
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 June 2022
Sec. Parasitology
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.917389
This article is part of the Research Topic Vaccines Against Parasitic Infections in Domestic Animals View all 8 articles
 Amarin Rittipornlertrak1,2
Amarin Rittipornlertrak1,2 Boondarika Nambooppha1,3
Boondarika Nambooppha1,3 Anucha Muenthaisong1
Anucha Muenthaisong1 Nisachon Apinda1
Nisachon Apinda1 Pongpisid Koonyosying1
Pongpisid Koonyosying1 Wanwisa Srisawat1
Wanwisa Srisawat1 Paweena Chomjit1
Paweena Chomjit1 Kanokwan Sangkakam1
Kanokwan Sangkakam1 Veerasak Punyapornwithaya2
Veerasak Punyapornwithaya2 Saruda Tiwananthagorn3
Saruda Tiwananthagorn3 Naoaki Yokoyama4
Naoaki Yokoyama4 Nattawooti Sthitmatee1,3,5*
Nattawooti Sthitmatee1,3,5*Both strong innate and adaptive immune responses are an important component of protection against intraerythrocytic protozoan parasites. Resistance to bovine babesiosis is associated with interferon (IFN)-γ mediated responses. CD4+ T cells and macrophages have been identified as major effector cells mediating the clearance of pathogens. Previously, the apical membrane antigen 1 (AMA-1) was found to significantly induce the immune response inhibiting B. bovis merozoite growth and invasion. However, a detailed characterization of both humoral and cellular immune responses against the structure of B. bovis AMA-1 (BbAMA-1) has not yet been established. Herein, the present study aimed to express the recombinant BbAMA-1 domain I+II protein [rBbAMA-1(I/II)], which is the most predominant immune response region, and to characterize its immune response. As a result, cattle vaccinated with BbAMA-1(I/II) significantly developed high titters of total immunoglobulin (Ig) G antibodies and a high ratio of IgG2/IgG1 when compared to control groups. Interestingly, the BbAMA-1(I/II)-based formulations produced in our study could elicit CD4+ T cells and CD8+ T cells producing IFN-γ and tumor necrosis factor (TNF)-α. Collectively, the results indicate that immunization of cattle with BbAMA-1(I/II) could induce strong Th1 cell responses. In support of this, we observed the up-regulation of Th1 cytokine mRNA transcripts, including IFN-γ, TNF-α, Interleukin (IL)-2 and IL-12, in contrast to down regulation of IL-4, IL-6 and IL-10, which would be indicative of a Th2 cytokine response. Moreover, the up-regulation of inducible nitric oxide synthase (iNOS) was observed. In conclusion, this is the first report on the in-depth immunological characterization of the response to BbAMA-1. According to our results, BbAMA-1 is recognized as a potential candidate vaccine against B. bovis infection. As evidenced by the Th1 cell response, it could potentially provide protective immunity. However, further challenge-exposure with virulent B. bovis strain in immunized cattle would be needed to determine its protective efficacy.
Most apicomplexans are obligate intracellular parasites. Babesia, an apicomplexan parasite, is a tick-transmitted hemoprotozooan. The most economically relevant bovine babesias are Babesia bovis (B. bovis) and Babesia bigemina (B. bigemina). Presently, a large number of cattle are at risk of exposure to bovine babesiosis. Bovine babesiosis is recognized as a global disease that has been the cause of significant economic losses to the livestock industry (1). Climate change is one of the factors that can influence the control of bovine babesiosis (2). Pathologically, a cyclical asexual replication of Babesia in red blood cells (RBC) that is associated with an excessive immune response has been known to lead to the development of several clinical manifestations. In general, bovine babesiosis can be treated with chemotherapy (1). However, the emergence of drug resistance in B. bovis has been recently documented (3). Although live attenuated vaccines are available in some endemic countries, the applications for wide use of these vaccines are limited for a number of reasons. At present, there are no safe and effective vaccines that protect cattle against the virulent pathogens (4). Thus, novel vaccines are increasingly desirable and urgently needed to control this disease (2).
The host-parasite interaction is of major importance for parasite survival. Without host cells, which supply environment and nutrition, the parasitic protozoa cannot grow and survive. The formation of a moving junction (MJ) between the parasite and host cell membranes is an important conserved mechanism that facilitates the parasitic invasion into host cells during the asexual growth cycle of apicomplexan parasites (5). The apical membrane antigen 1 (AMA-1) has been well characterized and reported to be involved in MJ formation in coordination with the rhoptry neck 2 (RON2) protein in Toxoplasma and Plasmodium (6, 7). It is likely directly responsible for reorientation and initiates the junctional contact. AMA-1, a microneme protein (MIC), is a type I integral membrane protein that is composed of three distinct structures (8). This protein is not only well conserved among Plasmodium species (9) but also other Apicomplexa species including Toxoplasma (10), Neospora (11) and Babesia (12–14). Unfortunately, little is known about the biological function of AMA-1. Several lines of evidence have demonstrated that AMA-1 plays an essential role in the invasion process (6, 7, 10, 15) and is secreted onto the surface at or around the time of merozoite egression (16). Attempts to inactivate ama-1 of Plasmodium falciparum (P. falciparum) (17) and Toxoplasma gondii (T. gondii) (18) have strongly supported the contention that AMA-1 plays a central role in merozoite invasion. Moreover, it either directly or indirectly plays a role in the resealing of the red blood cells at the posterior end of the invasion event, which would indicate that this protein may be essential to the survival of the parasite. However, it has been argued that AMA-1 plays a role in host cell attachment rather than MJ formation as a way of facilitating the invasion (19). Therefore, the importance of this protein continues to be debated.
Interestingly, AMA-1 is not only capable of involvement in parasitic invasion, it is also potentially an immunogen. Immunological studies in animal models have revealed that immunization with AMA-1 was able to induce significant protection against homologous but not heterologous malaria parasites (20–22). Accordingly, the molecules that are involved in immune system recognition and the steps of invasion are of great interest for the development of prophylaxis. For this reason, AMA-1 is a candidate antigen of significant interest for vaccine development against apicomplexan parasite infection. Given its important role and the immune response data, AMA-1 has been extensively studied as a potential long-standing malaria vaccine candidate (23). Genetically, a high degree of sequence polymorphisms was observed in domain I of the extracellular domain of Plasmodium ama-1. It has been suggested that the presence of the polymorphism is presumably due to host immune pressure (24, 25). The crystal structure reveals AMA-1 has a hydrophobic cleft that runs across domains I and II (26). This hydrophobic cleft was thought to be an AMA-1 ligand-binding site and a major target of protective immunity (27). Interestingly, it has been previously identified as the major target of the invasion-inhibitory monoclonal antibody (25, 27–29) and the invasion-inhibitory peptide (30, 31). Therefore, it is likely that domains I and II are the predominant targets for inhibitory responses and immunogenicity (32–34).
In consideration of the fact that domains I and II of AMA-1 are of vital importance to the apicomplexan parasite, this protein is of great interest as a candidate for subunit vaccine development against bovine babesiosis. Among B. bovis strain, ama-1 has been highly conserved. Importantly, a strong negative or purifying selection across the whole of the gene was established, especially in domain I, indicating strong functional constraints on this gene (35). Interestingly, B. bovis AMA-1 (BbAMA-1) is recognized by the antibodies against the epitopes that are mainly situated within domain I (12, 36) and domain II (37). These antibodies could inhibit the in vitro growth and invasion of erythrocytes by B. bovis merozoites. Collectively, the results indicate that BbAMA-1 domains I and II are potential targets in the development of a promising bovine babesiosis vaccine. To the best of our knowledge, detailed characterizations of both humoral and cellular immune responses against the structure of BbAMA-1 have not yet been established. To gain a more comprehensive understanding of the immune response elicited by BbAMA-1, we have herein expressed the structure of domains I and II of BbAMA-1 (Ser40-Glu438) and characterized their immunological functions.
Twenty healthy 10–16-month-old Holstein Friesian dairy cows were included in this study. Dairy cattle were kept in a free-stall barn. All cattle were acquired from farm members under the Mae Wang Dairy Cooperative, Mae Wang District, Chiang Mai Province, Thailand. Each cow was screened for B. bovis infection by PCR, as has been previously described (35), and then subjected to the immunofluorescence antibody test (IFAT) (38).
The sequence of ectodomains I and II B. bovis ama-1 encoding Ser40-Glu438 was selected from GenBank accession number KY575957. Codons were optimized, synthesized (Genscript, Piscataway, NJ, United States) and inserted into pQE-32 vector (Qiagen GmbH, Hilden, Germany) with N-terminal SacI and C-terminal HindIII enzyme sites. Accordingly, pQE-32/BbAMA-1(I/II) plasmid was transformed into Escherichia coli (E. coli) M15 strain using the heat shock method at 42°C for 45 s and then spread on Luria-Bertani (LB; Difco™, Sparks, MD, United States) agar containing 100 μg/ml ampicillin and 25 μg/ml kanamycin and incubated at 37°C for 16 h. A positive single colony detected by pQE specific vector primer (Primer-Type III/IV-pQE forward: CGGATAACAATTTCACACAG; Primer-pQE Reverse: GTTCTGAGGTCATTACTGG) was inoculated in LB broth (Difco™) containing 100 μg/ml ampicillin and 25 μg/ml kanamycin. It was then incubated in a shaking incubator at 250 rpm at a temperature of 37°C for 16 h for the purpose of starter culture preparation. One-liter of LB broth medium (LB broth, 100 μg/ml ampicillin and 25 μg/ml kanamycin) was inoculated in a ratio of 1:50 with the starter culture and continuously grown in the shaking incubator under the same growth conditions until an OD600 of 0.6 was reached. The recombinant protein expression was subsequently induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG; Amresco, Solon, OH, United States) to a final concentration of 0.5 mM, while the culture was incubated for an additional 12 h. Finally, cells were harvested by centrifugation at 4,000 × g for 20 min at 4°C and kept at −20°C for purification.
The cell pellets were lysed in lysis buffer (Denaturing conditions; 100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea; pH 8.0) with gentle shaking at 4°C for 1 h. The suspension was then centrifuged at 10,000 rpm at 4°C for 30 min. The supernatant was transferred to new tubes and maintained at −80°C. The purification process of the recombinant 6xHis-tagged proteins in this study was conducted by employing Ni-NTA affinity chromatography according to the manufacturer's instructions (Qiagen GmbH, Hilden, Germany). The purified recombinant protein was dialyzed by gradually decreasing the urea concentration. The rBbAMA-1(I/II) protein was concentrated using Amicon® centrifugation filters (30 kDa MWCO; Merck KGaA, Darmstadt, Germany) and concentration was then measured using the BCA protein assay kit (Pierce®, Rockford, IL, United States) according to the manufacturer's instructions.
Either lysate or purified protein was separated by 12.5% SDS-PAGE gel in a mini-slab apparatus (Bio-Rad Laboratories, Hercules, CA, United States). The process was run with 100 V for 1 h. The SDS-PAGE slab gels were then stained with Coomassie blue R-250 (Sigma-Aldrich, St. Louis, MO, United States) for protein band detection. Moreover, the proteins obtained from the SDS-PAGE gel were electrically transferred onto a nitrocellulose membrane (Merck Millipore™, Merck KGaA, Darmstadt, DEU) at 15 V for 1 h. The membrane was then incubated with previously produced rabbit polyclonal anti-sBbAMA-1 serum (1:50 dilution) at 4°C overnight (36). Subsequently, the membrane was probed with horseradish peroxidase (HRP)-conjugated goat anti-bovine immunoglobulin (Ig) G antibody (1:2,000 dilution; KPL, Gaithersburg, MD, United States). The membrane was then incubated with gentle shaking at room temperature for 1 h and then washed three times with washing buffer. Finally, the reactions were visualized using a solution containing 3,3'-diaminobenzidine (DAB; Invitrogen, Carlsbad, CA, United States) and hydrogen peroxide (H2O2; Merck, Germany).
Vaccine formulations were prepared by mixing different rBbAMA-1(I/II) concentrations (50 and 100 μg) with the Montanide™ ISA 206 VG adjuvant (1:1 V/V, SEPPIC, Paris, France) to a total volume of 1 ml/dose. The phosphate-buffered saline (PBS) formulated with only the adjuvant was used as a control. The vaccines were freshly prepared and stored at 4°C until being used. Cattle were divided into four groups based on the vaccine formulations (Table 1). All groups were intramuscularly immunized four times at 2-week intervals with the exception of the non-immunized group. Blood samples were collected before immunization and every 2 weeks for 10 weeks in order to determine the immunological responses. Adverse events, including pain, swelling at the injection site and behavioral changes, were monitored throughout the course of the experiment.
The immunoplate (Nunc-immunoTM plate, Denmark) was coated with 100 μg of rBbAMA-1(I/II) protein in a coating buffer (0.05 M carbonate bicarbonate buffer, pH 9.6) and then incubated overnight at 4°C. Unbound antigens were washed three times with a washing buffer [PBS (pH 7.2) containing 0.05% Tween-20; PBST]. Non-specific bindings were blocked with a blocking buffer (1% skim milk in PBS) and the plate was incubated at room temperature for 1 h. After thrice washing, cattle serum samples at a dilution of 1:50 in the blocking buffer were added to the wells in duplicate and they were then incubated at room temperature for 1 h. After being washed thrice with the washing buffer, HRP-conjugated goat anti-bovine IgG antibody (KPL, Gaithersburg, MD, United States) was used as the secondary antibody at a dilution of 1:2000 in the blocking buffer and incubated at room temperature for 1 h. After being washed three times, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (SeraCare Life Sciences, Gaithersburg, MD, United States) was added and the samples were incubated at room temperature in the dark for 15 min. The reaction was terminated by the addition of 2 M H2SO4. The optical density at 450 nm (OD450) was measured using an automatic ELISA plate reader (AccuReader, Metertech, Taipei, Taiwan R.O.C.). All samples and controls were run in duplicate.
To determine the IgG subclass responses (IgG1 and IgG2), dilutions of the specific antibodies were used as is shown in Table 2. The indirect ELISA protocol has been described above.
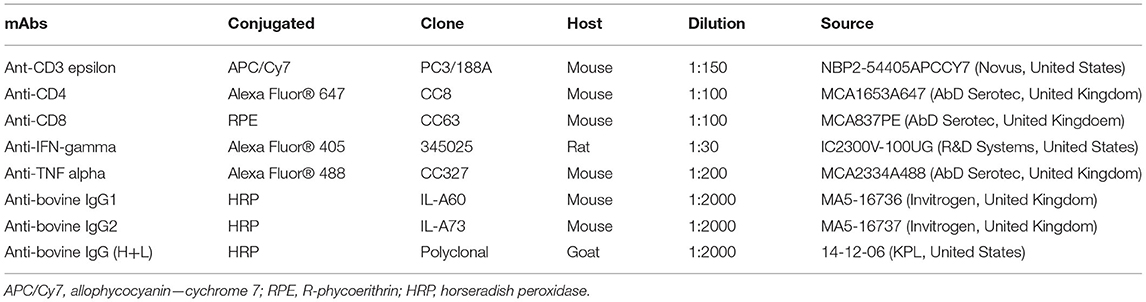
Table 2. Monoclonal and polyclonal antibodies (mAbs) used in this study for flow cytometry analysis and immunological assay.
Peripheral blood mononuclear cell (PBMC) isolation was performed according to the method described in a previous study (39). Jugular or tail blood samples collected at week 10 of the experiment, that had been kept in ethylene diamine tetraacetic acid (EDTA) tubes (BD Vacutainer, Plymouth, United Kingdom), were diluted with sterilized PBS (pH 7.2; 1:1 dilution) and overlaid on 5 ml of Lymphoprep™ (STEMCELL Technologies, Vancouver, Canada) in 15 ml conical tubes. They were then centrifuged using a swinging bucket rotor at 400 × g at a temperature of 25°C and an acceleration of nine with no break for 30 min. The PBMC layer was carefully collected from the tube and transferred to a new 50 ml conical tube. The contaminated red blood cells were lysed by the 1 × red blood cell lysis buffer for 3 min at room temperature. PBMCs were then washed twice with PBS by centrifugation at 700 × g for 5 min at 25°C. The pellets were then resuspended with complete RPMI medium [RPMI 1640 medium supplemented with 1X antibiotic-antimycotic (100 units/ml of penicillin, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B; Gibco™, Life Technologies Waltham, MA, United States) and 10% fetal calf serum (FCS, Gibco™)]. Cell viability was assessed by employing the Trypan Blue exclusion test carried out in a 0.1 mm Bürker chamber. Cells were then cultured in 24-well cell culture plates (2 × 106 cells/well). PBMC cells were re-stimulated with rBbAMA-1(I/II) at a concentration of 30 μg/ml that had been obtained from the previous step. Concanavalin A (2.5 μg/ml, eBioscience, Carlsbad, CA, USA) and RPMI medium were used as positive and negative controls, respectively. Plates were incubated for 72 h at 37°C in an atmosphere containing 5% CO2.
To investigate the effects of rBbAMA-1(I/II) on cytokine expression, total RNA was extracted from cultured PBMCs, which were then stimulated with antigen using a PureLink™ RNA Mini Kit (Invitrogen, San Diego, CA, United States) according to the manufacturer's instructions. The concentration and quality of RNA were determined using a spectrophotometer. Total RNA (1 μg) was reverse-transcribed to synthesize cDNA using a Tetro™ cDNA Synthesis Kit (Bioline, London, United Kingdom) according to the manufacturer's instructions. The cDNA obtained from each sample was then used as a template to measure the expression of some genes involved in the inflammatory response. Real-Time quantitative PCR was carried out in the CFX96 Touch™Real-Time PCR (Bio-Rad, Hercules, CA, United States). A SensiFAST™ SYBR® Lo-ROX Kit (Bioline, London, United Kingdom) was used in an optimized 20 μl reaction volume according to the manufacturer's instructions. The reaction mixture contained 10 μl of 2x SensiFAST SYBR Lo-ROX Mix, 0.8 μl of 10 μM each primer, 2 μl of cDNA and 6.4 μl of RNase-free water. The real-time PCR conditions were as follows: an initial denaturation step at 95°C for 2 min followed by 40 cycles of 95°C for 10 s and an annealing step at 58°C for 30 s. The expression of each gene was normalized relative to the expression of Glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Genes involved in the inflammatory response, including inducible nitric oxide synthase (iNOS), Interleukin (IL)-2, IL-4, IL-6, IL-10, interferon (IFN)-γ and tumor necrosis factor (TNF)-α were analyzed. Sequences of the primers used in this Real-Time PCR are presented in Table 3. The expression levels (fold-difference) of each gene were calculated using the 2−ΔΔCT method (45).
For functional characterizations of the CD4+ and CD8+ cells, PBMC cells were stimulated in vitro as has been mentioned above. Brefeldin A (2 μg/ml; Sigma, St. Louis, MO, USA) was added for the last 6 h to facilitate intracellular cytokine accumulation. Following incubation, the PBMC cells were washed twice with staining buffer (1X PBS, 0.01% NaN3, 10% FBS) and were subsequently incubated with the respective anti-bovine monoclonal antibodies (CD3- APC/Cy7, CD4- Alexa Fluor® 647, CD8-RPE; Table 2). Cells were then fixed and permeabilised with Intracellular Fixation & Permeabilization Buffer Set (eBioscience, Carlsbad, CA, USA) and stained intracellularly with anti-bovine IFN-γ-Alexa Fluor® 405 and TNFα- Alexa Fluor® 488 antibodies (Table 2). Flow cytometry was performed by acquiring 10,000 events in the live lymphocyte gate using DxFLEX Flow Cytometer (Beckman Coulter, Brea, CA, United States) and further analyzed using CytExpert for DxFLEX Software.
Statistical analysis was carried out using Graph Pad Prism 8.0.2 (GraphPad Software, Inc., San Diego, CA, United States). Statistical significance between groups was determined by one-way ANOVA or non-parametric ANOVA. Values of P < 0.05 were taken to be statistically significant. The results of the assumption test were analyzed before statistical analysis was performed.
The expression of the rBbAMA-1(I/II) protein was analyzed by SDS-PAGE (Figure 1A) and western blotting analysis (Figure 1B). The overexpression of the target protein band was observed to be ~48 kDa on SDS-PAGE gel. Western blotting analysis confirmed that the rBbAMA-1(I/II) protein was successfully expressed in this study, as was established through the use of the specific-sBbAMA-1 antibody.
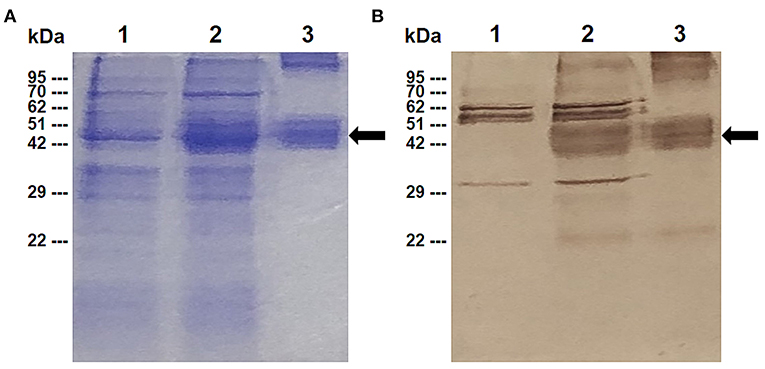
Figure 1. Expression of rBbAMA-1(I/II) analyzed using SDS-PAGE stained with Coomassie brilliant blue (A) and western blotting analysis (B). Lane 1: Non-induced E. coli strain M15 containing pQE-32/BbAMA-1(I/II) plasmid; Lane 2: Induced E. coli strain M15 containing pQE-32/BbAMA-1(I/II) plasmid; Lane 3: Purified rBbAMA-1(I/II) protein. Black arrows indicate the target protein band of rBAMA-1(I/II).
In order to characterize the antibody response elicited by rBbAMA-1(I/II), the cattle sera that were immunized with rBbAMA-1(I/II) were evaluated using western blotting analysis and indirect ELISA. The results revealed that the pooled sera of the rBbAMA-1(I/II)-immunized groups collected from week 10 specifically reacted to the purified rBbAMA-1(I/II) at ~48 kDa on the nitrocellulose membrane (Figure 2A).In consideration of the antibody response, the specific IgG to purified rBbAMA-1(I/II) was detected among the rBbAMA-1(I/II)-immunized groups. Remarkably, the antibody titers continuously increased throughout the experiment when compared to those of the control groups (Figure 2B).
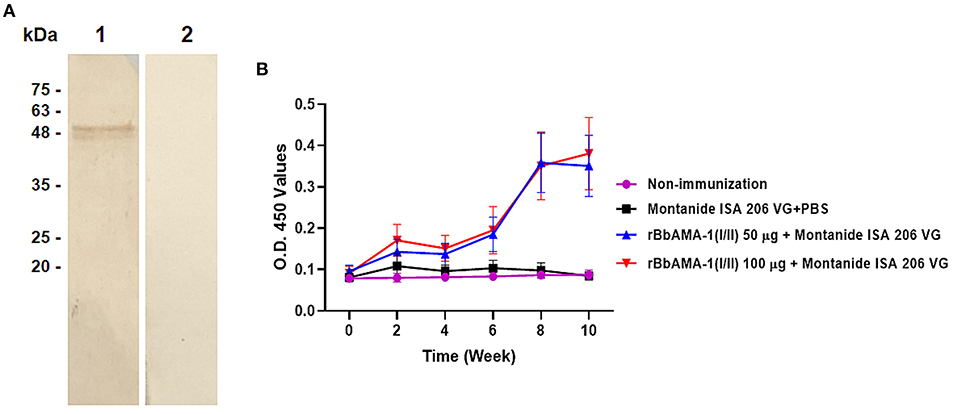
Figure 2. Determination of rBbAMA-1(I/II)-specific antibody response. (A) Western blotting analysis demonstrated that pooled cattle serum immunized with rBbAMA-1(I/II) reacted with the purified recombinant protein (lane 1) when compared to the pre-immunization period (lane 2). (B) Indirect ELISA test revealed that the antibody response continuously increased after the first immunization with rBbAMA-1(I/II).
Furthermore, an indication of the bias toward the T helper type 1 (Th1) cell response was also characterized by measuring the IgG1 and IgG2 bovine isotypes in the present study. The IgG2/IgG1 ratio >1 was an indicator of a Th1 type response. As a result, the ratio of IgG2/IgG1 in cattle immunized with 100 μg of the rBbAMA-1(I/II) group was higher than for the cattle immunized with 50 μg of rBbAMA-1(I/II) and the control groups. As observed, this ratio increased in the vaccinated groups throughout the course of the experiment. However, there was no statistical significance in the IgG2/IgG1 ratio among the rBbAMA-1(I/II)-immunized groups (Figure 3). Additionally, statistically significant differences were observed over the last 3 weeks, especially in cattle immunized with 100 μg of the rBbAMA-1(I/II) when compared with those in the control groups.
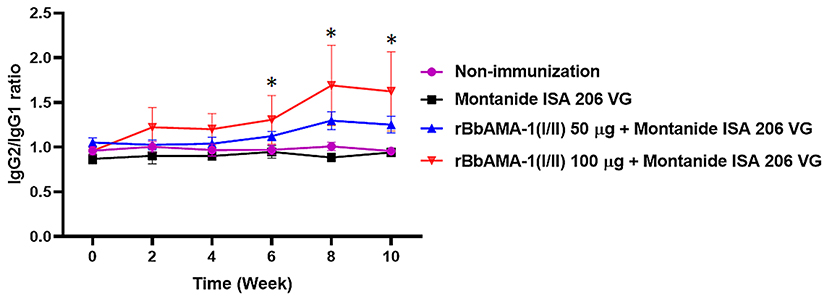
Figure 3. Time course of IgG2/IgG1 ratio response to rBbAMA-1(I/II) assessed by indirect ELISA. Each time point depicts the group mean value ± SD of the IgG2/IgG1 ratio. Asterisks indicate a significant difference (P < 0.05).
To determine the cellular immune responses induced by rBbAMA-1(I/II), peripheral blood mononuclear cells (PBMCs) isolated from animals in each group were cultured in vitro and re-stimulated with rBbAMA-1(I/II). Subsequently, flow cytometry analysis was performed to characterize the CD4+ and CD8+ T cell response by producing pro-inflammatory cytokines (IFN-γ and TNF-α). The present findings demonstrate that rBbAMA-1(I/II) could trigger bovine PBMCs to elevate the proportion of antigen-specific CD4+ T cell secreting IFN-γ with statistical significance in both groups of cattle immunized with 50 μg (P < 0.05) and 100 μg of rBbAMA-1(I/II; P < 0.01), as is shown in Figure 4A. Furthermore, 100 μg of rBbAMA-1(I/II) significantly produced higher percentages of CD4+ T cell secreting TNF-α (P < 0.01) in comparison with the control groups. With regard to the CD8+ T cells, the present study found that both rBbAMA-1(I/II) immunized groups elicited high frequencies of CD8+ T cell secreting IFN-γ with statistical significance (P < 0.05 and P < 0.0001 for 50 and 100 μg of the rBbAMA-1(I/II), respectively). Meanwhile, high proportions of antigen-specific CD8+ T cell secreting TNF-α were detected in cattle immunized with 100 μg of the rBbAMA-1(I/II) with statistical significance (P < 0.0001) (Figure 4B).
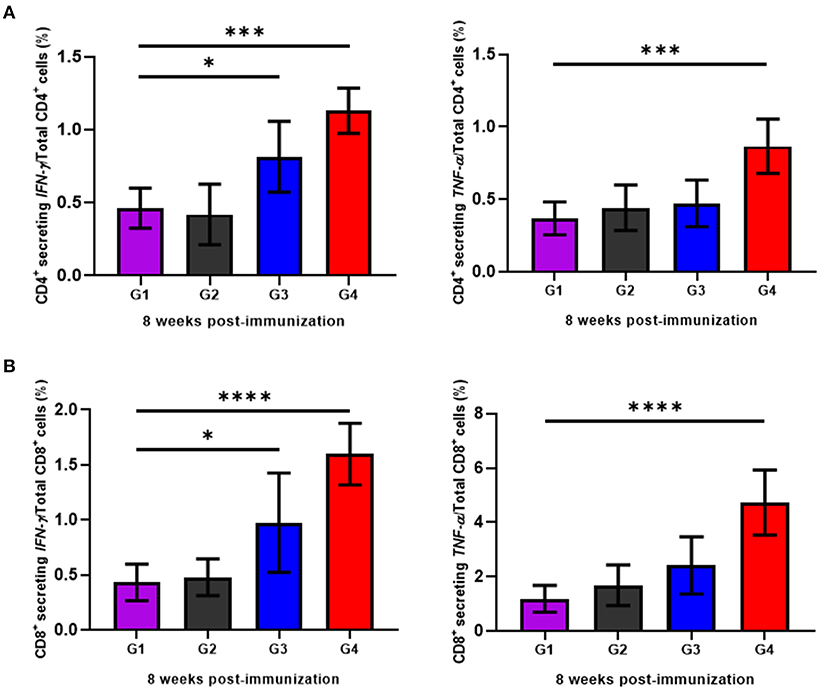
Figure 4. Determination of cellular immune response of cattle immunized with rBbAMA-1(I/II) analyzed by flow cytometry analysis. (A) Frequency of IFN-γ and TNF-α secreting CD4+ T cells. (B) Frequency of IFN-γ and TNF-α secreting CD8+ T cells. The results are expressed as group mean values ± SD. Asterisks indicate a significant difference (* < 0.05; *** < 0.001; **** < 0.0001). Experimental groups G1 = non-immunization, G2 = Montanide ISA 206 VG + PBS, G3 = rBbAMA-1(I/II) 50 μg + Montanide ISA 206 VG and G4 = rBbAMA-1(I/II) 100 μg + Montanide ISA 206 VG.
In particular, cytokine gene expression levels were also investigated in order to gain an insight into cellular response at the gene level. A significant up-regulation among cytokine genes, including IFN-γ (P < 0.05), TNF-α (P < 0.01), IL-2 (P < 0.001), IL-12 (P < 0.01) and iNOS (P < 0.05) in cattle immunized with 100 μg of the rBbAMA-1(I/II), were observed after the rBbAMA-1(I/II)-stimulated cultures were compared with the control groups (Figure 5). Higher expression levels of pro-inflammatory cytokines IFN-γ and IL-12 were observed. Although cattle immunized with 50 μg of rBbAMA-1(I/II) induced cytokine gene expression, no significant differences were observed with the exception of IL-2 (P < 0.01) and IFN-γ (P < 0.05). In contrast, mRNA transcription of IL-4, IL-6 and IL-10 were down-regulated when compared with the control groups. In addition, IL-10 gene expression was significantly down-regulated in cattle immunized with 100 μg of the rBbAMA-1(I/II) group (P < 0.01).
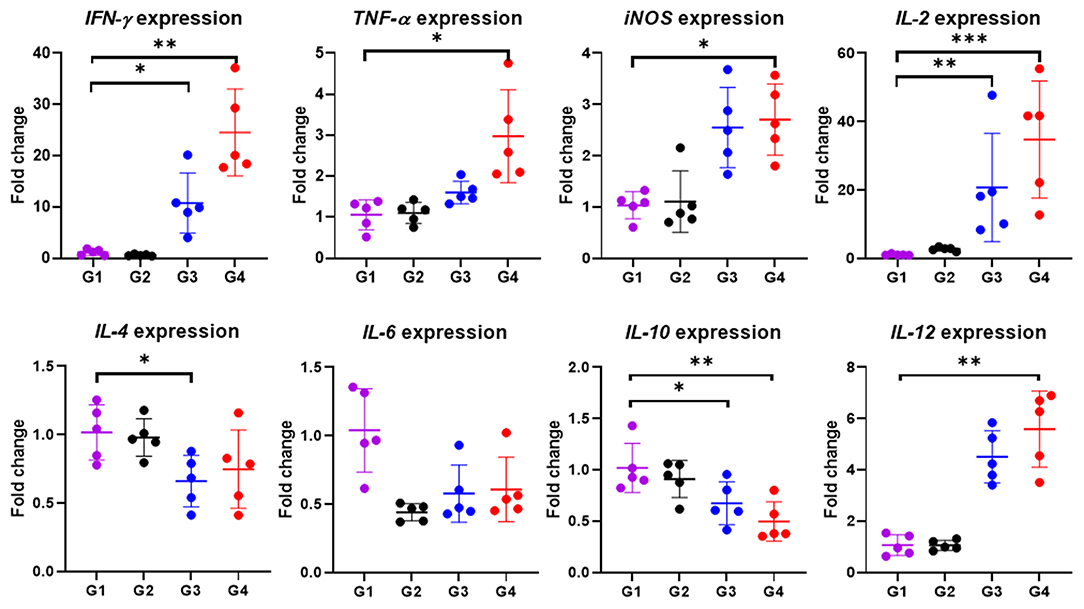
Figure 5. Inflammatory cytokine gene expression profiles induced by rBbAMA-1(I/II). The results are expressed as mean values ± SD. Asterisks indicate a significant difference (* < 0.05; ** < 0.01; *** < 0.001). Experimental groups G1 = non-immunization, G2 = Montanide ISA 206 VG + PBS, G3 = rBbAMA-1(I/II) 50 μg + Montanide ISA 206 VG and G4 = rBbAMA-1(I/II) 100 μg + Montanide ISA 206 VG.
Babesia bovis is an economically important pathogen known to cause bovine babesiosis worldwide. Due to the fact that the vaccines currently being administered are neither fully safe nor effective, researchers have extensively searched for a candidate vaccine antigen that could be used to develop an effective vaccine against B. bovis (4). As has been found with BbAMA-1, studies of its immunogenicity demonstrated its vaccine potential (12, 36, 37). However, a lack of in-depth knowledge and understanding of the immunological characterization of the response may have implications for the design and assessment of AMA-1-based vaccines for bovine babesiosis. Therefore, basic research efforts involving the identification and characterization of the immunological function would be very important in the development of recombinant vaccines. In the present study, the ectodomains I and II region of BbAMA-1 has been successfully expressed as a doublet band corresponding to around 48 kDa. Because BbAMA-1 encompasses the extracellular cysteine-rich domains I and II (12, 36, 37). It is likely that cysteine oxidation and formation of intra-molecular disulfide bonds is responsible for the doublet formation in Laemmli SDS-PAGE. Moreover, doublet in SDS-PAGE may result from proteolytic degradation (46, 47). However, the doublet bands were identical as the same protein identified by anti-sBbAMA-1 serum, which is specific to 50 amino acid residues of recombinant BbAMA-1(I/II) protein. Therefore, it was used for further immunization.
Determining the functional regions of the B. bovis antigens could be one of the best strategies for blocking this parasite's biological functions during host cell entry (48). It has been suggested that AMA-1 participates in the invasion stage, which is conserved for most apicomplexa. Immunological investigations in the present study showed that rBbAMA-1(I/II) could induce a humoral immune response, for which the IgG2 isotype response was predominantly higher than IgG1 isotype. Enhancement of IgG2 over the IgG1 antibody response indicates a bias toward the Th1 cell response (49). The evidence of IFN-γ detected in our present study is one of the justifications for the predominant IgG2 isotype (50). Generally, it has been suggested that the bovine IgG2 isotype possesses more functional opsonic characteristics than IgG1 in enhancing phagocytic activity and NO production (51). Interestingly, it has been reported that a higher IgG2a/IgG1 ratio is associated with a protective immune response against the intracellular pathogen (52). Therefore, evidence of the fact that IgG2 response in this study was higher than IgG1 indicated the possibility that rBbAMA-1(I/II) was capable of providing protective immunity against intracellular protozoan B. bovis infection.
To defeat the Babesia pathogen, not only is a humoral immune response important in eliminating the parasite, but so is a cellular immune response (1). Therefore, a cellular immune response has been studied and described in the present study in an attempt to gain a more comprehensive understanding of rBbAMA-1(I/II) immunological characteristics so as to evaluate its vaccine performance. In Plasmodium, CD4+ T cells displayed a central role against parasitized erythrocytes after being activated by antigen-presenting cells during blood-stage malaria (53). Interestingly, IFN-γ and TNF-α producing CD4+ T cells play a crucial role in the protective immune response to the blood-stages of the malaria parasite in humans (54, 55). Remarkably, the secretion of both IFN-γ and TNF-α from the same T cell would be more effective than cytokine alone for killing (56). Previous studies have also suggested that CD4+ T cells and IFN-γ are required to establish protective immunity against infection with Babesia parasites (57, 58). It has been determined that IFN-γ is involved in the protective innate immune response to B. bovis in calves (59–61). Therefore, it has been proposed that vaccines that prime CD4+ T cells to produce IFN-γ could induce and provide protective immunity against Babesia infection (4). Previously, the search for B. bovis candidate antigens as subunit vaccines has determined that some antigens have the potential to elicit CD4+ T cells in producing IFN-γ or TNF-α such as Bb-1 (62), Bbo20 (63), rhoptry-associated protein 1 (RAP-1) (64), small heat shock protein (Hsp20) (65) and merozoite surface antigens (MSAs) (66). Likewise, the flow cytometry analysis conducted in our study found that CD4+ secreting IFN-γ/TNF-α T cells was detected in cattle that had been immunized with rBbAMA-1(I/II). In support of this result, the up-regulation of IFN-γ and TNF-α mRNA transcription was noted. In this regard, the presence of IFN-γ cytokine indicates that the Th1 cell response is required to mediate protection against a variety of intracellular infections (67). In accordance with the antibody response obtained from our study, we have verified the immunological role of BbAMA-1(I/II) in induction of Th1 response against B. bovis. Therefore, it can be concluded that the rBbAMA-1(I/II)-based vaccine formulated in this study may provide effective protective antibodies against B. bovis infection.
Apart from the CD4+ T cell response, CD8+ T cells are vital in offering protection against liver-stage malaria (53). Unlike Plasmodium parasites, the pre-erythrocytic stage is not found in the Babesia parasite (68). Therefore, the majority of the immune responses against Babesia is dependent upon the CD4+ T cell response rather than the CD8+ T cell response (4). However, CD8+ T cells still play an important role in contributing to the clearance and immune memory against many intracellular pathogens, as has been observed in many incidences of malaria parasite infection (53). Currently, a number of studies have reported that CD8+ T cells are associated with the control of the Babesia parasite (60, 69). Interestingly, high percentages of antigen-specific CD8+ secreting IFN-γ/TNF-α T cells have been detected in our study. The CD8+ T cells secreting IFN-γ/TNF-α may have enhanced cytolytic activity (56). In this case, the generation of robust CD8+ responses would likely be relevant in vaccinations. This would likely be due to the fact that Th1 CD4+ T cells provide assistance in establishing optimal CD8+ T cell effector activity (70). Therefore, if sufficient antigen-specific CD8+ T cell responses are generated and provided against the vaccine, disease can be prevented and controlled.
In addition to T cell lymphocytes, macrophages are the key effector cells that mediate the clearance of pathogens by killing the organisms associated with phagocytosis in innate immune response and by regulating the consequent adaptive immune responses (4, 68). Previous studies have demonstrated that macrophages are critical for establishing protective immunity and resistance to Babesia infection in mice (71). Furthermore, they are responsible for producing an inflammatory cytokine of nitric oxide (NO) via the co-stimulation of biologically active IFN-γ and TNF-α or the presence of parasite derived products (72). Interestingly, it has been determined that NO has been shown to exhibit babesiacidal activity (73). Therefore, the resistance to babesiosis appears to correlate with an increase in NO production (58). Although, active NO levels were not determined in our study, the in vitro up-regulation of mRNA expression by iNOS, a key enzyme in the macrophage inflammatory response, was observed among cattle that had been immunized with rBbAMA-1(I/II) groups when compared with the control groups. This observation is likely related to the presence of the CD4+ secreting IFN-γ/TNF-α T cells or the indication that rBbAMA-1(I/II) may contribute to the enhancement of the transcription of iNOS. Consequently, this would imply that active NO production could possibly be induced by the rBbAMA-1(I/II)-based vaccine.
In addition to NO, other pro-inflammatory cytokines are involved in the defense against intracellular pathogens and are important in activating an innate and acquired immune response. Using real-time PCR, our results showed that not only were IFN-γ and TNF-α responsible for the Th1 response when up-regulated in cattle receiving rBbAMA-1(I/II), but that IL-2 and IL-12 were also directly involved. Interestingly, our results were similar to those of the study involving the AMA-1 of Plasmodium yoelii (P. yoelii), for which significantly higher levels of bioactive IFN-γ, TNF-α and IL-2 were noted in the immunized group (22). Notably, previous studies have reported that the multifunctional Th1 cells simultaneously expressing IFN-γ, TNF-α and IL-2 correlated best with the degree of protection of vaccine models against parasite infection (55, 74). These cells were classified as effector memory cells (74). With regard to IL-2, it was found to exhibit little direct effector function; however, it appears to be a promoter of CD4+ and CD8+ T cells expansion in serving the effector T-cell responses. Furthermore, it could enhance the memory capacity and effector function of CD8+ T cells and natural killer (NK)-cell activity. Therefore, cytokine markers of IL-2, TNF-α and IFN-γ can provide a relatively simple set of cytokines that could be used to define a vaccine-elicited response against specific infections that require T cells for protection (56). During the acute stage of babesiosis, the resistance is associated with early transcriptional up-regulation of IL-12 and IFN-γ (59, 75). IL-12 is a Th1 cytokine produced by activated monocytes and macrophages. Bioactive IL-12 develops and maintains the Th1 cells by activating Th1 cells and NK cells to produce IFN-γ (76, 77). Importantly, IL-12 plays an important role in the inhibition of Th2 differentiation (76). Therefore, the significant down-regulation of IL-4 and IL-10 transcription among cattle immunized with rBbAMA-1(I/II) in our study was likely due to IL-12 cytokine suppression.
Given that the structural BbAMA-1 domains I and II could elicit both humoral and cellular immune responses along with predominantly Th1 responses, it is interesting to employ this antigen as a vaccine candidate against bovine babesiosis. Regarding strain variations that dramatically interfere with the live vaccine efficacy and are an obstacle in the development of recombinant vaccines, BbAMA-1 which is highly conserved and distributed throughout B. bovis would help to address issues of ethnicity by providing protective immunity across B. bovis strains in the field condition. To complete the evaluation of its vaccine potential, this vaccine formulation will be used in future work in order to determine protection efficacy against challenge infection with a virulent strain. The findings reported here should be useful in better understanding and improving the design of BbAMA-1-based vaccines against B. bovis infections.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, KY575957.
The animal study was reviewed and approved by the Faculty of Veterinary Medicine, Chiang Mai University Animal Care and Use Committee (FVM – ACUC). Written informed consent was obtained from the owners for the participation of their animals in this study.
AR, VP, ST, and NS conceived and designed the experiments and performed the data analysis. AR, BN, AM, NA, PK, WS, PC, and KS collected samples and performed the experiments. AR and NS wrote the manuscript. AR, VP, ST, NY, and NS directed the analyses and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Royal Golden Jubilee Ph.D. Programme Scholarship, Grant No. PHD/0224/2560. This work was also supported by the National Research Council of Thailand, Grant No. FF65/089. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to express our deep appreciation to all members of the Laboratory of Veterinary Vaccine and Biological Products for their technical support and constructive suggestions. We are also grateful to all of the farmers who kindly provided us with access to the animals included in this study.
1. Bock R, Jackson L, De Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. (2004) 129:S247–69. doi: 10.1017/S0031182004005190
2. Suarez CE, Alzan HF, Silva MG, Rathinasamy V, Poole WA, Cooke BM. Unraveling the cellular and molecular pathogenesis of bovine babesiosis: is the sky the limit? Int J Parasitol. (2019) 49:183–97. doi: 10.1016/j.ijpara.2018.11.002
3. Tuvshintulga B, Sivakumar T, Yokoyama N, Igarashi I. Development of unstable resistance to diminazene aceturate in Babesia bovis. Int J Parasitol Drugs Drug Resist. (2019) 9:87–92. doi: 10.1016/j.ijpddr.2019.02.001
4. Brown W, Norimine J, Goff W, Suarez C, McElwain T. Prospects for recombinant vaccines against Babesia bovis and related parasites. Parasite Immunol. (2006) 28:315–27. doi: 10.1111/j.1365-3024.2006.00849.x
5. Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. (1978) 77:72–82. doi: 10.1083/jcb.77.1.72
6. Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. (2011) 7:e1001276. doi: 10.1371/journal.ppat.1001276
7. Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci USA. (2011) 108:13275–80. doi: 10.1073/pnas.1110303108
8. Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, et al. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem. (1996) 271:29446–52. doi: 10.1074/jbc.271.46.29446
9. Waters AP, Thomas A, Deans J, Mitchell G, Hudson DE, Miller LH, et al. A merozoite receptor protein from Plasmodium knowlesi is highly conserved and distributed throughout plasmodium. J Biol Chem. (1990) 265:17974–9. doi: 10.1016/S0021-9258(18)38259-0
10. Hehl AB, Lekutis C, Grigg ME, Bradley PJ, Dubremetz J-F, Ortega-Barria E, et al. Toxoplasma gondii homolog of Plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect Immun. (2000) 68:7078–86. doi: 10.1128/IAI.68.12.7078-7086.2000
11. Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ. Babesia divergens and Neospora caninum apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion. Protein Sci. (2013) 22:114–27. doi: 10.1002/pro.2193
12. Gaffar FR, Yatsuda AP, Franssen FF, de Vries E. Erythrocyte invasion by Babesia bovis merozoites is inhibited by polyclonal antisera directed against peptides derived from a homolog of Plasmodium falciparum apical membrane antigen 1. Infect Immun. (2004) 72:2947–55. doi: 10.1128/IAI.72.5.2947-2955.2004
13. Montero E, Rodriguez M, Oksov Y, Lobo CA. Babesia divergens apical membrane antigen 1 and its interaction with the human red blood cell. Infect Immun. (2009) 77:4783–93. doi: 10.1128/IAI.00969-08
14. Moitra P, Zheng H, Anantharaman V, Banerjee R, Takeda K, Kozakai Y, et al. Expression, purification, and biological characterization of Babesia microti apical membrane antigen 1. Infect Immun. (2015) 83:3890–901. doi: 10.1128/IAI.00168-15
15. Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, et al. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. (2000) 38:706–18. doi: 10.1046/j.1365-2958.2000.02175.x
16. Narum DL, Thomas AW. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol Biochem Parasitol. (1994) 67:59–68. doi: 10.1016/0166-6851(94)90096-5
17. Yap A, Azevedo MF, Gilson PR, Weiss GE, O'Neill MT, Wilson DW, et al. Conditional expression of apical membrane antigen 1 in Plasmodium falciparum shows it is required for erythrocyte invasion by merozoites. Cell Microbiol. (2014) 16:642–56. doi: 10.1111/cmi.12287
18. Mital J, Meissner M, Soldati D, Ward GE. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol Biol Cell. (2005) 16:4341–9. doi: 10.1091/mbc.e05-04-0281
19. Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, et al. Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat Commun. (2013) 4:1–13. doi: 10.1038/ncomms3552
20. Collins WE, Pye D, Crewther PE, Vandenberg KL, Galland GG, Sulzer AJ, et al. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am J Trop Med Hyg. (1994) 51:711–9. doi: 10.4269/ajtmh.1994.51.711
21. Srinivasan P, Baldeviano GC, Miura K, Diouf A, Ventocilla JA, Leiva KP, et al. A malaria vaccine protects Aotus monkeys against virulent Plasmodium falciparum infection. NPJ Vaccines. (2017) 2:1–10. doi: 10.1038/s41541-017-0015-7
22. Schussek S, Trieu A, Apte SH, Sidney J, Sette A, Doolan DL. Immunization with apical membrane antigen 1 confers sterile infection-blocking immunity against Plasmodium sporozoite challenge in a rodent model. Infect Immun. (2013) 81:3586–99. doi: 10.1128/IAI.00544-13
23. Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. (2008) 24:74–84. doi: 10.1016/j.pt.2007.12.002
24. Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. (2001) 158:1505–12. doi: 10.1093/genetics/158.4.1505
25. Coley A, Parisi K, Masciantonio R, Hoeck J, Casey J, Murphy V, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. (2006) 74:2628–36. doi: 10.1128/IAI.74.5.2628-2636.2006
26. Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. (2005) 102:12736–41. doi: 10.1073/pnas.0501808102
27. Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Anders RF, et al. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. (2007) 3:e138. doi: 10.1371/journal.ppat.0030138
28. Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J Biol Chem. (2007) 282:7431–41. doi: 10.1074/jbc.M610562200
29. Maskus DJ, Królik M, Bethke S, Spiegel H, Kapelski S, Seidel M, et al. Characterization of a novel inhibitory human monoclonal antibody directed against Plasmodium falciparum apical membrane antigen 1. Sci Rep. (2016) 6:1–14. doi: 10.1038/srep39462
30. Harris KS, Casey JL, Coley AM, Masciantonio R, Sabo JK, Keizer DW, et al. Binding hot spot for invasion inhibitory molecules on Plasmodium falciparum apical membrane antigen 1. Infect Immun. (2005) 73:6981–9. doi: 10.1128/IAI.73.10.6981-6989.2005
31. Wang G, Drinkwater N, Drew DR, MacRaild CA, Chalmers DK, Mohanty B, et al. Structure–activity studies of β-hairpin peptide inhibitors of the Plasmodium falciparum AMA1–RON2 interaction. J Mol Biol. (2016) 428:3986–98. doi: 10.1016/j.jmb.2016.07.001
32. Lalitha P, Biswas S, Pillai C, Saxena R. Immunogenicity of a recombinant malaria vaccine candidate, domain I+ II of AMA-1 ectodomain, from Indian P. falciparum alleles. Vaccine. (2008) 26:4526–35. doi: 10.1016/j.vaccine.2008.06.031
33. Healer J, Triglia T, Hodder AN, Gemmill AW, Cowman AF. Functional analysis of Plasmodium falciparum apical membrane antigen 1 utilizing interspecies domains. Infect Immun. (2005) 73:2444–51. doi: 10.1128/IAI.73.4.2444-2451.2005
34. Múfalo BC, Gentil F, Bargieri DY, Costa FT, Rodrigues MM, Soares IS. Plasmodium vivax apical membrane antigen-1: comparative recognition of different domains by antibodies induced during natural human infection. Microbes Infect. (2008) 10:1266–73. doi: 10.1016/j.micinf.2008.07.023
35. Rittipornlertrak A, Nambooppha B, Simking P, Punyapornwithaya V, Tiwananthagorn S, Jittapalapong S, et al. Low levels of genetic diversity associated with evidence of negative selection on the Babesia bovis apical membrane antigen 1 from parasite populations in Thailand. Infect Genet Evol. (2017) 54:447–54. doi: 10.1016/j.meegid.2017.08.009
36. Rittipornlertrak A, Nambooppha B, Muenthaisong A, Punyapornwithaya V, Tiwananthagorn S, Chung Y-T, et al. Structural and immunological characterization of an epitope within the PAN motif of ectodomain I in Babesia bovis apical membrane antigen 1 for vaccine development. PeerJ. (2021) 9:e11765. doi: 10.7717/peerj.11765
37. Salama AA, Terkawi MA, Kawai S, AbouLaila M, Nayel M, Mousa A, et al. Specific antibody to a conserved region of Babesia apical membrane antigen-1 inhibited the invasion of B. bovis into the erythrocyte. Exp Parasitol. (2013) 135:623–8. doi: 10.1016/j.exppara.2013.09.017
38. World Organization for Animal Health (OIE). Bovine babesiosis. (2021). Available online at: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.02_BABESIOSIS.pdf (accessed March 1, 2022).
39. Muenthaisong A, Rittipornlertrak A, Nambooppha B, Tankaew P, Varinrak T, Pumpuang M, et al. Immune response in dairy cattle against combined foot and mouth disease and haemorrhagic septicemia vaccine under field conditions. BMC Vet Res. (2021) 17:1–12. doi: 10.1186/s12917-021-02889-8
40. Amadori M, Soares-Filipe JF, Riva F, Vitali A, Ruggeri J, Lacetera N. Characterization of the blastogenic response to LPS of bovine peripheral blood mononuclear cells. PLoS ONE. (2018) 13:e0204827. doi: 10.1371/journal.pone.0204827
41. Okagawa T, Konnai S, Mekata H, Githaka N, Suzuki S, Kariuki E, et al. Transcriptional profiling of inflammatory cytokine genes in African buffaloes (Syncerus caffer) infected with Theileria parva. Vet Immunol Immunopathol. (2012) 148:373–9. doi: 10.1016/j.vetimm.2012.06.015
42. Yang L, Liu Z, Li J, He K, Kong L, Guo R, et al. Association of the expression of Th cytokines with peripheral CD4 and CD8 lymphocyte subsets after vaccination with FMD vaccine in Holstein young sires. Res Vet Sci. (2018) 119:79–84. doi: 10.1016/j.rvsc.2018.05.017
43. Leutenegger CM, Alluwaimi AM, Smith WL, Perani L, Cullor JS. Quantitation of bovine cytokine mRNA in milk cells of healthy cattle by real-time TaqMan® polymerase chain reaction. Vet Immunol Immunopathol. (2000) 77:275–87. doi: 10.1016/S0165-2427(00)00243-9
44. Gonzalez DD, Rimondi A, Aguirreburualde MP, Mozgovoj M, Bellido D, Wigdorovitz A, et al. Quantitation of cytokine gene expression by real time PCR in bovine milk and colostrum cells from cows immunized with a bovine rotavirus VP6 experimental vaccine. Res Vet Sci. (2013) 95:703–8. doi: 10.1016/j.rvsc.2013.03.016
45. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
46. Crow MK, Karasavvas N, Sarris AH. Protein aggregation mediated by cysteine oxidation during the stacking phase of discontinuous buffer SDS-PAGE. Biotechniques. (2001) 30:311–6. doi: 10.2144/01302st04
47. Grabski AC, Novagen RRB. Preparation of protein samples for SDS-polyacrylamide gelelectrophoresis: procedures and tips. Innovations. (2001) 13:1–12.
48. Arévalo-Pinzón G, Bermúdez M, Hernández D, Curtidor H, Patarroyo MA. Plasmodium vivax ligand-receptor interaction: PvAMA-1 domain I contains the minimal regions for specific interaction with CD71+ reticulocytes. Sci Rep. (2017) 7:1–13. doi: 10.1038/s41598-017-10025-6
49. Ortiz JMJ, Zajac MPDM, Zanetti FA, Molinari MP, Gravisaco MJ, Calamante G, et al. Vaccine strategies against Babesia bovis based on prime-boost immunizations in mice with modified vaccinia Ankara vector and recombinant proteins. Vaccine. (2014) 32:4625–32. doi: 10.1016/j.vaccine.2014.06.075
50. Brown W, Palmer G. Designing blood-stage vaccines against Babesia bovis and B. bigemina. Parasitol Today. (1999) 15:275–81. doi: 10.1016/S0169-4758(99)01471-4
51. McGuire T, Musoke A. Biologic activities of bovine IgG subclasses. Adv Exp Med Biol. (1981) 137:359–66.
52. Rostamian M, Sohrabi S, Kavosifard H, Niknam HM. Lower levels of IgG1 in comparison with IgG2a are associated with protective immunity against Leishmania tropica infection in BALB/c mice. J Microbiol Immunol Infect. (2017) 50:160–6. doi: 10.1016/j.jmii.2015.05.007
53. Kurup SP, Butler NS, Harty JT. T cell-mediated immunity to malaria. Nat Rev Immunol. (2019) 19:457–71. doi: 10.1038/s41577-019-0158-z
54. Seder RA, Chang L-J, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, et al. Protection against malaria by intravenous immunization with a non-replicating sporozoite vaccine. Science. (2013) 341:1359–65. doi: 10.1126/science.1241800
55. Mordmüller B, Surat G, Lagler H, Chakravarty S, Ishizuka AS, Lalremruata A, et al. Sterile protection against human malaria by chemo-attenuated PfSPZ vaccine. Nature. (2017) 542:445–9. doi: 10.1038/nature21060
56. Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. (2008) 8:247–58. doi: 10.1038/nri2274
57. Igarashi I, Suzuki R, Waki S, Tagawa Y-I, Seng S, Tum S, et al. Roles of CD4+ T cells and gamma interferon in protective immunity against Babesia microti infection in mice. Infect Immun. (1999) 67:4143–8. doi: 10.1128/IAI.67.8.4143-4148.1999
58. Aguilar-Delfin I, Wettstein PJ, Persing DH. Resistance to acute babesiosis is associated with interleukin-12-and gamma interferon-mediated responses and requires macrophages and natural killer cells. Infect Immun. (2003) 71:2002–8. doi: 10.1128/IAI.71.4.2002-2008.2003
59. Goff WL, Johnson WC, Parish SM, Barrington GM, Tuo W, Valdez RA. The age-related immunity in cattle to Babesia bovis infection involves the rapid induction of interleukin-12, interferon-γ and inducible nitric oxide synthase mRNA expression in the spleen. Parasite Immunol. (2001) 23:463–71. doi: 10.1046/j.1365-3024.2001.00402.x
60. Goff W, Johnson WC, Horn R, Barrington G, Knowles D. The innate immune response in calves to Boophilus microplus tick transmitted Babesia bovis involves type-1 cytokine induction and NK-like cells in the spleen. Parasite Immunol. (2003) 25:185–8. doi: 10.1046/j.1365-3024.2003.00625.x
61. Goff WL, Storset AK, Johnson WC, Brown WC. Bovine splenic NK cells synthesize IFN-γ in response to IL-12-containing supernatants from Babesia bovis-exposed monocyte cultures. Parasite Immunol. (2006) 28:221–8. doi: 10.1111/j.1365-3024.2006.00830.x
62. Brown W, Zhao S, Woods V, Dobbelaere D, Rice-Ficht A. Babesia bovis-specific CD4+ T cell clones from immune cattle express either the Th0 or Th1 profile of cytokines. Rev Elev Med Vet Pays Trop. (1993) 46:65–9. doi: 10.19182/remvt.9400
63. Brown WC, Ruef BJ, Norimine J, Kegerreis KA, Suarez CE, Conley PG, et al. A novel 20-kilodalton protein conserved in Babesia bovis and B. bigemina stimulates memory CD4+ T lymphocyte responses in B bovis-immune cattle. Mol Biochem Parasitol. (2001) 118:97–109. doi: 10.1016/S0166-6851(01)00375-9
64. Brown WC, McElwain TF, Ruef BJ, Suarez CE, Shkap V, Chitko-McKown CG, et al. Babesia bovis rhoptry-associated protein 1 is immunodominant for T helper cells of immune cattle and contains T-cell epitopes conserved among geographically distant B. bovis strains. Infect Immun. (1996) 64:3341–50. doi: 10.1128/iai.64.8.3341-3350.1996
65. Norimine J, Mosqueda J, Palmer GH, Lewin HA, Brown WC. Conservation of Babesia bovis small heat shock protein (Hsp20) among strains and definition of T helper cell epitopes recognized by cattle with diverse major histocompatibility complex class II haplotypes. Infect Immun. (2004) 72:1096–106. doi: 10.1128/IAI.72.2.1096-1106.2003
66. Gimenez AM, Françoso KS, Ersching J, Icimoto MY, Oliveira V, Rodriguez AE, et al. A recombinant multi-antigen vaccine formulation containing Babesia bovis merozoite surface antigens MSA-2a 1, MSA-2b and MSA-2c elicits invasion-inhibitory antibodies and IFN-γ producing cells. Parasit Vectors. (2016) 9:1–13. doi: 10.1186/s13071-016-1862-1
67. Jankovic D, Liu Z, Gause WC. Th1-and Th2-cell commitment during infectious disease: asymmetry in divergent pathways. Trends Immunol. (2001) 22:450–7. doi: 10.1016/S1471-4906(01)01975-5
68. Brown WC. Molecular approaches to elucidating innate and acquired immune responses to Babesia bovis, a protozoan parasite that causes persistent infection. Vet Parasitol. (2001) 101:233–48. doi: 10.1016/S0304-4017(01)00569-6
69. Rauf U, Suleman M, Abid A, Jamil H, Menghwar H, Durrani AZ, et al. Humoral and cell-mediated immune response validation in calves after a live attenuated vaccine of Babesia bigemina. Pathogens. (2020) 9:936. doi: 10.3390/pathogens9110936
70. Doolan D, Martinez-Alier N. Immune response to pre-erythrocytic stages of malaria parasites. Curr Mol Med. (2006) 6:169–85. doi: 10.2174/156652406776055249
71. Li Y, Terkawi MA, Nishikawa Y, Aboge GO, Luo Y, Ooka H, et al. Macrophages are critical for cross-protective immunity conferred by Babesia microti against Babesia rodhaini infection in mice. Infect Immunity. (2012) 80:311–20. doi: 10.1128/IAI.05900-11
72. Stich RW, Shoda LK, Dreewes M, Adler B, Jungi TW, Brown WC. Stimulation of nitric oxide production in macrophages by Babesia bovis. Infect Immun. (1998) 66:4130–6. doi: 10.1128/IAI.66.9.4130-4136.1998
73. Johnson WC, Cluff C, Goff W, Wyatt C. Reactive oxygen and nitrogen intermediates and products from polyamine degradation are babesiacidal in vitro. Ann N Y Acad Sci. (1996) 791:136–47. doi: 10.1111/j.1749-6632.1996.tb53520.x
74. Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, et al. Multifunctional TH 1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. (2007) 13:843–50. doi: 10.1038/nm1592
75. Shoda LK, Palmer GH, Florin-Christensen J, Florin-Christensen M, Godson DL, Brown WC. Babesia bovis-stimulated macrophages express interleukin-1β, interleukin-12, tumor necrosis factor alpha, and nitric oxide and inhibit parasite replication in vitro. Infect Immun. (2000) 68:5139–45. doi: 10.1128/IAI.68.9.5139-5145.2000
76. Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. (2000) 14:1693–711. doi: 10.1101/gad.14.14.1693
Keywords: Babesia bovis, AMA-1, recombinant protein, Th1 immune response, cattle, vaccine
Citation: Rittipornlertrak A, Nambooppha B, Muenthaisong A, Apinda N, Koonyosying P, Srisawat W, Chomjit P, Sangkakam K, Punyapornwithaya V, Tiwananthagorn S, Yokoyama N and Sthitmatee N (2022) Immunization of Cattle With Recombinant Structural Ectodomains I and II of Babesia bovis Apical Membrane Antigen 1 [BbAMA-1(I/II)] Induces Strong Th1 Immune Response. Front. Vet. Sci. 9:917389. doi: 10.3389/fvets.2022.917389
Received: 11 April 2022; Accepted: 23 May 2022;
Published: 23 June 2022.
Edited by:
Luiz Daniel De Barros, State University of Londrina, BrazilReviewed by:
Siju Susan Jacob, National Institute of Veterinary Epidemiology and Disease Informatics (ICAR), IndiaCopyright © 2022 Rittipornlertrak, Nambooppha, Muenthaisong, Apinda, Koonyosying, Srisawat, Chomjit, Sangkakam, Punyapornwithaya, Tiwananthagorn, Yokoyama and Sthitmatee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nattawooti Sthitmatee, ZHJuZWF3QGdtYWlsLmNvbQ==; bmF0dGF3b290aS5zQGNtdS5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.