coding sequence of ORF25; dotted line: non-coding sequence).
According to the species demarcation criteria of the International Committee on Taxonomy of Viruses (ICTV), the species designation of FAdVs depends on the distance matrix analysis of the DNA polymerase amino acid sequence. Phylogenetic tree based on amino acid sequence of DNA polymerase showed that GX01 was clustered within the species FAdV-D (Figure 4A). In addition, the hexon loop-1 gene is recognized as a gene that distinguishes serotypes of FAdVs (4). Phylogenetic tree based on hexon loop-1 gene showed that strain GX01 was most closely related to FAdV-2 strain 685 (Figure 4B).
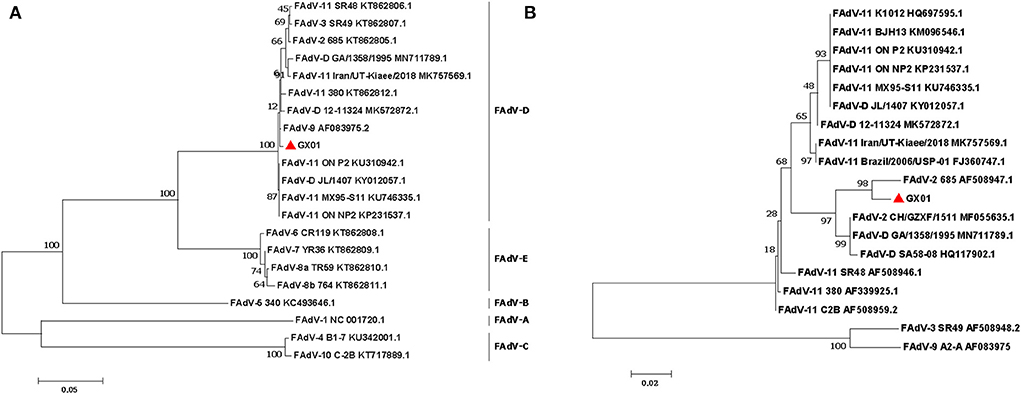
Figure 4. Phylogenetic analysis of strain GX01. The phylogenetic trees were constructed by the maximum likelihood method in MEGA 7.0. Bootstrap majority consensus values based on 1,000 replicates are indicated at each branch point as a percentage. (A) Phylogenetic tree based on amino acid sequence of DAN polymerase. (B) Phylogenetic tree based on the nucleotides of hexon loop-1 gene.
Pairwise comparisons were performed to determine the sequence identities. The results showed that GX01 shared of 90.5–97.9% identify in the complete genome sequence with other FAdV-D strains (Table 1). The homology analysis based on hexon loop-1 gene showed that GX01 shared 97.7% identity with FAdV-2 strain 685. The sequence identities at the amino acid level were 89.5–98.6% (hexon), 78.1–95.9% (fiber), and 90.1–99.3% (DNA polymerase) between GX01 and other FAdV-D strains.
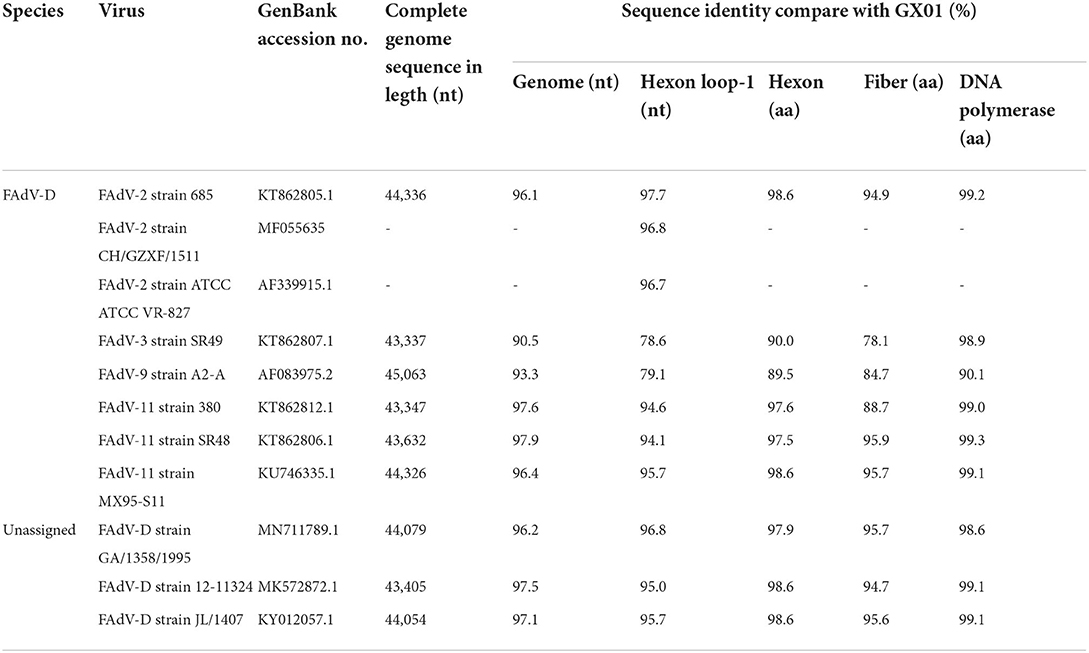
Table 1. Comparison of complete genome sequences, hexon loop-1 gene and amino acid sequences of the three ORFs of GX01 with other FAdV-D strains.
In order to assess the pathogenicity of GX01 to chickens, we used GX01 to challenge chickens. All of chickens in the challenge groups showed clinical signs of depression and anorexia at 1 to 3 dpi. The chickens in the control groups did not show any clinical signs. Moreover, although none of the chickens died through the study, the weight gain of infected 3-days-old chickens was significantly inhibited than that of the control group at 7, 14, and 21 dpi. The average weight of control chickens reached 245 g at 21 days, while the average weight of infected chickens was only 159 g (Figure 5A). The weight gain of infected 10-day-old chickens was significantly inhibited than that of the control group at 14 and 21 dpi. The average weight of control chickens reached 334 g at 21 days, while the average weight of infected chickens was only 261 g (Figure 5B).
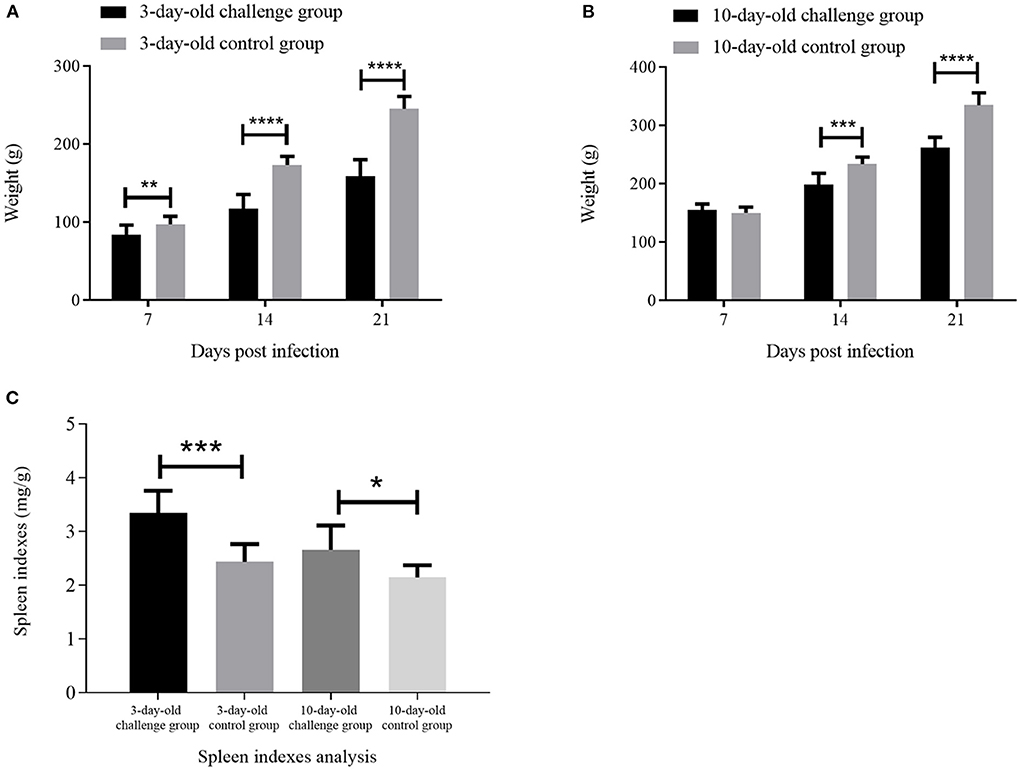
Figure 5. The weight analysis and spleen indexes analysis. (A) and (B) are the weight analysis of the challenge groups and control groups at 7, 14, and 21 days post infection (dpi); (C) is the spleen indexes analysis of challenge groups and control groups at 21 dpi. * represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001; **** represents p < 0.0001.
At 21 dpi, necropsy results showed that livers of challenge groups exhibited obvious petechial hemorrhage (Figures 6A,B). And the pericardium showed yellowish effusion (Figures 6D,E). In addition, the spleen indexes showed that the spleens in challenge groups were significantly larger than that of the control groups (Figure 5C). Histological examination showed that massive infiltration of inflammatory cells was observed in liver and kidney of infected chickens (Figure 7A). Typically IBH was observed in livers of 3-days-old challenge and 10-days-old challenge groups (Figure 7B). Lesions in the spleen were characterized as massive lymphocyte necrosis in red-white pith. Absolute quantification of FAdV-2 in the organs of the euthanized chickens showed that the FAdV-2 was detectable in the heart, liver, spleen, and kidneys. The viral loads were still remained 60 to 4.67 × 102 copies (Supplementary Figure 1). No FAdV-2 was detected in the negative control groups.
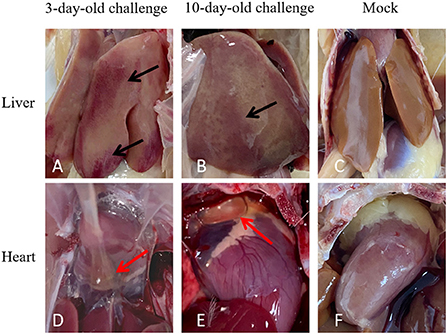
Figure 6. Pathogenicity analysis of FAdV-2 strain GX01. Clinical signs were observed in the challenge and negative control groups at 21 days post infection: livers enlargement and hemorrhage (Black arrow) and hearts mild hydropericardium (Red arrow). (A,B) Abnormal liver in 3-day-old challenge group and 10-day-old challenge group respectively. (C) Normal liver in mock group. (D,E) Abnormal heart in 3-day-old challenge group and 10-day-old challenge group, respectively. (F) Normal heart in mock group.
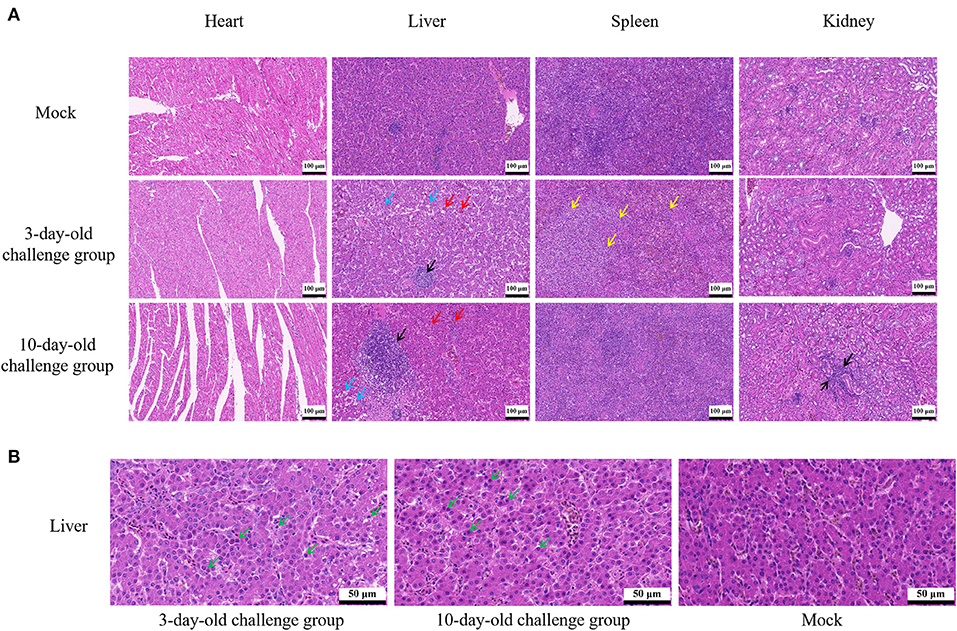
Figure 7. Histopathological analysis of FAdV-2 strain GX01. (A) Histopathological analysis showed massive infiltration of inflammatory cells was observed in liver and kidney of infected chickens (black arrow); Congested in liver sinuses (red arrow); Hepatocyte vacuolar degeneration with small vacuoles visible in the cytoplasm (blue arrow); Massive lymphocyte necrosis in red-white pith of spleen (yellow arrow). (B) Typically inclusion body was observed in liver (green arrow).
To determine viral shedding of GX01 in the 3-days-old and 10-days-old SPF chickens, we collected the anal swabs of all chickens from 0 to 21dpi. The virus genome was detected in anal swabs by quantitative real-time PCR assays (Figure 8). At 1 dpi, FAdV-2 in infected 3-days-old and 10-days-old groups has been detected about 1.26 × 102 copies and 1.93 × 102 copies, respectively. The viral shedding of infected 3-days-old chickens increased sharply at 3dpi and peaked at 5dpi (5.42 × 106 copies), then it dramatically declined. The viral shedding of infected 10-days-old chickens increased sharply at 3dpi and peaked at 4dpi (1.82 × 106 copies), then it declined slowly. At 21 dpi, FAdV-2 in infected 3-days-old and 10-days-old groups was still detectable approximately 2.64 × 102 copies and 1.26 × 102 copies, respectively. No FAdV-2 was detected in the negative control chickens throughout the experiment.
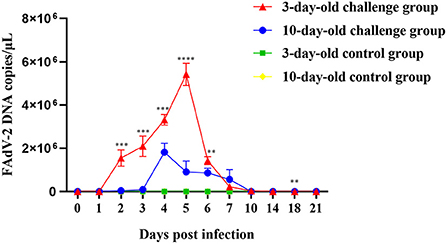
Figure 8. Viral shedding of FAdV-2 strain GX01. The viral shedding of challenge and negative control groups at 0, 1, 2, 3, 4, 5, 6, 7, 10, 14, 18, and 21 days post infection (Asterisk represents the significant difference in virus shedding between the 3-day-old challenge group and the 10-day-old challenge group on the same day).
In recent years, FAdVs have been frequently reported worldwide, causing huge economic loss to the poultry industry (34). Co-infection of multiple serotypes was observed in different regions, such as FAdV-2 and 8b in South Africa (18, 19), FAdV-2, 8a, 8b, and 11 in Asia, Europe and North America (2, 15, 21, 35–37). In China, FAdVs are transmitted both vertically and horizontally. The prevalence is dominated by serotype 4, 8a, 8b and 11 (23). Co-infection has also been reported in southern China (23). In this work, we successfully isolated a FAdV-2 strain GX01 and obtained the complete genome sequence of the virus. Moreover, the pathogenicity of FAdV-2 strain GX01 was deeply evaluated in 3-old-days and 10-old-day SPF chickens.
Up till now, only two complete genome sequences of FAdV-2 are documented in GenBank. They are FAdV-2 strain 685 and FAdV-2 strain SR48. FAdV-2 strain 685 was isolated from United Kingdom. FAdV-2 strain SR48 has already been confirmed to be FAdV-11 (2). Besides, our study confirmed the unassigned species FAdV-D strain GA/1358/1995 to be a FAdV-2, based on phylogenetic analysis of hexon loop-1 gene. The hexon gene of FAdVs was related to the classification of serotypes, especially the hexon loop-1 gene (4, 26, 35). This gene is sufficiently variable to ensure species identification and additional differentiation of the currently recognized 12 serotypes (38–40). Moreover, the ORFs of GX01 were almost identical to the other reported FAdV-2 strains by alignment with reference sequences, suggesting that the genome sequence of FAdV-2 was relatively conserved. However, there are some differences in ORF25 gene of GX01. The ORF25 gene of other strains in species FAdV-D is composed of two discontinuous CDS (5). In our study, the two CDS of GX01 ORF25 gene were contiguous, indicating that some non-coding sequences of GX01 were deleted in natural evolution. Moreover, the sequence of the first CDS of GX01 ORF25 gene was quite different from the other known FAdV-2 strains. To date, the function of FAdVs ORF25 gene is still unclear (25, 41, 42). It may be associated with evolution and mutation of the virus.
The pathogenicity of FAdV-2 is not well–understood, especially with regard to the effects on weight gain and viral shedding. In our study, although none of the chickens died throughout the experiment, we found the weight gain of infected chickens was significantly inhibited than that of control groups. It means that the growth of chickens is affected by FAdV-2 strain GX01, which will cause significant economic losses to poultry industry. Moreover, the viral shedding increased sharply, indicating strain GX01 has a strong ability to proliferate in chickens. Furthermore, FAdV-2 was still detectable in anal swabs at 21 dpi, indicating viral shedding lasted for a long period of time after infection, which will produce significant cross-infection in production and endanger healthy flocks.
FAdV-2 has only been reported to cause IBH (16). HHS was related with FAdV-4 and 11 (12, 43). Interestingly, in addition to severe hepatitis, hydropericardium was also observed in the infected chickens in our study. Thus, GX01 was able to cause HHS in both clinical and experimental cases. To our knowledge, this is the first report in which hydropericardium were associated with the FAdV-2. The volume of hydropericardium caused by FAdV-4 can reach as much as 15 ml, while the volume of that caused by FAdV-2 strain GX01 and FAdV-11 was 1 to 2 ml (43, 44). We suggested that the pathogenicity of FAdV-2 strain GX01 may be diverse.
In recent years, FAdV-4 has been proven to have immunosuppressive potential, which caused structural and functional damage of immune organs via apoptosis along with induction of severe inflammatory responses (45–47). Other serotypes of FAdVs have also been reported to cause immune system damage in chickens, such as FAdV-8b, and FAdV-11 (48–50). In our study, not only the spleen indexes of challenge groups were significantly higher than that of the control groups, but also massive necrosis of lymphocyte was observed in spleen of infected 3-days-old chickens. These result revealed that GX01 may also be an immunosuppressive pathogen. The immunosuppression potential of FAdV-2 is required for further investigation.
In our study, a FAdV-2 strain GX01 was successfully isolated from commercial broiler with HHS in Guangxi Province, China. The first complete genome of FAdV-2 in China was determined and characterized, which not only increased the knowledge of the molecular characteristics, but also enriched the understanding of FAdV-2 diversity. Moreover, the Pathogenicity of GX01 in SPF chickens showed that GX01 significantly inhibited weight gain in infected chickens and caused viral shedding lasted for at least 21 dpi. Furthermore, FAdV-2 strain GX01 is capable of causing HHS. We concluded that this virus may become a severe threat to poultry industry. Therefore, further studies of pathogenic mechanism and vaccine development of FAdV-2 are needed to obtain more insights for the prevention and control of this disease.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments of Institute of Animal Health, Guangdong Academy of Agricultural Sciences Experimental Animal Welfare Ethics Committee on 8 November, 2021 (Approve ID: SPF2021027).
ZX and JZ designed this study and critically revised the manuscript and performed the experiments, data analysis, and drafted the manuscript. ZX, MS, QZ, YH, JD, and LL participated in sample collection, virus isolation, and animal experiment. SH and ML participated in the coordination and manuscript revision. All authors read and approved the final manuscript.
This work was funded by Special Fund for Scientific Innovation Strategy-Construction of High Level Academy of Agriculture Science (202110TD, R2020PY-JX014, R2020QD-049, and R2020PY-JC001), Guangzhou Basic and Applied Basic Research Foundation (202102021162), Guangdong Laboratory for Lingnan Modern Agriculture Project (NZ2021015), Guangdong Basic and Applied Basic Research Foundation (2018A0303130003), Scientific Research Plan of Guangzhou Science and Technology Project (201904010379), Collaborative Innovation Center of GDAAS (XTXM202202), the Science and Technology Planning Project of Jiangmen (2020030103310009038), Guangdong Basic and Applied Basic Research Foundation (2019B1515210008), Key Research and Development Program of Guangdong Province (2020B0202090004), Yunnan Liao Ming Expert Workstation (202105AF150077), Science and Technology Program of Guangdong Province (2021B1212030015), and Zengcheng District Entrepreneurial Leading Team Project (2021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.951554/full#supplementary-material
1. Harrach B, Benko M, Both GW, Brown M, Wadell G. Family Adenoviridae. New York, NY; Oxford: Elsevier (2011).
2. Marek A, Kajan GL, Kosiol C, Benko M, Schachner A, Hess M. Genetic diversity of species fowl aviadenovirus D and fowl aviadenovirus E. J Gen Virol. (2016) 97:2323–32. doi: 10.1099/jgv.0.000519
3. Kaján GL, Affranio I, Tóthné Bistyák A, Kecskeméti S, Benko M. An emerging new fowl adenovirus genotype. Heliyon. (2019) 5:e01732. doi: 10.1016/j.heliyon.2019.e01732
4. Hess M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. (2000) 29:195–206. doi: 10.1080/03079450050045440
5. Marek A, Günes A, Schulz E, Hess M. Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the Hexon L1 gene region. J Virol Methods. (2010) 170:147–54. doi: 10.1016/j.jviromet.2010.09.019
6. Schachner A, Matos M, Grafl B, Hess M. Fowl adenovirus-induced diseases and strategies for their control - a review on the current global situation. Avian Pathol. (2018) 47:111–26. doi: 10.1080/03079457.2017.1385724
7. Gjevre AG, Kaldhusdal M, Eriksen GS. Gizzard erosion and ulceration syndrome in chickens and turkeys: a review of causal or predisposing factors. Avian Pathol. (2013) 42:297–303. doi: 10.1080/03079457.2013.817665
8. Shah MS, Ashraf A, Khan MI, Rahman M, Habib M, Chughtai MI, et al. Fowl adenovirus: history, emergence, biology and development of a vaccine against hydropericardium syndrome. Arch Virol. (2017) 162:1833–43. doi: 10.1007/s00705-017-3313-5
9. Mittal D, Jindal N, Tiwari AK, Khokhar RS. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virusdisease. (2014) 25:114–9. doi: 10.1007/s13337-013-0183-7
10. Mohamed MHA, El-Sabagh IM, Abdelaziz AM, Al-Ali AM, Alramadan M, Lebdah MA, et al. Molecular characterization of fowl aviadenoviruses species D and E associated with inclusion body hepatitis in chickens and falcons indicates possible cross-species transmission. Avian Pathol. (2018) 47:384–90. doi: 10.1080/03079457.2018.1457769
11. Lim TH, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, et al. Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis. (2011) 55:554–60. doi: 10.1637/9730-032011-Reg.1
12. Sun J, Zhang Y, Gao S, Yang J, Tang Y, Diao Y. Pathogenicity of fowl adenovirus serotype 4 (Fadv-4) in chickens. Infect Genet Evol. (2019) 75:104017. doi: 10.1016/j.meegid.2019.104017
13. Okuda Y, Ono M, Shibata I, Sato S, Akashi H. Comparison of the polymerase chain reaction-restriction fragment length polymorphism pattern of the fiber gene and pathogenicity of serotype-1 fowl adenovirus isolates from gizzard erosions and from feces of clinically healthy chickens in Japan. J Vat Diagn Invest. (2006) 18:162–7. doi: 10.1177/104063870601800204
14. Okuda Y, Ono M, Shibata I, Sato S. Pathogenicity of serotype 8 fowl adenovirus isolated from gizzard erosions of slaughtered broiler chickens. J Vet Med Sci. (2004) 66:1561–6. doi: 10.1292/jvms.66.1561
15. Kiss I, Homonnay ZG, Mato T, Banyai K, Palya V. Research note: an overview on distribution of fowl adenoviruses. Poult Sci. (2021) 100:101052. doi: 10.1016/j.psj.2021.101052
16. Xia J, Yao KC, Liu YY, You GJ Li SY, Liu P, et al. Isolation and molecular characterization of prevalent fowl adenovirus strains in southwestern China during 2015-2016 for the development of a control strategy. Emerg Microbes Infect. (2017) 6:e103. doi: 10.1038/emi.2017.91
17. Cui J, Xu Y, Zhou Z, Xu Q, Wang J, Xiao Y, et al. Pathogenicity and molecular typing of fowl adenovirus-associated with hepatitis/hydropericardium syndrome in central China (2015-2018). Front Vet Sci. (2020) 7:190. doi: 10.3389/fvets.2020.00190
18. Maartens LH, Joubert HW, Aitchison H, Venter EH. Inclusion body hepatitis associated with an outbreak of fowl adenovirus type 2 and type 8b in broiler flocks in South Africa. J S Afr Vet Assoc. (2014) 85:e1–5. doi: 10.4102/jsava.v85i1.1146
19. Joubert HW, Aitchison H, Maartens LH, Venter EH. Molecular differentiation and pathogenicity of aviadenoviruses isolated during an outbreak of inclusion body hepatitis in South Africa. J S Afr Vet Assoc. (2014) 85:1058. doi: 10.4102/jsava.v85i1.1058
20. Nakamura K, Mase M, Yamamoto Y, Takizawa K, Kabeya M, Wakuda T, et al. Inclusion body hepatitis caused by fowl adenovirus in broiler chickens in Japan, 2009-2010. Avian Dis. (2011) 55:719–23. doi: 10.1637/9813-052511-Case.1
21. Niczyporuk JS. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch Virol. (2016) 161:33–42. doi: 10.1007/s00705-015-2635-4
22. Niczyporuk JS, Kozdrun W, Czekaj H, Stys-Fijol N, Piekarska K. Detection of fowl adenovirus D strains in wild birds in Poland by loop-mediated isothermal amplification (Lamp). BMC Vet Res. (2020) 16:58. doi: 10.1186/s12917-020-2271-4
23. Chen L, Yin L, Zhou Q, Peng P, Du Y, Liu L, et al. Epidemiological investigation of fowl adenovirus infections in poultry in China during 2015-2018. BMC Vet Res. (2019) 15:271. doi: 10.1186/s12917-019-1969-7
24. Schrenzel M, Oaks JL, Rotstein D, Maalouf G, Snook E, Sandfort C, et al. Characterization of a new species of adenovirus in falcons. J Clin Microbiol. (2005) 43:3402–13. doi: 10.1128/JCM.43.7.3402-3413.2005
25. Schachner A, Gonzalez G, Endler L, Ito K, Hess M. Fowl adenovirus (Fadv) recombination with intertypic crossovers in genomes of Fadv-D and Fadv-E, displaying hybrid serological phenotypes. Viruses. (2019) 11:1094. doi: 10.3390/v11121094
26. Meulemans G, Boschmans M, Berg TP, Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. (2001) 30:655–60. doi: 10.1080/03079450120092143
27. Chen L, Yin L, Peng P, Zhou Q, Du Y, Zhang Y, et al. Isolation and characterization of a novel fowl adenovirus serotype 8a strain from China. Virol Sin. (2020) 35:517–27. doi: 10.1007/s12250-019-00172-7
28. Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, Lmh. Cancer Res. (1987) 47:4460–4.
29. Steer PA, Sandy JR, O'Rourke D, Scott PC, Browning GF, Noormohammadi AH. Chronological analysis of gross and histological lesions induced by field strains of fowl adenovirus serotypes 1, 8b and 11 in one-day-old chickens. Avian Pathol. (2015) 44:106–13. doi: 10.1080/03079457.2015.1007919
30. Pizzi M. Sampling variation of the fifty percent end-point, determined by the reed-muench (behrens) method. Hum Biol. (1950) 22:151–90.
31. Alexander HS, Huber P, Cao J, Krell PJ, Nagy E. Growth Characteristics of Fowl Adenovirus Type 8 in a Chicken Hepatoma Cell Line. J Virol Methods. (1998) 74:9–14.
32. Kumar S, Stecher G. Tamura K. Mega7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
33. Günes A, Marek A, Grafl B, Berger E, Hess M. Real-Time Pcr Assay for Universal Detection and Quantitation of All Five Species of Fowl Adenoviruses (Fadv-a to Fadv-E). J Virol Methods. (2012) 183:147–53. doi: 10.1016/j.jviromet.2012.04.005
34. Hess M. Commensal or Pathogen - a Challenge to Fulfil Koch's Postulates. Br Poult Sci. (2017) 58:1–12. doi: 10.1080/00071668.2016.1245849
35. Schachner A, Marek A, Grafl B, Hess M. Detailed Molecular Analyses of the Hexon Loop-1 and Fibers of Fowl Aviadenoviruses Reveal New Insights into the Antigenic Relationship and Confirm That Specific Genotypes Are Involved in Field Outbreaks of Inclusion Body Hepatitis. Vet Microbiol. (2016) 186:13–20. doi: 10.1016/j.vetmic.2016.02.008
36. Grgić H, Yang DH, Nagy E. Pathogenicity and Complete Genome Sequence of a Fowl Adenovirus Serotype 8 Isolate. Virus Res. (2011) 156:91–7. doi: 10.1016/j.virusres.2011.01.002
37. Niu Y, Sun Q, Zhang G, Sun W, Liu X, Xiao Y, et al. Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transbound Emerg Dis. (2018) 65:e121–6. doi: 10.1111/tbed.12691
38. Hosseini H, Langeroudi AG, FallahMehrabadi MH, Ziafati Kafi Z, Dizaji RE, Ghafouri SA, et al. The fowl adenovirus (Fadv-11) outbreak in iranian broiler chicken farms: the first full genome characterization and phylogenetic analysis. Comp Immunol Microbiol Infect Dis. (2020) 70:101365. doi: 10.1016/j.cimid.2019.101365
39. Steer PA, Kirkpatrick NC, O'Rourke D, Noormohammadi AH. Classification of fowl adenovirus serotypes by use of high-resolution melting-curve analysis of the hexon gene region. J Clin Microbiol. (2009) 47:311–21. doi: 10.1128/JCM.01567-08
40. Niczyporuk JS, Kozdrun W, Czekaj H, Piekarska K, Stys-Fijol N. Isolation and molecular characterization of fowl adenovirus strains in black grouse: first reported case in Poland. PLoS ONE. (2020) 15:e0234532. doi: 10.1371/journal.pone.0234532
41. Mat Isa N, Mohd Ayob J, Ravi S, Mustapha NA, Ashari KS, Bejo MH, et al. Complete genome sequence of fowl adenovirus-8b Upm04217 isolate associated with the inclusion body hepatitis disease in commercial broiler chickens in malaysia reveals intermediate evolution. Virusdisease. (2019) 30:426–32. doi: 10.1007/s13337-019-00530-9
42. Marek A, Kaján GL, Kosiol C, Harrach B, Schlötterer C, Hess M. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus aviadenovirus. Virology. (2014) 462-463:107–14. doi: 10.1016/j.virol.2014.04.033
43. Zhao J, Zhong Q, Zhao Y, Hu YX, Zhang GZ. Pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS One. (2015) 10:e0133073. doi: 10.1371/journal.pone.0133073
44. Kumar R, Chandra R, Shukla SK, Agrawal DK, Kumar M. Hydropericardium syndrome (Hps) in India: a preliminary study on the causative agent and control of the disease by inactivated autogenous vaccine. Trop Anim Health Prod. (1997) 29:158–64. doi: 10.1007/BF02633014
45. Niu Y, Sun Q, Shi Y, Ding Y, Li Z, Sun Y, et al. Immunosuppressive potential of fowl adenovirus serotype 4. Poult Sci. (2019) 98:3514–22. doi: 10.3382/ps/pez179
46. Schonewille E, Singh A, Gobel TW, Gerner W, Saalmuller A, Hess M. Fowl adenovirus (Fadv) serotype 4 causes depletion of B and T cells in lymphoid organs in specific pathogen-free chickens following experimental infection. Vet Immunol Immunopathol. (2008) 121:130–9. doi: 10.1016/j.vetimm.2007.09.017
47. Niu Y, Sun Q, Zhang G, Sun W, Liu X, Xiao Y, et al. Pathogenicity and immunosuppressive potential of fowl adenovirus in specific pathogen free chickens. Poult Sci. (2017) 96:3885–92. doi: 10.3382/ps/pex206
48. Cizmecigil UY, Umar S, Yilmaz A, Bayraktar E, Turan N, Tali B, et al. Characterisation of fowl adenovirus (fadv-8b) strain concerning the geographic analysis and pathological lesions associated with inclusion body hepatitis in broiler flocks in Turkey. J Vet Res. (2020) 64:231–7. doi: 10.2478/jvetres-2020-0026
49. Wang J, Zaheer I, Saleemi MK Qi X, Gao Y, Cui H, et al. The first complete genome sequence and pathogenicity characterization of fowl adenovirus 11 from chickens with inclusion body hepatitis in Pakistan. Vet Microbiol. (2020) 244:108670. doi: 10.1016/j.vetmic.2020.108670
Keywords: fowl adenovirus serotype 2, complete genome, pathogenicity, weight gain, viral shedding
Citation: Xie Z, Zhang J, Sun M, Zeng Q, Huang Y, Dong J, Li L, Huang S and Liao M (2022) The first complete genome sequence and pathogenicity characterization of fowl adenovirus serotype 2 with inclusion body hepatitis and hydropericardium in China. Front. Vet. Sci. 9:951554. doi: 10.3389/fvets.2022.951554
Received: 24 May 2022; Accepted: 27 July 2022;
Published: 15 August 2022.
Edited by:
José Manuel Rojas, Centro de Investigación en Sanidad Animal (CISA), SpainReviewed by:
Qi Su, The Chinese University of Hong Kong, ChinaCopyright © 2022 Xie, Zhang, Sun, Zeng, Huang, Dong, Li, Huang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shujian Huang, c2podWFuZy5mb3NoYW5AMTYzLmNvbQ==; Ming Liao, bWxpYW9Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.