- State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
Porcine reproductive and respiratory syndrome (PRRS) is an infectious viral disease that causes great harm to the pig industry. PRRS virus (PRRSV), the causative agent of PRRS, is characterized by severe reproductive failure and respiratory confusion. This study performed a cross-sectional investigation of PRRSV seroprevalence and collected 14,134 serum samples in pig farms without PRRSV vaccination from 12 provinces and two cities in China from 2017 to 2021 to detect PRRSV antibodies by enzyme-linked immunosorbent assay (ELISA). The apparent and true PRRSV antibody prevalence was estimated and compared based on the Clopper-Pearson method and Pearson chi-square test, respectively. Risk factors associated with the PRRSV serological status of pig farms were analyzed through univariate and multivariable logistic regression analysis. An automatic autoregressive integrated moving average (ARIMA) model procedure was used for time-series analysis for PRRSV seroprevalence. Spatial clusters of high PRRSV seroprevalence were detected by SaTScan software. The total true PRRSV seroprevalence of the animal level was 62.56% (95% confidence interval [CI]: 61.74–63.37%). Additionally, 286 out of 316 pig farms were positive for PRRSV antibodies at the herd level. Pig farms without pseudorabies virus (PRV) infection were 5.413 (95% CI: 1.977–17.435) times more likely to be PRRSV antibody positive than those with PRV. Identically, the possibility of pig farms being PRRSV antibody positive before an African swine fever (ASF) outbreak was 3.104 (95% CI: 1.122–10.326) times more than after ASF. The odd ratio values of medium and large pig farms with PRRSV infection are 3.076 (95% CI: 1.005–9.498) and 6.098 (95% CI: 1.814–21.290). A fluctuant decline pattern for PRRSV prevalence was observed in the temporal analysis. Three significant clusters of high PRRSV seroprevalence were first detected in China, covering a time frame from January 2018 to September 2018, which reveals high PRRSV prevalence before the outbreak of ASF. These findings show the epidemic situation and spatial-temporal distribution of PRRSV infection in China in recent years and could help develop reasonable measures to prevent PRRSV infection.
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is an infectious viral disease that is caused by PRRSV and characterized by reproductive failure in sows and severe respiratory confusion and mortality in young pigs. It is responsible for substantial economic losses in the global swine industry (1–3). PRRSV is a single-stranded, positive-sense RNA virus from Arteriviridae, genus Arterivirus, that contains ≈15 kb nucleotides and encodes >10 open reading frames (ORFs) (4, 5). PRRSV had two main genotypes, namely European type (type 1) and North American type (type 2), of which North American type is the major epidemic genotype in China (6–9).
The first PRRSV strain of China was isolated and identified from aborted fetuses in 1996 that belonged to the North American type (8, 10). A highly pathogenic PRRSV (HP-PRRSV) strain that was featured with the discontinuous deletion of 30 amino acids in the Nsp2 gene and 20% mortality in pigs had occurred and resulted in the deaths of one million pigs in China in 2006 (11, 12). In recent years, new PRRSV field isolates with low mortality show high similarity in gene sequences with NADC30 strains isolated in America in 2008 and have been successively reported and defined as NADC30-like strains in China since 2014 (13, 14). The HP-PRRSV and NADC30-like strains are the primary epidemic strains that circulate in pig farms in China (9). Three hundred and sixty-five PRRSV strains were isolated by Jiang et al. (15) from 1996 to 2017 in China and used to analyze evolution and genome, which demonstrated that the HP-PRRSV, NADC30-like, and intermediated PRRSV were the major epidemic strains.
Infection and transmission of PRRSV from infected pigs to susceptible pigs can horizontally or vertically emerge through direct or indirect contact within the herd (6). Pitkin et al. even reported that PRRSV could be disseminated by aerosol with a distance of ≈120 m (16). The high variability in the PRRSV genome and diversity in PRRSV transmission modes causes tremendous challenges in controlling PRRSV infection in fields. Therefore, knowing the risk factors and spatial-temporal distribution of PRRSV infection is necessary to contain and eradicate PRRS illness.
Risks of PRRSV infection in pig herds increased with increased pig farm size, absence of purchased gilts quarantine, and semen purchased outside for artificial insemination (17). Fablet et al. (18) found that setting the temperature low in fattening rooms and with pigs, which coinfect with Mycoplasma hyopneumoniae and H1N2 swine influenza viruses, could raise herds' PRRSV seropositive possibility. Spatial analysis of PPRSV type 1 and 2 seroprevalences executed in Denmark statistically significantly detected clusters from 2007 to 2010 with higher PRRSV type 1 seroprevalence (19). However, related research on risk factors and spatial-temporal distribution of PRRSV infection are highly inadequate in China. Meanwhile, the ASF outbreak across China has caused a massive loss of pig populations, threatened the stability of the meat supply chain, and changed the feeding and management mode of pig farms, which is important for China's pig industry (20, 21). Therefore, knowing about the epidemic situation of PRRSV infection and providing reference information for policymakers related to future PRRSV control in China is urgently demanded after the ASF outbreak in 2018 (22).
Materials and Methods
Sampling Region and Population
This study included 14,134 serum samples from 316 farms without PRRSV vaccination located in 12 provinces and two cities in China from 2017 to 2021. Collated serum samples were tested for PRRSV antibodies using an enzyme-linked immunosorbent assay (ELISA). Then, attained data were used to seek risk factors associated with the PRRSV serological status of pig farms and investigate spatial-temporal PRRSV seroprevalence clusters in China before and after the ASF outbreak. The sampling regions successively covered all seven geographical areas of China, namely central (Henan, Hubei, and Hunan provinces), eastern (Shandong, Jiangsu, Anhui, Jiangxi, Fujian provinces, and Shanghai city), northeast China (Liaoning province), south (Guangdong province), southwest (Sichuan province), northwest (Shaanxi province), and northern (Tianjin city) districts. These areas are ≈2.3 million square kilometers located in longitudes of 97°20′ E to 126°00′ E and latitudes of 18°10′ N to 43°30′ N with high pig feeding density. There are various monsoon climates with 3–28°C of annual average temperature and multiple geographic patterns, including plateaus, mountains, plains, hills, and basins. Pigs were divided into six categories according to different age and usage as follows: piglets (from birth to 21 days), weaned piglets (age of 22–70 days), growing-finishing pigs (above 70 days), replacement gilts, multiparous sows (at least one party), and boars (23). All the selected pig farms were without PRRSV vaccine immunization. Moreover, location coordinates of pig farms were acquired from Baidu Map (https://map.baidu.com/).
Sampling Design
Biosecurity measures were enhanced in pig farms after the ASF outbreak, thereby increasing the difficulty of collecting serum. Meanwhile, an accurate estimate for the number of pigs in the study region was inadequate. Therefore, a convenience sampling plan was conducted in this investigation. Detailed variable information on animal and farm levels was obtained by face-to-face interviews with pig farm owners. The documented variable information primarily included sampling time, sampling position, number of samples, season, farm size, pig farm topography, the background of pigs, ASF outbreak, and pseudorabies virus (PRV) purification in pig farms. The ASF outbreak, PRV purification, and pig farms topography are binary variables. Farm size is defined as small (≤100 sows), medium (100–500 sows), and large (≥500 sows) according to the number of sows in herds.
Sampling Method
Randomly selected pigs had blood extracted from the precaval vein using sterile vacutainer tubes without decoagulant. The gathered whole blood was sent to a third-party laboratory, “Wuhan Keweichuang Biotechnology Co, Ltd,” in a cold chain and centrifuged to obtain serum under 3,000 rpm for 5 min. The acquired serum was stored at −20°C until use.
Detection of Serum for PRRSV Antibody by ELISA
PRRSV antibodies were detected by the PRRS X3 Ab Test kit with a sensitivity of 98.8% and specificity of 99.9% (IDEXX, USA) according to instruction (24). The operating procedures were described as follows. First, the serum was diluted in a ratio of 1:40 by sample diluent. Then 100 μl of diluted serum was added to the coated plates and incubated for 30 min at 18–26°C. Secondly, the plates' solution was discarded, and the plates were washed three times using the wash solution. Subsequently, 100 μl of the conjugate was added to plates to incubate for 30 min at 18–26°C again. The above-mentioned wash process was repeated. Thirdly, 100 μl of the substrate was dispensed to test the well for 15 min of incubation in a dark place. Then 100 μl of stop solution was added to the wells. Finally, the absorbance of each well in plates was measured at 650 nm by Multiskan FC (Thermo scientific, USA). The S/P value of PRRSV antibodies was calculated according to the formula: (absorbance of sample-absorbance of negative control)/(absorbance of positive control-absorbance of negative control). S/P values ≥ 0.4 were considered PRRSV antibody positive. Otherwise, the serum was negative. A pig farm was deemed as PRRSV infection with at least a PRRSV antibody-positive sample.
Data Analysis
Obtained data were entered and organized in Excel (Microsoft Excel 2007, USA). The apparent and true PRRSV prevalence of animal levels was estimated using the EpiR package (version 2.0.43) based on the Clopper-Pearson method (25). Simultaneously, the Pearson chi-square test was used to analyze the differences in PRRSV seroprevalence among provinces and pig categories (26).
The serological status of pig farms was registered as a dichotomous variable (positive or negative). The potential risk factors associated with the serological status of pig farms were explored among putative variables using univariate logistic regression analysis. Variables with a p-value of <0.1 in the univariate analysis were selected for multivariable logistic regression analysis (27). The variance inflation factor (VIF) was used to identify multicollinearity (28). Variables relevant to biological meaning are retained in the model in presence of multicollinearity between variables.
Samples were not collected because of coronavirus disease 2019, which caused missed PRRSV seroprevalence data in February and March 2020. Hence, the missed data were filled using the mice package (version 3.14.0) through multiple interpolation methods (29). Time-series of PRRSV seroprevalence were analyzed using an automatic (ARIMA) procedure. The parameters p (the number of autoregressive terms), d (the number of non-seasonal differences), and q (the number of moving average terms) were determined to forecast PRRSV seroprevalence with 95 and 80% CI in the 19 months following the study period (May 2021).
SaTScan software release 9.6 version was employed to analyze spatial and temporal clusters of high PRRSV seroprevalence (30). The numbers of PRRSV seropositive and seronegative samples from each pig farm were respectively treated as case and control groups. We used the month level for time aggregation to cover all PRRSV seropositive samples from December 2017 to May 2021. All data analyses were achieved utilizing R software (31). Maps were plotted using ArcGIS 10.7 (ESRI, USA).
Result
PRRSV Seroprevalence of Animal and Herd Levels
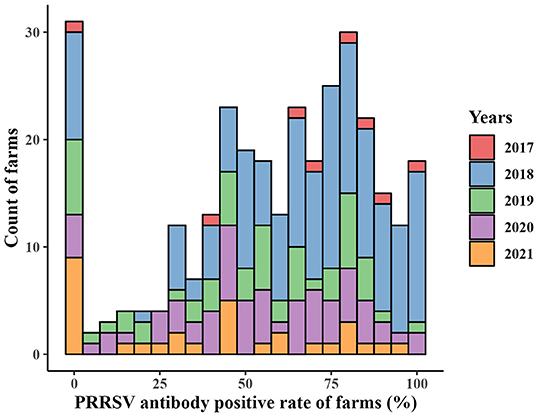
The numbers of collected samples in each province and locations of pig farms are shown and labeled in Figure 1. A total of 14,134 samples were obtained with an overall 62.56% (95% CI: 61.74–63.37%) true prevalence of PRRSV infection. PRRSV seroprevalences of Tianjin city and Liaoning province have the highest (>80%). Hubei and Shaanxi provinces have the lowest PRRSV prevalence in all sampling regions (<50%). The Pearson chi-square test results revealed statistically significant differences in PRRSV prevalence in various provinces and pig categories (Tables 1, 2). The true prevalence of replacement gilts is the highest (75.466% 95% CI: 73.000–77.821%). The lowest prevalence of PRRSV (43.304%, 95% CI: 41.400–45.228%) appears in weaned-piglets. We selected 316 pig farms to sample serum in this study, of which 286 are PRRSV antibody-positive and 30 pig farms PRRSV antibody-negative (data not shown). Figure 2 shows the histogram of PRRSV antibody positive rate of pig farms (mean: 58.12%; median: 64.69%; range: 0–100%).
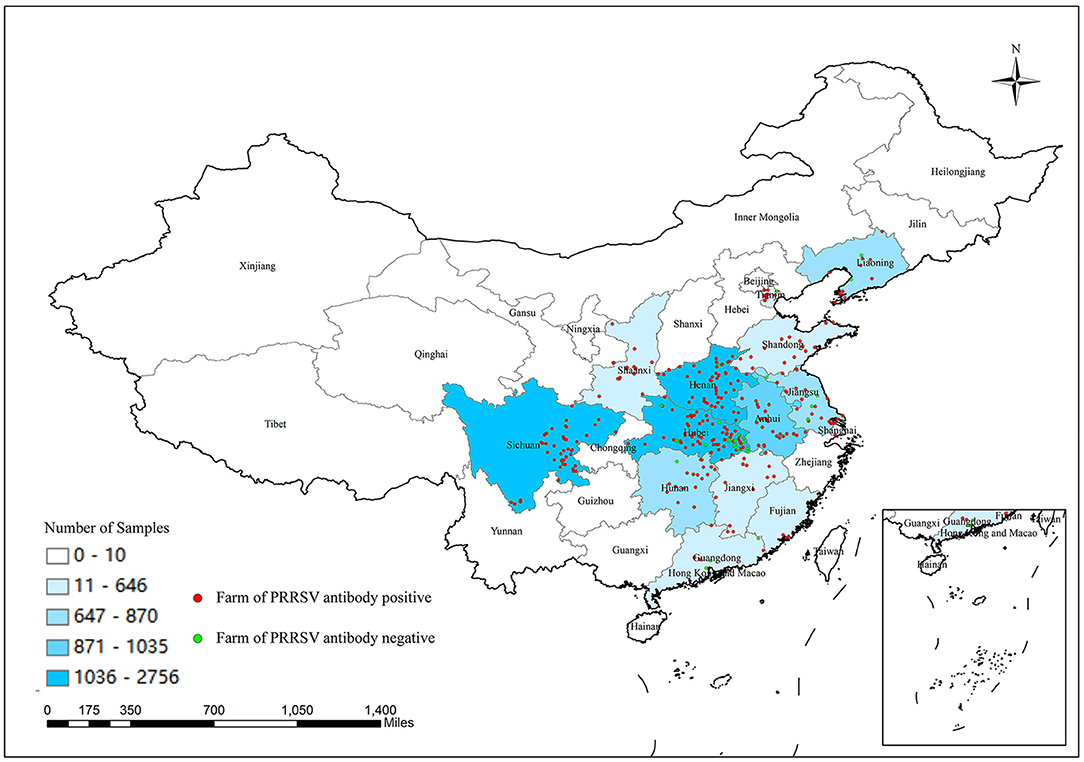
Figure 1. Number of samples collected from different provinces or cities in China and location of selected pig farms.
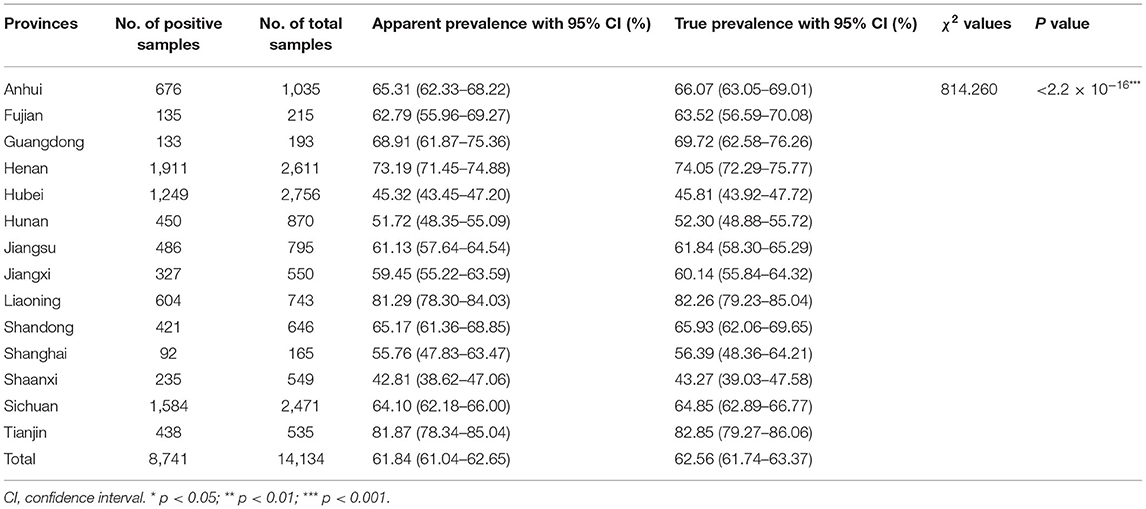
Table 1. The apparent and true prevalence of PRRSV antibody with Pearson chi-square test in 12 provinces and two cities in China.
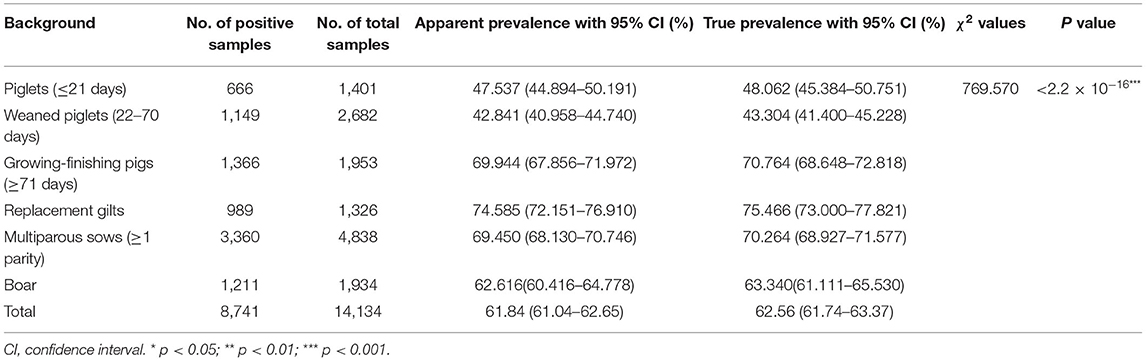
Table 2. The apparent and true prevalence of PRRSV antibody with Pearson chi-square test in different stages of pigs.
Risk Factors Analysis Related to PRRSV Serological Status of Pig Farms
Season and topography variables are not significantly associated with the PRRSV serological status of pig farms (p-value > 0.1) in the univariate logistic analysis presented in Table 3, which are removed from the subsequent multivariable logistic analysis. The identified risk factors associated with the PRRSV serological status of pig farms are shown in Table 4 through multivariable logistic analysis. Medium and large pig farms are 3.076 (95% CI: 1.005–9.498) and 6.098 (95% CI: 1.814–21.290) times more likely to be PRRSV antibody-positive compared with small pig farms, respectively. Similarly, the possibility of pig farms being PRRSV antibody-positive before the ASF outbreak is 3.104 (95% CI: 1.122–10.326) times that after the ASF outbreak. The odds ratio (OR) value of pig farms with PRV purification is 5.414 (95% CI: 1.977–17.435), implying that PRV purification may decrease PRRSV infection pressure.
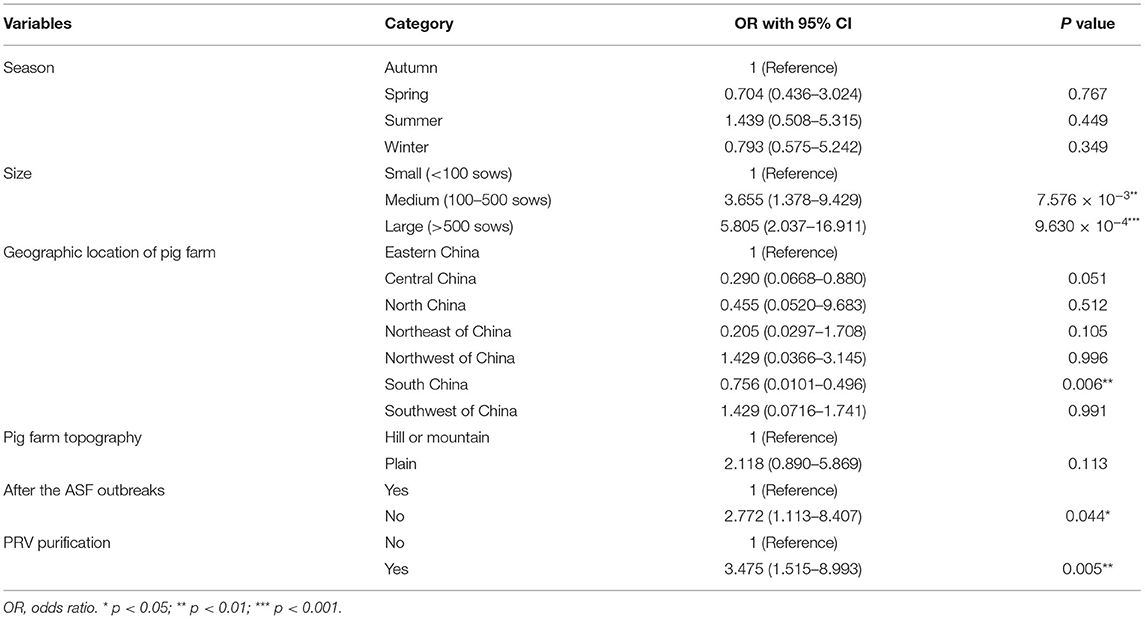
Table 3. Univariate logistic analysis of risk factors associated with PRRSV serological status of pig farm levels.
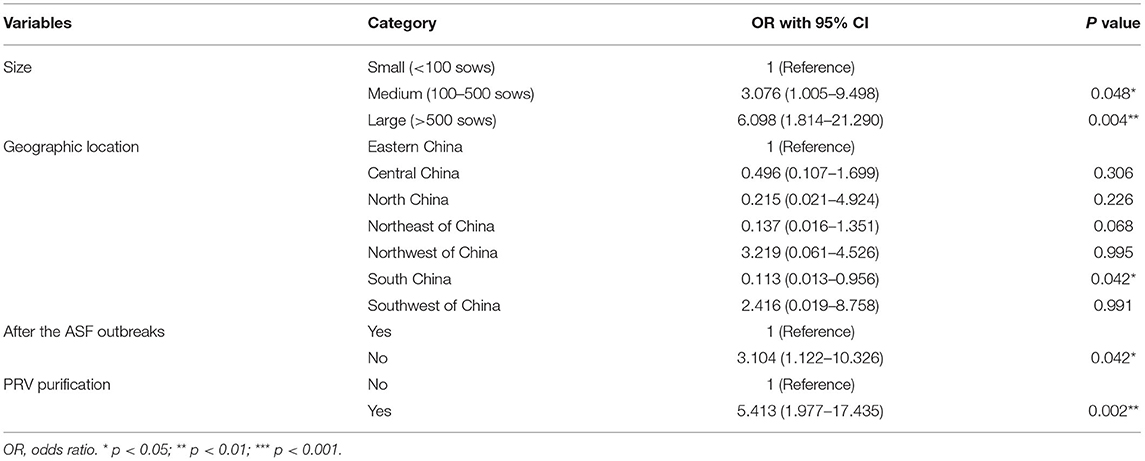
Table 4. Multivariable logistic analysis of risk factors associated with PRRSV serological status of pig farm levels.
Temporal Analysis of PRRSV Seroprevalence
Figure 3A reveals an epidemic curve of PRRSV seroprevalence by month. The highest PRRSV prevalence appeared in February 2018 and showed a comprehensive fluctuant decline trend since November 2018. Meanwhile, an ARIMA model has been established and used to forecast PRRSV prevalence with predictive limits of 95 and 80% CI in the 19 months following the current research date (May 2021). No change pattern of autocorrelation residuals was observed, which indicated the applicability of the established model and forecasts of PRRSV prevalence in this study (Figure 3B).
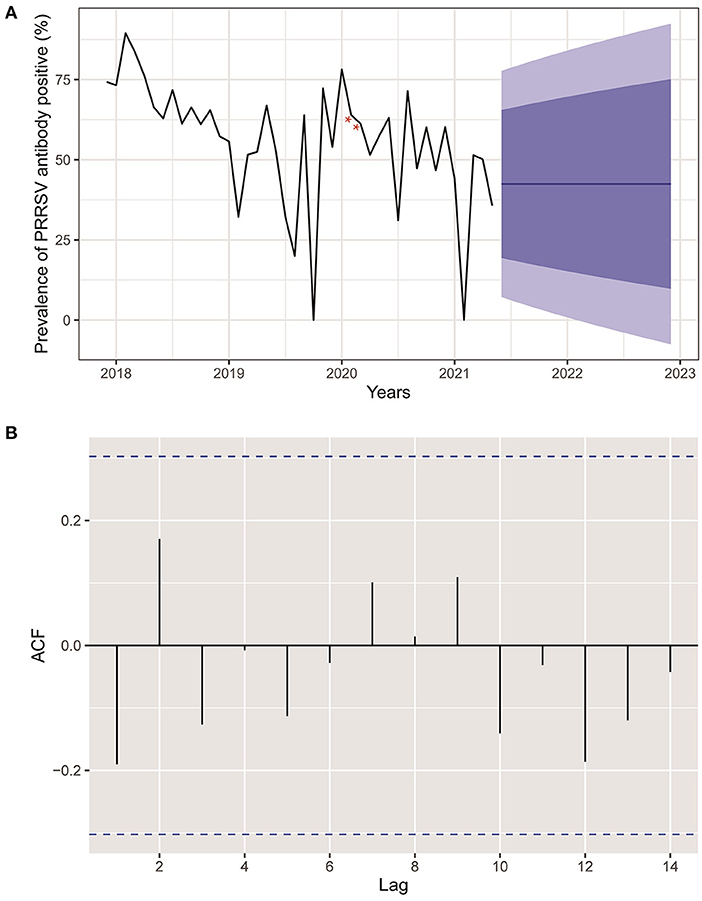
Figure 3. (A) Actual and forecasted PRRSV prevalence. Black line was the actual PRRSV prevalence from December 2017 to May 2021. The blue line represented the forecasted PRRSV prevalence in the next 19 months. The dark and light blue areas were 80 and 95% CI of forecasted PRRSV prevalence, respectively. Red stars indicated interpolated values for PRRSV prevalence of February and March 2020 deficiency because of COVID-2019. (B) Autocorrelation plots of PRRSV prevalence.
Spatial Analysis of PRRSV Seroprevalence
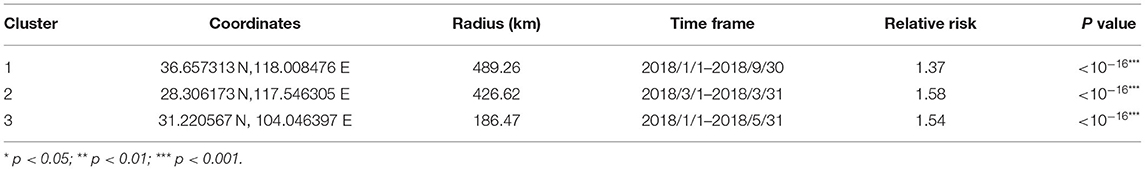
Three significant clusters of high PRRSV seroprevalence are detected in China through spatial-temporal analysis from December 2017 to May 2021 (Figure 4, Table 5). The time frame of three clusters of high PRRSV seroprevalence all occurred between January and September 2018, before the ASF outbreak. The coordinate of the largest cluster is 36.657313 N, 118.008476 E, with a radius of 489.26 km. Its time frame and relative risk are from January 1, 2018, to September 30, 2018, and 1.37, respectively. The second cluster locates at 28.306173 N, 117.546305 E, with a radius of 426.62 km. This cluster's time frame and relative risk are from March 1, 2018, to March 31, 2018, and 1.58, respectively. The coordinate of the smallest cluster is 31.220567 N, 104.046397 E, with a radius of 186.47 km. The time frame and relative risk are January 1, 2018, to May 31, 2018, and 1.54, respectively.
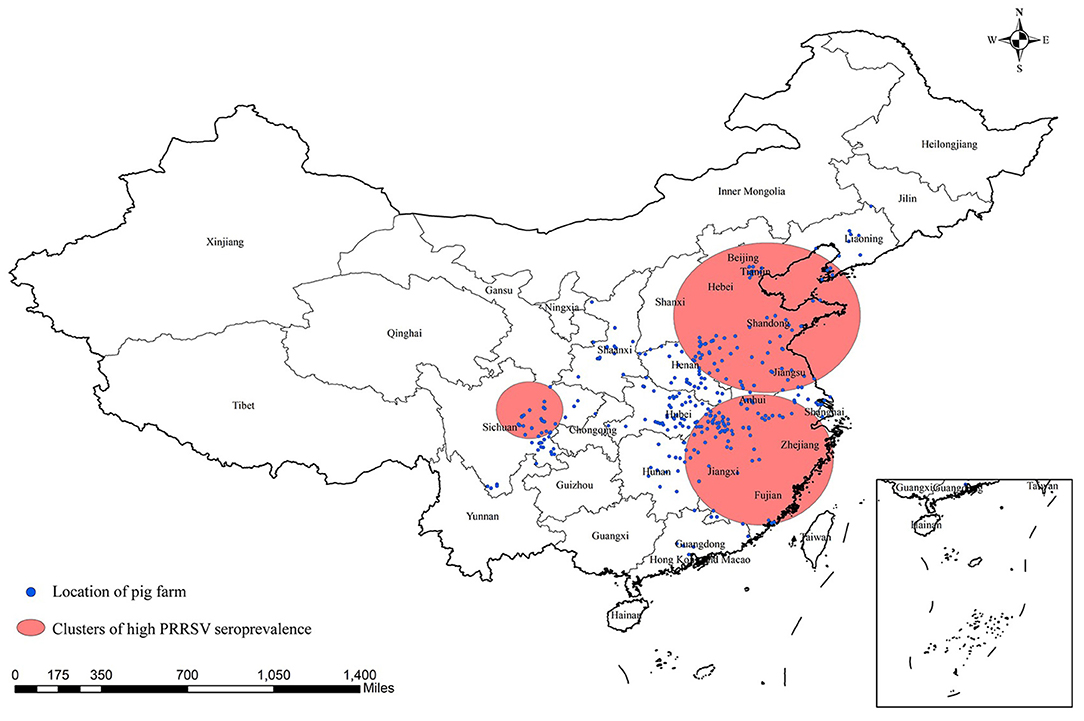
Figure 4. Significant spatial clusters (p-value < 0.05) of high PRRSV seroprevalence in China from December 2017 to May 2021 with a maximum window size of 25% of the population at risk.
Discussion
PRRSV is the causative agent of PRRS and is an influential pig disease that causes great harm to the pig industry. Meanwhile, China is the largest pork producer and consumer worldwide (32), thus, investigating the epidemic status of PRRSV infection in China is necessary. However, most studies about PRRSV epidemiology focused more on molecular genetic evolutionary analysis in China, not on serological prevalence (15, 33–35). Consequently, we performed a widespread cross-sectional study to collect serum samples in pig farms without PRRSV vaccination from 12 provinces and two cities in China from 2017 to 2021. Implementing a complete sampling plan, including most pig farms in China, is impracticable because of the inability to know accurate numbers of pigs fed in China and the expense limit. Therefore, convenience sampling was adopted in our study.
A total of 14,134 serum samples were collected from 316 pig farms to detect antibodies by ELISA as PRRSV infection diagnosis. The PRRSV seroprevalence of animal and herd levels are, respectively, 61.84 and 90.51% (286 pig farms of PRRSV seropositive), implying a high PRRSV infection in China, which follows a previous report performed by Guo et al. (9), wherein >80% of pig farms were PRRSV seropositive. The apparent and true prevalence of PRRSV infection in different provinces and pig categories show subtle differences, which indicates our results can reflect real PRRSV infection situations. Meanwhile, a significant difference was found in PRRSV seroprevalence for diverse provinces or cities by the Pearson chi-square test, which imply a probable existence of different spatial risks of PRRSV infection. The top three highest PRRSV seroprevalences are Tianjin, Liaoning, and Henan, all located to the north of China. The reason for this might be the low temperature that contributes to PRRSV survival and increases infection risk (36). The PRRSV prevalence in growing-finishing pigs, replacement gilts, boars, and multiparous sows are all >60%, which signifies that they could be a major source of PRRSV infection in herds. The replacement gilts had the highest PRRSV prevalence of 75.466% and are a dangerous signal for comprehensive control of PRRSV in herds because gradually replacing multiparous sows with negative gilts has been an important measure for PRRSV eradication (37). Previous research has also reported that quarantining new incoming sows can reduce the risks of PRRSV infection (38). Maternal antibodies of PRRSV obtained from sows can maintain for ≈2–4 weeks in piglets and start to decline at 4–10 weeks of age (39). Hence, the PRRSV prevalence of piglets and weaned-piglets might be overestimated because of the existence of maternal antibodies. Meanwhile, we can infer that the PRRSV positive antibodies of growing-finishing are induced by field PRRSV infection because the maternal antibodies have disappeared at this stage.
The possibility of pig farms being PRRSV seropositive increased with the size of pig farms, as already described by Firkins et al. (17), who revealed an association between larger herd size with increased risks of PRRSV infection. The farmers of pig farms without PRRSV vaccination often prefer to control disease through enhanced biosecurity measures whether on large or small farms, which implies that biosecurity levels might be similar for pig farms of different sizes (private communication). However, the high probability of PRRSV seropositivity in large pig farms might be due to the following reasons. More frequent contact between pigs in large pig farms might enhance the chances of virus spread. Furthermore, pigs on small pig farms have a simpler herd structure and can receive more care from breeders. A pig farm with PRV infection was 3.104 times more likely to be PRRSV-positive than one without PRV infection, which implies that PRV infection in pig herds was associated with a PRRSV seropositive status, playing a similar role in PRRSV infection as Mycoplasma hyopneumoniae and H1N2 swine influenza A virus found by Fablet et al. (18). The underlying mechanisms and causal relationship of these viral interactions remained murky, hinting that control of PRRSV infection needed to simultaneously adopt collaborative measures for multiple viruses in herds. The OR value of pig farms being PRRSV seropositive before the ASF outbreak was remarkably higher than that after the ASF outbreak, which was potentially caused by implementation of strengthened biosafety measures in pig farms after the ASF outbreak (40). Only the OR value of pig farms located in south China was statistically significant compared to the reference pig farms located in eastern China in the geographic location variable. However, considering the large p-value (0.042) and possible sampling error, the geographic location variable might not be the risk factor related to the PRRSV serological status of farms.
Little information is available about the spatial-temporal distribution of PRRSV infection in China due to the weak development of veterinary epidemiology, not to mention the comparation of the PRRSV epidemic before and after the ASF outbreak. Additionally, spatial-temporal analysis of PRRSV infection can contribute to detecting clusters of high PRRSV prevalence and exploring variation trends of PRRSV infection helping policymakers to design more precise and cost-effective intervention policies related to future PRRSV control in China. This study was the first to perform a spatial-temporal analysis of PRRSV seroprevalence in China after the ASF outbreak. The prevalence of PRRSV began a gradual decline in November 2018, when ASF entered China. The results of temporal analysis of PRRSV prevalence in our study exhibited a fluctuant decline pattern without obvious seasonal or periodic trends, which might be caused by the disadvantage of convenience sampling method with imperfect sample representativeness, although the incidence of PRRSV infection in autumn and winter is usually higher than that in spring and summer according to expert opinions (private communication) (41). Additionally, the PRRSV prevalence by month in Figure 3 showed a general decline after the ASF outbreak similar to the risk factor analysis described above. The forecasted values of PRRSV prevalence in the next 19 months (until December 2022) remained unchanged, which indicated that the PRRSV prevalence would tend toward a stable epidemic in China in the future. Three significant-high PRRSV seroprevalence clusters were first detected in China: two large clusters located in eastern China and a small cluster located in southwest China close to Sichuan province. Interestingly, the time frames of the three clusters of high PRRSV seroprevalence were all between January 2018 and September 2018 before the massive ASF outbreak in China (42), further demonstrating reduced PRRSV infection after the ASF outbreak. The detailed spatial epidemiology of PRRSV infection in China remained unknown, which demands another example to study, although we detected three clusters of high PRRSV prevalence in this study. We all demonstrate that PRRSV seroprevalence after the ASF outbreak displays an apparent decrease compared to that before the ASF outbreak through risk factors and spatial-temporal analysis.
We collected 14,134 samples from 316 pig farms without PRRSV vaccination located in 12 provinces and two cities in China from 2017 to 2021 to detect PRRSV antibodies using the ELISA method. The total true prevalence rate of PRRSV was 62.56% (95% CI: 61.74–63.37%) for pig level, and 286 pig farms (90.51%) were PRRSV antibody-positive, showing a widespread PRRSV epidemic in China. Additionally, we found that farm size, the ASF outbreak, and PRV purification variables were risk factors associated with the PRRSV serological status of pig herd using the multivariable logistic analysis. Temporal analysis for PRRSV seroprevalence showed a fluctuant declining trend. Three significant clusters of high PRRSV seroprevalence that occurred before the ASF outbreak were first detected in China through spatial-temporal analysis. The research findings obtained in our study fill in the knowledge gap of the epidemic situation, risk factors, and spatial-temporal distribution of PRRSV infection in China in recent years and could help form policies for PRRSV prevention in the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Ethical Committee of Huazhong Agricultural University Huazhong Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
Conceptualization, experimental design, and project guide: JZ. Investigation, sampling, data analysis, and writing draft: PZ. Investigation and sampling: CW and WC. Supervision: RF. All authors have read and consented to publish the manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities in China (Project number: 2662020DKPY016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Goyal SM. Porcine reproductive and respiratory syndrome. J Vet Diag Invest. (1993) 5:656–64. doi: 10.1177/104063879300500435
2. Wensvoort G, de Kluyver EP, Pol JM, Wagenaar F, Moormann RJ, Hulst MM, et al. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at Lelystad. Vet Microbiol. (1992) 33:185–93. doi: 10.1016/0378-1135(92)90046-V
3. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. (2005) 227:385–92. doi: 10.2460/javma.2005.227.385
4. Dea S, Gagnon C, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. (2000) 145:659–88. doi: 10.1007/s007050050662
5. Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, et al. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Ann Rev Anim Biosci. (2016) 4:129–54. doi: 10.1146/annurev-animal-022114-111025
6. Wensvoort G, Terpstra C, Pol J, Ter Laak E, Bloemraad M, De Kluyver E, et al. Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet Q. (1991) 13:121–30. doi: 10.1080/01652176.1991.9694296
7. Mardassi H, Mounir S, Dea S. Identification of major differences in the nucleocapsid protein genes of a Quebec strain and European strains of porcine reproductive and respiratory syndrome virus. J Gen Virol. (1994) 75:681–5. doi: 10.1099/0022-1317-75-3-681
8. Valicek L, Psikal I, Smid B, Rodák L, Kubalikova R, Kosinova E. Isolation and identification of porcine reproductive and respiratory syndrome virus in cell cultures. Vet Med. (1997) 42:281–7.
9. Guo Z. Chen X-x, Li R, Qiao S, Zhang G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J. (2018) 15:1–14. doi: 10.1186/s12985-017-0910-6
10. Guo B, Chen Z, Liu W, Cui Y. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin J Anim Poult Infect Dis. (1996) 2:117–24.
11. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. (2007) 2:e526. doi: 10.1371/journal.pone.0000526
12. Karniychuk UU, Geldhof M, Vanhee M, Van Doorsselaere J, Saveleva TA, Nauwynck HJ. Pathogenesis and antigenic characterization of a new east European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res. (2010) 6:30. doi: 10.1186/1746-6148-6-30
13. Brockmeier SL, Loving CL, Vorwald AC, Kehrli ME Jr, Baker RB, Nicholson TL, et al. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. (2012) 169:212–21. doi: 10.1016/j.virusres.2012.07.030
14. Zhao K, Ye C, Chang X-B, Jiang C-G, Wang S-J, Cai X-H, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol. (2015) 89:10712–6. doi: 10.1128/JVI.01446-15
15. Jiang Y, Li G, Yu L, Li L, Zhang Y, Zhou Y, et al. Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) from 1996 to 2017 in China. Front Microbiol. (2020) 11:618. doi: 10.3389/fmicb.2020.00618
16. Pitkin A, Deen J, Dee S. Use of a production region model to assess the airborne spread of porcine reproductive and respiratory syndrome virus. Vet Microbiol. (2009) 136:1–7. doi: 10.1016/j.vetmic.2008.10.013
17. Firkins LD, Weigel RM. A retrospective study of risk factors for porcine reproductive and respiratory syndrome virus infection and clinical disease in swine herds in illinois during the early years of the pandemic. J Swine Health Prod. (2004) 12:23–8.
18. Fablet C, Marois-Crehan C, Grasland B, Simon G, Rose N. Factors associated with herd-level PRRSV infection and age-time to seroconversion in farrow-to-finish herds. Vet Microbiol. (2016) 192:10–20. doi: 10.1016/j.vetmic.2016.06.006
19. Lopes Antunes AC, Halasa T, Lauritsen KT, Kristensen CS, Larsen LE, Toft N. Spatial analysis and temporal trends of porcine reproductive and respiratory syndrome in Denmark from 2007 to 2010 based on laboratory submission data. BMC Vet Res. (2015) 11:1–11. doi: 10.1186/s12917-015-0617-0
20. Woonwong Y, Do Tien D, Thanawongnuwech R. The future of the pig industry after the introduction of African swine fever into Asia. Anim Front. (2020) 10:30–7. doi: 10.1093/af/vfaa037
21. Wang T, Sun Y, Qiu H-J. African swine fever: an unprecedented disaster and challenge to China. Infect Dis Poverty. (2018) 7:111. doi: 10.1186/s40249-018-0495-3
23. Zhao P, Wang C, Xia Y, Hu Y, Fang R, Zhao J. Seroprevalence investigation of classic swine fever virus before, during, and after african swine fever virus outbreak in some provinces of China from 2017 to 2021. Viral Immunol. (2022) 35:33–40. doi: 10.1089/vim.2021.0109
24. Biernacka K, Podgórska K, Tyszka A, Stadejek T. Comparison of six commercial elisas for the detection of antibodies against porcine reproductive and respiratory syndrome virus (PRRSV) in field serum samples. Res Vet Sci. (2018) 121:40–5. doi: 10.1016/j.rvsc.2018.10.005
25. Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. (1934) 26:404–13. doi: 10.1093/biomet/26.4.404
26. Curtis K, Youngquist ST. Part 21: categoric analysis: pearson chi-square test. Air Med J. (2013) 32:179–80. doi: 10.1016/j.amj.2013.04.007
27. Venables WN, Ripley BD. Modern Applied Statistics with S-Plus. Berlin: Springer Science & Business Media (2013).
28. Mansfield ER, Helms BP. Detecting multicollinearity. Am Stat. (1982) 36:158–60. doi: 10.1080/00031305.1982.10482818
29. Van Buuren S, Boshuizen HC, Knook DL. Multiple imputation of missing blood pressure covariates in survival analysis. Stat Med. (1999) 18:681–94. doi: 10.1002/(sici)1097-0258(19990330)18:6<681::aid-sim71>3.0.co;2-r
30. Kulldorff M, Heffernan R, Hartman J, Assunçao R, Mostashari F. A space–time permutation scan statistic for disease outbreak detection. PLoS Med. (2005) 2:e59. doi: 10.1371/journal.pmed.0020059
31. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna (2020). Available online at: https://www.R-project.org/
32. Oh S-H, Whitley NC. Pork production in China, Japan and South Korea. Asian-Aust J Anim Sci. (2011) 24:1629–36. doi: 10.5713/ajas.2011.11155
33. Gao J-C, Xiong J-Y, Ye C, Chang X-B, Guo J-C, Jiang C-G, et al. Genotypic and geographical distribution of porcine reproductive and respiratory syndrome viruses in Mainland China in 1996–2016. Vet Microbiol. (2017) 208:164–72. doi: 10.1016/j.vetmic.2017.08.003
34. Guo Z, Chen X-X, Li X, Qiao S, Deng R, Zhang G. Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central china during 2016–2017: Nadc30-like PRRSVS are predominant. Microbial Pathogen. (2019) 135:103657. doi: 10.1016/j.micpath.2019.103657
35. Fang K, Liu S, Li X, Chen H, Qian P. Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus in South China between 2017 and 2021. Front Vet Sci. (2022) 9:853044. doi: 10.3389/fvets.2022.853044
36. Loza-Rubio E, Rojas-Anaya E. “Porcine reproductive and respiratory syndrome (Prrs),” in Prospects of Plant-Based Vaccines in Veterinary Medicine, ed MacDonald J (Cham: Springer International Publishing) (2018). p. 267–81. doi: 10.1007/978-3-319-90137-4_13
37. Fano E, Olea L, Pijoan C. Eradication of porcine reproductive and respiratory syndrome virus by serum inoculation of Naïve Gilts. Can J Vet Res-revue Canadienne De Recherche Veterinaire. (2005) 69:71–4.
38. Evans CM, Medley GF, Green LE. Porcine reproductive and respiratory syndrome virus (PRRSV) in Gb pig herds: farm characteristics associated with heterogeneity in seroprevalence. BMC Vet Res. (2008) 4:1–11. doi: 10.1186/1746-6148-4-48
39. Nodelijk G, Van Leengoed L, Schoevers E, Kroese A, De Jong M, Wensvoort G, et al. Seroprevalence of porcine reproductive and respiratory syndrome virus in Dutch weaning pigs. Vet Microbiol. (1997) 56:21–32. doi: 10.1016/S0378-1135(96)01349-1
40. Ding Y, Wang Y. Big Government: the fight against the African swine fever in China. J Biosaf Biosecu. (2020) 2:44–9. doi: 10.1016/j.jobb.2020.04.001
41. Stratton SJ. Population research: convenience sampling strategies. Prehosp Disaster Med. (2021) 36:373–4. doi: 10.1017/S1049023X21000649
Keywords: risk factors analysis, spatial-temporal analysis, seroprevalence, PRRSV, ASF outbreak
Citation: Zhao P, Wang C, Cao W, Fang R and Zhao J (2022) Risk Factors and Spatial-Temporal Analysis of Porcine Reproductive and Respiratory Syndrome Seroprevalence in China Before and After African Swine Fever Outbreak. Front. Vet. Sci. 9:929596. doi: 10.3389/fvets.2022.929596
Received: 27 April 2022; Accepted: 08 June 2022;
Published: 02 August 2022.
Edited by:
Ioannis Magouras, City University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Shankar Yadav, European Commission for the Control of Foot and Mouth Disease (EuFMD), ItalyKaichuang Shi, Guangxi Center for Animal Disease Control and Prevention, China
Copyright © 2022 Zhao, Wang, Cao, Fang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junlong Zhao, emhhb2p1bmxvbmdAbWFpbC5oemF1LmVkdS5jbg==
 Pengfei Zhao
Pengfei Zhao Chaofei Wang
Chaofei Wang Rui Fang
Rui Fang Junlong Zhao
Junlong Zhao