
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 05 July 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.928375
 Panpan Cui1
Panpan Cui1 Weimin Wang1,2
Weimin Wang1,2 Deyin Zhang2
Deyin Zhang2 Chong Li1
Chong Li1 Yongliang Huang1
Yongliang Huang1 Zongwu Ma1
Zongwu Ma1 Xiaojuan Wang1
Xiaojuan Wang1 Liming Zhao1
Liming Zhao1 Yukun Zhang1
Yukun Zhang1 Xiaobin Yang1
Xiaobin Yang1 Dan Xu1
Dan Xu1 Jiangbo Cheng1
Jiangbo Cheng1 Xiaolong Li1
Xiaolong Li1 Xiwen Zeng1
Xiwen Zeng1 Yuan Zhao1
Yuan Zhao1 Wenxin Li1
Wenxin Li1 Jianghui Wang1
Jianghui Wang1 Changchun Lin1
Changchun Lin1 Bubo Zhou1
Bubo Zhou1 Jia Liu1
Jia Liu1 Rui Zhai1
Rui Zhai1 Xiaoxue Zhang1*
Xiaoxue Zhang1*Fat deposition is an important economic trait that is closely related to feed efficiency and carcass performance in livestock. In this study, the fat deposition-related traits of 1,293 Hu sheep were measured and descriptive statistical analysis was conducted. The results showed that the coefficient of variation of all fat deposition-related traits was higher than 24%. In addition, single nucleotide polymorphisms and the expression characteristics of TRAPPC9 (encoding trafficking protein particle complex subunit 9) and BAIAP2 (encoding brain-specific Angiogenesis inhibitor 1-associated protein 2) genes in Hu sheep were detected using PCR amplification, Sanger sequencing, KASPar genotyping, and quantitative real-time reverse transcription PCR (qRT-PCR). The associations between SNPs and fat deposition-related traits were also analyzed. Two intronic mutations, TRAPPC9 g.57654 A > G and BAIAP2 g.46061 C > T, were identified in Hu sheep. The result of association analysis showed that TRAPPC9 g.57654 A > G and BAIAP2 g.46061 C > T were both significantly associated with the weight of tail fat, tail fat relative weight (body weight), and tail fat relative weight (carcass) (P < 0.05). Comprehensive effects analysis showed that there were significant differences between the combined genotypes and tail fat and perirenal fat deposition. Moreover, qRT-PCR analysis showed that TRAPPC9 and BAIAP2 are widely expressed, and their expression levels were significantly higher in the small-tail group compared with those in the big-tail group (P < 0.01). These results provided important candidate molecular markers that could be used in strategies to reduce tail fat deposition in Hu sheep.
Hu sheep are one of the important livestock breeds in Taihu Lake Basin region of China. They have the advantages of high reproductive performance (long estrus period, average litter size 2.06), fast growth and development performance, strong environmental adaptability, and good lactation performance (1, 2). Nowadays, sheep can be classified into five types on the basis of their tail size: short fat-tailed sheep, long fat-tailed sheep, short thin-tailed sheep, long thin-tailed sheep, and fat-rumped sheep (3). The important fat-tailed breed of sheep was first recorded 5,000 years ago (4). Hu sheep belong to short fat-tailed sheep (3). Adipose tissue plays a vital role in maintaining the balance of homeostatic metabolic processes in domestic animals. And it is found in various parts of the sheep body, including the perirenal, mesentery, and tail. Tail fat is the most typical type of deposited fat (5). During severe conditions, such as food scarcity resulting from migration, drought, and winter, tail fat can provide energy (6). Fat has added value to humans as it can provide high-energy food during droughts and famines (4). Currently, with the improvement of people's living standards and diet structure, consumers are paying increased attention to their own health and meat quality. However, for fat-tailed sheep, most of the fat is deposited in the tail, leading to the reduction of fat deposition in other parts of the body, which affects meat quality (7). In modern mutton sheep production systems, tail fat deposition requires higher energy costs. In addition, tail fat accounts for 20% of the carcass weight, which greatly reduces the economic value of the carcass and increases the feeding cost (8). Thus, reducing tail fat deposition has become a research hotspot in sheep genetic improvement.
NIK- and IKKβ-binding protein (NIBP), also known as trafficking protein particle complex 9 (TRAPPC9), is a nuclear factor kappa B (NF-κB) signaling pathway regulating factor that has been detected in human nerve cells. It has become clear that the TRAPP complex might exist in different forms depending on its specific functions (9). In addition, the NF-κB signaling pathway is the key mediator of cell proliferation, apoptosis, and physiological and pathological events in tumorigenesis (10). Wang et al., conducted genome-wide association studies on Chinese Holstein cows, and the results supported the presence of significant single nucleotide polymorphisms (SNPs), mainly located in Bos taurus autosome (BTA) 14 of Chinese Holstein cows, revealing a new candidate gene, TRAPPC9, gene related to cow mastitis resistance (11). Another study showed that microcephaly and obesity are common features of TRAPPC9-deficient patients (12/23 cases) and summarized this phenotype in a TRAPPC9-deficient mouse model (12). Briollais et al. showed that the mean effect of exclusive breastfeeding (EBF) was associated with a 0.06 reduction in the M value of the TRAPPC9 CpG locus in the first 2 years of life, which resulted in a 0.20 kg/m2 reduction in body mass index (BMI) (13). In a study by Liang et al., deleting TRAPPC9 in mice resulted in the weight of mice increasing significantly, and it was concluded that the loss of TRAPPC9 function led to the weight gain (14). Liu et al. showed that TRAPPC9 is related to body shape traits in pigs, in which it participates in the regulation of bone growth and development and nutrient absorption, and is associated with obesity (15). Insulin receptor substrate p53 (IRSp53), also known as brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2), is a multi-domain adapter protein originally identified as a tyrosine protein phosphorylated by the insulin receptor and insulin-like growth factor 1 (IGF-1) receptor (16). Lakshman et al. found that BAIAP2 was significantly associated with weight loss in participants with chronic obstructive pulmonary disease (COPD) (17). Al-Dokhi showed that adults tend to gain weight as they get older, caused by fat deposits (18). In addition, Lee and Shin reported that TRAPPC9 and BAIAP2 were related to fat accumulation of pigs using a genome-wide association study (GWAS) (19). However, to the best of our knowledge, there have been no reports on the association of polymorphisms in TRAPPC9 and BAIAP2 with fat deposition related-traits in Hu sheep.
Therefore, in the present study, we analyzed the relationship between the single SNPs of TRAPPC9 and BAIAP2 and fat deposition-related traits in Hu sheep. In addition, the expression levels of TRAPPC9 and BAIAP2 mRNAs in ten different tissues of Hu sheep and tail adipose tissue of small-tail and big-tail Hu sheep were also investigated. This study provided valuable molecular markers for Hu sheep breeding.
A total of 1,293 Male Hu sheep were purchased from Jinchang Zhongtian Sheep Industry Co., Ltd., Gansu Zhongsheng Huamei Sheep Industry Development Co., Ltd., Gansu Sanyang Jinyuan Animal Husbandry Co., Ltd., Shandong Runlin Sheep Industry Co., Ltd., and Wuwei Pukang Sheep Industry Co., Ltd. Lambs were immunized according to standard procedures before weaning at 56 days of age. All weaned lambs were raised at Minqin Defu Agriculture Co., Ltd. (Gansu, China). The lamb acclimation period was 14 days, the pre-experiment period was 10 days, and the experimental period was 100 days. The feeding conditions were consistent, including housing, feeding, and drinking water. Hu sheep were fed with pellet feed purchased from Gansu Sanyang Jinyuan Animal Husbandry Co., Ltd. All experimental animals were weighed and slaughtered at 180 days. After slaughter, the weight of tail fat, mesenteric fat, and perirenal fat were measured, and collected, and then stored at −80°C for subsequent RNA extraction. DNA was extracted from blood of 1,293 adult sheep (6 months old) using an EASYPURE Blood Genomic DNA Kit (Transgen Biotech, Beijing, China). DNA was stored in elution buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) at −20°C.
PCR primers were designed according to the gene sequences to conduct PCR amplification of TRAPPC9 (NC_040260.1 Chromosome 9 Reference Oar_rambouillet_v1.0 Primary Assembly) and BAIAP2 (NC_040262.1 Chromosome 11 Reference Oar_rambouillet_v1.0 Primary Assembly) sequences (Table 1). Using mixed DNA (n = 20), the PCR products were sequenced using Sanger sequencing to determine the SNPs of TRAPPC9 and BAIAP2. The PCR reaction (35 μL) included 17.5 μL of 2 × Easy Taq PCR Super Mix (Transgen), 1.12 μL of each primer (forward and reverse), 1.4 μL of dNTPs, and 14 μL ddH2O. The thermal cycling procedure for the TRAPPC9 gene included 5 min at 94°C; followed by 30 s at 94°C, 30 s at 54°C, and 30 s at 72°C for 35 cycles; with a final extension for 5 min at 72°C. The thermal cycling procedure for the BAIAP2 gene included 5 min at 94°C; 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C for 35 cycles; with a final extension for 5 min at 72°C. According to previous studies, genotyping was performed using competitive allele-specific fluorescence resonance energy transfer (FRET)-based PCR (KASPar) assays (LGC Genomics, Hoddesdon, UK) (20). The primers used for genotyping are shown in Table 2. In this experiment, the TRAPPC9 and BAIAP2 genes of 1,162 and 1,046 individuals, respectively, were successfully genotyped, and 970 individuals were successfully genotyped for both genes.
The total RNA of each tissue was extracted using Transzol (Transgen) and reverse transcribed into cDNA using an Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate Biotechnology Co., Ltd, Hunan, China) following the manufacturer's protocols. Six 180-day-old Hu sheep were randomly selected to analyze the expression levels of TRAPPC9 and BAIAP2 mRNA in ten tissues (heart, liver, spleen, lung, kidney, rumen, duodenum, muscle, lymph, and tail fat, n = 4 for each tissue). In addition, the mRNA expression levels of TRAPPC9 and BAIAP2 in the tail adipose tissue of six small-tail and six big-tail Hu sheep were detected. We assessed the tail fat deposition traits of the sheep, which are shown in Table 3. The mRNA sequences of sheep TRAPPC9 (XM_042254050.1 Chromosome 9 Reference Oar_rambouillet_v1.0 Primary Assembly) and BAIAP2 (XM_024450535.2 Chromosome 11 Reference Oar_rambouillet_v1.0 Primary Assembly), were used as templates. Specific primer pairs used for detecting TRAPPC9 and BAIAP2 expression were designed using Oligo 7.0 software, which the expected band sizes are 197 bp and 152 bp, respectively. β-actin (GenBank Accession no. NM_001009784.3) as reference gene (Table 1). The quantitative real-time PCR (qPCR) step of the qRT-PCR protocol was carried out at 94°C for 3 min; followed by 40 cycles of 15 s at 94°C, the optimum annealing temperature for 15 s, and 72°C for 20 s; with a final extension at 72°C for 5 min. The 2−ΔΔCT method was used to analyze the data (21).
The association analysis between genotypes and the fat deposition-related traits was performed using a general linear model program, which was defined as:
Where, Yijk and Yimjkn are the phenotypic observation value of the tail fat deposition traits, μ is the mean, Gi and Gm is the effect of the ith and mth genotypes, Fj represents the farm effect (j = 1, 2……5), Cn refers to the effect of combination, and εijk and εimjkn are the residuals corresponding to the observed trait values. A P-value < 0.05 or a P-value < 0.01 were regarded as statistically significant and highly significant, respectively. The genotypic frequency and allele frequency were calculated. SPSS v.23 software was used for all statistical analyses (IBM Corp., Armonk, NY, USA).
In the present study, the fat deposition-related traits of all lambs (n = 1,293) were measured after slaughter at 180 days of age, and the descriptive statistics for the phenotypes of all the traits are shown in Table 4. The coefficient of variation for all traits was great than 24%, among which the variation coefficient of tail fat weight, perirenal fat weight, and mesenteric fat weight were 30.28, 47.43, and 38.34%, respectively. These results suggested the fat deposition traits have marked phenotypic variation in the experimental population.
The 988 bp fragment of TRAPPC9 and the 604 bp fragment of BAIAP2 were amplified using the primers shown in Table 2 (Figure 1). The amplified PCR products were sequenced by Tsingke Ltd. (Xi'an, China). A new mutation was found in TRAPPC9, located in intron 10 (g.57654 A > G), and a new mutation was also found in BAIAP2, located in intron 6 (g.46061 C > T, Figure 2).
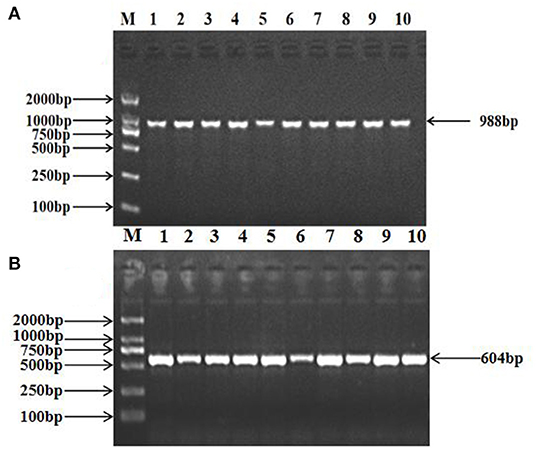
Figure 1. PCR amplification of the target fragments of the ovine TRAPPC9 (A) and BAIAP2 (B) genes. M: DL2000 DNA Marker; 1–10: PCR products.
KASPar analysis was used to genotype the two SNPs, and three genotypes of the two genes were determined: AA, AG, and GG (TRAPPC9) and CC, CT, and TT (BAIAP2) (Figure 3). The genetic parameters of the SNPs recognized in Hu sheep at TRAPPC9 g.57654 A > G and BAIAP2 g.46061 C > T loci were calculated (Table 5). For the TRAPPC9 g.57654 A > G locus, the genotype frequencies were 0.45, 0.44, and 0.11, respectively. The results of allele frequency analysis showed that the frequency of the A allele was 0.67, which accounted for the highest proportion in the population. For the BAIAP2 g.46061 C > T locus, the genotype frequencies were 0.58, 0.29, and 0.13, respectively. The results of allele frequency analysis showed that the frequency of the C allele was 0.73, which accounted for the highest proportion in the population. The polymorphism information content (PIC), effective allele number (Ne), expected homozygosity (Ho), expected heterozygosity (He), and the P-value of the Hardy-Weinberg equilibrium (PHWE) of TRAPPC9 were 0.34, 1.79, 0.56, 0.44, and 0.84, respectively, and the PIC, Ne, Ho, He, and PHWE of BAIAP2 were 0.31, 1.64, 0.61, 0.39, and 0, respectively (Table 5).
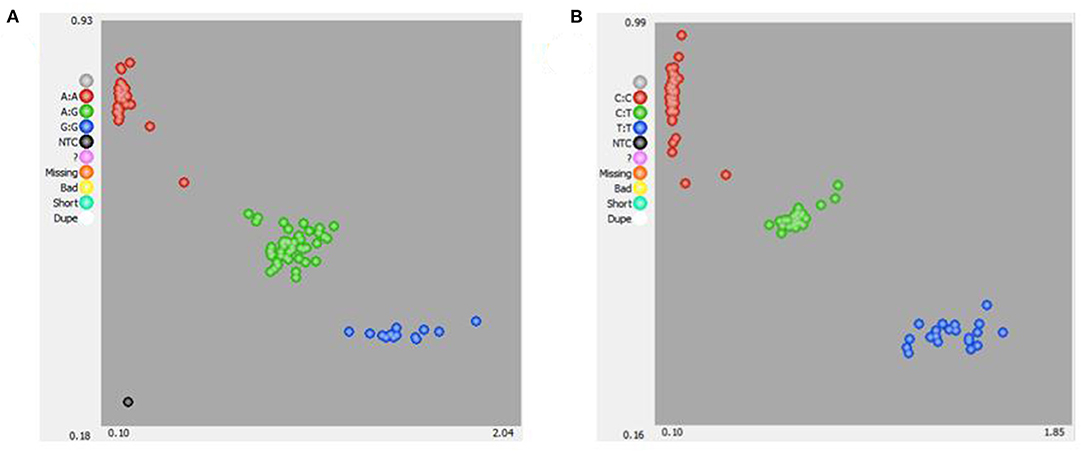
Figure 3. KASPar-based single nucleotide polymorphism (SNP) genotyping of sheep TRAPPC9 g.57654 A > G (A) and BAIAP2 g.460 C > T (B).
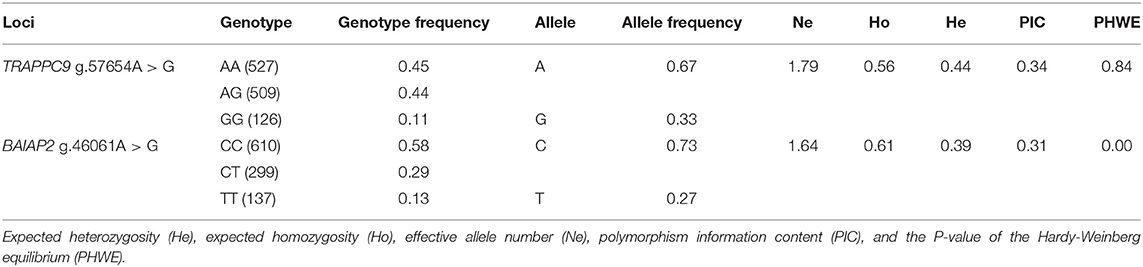
Table 5. The genotype frequency, allele frequency, and genetic diversity of the TRAPPC9 and BAIAP2 SNP sites.
The association analysis showed that the TRAPPC9 g.57654 A > G gene polymorphism correlated significantly with tail fat deposition traits (P < 0.05). The tail fat weight, relative tail fat weight (body weight), and relative tail fat weight (carcass) of the GG genotype were significantly lower than those of the AA genotype (P < 0.05). Therefore, GG was a significant genotype associated with tail fat deposition in Hu sheep. The BAIAP2 g.46061 C > T gene polymorphism also correlated significantly with tail fat deposition traits (P < 0.05). The tail fat weight, relative tail fat weight (body weight), and relative tail fat weight (carcass) of the CT genotype were significantly lower than those of the TT genotype (P < 0.05). However, no significant association was observed between the genes and perirenal fat weight and mesenteric fat weight (P > 0.05) (Table 6).
The comprehensive effects of TRAPPC9 g.57654 A > G and BAIAP2 g.46061 C > T on tail fat deposition traits were analyzed (Table 7). The results showed that tail fat weight, tail fat relative weight (body weight), and tail fat relative weight (carcass) of the GGTRAPPC9/CTBAIAP2, AGTRAPPC9/CCBAIAP2, GGTRAPPC9/CCBAIAP2, and AATRAPPC9/CCBAIAP2 genotype were significantly lower than those of the AATRAPPC9/TTBAIAP2, and AATRAPPC9/CTBAIAP2 combined genotypes (P < 0.05). The perirenal fat weight, perirenal fat relative weight (body weight), and perirenal fat relative weight (carcass) of the GGTRAPPC9/CTBAIAP2 genotype were significantly lower than those of the AATRAPPC9/TTBAIAP2, AATRAPPC9/CTBAIAP2, AGTRAPPC9/TTBAIAP2, and GGTRAPPC9/CCBAIAP2 combined genotypes (P < 0.05). The mesenteric fat weight of the AGTRAPPC9/CTBAIAP2 genotype was significantly lower than that of the AATRAPPC9/CTBAIAP2 genotype (P < 0.05), but not significantly different compared with other genotypes, and there was no significant difference between the mesenteric fat relative weight (body weight) and the mesenteric fat relative weight (carcass) among the genotypes (P > 0.05).
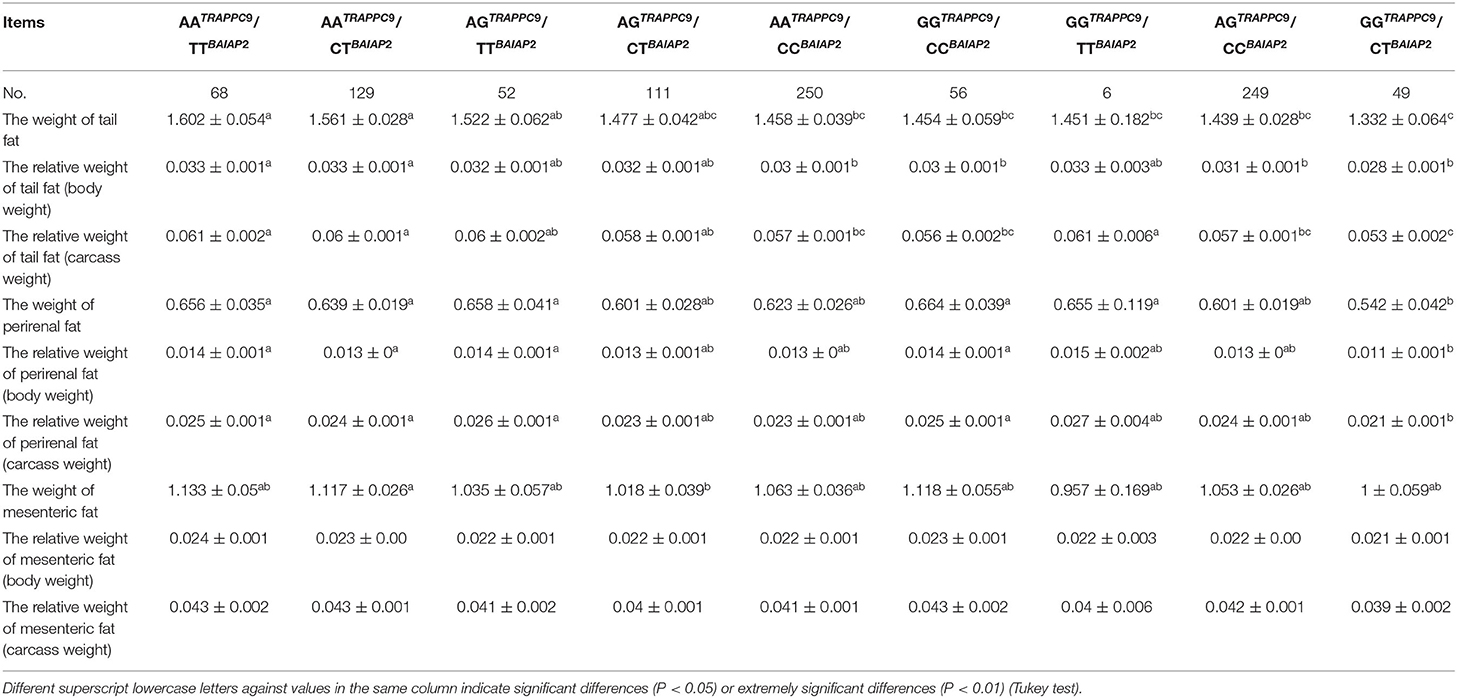
Table 7. Analysis of the associations of combined genotypes at the TRAPPC9 and BAIAP2 loci and sheep tail fat deposition traits.
The expression levels of TRAPPC9 and BAIAP2 in the tail fat, lymph, muscle, duodenum, rumen, kidney, lung, spleen, liver, and heart were detected using qRT-PCR. The results showed that TRAPPC9 and BAIAP2 were expressed in these ten tissues. Moreover, the expression levels of TRAPPC9 in the tail fat and rumen were significantly higher than those in the other tissues (P < 0.05), and the expression levels of BAIAP2 in the heart, liver, kidney, muscle, and tail fat were significantly higher than those in the other tissues (P < 0.05; Figure 4).
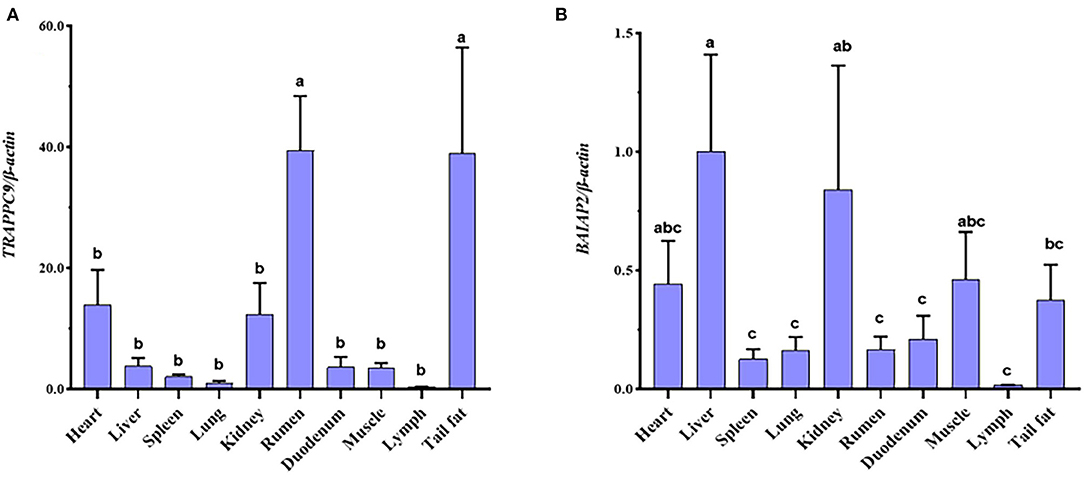
Figure 4. TRAPPC9 mRNA expression profile in sheep tissues (A). BAIAP2 mRNA expression profile in sheep tissues (B). Different lowercase letters indicate a significant difference (P < 0.05).
The mRNA expression levels of TRAPPC9 and BAIAP2 between small-tailed sheep and big-tailed sheep were analyzed quantitatively using qRT-PCR. The results showed that the expression levels of TRAPPC9 and BAIAP2 in the small-tailed group were significantly higher than those in the big-tailed group (P < 0.01; Figure 5).
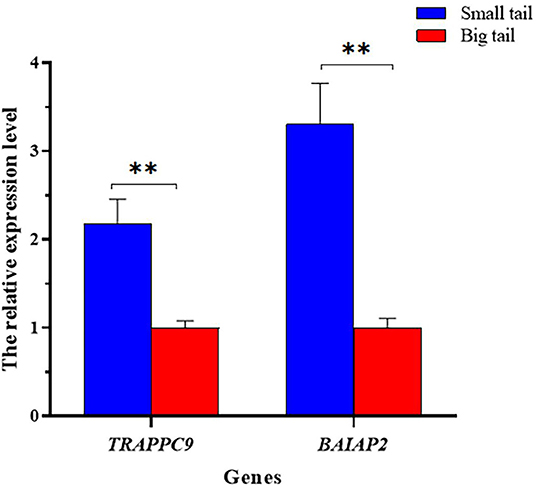
Figure 5. The relative TRAPPC9 and BAIAP2 mRNA expression levels between the small-tail and big-tail groups. Asterisks indicate a significant differences between the small- and big-tail groups (P < 0.05), double asterisks indicate a very significant differences between the small-tail and big-tail groups (P < 0.01).
Fat tails in sheep are perceived to have developed following domestication and are a valuable energy reserve for the animals during migration and winter (4, 22). However, fat tails might affect reproduction and fattening, thereby increasing the cost of raising sheep and reducing their economic value (23, 24). Fat deposition in sheep tails is the result of a complex mechanism. Previous studies have conducted several investigations into the inheritance of fat tails (25); nevertheless, the mechanisms of the genes affecting fat deposition in fat tail sheep remain unknown. TRAPPC9 is a subunit of the highly conserved protein complex called the transport protein particle (TRAPP), a guanine nucleotide exchange factor for rab proteins that operates in secretory, endocytic, and autophagic pathways (26). Over half of the patients with TRAPPC9 mutations are reported to present different degrees of obesity (27). Hnoonual et al. (12) found that the deletion of the TRAPPC9 gene leads to obesity. BAIAP2, which is located on 17q25 and encodes brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2), has been suggested to be involved in cerebral asymmetry (28). In a study by Lakshman et al. (17), the BAIAP2 gene was significantly associated with weight change in patients with chronic obstructive pulmonary disease (COPD). TRAPPC9 and BAIAP2 were implicated in fat accumulation by a GWAS (19). In this study, Two intronic mutations were detected in the TRAPPC9 and BAIAP2 genes, respectively. Recent studies suggest intronic mutations can change protein levels or protein conformation by altering regulatory splice sites, mRNA stability, miRNA binding sites, or translation efficiency (29–33). To further investigate the effect of these two intronic mutations on sheep phenotype, we analyzed the potential association between genotype and fat deposition. The results showed that TRAPPC9 g.57654 A > G and BAIAP2 g.46061 C > T were both associated significantly with tail fat deposition.
We detected that TRAPPC9 and BAIAP2 were expressed in all ten tissues of Hu sheep tested. Zhang et al. showed that TRAPPC9 was highly expressed in muscles and kidneys of human tissues; showed low expression in the heart, brain, and placenta; and was weakly expressed in the thymus and spleen (34). In Abbott et al.'s study, the tissue distribution of BAIAP2 mRNA was determined by northern blotting, appearing mainly in the heart, brain, spleen, lung, liver, kidney, and testis (35). Our results are consistent with those of previous studies. TRAPPC9 was significantly expressed in the rumen and tail fat (P < 0.05). However, studies on the TRAPPC9 gene in the rumen are rarely reported. This gene may be related to the growth and development of the rumen, but the specific mechanism still needs further study. Usman et al. found through research that mouse TRAPPC9 gene deficiency affects the proliferation and differentiation ability of adipose stem cells (ASCs) (36). Our qRT-PCR results also confirmed this view. Therefore, we speculate that TRAPPC9 has the same function in sheep.
The expression of BAIAP2 in the liver was significantly higher than that in other tissues (P < 0.05). Previous studies have found that the BAIAP2 is significantly expressed in the liver (37–39). The liver plays a pivotal role in regulating the metabolism of fatty acids (FA) and their neutral storage form, triglycerides (TGs) (40, 41). A study identified that interactions between adipose tissue and the liver might play a role in the development of non-alcoholic fatty liver disease (42). Therefore, we speculated that BAIAP2 might affect tail fat deposition by regulating FA synthesis.
To verify the potential roles of TRAPPC9 and BAIAP2 in ovine tail fat deposition, we analyzed the mRNA expression levels of TRAPPC9 and BAIAP2 between small-tail and big-tail sheep. The results showed that the expression levels of TRAPPC9 and BAIAP2 in the small-tailed group were significantly higher than those in the large-tailed group (P < 0.01). Therefore, we speculated the polymorphic loci in TRAPPC9 and BAIAP2 might represent important genetic markers in studies designed to reduce tail fat deposition in Hu sheep, and also provide clues for further search for causal mutations of tail fat deposition. However, the regulatory mechanism of TRAPPC9 and BAIAP2 on tail fat deposition in Hu sheep require further study.
In the present study, two novel SNPs in the TRAPPC9 and BAIAP2 genes were identified. Association analysis indicated that the two SNPs were significantly related to tail fat weight-related traits of Hu sheep. In addition, the two genes were expressed widely in ten tissues of Hu sheep, and the expression levels of TRAPPC9 and BAIAP2 in the small-tail group were significantly higher than those in the big-tail group. Thus, we speculated that the polymorphic loci in TRAPPC9 and BAIAP2 might be used as genetic markers of tail fat deposition in Hu sheep.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All experiments in this study were authorized and approved by the Animal Welfare and Ethics Committee of Gansu Agricultural University, and carried out in accordance with the regulations of the Standing Committee of the People's Congress of Gansu Province. License No. 2012-2-159.
WW, XZh, and PC were involved in the design and design of the experiment. CLi, LZ, DX, JC, YZhan, XL, XZe, YZhao, WL, JW, RZ, CLin, JL, and BZ collected the experimental samples. DZ, YH, ZM, XW, XY, and PC conducted the data analysis. PC wrote the paper. XZh revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (No. 2021YFD1300901), the National Natural Science Foundation of China (31960653), and the Key Research and Development Project of Gansu Province, China (20YF3NA012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.928375/full#supplementary-material
1. Feng X, Li F, Wang F, Zhang G, Pang J, Ren C, et al. Genome-wide differential expression profiling of mrnas and lncrnas associated with prolificacy in Hu sheep. Bioscience reports. (2018) 38: BSR20171350. doi: 10.1042/BSR20171350
2. H EE, Ma L, Xie X, Ma J, Ma X, Yue C, et al. Genetic polymorphism association analysis of SNPs on the species conservation genes of tan sheep and Hu sheep. Trop Anim Health Prod. (2020) 52:915–26. doi: 10.1007/s11250-019-02063-1
3. Wang J, Zhang X, Wang X, Li F, Zhang D, Li X, et al. Polymorphism and expression of the HMGA1 gene and association with tail fat deposition in Hu sheep. Anim Biotechnol. (2021) 1–9. doi: 10.1080/10495398.2021.1998093
4. Moradi MH, Nejati-Javaremi A, Moradi-Shahrbabak M, Dodds KG, McEwan JC. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. (2012) 13:10. doi: 10.1186/1471-2156-13-10
5. He X, Wu R, Yun Y, Qin X, Chen L, Han Y, et al. Transcriptome analysis of messenger RNA and long noncoding RNA related to different developmental stages of tail adipose tissues of sunite sheep. Food Sci Nutr. (2021) 9:5722–34. doi: 10.1002/fsn3.2537
6. Ben Sassi-Zaidy Y, Maretto F, Charfi-Cheikhrouha F, Mohamed-Brahmi A, Cassandro M. Contribution of microsatellites markers in the clarification of the origin, genetic risk factors, and implications for conservation of tunisian native sheep breeds. Genet Mol Res. (2016) 15:15017059. doi: 10.4238/gmr.15017059
7. Zhu C, Li N, Cheng H, Ma Y. Genome wide association study for the identification of genes associated with tail fat deposition in Chinese sheep breeds. Biol Open. (2021) 10:bio054932. doi: 10.1242/bio.054932
8. Bakhtiarizadeh MR, Alamouti AA. RNA-seq based genetic variant discovery provides new insights into controlling fat deposition in the tail of sheep. Sci Rep. (2020) 10:13525. doi: 10.1038/s41598-020-70527-8
9. Zong M, Wu XG, Chan CW, Choi MY, Chan HC, Tanner JA, et al. The adaptor function of TRAPPC2 in mammalian trapps explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS ONE. (2011) 6:e23350. doi: 10.1371/journal.pone.0023350
10. Fu ZH, Liu SQ, Qin MB, Huang JA, Xu CY, Wu WH, et al. NIK- and IKK?-binding protein contributes to gastric cancer chemoresistance by promoting epithelial-mesenchymal transition through the NF-?b signaling pathway. Oncol Rep. (2018) 39:2721–30. doi: 10.3892/or.2018.6348
11. Wang X, Ma P, Liu J, Zhang Q, Zhang Y, Ding X, et al. Genome-wide association study in Chinese holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. (2015) 16:111. doi: 10.1186/s12863-015-0263-3
12. Hnoonual A, Graidist P, Kritsaneepaiboon S, Limprasert P. Novel compound heterozygous mutations in the trappc9 gene in two siblings with autism and intellectual disability. Front Genet. (2019) 10:61. doi: 10.3389/fgene.2019.00061
13. Briollais L, Rustand D, Allard C, Wu Y, Xu J, Rajan SG, et al. DNA methylation mediates the association between breastfeeding and early-life growth trajectories. Clin Epigenet. (2021) 13:231. doi: 10.1186/s13148-021-01209-z
14. Liang ZS, Cimino I, Yalcin B, Raghupathy N, Vancollie VE, Ibarra-Soria X, et al. TRAPPC9 deficiency causes parent-of-origin dependent microcephaly and obesity. PLoS Genet. (2020) 16:e1008916. doi: 10.1371/journal.pgen.1008916
15. Liu H, Song H, Jiang Y, Jiang Y, Zhang F, Liu Y, et al. A single-step genome wide association study on body size traits using imputation-based whole-genome sequence data in yorkshire pigs. Front Genet. (2021) 12:629049. doi: 10.3389/fgene.2021.629049
16. Yeh TC, Ogawa W, Danielsen AG, Roth RA. Characterization and cloning of a 58/53-KDA substrate of the insulin receptor tyrosine kinase. J Biol Chem. (1996) 271:2921–8. doi: 10.1074/jbc.271.6.2921
17. Lakshman Kumar P, Wilson AC, Rocco A, Cho MH, Wan E, Hobbs BD, et al. Genetic variation in genes regulating skeletal muscle regeneration and tissue remodelling associated with weight loss in chronic obstructive pulmonary disease. J Cachexia Sarcopenia Muscle. (2021) 12:1803–17. doi: 10.1002/jcsm.12782
18. Al-Dokhi L. Association of the new index of sarcopenic obesity with physical fitness in healthy Saudi men and women. Eur Rev Med Pharmacol Sci. (2015) 19:328–33.
19. Lee YS, Shin D. Genome-wide association studies associated with backfat thickness in landrace and yorkshire pigs. Genomics Inform. (2018) 16:59–64. doi: 10.5808/GI.2018.16.3.59
20. Smith SM, Maughan PJ. SNP genotyping using kaspar assays. Methods Mol Biol. (2015) 1245:243–56. doi: 10.1007/978-1-4939-1966-6_18
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
22. Ermias E, Yami A, Rege J. Fat deposition in tropical sheep as adaptive attribute to periodic feed fluctuation. Journal of Animal Breeding Genet. (2002) 119:235–46. doi: 10.1046/j.1439-0388.2002.00344.x
23. Kilminster TF, Greeff JC. A note on the reproductive performance of damara, dorper and merino sheep under optimum management and nutrition for Merino Ewes in the Eastern Wheatbelt of Western Australia. Trop Anim Health Prod. (2011) 43:1459–64. doi: 10.1007/s11250-011-9871-8
24. Frisch RE. Body fat, menarche, fitness and fertility. Hum Reprod. (1987) 2:521–33. doi: 10.1093/oxfordjournals.humrep.a136582
25. Kashan N, Azar G, Afzalzadeh A, Salehi A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Ruminant Res. (2005) 60:267–71. doi: 10.1016/j.smallrumres.2005.01.001
26. Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. Trapp complexes in membrane traffic: convergence through a common rab. Nat Rev Mol Cell Biol. (2010) 11:759–63. doi: 10.1038/nrm2999
27. Radenkovic S, Martinelli D, Zhang Y, Preston GJ, Maiorana A, Terracciano A, et al. TRAPPC9-CDG: a novel congenital disorder of glycosylation with dysmorphic features and intellectual disability. Genet Med. (2022) 24:894–904. doi: 10.1016/j.gim.2021.12.012
28. Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, et al. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. (2005) 308:1794–8. doi: 10.1126/science.1110324
29. Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Groß M, et al. A pan-cancer analysis of synonymous mutations. Nat Commun. (2019) 10:2569. doi: 10.1038/s41467-019-10489-2
30. Gupta SK, Carmi S, Waldman Ben-Asher H, Tkacz ID, Naboishchikov I, Michaeli S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J Biol Chem. (2013) 288:4991–5006. doi: 10.1074/jbc.M112.416578
31. Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. (2007) 17:682–90. doi: 10.1101/gr.6525007
32. Reis EM, Louro R, Nakaya HI, Verjovski-Almeida S. As antisense RNA gets intronic. Omics. (2005) 9:2–12. doi: 10.1089/omi.2005.9.2
33. Chorev M, Carmel L. The function of introns. Front Genet. (2012) 3:55. doi: 10.3389/fgene.2012.00055
34. Zhang Y, Bitner D, Pontes Filho AA, Li F, Liu S, Wang H, et al. Expression and function of NIK- and IKK2-binding protein (NIBP) in mouse enteric nervous system. Neurogastroenterol Motility. (2014) 26:77–97. doi: 10.1111/nmo.12234
35. Abbott MA, Wells DG, Fallon JR. The insulin receptor tyrosine kinase substrate P58/53 and the insulin receptor are components of CNS synapses. J Neurosci. (1999) 19:7300–8. doi: 10.1523/JNEUROSCI.19-17-07300.1999
36. Usman M, Li Y, Ke Y, Chhetri G, Islam MA, Wang Z, et al. TRAPPC9 deficiency impairs the plasticity of stem cells. Int J Mol Sci. (2022) 23:4900. doi: 10.3390/ijms23094900
37. Yang Y, Ge H, Li DQ, Xu AX. E2F1-induced lncRNA BAIAP2-AS1 overexpression contributes to the malignant progression of hepatocellular carcinoma via miR-361-3p/SOX4 axis. Dis Markers. (2021) 2021:6256369. doi: 10.1155/2021/6256369
38. Solís-Fernández G, Montero-Calle A, Martínez-Useros J, López-Janeiro Á, de Los Ríos V, Sanz R, et al. Spatial proteomic analysis of isogenic metastatic colorectal cancer cells reveals key dysregulated proteins associated with lymph node, liver, and lung metastasis. Cells. (2022) 11:447. doi: 10.3390/cells11030447
39. Chen Q, Jia A, Snyder SA, Gong Z, Lam SH. Glucocorticoid activity detected by in vivo zebrafish assay and in vitro glucocorticoid receptor bioassay at environmental relevant concentrations. Chemosphere. (2016) 144:1162–9. doi: 10.1016/j.chemosphere.2015.09.089
40. Seebacher F, Zeigerer A, Kory N, Krahmer N. hepatic lipid droplet homeostasis and fatty liver disease. Semin Cell Dev Biol. (2020) 108:72–81. doi: 10.1016/j.semcdb.2020.04.011
41. Alves-Bezerra M, Cohen DE. Triglyceride metabolism in the liver. Compr Physiol. (2017) 8:1–8. doi: 10.1002/cphy.c170012
Keywords: Hu sheep, TRAPPC9, BAIAP2, fat deposition related traits, qRT-PCR
Citation: Cui P, Wang W, Zhang D, Li C, Huang Y, Ma Z, Wang X, Zhao L, Zhang Y, Yang X, Xu D, Cheng J, Li X, Zeng X, Zhao Y, Li W, Wang J, Lin C, Zhou B, Liu J, Zhai R and Zhang X (2022) Identification of TRAPPC9 and BAIAP2 Gene Polymorphisms and Their Association With Fat Deposition-Related Traits in Hu Sheep. Front. Vet. Sci. 9:928375. doi: 10.3389/fvets.2022.928375
Received: 25 April 2022; Accepted: 14 June 2022;
Published: 05 July 2022.
Edited by:
Ran Di, Institute of Animal Sciences (CAAS), ChinaReviewed by:
Yongfu La, Lanzhou Institute of Husbandry and Pharmaceutical Sciences (CAAS), ChinaCopyright © 2022 Cui, Wang, Zhang, Li, Huang, Ma, Wang, Zhao, Zhang, Yang, Xu, Cheng, Li, Zeng, Zhao, Li, Wang, Lin, Zhou, Liu, Zhai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxue Zhang, emhhbmd4eEBnc2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.