- 1Laboratory of Small Animal Internal Medicine, School of Veterinary Medicine, Azabu University, Sagamihara, Japan
- 2Research and Development Section, Anicom Specialty Medical Institute Inc., Yokohama, Japan
- 3Laboratory of Clinical Diagnosis, School of Veterinary Medicine, Azabu University, Sagamihara, Japan
Haematopoietic stem and progenitor cells (HSPCs) are used for transplantation to reconstruct the haematopoietic pathways in humans receiving severe chemotherapy. However, the characteristics of canine HSPCs, such as specific surface antigens and gene expression profiles, are still unclear. This study aimed to characterise the haematopoietic ability and gene expression profiles of canine bone marrow HSPCs in healthy dogs. In this study, the CD34 positive (CD34+) cells were defined as classical HSPCs, CD34+/CD45 diminished (CD45dim) cells as more enriched HSPCs, and whole viable cells as controls. Haematopoietic abilities and gene expression profiles were evaluated using a colony-forming unit assay and RNA-sequencing analysis. Canine CD34+/CD45dim cells exhibited a significantly higher haematopoietic colony formation ability and expressed more similarity in the gene expression profiles to human and mouse HSPCs than those of the other cell fractions. Furthermore, the canine CD34+/CD45dim cells expressed candidate cell surface antigens necessary to define the canine haematopoietic hierarchy roadmap. These results indicate that the canine CD34+/CD45dim cells express the HSPC characteristics more than the other cell fractions, thereby suggesting that these cells have the potential to be used for studying haematopoietic stem cells in dogs.
Introduction
Haematopoietic stem and progenitor cells (HSPCs), present in the bone marrow, are multipotent cells with the ability to differentiate into mature blood cells. The haematopoietic stem cells (HSCs) have the ability to self-renew and reconstruct the haematopoietic system post-transplantation, whereas the haematopoietic progenitor cells (HPCs) have no detectable ability for self-renewal. In the classical haematopoietic hierarchy roadmap, HSCs are classified into two populations, namely, long-term (LT) HSCs and short-term (ST) HSCs, while HPCs are classified into five populations, namely, multipotent progenitors (MPPs), common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), megakaryocyte/erythrocyte progenitors (MEPs), and granulocyte/macrophage progenitors (GMPs) (1, 2). In human medicine, these HSPCs are used for haematopoietic stem cell transplantation (HSCT), and hence, they are administered to patients with haematopoietic tumour diseases after high-dose chemotherapy (3).
In veterinary medicine, autologous HSCT using peripheral blood stem cells (PBSCs) has been performed successfully in cases of canine B-cell and T-cell lymphoma; therefore, it may be considered as a new potential treatment option for canine haematopoietic tumours (4, 5). However, due to the low HSPC population in the bone marrow, it is difficult to obtain the number of HSPCs necessary for successful transplantation; hence, HSPC transplantation is uncommon in small animal practise. In human medicine, the isolation of HSPCs using cell surface antigens is critical for a successful PBSC transplantation since a high number of HSPCs contributes to an increase in the engraftment ratio post-HSCT (6). Previously, the canine CD34-positive (CD34+) cells were considered to be a definitive marker of HSCs; however, a study has revealed that the CD34+ cell population isolated from canine peripheral blood mostly consists of CD34+/CD45-high (CD45hi) cells and rarely the presence of CD34+/CD45-low (CD45lo) cells (7). Furthermore, 98% of CD34+/CD45hi cells exhibited CD21-positive, thereby suggesting the presence of differentiated and maturated B-cells. Therefore, the characterisation of other cell surface antigens for the identification of canine HSPCs is necessary to develop successful HSCT in these animals.
Even though human and mouse studies have extensively reported about the cell surface protein and gene expression markers of HSPC populations, there are only a few reports on similar analysis in dogs (7–10). In general, human CD34+ cells and CD34-positive/CD45-diminished (CD34+/CD45dim) cells are counted to evaluate the number of HSPCs prior to transplantation. In fact, the cell surface antigens of HSPCs that are centred on CD34+ cells, such as Lineage-negative (Lin-)/CD34+/CD38– and CD34+/CD45dim, have generally been used in human medicine (11–14). In veterinary medicine, the CD34 antigen has been considered as a marker for HSCs, and hence, it has been used in HSCT; however, although most of the CD34+ cells were reported to be composed of differentiated B-cells, bone marrow and PBSC transplants using CD34+/CD45dim have not yet been performed in dogs and cats with spontaneous haematopoietic tumors. Therefore, using CD34+/CD45dim cells or hematopoietic Lin-/CD34+ cells is necessary to accurately count the canine HSPCs.
Incidentally, HSPCs have been widely studied in haematopoietic research in humans, mice, and zebrafish, and they are required as a positive control while studying haematopoietic differentiation, haematopoietic insufficiency diseases, and haematopoietic tumors. Additionally, there are several reports of canine HSPC surface antigens other than CD34, including KIT proto-oncogene receptor tyrosine kinase (KIT; CD117), FMS-like tyrosine kinase 3 (Flt-3), major histocompatibility complex (MHC) class II, and wheat germ agglutinin (WGA) (9, 15, 16); however, the functions and gene expression profiles that characterise each cell fraction are unclear with respect to veterinary medicine. Therefore, it is important to characterise the other HSPC surface antigens for the successful detection of these cells in dogs.
In this study, we analysed the haematopoietic colony forming ability and the transcriptome of canine CD34+/CD45dim cells to identify potential markers for canine HSPCs.
Materials and methods
Animals
Clinically healthy four Beagle dogs (mean age, 5.75 ± 0.5 years) with clean medical histories were used for this study. They were kept at a small animal breeding facility in the Azabu University Veterinary Clinical Centre. All animal experiments were conducted according to the laboratory dynamic experimental guidelines of Azabu University (No. 190927-2). The dogs were not administered any drugs from 30 days prior to the beginning of the experiments.
Preparation of canine bone marrow-derived mononuclear cells (BM-MNCs)
The dogs were sedated with medetomidine hydrochloride (0.04 mg/kg IV), butorphanol tartrate (0.02 mg/kg IV), and buprenorphine hydrochloride (0.02 mg/kg IV). Thereafter, bone marrow specimens were collected from the ilium and humerus/femur using a bone marrow aspiration needle attached to a syringe containing 1 ml heparin. The collected specimens were quickly diluted with Iscove's modified Dulbecco's medium (IMDM; Gibco, MA, USA). Subsequently, the diluted specimens were separated by centrifugation with 50 mL Leucosep (Greiner Bio-One, Frickenhausen, Germany) at 1,000 × g for 10 min, and the supernatant containing the BM-MNCs was transferred to another centrifuge tube, diluted with phosphate-buffered saline (PBS; Gibco), and further separated by centrifugation at 250 × g for 10 min. Finally, the supernatant was removed, the cell sediment was washed twice with PBS, and stored in a cell cryopreservation solution using HSC-Banker GMP grade (Zenogen Pharma, Fukushima, Japan), according to the manufacturer's protocol. The cryopreserved cells were used for further experiments.
Flow cytometry analysis of cell surface antigen and cell sorting of canine BM-MNCs
To isolate the HSPCs, each cell fraction was assessed according to the clinical guidelines for CD34+ cell quantification in mobilised PBSCs using the sequential gating strategy of the International Society of Hematotherapy and Graft Engineering (ISHAGE) (11, 17). Our analysis was based upon the protocol described in a previous study with some modifications (18). Non-specific binding of antibodies was blocked using the Fc receptor binding inhibitor polyclonal antibody (Thermo Fisher Scientific, diluted 1:10). Phycoerythrin-conjugated mouse anti-canine CD34 (clone 1H6, BD Bioscience, diluted 1:20) and fluorescein-isothiocyanate-conjugated rat anti-canine CD45 (clone YKIX716.13, Bio-Rad, Hercules, CA, diluted 1:20) monoclonal antibodies were used for flow cytometry analysis. Additionally, phycoerythrin-conjugated mouse immunoglobulin G1 (IgG1) kappa isotype control (clone P3.6.2.8.1, Thermo Fisher Scientific, diluted 1:20) and fluorescein-isothiocyanate-conjugated rat IgG2b kappa isotype control (clone eB149/10H5, Thermo Fisher Scientific, diluted 1:20) were used as the isotype controls during the flow cytometry process. The 7-aminoactinomycin D (7-AAD; BD Bioscience, diluted 1:20) was used as the viability dye. Fluorescence-activated single cell sorting (FACS) melody (BD Bioscience, San Jose, CA, USA) was used for CD34 and CD45-based cell sorting of the canine BM-MNCs, while FlowJo (Tree Star, Ashland, OR) was used for data analysis. The HSPCs were defined as CD34+ or CD34+/CD45dim cells, and they were sorted for RNA extraction and colony-forming unit assay.
Colony-forming unit assay (CFU-A)
Haematopoietic differential ability of the sorted cells was evaluated using CFU-A. Approximately 1,000–10,000 canine BM-MNCs were sorted into three fractions, namely, whole viable cells, CD34+ cells, and CD34+/CD45dim cells, according to be said cell sorting strategy. The CFU-A was performed using MethoCult H4435 enriched medium (MethoCult; Stem Cell Technologies, Vancouver, BC, Canada), which is composed of a semi-solid matrix containing supplements necessary for differentiating into mature blood cells. Each cell fraction was added to MethoCult, mixed by vigorous vortexing, and incubated at 37°C and 5% CO2. After 7–14 days, haematopoietic colonies, such as the burst-forming unit erythroid (BFU-E), colony-forming unit erythroid (CFU-E), colony-forming unit granulocyte (CFU-G), colony-forming unit macrophage (CFU-M), and colony-forming unit granulocyte-macrophage (CFU-GM), were observed with a phase-contrast microscope. The proportion of colony formation was calculated using the following formula: proportion of colony formation (%) = (total colony count / count of seeding cells) ×100. Statistical significance (*p < 0.05, ***p < 0.001) was assessed by one-way analysis of variance (ANOVA) using EZR (version 1.54; Jichi Medical University, Saitama, Japan) (19).
Total RNA extraction
All four canine BM-MNC samples were subjected to RNA-sequencing (RNA-seq) analysis. The 1,000–10,000 canine BM-MNCs that had been sorted using a previously described cell sorting strategy were subsequently treated with TRIzol LS Reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the total RNA was extracted using Direct-zol RNA Miniprep Plus kit (Zymo Research, Irvine, CA, USA). The RNA integrity number (RIN) was evaluated using Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA), and samples with RIN ≥ 7 were subjected to further gene expression analysis.
Gene expression profiling by RNA-seq
A complementary DNA (cDNA) library was constructed from 250 pg of total RNA for each sample using the SMART-Seq v4 PLUS kit (TaKaRa Bio, Shiga, Japan). The cDNA library was sequenced on an Illumina NextSeq 500 (Illumina, San Diego, USA) using the NextSeq 500/550 High Output Kit v2.0 (Illumina), according to the manufacturer's protocol (Supplementary Table 1). The read length was 150 bp single-end reads. Low-quality sequenced reads were trimmed using Trimmomatic (Version 0.39) with the following parameter settings: HEADCROP: 3, CROP: 145, LEADING: 30, TRAILING: 30, SLIDEWINDOW: 10:30, MINLEN: 50, and AVGQUAL: 31. Finally, the read length was a range of 50–145 bp single-end reads. The quality of the sequenced reads was assessed using FastQC (version 0.11.9). Moreover, transcripts per million (TPM) were used for RNA quantification as well as subsequent statistical analysis. The reference cDNA datasets were CanFam3.1-based annotations deposited in the Ensembl genome browser (release 104). Gene expression was quantified using Kallisto/Sleuth pipeline. Incidentally, Kallisto (v0.46.1) is a pseudoalignment-based method used to quantify RNA abundance at the transcriptional level (20). The TPM was calculated using the quant option implemented in Kallisto. Thereafter, the downstream differential gene expression analysis was analysed by Sleuth (v0.30.0) using the TPM data (21). All statistical tests were corrected by the Benjamini–Hochberg method, and the statistical significance level was set as false discovery rate (FDR) <0.1 in the Wald test. Gene ontology (GO) analysis, a key bioinformatics tool for integrating the representation of genes and gene product attributes across all species, and Kyoto Encyclopaedia Encyclopaedia of Genes and Genomes (KEGG) pathway analysis were performed using the g:GOSt (version e104_eg51_p15_3922dba), which is a web server for performing functional enrichment analyses (22).
Results
Characterisation of CD34+ and CD34+/CD45dim cells among canine BM-MNCs
In this study, canine HSPCs were collected using a method similar to the ISHAGE sequential gating strategy. Typical flow cytometry plots of the canine BM-MNCs are shown in Figure 1. The proportions of whole viable cells, CD34+ cells, and CD34+/CD45dim cells are 49.42 ± 3.67%, 0.36 ± 0.06%, and 0.16 ± 0.03%, respectively, among 100,000 canine BM-MNCs.
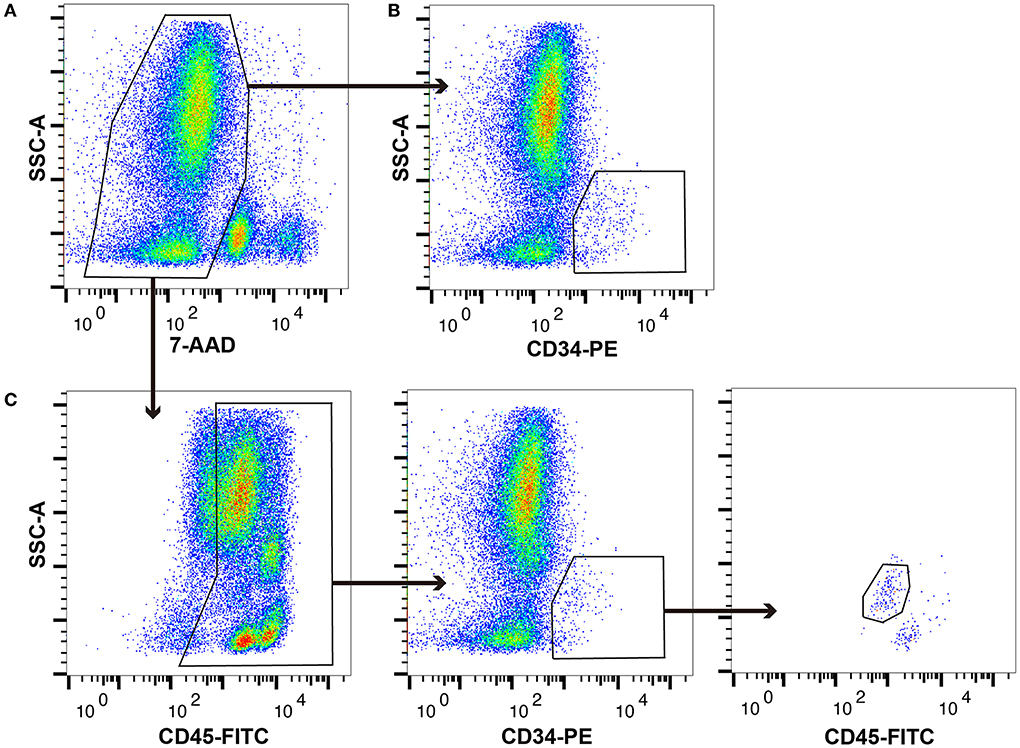
Figure 1. Typical plot profiles of canine haematopoietic stem and progenitor cells (HSPCs). Canine bone marrow-derived mononuclear cells (cBM-MNCs) were stained with 7-aminoactinomycin D (7-AAD), which is a viability dye, phycoerythrin (PE)-labelled anti-canine CD34 monoclonal antibody (mAb), and fluorescein isothiocyanate (FITC)-labelled anti-canine CD45 mAb. (A) Whole viable cells, (B) CD34+ cells, and (C) CD34+/CD45 diminished (CD45dim) cells as analysed by flow cytometry.
Haematopoietic colony forming ability among canine HSPC phenotypes
To evaluate the haematopoietic multipotency of the canine HSPC phenotypes arising due to different cell surface antigens, we examined their colony forming abilities using CFU-A. In fact, each cell fraction collected during cell sorting was evaluated for its colony-forming ability. All cell fractions showed blood cell colony formation (Figure 2A). For every 1,000 canine BM-MNCs, the number of colonies formed by whole viable cells, CD34+ cells, and CD34+/CD45dim cells are 0.6 ± 0.05, 19.5 ± 6.0, and 67 ± 1.85, respectively (Figure 2B). Moreover, the ability of the CD34+/CD45dim cells to differentiate into haematopoietic colonies is significantly higher as compared to that of the other cell fractions. Incidentally, the CD34+/CD45bright/side scatter low (SSClow) cells were unable to form haematopoietic colonies (data not shown). Notably, there was no significant difference in the individual haematopoietic colony construction abilities of the CD34+ and CD34+/CD45dim cells.
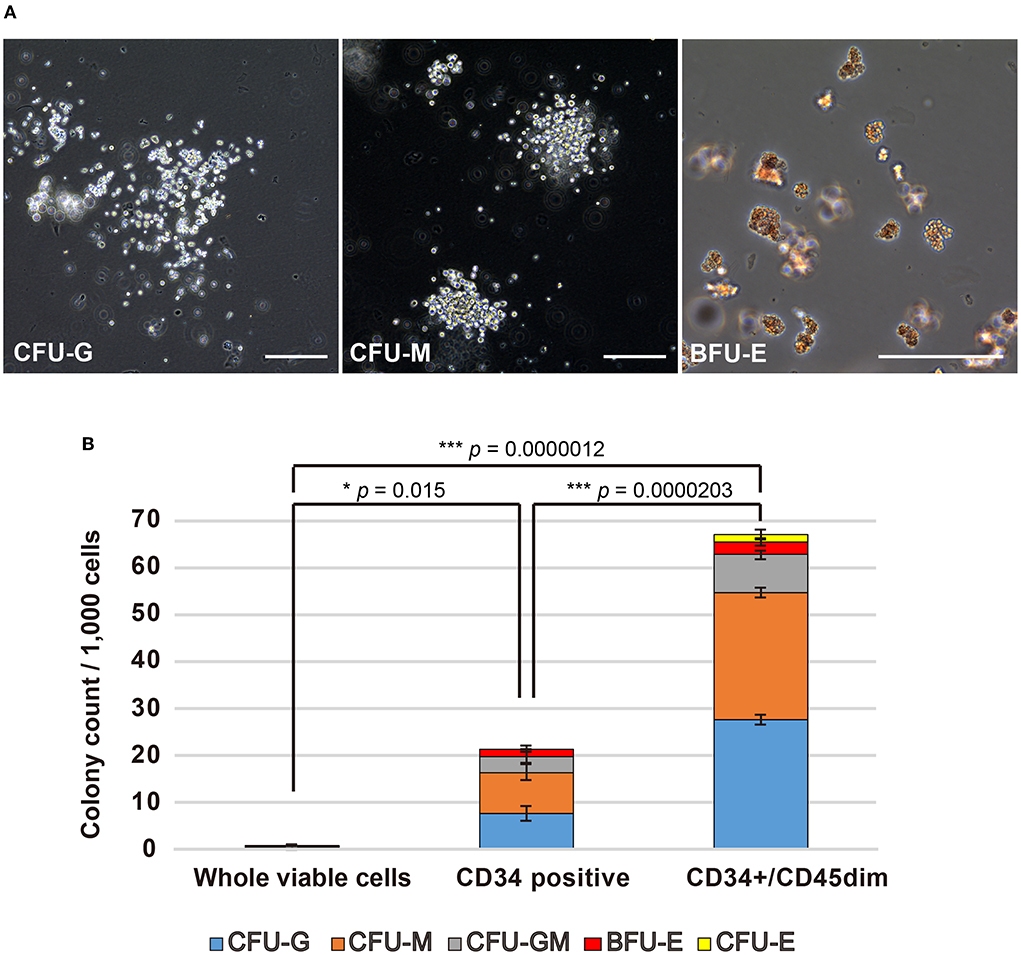
Figure 2. Haematopoietic colony formation by canine haematopoietic stem and progenitor cells (HSPCs). (A) Microscopic observation of canine haematopoietic colonies (Scale bars: 200 μm). (B) Haematopoietic colonies for every 1,000 cells, as counted for whole viable cells, CD34+ cells, and CD34+/CD45diminished (CD45dim) cells. Experiments were performed four times independently. Results are presented as mean ± standard error of the mean (SEM). Statistical significance of total colony counts was assessed by one-way analysis of variance (ANOVA). *p < 0.05, ***p < 0.001.
RNA-seq analysis of canine HSPC phenotypes
To investigate the similarities and differences between the CD34+ and CD34+/CD45dim cells, we performed whole transcriptome analysis using RNA-seq. Principal component analysis demonstrates a clear separation of the whole viable cells (blue dots) from the canine HSPCs, including the CD34+ cells (red dots) and CD34+/CD45dim cells (green dots) in PC1 (Figure 3A). Furthermore, the CD34+/CD45dim cells are convergent, as compared to the CD34+ cells in PC2. The contribution ratio of PC1 is 93.96 % and that of PC2 is 3.46 %. Additionally, we detected 148 differentially expressed genes (DEGs) with FDR <0.1 in the canine CD34+/CD45dim cells, as compared to the corresponding gene expressions in the CD34+ cells; among these 148 DEGs, 7 are upregulated and 141 are downregulated (Figure 3B). The genes that are upregulated in CD34+/CD45dim cells include the ones encoding phospholipase A2 group IVA (PLA2G4A), integrin subunit alpha 8 (ITGA8), chondroitin sulfate N-acetylgalactosaminyltransferase 1 (CSGALNACT1), germinal centre associated signalling and motility like protein (GCSAML), cystathionine beta-synthase (CBS), TNF superfamily member 8 (TNFSF8), and retinoic acid receptor responder 2 (RARRES2). Moreover, the GO analysis of the 131 downregulated DEGs in the CD34+/CD45dim cells revealed that the majority of the GO terms are associated with cell cycle and cell division, and the KEGG pathway analysis revealed that the downregulated DEGs are significantly enriched in B cell receptor signalling pathways (adjusted p value = 1.699 ×10−2) (Figure 3C). On the contrary, there are 2,286 upregulated DEGs and 2,295 downregulated DEGs in the CD34+/CD45dim cells, as compared to the corresponding gene expressions in the whole viable cells (Figure 3D). The upregulated DEGs associated with the major surface antigens of HSPCs are KIT proto-oncogene (KIT; CD117), CD34, FMS-like tyrosine kinase 3 (FLT3; CD135), endothelial cell adhesion molecule (ESAM), TEK Receptor Tyrosine Kinase (TEK; Tie2), and endoglin (ENG; CD105), while the downregulated DEGs are integrin alpha M (ITGAM; CD11b), protein tyrosine phosphatase receptor type C (PTPRC; CD45RA), CD48, Thy-1 cell surface antigen (THY1; CD90), CXC chemokine receptor type 4 (CXCR4), and CD38. Additionally, MYB Proto-Oncogene (MYB), ETS variant transcription factor 6 (ETV6), GATA binding factor 2 (GATA2), homeobox A5 (HOXA5), HOXA7, HOXA9, and HOXA10, which are highly expressed in HSCs, are upregulated in the CD34+/CD45dim cells.
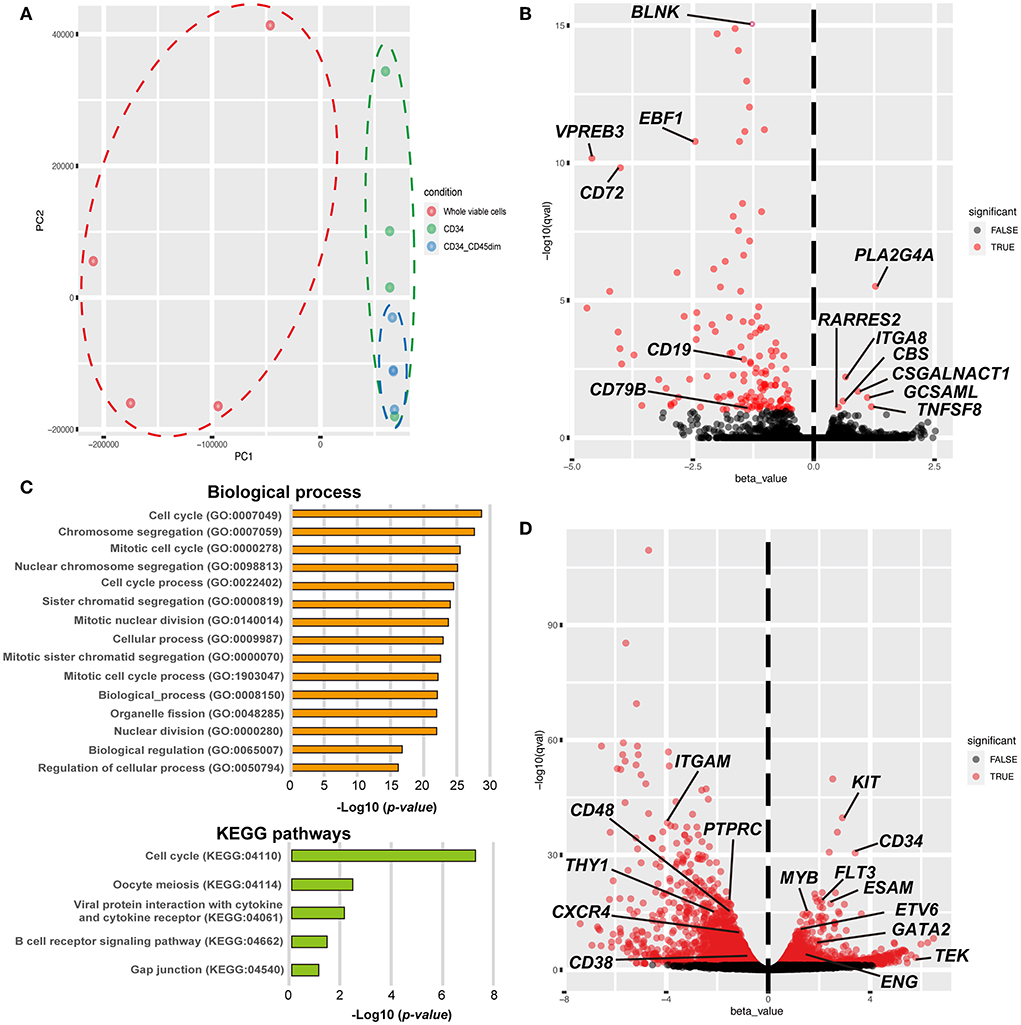
Figure 3. Global gene expression profiling of canine haematopoietic stem and progenitor cells (HSPCs). (A) Principal component analysis of the global gene expression of canine HSPCs. Gene expression is demonstrated by dots and circles of red (whole viable cells), green (CD34+ cells), and blue [CD34+/CD45 diminished (CD45dim) cells]. (B) The volcano plot shows the results of differentially expressed gene (DEG) analysis between CD34+/CD45dim cells and CD34+ cells. Statistical significance was set as false discovery rate (FDR) <0.1. (C) Gene ontology (GO) and Kyoto Encyclopaedia Gene and Genomes (KEGG)-based pathway enrichment analysis of the downregulated genes of CD34+/CD45dim cells. The candidate terms of biological processes are illustrated with adjusted p-values < −Log1016 (top). The candidate terms revealed by KEGG pathway analysis are demonstrated (bottom). (D) The volcano plot shows the results of DEG analysis between CD34+/CD45dim cells and whole viable cells. Statistical significance was set as FDR <0.1.
Discussion
This study demonstrates that the canine CD34+/CD45dim cells have a high haematopoietic colony forming ability and a high degree of HSPC-associated gene expression pattern, indicating their potential to function as canine HSPCs. Generally, HSPCs are collected directly from the bone marrow by aspiration, or they are mobilised into peripheral blood and collected by apheresis (1). Moreover, low population cells, like these HSPCs, are generally analysed after enrichment or by single-cell analysis using flow cytometric cell sorting (23, 24). In this study, the canine CD34+ and CD34+/CD45dim cells had a very low population in the bone marrow; therefore, they were subjected to cell sorting enrichment prior to any experiments (Figure 1). With respect to veterinary medicine, Faldyna et al. reported the presence of 1%−3% CD34+ cells (15), and Tsumagari et al. reported the presence of 0.30 ± 0.07% CD34+/CD45dim cells in the adult canine bone marrow (7). Interestingly, the population of CD34+/CD45dim cells (0.16 ± 0.03%) recorded in this study is lower than that reported in the previous studies. However, the reason for this striking difference is unknown. We believe that this low population of CD34+/CD45dim cells in this study might be due to the exclusion of doublets, dead cells, and surrounding fraction cells.
In this study, the canine CD34+/CD45dim cells exhibited a high cell differentiation ability, i.e., their colony forming ability was significantly greater than that of CD34+ cells and whole viable cells (Figure 2B). Interestingly, a study related to human medicine reports that the CD45 antigen expression is lower in the immature cells of the early differentiation stages than that in the mature lymphocytes and monocytes (17). Furthermore, the expression level of CD45 antigen is low to moderate, even in the CD34+ HSPCs, and the endothelial cells, the other cell types of CD34+ cells, exhibit negative expression of CD45 antigens. Therefore, the combination of CD34 and CD45 makes it possible to distinctly select the HSPCs. Moreover, we did not observe any differences between the CD34+/CD45dim and CD34+ cell fractions with respect to the BFU-E, CFU-E, CFU-G, CFU-M, and CFU-GM types of haematopoietic colony formations. This suggests that CD34+/CD45dim cells did not inhibit the differentiation of any specific cell lineage of the canine HSPCs. On the contrary, CFU-A of canine CD34+/CD45bright/SSClow cells demonstrated their inability to form haematopoietic colonies. A previous study had revealed that the canine CD34+ cell fraction mostly consisted of B-cells and exhibited non-specific binding to anti-CD34 antibodies (7). This suggests that the CD34+/CD45bright/SSClow cells are non-HSPCs, including other haematopoietic cell lineages.
Based on transcriptome analysis, we showed that the canine CD34+/CD45dim cells have a gene expression profile similar to that of the HSPCs. Moreover, the principal component of the canine CD34+/CD45dim cells gene expression was concentrated with similar components compared to whole viable cells and CD34+ cells. Additionally, a DEG analysis between the canine CD34+/CD45dim cells and the CD34+ cells revealed that four out of the seven upregulated DEGs in the CD34+/CD45dim cells, (PLA2G4A, CSGALNACT1, CBS, and RARRES2) were reportedly associated with HSPCs (25–30), and the remaining three (ITGA8, GCSAML, and TNFSF8) with haematopoietic lineage cells (31–37). The PLA2G4A encodes a key enzyme that catalyses the hydrolysis of membrane phospholipids to release arachidonic acid (38). The CSGALNACT1 encodes a key enzyme in chondroitin sulphate biosynthesis (27), and in CSGALNACT1-knockout mice, there is a reported disruption of HSC maintenance and early T-cell development (28). Previous studies have demonstrated a higher expression of these two genes in HSPCs, as compared to that in myelocytes or lymphocytes (26, 27), and in this study, these two gene expressions were significantly increased in the canine CD34+/CD45dim cells than in the CD34+ cells. The CBS is a key enzyme for hydrogen sulphide (H2S) production from L-cysteine, and CBS deficiency induces a disruption of erythropoiesis by interfering with the expression of haem-biosynthetic enzymes and haem-transporters (29). The RARRES2, which encodes a protein named chemerin, is secreted from Lin-/Sca-1+/c-kit+/CD34- LT-HSC in mice, and RARRES2 neutralisation blocks osteoclastogenesis of HSCs (30). These reports demonstrate that the abovementioned genes play an important role in HSPC functions and haematopoiesis, and the upregulation of these genes characterises canine CD34+/CD45dim cells as HSPCs.
Furthermore, we detected 141 downregulated DEGs in the canine CD34+/CD45dim cells as compared to the corresponding gene expressions in the CD34+ cells, and a GO analysis of 131 of these DEGs revealed numerous GO annotations associated with cell cycle and cell division. This suggests that the canine CD34+/CD45dim cells have a longer quiescent phase of the cell cycle than the CD34+ cells (Figure 3C). Under normal conditions, the bone marrow is in a hypoxic environment, and majority of the HSPCs are maintained in the quiescent G0 phase by hypoxia-inducible factor 1-alpha (HIF-1α) (39). Additionally, the HSPCs are tightly regulated with respect to quiescence, proliferation, and differentiation in the bone marrow microenvironment or niche (40). Another study has reported that HSPCs are maintained by CXC chemokine ligand (CXCL)12-abundant reticular (CAR) cells, and they form bone marrow microenvironments known as haematopoietic stem cell niches (41). Therefore, it is possible that the canine bone marrow HSCs contain quiescent phase cells. This supports the presence of a high number of quiescent phase cells among the canine CD34+/CD45dim cells.
The KEGG pathway enrichment analysis revealed that the downregulated DEGs are associated with the B cell receptor signalling pathway (Figure 3C). A previous report revealed that canine CD34+ cell fraction contained B-cells that exhibited non-specific binding to anti-CD34 antibodies (7). Therefore, in this study, the canine CD34+ cell fraction also included other kinds of HSPCs. We have demonstrated that the major surface antigen genes associated with HSPCs are differentially expressed in CD34+/CD45dim cells than in the whole viable cells (Figure 3D). In humans and in mice, KIT, CD34, FLT3, Tie2, CD105, CD45RA, and CD90 are generally used as surface antigens to define the haematopoietic hierarchy roadmap (13, 42–45). Moreover, CD38, which is expressed in many cell types, including plasma cells, is generally used as a negative HSC marker in humans (12, 46). The ESAM is a novel, human primitive HSC marker that plays an important role in definitive haematopoiesis development (47). These antigen expressions are common to humans and mice; therefore, they can be considered as candidates for determining the haematopoietic hierarchy roadmap in dogs, as well. Additionally, MYB, ETV6, GATA2, HOXA5, HOXA7, HOXA9, and HOXA10, which are reportedly HSPC markers (48–51), were upregulated in the canine CD34+/CD45dim cells, thereby supporting the characterisation of canine CD34+/CD45dim cells as HSPCs.
According to the observations of this study, CD34+/CD45dim seems to have the potential to function as a surface antigen marker for the identification of canine HSPCs. Canine HSPC surface antigens have not been elucidated clearly, and the definition of the haematopoietic hierarchy roadmap has been incomplete in veterinary medicine. McSweeney et al. have reported that CD34 is expressed in the canine bone marrow cells, and CD34+ cells have a haematopoietic colony forming ability, thereby suggesting their probable characterisation as HSPCs (52). Furthermore, cryopreserved CD34+ cells can retain their colony formation ability (53). Kim et al. demonstrated that the comparative study regarding the safety of PBSC collection methods by apheresis in healthy dogs using CD34+/CD45dim cells (18). Incidentally, the molecular-level characteristics of CD34+/CD45dim cells were not assessed in that study; however, it is necessary to analyse whether the CD34+/CD45dim cells have the cell differentiation potential and haematopoietic stem cell characteristics suitable for induction in transplantation therapy. In this study, we have successfully demonstrated that canine CD34+/CD45dim cells exhibit more HSPC characteristics than the CD34+ cells.
In the clinical practise of human medicine, HSPCs have been extensively used as a tumour treatment option. In fact, HSCT is performed by autologous and allogeneic methods, both of which generally use PBSCs collected by apheresis. Autologous transplantation has the advantage of avoiding graft-vs.-host disease (GvHD), and it has a reduced relapse risk (RR) over allogeneic transplantation (54). Even though allogeneic transplantation has an increased GvHD risk in some recipients, it has been widely used to attack and eradicate malignant cells, in which the donor immune cells are capable of eradicating chemotherapy-resistant tumour cells by the graft-versus-tumour (GVT) effect (55). Meanwhile, pluripotent stem cells, such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), can differentiate into various types of cells and tissues, and many studies have reported haematopoietic differentiation methods via HSPCs using pluripotent stem cells in humans and mice (56–59). Therefore, it is expected that allogeneic HSCTs will be performed increasingly in the future, and the analysis of immunogenicity and cell surface markers will be important for determining the safety of transplantation in canines and humans.
There are a few limitations in this study. First, we did not confirm the haematopoietic reconstruction ability in vivo by transplantation of CD34+/CD45dim cells into immune-deficient mice. Second, the CD34+/CD45dim cells are not sufficient to exclusively confirm the presence of HSCs; therefore, the location of the HSPCs among the canine CD34+/CD45dim cells is still unclear with respect to the classical haematopoietic hierarchy Roadmap. Additional Markers Need to be Determined for This Purpose.
Conclusion
These results indicate that the canine CD34+/CD45dim cells conformed to the characteristics of HSPCs based upon the ISHAGE sequential gating strategy, especially in terms of haematopoietic colony forming ability and gene expression profile. Moreover, it contributes to the determination of candidate surface antigens that define the canine haematopoietic hierarchy roadmap. We believe that these results will lead to long-term haematopoietic reconstructions in HSCT and efficient haematopoietic differentiation from pluripotent stem cells in dogs. However, further research is needed for the exclusive identification of canine HSPCs in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Submission: DRA014248, Run: DRR378823-DRR378834. BioProject: PRJDB13629 (PSUB017583), BioSample: SAMD00493090-SAMD00493092 (SSUB021958), SAMD00493093-SAMD00493095 (SSUB021989), SAMD00493096-SAMD00493098 (SSUB021990), and SAMD00493099-SAMD00493101 (SSUB021991).
Ethics statement
The animal study was reviewed and approved by Animal experiment Committee of Azanu University.
Author contributions
TA, MH, and KKa contributed to conception and design of the study. YM and RH organised the database. TA, SNi, YY, SNe, and KKi performed the experiment. TA and MH wrote the first draught of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the Grant and Academic Consulting Fee of Laboratory of Small Animal Internal Medicine, Azabu University, and Research Foundation of Anicom Specialty Medical Institute Inc. (Japan).
Acknowledgments
We would like to express our deep gratitude to the students and staff of Azabu University for their cooperation in the research. Academic Consulting Fee was provided by Sainomori Animal Hospital, Meguro Senzoku Animal Hospital, and QIX Co., Ltd. Also, we would like to thank Editage (www.editage.com) for English language editing.
Conflict of interest
Authors TA, YM, and KKa were employed by Anicom Specialty Medical Institute Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.936623/full#supplementary-material
Abbreviations
HSPC, haematopoietic stem and progenitor cell; HSC, haematopoietic stem cell; HPC, haematopoietic progenitor cell; HSCT, haematopoietic stem cell transplantation; LT-HSC, long-term HSC; ST-HSC, short-term HSC; MPP, multipotent progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; GMP, granulocyte/macrophage progenitor; BM-MNC, bone marrow-derived mononuclear cells; Flt-3, FMS-like tyrosine kinase 3; MHC, major histocompatibility complex; CFU-A, colony forming unit assay; BFU-E, burst forming unit-erythroid; CFU-E, colony forming unit-erythroid; CFU-G, colony forming unit granulocyte; CFU-M, colony forming unit macrophage; CFU-GM, colony forming unit granulocyte-macrophage; TPM, transcripts per million; PLA2G4A, phospholipase A2 group IVA; ITGA8, integrin subunit alpha 8; CSGALNACT1, chondroitin sulphate N-acetylgalactosaminyltransferase 1; GCSAML, germinal centre associated signalling and motility like; CBS, cystathionine beta-synthase; TNFSF8, TNF superfamily member 8; RARRES2, retinoic acid receptor responder 2; ITGAM, integrin alpha M; PTPRC, protein tyrosine phosphatase receptor type C; CXCR4, CXC chemokine receptor type 4; ETV6, ETS variant transcription factor 6; GATA2, GATA binding factor 2; HOXA5, homeobox A5; ESAM, endothelial cell adhesion molecule; ENG, endoglin; DEG, differentially expressed gene; GvHD, graft-vs.-host disease; GvT, graft-vs.-tumor.
References
1. Yang L, Bryder D, Adolfsson J, Nygren J, Månsson R, Sigvardsson M, et al. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. (2005) 105:2717–23. doi: 10.1182/blood-2004-06-2159
2. Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. (1997) 124:1929–39. doi: 10.1242/dev.124.10.1929
3. Hequet O. Hematopoietic stem and progenitor cell harvesting: technical advances and clinical utility. J Blood Med. (2015) 6:55–67. doi: 10.2147/JBM.S52783
4. Willcox JL, Pruitt A, Suter SE. Autologous peripheral blood hematopoietic cell transplantation in dogs with B-cell lymphoma. J Vet Intern Med. (2012) 26:1155–63. doi: 10.1111/j.1939-1676.2012.00980.x
5. Warry EE, Willcox JL, Suter SE. Autologous peripheral blood hematopoietic cell transplantation in dogs with T-cell lymphoma. J Vet Intern Med. (2014) 28:529–37. doi: 10.1111/jvim.12302
6. Singhal S, Powles R, Treleaven J, Kulkarni S, Sirohi B, Horton C, et al. A low CD34+ cell dose results in higher mortality and poorer survival after blood or marrow stem cell transplantation from HLA-identical siblings: Should 2 x 10(6) CD34+ cells/kg be considered the minimum threshold? Bone Marrow Trans. (2000) 26:489–96. doi: 10.1038/sj.bmt.1702542
7. Tsumagari S, Otani I, Tanemura K, Namba S, Ohtaki T, Kamata H, et al. Characterization of CD34+ cells from canine umbilical cord blood, bone marrow leukocytes, and peripheral blood by flow cytometric analysis. J Vet Med Sci. (2007) 69:1207–9. doi: 10.1292/jvms.69.1207
8. Neuner E, Schumm M, Schneider EM, Guenther W, Ellwart J, Kremmer E, et al. Immune phenotype of canine hematopoietic progenitor cells. Tissue Antigens. (1997) 50:466–74. doi: 10.1111/j.1399-0039.1997.tb02901.x
9. Wijewardana V, Sugiura K, Shigeyama N, Moriguchi M, Tsunoda S, Ikehara S, et al. Isolation and characterization of hematopoietic progenitor cells in canine bone marrow. Vet Immunol Immunopathol. (2007) 115:230–8. doi: 10.1016/j.vetimm.2006.11.007
10. Ide K, Goto-Koshino Y, Momoi Y, Fujino Y, Ohno K, Tsujimoto H. Quantitative analysis of mRNA transcripts of hox, SHH, PTCH, Wnt, and Fzd genes in canine hematopoietic progenitor cells and various in vitro colonies differentiated from the cells. J Vet Med Sci. (2009) 71:69–77. doi: 10.1292/jvms.71.69
11. Barnett D, Janossy G, Lubenko A, Matutes E, Newland A, Reilly JT. Guideline for the flow cytometric enumeration of CD34+ haematopoietic stem cells. Prepared by the CD34+ haematopoietic stem cell working party general haematology task force of the British committee for standards in haematology. Clin Lab Haematol. (1999) 21:301–8. doi: 10.1046/j.1365-2257.1999.00253.x
12. Kerre TCC, De Smet G, De Smedt M, Offner F, De Bosscher J, Plum J, et al. Both CD34+38+ and CD34+38- cells home specifically to the bone marrow of NOD/LtSZ scid/scid mice but show different kinetics in expansion. J Immunol. (2001) 167:3692–8. doi: 10.4049/jimmunol.167.7.3692
13. Yahata T, Muguruma Y, Yumino S, Sheng Y, Uno T, Matsuzawa H, et al. Quiescent human hematopoietic stem cells in the bone marrow niches organize the hierarchical structure of hematopoiesis. Stem Cells. (2008) 26:3228–36. doi: 10.1634/stemcells.2008-0552
14. Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. (2014) 32:1380–9. doi: 10.1002/stem.1661
15. Faldyna M, Sinkora J, Knotigova P, Rehakova Z, Moravkova A, Toman M. Flow cytometric analysis of bone marrow leukocytes in neonatal dogs. Vet Immunol Immunopathol. (2003) 95:165–76. doi: 10.1016/S0165-2427(03)00135-1
16. Niemeyer GP, Hudson J, Bridgman R, Spano J, Nash RA, Lothrop CD. Isolation and characterization of canine hematopoietic progenitor cells. Exp Hematol. (2001) 29:686–93. doi: 10.1016/S0301-472X(01)00638-5
17. Sutherland DR, Anderson L, Keeney M, Nayar R, Chin-Yee I. The ISHAGE guidelines for CD34+ cell determination by flow cytometry. International Society of Hematotherapy and Graft Engineering. J Hematother. (1996) 5:213–26. doi: 10.1089/scd.1.1996.5.213
18. Kim S, Hosoya K, Kobayashi A, Okumura M. Comparison of three mobilization protocols for peripheral blood stem cell apheresis with Spectra Optia continuous mononuclear cell protocol in healthy dogs. Vet Comp Oncol. (2019) 17:61–8. doi: 10.1111/vco.12446
19. Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Trans. (2013) 48:452–8. doi: 10.1038/bmt.2012.244
20. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. (2016) 34:525–7. doi: 10.1038/nbt.3519
21. Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. (2017) 14:687–90. doi: 10.1038/nmeth.4324
22. Reimand J, Arak T, Vilo J. G:profiler - A web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. (2011) 39:W307–15. doi: 10.1093/nar/gkr378
23. Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ Li P, Yzaguirre AD, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. (2015) 160:241–52. doi: 10.1016/j.cell.2014.12.032
24. Watcham S, Kucinski I, Gottgens B. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood. (2019) 133:1415–26. doi: 10.1182/blood-2018-08-835355
25. Patel MI, Singh J, Niknami M, et al. Cytosolic phospholipase A2-alpha: a potential therapeutic target for prostate cancer. Clin Cancer Res. (2008) 14:8070–9. doi: 10.1158/1078-0432.CCR-08-0566
26. McGraw KL, Cheng CH, Chen YA, Hou HA, Nilsson B, Genovese G, et al. Non-del(5q) myelodysplastic syndromes–associated loci detected by SNP-array genome-wide association meta-analysis. Blood Adv. (2019) 3:3579–89. doi: 10.1182/bloodadvances.2019000922
27. Katagiri T, Uemura S, Ushiki T, Nakajima-Takagi Y, Oshima M, Mikami T, et al. Distinct effects of chondroitin sulfate on hematopoietic cells and the stromal microenvironment in bone marrow hematopoiesis. Exp Hematol. (2021) 96:52–62.e5. doi: 10.1016/j.exphem.2021.02.003
28. Abramowitz LK, Harly C, Das A, Bhandoola A, Hanover JA. Blocked O-GlcNAc cycling disrupts mouse hematopoeitic stem cell maintenance and early T cell development. Sci Rep. (2019) 9:12569. doi: 10.1038/s41598-019-48991-8
29. Zhao P, Qian C, Chen YJ, Sheng Y, Ke Y, Qian ZM. Cystathionine β-synthase (CBS) deficiency suppresses erythropoiesis by disrupting expression of heme biosynthetic enzymes and transporter. Cell Death Dis. (2019) 10:708. doi: 10.1038/s41419-019-1951-0
30. Muruganandan S, Dranse HJ, Rourke JL, Mcmullen NM, Sinal CJ. Chemerin neutralization blocks hematopoietic stem cell osteoclastogenesis. Stem Cells. (2013) 31:2172–82. doi: 10.1002/stem.1450
31. Schnapp LM, Hatch N, Ramos DM, Klimanskaya IV, Sheppard D, Pytela R. The human integrin α8β1 functions as a receptor for tenascin, fibronectin, and vitronectin. J Biol Chem. (1995) 270:23196–202. doi: 10.1074/jbc.270.39.23196
32. Ryu J, Koh Y, Park H, Kim DY, Kim DC, Byun JM, et al. Highly expressed integrin-α8 induces epithelial to mesenchymal transition-like features in multiple myeloma with early relapse. Mol Cells. (2016) 39:898–908. doi: 10.14348/molcells.2016.0210
33. Eicher JD, Chami N, Kacprowski T, Nomura A, Chen MH, Yanek LR, et al. Platelet-related variants identified by Exomechip meta-analysis in 157,293 individuals. Am J Hum Genet. (2016) 99:40–55. doi: 10.1016/j.ajhg.2016.05.005
34. Gattei V, Degan M, Gloghini A, De Iuliis A, Improta S, Rossi FM, et al. CD30 ligand is frequently expressed in human hematopoietic malignancies of myeloid and lymphoid origin. Blood. (1997) 89:2048–59. doi: 10.1182/blood.V89.6.2048
35. Nicod LP, Isler P. Alveolar macrophages in sarcoidosis coexpress high levels of CD86 (B72), CD40, and CD30L Am J Respir Cell Mol Biol. (1997) 17:91–6. doi: 10.1165/ajrcmb.17.1.2781
36. Shimozato O, Takeda K, Yagita H, Okumura K. Expression of CD30 ligand (CD153) on murine activated T cells. Biochem Biophys Res Commun. (1999) 256:519–26. doi: 10.1006/bbrc.1999.0336
37. Pinto A, Aldinucci D, Gloghini A, Zagonel V, Degan M, Improta S, et al. Human eosinophils express functional CD30 ligand and stimulate proliferation of a Hodgkin's disease cell line. Blood. (1996) 88:3299–305. doi: 10.1182/blood.V88.9.3299.bloodjournal8893299
38. Murakami M, Kudo I. Phospholipase A2. J Biochem. (2002) 131:285–92. doi: 10.1093/oxfordjournals.jbchem.a003101
39. Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, et al. Regulation of the HIF-1α level is essential for hematopoietic stem cells. Cell Stem Cell. (2010) 7:391–402. doi: 10.1016/j.stem.2010.06.020
40. Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. (2014) 20:833–46. doi: 10.1038/nm.3647
41. Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. (2014) 508:536–40. doi: 10.1038/nature13071
42. Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. (2011) 333:218–21. doi: 10.1126/science.1201219
43. Hsu HC, Ema H, Osawa M, Nakamura Y, Suda T, Nakauchi H. Hematopoietic stem cells express Tie-2 receptor in the murine fetal liver. Blood. (2000) 96:3757–62. doi: 10.1182/blood.V96.12.3757
44. Kays SK, Kaufmann KB, Abel T, Brendel C, Bonig H, Grez M, et al. CD105 is a surface marker for receptor-targeted gene transfer into human long-term repopulating hematopoietic stem cells. Stem Cells Dev. (2015) 24:714–23. doi: 10.1089/scd.2014.0455
45. Ito T, Kawai Y, Yasui Y, et al. The therapeutic potential of multiclonal tumoricidal T cells derived from tumor infiltrating lymphocyte-1derived iPS cells. Commun Biol. (2021) 4:694. doi: 10.1038/s42003-021-02195-x
46. Terstappen LWMM, Huang S, Safford M, Lansdorp PM, Loken MR. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38- progenitor cells. Blood. (1991) 77:1218–27. doi: 10.1182/blood.V77.6.1218.1218
47. Ueda T, Yokota T, Okuzaki D, Uno Y, Mashimo T, Kubota Y, et al. Endothelial cell-selective adhesion molecule contributes to the development of definitive hematopoiesis in the fetal liver. Stem Cell Rep. (2019) 13:992–1005. doi: 10.1016/j.stemcr.2019.11.002
48. Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci U S A. (2004) 101:16240–5. doi: 10.1073/pnas.0407241101
49. Crooks GM, Fuller J, Petersen D, Izadi P, Malik P, Pattengale PK, et al. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood. (1999) 94:519–28. doi: 10.1182/blood.V94.2.519
50. Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. (2005) 106:3988–94. doi: 10.1182/blood-2005-05-2003
51. Bhatlekar S, Fields JZ, Boman BM. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. (2018) 2018:3569493. doi: 10.1155/2018/3569493
52. McSweeney PA, Rouleau KA, Storb R, Bolles L, Wallace PM, Beauchamp M, et al. Canine CD34: cloning of the cDNA and evaluation of an antiserum to recombinant protein. Blood. (1996) 88:1992–2003. doi: 10.1182/blood.V88.6.1992.bloodjournal8861992
53. Ide K, Matsuura S, Fujino Y, Ohno K, Tsujimoto H. Investigation of various methods for the cryopreservation of canine bone marrow-derived CD34(+) cells. J Vet Med Sci. (2008) 70:1211–7. doi: 10.1292/jvms.70.1211
54. Zhao Y, Chen X, Feng S. Autologous hematopoietic stem cell transplantation in acute myelogenous leukemia. Biol Blood Marrow Transplant. (2019) 25:e285–92. doi: 10.1016/j.bbmt.2019.04.027
55. Copelan EA, Chojecki A, Lazarus HM, Avalos BR. Allogeneic hematopoietic cell transplantation; the current renaissance. Blood Rev. (2019) 34:34–44. doi: 10.1016/j.blre.2018.11.001
56. Kobari L, Yates F, Oudrhiri N, Francina A, Kiger L, Mazurier C, et al. Human induced pluripotent stem cells can reach complete terminal maturation: in vivo and in vitro evidence in the erythropoietic differentiation model. Haematologica. (2012) 97:1795–803. doi: 10.3324/haematol.2011.055566
57. Ferreira AF, Calin GA, Picanço-Castro V, Kashima S, Covas DT, de Castro FA. Hematopoietic stem cells from induced pluripotent stem cells - considering the role of microRNA as a cell differentiation regulator. J Cell Sci. (2018) 131:203018. doi: 10.1242/jcs.203018
58. Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. (2017) 545:432–8. doi: 10.1038/nature22370
Keywords: surface antigen, stem cell, dog, CD34, bone marrow
Citation: Ayabe T, Hisasue M, Yamada Y, Nitta S, Kikuchi K, Neo S, Matsumoto Y, Horie R and Kawamoto K (2022) Characterisation of canine CD34+/CD45 diminished cells by colony-forming unit assay and transcriptome analysis. Front. Vet. Sci. 9:936623. doi: 10.3389/fvets.2022.936623
Received: 12 May 2022; Accepted: 26 July 2022;
Published: 12 September 2022.
Edited by:
Marxa L. Figueiredo, Purdue University, United StatesReviewed by:
Andrea Pires Dos Santos, Purdue University, United StatesCarlos Alberto Antunes Viegas, University of Trás-os-Montes and Alto Douro, Portugal
Copyright © 2022 Ayabe, Hisasue, Yamada, Nitta, Kikuchi, Neo, Matsumoto, Horie and Kawamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaharu Hisasue, aGlzYXN1ZUBhemFidS11LmFjLmpw
 Taro Ayabe
Taro Ayabe Masaharu Hisasue
Masaharu Hisasue Yoko Yamada1
Yoko Yamada1 Sakurako Neo
Sakurako Neo