- 1Department of Veterinary Medicine, University of Bari, Bari, Italy
- 2Department of Veterinary Microbiology, College of Veterinary Medicine, Michael Okpara University of Agriculture, Umudike, Nigeria
- 3Department of Microbiology and Immunology, National Research Centre, Veterinary Research Institute, Giza, Egypt
Canine parvovirus (CPV) and feline panleukopenia virus (FPV), now included in the unique species Carnivore protoparvovirus 1 (CPPV1), have been circulating in dogs and cats for several decades and are considered the causes of clinically important diseases, especially in young animals. While genetic evidence of the circulation of parvoviruses in Egyptian domestic carnivores has been provided since 2016, to date, all available data are based on partial fragments of the VP2 gene. This study reports the molecular characterization of CPPV strains from Egypt based on the full VP2 gene. Overall, 196 blood samples were collected from dogs and cats presented at veterinary clinics for routine medical assessment in 2019 in Egypt. DNA extracts were screened and characterized by real-time PCR. Positive samples were amplified by conventional PCR and then were sequenced. Nucleotide and amino acid changes in the sequences were investigated and phylogeny was inferred. Carnivore protoparvovirus DNA was detected in 18 out of 96 dogs (18.8%) and 7 of 100 cats (7%). Phylogenetic analyses based on the full VP2 gene revealed that 9 sequenced strains clustered with different CPV clades (5 with 2c, 2 with 2a, 1 with 2b, and 1 with 2) and 1 strain with the FPV clade. All three CPV variants were detected in dog and cat populations with a predominance of CPV-2c strains (7 of 18, 38.9%) in dog samples, thus mirroring the circulation of this variant in African, European, and Asian countries. Deduced amino acid sequence alignment revealed the presence of the previously unreported unique mutations S542L, H543Q, Q549H, and N557T in the Egyptian CPV-2c strains.
Introduction
Canine parvovirus (CPV) and feline panleukopenia virus (FPV) [species Carnivore protoparvovirus 1 (CPPV 1), genus Protoparvovirus, family Parvoviridae] have been circulating globally in domestic dog and cat populations for several decades (1, 2). These viruses are highly pathogenic in their hosts, especially in young puppies and kittens, causing a severe enteric disease and requiring systematic vaccination for its prevention (3). Parvoviruses are small (diameter of 25 nm), non-enveloped viruses infecting vertebrates and insects. The virion consists of a spherical capsid, which is composed by three proteins (VP1, VP2, and VP3), with VP2 forming two-thirds of the capsid and being responsible for host range and immune response (4). The genome is a positive-sense, single-stranded DNA (4.5–5.5 kb), with complex hairpin-like structures at the 5′ and 3′ ends. The coding region of the genome contains two major expression cassettes, with open reading frames (ORFs) on the left-hand side giving rise to non-structural (NS) proteins (ORF1), whereas mRNA populations responsible for translating structural protein (viral proteins; VPs) are transcribed from the right-hand cassette (ORF2) (1, 2, 5).
Feline panleukopenia virus has been known since 1928 (6, 7) and is genetically and antigenically similar to CPV (8). CPV was first identified in the late 1970s when severe hemorrhagic gastroenteritis and myocarditis were reported in puppies (9). The virus was initially designated CPV-2 to distinguish it from the genetically unrelated CPV type 1 (currently known as Carnivore bocaparvovirus 1), but nowadays, CPV-2 generally refers to the original strain (3). It is speculated that CPV-2 has evolved from FPV after crossing the species barriers by acquiring a few amino acid mutations in the VP2 protein (10, 11). Shortly after its emergence, CPV-2 started evolving, thereby generating three antigenic variants, namely, CPV-2a, 2b, and 2c, which spread and substituted the original strain (12–16). CPV has shown a higher mutation rate than FPV, with the capsid protein gene mutating faster than the NS regions of the genome (17, 18). Consequently, the hypervariable VP2 protein remains the focus in CPV characterization. The numerous mutations reported over the years are postulated to provide advantages to the virus in the form of antigenic variation, capsid stability, and improved receptor-binding capacity, thus extending the host range and increasing the pathogenicity of new variants of the virus (13, 15, 19). The ability of these viruses to infect several wild carnivores further complicates their control as spillover infections occur from these animals to domestic pets and vice versa (20–22). CPV has also been shown to have gained the ability to infect domestic cats (23), highlighting the need for constant surveillance of these carnivores alongside the canine species.
The first report on CPV in Egypt dates back to 1982 (24). Since then, all three variants of the virus have been reported along with FPV (25–28). However, the presence of these viruses was assessed by serological tools or by molecular amplification of short fragments of the VP2 gene, providing limited genetic information on the circulation of CPPVs in Egypt. In the present paper, we report the characterization of CPPV strains from Egyptian domestic carnivores on the basis of the analysis of the full VP2 gene.
Materials and methods
Sample collection
A total of 196 blood samples previously collected from dogs (n = 96) and cats (n = 100) presented at the veterinary clinics for bacteriological surveillance from August to September 2019 were included in this study.
Samples were briefly stored at −20°C at the collection points and subsequently transported under a cold chain to the Infectious Diseases Unit of the Department of Veterinary Medicine, University of Bari for analysis. The samples were obtained from Cairo (n = 125) and Giza (n = 71), Egypt (Figure 1), and general information regarding the animals were collected.
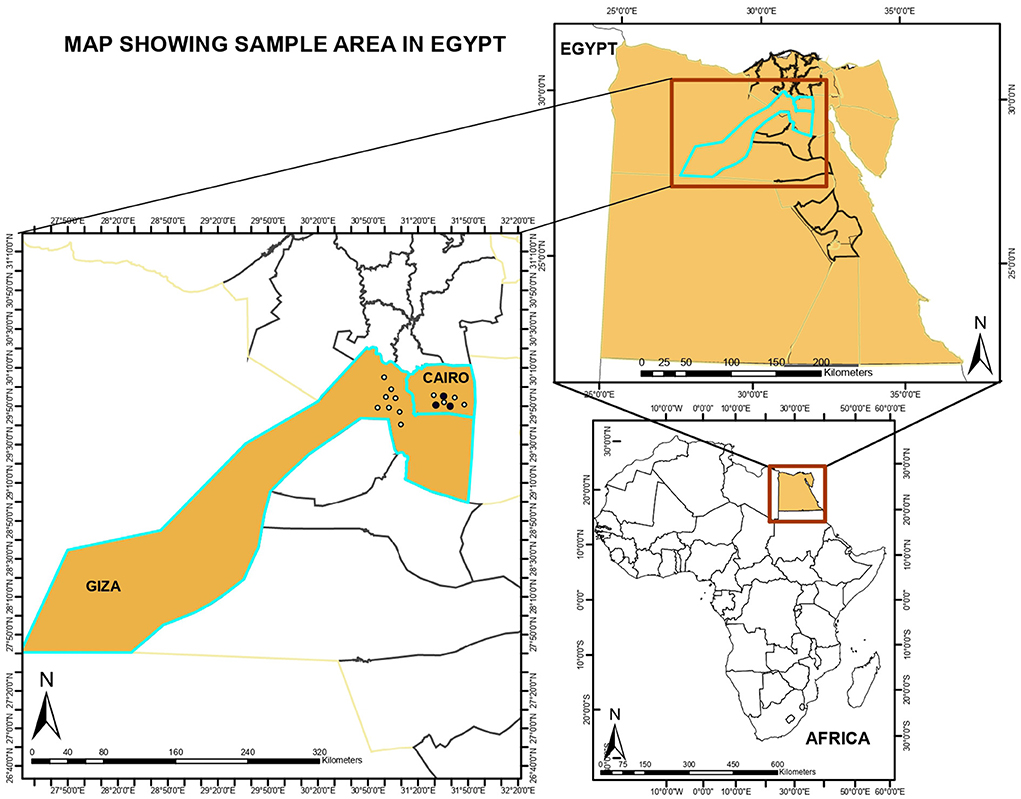
Figure 1. Shape files of study locations were obtained from the ArcGIS online map tools and imported for visualization into ArcGIS version 10.8.1 Redlands, CA: Environmental Systems Research Institute, Inc., 2020.
DNA extraction and screening for CPPV DNA
Viral DNA was extracted from the samples using the IndiSpin Pathogen Kit (Indical Bioscience GmbH, Leipzig, Germany), following the instructions of the manufacturer, and stored at −80°C until use. Quantification of extracted DNA was performed with the Fluorometric Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The quality of the extracted DNA was compared by measuring the concentration and purity using a UV Spectrophotometer (NanoDrop™ 1000, Thermo Fisher Scientific). Testing of samples was done in three steps, the first of which involved the screening for the presence of CPV/FPV DNA using a real-time PCR (qPCR) assay based on TaqMan technology (29). This was followed by the characterization of positive samples by minor groove binder (MGB) probe-based qPCR assays to differentiate CPV types 2a/2b and 2b/2c and to discriminate between CPV and FPV (30) (Supplementary Table 1). Real-time PCR assays were performed using iTaq™ Universal Probes Supermix (Bio-Rad Laboratories SRL, Segrate, Italy). To evaluate the sensitivity of qPCR assays, 10-fold serial plasmid dilutions (108-100 DNA copy numbers per μl) containing the VP2 portion targeted by qPCR assays were used as templates. The repeatability was determined using four different DNA concentrations of CPPV tested. Concentrations of the DNA standard 102, 103, 105, and 107 DNA copy numbers per μl were tested and analyzed by qPCR. A total of 10 samples per reaction were tested to estimate the intra- and inter-assay coefficients. For intra-assay variability, each dilution was analyzed in triplicate. To evaluate the inter-assay precision of the assay, each dilution was analyzed in different runs performed by two different laboratory technicians on different days. The coefficient of variation (CV) was determined following the formula: CV = [SD (Ct-value)/overall mean (Ct-value)] × 100.
Variable categorization and analyses
Data collated regarding clinical signs, age, and sampling location were sorted and inputted into Microsoft Excel (Microsoft Corporation, Redmond, WA, USA). Further, data were exported into SPSS (version 22; IBM Corp., Armonk, NY, USA), where descriptive and inferential statistical analyses were conducted. Age, clinical signs, and sampling location were categorized with corresponding cell values assigned in a “2 × 2” contingency matrix and the association between CPPV positivity and categorized variables was assessed by the chi-square test. p < 0.05 was considered statistically significant.
Mapping
Shape files of study locations were obtained from the ArcGIS online map tools and imported for visualization into ArcGIS version 10.8.1 Redlands, CA: Environmental Systems Research Institute, Inc., 2020 (Figure 1).
VP2 gene amplification and sequencing
VP2 gene was amplified in two overlapping fragments from samples that tested positive by qPCR, using primer sets previously described (17, 31) (Supplementary Table 1). Each 50 μl PCR reaction contained 5 μl of DNA extract, TaKaRa LA Taq TM Kit (Takara Bio Europe S.A.S. Saint-Germain-en-Laye, France), consisting of 24.5 μl of PCR grade water, 5 μl of 10× buffer, 5 μl of MgCl2 (25 mM), 1 μl of forward and reverse primers (50 μM), 8 μl of deoxynucleotide triphosphates (dNTPs) (2.5 mM), and 0.5 μl of Takara La Taq polymerase (5 U/μl). Initial denaturation was set at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 1 min, 30 s annealing at 59°C for 1 min and extension at 68°C for 2 min, and a final extension at 68°C for 10 min. A DNA extract of an FPV vaccine (Vanguard® Feline RCP, Zoetis) and nuclease-free water were included as positive control and blank, respectively. The PCR products were electrophoresed in a 1.5% agarose gel at 80 V for 40 min and the amplification bands were visualized on a Gel Doc™ EZ (Bio-Rad Laboratories SRL, Segrate, Italy), using Image LabTM software.
Sequencing and sequence analysis
Purification of PCR products was performed by QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany), followed by nucleotide (nt) sequencing in both directions by the Sanger method using BigDye 3.1 Ready Reaction Mix (Applied Biosystems), according to the instructions of the manufacturer. Generated reads were edited, and contigs were assembled using Geneious Prime version 2021.1 (Biomatters, Auckland, New Zealand). Related sequences were explored using web-based tools Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch) and FAST-All (FASTA; https://www.ebi.ac.uk/Tools/sss/fasta/nucleotide.html). The obtained sequences were aligned with reference CPPV sequences retrieved from the GenBank database by the multiple alignment using fast Fourier transform (MAFFT) algorithm (32).
Sequence submission
Nucleotide sequences of strains EGY/2019/39-122, EGY/2019/39-134, EGY/2019/39-161, EGY/2019/39-167, EGY/2019/39-168, EGY/2019/39-178, EGY/2019/39-200, EGY/2019/39-517, EGY/2019/39-549, and EGY/2019/39-566 used for phylogeny were deposited in the GenBank under the accession nos. OM937907, OM937916.
Phylogenetic analysis
The most appropriate model of evolution for phylogenetic analysis on the full VP2 gene of CPPV strains was evaluated using a jModelTest software (http://evomics.org/resources/software/molecular-evolution-software/modeltest/). The identified program settings for all partitions, under the Bayesian Information Criteria, included 5-character states (general time-reversible model), a proportion of invariant sites, and a discrete gamma distribution (6 categories) of rate variation across sites. Phylogenetic analyses were conducted using MrBayes 4 chains run for >1 million generations (33, 34) and a bootstrap analysis with 1,000 pseudoreplicated datasets.
Phylogenetic analyses using other evolutionary models (maximum likelihood and neighbor joining) were performed to compare the topology of the phylogenetic trees. Similar topologies with slight differences in bootstrap values at the nodes of the tree were observed. Accordingly, the Bayesian tree was retained.
Results
CPV diagnosis and statistical analysis
In the screening performed using a qPCR assay based on TaqMan technology, CPPV DNA was detected in a total of 25 animals consisting of 18 of 96 dogs (18.8%) and 7 of 100 cats (7%) (Table 1). The detection limit of the CPPV qPCR was 101 DNA copy numbers per μl. The qPCR assay expressed a high repeatability with CV within runs (intra-assay variability) and between runs (inter-assay variability) that ranged from 0.73 to 1.69% and 0.97 to 2.18%, respectively. Overall, the viral load of CPPV in samples ranged from 1.6 × 101 to 1.1 × 105 DNA copy numbers per μl (mean 7.3 × 103 DNA copy numbers per μl, median 2.1 × 103 DNA copy numbers per ml). In the MGB probe-based qPCR assays, the most frequently detected variant in dogs was CPV-2c (n = 7/18, 38.9%), followed by CPV-2a (n = 2/18, 11.1%), CPV-2b (1/18, 5.6%), and CPV-2 1/18 dog (5.6%). Due to lower viral load (mean 2.1 × 102 DNA copy numbers per μl, median 1.1 × 102 DNA copy numbers per μl), 7 of 18 CPV-positive samples (38.9%) identified in dogs could not be characterized by MGB probe-based qPCR assays. Out of a total of 7 cats, FPV and CPV-2a were identified in 3 (42.9%) and 2 (28.5%) cats, respectively. One cat (14.3%) tested positive for CPV-2b, while in another cat (14.3%), the concurrent presence of CPV-2a and−2c DNA was observed.
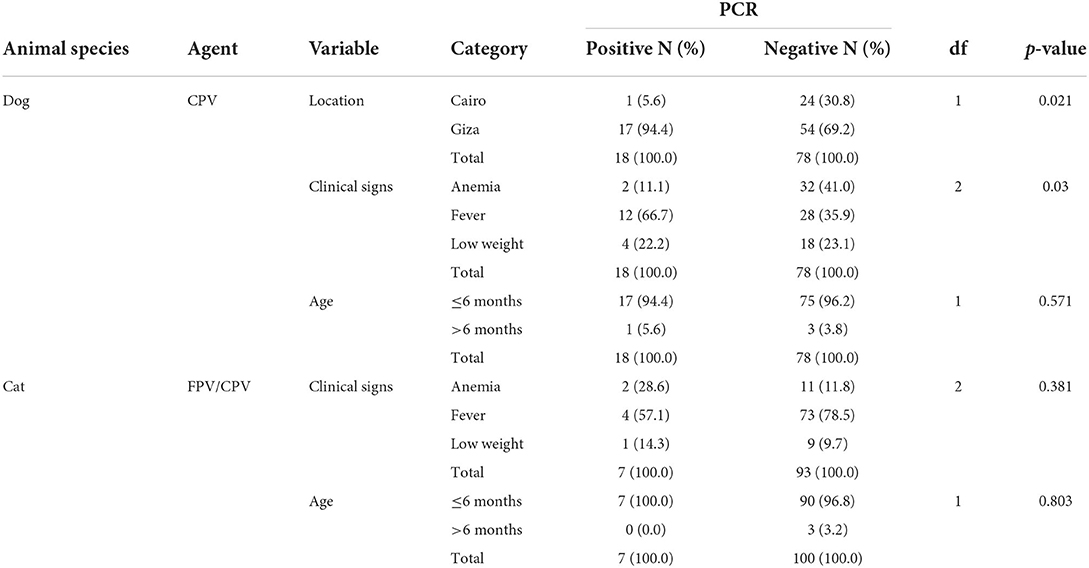
Table 1. Inferential statistics testing the association between socio-demographics, clinical signs, and disease outcome in the sampled dogs and cats.
The total mean age in weeks of the 100 cats and 96 dogs tested was 13.54 ± 0.5 and 13.69 ± 4.92, respectively. While all the cat samples tested were collected only from Cairo, a significantly higher number of CPV-positive dogs were reported for Giza in comparison to Cairo (p = 0.021). In addition, a significant association was found between the presence of CPPV-1 DNA and clinical signs, with fever identified as the most observed clinical sign in disease outcomes (p = 0.03). Similarly, fever was more consistently associated with feline FPV and CPV than anemia or low weight, although this association was not statistically significant (p > 0.05). All positive cat cases occurred in kittens of 6 months of age or under (n = 7), although without statistical significance (Table 1).
VP2 sequence and phylogenetic analyses
Out of 25 CPPV strains detected by qPCR, 10 full VP2 sequences (7 from dog samples and 3 from cat samples) were successfully amplified by conventional PCR and sequenced. BLAST and FASTA analyses revealed a high nt identity with other reference sequences from the GenBank database (99.6–100%) while identity within the sequences from this study was 97.8–100%. By sequence comparison of amino acid (aa) residues of FPV strains identified in the present study with cognate reference sequences, no aa substitutions were observed (Table 2). Conversely, eleven non-synonymous mutations were found in the CPV sequences from this study, compared to cognate reference sequences used in the phylogenetic tree (Table 2). CPV-2 strain EGY/2019/39-122 (OM937907) displayed three aa substitutions at positions 219 (L → V), 386 (Q → K), and 418 (I → T), while CPV2a strain EGY/2019/39-200 (OM937913) exhibited two aa substitutions at positions 13 (P → S) and 440 (T → A). CPV-2c strain EGY/2019/39-167 (OM937910) displayed two aa substitutions at positions 549 (Q → H) and 577 (N → T). Unique aa substitutions were observed in the CPV-2b strain EGY/2019/39-517 (OM937914) at position 440 (T → A), in the CPV-2c strain EGY/2019/dog/39-134 (OM937908) at position 542 (S → L), and in the CPV-2c strain EGY/2019/39-168 at position 543 (H → Q). All the CPV variants identified in Egyptian samples displayed aa substitution S → A at position 297.

Table 2. Summary table of nucleotide and amino acid substitutions in the VP2 region of carnivore protoparvoviruses (canine parvovirus 2, CPV-2 and feline panleukopenia virus, FPV) detected in blood samples of dogs and cats in Egypt as compared to reference strains used for the phylogeny (Figure 2).
Phylogenetic analysis revealed that 5 strains (EGY/2021/39-134, EGY/2021/39-168, EGY/2021/39-161, EGY/2021/39-167, and EGY/2021/39-178) segregated into the CPV-2c clade together with European, Asian, and Nigerian strains. Strain EGY/2021/39-517 clustered with Turkish and Thai CPV-2b strain while with strain EGY/2021/39-200, characterized as CPV-2a, was immediately basal to the clade. Strain EGY/2021/39-549 segregated into CPV-2a clade together with other strains identified in China, Middle East, and Uruguay. Strain EGY/2021/39-122 clustered with CPV-2 strains retrieved from the USA, Italy, China, and India, while strain EGY/2021/39-566 segregated with FPV strains identified in Thailand (Figure 2).
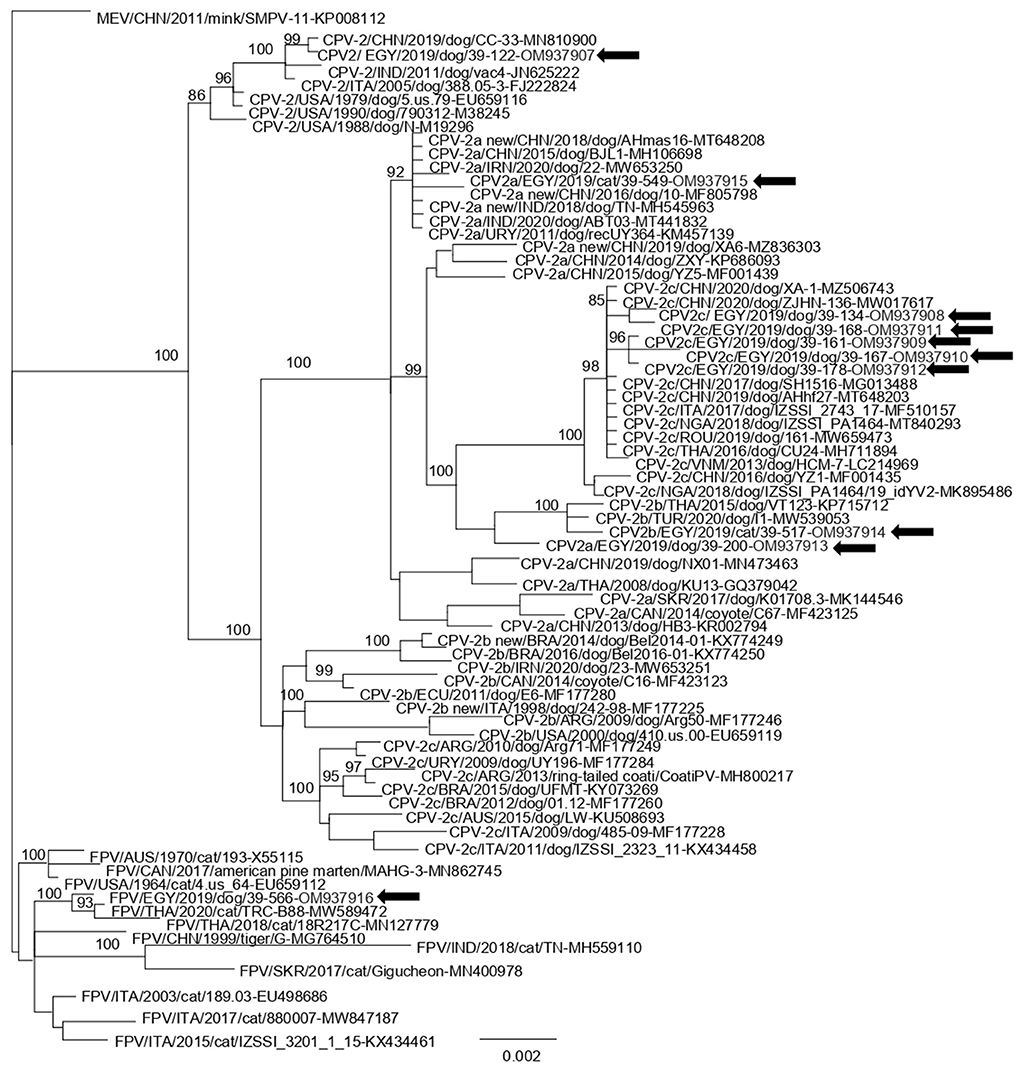
Figure 2. Bayesian open reading frame (ORF)2-based phylogenetic tree of Carnivore protoparvovirus 1. The tree was elaborated using a 1,755 nt long alignment of the ORF2 sequence of the Egyptian canine parvovirus (CPV) and feline panleukopenia virus (FPV) strains identified in this study and the cognate sequences of Carnivore protoparvovirus 1 strains retrieved from the GenBank database. The posterior output of the tree was derived using a general time-reversible model, a proportion of invariable sites, a gamma distribution of rate variation across sites, and a subsampling frequency of 1,000. Posterior probability values >95% are indicated at the tree nodes. The black arrows indicate the Egyptian strains generated in this study. The scale bar indicates the number of nt substitutions per site.
Discussion
Carnivore protoparvoviruses have maintained their status as a major cause of mortality, especially in juvenile dogs and cats globally, despite decades of vaccine use. Mutations in the hypervariable capsid gene VP2 are of importance as they are known to influence virus/receptor binding, thereby playing key roles as determinants of host range and antigenicity (10, 35–38). These mutant viruses also tend to acquire evolutive advantages, thriving, spreading, and replacing existing variants, thus, driving the dynamic epidemiology of CPV across countries. The use of blood samples in this study rather than feces cannot account for the overall low detection rate, as in infected animals, viremia is longer lasting than fecal shedding (39), and CPPVs have been frequently detected also in the blood of healthy dogs and cats (40). Previous studies in Egypt based their selection criteria on the development of diarrhea, followed by positivity to rapid tests, resulting in the prevalence of 84% and higher (28, 41).
This study confirms the circulation of all three CPV variants in domestic dog and cat populations from Egypt. CPV is known to infect cats and has been suggested to contribute to genetic diversity of CPPV as a consequence of infecting both dogs and cats (23, 42). CPV infections in cats are usually mild (42) but clinical cases resembling to feline panleukopenia have been reported (23). Cats have been also found to shed CPV without clinical signs (43, 44), serving as reservoir hosts. The present study, however, cannot rule out the role of other blood-borne pathogens in the induction of fever in the animals tested. A lack of vaccination and of history of gastroenteric disease in the animals limits the interpretation of the clinical significance of this study.
A significantly higher number of CPV-positive dogs were reported for Giza (17/71, 23.9%) as compared to Cairo (1/25, 0.04%) (Table 1). While Cairo is more densely human-populated, Giza is home to the popular pyramids and attracts a huge number of tourists from around the world throughout the year. The intensive movements of people (and their pets) from diverse origins might favor the introduction of new pathogens into the region, possibly through fomites. In addition, the complexity of the host immunity response against the CPPV vaccine cannot rule out the possibility of a vaccinated animal getting infected following vaccination or shedding wild-type virus without clinical signs.
A total of 10 CPPV strains have been characterized in this study. The CPV-2 strain EGY/2019/39-122 was 100% identical to isolate CC-33 (MN810900) identified from a dog in China in 2019. CPV-2 has been sporadically detected in other studies, usually as a consequence of recent vaccination (45, 46), since the original strain is no longer circulating in the field, but it is still contained in a number of vaccines (3). The CPV-2 strain displayed the presence of Val-219 and Lys-386 as also observed in the VP2 of the Nobivac® vaccine (C3) (MG264079) (47), and Thr-418 has been previously described in CPPV strains from domestic and wild carnivores (20, 48).
CPV-2a strain EGY/2019/dog/39-200 displayed the mutation S13P consistently reported in Italy in the last decades (48–50). Both CPV-2a strain EGY/2019/dog/39-200 (OM937913) and CPV-2b strain EGY/2019/cat/39-517 (OM937914) displayed the mutation T440A and were closely related to the isolates from Turkey and Thailand (51, 52). The T440A mutation was also prevalent in the CPV-2a strains previously identified in Egypt (53). CPV variants displaying VP2 changes F267Y, Y342I, and T440A are considered immune escape mutants, which are likely emerged due to vaccine pressure, with the role of the 267 mutation still unclear although it is an unexposed residue (37, 46). Residues 324 and 440 are located next to the spike residues 423 and 427, respectively (37).
CPV-2c was the predominant variant circulating in domestic dogs in Egypt, in contrast to earlier reports that accounted for a limited circulation of CPV-2c in this country (28, 53). The CPV-2c mutant detected has been also reportedly spreading in Europe, Asia, and Africa (54–63). This variant was recently detected in Nigeria (64) and is widespread in this country (63, 64). Considering the rapid spreading of this CPV-2c mutant, and Egypt being a touristic country, a predominance of this variant is expected in the next few years.
Amino acid substitutions observed in the CPV2c strains EGY/2019/39-21-134 (S542L), EGY/2019/39-167 (Q549H, N557T), and EGY/2019/39-168 (H543Q) were unique and have not been previously reported.
All CPV variants identified in this study displayed a VP2 with Ala-297, a recent widespread mutation (S → A) due to host adaptation (31, 65–70). The residue 297 is under strong positive selection pressure (68) and mutants displaying such a change have been considered a subvariant of CPV-2a/2b (46).
The FPV sequence from this study was 99.7% identical to FPV TRC-B88/TH/2020, which had been detected in the brain of a cat in Thailand. Overall, the FPV genome has a lower mutation rate than CPV (18), hence the observation of nt changes with no effect on the VP2 sequence is more frequent in the former.
A more extensive epidemiological surveillance is needed in domestic carnivores from Egypt and other African countries in order to better understand the evolution and variability of CPPV in geographic areas where the epidemiological data of these viruses are still scarce.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was reviewed and approved by Medical research Ethics Committee of the National Research Centre, Cairo, Egypt (approval number 6211022021). Written informed consent was obtained from the owners for the participation of their animals to this study.
Author contributions
ND, CB, and GL designed the experiment. LN, CD, EO, AZ, and PC carried out the experiment. LN, GL, and PC wrote the manuscript. LN, GL, FE, and GG participated in the data analysis. ND and CB reviewed the manuscript. All authors read and approved the manuscript before submission.
Funding
This study was supported by grants from the Italian Ministry of Health: Ricerca Corrente 2019 NGS e diagnostica molecolare in Sanità Animale: Fast D2, recipient ND.
Acknowledgments
The authors thank Carlo Armenise and Arturo Gentile for their excellent technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.932247/full#supplementary-material
References
1. Chung HC, Kim SJ, Nguyen VG, Shin S, Kim JY, Lim SK, et al. New genotype classification and molecular characterization of canine and feline parvoviruses. J Vet Sci. (2020) 21:e43. doi: 10.4142/jvs.2020.21.e43
2. Penzes JJ, Soderlund-Venermo M, Canuti M, Eis-Hubinger AM, Hughes J, Cotmore SF, et al. Reorganizing the family parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Arch Virol. (2020) 165:2133–46. doi: 10.1007/s00705-020-04632-4
3. Decaro N, Buonavoglia C, Barrs V. Canine parvovirus vaccination and immunisation failures: are we far from disease eradication? Vet Microbiol. (2020) 247:108760. doi: 10.1016/j.vetmic.2020.108760
4. Muzyczka N, Berns KI. Parvoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology, 4th Edn. Philadephia, PA: Lippincott Williams & Wilkins (2001). p. 2327–59.
5. Reed AP, Jones EV, Miller TJ. Nucleotide sequence and genome organization of canine parvovirus. J Virol. (1988) 62:266–76. doi: 10.1128/jvi.62.1.266-276.1988
6. Hindle E, Findlay G. Studies on feline distemper. J Comp Pathol Ther. (1932) 45:11–26. doi: 10.1016/S0368-1742(32)80002-0
7. Verge J, Christoforoni N. La gastroenterite infectieuse des chats; Est-Elle Due À Un virus filtrable. CR Seances Soc Biol Fil. (1928) 99:312.
8. Stuetzer B, Hartmann K. Feline parvovirus infection and associated diseases. Vet J. (2014) 201:150–5. doi: 10.1016/j.tvjl.2014.05.027
9. Appel MJ, Scott FW, Carmichael LE. Isolation and immunisation studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Vet Rec. (1979) 105:156–9. doi: 10.1136/vr.105.8.156
10. Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR. the natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. (2003) 77:1718–26. doi: 10.1128/JVI.77.3.1718-1726.2003
11. Truyen U. Emergence and recent evolution of canine parvovirus. Vet Microbiol. (1999) 69:47–50. doi: 10.1016/S0378-1135(99)00086-3
12. Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, et al. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. (2001) 82 (Pt. 12):3021–5. doi: 10.1099/0022-1317-82-12-3021
13. Carmichael LE. An annotated historical account of canine parvovirus. J Vet Med B Infect Dis Vet Public Health. (2005) 52:303–11. doi: 10.1111/j.1439-0450.2005.00868.x
14. Decaro N, Buonavoglia C. Canine parvovirus—a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol. (2012) 155:1–12. doi: 10.1016/j.vetmic.2011.09.007
15. Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol. (1991) 65:6544–52. doi: 10.1128/jvi.65.12.6544-6552.1991
16. Parrish CR, O'Connell PH, Evermann JF, Carmichael LE. Natural variation of canine parvovirus. Science. (1985) 230:1046–8. doi: 10.1126/science.4059921
17. Decaro N, Desario C, Miccolupo A, Campolo M, Parisi A, Martella V, et al. Genetic analysis of feline panleukopenia viruses from cats with gastroenteritis. J Gen Virol. (2008) 89 (Pt 9):2290–8. doi: 10.1099/vir.0.2008/001503-0
18. Hoelzer K, Parrish CR. The emergence of parvoviruses of carnivores. Vet Res. (2010) 41:39. doi: 10.1051/vetres/2010011
19. Wang J, Lin P, Zhao H, Cheng Y, Jiang Z, Zhu H, et al. Continuing evolution of canine parvovirus in China: isolation of novel variants with an Ala5gly mutation in the Vp2 Ptein. Infect Genet Evol. (2016) 38:73–8. doi: 10.1016/j.meegid.2015.12.009
20. Ndiana LA, Lanave G, Desario C, Berjaoui S, Alfano F, Puglia I, et al. Circulation of diverse protoparvoviruses in wild carnivores, Italy. Transbound Emerg Dis. (2021) 68:2489–502. doi: 10.1111/tbed.13917
21. Vieira FV, Hoffmann DJ, Fabri CU, Bresciani KD, Gameiro R, Flores EF, et al. Circulation of canine parvovirus among dogs living in human-wildlife interface in the atlantic forest biome, Brazil. Heliyon. (2017) 3:e00491. doi: 10.1016/j.heliyon.2017.e00491
22. Zaccaria G, Malatesta D, Scipioni G, Di Felice E, Campolo M, Casaccia C, et al. Circovirus in domestic and wild carnivores: an important opportunistic agent? Virology. (2016) 490:69–74. doi: 10.1016/j.virol.2016.01.007
23. Decaro N, Buonavoglia D, Desario C, Amorisco F, Colaianni ML, Parisi A, et al. Characterisation of canine parvovirus strains isolated from cats with feline panleukopenia. Res Vet Sci. (2010) 89:275–8. doi: 10.1016/j.rvsc.2010.03.001
24. Bucci T, Botros B, El Molla M. Canine parvovirus infection: a brief review and report of first cases in Egypt. J Egypt Vet Med Assoc. (1982) 41:21–5.
25. Abdel-Rhman A, El Balkemy F, Abouzeid N, Edries S. Canine parvo enteritis infection in Egypt: isolation, molecular characterization and sequencing. Adv Anim Vet Sci. (2019) 7:117–22. doi: 10.17582/journal.aavs/2019/7.s2.117.122
26. AL-Hosary AA. Detection and molecular characterization of parvovirus serotypes in Egypt. J Adv Vet Res. (2018) 8:79–83. Available online at: https://advetresearch.com/index.php/AVR/article/view/322 (accessed July, 2022).
27. El-Neshwy W, El-Zahar H, Morsi A, Shety T. Molecular and phylogenetic analysis of canine parvovirus variants (2a-2b-2c) in Egypt. Res J Vet Pract. (2019) 7:74–82.
28. Elbaz E, El-Tholoth M, Abo Elfadl EA, Mosad SM. Molecular investigation on the presence of canine parvovirus in Egypt. Comp Immunol Microbiol Infect Dis. (2021) 74:101576. doi: 10.1016/j.cimid.2020.101576
29. Decaro N, Elia G, Martella V, Desario C, Campolo M, Trani LD, et al. A real-time Pcr assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet Microbiol. (2005) 105:19–28. doi: 10.1016/j.vetmic.2004.09.018
30. Decaro N, Elia G, Martella V, Campolo M, Desario C, Camero M, et al. Characterisation of the canine parvovirus type 2 variants using minor groove binder Pbe technology. J Virol Methods. (2006) 133:92–9. doi: 10.1016/j.jviromet.2005.10.026
31. Ikeda Y, Mochizuki M, Naito R, Nakamura K, Miyazawa T, Mikami T, et al. Predominance of canine parvovirus (Cpv) in unvaccinated cat populations and emergence of new antigenic types of Cpvs in cats. Virology. (2000) 278:13–9. doi: 10.1006/viro.2000.0653
32. Katoh K, Misawa K, Kuma K, Miyata T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. (2002) 30:3059–66. doi: 10.1093/nar/gkf436
33. Huelsenbeck JP, Ronquist F. Mrbayes: bayesian inference of phylogenetic trees. Bioinformatics. (2001) 17:754–5. doi: 10.1093/bioinformatics/17.8.754
34. Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. (2003) 19:1572–4. doi: 10.1093/bioinformatics/btg180
35. Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. Structure determination of feline panleukopenia virus empty particles. Proteins. (1993) 16:155–71. doi: 10.1002/prot.340160204
36. Chang SF, Sgro JY, Parrish CR. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol. (1992) 66:6858–67. doi: 10.1128/jvi.66.12.6858-6867.1992
37. Goetschius DJ, Hartmann SR, Organtini LJ, Callaway H, Huang K, Bator CM, et al. High-Resolution asymmetric structure of a fab-virus complex reveals overlap with the receptor binding site. Proc Natl Acad Sci USA. (2021) 118:e2025452118. doi: 10.1073/pnas.2025452118
38. Llamas-Saiz AL, Agbandje-Mckenna M, Parker JS, Wahid A, Parrish CR, Rossmann MG. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology. (1996) 225:65–71. doi: 10.1006/viro.1996.0575
39. Decaro N, Buonavoglia C. Canine parvovirus post-vaccination shedding: interference with diagnostic assays and correlation with host immune status. Vet J. (2017) 221:23–4. doi: 10.1016/j.tvjl.2017.01.020
40. Marenzoni ML, Antognoni MT, Baldelli F, Miglio A, Stefanetti V, Desario C, et al. Detection of parvovirus and herpesvirus DNA in the blood of feline and canine blood donors. Vet Microbiol. (2018) 224:66–9. doi: 10.1016/j.vetmic.2018.08.030
41. Zaghawa A, Abualkhier M. Genetic and molecular typing of canine parvovirus strains circulating in symptomatic dogs in Egypt. J Curr Vet Res. (2019) 1:75–85. doi: 10.21608/jcvr.2019.36556
42. Battilani M, Balboni A, Ustulin M, Giunti M, Scagliarini A, Prosperi S. Genetic complexity and multiple infections with more parvovirus species in naturally infected cats. Vet Res. (2011) 42:43. doi: 10.1186/1297-9716-42-43
43. Balboni A, Bassi F, De Arcangeli S, Zobba R, Dedola C, Alberti A, et al. Molecular analysis of carnivore protoparvovirus detected in white blood cells of naturally infected cats. BMC Vet Res. (2018) 14:41. doi: 10.1186/s12917-018-1356-9
44. Carrai M, Decaro N, Van Brussel K., Dall'Ara P, Desario C, Fracasso M, et al. Canine parvovirus is shed infrequently by cats without diarrhoea in multi-cat environments. Vet Microbiol. (2021) 261:109204. doi: 10.1016/j.vetmic.2021.109204
45. Decaro N, Desario C, Elia G, Campolo M, Lorusso A, Mari V, et al. Occurrence of severe gastroenteritis in pups after canine parvovirus vaccine administration: a clinical and laboratory diagnostic dilemma. Vaccine. (2007) 25:1161–6. doi: 10.1016/j.vaccine.2006.10.020
46. Zhou P, Zeng W, Zhang X, Li S. The genetic evolution of canine parvovirus–a new perspective. PLoS ONE. (2017) 12:e0175035. doi: 10.1371/journal.pone.0175035
47. De la Torre D, Mafla E, Puga B, Erazo L, Astolfi-Ferreira C, Ferreira AP. Molecular characterization of canine parvovirus variants (CPV-2a, CPV-2b, and CPV-2c) based on the VP2 gene in affected domestic dogs in Ecuador. Vet World. (2018) 11:480. doi: 10.14202/vetworld.2018.480-487
48. Dei Giudici S, Cubeddu T, Giagu A, Sanna G, Rocca S, Oggiano A. First molecular characterization of canine parvovirus strains in Sardinia, Italy. Arch Virol. (2017) 162:3481–6. doi: 10.1007/s00705-017-3457-3
49. Tucciarone CM, Franzo G, Mazzetto E, Legnardi M, Caldin M, Furlanello T, et al. Molecular insight into Italian canine parvovirus heterogeneity and comparison with the worldwide scenario. Infect Genet Evol. (2018) 66:171–9. doi: 10.1016/j.meegid.2018.09.021
50. Battilani M, Modugno F, Mira F, Purpari G, Di Bella S, Guercio A, et al. Molecular epidemiology of canine parvovirus type 2 in Italy from 1994 to 2017: recurrence of the CPV-2b variant. BMC Vet Res. (2019) 15:393. doi: 10.1186/s12917-019-2096-1
51. Temizkan MC, Temizkan SS. The first whole CPV genome and accidentally found CPV-2c strain in Turkey. In: Proceedings of 3rd Euroscicon Conference on Virology. Edinburgh (2021). doi: 10.17582/journal.rjvp/2019/7.4.74.82
52. Inthong N, Kaewmongkol S, Meekhanon N, Sirinarumitr K, Sirinarumitr T. Dynamic evolution of canine parvovirus in Thailand. Vet World. (2020) 13, 245. doi: 10.14202/vetworld.2020.245-255
53. Etman R, Safwat M, Khodeir H. Molecular characterization of canine parvovirus-2 (Cpv-2) in dogs in Egypt. Adv Anim Vet Sci. (2021) 9:933–40. doi: 10.17582/journal.aavs/2021/9.6.933.940
54. Balboni A, Niculae M, Di Vito S, Urbani L, Terrusi A, Muresan C, et al. The detection of canine parvovirus type 2c of asian origin in dogs in romania evidenced its progressive worldwide diffusion. BMC Vet Res. (2021) 17:206. doi: 10.1186/s12917-021-02918-6
55. Charoenkul K, Tangwangvivat R, Janetanakit T, Boonyapisitsopa S, Bunpapong N, Chaiyawong S, et al. Emergence of canine parvovirus type 2c in domestic dogs and cats from Thailand. Transbound Emerg Dis. (2019) 66:1518–28. doi: 10.1111/tbed.13177
56. Hao X, He Y, Wang C, Xiao W, Liu R, Xiao X, et al. The increasing prevalence of CPV-2c in domestic dogs in China. PeerJ. (2020) 8:e9869. doi: 10.7717/peerj.9869
57. Hu W, Zheng L, Xu X, Liu Q, Ji J, Yao L, et al. Genetic Characterisation and Local genotypes of canine parvovirus strains collected from pet dogs in central and eastern China during 2018-2019. J Vet Res. (2020) 64:477–86. doi: 10.2478/jvetres-2020-0076
58. Mira F, Purpari G, Lorusso E, Di Bella S, Gucciardi F, Desario C, et al. Introduction of Asian canine parvovirus in Europe through dog importation. Transbound Emerg Dis. (2018) 65:16–21. doi: 10.1111/tbed.12747
59. Mira F, Purpari G, Di Bella S, Colaianni ML, Schiro G, Chiaramonte G, et al. Spreading of canine parvovirus type 2c mutants of asian origin in southern Italy. Transbound Emerg Dis. (2019) 66:2297–304. doi: 10.1111/tbed.13283
60. Moon BY, Jang J, Kim SH, Kim YH, Lee HK, So B, et al. Genetic characterization of canine parvovirus type 2c from domestic dogs in Korea. Transbound Emerg Dis. (2020) 67:1645–53. doi: 10.1111/tbed.13501
61. Nguyen Manh T, Piewbang C, Rungsipipat A, Techangamsuwan S. Molecular and phylogenetic analysis of Vietnamese canine parvovirus 2C originated from dogs reveals a new Asia-IV clade. Transbound Emerg Dis. (2021) 68:1445–53. doi: 10.1111/tbed.13811
62. Qi S, Zhao J, Guo D, Sun D. A Mini-Review on the epidemiology of canine parvovirus in China. Front Vet Sci. (2020) 7:5. doi: 10.3389/fvets.2020.00005
63. Ogbu KI, Mira F, Purpari G, Nwosuh C, Loria GR, Schirò G, et al. nearly full-length genome characterization of canine parvovirus strains circulating in Nigeria. Transbound Emerg Dis. (2020) 67:635–47. doi: 10.1111/tbed.13379
64. Ndiana L, Odaibo G, Olaleye D. Molecular characterization of canine parvovirus from domestic dogs in Nigeria: introduction and spread of a CPV-2c mutant and replacement of older CPV-2a by the “new CPV-2a” strain. Virus Dis. (2021) 32:361–8. doi: 10.1007/s13337-021-00689-0
65. Battilani M, Scagliarini A, Tisato E, Turilli C, Jacoboni I, Casadio R, et al. Analysis of Canine parvovirus sequences from wolves and dogs isolated in Italy. J Gen Virol. (2001) 82 (Pt. 7):1555–60. doi: 10.1099/0022-1317-82-7-1555
66. Geng Y, Guo D, Li C, Wang E, Wei S, Wang Z, et al. Co-Circulation of the rare Cpv-2c with unique Gln370arg substitution, new Cpv-2b with unique Thr440ala substitution, and New Cpv-2a with high prevalence and variation in Heilongjiang Province, Northeast China. PLoS ONE. (2015) 10:e0137288. doi: 10.1371/journal.pone.0137288
67. Lin CN, Chien CH, Chiou MT, Chueh LL, Hung MY, Hsu HS. Genetic characterization of type 2a canine parvoviruses from Taiwan reveals the emergence of an Ile324 mutation in Vp2. Virol J. (2014) 11:39. doi: 10.1186/1743-422X-11-39
68. Pereira CA, Leal ES, Durigon EL. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect Genet Evol. (2007) 7:399–409. doi: 10.1016/j.meegid.2006.03.007
69. Steinel A, Munson L, Van Vuuren M, Truyen U. Genetic characterization of feline parvovirus sequences from various carnivores. J Gen Virol. (2000) 81:345–50. doi: 10.1099/0022-1317-81-2-345
Keywords: Carnivore protoparvovirus 1, dogs, cats, Egypt, molecular characterization, phylogeny
Citation: Ndiana LA, Lanave G, Zarea AAK, Desario C, Odigie EA, Ehab FA, Capozza P, Greco G, Buonavoglia C and Decaro N (2022) Molecular characterization of carnivore protoparvovirus 1 circulating in domestic carnivores in Egypt. Front. Vet. Sci. 9:932247. doi: 10.3389/fvets.2022.932247
Received: 29 April 2022; Accepted: 27 June 2022;
Published: 22 July 2022.
Edited by:
Lester J. Perez, Abbott, United StatesReviewed by:
Chutchai Piewbang, Chulalongkorn University, ThailandAhsan Naveed, University of Kentucky, United States
Copyright © 2022 Ndiana, Lanave, Zarea, Desario, Odigie, Ehab, Capozza, Greco, Buonavoglia and Decaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianvito Lanave, Z2lhbnZpdG8ubGFuYXZlQHVuaWJhLml0
 Linda A. Ndiana
Linda A. Ndiana Gianvito Lanave
Gianvito Lanave Aya A. K. Zarea
Aya A. K. Zarea Costantina Desario
Costantina Desario Eugene A. Odigie1
Eugene A. Odigie1 Grazia Greco
Grazia Greco Canio Buonavoglia
Canio Buonavoglia Nicola Decaro
Nicola Decaro