
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 06 December 2022
Sec. Veterinary Epidemiology and Economics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.947154
This article is part of the Research Topic Women in Veterinary Epidemiology and Economics View all 11 articles
Methicillin-resistant Staphylococcus aureus (MRSA) is an opportunistic bacterium that causes many human and animal infections worldwide. MRSA infections are classified as priority infections owing to their high morbidity and mortality, with a significant risk of zoonotic transmission. This study aimed to determine the pooled prevalence of MRSA in dairy cattle farms and its heterogeneity. Relevant studies were retrieved from three databases: PubMed, Web of Science, and Scopus. The pooled prevalence of MRSA in dairy farms was estimated using a random-effects model. Subgroup and meta-regression analyses were used to assess the probable sources of heterogeneity. Sensitivity and publication bias analyses were also performed. A total of 94 articles were eligible for inclusion in this meta-analysis. The pooled prevalence of MRSA was estimated to be 3.81% [95% confidence interval (95% CI) = 2.61–5.20] with significantly high heterogeneity (I2 = 96.6%, p = 0.00). For the subgroup analysis among continents, the prevalence was highest in Asia (4.89%; 95% CI = 2.88–7.35) and lowest in South America (1.33%, 95% CI = 0.00–5.49). As for the year of publication, MRSA prevalence was highest in reports published from 2015 to 2018 (4.36%, 95% CI = 2.41–6.80) and lowest in reports published before 2015 (2.65%, 95% CI = 0.75–5.52). As for sample type, the prevalence of MRSA in cattle milk (3.91%, 95% CI = 2.64–5.39) was higher than that in other sample types (1.19%, 95% CI = 0.05–3.24). These three factors were not significantly associated with the pooled prevalence of MRSA (p > 0.05). Therefore, the findings of this study indicate that the prevalence of MRSA has been minimal and consistent in dairy cattle farms over time.
Staphylococcus aureus is a commensal bacterium that can be found on the skin, mucous membranes, and upper respiratory tracts of both animals and humans (1). However, it can be an opportunistic pathogen that causes various infectious illnesses in humans and animals (2). S. aureus is associated with many human disorders, from skin and soft tissue infections to life-threatening septicemia (3). In veterinary medicine, it is a common cause of bovine mastitis in dairy cattle, resulting in high economic losses worldwide (4).
Methicillin-resistant S. aureus (MRSA) was first documented in 1961 (5). MRSA strains were phenotypically identified using cefoxitin and oxacillin susceptibility assays (6). The gold standard for detecting MRSA is through the detection of the mecA gene, which encodes a protein called PBP2a, that has a poor affinity for β-lactam drugs, resulting in resistance to methicillin (7, 8). According to the recorded data, methicillin resistance has been identified in 50–70% of S. aureus strains isolated from the hospital environment, causing ~100,000 infections in the United States each year, with a 20% mortality rate (9). In 1972, MRSA was first reported in domestic animals as a pathogen causing bovine mastitis in dairy cattle in Belgium (10). Since then, various studies reported the zoonotic transmission of MRSA from livestock, especially pigs, poultry, and cattle, to farm workers and exposed people, which has been known as livestock-associated MRSA (LA-MRSA) (11–13). The majority of LA-MRSA isolates lack toxins such as PVL and enterotoxins (14) and are reported to have multiple antimicrobial resistance (15).
In the past two decades, numerous studies have reported different prevalence rates of MRSA on dairy cattle farms in different regions. These variations might be associated with isolation protocols, farm management systems, sample sizes, sample sources, and other factors (16). Most studies have detected MRSA in bovine mastitis cases. However, several studies have demonstrated the presence of MRSA in raw milk, farm workers, and dairy cattle farms, indicating the possible risk of MRSA transmission within dairy cattle farms and across the dairy supply chain to the general public (17–19). Hence, the objective of this study was to estimate the global prevalence of MRSA isolated from various sample sources in dairy cattle farms through a systematic review and meta-analysis of published articles.
The Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) guidelines were adopted for this study. Relevant studies published until 31 December 2021 were retrieved from three online databases: PubMed, Scopus, and Web of Science. The search was limited to original articles published in English. The keywords used for searching the relevant studies were “MRSA” OR “Methicillin-resistant Staphylococcus aureus” AND “dairy cattle” OR “dairy cow.”
All original publications reporting the prevalence of MRSA, as determined by the detection of mecA and/or mecC genes, in dairy cattle farms were considered for analysis. The inclusion criteria were observational, cross-sectional, and case–control studies that determined the prevalence of MRSA from any sample source in dairy cattle farms. Studies were excluded from the analysis if they were (1) review articles, (2) experimental studies, (3) not written in English, (4) lack of a clear report on the prevalence of MRSA from any sample sources in dairy cattle farms, (5) lack of clear sample size, (6) performed on archived isolates, (7) no full text available, and (8) studies that used only phenotypic tests to detect MRSA. The titles and abstracts of the retrieved studies were evaluated for eligibility. After title and abstract screening, the full text of each article was thoroughly reviewed for inclusion. Two authors, SK and SB, independently performed study screening and selection. Disagreements were resolved through discussion.
Two authors individually extracted data from all included studies. Discrepancies between the data obtained by these two authors were discussed with a third author for consensus to avoid bias. The extracted data included (1) the name of the author and year of publication, (2) the continent where the study was conducted, (3) sample size, (4) sources of samples, (5) the number of S. aureus isolates, (6) the number of MRSA isolates, and (7) the detection method used.
The quality assessment criteria derived from Ding et al. (20) were used to evaluate the quality of the included studies. The checklist for determining the quality of studies consisted of these five questions: (1) Was the research objective clearly stated? (2) Was the sampling method described? (3) Was the study period and location clearly stated? (4) Were the examination methods and procedures for MRSA detection described clearly? (5) Were the samples clearly classified into different subgroups? The answers to each question were scored as “2” for yes, “0” for no, or “1” for unsure. A summation of the scores from all five questions was calculated, and the overall quality of each study was evaluated.
Meta-analysis was performed using the R package meta in the statistics software R (21, 22). The prevalence of MRSA in dairy cattle farms was determined by dividing the number of MRSA isolates by the total sample size. Because several studies reported zero prevalence of MRSA, Freeman–Tukey double arcsine transformation was performed for all raw proportions before conducting the meta-analyses to avoid excluding these studies (23). The classic meta-analysis model utilizing logit-transformed proportions and the corresponding standard errors in the inverse variance method was used to pool studies (24). Back-transformation of all estimated pooled prevalence was performed for ease of interpretation.
A random-effects model was used to estimate the overall pooled prevalence of MRSA in dairy cattle farms, together with its 95% confidence interval (95% CI). Cochran's Q-test was used to determine the heterogeneity of the pooled prevalence. Furthermore, the I2 statistic was used to characterize the degree of heterogeneity across studies, with values of 25, 50, and 75% indicating low, medium, and high heterogeneity, respectively (25).
The subgroups in each study were used as the unit of analysis for all subgroup meta-analyses. Subgroup analyses were performed to investigate the heterogeneity between three variables: year of publication, continent, and sample type. The year of publication for each study was categorized into three groups consisting of “before 2015,” “2015–2018,” and “after 2018.” Each study was classified into five continents: “Asia,” “Africa,” “Europe,” “South America,” and “North America.” Sample type, referring to the sources of samples, was analyzed as two subgroups: “cattle milk” and “others.” The “cattle milk” category included quarter milk, composite milk, bulk tank milk, and milk from clinical and subclinical mastitis cases. Other sources of samples, such as cattle nasal swabs, human samples, and environmental samples collected from dairy cattle farms, were included in the “others” category.
Meta-regression analyses were performed to investigate the significance of the between-study heterogeneity associated with three independent variables: year of publication, continent, and sample type. Levels within each independent variable were similar to those described for the subgroup meta-analyses. A univariate meta-regression model was created to determine the association between each independent variable and the prevalence of MRSA in dairy cattle farms. Furthermore, variables with p ≤ 0.25 in the univariable meta-regression analysis were introduced to the random-effects multivariable meta-regression model.
Publication bias was examined using a funnel plot and Egger's test, where a p < 0.05 indicates statistical significance (26). The robustness of the results was evaluated using two sensitivity analyses. The first is a comparison of the results obtained from the random-effects and fixed-effects models. In addition, a leave-one-out meta-analysis was performed to evaluate whether any single study affected the results.
A total of 2,601 records were identified from the three databases searched. These records consisted of 69 from PubMed, 2,446 from Scopus, and 86 from Web of Science. Of these, 155 records were duplicates and were removed before screening the titles and abstracts. After the screening process, 272 articles were included in the full-text review for inclusion criteria. Finally, 94 studies met the inclusion criteria and were included in the meta-analysis (16, 27–118). The remaining 178 articles were excluded for the following reasons: lack of clear prevalence of MRSA from any sample sources in dairy cattle farms (n = 47), lack of clear sample size (n = 13), archived isolates (n = 56), not written in English (n = 9), and studies that used only phenotypic tests to detect MRSA (n = 53), as shown in Figure 1.
The 94 studies considered in this review were published between 2003 and 2021, with the majority published after 2018 (n = 38). These studies reported the prevalence of MRSA in 30 countries across five continents. Most studies were conducted in Asia (n = 43), followed by Africa (n = 20) and Europe (n = 20). The majority of studies reported MRSA detection in milk samples (n = 90), whereas only 22 studies reported the presence of MRSA from other sample types. The mean ± standard deviation of quality scores of all included studies was 7.91 ± 1.62, with a range from 4 to 10. The characteristics of the selected studies are shown in Supplementary Table 1.
After data extraction, a total of 1,251 MRSA strains isolated from 47,236 samples collected from dairy cattle farms worldwide were included in the meta-analysis. As estimated from the random-effects model, the overall pooled prevalence of MRSA in dairy cattle farms was 3.81% (95% CI = 2.61–5.20), with high heterogeneity (Q = 2773.64; I2 = 96.6%; p = 0.00). These data are shown in Table 1 and Figure 2.
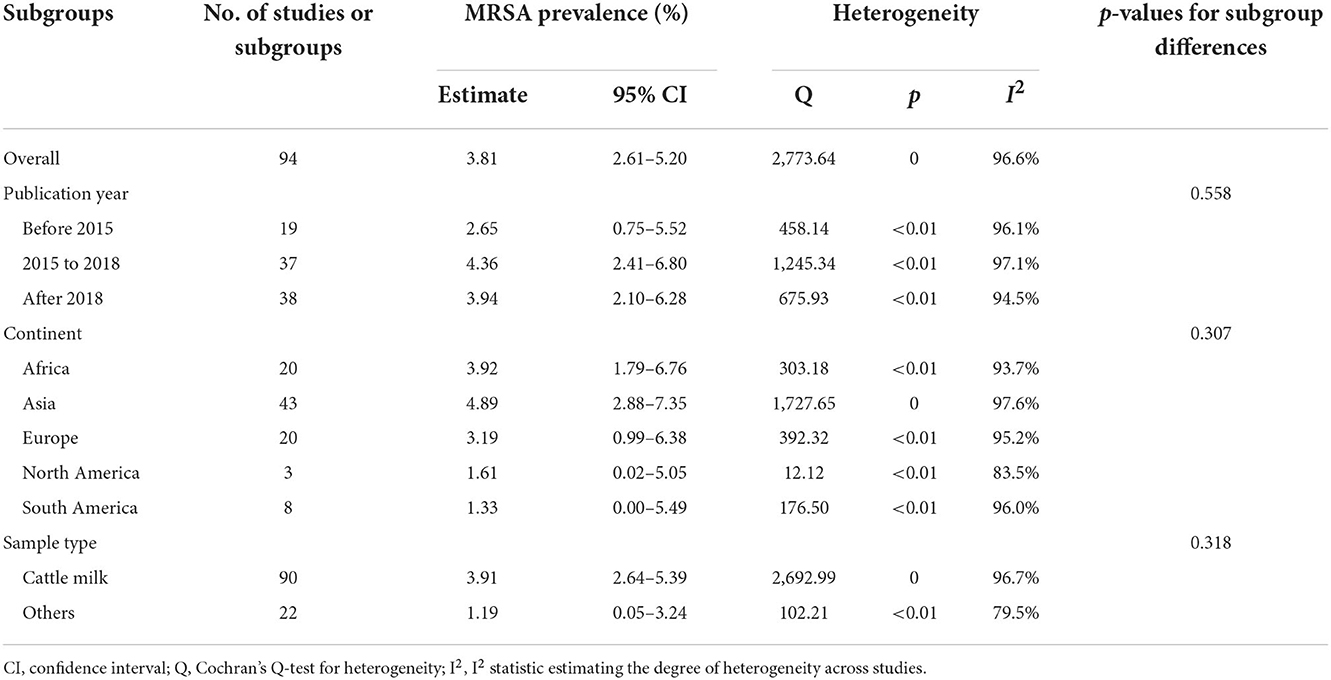
Table 1. Meta-analysis of methicillin-resistant Staphylococcus aureus (MRSA) prevalence from dairy cattle farms.
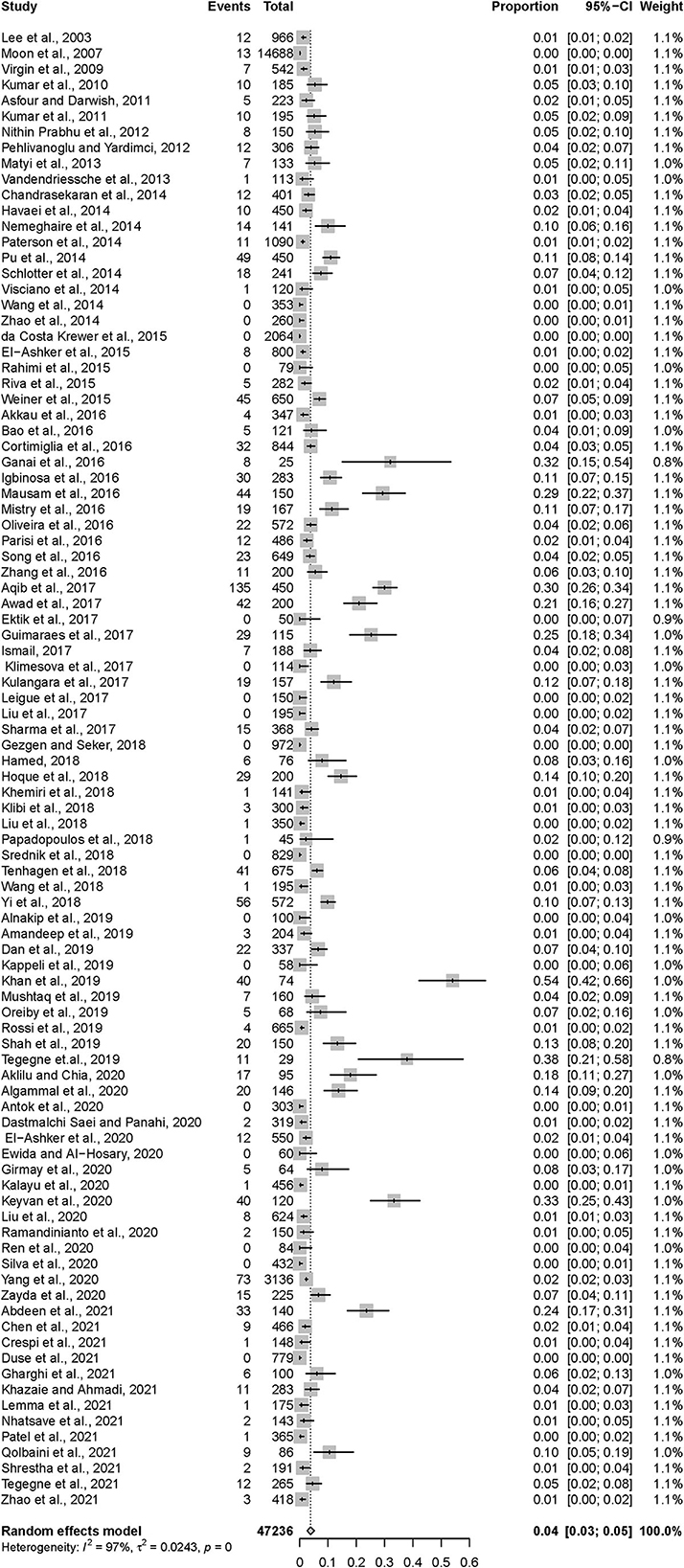
Figure 2. Forest plot demonstrating the pooled prevalence of MRSA in dairy cattle farms and its 95% confidence interval estimated by a random-effects model.
The pooled prevalence of MRSA and the 95% CI for different subgroups of the year of publication, continent, and sample type are shown in Table 1. High heterogeneity was observed among all the tested subgroups. However, no statistically significant differences were detected between these subgroups. According to the year of publication, no significant trend in MRSA prevalence was observed, but the highest prevalence was observed among studies published between 2015 and 2018 (4.36%, 95% CI = 2.41–6.80). The pooled prevalence of MRSA in Asia appeared to be highest (4.89%, 95% CI = 2.88–7.35), followed by Africa (3.92%, 95% CI = 1.79–6.76) and Europe (3.19%, 95% CI = 0.99–6.38). The estimated prevalence of MRSA was lowest in South America (1.33%, 95% CI = 0.00–5.49). The pooled prevalence of MRSA in cattle milk (3.91%, 95% CI = 2.64–5.39) was higher than, but not statistically significantly different from, those in other sample types from dairy cattle farms (1.19%, 95% CI = 0.05–3.24). When the meta-regression models were analyzed for all three variables, no significant variable was associated with the heterogeneity of the overall pooled prevalence of MRSA in dairy cattle farms (Supplementary Tables 2, 3).
The funnel plot created from the data obtained from the selected studies demonstrated asymmetry of distribution, as shown in Figure 3, indicating a publication bias among the selected studies. To investigate the sources of funnel plot asymmetry, the results from Egger's test showed a statistically significant coefficient bias (5.30 ± 0.77, p < 0.0001), revealing evidence of small-study effects. Furthermore, sensitivity analysis was performed to assess the robustness of the models used to estimate the pooled prevalence of MRSA. The overall pooled prevalence of MRSA in dairy cattle farms using a fixed-effects model was much lower than that using a random-effects model, as shown in Table 2. In addition, a leave-one-out meta-analysis was performed to investigate the impact of each study on the pooled prevalence of MRSA in dairy cattle farms. Removing the studies with the lowest or highest prevalence did not significantly influence the overall pooled prevalence of MRSA in dairy cattle farms, as shown in Table 2.
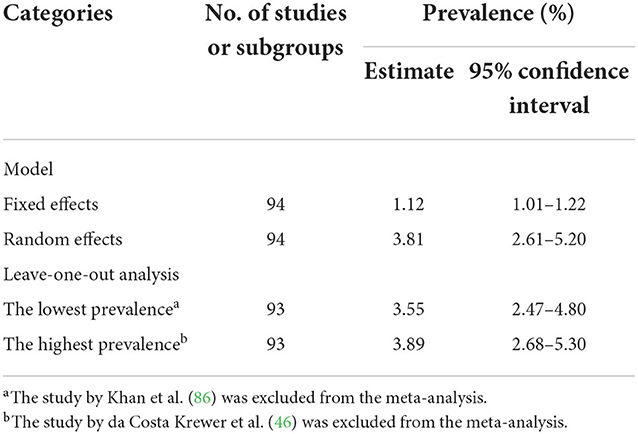
Table 2. Sensitivity analysis to determine the robustness of the results obtained from the models used.
The current study revealed that the global prevalence of MRSA isolated from various sample sources in dairy cattle farms, using a random-effects meta-analysis model, was 3.81%. Recently, Zaatout and Hezil (119) reported the global prevalence of MRSA isolated from bovine mastitis cases using a meta-analysis. Their reported prevalence was 4.30%, which was similar to our findings. The small variation in the estimated prevalence could be due to the fact that the current study included data from various sample types presented in dairy cattle farms, while the study by Zaatout and Hezil only selected reports from MRSA in the milk of clinical and subclinical bovine mastitis cases. We included data from a broad range of sample types to demonstrate the overall pooled prevalence of MRSA in dairy cattle farms, which can be used to determine the risk of MRSA transmission and contamination between cattle, humans, and the environment within the farms, and between the farms and other population at risk, especially the dairy consumers.
Subgroup analyses were carried out depending on the year of publication, continent, and sample type. We observed that the number of selected articles published before 2015 was limited (19/94). Increased numbers of studies were observed from 2015 to 2018 (37/94) and after 2018 (38/94). The highest pooled prevalence of MRSA was observed from 2015 to 2018 (4.36%) but not statistically different from that before 2015 and after 2018. In contrast, a recent meta-analysis on MRSA associated with bovine mastitis reported a significantly increasing trend in prevalence by the year of publication and suggested that it might be influenced by the advancements in the detection methods used (119). This contrast can be explained by the difference in included studies, the different categorization used to create levels for the subgroup analysis of the publication year, and the different sample types to be included in both studies. All of these differences might be resulted in narrower CIs of the reported prevalence of each level of year of publication in the previous study, compared to those of reported prevalence in the current study. The narrower CIs could be potentially associated with the statistical significance observed in the previous study. The changes in prevalence emphasize the importance of monitoring MRSA in dairy cattle farms to assess the progress or success of any implemented antimicrobial resistance control program.
Although we could not demonstrate a statistically significant difference in pooled prevalence among subgroups, our results showed a substantially higher prevalence of MRSA in dairy cattle farms in Asia (4.89%) than in South America (1.33%). The milk production and dairy animal population in Asia have been increasing (120). During the period from 2010 to 2020, cow milk production in Asia increased up to 4.2%, which was the highest growth compared to other regions of the world (121). This could be associated with the high number of research studies investigating the presence of any zoonotic pathogens, especially MRSA, in dairy cattle farms. The limited number of publications in South and North America can potentially lead to an underestimation of the prevalence of MRSA in dairy cattle farms in these regions and should be noted. Moreover, the higher prevalence of MRSA in Asia compared to other regions might be due to the high consumption of antimicrobial agents in food animals (122), which could be related to the increased dairy cattle population in this region and the available antimicrobial agents used in the region as they have a different selective pressure on MRSA. This phenomenon can also be attributed to the unethical use of antibiotics, especially in developing countries, where drugs are administered on the spur of the moment and without veterinarian monitoring (123). Another concern is poor farm sanitation and water management, both of which can facilitate MRSA transmission from animals to humans and vice versa and the development of antimicrobial resistance (124).
In addition to cattle milk, MRSA has been isolated from farm workers, farm environments, and other cattle organs. The pooled prevalence of MRSA in milk samples was lower than that in other sample sources from dairy cattle farms. Most milk samples reported in the selected studies were quarter milk samples collected aseptically; therefore, MRSA detected in cattle milk is generally a representative pathogenic strain of MRSA associated with intramammary infection and/or mastitis in cattle. In contrast, MRSA isolated from other sample sources could be either pathogenic or non-pathogenic strains, or a mix of both. Our findings suggest that MRSA is higher prevalent among bovine mastitis-causing S. aureus than other pathogenic or non-pathogenic S. aureus found in other sources in dairy cattle farms. However, the difference in the prevalence of MRSA isolated from these two sample types was not significant. Moreover, MRSA transmission among cattle, humans, and the environment cannot be ruled out. Therefore, MRSA monitoring and prudent antimicrobial use in dairy cattle farms should be regularly implemented.
Regarding univariable and multivariable meta-regression, there was no significant association between the overall pooled prevalence of MRSA and any variable, suggesting that the source of heterogeneity could not be explained by the year of publication, continent, or sample type. It suggests that the heterogeneity of reported prevalence among included publications might be associated with other factors, such as the method of isolation, the sampling and sample handling procedure, and the history of MRSA infection or transmission in the farms. However, the information regarding those factors was not equally and well-explained in most of the included publications. Therefore, they were not extracted during the systematic review and included in the meta-analysis. A further study with a more specific hypothesis using different search strategies and inclusion and exclusion criteria should be performed to investigate the source of heterogeneity of the prevalence of MRSA in dairy cattle farms.
Analysis of publication bias performed using the funnel plot and Egger's test revealed the bias of publications with small-study effects. Small-study effects are generally defined as a phenomenon in which studies with smaller sample size show different, and often larger, effects than studies with a larger sample size. This phenomenon can be due to the publication bias, when small studies reporting larger effects are more likely to be published compared to those reporting smaller effects. A funnel plot, showing the reported effects from small studies which are usually associated with high standard errors and large studies which are usually associated with low standard errors, can be used to illustrate the publication bias. According to the present study, the funnel plot clearly shows that small studies reporting low prevalence are missing which is illustrated as an area without any dots in the bottom left corner of the plot. Even though the small-study effect is a potential limitation of this study, all included publications were of fair to high quality. Moreover, using a sensitivity analysis, we showed that our meta-analysis was robust and stable. Other study limitations should also be concerned. First, only articles that were written in English were included. Second, the included studies were obtained from only three distinct databases. Third, the year of publication of several studies was not identical to the year of MRSA isolation, which may have influenced the misclassification and misinterpretation of the subgroup meta-analysis. Even though we successfully revealed the global prevalence of MRSA in dairy cattle farms, other knowledge such as the risk factors associated with the presence of MRSA and the antimicrobial use in dairy cattle farms was not described. This gap in knowledge is critical in controlling and monitoring MRSA in dairy cattle farms and needed to be further investigated in a future study.
The global pooled prevalence of MRSA in dairy cattle farms has been minimal yet consistent over time. The pooled prevalence of MRSA in dairy cattle farms was the highest in Asia, followed by Africa and Europe. Cattle milk samples were found to harbor a higher prevalence of MRSA than other sample types. Therefore, following the results of this study, we recommend that appropriate levels of barn sanitation, personnel sanitation while handling animals and animal products, implementation of a continuous surveillance and monitoring program for evaluating animal health, and monitoring of antimicrobial resistance patterns at the farm level, be employed to control the spread of MRSA in dairy cattle farms.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
SB and NA designed this study. SB, SK, and NA performed the systematic review and meta-analysis. SB and SK prepared and revised the manuscript accordingly. All authors contributed to the article and approved the submitted version.
This study was funded by the Center of Excellence in Veterinary Public Health, Faculty of Veterinary Medicine, and CMU Presidential scholarship, Chiang Mai University.
We would like to thank Miss Chananthida Phasom for her technical guidance and support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.947154/full#supplementary-material
1. Gould D, Chamberlaine A. Staphylococcus aureus: a review of the literature. J Clin Nurs. (1995) 4:5–12. doi: 10.1111/j.1365-2702.1995.tb00004.x
2. Biedenbach DJ, Moet, GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY antimicrobial surveillance program (1997-2002). Diagn Microbiol Infect Dis. (2004) 50:59–69. doi: 10.1016/j.diagmicrobio.2004.05.003
3. Feng Y, Chen CJ, Su LH, Hu S, Yu J, Chiu CH. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev. (2008) 32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x
4. Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res. (2003) 34:475–91. doi: 10.1051/vetres:2003027
5. Pantosti A. Methicillin-Resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol. (2012) 3:127. doi: 10.3389/fmicb.2012.00127
6. Panda AM, Mallick B, Chayani N. Evaluation of genotypic and phenotypic methods for detection of methicillin resistant Staphylococcus aureus in a tertiary care hospital of eastern Odisha. J Clin Diagn Res. (2016) 10:DC19–21. doi: 10.7860/JCDR/2016/17476.7278
7. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (2001) 45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001
8. Koupahi H, Honarmand Jahromy S, Rahbar M. Evaluation of different phenotypic and genotypic methods for detection of methicillin resistant Staphylococcus aureus (MRSA). Iran J Pathol. (2016) 11:370–6.
9. Kavanagh KT. Control of MSSA and MRSA in the United States: protocols, policies, risk adjustment and excuses. Antimicrob Resist Infect Control. (2019) 8:103. doi: 10.1186/s13756-019-0550-2
10. Devriese LA, Hommez J. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res Vet Sci. (1975) 19:23–7. doi: 10.1016/S0034-5288(18)33549-5
11. Armand-Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis. (2005) 11:711–4. doi: 10.3201/eid1105.040866
12. Graveland H, Wagenaar JA, Heesterbeek H, Mevius D, van Duijkeren E, Heederik D. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS ONE. (2010) 5:e10990. doi: 10.1371/journal.pone.0010990
13. Nemati M, Hermans K, Lipinska U, Denis O, Deplano A, Struelens M, et al. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob Agents Chemother. (2008) 52:3817–9. doi: 10.1128/AAC.00613-08
14. Hallin M, De Mendonca R, Denis O, Lefort A, El Garch F, Butaye P, et al. Diversity of accessory genome of human and livestock-associated ST398 methicillin resistant Staphylococcus aureus strains. Infect Genet Evol. (2011) 11:290–9. doi: 10.1016/j.meegid.2010.10.021
15. Butaye P, Argudín MA, Smith-Butaye TC. Livestock-Associated MRSA and its current evolution. Curr Clin Microbiol Rep. (2016) 3:19–31. doi: 10.1007/s40588-016-0031-9
16. Girmay W, Gugsa G, Taddele H, Tsegaye Y, Awol N, Ahmed M, et al. Isolation and identification of methicillin-resistant Staphylococcus aureus (MRSA) from milk in shire dairy farms, Tigray, Ethiopia. Vet Med Int. (2020) 2020:8833973. doi: 10.1155/2020/8833973
17. Giovanni N, Elisa S, Marta C, Rosa F, Loredana C, Alessandra B, et al. Occurrence and characteristics of methicillin-resistant Staphylococcus aureus (MRSA) in buffalo bulk tank milk and the farm workers in Italy. Food Microbiol. (2020) 91:103509. doi: 10.1016/j.fm.2020.103509
18. Papadopoulos P, Angelidis AS, Papadopoulos T, Kotzamanidis C, Zdragas A, Papa A, et al. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in bulk tank milk, livestock and dairy-farm personnel in north-central and north-eastern Greece: prevalence, characterization and genetic relatedness. Food Microbiol. (2019) 84:103249. doi: 10.1016/j.fm.2019.103249
19. Parisi A, Caruso M, Normanno G, Latorre L, Sottili R, Miccolupo A, et al. Prevalence, antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus (MRSA) in bulk tank milk from southern Italy. Food Microbiol. (2016) 58:36–42. doi: 10.1016/j.fm.2016.03.004
20. Ding H, Gao YM, Deng Y, Lamberton PHL, Lu DB. A systematic review and meta-analysis of the seroprevalence of Toxoplasma gondii in cats in mainland China. Parasit Vectors. (2017) 10:27. doi: 10.1186/s13071-017-1970-6
22. Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. (2019) 22:153–60. doi: 10.1136/ebmental-2019-300117
23. Lin L, Xu C. Arcsine-based transformations for meta-analysis of proportions: pros, cons, and alternatives. Health Sci Rep. (2020) 3:e178. doi: 10.1002/hsr2.178
24. Borenstein M, Hedges LV, Higgins JP, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. (2010) 1:97–111. doi: 10.1002/jrsm.12
25. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:57–60. doi: 10.1136/bmj.327.7414.557
26. Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
27. Lee JH. Methicillin (Oxacillin)-Resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. (2003) 69:6489–94. doi: 10.1128/AEM.69.11.6489-6494.2003
28. Moon JS, Lee AR, Kang HM, Lee ES, Kim MN, Paik YH, et al. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J Dairy Sci. (2007) 90:1176–85. doi: 10.3168/jds.S0022-0302(07)71604-1
29. Virgin JE, Van Slyke TM, Lombard JE, Zadoks RN. Methicillin-resistant Staphylococcus aureus detection in US bulk tank milk. J Dairy Sci. (2009) 92:4988–91. doi: 10.3168/jds.2009-2290
30. Kumar R, Yadav BR, Singh RS. Genetic determinants of antibiotic resistance in Staphylococcus aureus isolates from milk of mastitic crossbred cattle. Curr Microbiol. (2010) 60:379–86. doi: 10.1007/s00284-009-9553-1
31. Asfour HAE, Darwish SF. Phenotypic and genotypic detection of both mecA- and blaZ-genes mediated β-lactam resistance in staphylococcus strains isolated from bovine mastitis. Glob Vet. (2011) 6:39–50.
32. Kumar R, Yadav BR, Singh RS. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic sahiwal cattle. J Biosci. (2011) 36:175–188. doi: 10.1007/s12038-011-9004-6
33. Nithin Prabhu K, Wilfred SR, Hegde R, Naveen Kumar GS. Methicillin resistance pattern of Staphylococcus aureus from mastitis milk in correlation to its possession of methicillin resistance gene. Milchwissenschaft. (2012) 67:151–4.
34. Pehlivanoglu F, Yardimci H. Detection of methicillin and vancomycin resistance in staphylococcus strains isolated from bovine milk samples with mastitis. Kafkas Uni Vet Fak Derg. (2012) 18:849–55. doi: 10.9775/kvfd.2012.6642
35. Matyi SA, Dupre JM, Johnson WL, Hoyt PR, White DG, Brody T, et al. Isolation and characterization of Staphylococcus aureus strains from a paso del norte dairy. J Dairy Sci. (2013) 96:3535–42. doi: 10.3168/jds.2013-6590
36. Vandendriessche S, Vanderhaeghen W, Soares FV, Hallin M, Catry B, Hermans K, et al. Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J Antimicrob Chemother. (2013) 68:1510–6. doi: 10.1093/jac/dkt047
37. Chandrasekaran D, Venkatesan P, Tirumurugaan KG, Nambi AP, Thirunavukkarasu PS, Kumanan K, et al. Pattern of antibiotic resistant mastitis in dairy cows. Vet World. (2014) 7:389–94. doi: 10.14202/vetworld.2014.389-394
38. Havaei SA, Esfahani BN, Hoseini N, Assadbeigi B. Investigation of antibiotic resistance pattern and detection of methicillin- resistant strains (MRSA) in Staphylococcus aureus isolates associated with bovine mastitis. J Isfahan Med Sch. (2014) 32:1319–29.
39. Nemeghaire S, Argudín MA, Haesebrouck F, Butaye P. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet Res. (2014) 10:153. doi: 10.1186/1746-6148-10-153
40. Paterson GK, Morgan FJ, Harrison EM, Peacock SJ, Parkhill J, Zadoks RN, et al. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother. (2014) 69:598–602. doi: 10.1093/jac/dkt417
41. Pu WX, Su Y, Li, JX, Li CH, Yang ZQ, et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS ONE. (2014) 9:e88134. doi: 10.1371/journal.pone.0088134
42. Schlotter K, Huber-Schlenstedt R, Gangl A, Hotzel H, Monecke S, Muller E, et al. Multiple cases of methicillin-resistant CC130 Staphylococcus aureus harboring mecC in milk and swab samples from a Bavarian dairy herd. J Dairy Sci. (2014) 97:2782–8. doi: 10.3168/jds.2013-7378
43. Visciano P, Pomilio F, Tofalo R, Sacchini L, Saletti MA, Tieri E, et al. Detection of methicillin-resistant Staphylococcus aureus in dairy cow farms. Food Control. (2014) 46:532–8. doi: 10.1016/j.foodcont.2014.06.022
44. Wang X, Wang X, Wang Y, Guo G, Usman T, Hao D, et al. Antimicrobial resistance and toxin gene profiles of Staphylococcus aureus strains from holstein milk. Lett Appl Microbiol. (2014) 58:527–34. doi: 10.1111/lam.12221
45. Zhao JL, Ding YX, Zhao HX, He XL, Li PF, Li ZF, et al. Presence of superantigen genes and antimicrobial resistance in Staphylococcus isolates obtained from the uterine of dairy cows with clinical endometritis. Vet Rec. (2014) 175:352. doi: 10.1136/vr.102302
46. da Costa Krewer C, Santos Amanso E, Veneroni Gouveia G, de Lima Souza R, da Costa MM, Aparecido Mota R. Resistance to antimicrobials and biofilm formation in Staphylococcus spp. isolated from bovine mastitis in the Northeast of Brazil. Trop Anim Health Prod. (2015) 47:511–8. doi: 10.1007/s11250-014-0752-9
47. El-Ashker M, Gwida M, Tomaso H, Monecke S, Ehricht R, El-Gohary F, et al. Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J Dairy Sci. (2015) 98:7450–9. doi: 10.3168/jds.2015-9432
48. Rahimi H, Saei HD, Ahmadi M. Nasal carriage of Staphylococcus aureus: frequency and antibiotic resistance in healthy ruminants. Jundishapur J Microbiol. (2015) 8:e22413. doi: 10.5812/jjm.22413
49. Riva A, Borghi E, Cirasola D, Colmegna S, Borgo F, Amato E, et al. Methicillin-resistant Staphylococcus aureus in raw milk: prevalence, SCCmec typing, enterotoxin characterization, and antimicrobial resistance patterns. J Food Prot. (2015) 78:1142–6. doi: 10.4315/0362-028X.JFP-14-531
50. Weiner M, Rózańska H, Kubajka M, Szulowski K, Krajewska M, Wasiński B. Occurrence and characterisation of MRSA and extended-spectrum ß-lactamases producing Escherichia coli isolated from mastitic cows' milk. Bull Vet Inst Pulawy. (2015) 59:191–5. doi: 10.1515/bvip-2015-0029
51. Akkou M, Antri K, Bachtarzi MA, Bes M, Tristan A, Dauwalder O, et al. Phenotypic and genotypic characterization of Staphylococcus aureus strains associated bovine mastitis and nasal carriage of workers in contact to animals in Algeria. Pak Vet J. (2016) 36:184–8.
52. Bao H, Zhang H, Zhou Y, Zhang L, Wang R. Prevalence, enterotoxin gene and antimicrobial resistance of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from clinical healthy dairy cows. Pak Vet J. (2016) 36:270–4.
53. Cortimiglia C, Luini M, Bianchini V, Marzagalli L, Vezzoli F, Avisani D, et al. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy). Epidemiol Infect. (2016) 144:3046–51. doi: 10.1017/S0950268816001576
54. Ganai AW, Kotwal SK, Wani N, Malik A, Jeelani R, et al. Detection of mecA gene of methicillin resistant Staphylococcus aureus by PCR assay from raw milk. Indian J Anim Sci. (2016) 86:508–11.
55. Igbinosa EO, Beshiru A, Akporehe LU, Ogofure AG. Detection of methicillin-resistant staphylococci isolated from food producing animals: a public health implication. Vet Sci. (2016) 3:14. doi: 10.3390/vetsci3030014
56. Mausam, Ray PK, Dey A, Mohanty S, Kaushik P, Anjay, et al. Isolation, identification and antibiotic sensitivity profiling of methicillin resistant Staphylococcus aureus from bovine milk in Bihar. J Pure Appl Microbiol. (2016) 10:3183–8. doi: 10.22207/JPAM.10.4.95
57. Mistry H, Sharma P, Mahato S, Saravanan R, Kumar PA, Bhandari V. Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in india. PLoS ONE. (2016) 11:e0162256. doi: 10.1371/journal.pone.0162256
58. Oliveira CJB, Tiao N, de Sousa FGC, de Moura JFP, Santos Filho L, Gebreyes WA. Methicillin-resistant Staphylococcus aureus from Brazilian dairy farms and identification of novel sequence types. Zoonoses Public Health. (2016) 63:97–105. doi: 10.1111/zph.12209
59. Song JW, Yang SJ, Shin S, Seo KS, Park YH, Park KT. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus isolated from bovine mastitic milk in Korea. J Food Prot. (2016) 79:1725–32. doi: 10.4315/0362-028X.JFP-16-067
60. Zhang L, Li Y, Bao H, Wei R, Zhou Y, Zhang H, et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb Pathog. (2016) 97:103–9. doi: 10.1016/j.micpath.2016.06.005
61. Aqib, AI, Ijaz M, Anjum AA, Malik MAR, Mehmood K, et al. Antibiotic susceptibilities and prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine milk in Pakistan. Acta Trop. (2017) 176:168–72. doi: 10.1016/j.actatropica.2017.08.008
62. Awad A, Ramadan H, Nasr S, Ateya A, Atwa S. Genetic characterization, antimicrobial resistance patterns and virulence determinants of Staphylococcus aureus isolated form bovine mastitis. Pak J Biol Sci. (2017) 20:298–305. doi: 10.3923/pjbs.2017.298.305
63. Ektik N, Gökmen M, Çibik R. The prevalence and antibiotic resistance of methicillin-resistant Staphylococcus aureus (MRSA) in milk and dairy products in Balikesir, Turkey. J Hell Vet Med. (2017) 68:613–20. doi: 10.12681/jhvms.16062
64. Guimarães, F, F, Manzi MP, Joaquim SF, Richini-Pereira VB, Langoni H. Outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-associated mastitis in a closed dairy herd. J Dairy Sci. (2017) 100:726–30. doi: 10.3168/jds.2016-11700
65. Ismail ZB. Molecular characteristics, antibiogram and prevalence of multi-drug resistant Staphylococcus aureus (MDRSA) isolated from milk obtained from culled dairy cows and from cows with acute clinical mastitis. Asian Pac J Trop Biomed. (2017) 7:694–7. doi: 10.1016/j.apjtb.2017.07.005
66. Klimešová M, Manga I, Nejeschlebová L, Horáček J, PoníŽil A, Vondrušková E. Occurrence of Staphylococcus aureus in cattle, sheep, goat, and pig rearing in the Czech Republic. Acta Vet Brno. (2017) 1:3–10. doi: 10.2754/avb201786010003
67. Kulangara V, Nair N, Sivasailam A, Sasidharan S, Kollannur JD, Syam R. Genotypic and phenotypic beta-lactam resistance and presence of PVL gene in Staphylococci from dry bovine udder. PLoS ONE. (2017) 12:e0187277. doi: 10.1371/journal.pone.0187277
68. Leigue L, Hilgert AR, Fiorini A, Santos MF, Vendruscolo ECG. Occurrence and genetic characterization of Staphylococcus aureus in milk samples of cattle with mastitis, and in the veterinary hospital personnel and dairy workers. Braz J Vet Res Anim Sci. (2017) 54:117–28. doi: 10.11606/issn.1678-4456.bjvras.2017.115947
69. Liu H, Li S, Meng L, Dong L, Zhao S, Lan X, et al. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. (2017) 100:8796–803. doi: 10.3168/jds.2017-13370
70. Sharma V, Sharma S, Dahiya DK, Khan A, Mathur M, Sharma A. Coagulase gene polymorphism, enterotoxigenecity, biofilm production, and antibiotic resistance in Staphylococcus aureus isolated from bovine raw milk in North West India. Ann Clin Microbiol Antimicrob. (2017) 16:65. doi: 10.1186/s12941-017-0242-9
71. Gezgen C, Seker E. Investigation of methicillin resistance and panton-valentine leukocidin in staphylococci isolated from bovine mastitis. Acta Sci Vet. (2018) 44:9. doi: 10.22456/1679-9216.81080
72. Hamed MI. Detection of methicillin-resistant and biofilm-producing Staphylococcus aureus in bovine mastitis. J Adv Vet Res. (2018) 8:95–100.
73. Hoque MN, Das ZC, Rahman ANMA, Haider MG, Islam MA. Molecular characterization of Staphylococcus aureus strains in bovine mastitis milk in Bangladesh. Int J Vet Sci Med. (2018) 6:53–60. doi: 10.1016/j.ijvsm.2018.03.008
74. Khemiri M, Abbassi MS, Couto N, Mansouri R, Hammami S, Pomba C. Genetic characterisation of Staphylococcus aureus isolated from milk and nasal samples of healthy cows in Tunisia: first report of ST97-t267-agrI-SCCmecV MRSA of bovine origin in Tunisia. J Glob Antimicrob Resist. (2018) 14:161–5. doi: 10.1016/j.jgar.2018.03.013
75. Klibi A, Jouini A, Gómez P, Slimene K, Ceballos S, Torres C, et al. Molecular characterization and clonal diversity of methicillin-resistant and-susceptible Staphylococcus aureus isolates of milk of cows with clinical mastitis in Tunisia. Microb Drug Resist. (2018) 24:1210–6. doi: 10.1089/mdr.2017.0278
76. Liu B, Sun H, Pan Y, Zhai Y, Cai T, Yuan X, et al. Prevalence, resistance pattern, and molecular characterization of Staphylococcus aureus isolates from healthy animals and sick populations in Henan Province, China. Gut Pathog. (2018) 10:31. doi: 10.1186/s13099-018-0254-9
77. Papadopoulos P, Papadopoulos T, Angelidis AS, Boukouvala E, Zdragas A, Papa A, et al. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. (2018) 69:43–50. doi: 10.1016/j.fm.2017.07.016
78. Srednik ME, Usongo V, Lépine S, Janvier X, Archambault M, Gentilini ER. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. J Dairy Res. (2018) 85:57–63. doi: 10.1017/S0022029917000851
79. Tenhagen BA, Alt K, Pfefferkorn B, Wiehle L, Käsbohrer A, Fetsch A. Methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J Dairy Sci. (2018) 101:3380–886. doi: 10.3168/jds.2017-12939
80. Wang W, Lin X, Jiang T, Peng Z, Xu J, Yi L, et al. Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front Microbiol. (2018) 9:1123. doi: 10.3389/fmicb.2018.01123
81. Yi Y, Su L, Li B, Li S, Zhang B, Su Y. Analysis of the genetic diversity in methicillin-resistant Staphylococcus aureus isolates from bovine subclinical mastitis case in Xinjiang, China. Foodborne Pathog Dis. (2018) 15:568–75. doi: 10.1089/fpd.2018.2424
82. Alnakip ME, Quintela-Baluja M, Böhme K, Caamaño-Antelo S, Bayoumi MA, Kamal RM, et al. Molecular characterisation and typing the methicillin resistance of Staphylococcus spp. isolated from raw milk and cheeses in northwest Spain: a mini survey. Int Dairy J. (2019) 89:68–76. doi: 10.1016/j.idairyj.2018.09.006
83. Amandeep Singh R, Kaur S, Gill JPS. Panton-Valentine leukocidin (PVL) positive methicillin resistant Staphylococcus aureus (MRSA) in raw milk in Punjab. Indian J Anim Sci. (2019) 89:9–14.
84. Dan M, Yehui W, Qingling M, Jun Q, Xingxing Z, Shuai M, et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, northwest China. J Glob Antimicrob Resist. (2019) 16:98–104. doi: 10.1016/j.jgar.2018.08.024
85. Käppeli N, Morach M, Corti S, Eicher C, Stephan R, Johler S. Staphylococcus aureus related to bovine mastitis in Switzerland: clonal diversity, virulence gene profiles, and antimicrobial resistance of isolates collected throughout 2017. J Dairy Sci. (2019) 102:3274–81. doi: 10.3168/jds.2018-15317
86. Khan A, Durrani AZ, Yousaf A, Khan JA, Chaudhry M, Fatima Z, et al. Epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus in cattle of pothohar region, Pakistan. Pak Vet J. (2019) 39:438–42. doi: 10.29261/pakvetj/2019.049
87. Mushtaq M, Agrawal R, Bhat MA, Singh R, Pande N. Antibiotic resistance gene typing in Staphylococcus aureus isolated from bovine mastitis. Indian J Anim Sci. (2019) 89:1188–91.
88. Oreiby A, Khalifa H, Eid A, Ahmed A, Shimamoto T, Shimamoto T. Staphylococcus aureus and bovine mastitis: molecular typing of methicillin resistance and clinical description of infected quarters. J Hell Vet Med Soc. (2019) 70:1511–6. doi: 10.12681/jhvms.20826
89. Rossi BF, Bonsaglia ECR, Castilho IG, Dantas STA, Salina A, Langoni H, et al. Genotyping of long term persistent Staphylococcus aureus in bovine subclinical mastitis. Microb Pathog. (2019) 132:45–50. doi: 10.1016/j.micpath.2019.04.031
90. Shah MS, Qureshi S, Kashoo Z, Farooq S, Wani SA, Hussain MI, et al. Methicillin resistance genes and in vitro biofilm formation among Staphylococcus aureus isolates from bovine mastitis in India. Comp Immunol Microbiol Infect Dis. (2019) 64:117–24. doi: 10.1016/j.cimid.2019.02.009
91. Tegegne HA, Florianová M, Gelbíčová T, Karpíšková R, Koláčková I. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from bulk tank milk of cows, sheep, and goats. Foodborne Pathog Dis. (2019) 16:68–73. doi: 10.1089/fpd.2018.2511
92. Aklilu E, Chia HY. First mecC and mecA positive livestock-associated methicillin resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from dairy cattle in Malaysia. Microorganisms. (2020) 8:147. doi: 10.3390/microorganisms8020147
93. Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens. (2020) 9:362. doi: 10.3390/pathogens9050362
94. Antók FI, Mayrhofer R, Marbach H, Masengesho JC, Keinprecht H, Nyirimbuga V, et al. Characterization of antibiotic and biocide resistance genes and virulence factors of Staphylococcus species associated with bovine mastitis in Rwanda. Antibiotics. (2020) 9:1. doi: 10.3390/antibiotics9010001
95. Dastmalchi Saei H, Panahi M. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from dairy ruminants: differences in the distribution of clonal types between cattle and small ruminants. Arch Microbiol. (2020) 202:115–25. doi: 10.1007/s00203-019-01722-z
96. El-Ashker M, Gwida M, Monecke S, El-Gohary F, Ehricht R, Elsayed M, et al. Antimicrobial resistance pattern and virulence profile of S. aureus isolated from household cattle and buffalo with mastitis in Egypt. Vet Microbiol. (2020) 240:108535. doi: 10.1016/j.vetmic.2019.108535
97. Ewida RM, Al-Hosary AAT. Prevalence of enterotoxins and other virulence genes of Staphylococcus aureus caused subclinical mastitis in dairy cows. Vet World. (2020) 13:1193–8. doi: 10.14202/vetworld.2020.1193-1198
98. Kalayu AA, Woldetsadik DA, Woldeamanuel Y, Wang SH, Gebreyes WA, Teferi T. Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle, Northern Ethiopia. BMC Vet Res. (2020) 16:20. doi: 10.1186/s12917-020-2235-8
99. Keyvan E, Yurdakul O, Demirtas A, Yalcin H, Bilgen N. Identification of methicillin-resistant Staphylococcus aureus in bulk tank milk. Food Sci Technol. (2020) 40:150–6. doi: 10.1590/fst.35818
100. Liu K, Tao L, Li J, Fang L, Cui L, Li J, et al. Characterization of Staphylococcus aureus isolates from cases of clinical bovine mastitis on large-scale chinese dairy farms. Front Vet Sci. (2020) 7:580129. doi: 10.3389/fvets.2020.580129
101. Ramandinianto SC, Khairullah AR, Effendi MH. Meca gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from dairy farms in East Java, Indonesia. Biodiversitas. (2020) 21:3562–8. doi: 10.13057/biodiv/d210819
102. Ren Q, Liao G, Wu Z, Lv J, Chen W. Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J Dairy Sci. (2020) 103:3368–80. doi: 10.3168/jds.2019-17420
103. Silva ATF, da Silva JG, Aragão BB, Peixoto RM, Mota RA. Occurrence of β-lactam-resistant Staphylococcus aureus in milk from primiparous dairy cows in the northeastern region of Brazil. Trop Anim Health Prod. (2020) 52:2303–7. doi: 10.1007/s11250-020-02259-w
104. Yang F, Zhang S, Shang X, Li H, Zhang H, Cui D, et al. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from subclinical bovine mastitis cases in China. J Dairy Sci. (2020) 103:840–5. doi: 10.3168/jds.2019-16317
105. Zayda MG, Masuda Y, Hammad AM, Honjoh KI, Elbagory AM, Miyamoto T. Molecular characterisation of methicillin-resistant (MRSA) and methicillin-susceptible (MSSA) Staphylococcus aureus isolated from bovine subclinical mastitis and Egyptian raw milk cheese. Int Dairy J. (2020) 104:104646. doi: 10.1016/j.idairyj.2020.104646
106. Abdeen EE, Mousa WS, Abdel-Tawab AA, El-Faramawy R, Abo-Shama UH. Phenotypic, genotypic and antibiogram among Staphylococcus aureus isolated from bovine subclinical mastitis. Pak Vet J. (2021) 41:289–93. doi: 10.29261/pakvetj/2021.008
107. Chen C, Sun C, Li J, Ji X, Wang Y, Song C, et al. Characterisation of Staphylococcus aureus isolates from bovine mastitis in Ningxia, Western China. J Glob Antimicrob Resist. (2021) 25:232–7. doi: 10.1016/j.jgar.2021.03.021
108. Crespi E, Pereyra AM, Puigdevall T, Rumi MV, Testorelli MF, Caggiano N, et al. Antimicrobial resistance studies in staphylococci and streptococci isolated from cows with mastitis in Argentina. J Vet Sci. (2021) 22:1–10. doi: 10.4142/jvs.2021.22.e82
109. Duse A, Persson-Waller K, Pedersen K. Microbial aetiology, antibiotic susceptibility and pathogen-specific risk factors for udder pathogens from clinical mastitis in dairy cows. Animals. (2021) 11:2113. doi: 10.3390/ani11072113
110. Gharghi M, Bahador N, Rowshan-Ghasrodashti A. Study on isolated Staphylococcus aureus from bovine milk with mastitis containing methicillin and panton-valentine leukocidin gene. JRIFST. (2021) 10:359–68. doi: 10.22101/JRIFST.2021.256513.1199
111. Khazaie F, Ahmadi E. Bovine subclinical mastitis-associated methicillin-resistant Staphylococcus aureus, selective genotyping and antimicrobial susceptibility profile of the isolates in Kurdistan province of Iran. Iran J Microbiol. (2021) 13:65–73. doi: 10.18502/ijm.v13i1.5494
112. Lemma F, Alemayehu H, Stringer A, Eguale T. Prevalence and antimicrobial susceptibility profile of Staphylococcus aureus in milk and traditionally processed dairy products in Addis Ababa, Ethiopia. Biomed Res Int. (2021) 2021:5576873. doi: 10.1155/2021/5576873
113. Nhatsave N, Garrine M, Messa A, Massinga AJ, Cossa A, Vaz R, et al. Molecular characterization of Staphylococcus aureus isolated from raw milk samples of dairy cows in Manhiça district, southern Mozambique. Microorganisms. (2021) 9:1684. doi: 10.3390/microorganisms9081684
114. Patel K, Godden SM, Royster EE, Crooker BA, Johnson TJ, Smith EA, et al. Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of US dairy herds. BMC Genomics. (2021) 22:367. doi: 10.1186/s12864-021-07603-4
115. Qolbaini EN, Khoeri MM, Salsabila K, Paramaiswari WT, Tafroji W, Made Artika I, et al. Identification and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus-associated subclinical mastitis isolated from dairy cows in Bogor, Indonesia. Vet World. (2021) 14:1180–4. doi: 10.14202/vetworld.2021.1180-1184
116. Shrestha A, Bhattarai RK, Luitel H, Karki S, Basnet HB. Prevalence of methicillin-resistant Staphylococcus aureus and pattern of antimicrobial resistance in mastitis milk of cattle in Chitwan, Nepal. BMC Vet Res. (2021) 17:239. doi: 10.1186/s12917-021-02942-6
117. Tegegne DT, Mamo G, Waktole H, Messele YE. Molecular characterization of virulence factors in Staphylococcus aureus isolated from bovine subclinical mastitis in central Ethiopia. Ann Microbiol. (2021) 71:28. doi: 10.1186/s13213-021-01639-3
118. Zhao X, Yuan X, Hu M, Zhang Y, Li L, Zhang Q, et al. Prevalence and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus isolated from bulk tank milk in Shandong dairy farms. Food Control. (2021) 125:107836. doi: 10.1016/j.foodcont.2020.107836
119. Zaatout N, Hezil D. A meta-analysis of the global prevalence of methicillin-resistant Staphylococcus aureus (MRSA) isolated from clinical and subclinical bovine mastitis. J Appl Microbiol. (2022) 132:140–54. doi: 10.1111/jam.15192
120. Oliveros MCR. The dairy industry in southeast asia: perspective, challenges and opportunities. IOP Conf Ser Earth Environ Sci. (2019) 372:012068. doi: 10.1088/1755-1315/372/1/012068
121. International Dairy Federation. The World Dairy Situation 2021. Brussels: Bulletin of the International Dairy Federation (2021).
122. Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. (2015) 112:5649–54. doi: 10.1073/pnas.1503141112
123. Kayitsinga J, Schewe RL, Contreras GA, Erskine RJ. Antimicrobial treatment of clinical mastitis in the eastern United States: the influence of dairy farmers' mastitis management and treatment behavior and attitudes. J Dairy Sci. (2017) 100:1388–407. doi: 10.3168/jds.2016-11708
Keywords: methicillin-resistant Staphylococcus aureus, MRSA, meta-analysis, systematic review, dairy cattle farm
Citation: Khanal S, Boonyayatra S and Awaiwanont N (2022) Prevalence of methicillin-resistant Staphylococcus aureus in dairy farms: A systematic review and meta-analysis. Front. Vet. Sci. 9:947154. doi: 10.3389/fvets.2022.947154
Received: 18 May 2022; Accepted: 11 November 2022;
Published: 06 December 2022.
Edited by:
Alejandra Victoria Capozzo, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Timothy Frana, Boehringer Ingelheim, United StatesCopyright © 2022 Khanal, Boonyayatra and Awaiwanont. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sukolrat Boonyayatra, U3Vrb2xyYXQuQm9vbnlheWF0cmFAbGl1LmVkdQ==
†Present address: Sukolrat Boonyayatra, Department of Veterinary Clinical Sciences, College of Veterinary Medicine, Long Island University, Brookville, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.