- 1Department of Botany, University of North Bengal, Siliguri, India
- 2Department of Biotechnology, University of North Bengal, Siliguri, India
Aeromonas is omnipresent in aquatic environments and cause disease within a wide host range. A total of thirty-four isolates from water samples of small fish farms were identified as Aeromonas based on biochemical characteristics and 16S rRNA gene sequence. A total of six virulent factors were analyzed which indicated 100% of isolates as beta-haemolytic and proteolytic, whereas 44.1, 38.2, and 70.6% of isolates produced DNAse, siderophore, and amylase, respectively. Studies on the occurrence of four genetic determinants of virulence factors revealed that aer/haem (haemolytic toxin) and flaA (polar flagella) genes were present in 44.1% of strains whereas ascV (type 3 secretion system) and aspA (serine protease) genes were detected in 21.5 and 8.82% of strains, respectively. Fish (Anabas testudineus) challenge studies showed that the isolate GP3 (Aeromonas veronii) bearing five virulent factors with the combination of aer/haem+/ascV+/fla+ genes induced severe lesions leading to 100% of mortality. In contrast, RB7 possessing four virulence factors and three genes (aer/haem+/ascV+/aspA+) could not produce severe lesions and any mortality indicating the absence of correlation between the virulence factors, its genes, and the pathogenicity in fishes. GP3 was cytotoxic to human liver cell line (WRL-68) in trypan blue dye exclusion assay. The 431 bp aer/haem gene of GP3 was transferable to E. coli Dh5α with a conjugational efficiency of 0.394 × 10–4 transconjugants per recipient cell. The transfer was confirmed by PCR and by the presence of 23-kb plasmids in both donor and transconjugants. Therefore, the occurrence of mobile genetic elements bearing virulence-associated genes in Aeromonas indicates the need for periodic monitoring of the aquatic habitat to prevent disease outbreaks.
Introduction
The genus Aeromonas is a pervasive group of microorganisms in the aquatic environment (1). It has the ability to induce diseases in fishes, mammals, reptiles, and amphibians (2). Aeromonas has been recognized as an etiological agent of several pathogenic conditions such as motile aeromonad syndrome (MAS), haemorrhagic septicaemia, fin rot, red sore disease, and scale protrusions in fish (3). Additionally, its role as an opportunistic pathogen in immunocompromised human and in infections such as septicaemia, bacterial endocarditis, gastroenteritis, wound sepsis, and traveller's diarrhea cannot be undermined (4). At present, Aeromonas is an important water-borne pathogen and has gained significance with its ability to induce multiple infections in a wide host range (5). A total of 75% of freshwater fish production in India is contributed by the state of West Bengal (6). About 251 species of freshwater fishes are found here in various freshwater resources such as tanks, ponds, creeks, beels, barrages, reservoirs, small river streams, major rivers, and an interlinked river drainage system (7). However, the yield is largely affected by mortality due to infections. Several species of highly pathogenic motile aeromonads including Aeromonas hydrophila, Aeromonas caviae, and Aeromonas sobria were found to be associated with diseases with mass mortalities in fishes (8). Pathogenicity in Aeromonas spp. has been reported to be due to multiple virulence factors that contribute to different mechanisms of the infection process. These factors include haemolysins, proteases, elastases, lipases, enterotoxins, phospholipases, chitinases, and deoxyribonucleases which can cause tissue damage and lethality (9). The genes encoding virulence factors determine the pathogenicity of a microorganism (10, 11). Pore-forming toxin genes aerA (encoding aerolysin), act (encoding cytotoxic toxins), and alt and ast (encoding cytotonic enterotoxins) are the primary elements of virulence within Aeromonas spp. (12). In addition, virulence genes encoding serine protease (ser, aspA), polar flagella (flaA), lateral flagella (lafA), DNase (exu), elastase (ahyB), glycerophospholipid: cholesterol acyltransferase (gCAT), and type III secretion system (ascV) have also been traced within Aeromonas (13). The invasiveness and adherence of Aeromonas hydrophila to HEpG2 cell monolayers, Vero cells, Chinese hamster ovarian cells, and erythrocytes have also been documented (14). Mobile genetic elements that carry virulence factors have been reported as widespread in the aquatic environment among different genera of bacteria such as Vibrio (15, 16), Photobacterium damselae (17), and Staphylococcus aureus (18). Some pieces of evidence show that Aeromonas can actively participate in processes of transfer of genetic material (via conjugation) with phylogenetically distant bacteria (19). In a study on the whole-genome sequence, Poole et al. (20) observed that A. hydrophila may be capable of transferring resistance and virulence genes to other related genera in the environment. This study aims at the isolation of Aeromonas spp. from fish farming environments of North Bengal, India for investigating the occurrence and distribution of molecular markers responsible for virulence. The current work provides experimental evidence of the conjugational spread of virulence genes to E. coli Dh5α.
Materials and Methods
Sample Collection and Isolation of Aeromonas sp.
The Aeromonas populations from water samples of 9 different fish farming sites across Darjeeling and Jalpaiguri districts of Sub-Himalayan West Bengal were investigated. Samples were collected in sterilized vials, kept at 4°C, and analyzed within 24 h of procuring. For isolation of the bacteria, 0.1 ml of ten-fold diluted samples prepared in sterile distilled water was spread plated on Aeromonas isolation medium (HiMedia Laboratories Ltd., Mumbai, India). The plates were incubated for 24 h at 30°C in the bacterial incubator. Altogether, 83 single colonies were selected based on the expected morphology from the spread plates, and pure cultures were maintained by streaking them on tryptone soya agar slants (21). Glycerol stocks of cultures were maintained in 10% glycerol at −20°C. All of the isolates were initially screened presumptively using a slightly modified key test called Aerokey II, described by Carnahan et al. (22), to identify the genus Aeromonas. The isolates were tested for Gram-staining, oxidase, catalase, glucose fermentation, resistance to 0/129 discs, and esculin hydrolysis (23). The strains were labeled as presumptive Aeromonas spp. depending on the tests.
Detection of Phenotypic Attributes of Virulence
Haemolytic activity of Aeromonas positive bacterial isolates was assessed by cultivating on tryptone soya agar (TSA) medium supplemented with 5% sheep blood (vol/vol) for 24 h at 30°C. Expression of haemolysin was depicted by a zone of clearance around bacterial colonies (24). Protease production was determined by the growth of bacterial isolates in nutrient agar containing 1.5% of skimmed milk and incubated for 24 h at 30°C. Casein-degrading bacteria showed clear zone around the colonies (24). DNase production was tested by inoculating the isolates in DNAse agar medium (HiMedia Laboratories, Mumbai) for 24 h at 30°C. The appearance of pale pink to white halos around the colonies indicated a positive result (25). Lipolytic activity was tested by growing the isolates in Tween-80 medium for 24 h at 30°C. Opaque zones around streaked cultures indicating crystal formation were considered positive for lipase (26). Amylase production was determined in starch agar plates following the method given by Barrow and Feltham (25). Siderophore production was determined using the Universal Chromazurol S (CAS) assay (27).
Molecular Identification of the Isolates
Sequence Similarity and Phylogenetic Analysis
The bacterial isolates were identified by 16S rRNA gene sequencing using universal primers. The total genomic DNAs were isolated from all the strains by CTAB method (28). Amplification of the 16S rRNA gene was performed in 25 μl reaction using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1498R (5′-GGTTCACTTGTTACGACTT-3′) (29) following specific cycling conditions: initial denaturation at 95°C for 3 min, 30 cycles of denaturation at 95°C for 30 s, annealing at the temperature of 52.2°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min on a thermal cycler (Applied Biosystems GeneAmp PCR 2400). The amplified product was purified using PCR purification kit (BR Biochem Life Sciences Pvt. Ltd) and sequenced at Eurofins sequencing services, Bangalore. All the obtained sequences were submitted to NCBI and GenBank accession numbers are given in the tree. The phylogenetic tree was constructed with the neighbor-joining method using MEGA 6.0 (30). Confidence in the tree topology was determined by bootstrap analysis using 1,000 resamplings of the sequences (31).
Pathogenicity Testing in Fishes
Healthy small-sized fishes (weight ~25–30 g) of Anabas testudineus were collected from fish farms in nearby areas of Sonapur and Gangarampur of Darjeeling district and maintained for 15 days in glass aquaria measuring 90 cm × 35 cm × 35 cm (10–12 fishes in each aquarium) for acclimatization. Water temperature was maintained at 25–30°C. For the fish pathogenicity testing, six strains (GP3, RB7, BP3, RJB1, MG8, and PP21) bearing two or more virulence properties and isolated from different locations were selected. The selected isolates were cultured in tryptic soy broth (HiMedia Laboratories, Mumbai) for 18 h at 30°C with constant shaking at 90 rpm. The cells were harvested as pellets following the centrifugation at 10,000 g for 10 min at 4°C and resuspended in 0.85% saline solution. The suspension was then adjusted to cell density of 1 × 107cfu/ml by measuring O.D. at 600 nm (O.D. = 0.8) in a spectrophotometer. Prior to infection, the fishes were anesthetized by keeping in benzocaine solution (25 mg L−1) for 1–2 min. Intramuscular injection was given at 0.4 ml per 25 g of body weight with each of the prepared bacterial cell suspensions. The fishes injected with individual bacterium were maintained in separate aquaria (10 fishes in each aquarium). The control set of fishes kept in a separate aquarium was administered with 0.85% saline solution at similar dose. A negative control set was additionally maintained under similar conditions in which the fishes did not receive any injection. Development of ulcers and damage to the surface tissue were monitored every 24 h postadministration for 1 week (32). The Kaplan–Meier survival analysis was performed using the GraphPad Prism software (version 9.3.1).
Cytotoxicity Test
Cytotoxicity of the Aeromonas isolate GP3 was tested in WRL-68 cell lines. GP3 was grown in LB broth at 30°C for 16 h under shaking at 90 rpm. The resultant culture was centrifuged at 10,000 g for 30 min at 4°C and the cell-free supernatant was collected carefully and filtered through a 0.45-mm filter. Cell-free filtrates of a non-pathogenic strain Lactobacillus sp. were similarly prepared and included in the experiment as the positive control, whereas sterile LB medium was used for negative control. WRL-68 cell line (human, liver, embryonic) obtained from National Centre for Cell Science (NCCS) Pune, Maharashtra, India was grown in Dulbeco's modified Eagle's medium (DMEM) with 10% foetal calf serum in an atmosphere containing 5% CO2. For morphological examination, cells were seeded in 60-mm polyvinyl-coated culture plates and incubated at 37°C for 24 h in CO2 incubator. Next day, 1.5 ml of the two cell-free filtrates and sterile LB medium was added to the cells taken in three separate sets and incubated for 1 h at 25°C. Changes in the cell morphology were recorded by observing under phase-contrast inverted microscope (Olympus CK40-SLP) at 200X magnification. Trypan blue dye exclusion assay was used to confirm cell death. By this method, nonviable cells are stained whereas viable cells exclude the dye (33). Percent viability was determined as follows: [total number of viable cells per ml of aliquot/ total number of cells per ml of aliquot] × 100. The experiment was repeated three times and data were averaged. Standard error was calculated using the statistical software OriginPro® version 9.9 freely available from https://www.originlab.com
Detection of Virulence Factor Encoding Genes by Polymerase Chain Reaction (PCR)
A total of 34 isolated strains were tested for the occurrence of four genetic determinants of virulence factors, aer/haem (haemolytic toxin), aspA (serine protease), ascV (type 3 secretion system), and flaA (polar flagella) by PCR using gene-specific primers (34). The primers have been listed in Supplementary Table 1. PCR amplification was performed with a 25 μl reaction volume containing 5 μl of 5X Flexi-Taq DNA polymerase buffer, 0.5 μl of 10 mM dNTP mix, 0.25 μl of the gene primers (100 mM-both forward and reverse), 2 μl of 25 mM MgCl2, 2 μl template DNA, and 0.25 μl of 5U Taq polymerase. PCRs were carried out in thermal cycler (Applied Biosystems GeneAmp PCR 2,400) with the following cycling conditions: initial denaturation at 95°C for 3 min, 30 cycles of denaturation at 95°C for 30 s, annealing at temperature of 50–55°C appropriate for each primer pair for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The amplicons were separated electrophoretically in 2% agarose gel stained with ethidium bromide (0.5 μg/ml). Electrophoresis was performed in a tank containing 1X TAE buffer for 1 h at 55V and bands viewed under UV-transilluminator (Bio-Rad Laboratories). PCR products were extracted using Gel/PCR DNA Fragments Extraction Kit (BR Biochem Life Sciences Pvt. Ltd.) according to the supplier's protocol. The products were cloned using pGEM–T easy vector kit (PROMEGA Corporation, U.S.A.) following the manufacturer's instructions. The amplicons were sequenced at Eurofins sequencing services, Bangalore, India. The obtained sequences were subjected to the similarity search using the BLAST search program of the National Center for Biotechnology Information (NCBI) (35). The sequences obtained were annotated and thereafter deposited in the NCBI GenBank through the Bankit tool.
Conjugation and Plasmid Detection
The potential donor GP3 strain bearing the haemolytic gene and nalidixic acid-resistant Escherichia coli-DH5α recipient strain which tested negative in PCR with aer/haem primers was incubated overnight in LB broth at 37°C. Both were grown to equal optical densities of 0.5 as measured by spectrophotometer (107 cells/ml). Broth conjugation was performed by mixing equal volumes of donor and recipient strains and incubating at 25°C for 24 h without shaking. A ten-fold serial dilution of each mating mixture was spread on LB agar plates supplemented with 5% sheep blood and 20 μg/ml nalidixic acid. Colonies growing on these double selective plates after 24–48 h of incubation at 37°C were treated as the putative transconjugants (32). The efficiency of conjugation was estimated as the number of transconjugants per initial number of recipients. Subsequently, boiling lysis of the selected transconjugants was carried out, and PCR was performed using aer/heam primers to detect the presence of the transferred gene. The donor and transconjugants as well as recipients prior to conjugation were also screened for the presence of plasmids as described by Birnboim and Doly (36).
Results
Occurrence and Presumptive Identification of Aeromonas
Out of the total 83 strains, 34 strains were morphologically and biochemically identified as Aeromonas sp. (Supplementary Table 2), and the identity was further confirmed by 16S rRNA gene sequencing. The genetic relationship among the sequences was established by a phylogenetic tree constructed by MEGA software (version 7) (Supplementary Figure 1). The neighbor-joining tree method showed that all the 34 isolates clustered with the reference strains of Aeromonas species: A. veronii (n = 19), A. hydrophila (n = 7), A. cavie (n = 3), and Aeromonas jandei (n = 5).
Virulence Characteristics
The virulence characteristics of all isolates were studied, and the results are summarized in Supplementary Table 3. Haemolytic and proteolytic activity was a common trait exhibited by all the 34 (100%) isolates. On the other hand, 15 (44.1%), 13 (38.2%), and 24 (70.6%) isolated strains displayed DNase, siderophore, and amylase production, respectively. Only 2 (5.8%) isolates showed lipolytic activity.
Pathogenicity in Fish
Virulence of six potential pathogenic Aeromonas strains isolated from different locations (GP3, RB7, BP3, RJB1, MG8, and PP21) was tested in A. testidineus using intramuscular injection, and the results are presented in Figure 1. Superficial lesions were observed initially at the site of injection in most fishes, which progressively became severe with time. Ulcers induced by GP3 were found to be most severe and all the fishes died by the 3rd day (Figure 1A). PP21-injected fishes also showed 100% mortality on the 3rd day with severe to moderate ulcers at the site of injection (Figure 1B). Fishes injected with BP3 and MG8 exhibited mild to moderate ulcers2 with 100% of mortality on 4th and 5th day, respectively (Figure 1D). RJB1-injected fishes produced superficial lesions in 30% of the fishes and recorded low mortality (Figure 1C). However, fishes injected with strain RB7 did not record any mortality. The fishes in the control group were healthy with no signs of ulcers or disease. The median survival time of fishes injected with PP21, MG8, BP3, and GP3 was found to be 1.5, 1, 2, and 2.5. For RJB1, RB7, and the control group, it was undefined because some fishes survived at the end of the observation period (Supplementary Figure 2). The results obtained were statistically significant with p < 0.0001 in the log-rank test.
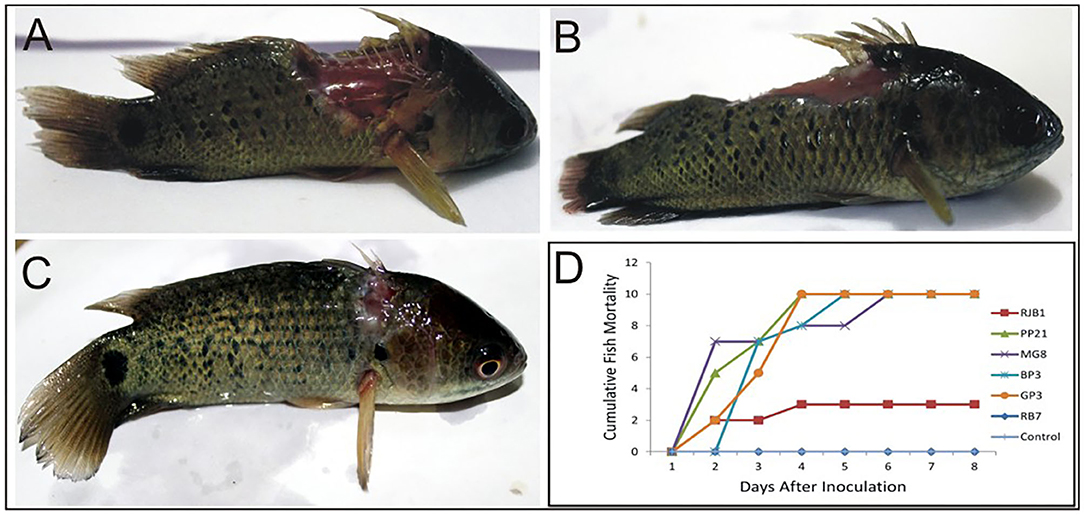
Figure 1. Pathogenicity testing of isolated Aeromonas strains in A. testudineus. (A) Extremely severe lesion in fish injected intramuscularly with cell suspension of GP3. (B) Fish injected with PP21 showing severe lesion. (C) Fish injected with RJB1showing superficial lesion. (D) Graph depicting cumulative mortality of fishes injected with bacterial cell suspensions within a 7-day observation period.
Cytotoxicity
The cell-free culture filtrate of isolate GP3 induced cytotoxic activity on WRL-68 human liver cell lines. Degenerative changes such as visible rounding off and adherence loss from the plate surface in cell cultures were observed (Figure 2A). However, no visible changes in morphology were induced in cell lines exposed to Lactobacillus cell-free culture filtrates (Figure 2B) as well as in control (Figure 2C). In the cell viability assay, a significant reduction in percent viability was observed in the GP3-exposed cells when compared to the control. GP3-exposed cells recorded a percent viability of only 0.48% which was much lower than Lactobacillus treated cells (66%) (Figure 2D).
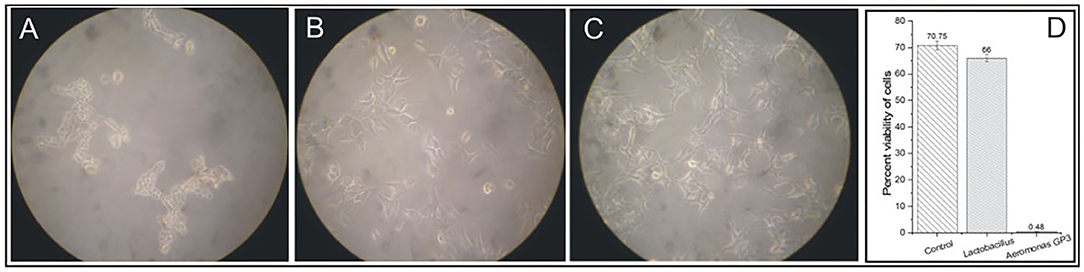
Figure 2. Cytotoxicity of isolated Aeromonas strain GP3 in WRL-68 cell line. (A) Treated with culture filtrate of GP3 showing visible rounding off and detachment of monolayers. (B) Treated with cell-free filtrate of Lactobacillus sp. showing minor morphological changes. (C) Control (treated with sterile LB medium) showing no change in cell morphology. (D) Graph representing the cell viability assay.
Virulence Genes
The virulence genes were detected in 23 of the 34 isolates which were identified as Aeromonas. The aerA/haem and flaA gene were amplified in 44.11% of the isolates (Supplementary Figures 3A,B). The ascV and aspA gene were detected in 23.5 and 8.82% of the isolates, respectively (Supplementary Figures 3C,D). The isolates could be classified into eight groups depending upon the occurrence of virulence genes (Table 1). A total of eleven isolates did not show the presence of any of the virulence genes. The GenBank accession numbers of the virulence genes are given as aer/haem (MT704303-MT704309, MT707932-MT707935, MH607886, MT591426, and MTT813045) aspA (MT909568-MT909570), ascV (MW001219-MW001222, MH607887-MH607890), and flaA (MT942623-MT942626, MT977537- MT977539).
Conjugation and Plasmid Isolation
Conjugation experiments revealed that determinants of haemolytic property of the isolated Aeromonas strain GP3 was transferable to recipient E. coli. GP3 contained a 23-kb plasmid which was also detectable in the transconjugants (Figure 3A). PCR analysis revealed the presence of the 431bp aer/haem gene in both donor and transconjugants (Figure 3B). The gene was not detected in recipient strains prior to conjugation. The frequency of conjugal transfer was recorded as 0.394 × 10−4 transconjugants per recipient cell in a mating mixture.
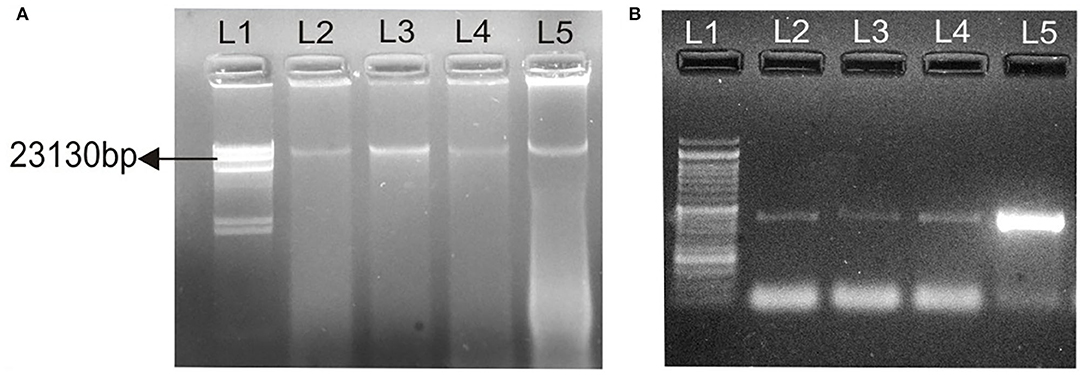
Figure 3. Transfer of virulence gene from isolated Aeromonas strain GP3 to E. coli DH5α. (A) lane 1, Lambda DNA/Hind III digest: molecular weight marker; lane 2, 23-kb plasmid (23,130 bp) of GP3 (donor); L3, L4, L5, 23 kb plasmids detected in transconjugants, (B) 431-bp PCR amplicons of the aerolysin/haemolysin (aer/haem) encoding gene from donor and transconjugants; lane 1, molecular weight marker as 50-bp DNA ladder; lanes 2-4, transconjugants and lane 5, donor GP3.
Discussion
In this study, several environmental strains of Aeromonas sp. harboring multiple virulence encoding genes were isolated from water samples of 10 fish-farming sites distributed across three districts of the Sub-Himalayan West Bengal. Furthermore, few isolates proved to be pathogenic in fish and cytotoxic to mammalian liver cells. Conjugational studies on the transfer of haemolysin-encoding gene from a virulent strain to E. coli Dh5α were also successfully demonstrated.
During the process of isolation, a lot of background microflora with similar cultural characteristics and morphology was obtained on Aeromonas isolation medium (37). Furthermore, we accurately identified the strains belonging to this genus using the aero key scheme proposed by Carnahan et al. (22) followed by 16S rRNA gene sequencing (38). Phylogenetic tree showed that all 34 isolates rearranged between four reference species with a highest number of species clustering to A. veronii followed by A. hydrophila. Few isolates formed distinct clades with A. jandaei and three isolates clustered with A. caviae. Such prevalence of variable species from the area of our study has not yet been reported elsewhere.
The foothill of Sub-Himalayan region is a geographically diverse region and serves as a habitat for a lot of endemic species of fish (39). A huge impact of bacterial diseases on fish productivity (40) and the role of Aeromonas in fish infections due to multiple virulence factors has been previously reported (41). In this study, all the strains isolated from water samples of small fish farms were found to be beta-haemolytic and proteolytic. This supports the theory of close association between the two factors since proteases have a prominent role in cleavage and activation of haemolysin (42). Additionally, 70.6% of the strains produced amylase whereas lesser isolates produced siderophore and DNase. Lower detection of siderophore-producing bacteria in this study may be due to alternate iron acquisition mechanisms found in Aeromonas for utilization in the process of host colonization (43). Likewise, DNase and amylase production are less as it depends on the nutritional requirement of the pathogen (42, 44).
In our study, four important virulence-related genes encoding haemolysin (aerA/haem), alkaline serine protease (aspA), inner component of type III secretion system (ascV), and polar flagella (flaA) had a heterogenous distribution among the isolates. Among these four genes, a prevalence of at least one virulence-related gene was observed in 67.7% of the total isolates. A higher percent (44.1%) of strains was found to harbor the aer/haem and flaA genes, which was similar to the findings by other authors (45, 46). However, lesser proportion of strains showed the presence of ascV (21.5%) and aspA (8.82%), indicating the possible involvement in proteases other than serine protease and secretion systems other than type III (47). Aeromonas isolates harboring at least one virulence gene were summarized into groups based on the presence of the type of gene. A total of eight different combinations were revealed, of which, the presence of single flaA gene, found in six isolates, was the most common. A total of five isolates showed the virulence gene pattern aer/haem+/ascV+/flaA+. The aerA (encoding aerolysin) and hlyA (encoding haemolysin) gene in Aeromonas hydrophila has been previously categorized as strong virulence determinant for pathogenicity (48, 49). Li et al. (12) indicated a strong link between bacterial virulence and the presence of aerA and ahp (encoding serine protease) gene. Hence, in Aeromonas, the presence of multiple virulence factors encoding genes either acting singly or in a synergistic pattern leads to disease development in the host (46).
In the fish challenge studies, all the six selected isolates of Aeromonas were found to be pathogenic to A. testudineus. Mortality was reported in 72.2% of fishes along with lesion development at the site of administration of bacteria. An isolate GP3 exhibiting five virulence characteristics and harboring three of the four tested virulence-related genes showed a highest pathogenicity with severe lesions leading to 100% of mortality. PP21, with only two virulent phenotype (protease and haemolysin) and genes aer/haem and flaA, also showed 100% of mortality on the 3rd day following injection. On the other hand, RB7 which exhibited four virulent phenotypes (haemolysin, protease, DNase, and amylase) and three genes (aer/haem+/ascV+/aspA+) did not record any mortality. Further, BP3 which showed four virulence traits and harbored only one of the four tested genes (ascV) was found to be strongly pathogenic to fishes. Therefore, no distinct correlation among the exhibition of virulence phenotypes, harboring of specific genes, and the mortality in fishes was evident. Similar results were obtained by Oliviera et al. (50) as they found no statistically significant relationship between the presence of virulence factors and mortality in Nile tilapia infected by A. hydrophila. Such absence of correlation between virulence factors and pathogenicity has also been reported by many other researchers (51, 52). However, several authors observed that the presence of specific virulence factors in aeromonads is closely related to mortality in injected fishes (9, 53).
The virulence of GP3 was further evaluated by assessing its ability to induce cytotoxic changes in liver cell lines in comparison with a non-virulent strain Lactobacillus. In our study, clear rounding off and detachment of monolayers were visible in liver cells exposed to cell-free GP3 filtrate. The cell viability reduced to only 0.48%, indicating extreme cytotoxicity. In similar studies, previous authors have reported rounding off, shriveling, and detachment of cell lines treated with bacterial filtrates of Aeromonas (54). The cytotoxicity in WRL-68 human liver cell lines indicated the possibility of disease-causing ability of Aeromonas in human, and similar reports suggesting its role in gastroenteritis and soft tissue infections of humans have been reported earlier (2).
Another significant finding of our study is the successful transfer of a component of haemolytic gene via conjugation from a virulent isolate GP3 to an E. coli recipient strain. A 23-kb plasmid was isolated from donor and transconjugants. Previous studies demonstrating the transfer of virulence genes from other genera such as Vibrio and S. aureus have been reported (15, 16, 18), but such reports involving aeromonads are rare. Most of the reports in Aeromonas sp. have their prime focus on the distribution of genetic determinants of several virulence factors, but there is very less evidence to suggest the transfer ability of these genes to non-pathogenic strains of bacteria belonging to a same or different genus. Majumdar et al. (55) observed that curing of a particular 21-kb plasmid detected in virulent Aeromonas isolate from environmental sources led to the loss in virulence. In this study, the 23-kb plasmid may be carrying the haemolysin gene as it was isolated from donor and transconjugants but not from the recipient strain prior to conjugation. Such mobile genetic elements contribute to the gene pool in aquatic reservoirs (56) and could be easily transferred to other microorganisms helping them thrive in nutrient-limiting conditions and increase their survivability within hosts (57).
In conclusion, this study revealed the occurrence of virulence factors and some of their genetic determinants in Aeromonas strains of aquatic origin. Further extensive studies on a greater number of virulence genes shall serve to accurately discriminate the various pathotypes of Aeromonas spp. We further studied the probability of horizontal spread of haemolytic property from the isolated Aeromonas to E. coli recipient strain. To our knowledge, this is the first report of the transfer of aer/haem gene encoding a haemolysin from a strain of virulent Aeromonas to any recipient strain. As the aquatic bodies provide a perfect environment for the interaction of microbes leading to gene transfer among cohabitant microflora, the occurrence of transferable virulence traits in bacteria especially in fish farming habitats is alarming. Being opportunistic pathogens, these bacteria may also infect humans as indicated by cytotoxicity studies. Future studies on the involvement of mobile genetic elements in virulence of Aeromonas spp. from aquaculture sources will allow to accurately assess the risk of spread of virulence traits to other non-pathogenic bacteria and the threat this poses to public health. Therefore, periodic monitoring of Aeromonas is required as these serve as the indicators of poor fish culture conditions and would possibly lead to zoonotic outbreaks.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval were not required as the study is in accordance with the institutional requirements and the institution has reviewed and approved the animal studies (Approval no. 161-(A)/R-2022). The fish used here is marketed in live condition commercially for purpose of human consumption. However, all fish handling and experimentation was done with maximum veterinary care.
Author Contributions
DS designed the study, helped in interpreting the study results, and critically revised the manuscript. PM and PB carried out the methodology, collected, and analysed the data. PM and DS drafted the manuscript. AS has helped in fish studies. AK has contributed in cytotoxicity studies. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funds from University Grants Commission (UGC), India (www.ugc.ac.in/) Grant No. F. No. 42-187/2013(SR) dated 02.07.2014. PM received fellowship from University Grants Commission (UGC), India (www.ugc.ac.in/) Grant No. F.25-1/2014-15(BSR)/7-132/2007(BSR) dated 07.10.2015. PB received fellowship from Department of Biotechnology, Govt. of India. The funders had no role in study design, data collection and analysis, decision to publish, or provide any funds for publishing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.887174/full#supplementary-material
References
1. Araujo RM, Pares R, Lucena F. The effect of terrestrial effluents on the incidence of Aeromonas spp. in coastal waters. J Appl Bacteriol. (1990) 69:439–44. doi: 10.1111/j.1365-2672.1990.tb01535.x
2. Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clinic Microbiol Rev. (2010) 23:35–73. doi: 10.1128/CMR.00039-09
3. Hoai TD, Trang TT, Van Tuyen N, Giang NT, Van Van K. Aeromonas veronii caused disease and mortality in channel catfish in Vietnam. Aquaculture. (2019) 513:734425. doi: 10.1016/j.aquaculture.2019.734425
4. Grim CJ, Kozlova EV, Sha J, Fitts EC, van Lier CJ, Kirtley ML, et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio. (2013) 4:e00064–13. doi: 10.1128/mBio.00064-13
5. Lamy B, Baron S, Barraud O. Aeromonas: the multifaceted middleman in the One Health world. Curr Opin Microbiol. (2022) 65:24–32. doi: 10.1016/j.mib.2021.09.012
6. Abraham TJ, Bharathkumar G. Distribution of motile aeromonads, pseudomonads and oxytetracycline resistant bacteria in freshwater catfish Pangasius pangasius Hatcheries of West Bengal, India. J Bio-Sci. (2009) 17:13–20. doi: 10.3329/jbs.v17i0.7094
7. Patra BC, Kar A, Bhattacharya M, Parua S, Shit PK. Freshwater fish resource mapping and conservation strategies of West Bengal, India. Spatial Inform Res. (2017) 25:635–45. doi: 10.1007/s41324-017-0129-z
8. Sahoo PK, Swaminathan TR, Abraham TJ, Kumar R, Pattanayak S, Mohapatra A, et al. Detection of goldfish haematopoietic necrosis herpes virus (Cyprinid herpesvirus-2) with multi-drug resistant Aeromonas hydrophila infection in goldfish: First evidence of any viral disease outbreak in ornamental freshwater aquaculture farms in India. Acta tropica. (2016) 161:8–17. doi: 10.1016/j.actatropica.2016.05.004
9. Mzula A, Wambura PN, Mdegela RH, Shirima GM. Virulence pattern of circulating aeromonads isolated from farmed Nile tilapia in Tanzania and novel antibiotic free attenuation of Aeromonas hydrophila strain TZR7-2018. Aquaculture Rep. (2020) 17:100300. doi: 10.1016/j.aqrep.2020.100300
10. Gao CX, Ren Y, Wang Q, Zeng WW, Li YY, Wang YY, et al. Isolation, identification antimicrobial susceptibility of pathogenic Aeromonas veronii isolated from grass carp. J Anhui Agr Univ. (2018) 45:409–415. doi: 10.13610/j.cnki.1672-352x.20180620.019
11. Kingombe CIB, Huys G, Tonolla M, Albert MJ, Swings J, Peduzzi, et al. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl Environ Microbiol. (1999) 65:5293–5302. doi: 10.1128/AEM.65.12.5293-5302.1999
12. Li J, Ni XD, Liu YJ, Lu CP. Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebra fish. J Appl Microbiol. (2011) 110:823–30. doi: 10.1111/j.1365-2672.2011.04944.x
13. Pattanayak S, Priyadarsini S, Paul A, Kumar PR, Sahoo PK. Diversity of virulence-associated genes in pathogenic Aeromonas hydrophila isolates and their in vivo modulation at varied water temperatures. Microb Pathogene. (2020) 147:104424. doi: 10.1016/j.micpath.2020.104424
14. Illanchezian S, Jayaraman S, Manoharan MS, Valsalam S. Virulence and cytotoxicity of seafood borne Aeromonas hydrophila. Brazil J Microbiol. (2010) 41:978–83. doi: 10.1590/S1517-83822010000400016
15. Castillo D, Kauffman K, Hussain F, Kalatzis P, Rørbo N, Polz MF, et al. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Scientific Rep. (2018) 8:1–9. doi: 10.1038/s41598-018-28326-9
16. Deng Y, Xu H, Su Y, Liu S, Xu L, Guo Z, et al. Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genom. (2019) 20:1–9. doi: 10.1186/s12864-019-6137-8
17. Osorio CR. Photobacterium damselae: how horizontal gene transfer shaped two different pathogenic lifestyles in a marine bacterium. Horizontal Gene Trans. (2019) 9:175–99. doi: 10.1007/978-3-030-21862-1_6
18. Alibayov B, Baba-Moussa L, Sina H, Zdenková K, Demnerová K. Staphylococcus aureus mobile genetic elements. Mol Biol Rep. (2014) 41:5005–18. doi: 10.1007/s11033-014-3367-3
19. Bello-López JM, Cabrero-Martínez OA, Ibáñez-Cervantes G, Hernández-Cortez C, Pelcastre-Rodríguez LI, Gonzalez-Avila LU, et al. Horizontal gene transfer and its association with antibiotic resistance in the genus Aeromonas spp. Microorganisms. (2019) 7:363. doi: 10.3390/microorganisms7090363
20. Poole TL, Schlosser WD, Anderson RC, Norman KN, Beier RC, Nisbet DJ. Whole-Genome sequence of Aeromonas hydrophila CVM861 isolated from diarrhetic neonatal swine. Microorganisms. (2020) 8:1648. doi: 10.3390/microorganisms8111648
21. Cipriano RC, Bullock GL. “Furunculosis and other diseases caused by Aeromonas salmonicida,” in National Fish Health Research Laboratory (2001).
22. Carnahan AM, Behram S, Joseph SW. Aerokey II: a flexible key for identifying clinical Aeromonas species. J Clinic Microbiol. (1991) 29:2843–9. doi: 10.1128/jcm.29.12.2843-2849.1991
23. Gerhardt P, Murray R, Wood W, Krieg N. Methods for general and molecular bacteriology. Am Soc Microbiol. (1994) 4:21–42. doi: 10.1002/food.19960400226
24. Aneja KR. Experiments in Microbiology Plant Pathology and Biotechnology. New Delhi: New Age International (P) Limited, Publishers (2003).
25. Barrow GI, Feltham RKA. Cowan and Steel's manual for identification of medical bacteria Great Britain: Cambridge University Press, p. 331 (1993).
26. Smibert RM, Krieg NR. “Phenotypic characterization,” In: editors Gerhardt P, et al. Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology, pp. 607–654 (1994).
27. Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Analytic biochemistr. (1987) 160:47–56. doi: 10.1016/0003-2697(87)90612-9
28. Gomes LH, Duarte KM, Andrino FG, Tavares FC. A simple method for DNA isolation from Xanthomonas spp. Scientia Agricola. (2000) 57:553–5. doi: 10.1590/S0103-90162000000300028
29. Srinivasan R, Karaoz U, Volegova M, MacKichan J, Kato-Maeda M, Miller S, et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PloS one. (2015) 10:e0117617. doi: 10.1371/journal.pone.0117617
30. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. (2011) 28:2731–9. doi: 10.1093/molbev/msr121
31. Felsenstein J. Confidence limits on phylogenies with a molecular clock. Systemat Zool. (1985) 34:152–61. doi: 10.2307/sysbio/34.2.152
32. Das A, Saha D, Pal J. Antimicrobial resistance and in vitro gene transfer in bacteria isolated from the ulcers of EUS-affected fish in India. Lett Appl Microbiol. (2009) 49:497–502. doi: 10.1111/j.1472-765X.2009.02700.x
33. Strober W. Trypan blue exclusion test of cell viability. Curr Protocols Immunol. (2001) 1:21. doi: 10.1002/0471142735.ima03bs21
34. Aravena-Román M, Inglis TJ, Riley TV, Chang BJ. Distribution of 13 virulence genes among clinical and environmental Aeromonas spp. in Western Australia. Euro J Clinic Microbiol Infect Dis. (2014) 33:1889–95. doi: 10.1007/s10096-014-2157-0
35. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
36. Bimboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. (1979) 7:1513–23. doi: 10.1093/nar/7.6.1513
37. Chaix G, Roger F, Berthe T, Lamy B, Jumas-Bilak E, Lafite R, et al. Distinct Aeromonas populations in water column and associated with copepods from estuarine environment (Seine, France). Front Microbiol. (2017) 8:1259. doi: 10.3389/fmicb.2017.01259
38. Pandove G, Sahota P, Vikal Y, Kaur B. Multiplex PCR water testing kit for rapid, economic and simultaneous detection of Escherichia coli, Yersinia enterocolitica and Aeromonas hydrophila from drinking water. Curr Sci. (2013) 10:352–8.
39. Panja S, Podder A, Chakrabarty M, Homechaudhuri S. Spatial pattern of freshwater habitats and their prioritization using additive partitions of beta diversity of inhabitant piscine assemblages in the Terai–Dooars ecoregion of Eastern Himalayas. Limnology. (2022) 23:57–72. doi: 10.1007/s10201-021-00666-y
40. Samayanpaulraj V, Velu V, Uthandakalaipandiyan R. Determination of lethal dose of Aeromonas hydrophila Ah17 strain in snake head fish Channa striata. Microbial Pathogene. (2019) 127:7–11. doi: 10.1016/j.micpath.2018.11.035
41. Shameena SS, Kumar K, Kumar S, Kumar S, Rathore G. Virulence characteristics of Aeromonas veronii biovars isolated from infected freshwater goldfish (Carassius auratus). Aquaculture. (2020) 518:734819. doi: 10.1016/j.aquaculture.2019.734819
42. Pemberton JM, Kidd SP, Schmidt R. Secreted enzymes of Aeromonas. FEMS Microbiol Lett. (1997) 152:1–0. doi: 10.1111/j.1574-6968.1997.tb10401.x
43. Maltz M, Levarge BL, Graf J. Identification of iron and heme utilization genes in Aeromonas and their role in the colonization of the leech digestive tract. Front Microbiol. (2015) 6:763. doi: 10.3389/fmicb.2015.00763
44. Campbell CM, Duncan D, Price NC, Stevens L. The secretion of amylase, phospholipase and protease from Aeromonas salmonicida, and the correlation with membrane-associated ribosomes. J Fish Dis. (1990) 13:463–74. doi: 10.1111/j.1365-2761.1990.tb00805.x
45. Hoel S, Vadstein O, Jakobsen AN. Species distribution and prevalence of putative virulence factors in mesophilic Aeromonas spp. isolated from fresh retail sushi. Front Microbiol. (2017) 8:931. doi: 10.3389/fmicb.2017.00931
46. Chen JS, Hsu GJ, Hsu BM, Yang PY, Kuo YJ, Wang JL, et al. Prevalence, virulence-gene profiles, antimicrobial resistance, and genetic diversity of human pathogenic Aeromonas spp. from shellfish and aquatic environments. Environ Pollut. (2021) 287:117361. doi: 10.1016/j.envpol.2021.117361
47. Hossain S, De Silva BC, Wickramanayake MV, Dahanayake PS, Wimalasena SH, Heo GJ. Incidence of antimicrobial resistance genes and class 1 integron gene cassettes in multidrug-resistant motile Aeromonas sp. isolated from ornamental guppy (Poecilia reticulata). Lett Appl Microbiol. (2019) 69:2–10. doi: 10.1111/lam.13162
48. Wang G, Clark CG, Liu C, Pucknell C, Munro CK, Kruk TM, et al. Detection and characterization of the hemolysin genes in Aeromonas hydrophila and Aeromonas sobria by multiplex PCR. J Clinic Microbiol. (2003) 41:1048–54. doi: 10.1128/JCM.41.3.1048-1054.2003
49. Heuzenroeder MW, Wong CY, Flower RL. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS Microbiol Lett. (1999) 174:131–6. doi: 10.1111/j.1574-6968.1999.tb13559.x
50. Oliveira ST, Veneroni-Gouveia G, Costa MM. Molecular characterization of virulence factors in Aeromonas hydrophila obtained from fish. PesquisaVeterináriaBrasileira. (2012) 32:701–6. doi: 10.1590/S0100-736X2012000800004
51. Wu CJ, Ko WC, Lee NY, Su SL, Li CW, Li MC, et al. Aeromonas isolates from fish and patients in Tainan City, Taiwan: Genotypic and phenotypic characteristics. Appl Environ Microbiol. (2019) 85:e01360–19. doi: 10.1128/AEM.01360-19
52. Roges EM, Gonçalves VD, Cardoso MD, Festivo ML, Siciliano S, Berto LH, et al. Virulence-associated genes and antimicrobial resistance of Aeromonashydrophilaisolates from animal, food, and human sources in Brazil. BioMed Res Int. (2020) 20:2607. doi: 10.1155/2020/1052607
53. Li T, Raza SH, Yang B, Sun Y, Wang G, Sun W, et al. Aeromonasveronii infection in commercial freshwater fish: a potential threat to public health. Animals. (2020) 10:608. doi: 10.3390/ani10040608
54. Kim KT, Lee SH, Lee KK, Han JE, Kwak D. Enhanced Virulence of Aeromonas hydrophila is induced by stress and serial passaging in mice. Animals. (2021) 11:508. doi: 10.3390/ani11020508
55. Majumdar T, Ghosh S, Pal J, Mazumder S. Possible role of a plasmid in the pathogenesis of a fish disease caused by Aeromonas hydrophila. Aquaculture. (2006) 256:95–104. doi: 10.1016/j.aquaculture.2006.02.042
56. Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha EP, et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nature Commun. (2020) 11:1–3. doi: 10.1038/s41467-020-17278-2
Keywords: Aeromonas, 16S rRNA gene, virulence related genes, Anabas testudineus, cytotoxicity, conjugational efficiency, plasmid
Citation: Mangar P, Barman P, Kumar A, Saha A and Saha D (2022) Detection of Virulence-Associated Genes and in vitro Gene Transfer From Aeromonas sp. Isolated From Aquatic Environments of Sub-himalayan West Bengal. Front. Vet. Sci. 9:887174. doi: 10.3389/fvets.2022.887174
Received: 01 March 2022; Accepted: 03 May 2022;
Published: 10 June 2022.
Edited by:
Huanying Pang, Guangdong Ocean University, ChinaReviewed by:
Guilherme Campos Tavares, Federal University of Minas Gerais, BrazilHiroshi Asakura, National Institute of Health Sciences (NIHS), Japan
Copyright © 2022 Mangar, Barman, Kumar, Saha and Saha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipanwita Saha, ZGlwYW53aXRhc2FoYUBuYnUuYWMuaW4=
 Preeti Mangar
Preeti Mangar Partha Barman2
Partha Barman2 Dipanwita Saha
Dipanwita Saha