
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 25 April 2022
Sec. Animal Nutrition and Metabolism
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.890585
The gut microbiota plays vital roles in metabolizing nutrient, maintaining the intestinal epithelial barrier but also in modulating immunity. Host genetics and the pig breed are implicated in shaping gut microbiota. Tibetan pig is a unique native Chinese breed and has evolved to manifest a strong disease resistance. However, the immunity and microbiota of growing Tibetan (TP) pigs were still rarely understood. The jejunal immunity phenotype and microbial composition of TP and Duroc × (Landrace × Yorkshire) (DLY) pigs were explored through immunohistochemistry and 16S rRNA sequencing. Higher scores of clusters of differentiation 4 (CD4+) and Toll-like receptor 9 (TLR9) were observed in TP pigs than those of DLY pigs (p < 0.05), as were Interleukin 10 (IL-10) and zonular occludens 1 (ZO-1) (p < 0.01). Similar levels of bacterial richness and diversity were found in the jejunal microbiota of the TP and DLY pigs. However, the TP pigs showed a significantly different microbiome compared to DLY pigs at the genus level (ANOSIM; p < 0.05). Pseudomonas, Stenotrophomonas, Phenylobacterium, and Sandaracinobacter were enriched in DLY pigs (p < 0.05), while the Lactobacillus and Solibacillus had higher abundances in TP pigs than DLY pigs (p < 0.05). Tibetan pigs have “healthier” intestinal microbial communities than DLY pigs. Close relationships were found between jejunal immune performance and the differential bacteria, Lactobacillus can enhance porcine jejunal immunity, while Stenotrophomonas will have a negative impact on porcine gut immunity.
The gastrointestinal tract of mammals comprises approximately 500–1,000 microbes (1) that constitute complicated and dynamic microbial communities mainly consist of bacteria (2). Indeed, gut microbiota are concerned with the regular physiology of the gastrointestinal tract, including degrading indigestible carbohydrate like plant polysaccharides and producing diverse metabolites like short chain fatty acids (SCFAs) and vitamins (3). Besides metabolic advantages, commensal gut microbiota provide the host with functions that promoting immune responses, immune homeostasis and defending against pathogen invasion (4, 5). Furthermore, alterations in microbial community can affect immune system development and modulate immune mediators (6, 7). The microbiota of small intestine also have a considerable influence on diverse aspects of the host's physiology, including metabolism, immunity and endocrine (8).
The previous studies have suggested that genetic factor was broadly conductive to the difference of the intestinal microbial composition (9) and pig breeds strongly affect the bacterial structure at the suckling period (10). The Duroc × (Landrace × Yorkshire) (DLY) pig is a worldwide commercial pig breed by means of high breeding selection to pursue a faster growth rate at the detriment of decreasing the capacity of disease resistance. On the contrary, the Tibetan pig is a local pig breed that is found primarily in the Tibetan highlands and whose specific phenotype and physiology that have allowed them to accommodate the extreme environments (11).
However, very little information is available regarding the jejunal immunity and microbial structure in growing-period Tibetan pigs. Exploring how host genetics and pig breed affect intestinal microbial diversity and composition is of great importance in the understanding of the host health. Besides, searching the relationships between gut microbiota and immunity may help us solve health problems in pig industry. Therefore, the present study studied the jejunal immunity and microbial differences in Tibetan and DLY pigs and investigated the correlations between immune indices and relative abundance of differential microbiota.
Twelve Tibetan pigs (4 months of age) and twelve DLY [Duroc × (Landrace × Yorkshire)] pigs (4 months of age) in the growth stage were selected and maintained in the same house with one pig per pen which equips with fully slatted floors, a feeder, and a nipple drinker. All pigs were housed at Yunnan Agricultural University and fed by the same NRC (NRC 2012) diet for 28 days (Table 1), then euthanized via exsanguination. Jejunal chyme samples were collected by advancing the lumenal contents into a cryotube. The jejunum samples (2–3 cm in middle section) were collected and fixed in 4% neutral paraformaldehyde fix solution. The treatment of the pigs was approved by the Institutional Animal Care and Use Committee of Yunnan Agricultural University (No. YNAU20201304).
Each jejunal sample was cut into 4 μm thick slides and embedded in paraffin after fixation. And then three to four transverse slides from each sample were chosen to stain with hematoxylin and eosin. For immunohistochemical staining, slides were heated in microwaves at 750 W for 15 min in citrate buffer (pH 6.0) for antigen repairing after deparaffinization and rehydration. Following washing with phosphate-buffered saline (PBS), slides were treated for 25 min with 3% H2O2 at 20–22°C and in a photophobic environment to block endogenous peroxidase activity, washed with PBS, and trated for 30 min at 20–22°C with 3% bovine serum albumin to block. Then the slides were incubated with the primary antibody including rabbit anti-CD3 (Servicebio Technology, Wuhan, CN, #GB13014, 1:50), rabbit anti-MYD88, mouse anti-TLR4 and rabbit anti-TLR9 (#GB11269 and #GB11266, 1:100), rabbit anti-ZO-1 and rabbit anti-CD4 (#GB11195 and #GB13064-2, 1:200), rabbit anti-TLR2 (#GB11554, 1:500), rabbit anti-IL-1β and rabbit anti-IL-6 (#GB11113 and #GB11117, 1:800), and rabbit anti-MUC-2 (#GB11344, 1:1,000) diluted in PBS and at 4°C overnight. Next, the slides were washed with PBS and incubated for 50 min at 20–22°C with a secondary antibody HRP conjugated goat anti-rabbit IgG (#GB23303, 1:200). The immunohistochemical reaction was revealed for visual inspection using diaminobenzidine (DAB) as a chromogen (Vector Laboratories). Slides were further counterstained with hematoxylin and coverslipped. A digital microscopy scanner Pannoramic P250 (3DHISTECH Ltd., Budapest, Hungary) with a 20 × microscope objective was used to scan the immunostained slides. The corresponding whole-slide scanned images were evaluated using Pannoramic Viewer 1.15.2 (3DHISTECH Ltd., Budapest, Hungary). The final immunohistochemistry results were expressed as H-score as reported by Yang et al. (12).
Following the manufacturer's instructions, total microbial DNA extraction of jejunal contents sample was carried out using the HiPure Stool DNA Kits (Magen, Guangzhou, China). Amplicon pooled libraries for sequencing on an Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) were prepared using primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′) targeted across the hypervariable V3-V4 regions of the 16S rRNA gene. The amplification was performed in the following steps: initial denaturation at 95°C for 2 min, followed by 27 cycles (98°C−10 s, 62°C−30 s, 68°C−30 s) and a final extension at 68°C for 10 min.
FASTP (https://github.com/OpenGene/fastp) was used to filter the raw sequences, and then with a minimal overlap of 10 bp and mismatch error ratio of 2%, paired-end reads were assembled as raw tags through FLSAH (version 1.2.11). QIIME (version 1.9.1) pipeline was used to filter noisy sequences of raw tags to obtain high-quality clean tags. Subsequently, with a similarity ≥97%, UPARSE (http://drive5.com/uparse/) was used to cluster the tags into OTUs (operational taxonomic units), while all chimeric tags were removed using Userach (version 7.0). Each representative sequence was assigned into organisms by RDP (Ribosomal Database Project) classifier (version 2.2) with a confidence threshold value of 0.8–1. Alpha diversity of observed species, Good's coverage, Chao1, Shannon, Simpson, PD whole tree, and beta diversity of principal component analysis (PCA) was determined using QIIME software (version 1.9.1). Based on high-quality reads, functional categories of KEGG ortholog were predicted by Tax4Fun (version 1.0).
The analysis of intestinal morphology and immune performance results was calculated by Welch's t-test using the SPSS 22.0 software. The Wilcoxon rank test was used to compute differential functional analysis among groups in R project (version 3.4.1). Correlations between variables were calculated using the Spearman rank correlation in GraphPad Prism 7.0.
In the jejunum, TP pigs had higher villus height and less deeper crypt depth than DLY pigs (p < 0.05). And TP pigs tended to have higher villus height to crypt depth ratio than DLY pigs (p = 0.162) (Table 2).
As shown in Table 3, at the protein level, the cluster of differentiation 4 (CD4+) and Toll-like receptor 9 (TLR9) were significantly higher in TP pigs than in DLY pigs (p < 0.05), as were the interleukin 10 (IL-10) and zonular occludens 1 (ZO-1) contents (p < 0.01).
We obtained 2,925,307 clean tags from the samples by using the IlluminaHiSeq2500 platform. Of these, 2,881,333 effective tags with an average length of 421 bp were selected after the quality control and filtering. This yielded an average of 120,055 effective reads per sample. The average effective tags to clean tags ratio was 94.68% (Table 4). We identified a total of 6,156 operational taxonomic units (OTUs) at a 97% similarity level. A Venn diagram (Figure 1) shows 249 OTUs are shared between TP and DLY groups, along with 188 and 276 unique OTUs in the TP and DLY groups, respectively.
Good's coverage for the two groups was >99.5%, indicating a satisfactory sequencing depth of the gut microbiota (Table 5). Alpha diversity indices were not significantly different between TP and DLY pigs (p > 0.05). For example, the Shannon index (p = 0.281) and simpson (p = 0.523) index of TP pigs were lower than those of DLY pigs.
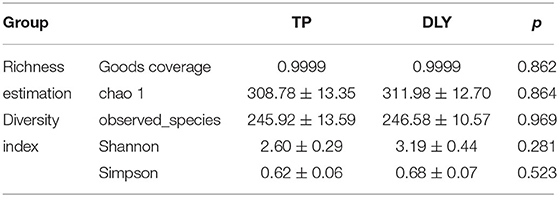
Table 5. Comparison of alpha diversity estimation of the 16S rRNA gene libraries for TP and DLY pigs at 97% similarity.
The principal component analysis (PCA) score plot (Figure 2A) from sequences at the OTU level showed that the microbial composition of TP pigs was like that of DLY pigs, and as were the analysis of similarity (ANOSIM) at the OTU and phylum levels (p > 0.05) (Figures 2B,C). However, the TP pigs showed a significant different microbiome compared to DLY pigs at the genus level, as shown in Figure 2D (ANOSIM; p < 0.05).
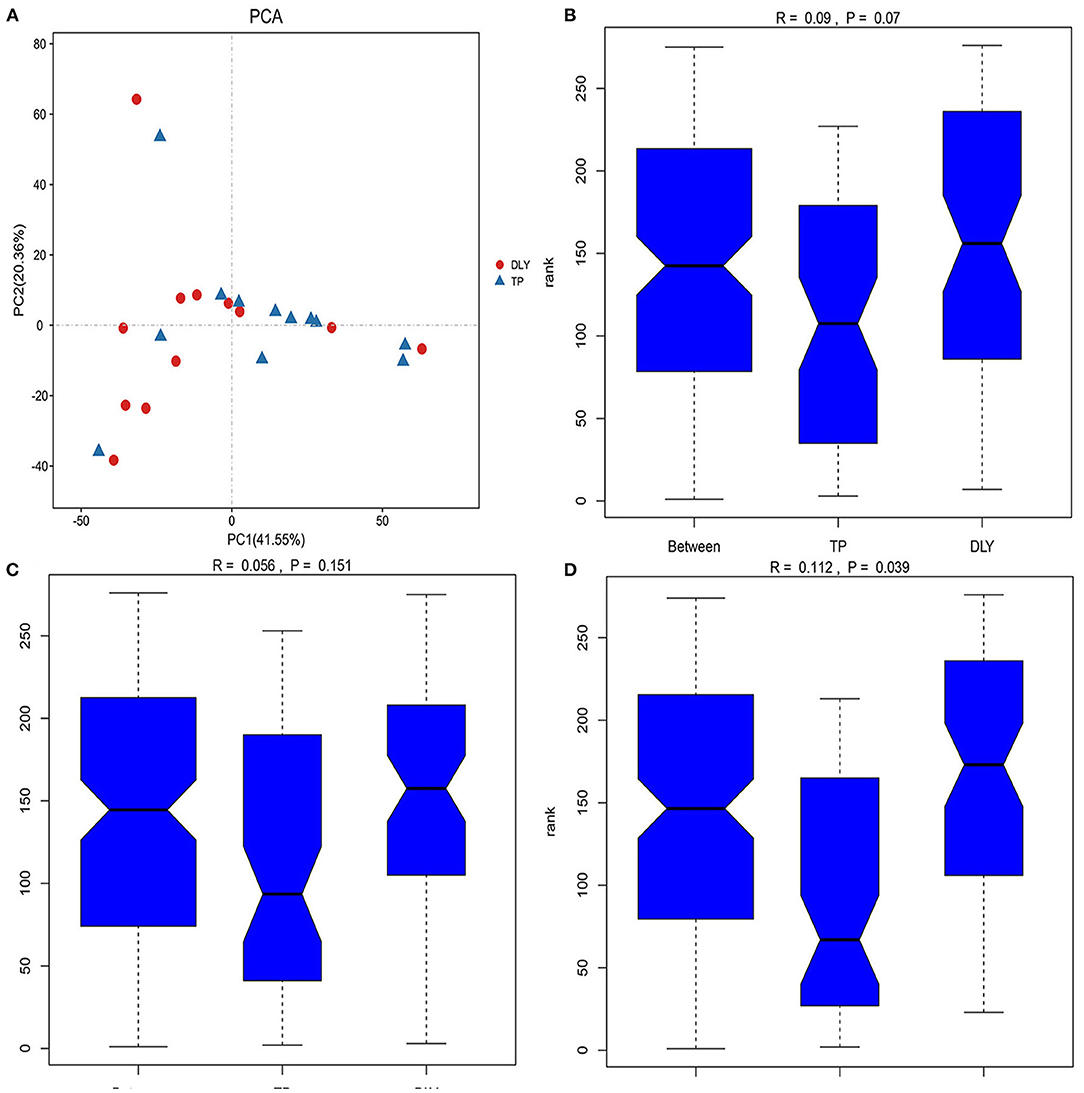
Figure 2. Beta-diversity analysis of jejunal microbiota between TP and DLY pigs. (A) PCA analysis; (B) ANOSIM analysis at the OTU level; (C) ANOSIM analysis at the phylum level; (D) ANOSIM analysis at the genus level.
The predominant phyla were Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes (Figure 3). The relative abundance of Tenericutes in TP (0.76%) pigs was significantly higher than in DLY (0.24%) pigs (p < 0.05). The TP pigs displayed a 23.53% increase in the relative abundances of Firmicutes compared with DLY pigs (p = 0.07), while the relative abundance of Proteobacteria (p = 0.24), Actinobacteria (p = 0.25), Bacteroidetes (p = 0.06), and Cyanobacteria (p = 0.16) tended to be lower in the TP pigs than in the DLY pigs (Table 6).
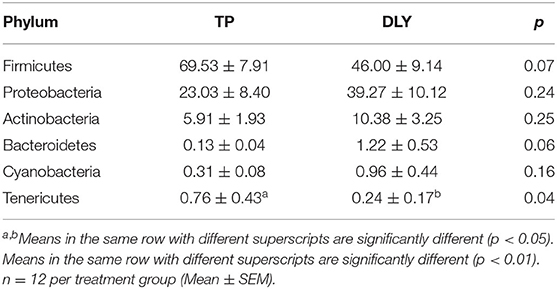
Table 6. Analysis of the jejunal composition and differences at the phylum level of growing DLY and TP pigs.
At the genus level, the most predominant was Lactobacillus, followed by Escherichia-Shigella, Megasphaera, Pantoea, Acinetobacter, Pseudoscardovia, Pseudomonas, Bifidobacterium, Streptococcus, Olsenella, and Clostridium_sensu_stricto_1 (Figure 4). As shown in Table 7, TP pigs manifested an increase of 29.05 and 0.47% increase in the relative abundances of Lactobacillus and Solibacillus compared with DLY pigs, while the relative abundances of Pseudomonas, Stenotrophomonas, Phenylobacterium, and Sandaracinobacter in TP pigs were significantly lower than those in DLY pigs (p < 0.05).
Spearman's rank correlations (Figure 5) between the jejunal morphology phenotype, immune performance and relative abundance of differential microbiota were assessed, the relative abundance of Lactobacillus positively correlated with villus height (R = 0.65, p = 0.049), TLR9 expression at the protein level (R = 0.66, p = 0.044) negatively correlated with crypt depth (R = −0.89, p = 0.0011). The relative abundance of Solibacillus negatively correlated with crypt depth (R = −0.65, p = 0.049). The relative abundance of Pseudomonas positively correlated crypt depth (R = 0.78, p = 0.011). The relative abundance of Stenotrophomonas negatively correlated with villus height (R = −0.72, p = 0.024), TLR2 (R = −0.73, p = 0.021) and TLR9 expression at the protein level (R = −0.65, p = 0.047). The relative abundance of Sandaracinobacter negatively correlated with IL-10 (R = −0.84, p = 0.040). The relative abundance of Phenylobacterium negatively correlated with CD4+(R = −0.70, p = 0.022) and IL-6 expression at the protein level (R = −0.70, p = 0.022).
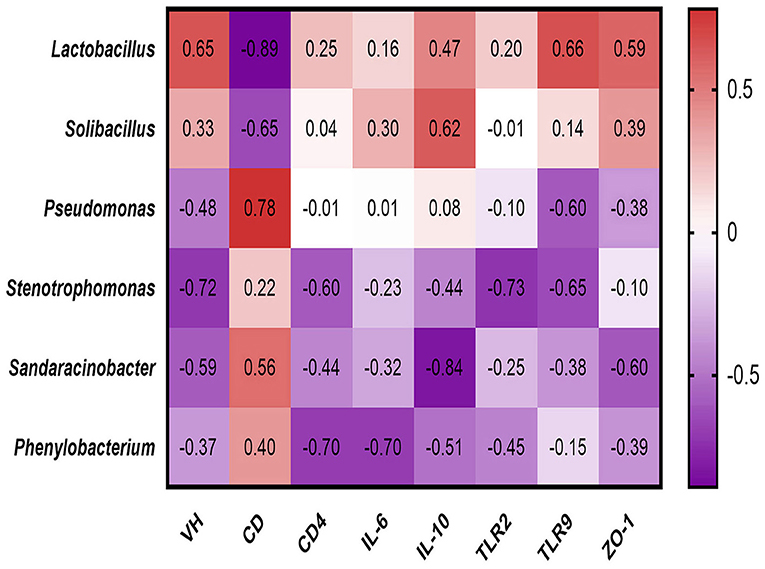
Figure 5. Spearman's rank correlations between differential jejunal microbiota and immune performance.
According to the Tax4Fun-based functional prediction, 16 differential KEGG pathways were found of the jejunal microbiota between the TP and DLY pigs (Figure 6). The pathways enriched in the TP pigs were mainly related to fructose and mannose metabolism, phosphotransferase system (PTS), glycerolipid metabolism, glycolysis/gluconeogenesis, RNA transport, glycosaminoglycan degradation, RIG-I-like receptor signaling pathway and flavonoid biosynthesis (p < 0.05). The pathways enriched in the DLY pigs were mainly related to Vitamin B6 metabolism, Toluene degradation, Meiosis – yeast, GABAergic synapse, Proximal tubule bicarbonate reclamation, Steroid biosynthesis, GnRH signaling pathway and Endocytosis (p < 0.05).
It is generally considered that the gastrointestinal tract is the largest immune organ in mammals and involves in modulating immunological homeostasis (13). In our study, the levels of jejunal IL-10, CD4+, TLR9, and ZO-1 were higher in Tibetan pigs than in DLY pigs. IL-10 is a cytokine with widely anti-inflammatory characteristics as well as a vital regulatory role of CD4+ T lymphocytes (14). CD4+ T lymphocytes are reported to be importantly involved in defending mammals against pathogens and are necessary for boosting cytokine production (15). ZO-1 is the principal component of the functional and constructional organization of tight junctions related to epithelial integrity (16). Porcine toll-like receptors are thought to be the frontline of pathogen monitoring and may be associated with disease tolerance (17). Our results indicate that Tibetan pigs may have better anti-inflammatory characteristics and intestinal barrier function than DLY pigs. Consistent with our research, the research of Gao et al. showed that Jinhua pigs showed lower immune activation and diarrhea frequency than Landrace pigs when challenging with an enterotoxigenic E. coli (ETEC) K88 species (18). Albin et al.' s research shows that resistance (decreasing conductance) increased in intestines of Yorkshire pigs in response to lipopolysaccharide (LPS), whereas resistance in Meishan intestines was unchanged with LPS (19). This indicates that the gut barrier functions are likely enhanced in the traditional pig breeds comparing that in the Yorkshire pigs. Furthermore, a previous study found that innate immunity in Tibetan pigs were stronger than Yorkshire pigs (20). These results indicate that TP pigs may have better anti-inflammatory properties and gut barrier functions than DLY pigs.
Consistent with our research, some researchers have found that small intestine of weanling piglets had the highest populations of Firmicutes and moderate levels of Proteobacteria and Actinobacteria (21). The most dominant genus was Lactobacillus, which is related to the fact that the small intestine is abounded in mono- and di-saccharides along with amino acids, supporting the proliferation of Lactobacillus (22). Recent studies have indicated that the pig breed and host genetics play fundamental roles in the microbial profiles of gastrointestinal tract (23). Our results show that the relative abundances of Lactobacillus and Solibacillus were higher in TP pigs than in DLY pigs, while Pseudomonas, Stenotrophomonas, Phenylobacterium, and Sandaracinobacter were lower in TP pigs than in DLY pigs, which indicated that the intestine microbial composition of different pig breeds was different. Consistent with our research, Pajarillo et al. found that feces of Duroc pigs had higher relative abundances of fecal Phascolarctobacterium, Catenibacterium, and Subdoligranulum than those in Landrace and Yorkshire pigs (10). Compared with Rongchang (RP) and TP pigs, Yorkshire pigs (YP) had a lower ratio of Firmicutes and Bacteroidetes, while higher abundances of Spirochaetes were found in TP in comparison with RP and YP (24). It is indicated that the host genetic factors have an influence on the structure and abundance of intestinal microbiota at the genus level. In addition, Lactobacillus are reported as probiotics in virtue of their anti-inflammatory ability (25), presence of three species of Solibacillus have been reported, of which Solibacillus silvestris can produce antineoplastic biomolecule (26). On the other hand, Stenotrophomonas species are multi-resistant bacteria with ability to cause opportunistic infections (27), and Pseudomonas has been reported as a known pathogen of mastitis microbiota in ruminants (28). Consequently, the intestinal microflora of TP pigs are more probably “healthier” than DLY pigs.
It is well-known that the intestinal microbiota composition influenced immune system development and modulated immune mediators. In this study, we found that Lactobacillus was positively correlated with IL-10 whereas the potential harmful Stenotrophomonas in this study were negatively with TLR2 and TLR9. The members within the genus Lactobacillus are reported to maintain the intestinal ecological equilibrium through avoiding the invasion of pathogens, modulating immunity, and enhancing mucosal barrier integrity (29). For example, treatment of porcine intestinal IPEC-1 epithelial cells with Lactobacillus sobrius repealed the upregulation in IL-1β and IL-8 levels and induced an increase of IL-10 (30). Stenotrophomonas is an multidrug-resistant global opportunistic pathogen, which is related to the increase of human and animal infections in recent years (31–33). In addition, researches shows that the reduction of pathogenic bacteria is helpful to enhance the immune performance of the body (32, 33). These results indicates that Lactobacillus play a vital role in enhancing porcine jejunal immunity while Stenotrophomonas have a negative effect on porcine jejunal immunity.
In summary, the present study revealed that the jejunal microbial diversity did not differ between TP pigs and DLY pigs, while jejunal immunity and microbial composition were different. TP pigs have better anti-inflammatory properties and gut barrier functions than DLY pigs, and TP pigs enhance the jejunal immunity through Lactobacillus, while Stenotrophomonas in the jejunum of DLY pigs has a negative impact on its jejunal immunity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by all animal research was approved by the Ethics Committee of Yunnan Agricultural University (Approval No: YNAU20201304). Written informed consent was obtained from the owners for the participation of their animals in this study.
HP designed the experiments. HP, YY, YL, YX, and LY performed the experiments. YY and YL analyzed the data and wrote the manuscript. SQ and HP revised this manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by Nature Science Foundation of China (Nos. U1802234 and 32160795).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. (2010) 10:159–69. doi: 10.1038/nri2710
2. Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. (2014) 10:416–24. doi: 10.1038/nchembio.1535
3. Sánchez B, Delgado S, Blanco-Míguez A, Lourenço A, Gueimonde M, Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res. (2017) 61. doi: 10.1002/mnfr.201600240
4. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, inflammatory disease. Immunol Rev. (2017) 279:70–89. doi: 10.1111/imr.12567
5. Wu S, Cong G, Zhang Q, Yao H, Wang Z, Kang K, et al. Infection heterogeneity and microbiota differences in chicks infected by Salmonella enteritidis. Microorganisms. (2021) 9:1705. doi: 10.3390/microorganisms9081705
6. Hu Y, Wang L, Shao D, Wang Q, Wu Y, Han Y, et al. Selectived and reshaped early dominant microbial community in the cecum with similar proportions and better homogenization and species diversity due to organic acids as AGP alternatives mediate their effects on broilers growth. Front Microbiol. (2020) 10:2948. doi: 10.3389/fmicb.2019.02948
7. Zhang S, Zhong G, Shao D, Wang Q, Hu Y, Wu T, et al. Dietary supplementation with Bacillus subtilis promotes growth performance of broilers by altering the dominant microbial community. Poult Sci. (2021) 100:100935. doi: 10.1016/j.psj.2020.12.032
8. El Aidy S, van den Bogert B, Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr Opin Biotechnol. (2015) 32:14–20. doi: 10.1016/j.copbio.2014.09.005
9. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U. S. A. (2010) 107:18933–8. doi: 10.1073/pnas.1007028107
10. Pajarillo EA, Chae JP, Balolong MP, Kim HB, Seo KS, Kang DK. Pyrosequencing-based analysis of fecal microbial communities in three purebred pig lines. J Microbiol. (2014) 52:646–51. doi: 10.1007/s12275-014-4270-2
11. Yang S, Zhang H, Mao H, Yan D, Lu S, Lian L, et al. The local origin of the Tibetan pig and additional insights into the origin of Asian pigs. PLoS ONE. (2011) 6:e28215. doi: 10.1371/journal.pone.0028215
12. Yang Y, Xiao M, Song Y, Tang Y, Luo T, Yang S, et al. H-score of 11β-hydroxylase and aldosterone synthase in the histopathological diagnosis of adrenocortical tumors. Endocrine. (2019) 65:683–91. doi: 10.1007/s12020-019-02022-8
13. Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. (2017) 5:e1373208. doi: 10.1080/21688370.2017.1373208
14. Tang W, Chen D, Yu B, He J, Huang Z, Zheng P, et al. Capsulized faecal microbiota transplantation ameliorates post-weaning diarrhoea by modulating the gut microbiota in piglets. Vet Res. (2020) 51:55. doi: 10.1186/s13567-020-00779-9
15. Jankovic D, Kugler DG, Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. (2010) 3:239–46. doi: 10.1038/mi.2010.8
16. Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. (1998) 273:29745–53. doi: 10.1074/jbc.273.45.29745
17. Uenishi H, Shinkai H. Porcine Toll-like receptors: the front line of pathogen monitoring and possible implications for disease resistance. Dev Comp Immunol. (2009) 33:353–61. doi: 10.1016/j.dci.2008.06.001
18. Gao Y, Han F, Huang X, Rong Y, Yi H, Wang Y. Changes in gut microbial populations, intestinal morphology, expression of tight junction proteins, and cytokine production between two pig breeds after challenge with Escherichia coli K88: a comparative study. J. Anim. Sci. (2013) 91:5614–25. doi: 10.2527/jas.2013-6528
19. Albin DM, Wubben JE, Rowlett JM, Tappenden KA, Nowak RA. Changes in small intestinal nutrient transport and barrier function after lipopolysaccharide exposure in two pig breeds. J Anim Sci. (2007) 85:2517–23. doi: 10.2527/jas.2006-237
20. Cheng C, Sun WK, Liu R, Wang RM, Chen YH, Wang Y, et al. Comparison of gene expression of Toll-like receptors and antimicrobial peptides in immune organs and tissues between Yorkshire and Tibetan pigs. Anim Genet. (2015) 46:272–9. doi: 10.1111/age.12286
21. Casas GA, Blavi L, Cross TL, Lee AH, Swanson KS, Stein HH. Inclusion of the direct-fed microbial Clostridium butyricum in diets for weanling pigs increases growth performance and tends to increase villus height and crypt depth, but does not change intestinal microbial abundance. J Anim Sci. (2020) 98:skz372. doi: 10.1093/jas/skz372
22. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. (2013) 13:321–35. doi: 10.1038/nri3430
23. Patil Y, Gooneratne R, Ju XH. Interactions between host and gut microbiota in domestic pigs: a review. Gut Microbes. (2020) 11:310–34. doi: 10.1080/19490976.2019.1690363
24. Diao H, Yan HL, Xiao Y, Yu B, Yu J, He J, et al. Intestinal microbiota could transfer host Gut characteristics from pigs to mice. BMC Microbiol. (2016) 16:238. doi: 10.1186/s12866-016-0851-z
25. Rieu A, Aoudia N, Jego G, Chluba J, Yousfi N, Briandet R, et al. The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell Microbiol. (2014) 16:1836–53. doi: 10.1111/cmi.12331
26. Markande AR, Vemuluri VR, Shouche YS, Nerurkar AS. Characterization of Solibacillus silvestris strain AM1 that produces amyloid bioemulsifier. J Basic Microbiol. (2018) 58:523–31. doi: 10.1002/jobm.201700685
27. Elufisan TO, Luna I, Oyedara OO, Varela AS, García VB, Oluyide BO, et al. Antimicrobial susceptibility pattern of Stenotrophomonas species isolated from Mexico. Afr Health Sci. (2020) 20:168–81. doi: 10.4314/ahs.v20i1.22
28. Catozzi C, Sanchez Bonastre A, Francino O, Lecchi C, De Carlo E, Vecchio D, et al. The microbiota of water buffalo milk during mastitis. PLoS ONE. (2017) 12:e0184710. doi: 10.1371/journal.pone.0184710
29. Zielińska D, Długosz E, Zawistowska-Deniziak A. Functional properties of food origin Lactobacillus in the gastrointestinal ecosystem-in vitro study. Probiotics Antimicrob Proteins. (2019) 11:820–9. doi: 10.1007/s12602-018-9458-z
30. Roselli M, Finamore A, Britti MS, Konstantinov SR, Smidt H, de Vos WM, et al. The novel porcine Lactobacillus sobrius strain protects intestinal cells from enterotoxigenic Escherichia coli K88 infection and prevents membrane barrier damage. J Nutr. (2007) 137:2709–16. doi: 10.1093/jn/137.12.2709
31. Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. (2012) 25:2–41. doi: 10.1128/CMR.00019-11
32. Shi S, Wu S, Shen Y, Zhang S, Xiao Y, He X, et al. Iron oxide nanozyme suppresses intracellular Salmonella Enteritidis growth and alleviates infection in vivo. Theranostics. (2018) 8:6149–62. doi: 10.7150/thno.29303
Keywords: Tibetan pigs, DLY pigs, jejunum, immune, microbial composition
Citation: Yang Y, Li Y, Xie Y, Qiao S, Yang L and Pan H (2022) Comparative Study on Jejunal Immunity and Microbial Composition of Growing-Period Tibetan Pigs and Duroc × (Landrace × Yorkshire) Pigs. Front. Vet. Sci. 9:890585. doi: 10.3389/fvets.2022.890585
Received: 06 March 2022; Accepted: 29 March 2022;
Published: 25 April 2022.
Edited by:
Shourong Shi, Poultry Institute (CAAS), ChinaReviewed by:
Zhi Peng Li, Institute of Special Animal and Plant Sciences (CAAS), ChinaCopyright © 2022 Yang, Li, Xie, Qiao, Yang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Pan, eW5zZHl6QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.