- 1Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Zhanjiang, China
- 3Guangdong Key Laboratory of Control for Diseases of Aquatic Economic Animals, Zhanjiang, China
Pyruvate kinase I (PykF) is one of the key enzymes of glycolysis and plays a crucial role in bacterial metabolism. Several acetylation sites of Vibrio alginolyticus PykF were reported in previous studies and then 11 sites were first verified in this study, however, the specific roles of PykF acetylation remains unclear. Overlap-PCR and homologous recombination were implied to delete V. alginolyticus pykF gene and constructed complementary strains of site-directed mutagenesis for the further research focus on the deacetylation regulation on PykF. The results showed that the pyruvate kinase activity was sharply suppressed in the deacetylation status of K52, K68, and K317 of PykF, as well as the extracellular protease activity was significantly decreased in the deacetylation status of K52 and K68, but not induced with K317. Moreover, the growth rates of V. alginolyticus were not influenced with these three deacetylation sites. The ΔpykF mutant exhibited a 6-fold reduction in virulence to zebrafish. Site-directed mutations of K52R and K68R also showed reduced virulence while mutations of K317R didn't. The in vitro experiments showed that PykF was acetylated by acetyl phosphate (AcP), with the increase of incubation time by AcP, the acetylation level of PykF increased while the enzyme activity of PykF decreased correspondingly. Besides, PykF was deacetylated by CobB deacetylase and in result that the deacetylation was significantly down-regulated while the pyruvate kinase activity of PykF increased. Moreover, deletion of cobB gene had no significant difference in pyruvate kinase activity. These results confirm that CobB can regulate the acetylation level and pyruvate kinase activity of PykF. In summary, the results of this study provide a theoretical basis for further understanding of the deacetylation modification of PykF. It provides a new idea for the prevention and cure of vibriosis.
Introduction
Vibrio alginolyticus is an opportunistic halophilic gram-negative bacterium, which has great harm to the development of aquaculture and human health (1–4). V. alginolyticus is associated with diseases of fish, shellfish, shrimp, and coral (5–7), and is also a human pathogen that causes gastrointestinal infections (8). Therefore, it is necessary to understand the pathogenic mechanism of V. alginolyticus to cure these diseases. Lysine acetylation is a reversible post-translational modification (PTMs) that are involved various cellular processes in organisms from eucaryote to prokaryote (9–12). Lysine acetylation has a wide implication on bacteria, and plays an important role in their physiology, metabolism and virulence (13–15). In bacteria, it is known that protein activity is regulated by site-specific acetylation. In addition, acetyltransferases and deacetylases regulate the activity of proteins by adding or removing charged acetyl groups (14). In contrast to the transcriptional regulation, PTMs such as acetylation usually finetune the activities of proteins rather than turn them on or off (16, 17). In some pathogens, lysine acetylation modification affects bacterial virulence by regulating the activity of key factors, such as acetylation regulating protein stability and DNA-binding ability of HilD to modulate Salmonella Typhimurium virulence (18), acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence (19), acetylation of PhoP K88 is involved in regulating Salmonella virulence (20), acetylation is involved in Salmonella enterica Serovar Typhimurium Virulence and so on (15, 21). Furthermore, lysine acetylation can regulate the activity of various metabolic enzymes, such as glyceraldehyde phosphate dehydrogenase, isocitrate lyase, isocitrate dehydrogenase kinase/phosphatase, isocitrate dehydrogenase, malate dehydrogenase citrate synthase (22–25). This suggests that acetylation modifications may play an important role in bacterial. But no studies have been reported about PTMs of vibrio key factors/proteins regulate vibrio virulence. In past studies, acetylome and succinylome of vibrio have shown that a key strategy of vibrio is to use PTMs to modulate essential factors for virulence. These essential factors are involved purine metabolism, ribosome, pyruvate metabolism, glycolysis/gluconeogenesis, the TCA cycle, and so on, include LuxR (Transcriptional activator protein LuxR), LuxO (Luminescence regulatory protein LuxO), LuxS (S-ribosylhomocysteine lyase), Tdh (L-threonine 3-dehydrogenase), SodB (Superoxide dismutase), PEPCK [Phosphoenolpyruvate carboxykinase (ATP)], ClpP (ATP-dependent Clp protease proteolytic subunit) and so on, they are very important to virulence systems of vibrio (26–29). At present, how acetylation affects V. alginolyticus metabolism as well as virulence is still uncovered.
Previous studies about global acetylome studies have revealed that PykF is acetylated at multiple sites (27, 30–32). In the last step of glycolysis, pyruvate kinase catalyzes the irreversible transfer of phosphate from phosphoenolpyruvate (PEP) to ADP resulting in the production of pyruvic acid and ATP. In mammals, two genes, respectively, encode two different PK (pyruvate kinase) isoforms. Pkrl gene encodes the PKL and PKR isoforms of PK, which expresses in the liver and red blood cells, respectively. Most tissues express the PKM1or PKM2 isoform encoded by Pkm gene (muscle form of PK) (33). Acetylation at Lys305 of the PKM decreases its activity and targets it for chaperone-mediated autophagy and subsequent lysosome degradation (34). Two isoenzymes of PK, PykF and PykA (pyruvate kinase II), have been identified in E. coli (35, 36). Acetylome studies have revealed that V. alginolyticus PykF is acetylated at multiple sites, but the effect of their acetylation sites on its biological functions is still unknown. In this study, 11 lysine acetylation sites of V. alginolyticus PykF were identified by specific antibody enrichment combined with high-resolution mass spectrometry analysis. We mimicked the effect of lysine deacetylation of V. alginolyticus PykF on its biological function by site-directed mutagenesis. In summary, the results in this study provide a theoretical basis for further understanding of the acetylation modification of PykF.
Materials and Methods
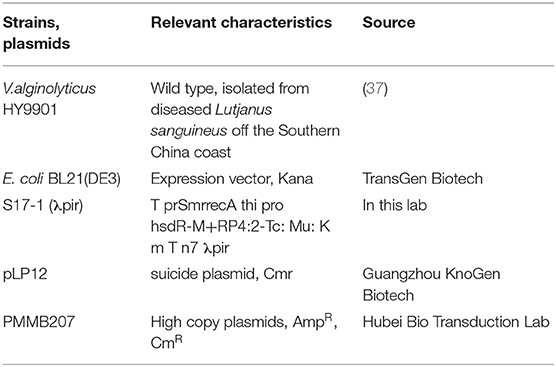
Strains and Plasmids
Vibrio alginolyticus HY9901 was isolated from a diseased red snapper (Lutjanus sanguineus) in Zhanjiang Port, Guangdong Province (37). The strains and plasmids used in this study are in Table 1. All primers were designed as shown in Table 2 according to the V. alginolyticus gene sequence (GenBank Number: GU074526.1) and the pykF gene (GenBank Number: OK376642). Healthy zebrafish (Danio rerio) were purchased from Zhanjiang Aquatic Market, with an average body length of 4 cm and weight of 0.2 g. The zebrafish were performed by bacteriological recovery tests and temporary rearing in seawater in a circulation system at 28°C for 2 weeks before the experiment.
Determining PykF Acetylation Sites by Mass Spectrometry
The protocol of LS-MS/MS analysis was performed by following the protocol described in previous studies (38, 39). Purified PykF proteins were fractionated on a 4–20% SDS-PAGE gel. For in-gel tryptic digestion, gel pieces were destained in 50 mM/L NH4HCO3 in 50% acetonitrile (v/v) until clear. Gel pieces were dehydrated with 100 μl of 100% acetonitrile for 5 min, the liquid removed, and the gel pieces rehydrated in 10 mM/L dithiothreitol and incubated at 56°C for 60 min. Gel pieces were again dehydrated in 100% acetonitrile, liquid was removed and gel pieces were rehydrated with 55 mM/L iodoacetamide. Samples were incubated at room temperature, in the dark for 45 min. Gel pieces were washed with 50 mM/L NH4HCO3 and dehydrated with 100% acetonitrile. Gel pieces were rehydrated with 10 ng/μl trypsin resuspended in 50 mM/L NH4HCO3 on ice for 1 h. Excess liquid was removed and gel pieces were digested with trypsin at 37°C overnight. Peptides were extracted with 50% acetonitrile /5% formic acid, followed by 100% acetonitrile. Peptides were dried to completion and resuspended in 2% acetonitrile/0.1% formic acid.
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a home-made reversed-phase analytical column (15-cm length, 75 μm i.d.). The gradient was comprised of an increase from 7 to 25% solvent B (0.1% formic acid in 98% acetonitrile) over 18 min, 25 to 38% in 6 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 450 nl/min on an EASY-nLC 1000 UPLC system.
The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in OrbitrapFusion (Thermo) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1,550 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 60,000. Peptides were then selected for MS/MS using NCE setting as 35 and the fragments were detected in the Orbitrap at a resolution of 15,000. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4. The peptides eluted from the HPLC column/electrospray source were subjected to MS survey scans. Raw MS/MS data were used to search a user-defined amino acid sequence database with the Proteome Discoverer 1.3 program. Cysteine Alkylation was used as a fixed modification, while lysine acetylation was set as the variable modification.
Expression and Purification of Lysine Deacetylated PykF Variants
The gene of pykF and their variants were cloned into the pET-28a plasmid with 6 xHis-tag and transformed into BL21 (DE3) cells (CD601, TransGen Biotech, Beijing, China) for expression and purified using BeyoGold™ His-tag Purification Resin (P2218, Beyotime, Shanghai, China) according to the manufacturer's recommended procedures. Site-directed mutagenesis of the pykF was performed using a Fast MultiSite Mutagenesis System kit (FM201-01, TransGen Biotech, Beijing, China) following manufacturer's protocol.
Construction of Deletion Mutants and Site-Directed Mutagenesis Complemented Strains
According to previous studies (40, 41), all deletion mutants and complemented strains were made. Primer pairs used for plasmid construction were included in Table 2. Overlap extension PCR was applied to generate an in-frame deletion of the pykF gene on the V. alginolyticus wild-type HY9901 chromosome. Two about 500 bp PCR fragments corresponding to genomic sequences flanking pykF for chromosomal in-frame deletions. Chromosomal in-frame deletions were cloned into a suicide plasmid (pLP12) by using standard cloning procedures followed by DNA sequencing. The resulting constructs were individually transformed into E. coli S17-1 λpir and introduced by conjugation into V. alginolyticus. Deletion mutants were selected on 10% sucrose TSA plates. Its presence was subsequently confirmed by PCR using primers located inside of the deleted sequence and subsequent sequencing of the PCR product.
For the construction experiments of the complemented strain, the pykF and its variants were cloned into expression vector pMMB207, which incorporates a C-terminal His-tag by PCR. The Fast MultiSite Mutagenesis System kit was used to perform site-directed mutagenesis of pykF with the corresponding primers (Table 2). These constructs were fully sequenced to check their inserts and then introduced by conjugation into the appropriate mutant strains. Then, all the resulting target mutations were confirmed by DNA sequencing. In addition to this, His Tag Mouse Monoclonal Antibody was used to confirm the normal expression by Western blot (described below).
PykF Activity Evaluation
Purified PykF and its variants were quantified using the detergent compatible Bradford protein assay kit (P0006, Beyotime, Shanghai, China). Pyruvate kinase enzymatic activity was measured using the pyruvate kinase Activity Detection Kit (BC0540, Solarbio, Beijing, China) according to the manufacturer's instruction. Pyruvate kinase catalyzes phosphoenolpyruvate and ADP to generate ATP and pyruvate, and lactate dehydrogenase further catalyzes NADH and pyruvate to generate lactate and NAD+. The decrease rate of NADH can be measured at OD340nm to reflect PykF activity.
PykF Deacetylation
For in vitro tests, the protocol of the deacetylation assay was performed by following described in previous studies (22–24), the reaction of the deacetylation was performed in the buffer containing 50 mM HEPES (pH 7.0), 5 mM MgCl2, 1 mM NAD+, 1 mM DTT, and 10% glycerol. The deacetylation reaction was initiated by mixing 5 μg PykF, 5 μg CobB proteins in a total volume of 100 μL and incubated at 37°C for 1 h. The acetylation level of the treated proteins was analyzed by Western blot (described below).
For in vivo tests, the acetylation level of the native PykF purified from the WT:pykF or ΔcobB:pykF of V. alginolyticus grown in TSB medium was determined by Western blot (described below).
PykF Acetylation
For in vitro tests, the protocol of the acetylation assay was performed by following described in previous studies (22–24), the reaction of the acetylation was performed in the buffer containing 50 mM HEPES (pH 7.0), 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 10 mM sodium butyrate. The acetylation reaction was initiated by mixing 5 μg PykF varied concentrations of AcP in a total volume of 100 μL and then incubated at 37°C for 1 h. The acetylation level of the treated proteins was analyzed by Western blot (described below).
Determination of Growth and Extracellular Protease Activity
The method of detection of growth was performed by following described in previous studies (41). All strains were incubated separately in TSB overnight and all diluted to (OD595 = 0.2), then inoculated at 28°C at a ratio of 1:100 and determined OD595 every hour. All measurements were repeated three times per group.
The method of detection of extracellular protease activity was performed by following described in previous studies (41). All strains were coated on TSA plates coated with sterile cellophane, and cultured at 28°C at 24 h, washed with sterile cooled PBS, centrifuged at 4°C for 30 min, and the supernatant filtered to obtain extracellular products. All measurements were repeated three times per group.
LD50 Assessment
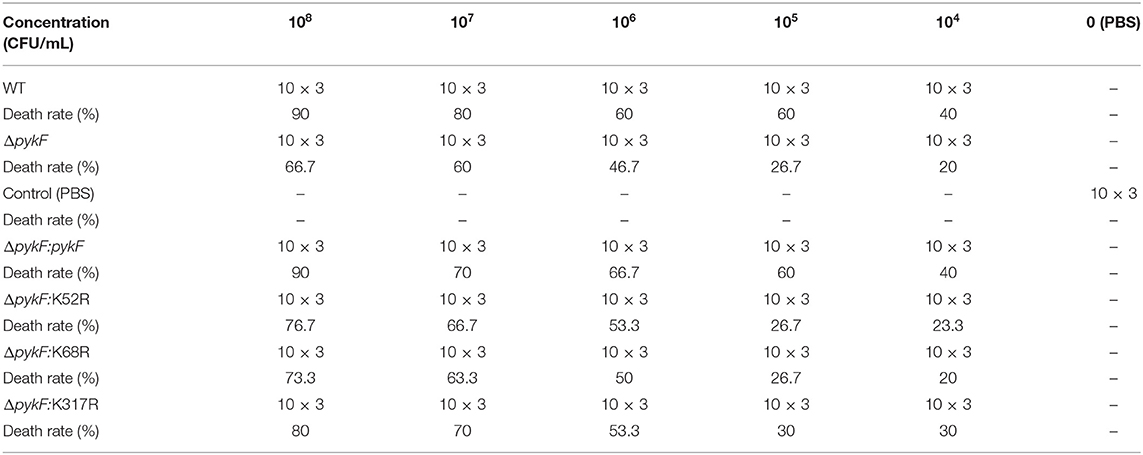
The LD50 evaluation was performed by following described in previous studies (41). The injection concentrations used for the dose response of wild-type strain HY9901, ΔpykF, and all complemented strains were 104, 105, 106, 107, and 108 CFU/mL. A total of 930 fish were randomly divided into seven groups (Table 3). The water temperature was adjusted to 28°C. The experiment was repeated three times. Five microliter of bacterial solution was injected into fish by intramuscular injection. The control group was injected with equivalent volumes of PBS. Fish were monitored for 14 days or until no morbidities occurred.
Western Blot
Protein concentrations were determined by Bradford Protein Assay (P0006C, Beyotime, Shanghai, China). For Western blot, purified PykF and its variants were fractionated on a 4–20% SDS-PAGE gel and transferred to the PVDF membrane by Trans-Blot Turbo (Bio-rad, USA). The samples were blocked with QuickBlock™ Blocking Buffer for Western blot (P0239, Beyotime, Shanghai, China) overnight at 4°C and then incubated with the horseradish peroxidase (HRP)-conjugated acetyl lysine antibody (9441S, Cell Signaling Technology, USA) as primary antibodies. The solutions were diluted at a ratio of 1: 2000 in QuickBlock™ Primary Antibody Dilution Buffer for Western blot (P0239, Beyotime, Shanghai, China) for 2 h at room temperature. Afterward, the membranes were washed three times in TBST and incubated with HRP-labeled goat anti-rabbit IgG (H+L) (A0208, Beyotime, Shanghai, China) at room temperature for 1 h. Antigen–antibody complexes were detected via the enhanced chemiluminescence method (P0018, Beyotime, Shanghai, China).
Statistical Analysis
Western blot images were quantified using Image J software. The experimental data were analyzed by single factor analysis of variance (ANOVA) with SPSS17.0 software and GraphPad Prism 8. The results in the figures are displayed as the mean ± standard deviation. Differences were considered statistically significant if the P-value was smaller than 0.05. Significance was indicated as *P ≤ 0.05; no *, P > 0.05, no significance.
Results
The Effect of Lysine Deacetylation and Acetylation Status of PykF on Pyruvate Kinase Activity
The acetylation signal of PykF was detected by Western blot, and then 11 acetylated lysine residues were identified via the previous acetylome profiling of V. alginolyticus and the mass spectrometry data (Supplementary Figures 1–11). Subsequently, 11 lysine residues, namely, K13, K19, K52, K59, K68, K145, K317, K319, K340, K368, and K382 were identified and analyzed to uncover the effects of deacetylated lysine residues on PykF.
Acetylation levels of PykF and its acetylation and deacetylated variants were evaluated. Compared to the acetylation levels of PykF, the acetylation mimicking status of those sites were not significantly different from PykF (Figure 1A top line). But the effect of deacetylation at different sites on acetylation level was different. Compared to the acetylation levels of PykF, the deacetylation mimicking status of K52, K68, K317, and K382 significantly decreased acetylation level, but other deacetylation mimicking status were no significant difference (Figure 1B top line).
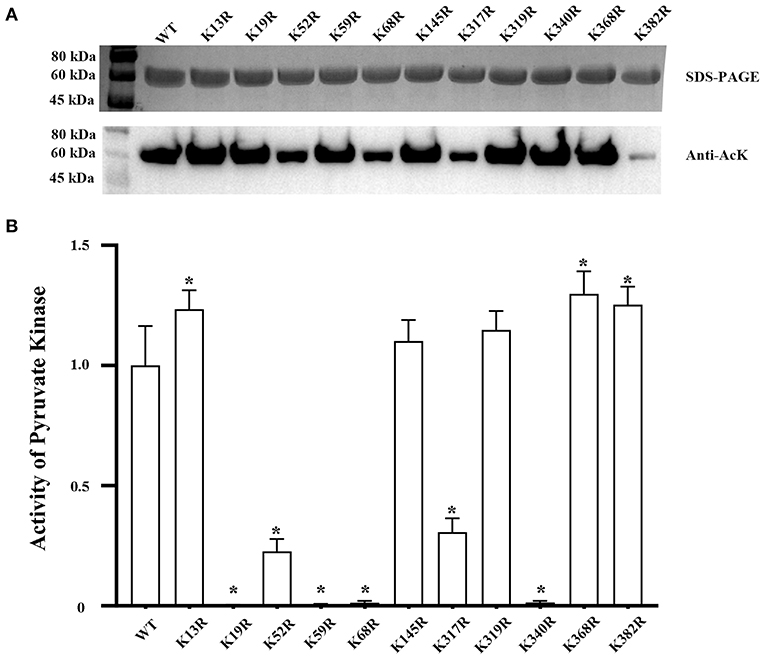
Figure 1. (A) Top line, SDS-PAGE and Western blot analysis of purified PykF and its acetylated variants from BL21 (DE3) cells. The acetylated variants with mutation of a single lysine site in PykF to glutamine. Lane 1, wild-type PykF; Lane 2, PykF-K13Q; Lane 3, PykF-K19Q; Lane 4, PykF-K52Q; Lane 5, PykF-K59Q; Lane 6, PykF-K68Q; Lane 7, PykF-145Q; Lane 8, PykF-K317Q; Lane 9, PykF-K319Q; Lane 10, PykF-K340Q; Lane 11, PykF-K368Q; Lane 12, PykF-K382Q. The same amount of protein was loaded. Anti-AcK: anti-acetyl lysine antibody. Bottom line, pyruvate kinase activity of PykF and its acetylated variants. Pyruvate kinase activity of the wild-type PykF was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). (B) Top line, SDS-PAGE and Western blot analysis of purified PykF and its deacetylated variants from BL21 (DE3) cells. The deacetylated variants with mutation of a single lysine site in PykF to arginine. Lane 1, wild-type PykF; Lane 2, PykF-K13R; Lane 3, PykF-K19R; Lane 4, PykF-K52R; Lane 5, PykF-K59R; Lane 6, PykF-K68R; Lane 7, PykF-145R; Lane 8, PykF-K317R; Lane 9, PykF-K319R; Lane 10, PykF-K340R; Lane 11, PykF-K368R; Lane 12, PykF-K382R. The same amount of protein was loaded. Anti-AcK, anti-acetyl lysine antibody. Bottom line, pyruvate kinase activity of PykF and its deacetylated variants. Pyruvate kinase activity of the wild-type PykF was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05. *p < 0.05.
Pyruvate kinase activities of PykF and its acetylation and deacetylated variants were evaluated. Compared to pyruvate kinase activities of PykF, the acetylation mimicking status of those sites were not significantly different from PykF (Figure 1A bottom line). But the effect of deacetylation at different sites on pyruvate kinase activities was different. Compared to the pyruvate kinase activities of PykF, the deacetylation mimicking status of K13, K368, or K382 significantly increased the activity, while acetylation at K145 or K319 only had no significantly change. Furthermore, the deacetylation mimicking status of K52 or K317 lost about 80% of pyruvate kinase activity, while the deacetylation mimicking status of K19, K59, K68, or K340 almost eliminated its activity (Figure 1B bottom line).
PykF Activity of the Deletion Mutants and Site-Directed Mutagenesis Complemented Strains
The principle of lysine site selection for the next experiments was that deacetylated variants had significantly lower acetylation levels and activity than PykF. According to the results 3.1, K52, K68, and K317 were selected to study the changes in pyruvate kinase activity. In this study, WT: pykF expression strain, ΔpykF mutant strain (Figure 6 of Supplementary Files), ΔpykF: K52R, ΔpykF: K68R, ΔpykF: K317R site-directed mutagenesis strains were successful constructed (Figure 2A). Pyruvate kinase activity of ΔpykF was decreased by about 60% compared to wild-type (Figure 2B). Site-directed mutagenesis complemented strains such as ΔpykF:K52R, ΔpykF:K68R and ΔpykF:K317R showed significantly decreased pyruvate kinase activity compared to ΔpykF:pykF complemented strain (Figure 2C). Therefore, the deacetylation status of K52, K68, and K317 was required for pyruvate kinase activity of V. alginolyticus.
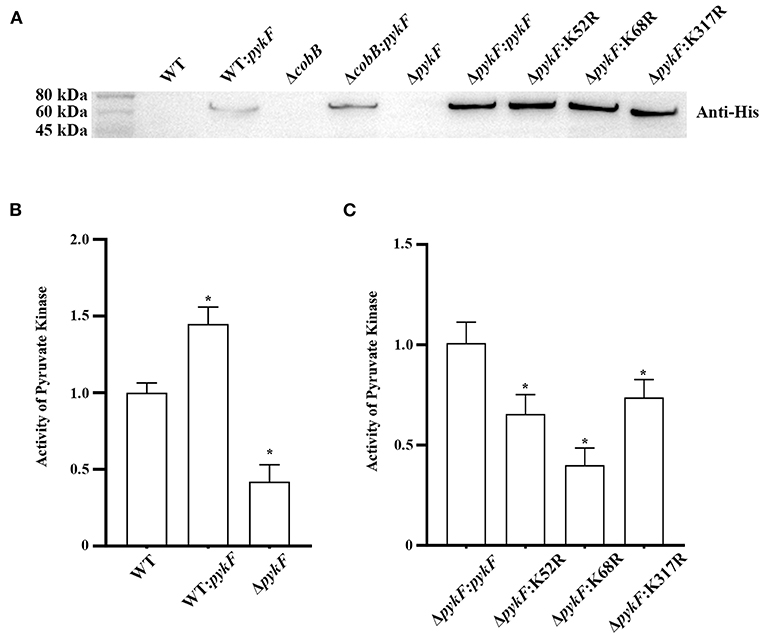
Figure 2. (A) Construction of complemented and overexpression strains. Lane 1, wild-type (WT); Lane 2, WT:pykF overexpression strain; Lane 3, deletion of cobB gene strain (ΔcobB); Lane 4, ΔcobB:pykF complemented strain; Lane 5, deletion of pykF gene strain (ΔpykF); Lane 6, ΔpykF:pykF complemented strain; Lane 7, ΔpykF:K52R complemented strain; Lane 8, ΔpykF:K68R complemented strain; Lane 9, ΔpykF:K317R complemented strain. Anti-His: His Tag Mouse Monoclonal Antibody. (B) Analysis of pyruvate kinase activity throughout the V. alginolyticus ΔpykF strain and WT:pykF overexpression strain. Pyruvate kinase activity of V. alginolyticus WT was set as 1. (C) Analysis pyruvate kinase activity of site-directed mutagenesis complemented strains. Pyruvate kinase activity of V. alginolyticus ΔpykF:pykF was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05. *p < 0.05.
Deacetylation of PykF by Deacetylase CobB
The acetyl on the ε-amino group of lysine residues is stable, and its removal needs a category of enzymes called lysine deacetylases (42). So far, only one lysine deacetylase has been identified in bacteria: the deacetylase CobB, a NAD+-dependent sirtuin class deacetylase (43, 44). we have verified the function of the CobB protein in V. alginolyticus (45). To confirm the effect of CobB on PykF, Western blot was used to detect the deacetylation of CobB to PykF, and then its enzyme activity was determined. The results showed that the CobB expressed and purified from E.coli BL21 (DE3) cells can deacetylate PykF with the participation of NAD+ (Figure 3A top line), and deacetylation of PykF significantly enhanced pyruvate kinase activity (Figure 3A bottom line). For in vivo tests, the acetylation level of the native PykF purified from the WT:pykF or ΔcobB:pykF was determined by Western blot. Deletion of the cobB gene increased the acetylation level of PykF (Figure 3B top line), but it was no significant difference in pyruvate kinase activity (Figure 3B bottom line). In conclusion, CobB can regulate the acetylation level of PykF and pyruvate kinase activity.
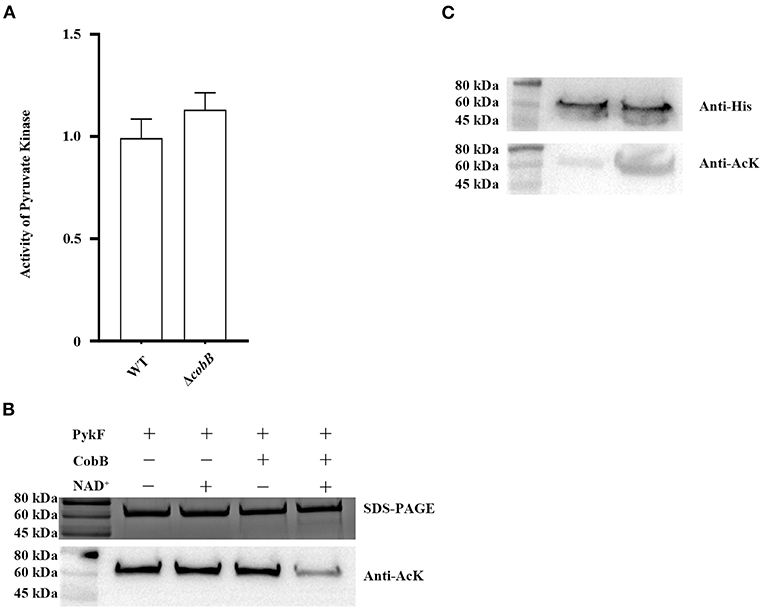
Figure 3. (A) CobB deacetylation PykF in vitro. Top line, Western blot was performed on PykF treated with CobB for 1 h. The same amount of protein was loaded in all lanes. Bottom line, pyruvate kinase activity was measured, and the enzyme activity of lane 1 sample was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05. (B) CobB deacetylates PykF in vivo. Top line, Western blot analysis of purified PykF from V. alginolyticus WT:pykF and ΔcobB:pykF. Lane 1, purified PykF from V. alginolyticus WT:pykF; Lane 2, purified PykF from V. alginolyticus ΔcobB:pykF. The same amount of protein was loaded in all lanes. Anti-His, His Tag Mouse Monoclonal Antibody. Anti-AcK: anti-acetyl lysine antibody. Bottom line, pyruvate kinase activity of V. alginolyticus WT and ΔcobB. Pyruvate kinase activity of V. alginolyticus WT was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05.
Acetylation of PykF by Acetyl Phosphate
So far, it is believed that there are two mechanisms of lysine acetylation in bacteria, the acetyl-CoA-dependent enzymatic process, and the AcP-dependent chemical reaction (11, 12, 14, 31, 46, 47). In order to study the acetylation mechanism of PykF, PykF expressed and purified from BL21 (DE3) cells was treated with AcP at concentrations of 200 μM, 3 mM, and 12 mM, corresponding to estimated intracellular AcP concentrations at the exponential phase, the stationary phase, and the ΔackA background, which accumulates AcP, respectively (30, 48, 49). The immunoblot with anti-AcK antibody and the detection of pyruvate kinase activity indicated that AcP can chemically acetylate PykF in a dose-dependent and time-dependent manner in vitro (Figure 4 top line). Furthermore, with the increase of incubation time, the acetylation level of PykF increased and its enzyme activity decreased correspondingly (Figure 4 bottom line). This suggested AcP can acetylate PykF, resulting in a decrease in enzyme activity of PykF.
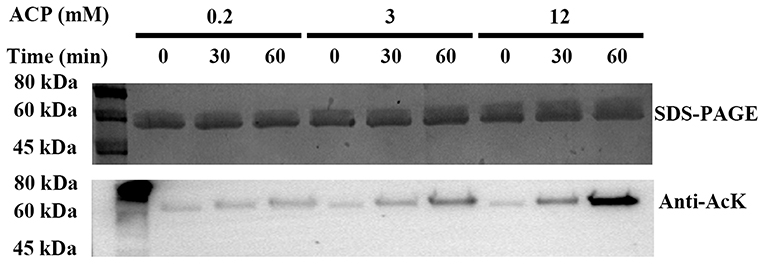
Figure 4. Top line, acetylation of PykF. SDS-PAGE and Western blot analysis were performed on purified PykF treated with AcP in different concentrations and incubation time. The same amount of protein was loaded. Anti-AcK, acetyl lysine antibody. Bottom line, pyruvate kinase activity was measured, and the enzyme activity of lane 1 sample was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05.
Growth and Extracellular Protease Activity
To determine the effect of delete or overexpress pykF gene and site-directed mutagenesis at the K7, K52, and K317 on the biological function of V. alginolyticus. WT, ΔpykF mutant strain, and WT:pykF, ΔpykF:pykF complemented strain, and ΔpykF:K52R, ΔpykF:K68R and ΔpykF:K317R site-directed mutagenesis complemented strains were subjected to determine growth and extracellular protease activity. The results showed that the growth rate of the WT:pykF overexpression strain was similar to that of the wild-type strain, while the growth rate of the ΔpykF mutant strain was significantly reduced (Figure 5A). In addition, the growth rate of site-directed mutagenesis complemented strains had no significant change compared to the ΔpykF:pykF complemented strain (Figure 5B). These results indicate that deletion of pykF gene significantly weakened the growth of V. alginolyticus, whereas overexpression of pykF gene and site-directed mutagenesis complemented strains had no significant effect on the growth of V. alginolyticus.
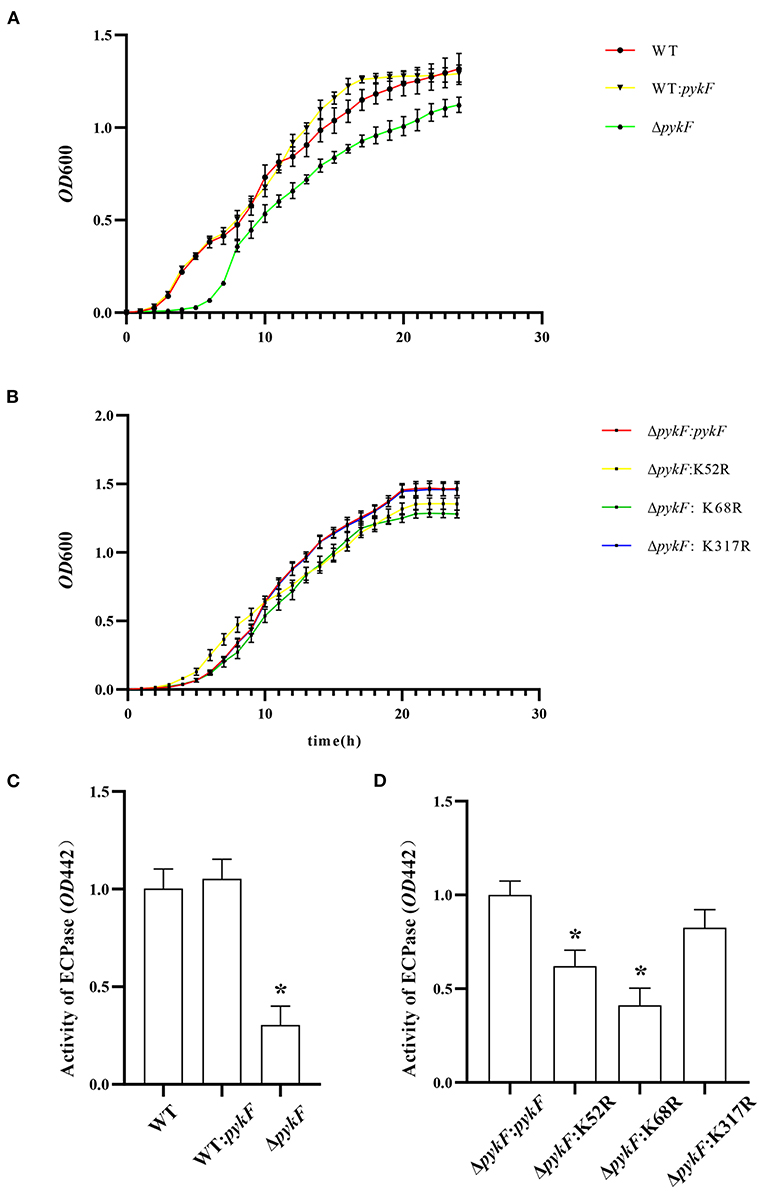
Figure 5. (A) Growth of V. alginolyticus. Analysis of growth throughout the V. alginolyticus ΔpykF strain and WT:pykF overexpression strain at 24 h. (B) Analysis of growth throughout V. alginolyticus ΔpykF:pykF, ΔpykF:K52R, ΔpykF: K68R and ΔpykF:K317R complemented strains at 24 h. (C) Analysis of extracellular protease activity throughout the V. alginolyticus ΔpykF mutant strain and WT:pykF overexpression strain. Extracellular protease activity of V. alginolyticus WT was set as 1. (D) Analysis site-directed mutagenesis complemented strains on extracellular protease activity. Extracellular protease activity of V. alginolyticus ΔpykF:pykF was set as 1. Mean values and standard deviations were calculated based on three replicates (n = 3). Two-tailed P-values were determined by the t-test, and the significance level is 0.05. *p < 0.05.
Extracellular protease, as metabolites of bacteria, play an important role as virulence factors in the process of infecting the host. Extracellular proteins have a variety of protease activities, including lecithin, amylase, lipase, and casein. The extracellular protease activity was reduced in the ΔpykF mutant strain compared to the wild-type strain (Figure 5C). Compared to the ΔpykF:pykF complemented strain, site-directed mutagenesis complemented strains ΔpykF:K52R and ΔpykF:K68R showed decreased extracellular protease activity, but extracellular protease activity of ΔpykF:K317R had no significant change (Figure 5D). Among them, site-directed mutagenesis complemented strains ΔpykF:K52R and ΔpykF:K68R showed about a 50% reduction in extracellular protease activity, indicating that deacetylation of these two sites has an important role in protease activity inhibition.
LD50
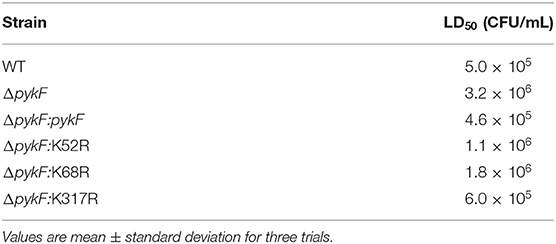
Zebrafish from quarantined stocks recognized as disease-free were used as models to assess the virulence of WT, ΔpykF, and all complemented strains. The results showed that LD50 of ΔpykF was 6 times higher than that of WT strain. Compared with ΔpykF: pykF complementary strains, ΔpykF: K52R and ΔpykF: K68R had about 3 and 4 times of LD50, while ΔpykF: K317R had no significant change (Tables 3, 4). The results showed that the deletion of pykF gene weakened the virulence of V. alginolyticus, and the deacetylation of K52 and K68 sites also weakened the virulence of V. alginolyticus, while the deacetylation of K317R did not significantly affect the virulence of V. alginolyticus.
Discussion
In recent years, more and more acetylome data have been published, showing that most acetylated proteins are mostly involved in cellular metabolism. However, no studies have been reported the effect of acetylation modifications on PykF in bacteria (13, 27, 50–52). In this study, we show the effect of deacetylation of lysine residues on PykF, and that deacetylases CobB can deacetylate PykF. As a classic approach, glutamine is commonly used as a mimic of acetylated lysine. Inconsistent with previous research is that PykF expressed in E.coli BL21 (DE3) cells had been detected acetylation signal (22–24). Glutamine was used to substitute for lysine to mimic acetylation, but acetylation mimicking status of those sites cannot significantly change their acetylation levels and pyruvate kinase activity. The reason for this may be that those lysine residues were already acetylated in prokaryotic expression system. On the other hand, arginine is commonly used as a mimic of deacetylated lysine, and then we used Q5 site-directed mutagenesis kit to mutate acetylated lysine residues to arginine, and measured pyruvate kinase activity of these variants to mimic the effect of lysine deacetylation on its activity. By site-specifically deacetylated from selected acetylated lysine of PykF, we provided a direct biochemical basis for the deacetylation study on PykF. We found that deacetylation status of K52, K68, and K317 sites decreased pyruvate kinase activity. Thus, the acetylation of these three lysine residues is essential for its activity.
CobB is a bacterial Sirtuins that regulates the function of its substrate by deacetylation at the active site of lysine. Although CobB is a predominant deacetylase in bacteria, it does not completely deacetylate PykF due to CobB only deacetylates a portion of acetyl lysine (30, 43). Our results showed that PykF can be deacetylated by the NAD+-dependent deacetylase CobB in vitro and that hyperacetylation of PykF occurs in the ΔcobB strain. These suggest that CobB can also regulate the deacetylation of PykF in vivo. We further found that pyruvate kinase activity has no significant difference in the ΔcobB strain compared to the wild-type strain. But in other studies, deletion of cobB gene increases the activity of some metabolic enzymes from other strains. CobB affects malate dehydrogenase activity by regulating the deacetylation of lysine residues at positions K301 and K314 of the non-protein structural domain of E. coli Malate Dehydrogenase (22). This regulatory mechanism has been more studied in eukaryotes, Sirtuin family deacetylation mediates nuclear localization of PKM2, protein kinase activity of pyruvate kinase M2, tetramerization and pyruvate kinase activity in vitro to influence oncogenic function, tumor growth and insect longevity (53–58). This indicates the importance of deacetylation for the regulation of PykF function, and the preference of CobB for acetyl groups may be a new ideal drug target.
Not all acetylations are reversed, and we obtained a protein that all the lysine sites may not have been acetylated. We found that PykF was acetylated in vitro in a time- and AcP dose-dependent manner, and with the increase of acetylation level, its enzyme activity decreases. This suggesting that there are still unidentified PykF acetylated lysine residues, or the known sites may be more acetylated. This has similarities with the studies in E. coli (22–24). Preliminary findings in E. coli revealed that most acetylation occurred at a low level and accumulated in growth-arrested cells in a manner that depended on the formation of AcP through glycolysis (30). The site-specificity of AcP-dependent protein acetylation has been studied, and these specificities depend on the surface accessibility, reactivity, and three-dimensional microenvironment of the target lysine. AcP affects the function of some key enzymes of bacteria as well as mediating bacterial virulence (15, 31, 59, 60). The study of AcP-dependent acetylation of substrates is important for understanding the mechanism of AcP action.
K52, K68, and K317 sites of V.alginolyticus were mutated to arginine to mimic the effect of deacetylation status on it. We verified the acetylation levels by Western blot and found that protein acetylation levels were reduced in mutation at K52, K68, and K317 sites, and the three site-directed mutagenesis complemented strains showed decreased pyruvate kinase activity. Lysine deacetylation at the K52 and K68 sites significantly reduced extracellular protease activity of V. alginolyticus. But lysine deacetylation at the K317 site had no significant effect on extracellular protease activity. Thus, protein deacetylation modification at these three sites has important effects. An important gene for carbon metabolism-related pyruvate kinase is encoded by pykF in many bacteria. In the present study, deletion of the pykF gene reduced the growth rate and extracellular protein activity of bacteria, which is consistent with the findings in other bacteria (61–64). However, overexpression of pykF gene had no significant effect on the growth rate and extracellular protease activity. We also found decreased rate of extracellular protein activity in complemented strains at the K52R and K68R sites. The two lysine residues are located in the A-domain of PykF, which consists of a (β/α)8-barrel structure characterized by three helices located at the top of the loop connecting the c-terminal chains of the β sheet, and these helices play a major role in catalytic and metastable regulation. The catalytic site of PyK is in the cleft between the A- and B-domains at the top of the barrel (65–69). These effects may be explained by that the deacetylation of the three sites changed the charged nature of the three lysine residues, the hydrophilicity, and the distance between the hydrogen bonds, thus affected the stability of the catalytic domain and the active site.
PykF is one of the key enzymes in glycolysis and plays an important role in V. alginolyticus virulence. Our results are like those of other pathogens, such as Brucella abortus (62). The deacetylation of K52 and K68 of PykF is virulent by reducing pyruvate kinase activity and extracellular protease activity of V. alginolyticus, but no similar studies have been conducted before. During the entry of intracellular pathogens into host cells, carbon metabolism may be directly or indirectly involved in regulating the expression of virulence genes in host cells, thus affecting the virulence of pathogens. In V. alginolyticus, silencing of the pykF gene reduces the expression level of some virulence genes, ndk (Nucleoside-diphosphate kinase encoding gene), eno (Enolase encoding gene), sdhB (succinate dehydrogenase iron-sulfur Subunit encoding Gene), glpF (glycerol uptake facilitator Protein-encoding gene) and cycH (phosphoadenosine phosphosulfate reductase encoding gene) (64), which in turn leads to reduced virulence. The deletion of pykF gene also showed a consistent situation, which proved that pykF is closely related to the regulation of virulence of V. alginolyticus.
Conclusion
In this study, we investigated the effect of lysine deacetylation of PykF protein on its biological function. The results showed that deacetylation status of three lysine residues in PykF, K52, K68, and K317, significantly reduced its activity. Deacetylated at the K52 and K68 sites significantly reduced extracellular protease activity and virulence of V. alginolyticus, but deacetylated at the K317 site had no significant difference on extracellular protease activity. Deacetylase CobB deacetylates PykF, and AcP catalyzes the acetylation of PykF. And with the acetylation level of PykF increased and its enzyme activity decreased. In summary, although deacetylation status of three lysine residues, K52, K68, and K317, all reduced pyruvate kinase activity. However, the mechanisms of acetylation and deacetylation of these sites are not clear and need to be further investigated. We are sure that the deacetylation modifications of these sites will become new drug targets that can better reduce the risk of vibriosis to the aquaculture industry.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Guangli Li and Guangdong Ocean University of Ethics Committee.
Author Contributions
ZX: conceptualization, methodology, and writing—original draft. LW, XW, MW, and MT: resources and investigation. YD: writing—review and editing and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Foundation for the High-level Talents in Higher Education of Guangdong and Graduate Education Innovation Program of Guangdong Ocean University (No. 201724).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.877067/full#supplementary-material
References
1. Thompson FL, Lida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. (2004) 68:403–31. doi: 10.1128/MMBR.68.3.403-431.2004
2. Scarano C, Spanu C, Ziino G, Pedonese F, Dalmasso A, Spanu V, et al. Antibiotic resistance of Vibrio species isolated from Sparus aurata reared in Italian mariculture. New Microbiol. (2014) 37:329–7. Available online at: http://www.newmicrobiologica.org/PUB/allegati_pdf/2014/3/329.pdf
3. Song X, Zang J, Yu W, Shi X, Wu Y. Occurrence and identification of pathogenic Vibrio contaminants in common seafood available in a Chinese traditional market in Qingdao, Shandong Province. Front Microbiol. (2020) 11:1488. doi: 10.3389/fmicb.2020.01488
4. Yen PTH, Linh NQ, Tram NDQ. The identification and determination of toxin genes of Vibrio strains causing hemorrhagic disease on red drum (Sciaenops ocellatus) using PCR. AMB Express. (2021) 11:4. doi: 10.1186/s13568-020-01161-w
5. Maugeri TL, Caccamo D, Gugliandolo C. Potentially pathogenic vibrios in brackish waters and mussels. J Appl Microbiol. (2000) 89:261–6. doi: 10.1046/j.1365-2672.2000.01096.x
6. Zhenyu X, Shaowen K, Chaoqun H, Zhixiong Z, Shifeng W, Yongcan Z. First characterization of bacterial pathogen, Vibrio alginolyticus, for Porites andrewsi White syndrome in the South China Sea. PLoS ONE. (2013) 8:e75425. doi: 10.1371/journal.pone.0075425
7. Arab S, Nalbone L, Giarratana F, Berbar A. Occurrence of Vibrio spp. along the Algerian Mediterranean coast in wild and farmed Sparus aurata and Dicentrarchus labrax. Vet World. (2020) 13:1199–208. doi: 10.14202/vetworld.2020.1199-1208
8. Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, et al. Vibrio spp. infections. Nat Rev Dis Primers. (2018) 4:8. doi: 10.1038/s41572-018-0005-8
9. Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem Sci. (2011) 36:211–20. doi: 10.1016/j.tibs.2010.10.001
10. Christensen DG, Baumgartner JT, Xie X, Jew KM, Basisty N, Schilling B, et al. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio. (2019) 10:e02708-18. doi: 10.1128/mBio.02708-18
11. Diallo I, Seve M, Cunin V, Minassian F, Poisson JF, Michelland S, et al. Current trends in protein acetylation analysis. Expert Rev Proteomics. (2019) 16:139–59. doi: 10.1080/14789450.2019.1559061
12. VanDrisse CM, Escalante-Semerena JC. Protein acetylation in bacteria. Annu Rev Microbiol. (2019) 73:111–32. doi: 10.1146/annurev-micro-020518-115526
13. Zhang K, Zheng S, Yang JS, Chen Y, Cheng Z. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J Proteome Res. (2013) 12:844–51. doi: 10.1021/pr300912q
14. Bernal V, Castaño-Cerezo S, Gallego-Jara J, Écija-Conesa A, de Diego T, Iborra JL, et al. Regulation of bacterial physiology by lysine acetylation of proteins. N Biotechnol. (2014) 31:586–95. doi: 10.1016/j.nbt.2014.03.002
15. Ren J, Sang Y, Qin R, Su Y, Cui Z, Mang Z, et al. Metabolic intermediate acetyl phosphate modulates bacterial virulence via acetylation. Emerg Microbes Infect. (2019) 8:55–69. doi: 10.1080/22221751.2018.1558963
16. Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. (2009) 1:a000950. doi: 10.1101/cshperspect.a000950
17. Muller MM. Post-translational modifications of protein backbones: unique functions, mechanisms, and challenges. Biochemistry. (2018) 57:177–85. doi: 10.1021/acs.biochem.7b00861
18. Sang Y, Ren J, Qin R, Liu S, Cui Z, Cheng S, et al. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J Infect Dis. (2017) 216:1018–26. doi: 10.1093/infdis/jix102
19. Ren J, Sang Y, Tan Y, Tao J, Ni J, Liu S, et al. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. (2016) 12:e1005458. doi: 10.1371/journal.ppat.1005458
20. Li J, Liu S, Su Y, Ren J, Sang Y, Ni J, et al. Acetylation of PhoP K88 is involved in regulating Salmonella virulence. Infect Immun. (2021) 89:e00588-20. doi: 10.1128/IAI.00588-20
21. Sang Y, Ren J, Ni J, Tao J, Lu J, Yao YF. Protein acetylation is involved in Salmonella enterica Serovar Typhimurium virulence. J Infect Dis. (2016) 213:1836–845. doi: 10.1093/infdis/jiw028
22. Venkat S, Gregory C, Sturges J, Gan Q, Fan C. Studying the lysine acetylation of malate dehydrogenase. J Mol Biol. (2017) 429:1396–405. doi: 10.1016/j.jmb.2017.03.027
23. Venkat S, Chen H, Stahman A, Hudson D, McGuire P, Gan Q, et al. Characterizing lysine acetylation of isocitrate dehydrogenase in Escherichia coli. J Mol Biol. (2018) 430:1901–11. doi: 10.1016/j.jmb.2018.04.031
24. Venkat S, Chen H, McGuire P, Stahman A, Gan Q, Fan C. Characterizing lysine acetylation of Escherichia coli type II citrate synthase. FEBS J. (2019) 286:2799–808. doi: 10.1111/febs.14845
25. Liu M, Guo L, Fu Y, Huo M, Qi Q, Zhao G. Bacterial protein acetylation and its role in cellular physiology and metabolic regulation. Biotechnol Adv. (2021) 53:107842. doi: 10.1016/j.biotechadv.2021.107842
26. Zeng F, Pang H, Chen Y, Zheng H, Li W, Ramanathan S, et al. First succinylome profiling of Vibrio alginolyticus reveals key role of lysine succinylation in cellular metabolism and virulence. Front Cell Infect Microbiol. (2021) 10:626574. doi: 10.3389/fcimb.2020.626574
27. Pang H, Li W, Zhang W, Zhou W, Hoare S, Monaghan R, et al. Acetylome profiling of Vibrio alginolyticus reveals its role in bacterial virulence. J Proteomics. (2020) 211:103543. doi: 10.1016/j.jprot.2019.103543
28. Wang J, Pang H, Yin L, Zeng F, Wang N, Hoare R, et al. A comprehensive analysis of the lysine acetylome in the aquatic animals pathogenic bacterium Vibrio mimicus. Front Microbiol. (2022) 13:816968. doi: 10.3389/fmicb.2022.816968
29. Pang R, Li Y, Liao K, Guo P, Li Y, Yang X, et al. Genome- and proteome-wide analysis of lysine acetylation in Vibrio vulnificus Vv180806 reveals its regulatory roles in virulence and antibiotic resistance. Front Microbiol. (2020) 11:591287. doi: 10.3389/fmicb.2020.591287
30. Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. (2013) 51:265–72. doi: 10.1016/j.molcel.2013.06.003
31. Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS ONE. (2014) 9:e94816. doi: 10.1371/journal.pone.0094816
32. Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, et al. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol. (2015) 98:847–63. doi: 10.1111/mmi.13161
33. Anastasiou D Yu Y Israelsen WJ . Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. (2012) 8:839–47. doi: 10.1038/nchembio.1060
34. Xiong Y, Lei QY, Zhao S, Guan KL. Regulation of glycolysis and gluconeogenesis by acetylation of PKM and PEPCK. Cold Spring Harb Symp Quant Biol. (2011) 76:285–9. doi: 10.1101/sqb.2011.76.010942
35. Ponce E, Flores N, Martinez A, Valle F, Bolívar F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J Bacteriol. (1995) 177:5719–22. doi: 10.1128/jb.177.19.5719-5722.1995
36. Meza E, Becker J, Bolivar F, Gosset G, Wittmann C. Consequences of phosphoenolpyruvate:sugar phosphotranferase system and pyruvate kinase isozymes inactivation in central carbon metabolism flux distribution in Escherichia coli. Microb Cell Fact. (2012) 11:127. doi: 10.1186/1475-2859-11-127
37. Cai SH, Wu ZH, Jian JC, Lu YS. Cloning and expression of the gene encoding an extracellular alkaline serine protease from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis in Lutjanus erythopterus (Bloch). J Fish Dis. (2007) 30:493–500. doi: 10.1111/j.1365-2761.2007.00835.x
38. Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. (2010) 22:4084–97. doi: 10.1105/tpc.110.079947
39. Wang ZQ, Wang L, Cui J. Proteomic analysis of Trichinella spiralis proteins in intestinal epithelial cells after culture with their larvae by shotgun LC-MS/MS approach. J Proteomics. (2012) 75:2375–83. doi: 10.1016/j.jprot.2012.02.005
40. Zhao Z, Liu J, Deng Y, Huang W, Ren C, Call DR, et al. The Vibrio alginolyticus T3SS effectors, Val1686 and Val1680, induce cell rounding, apoptosis and lysis of fish epithelial cells. Virulence. (2018) 9:318–30. doi: 10.1080/21505594.2017.1414134
41. Zhou S, Tu X, Pang H, Hoare R, Monaghan SJ, Luo J, et al. A T3SS regulator mutant of Vibrio alginolyticus affects antibiotic susceptibilities and provides significant protection to danio rerio as a live attenuated vaccine. Front Cell Infect Microbiol. (2020) 10:183. doi: 10.3389/fcimb.2020.00183
42. Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. (2004) 338:17–31. doi: 10.1016/j.jmb.2004.02.006
43. Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol. (2004) 337:731–41. doi: 10.1016/j.jmb.2004.01.060
44. Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Z, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. (2013) 12:3509–20. doi: 10.1074/mcp.M113.031567
45. Chenlong F, Yu D. Molecular cloning and functional verification of histone deacetylase gene cobB in Vibrio alginolyticus. Biotechnol Bull. (2021) 37:195–202. doi: 10.13560/j.cnki.biotech.bull.1985.2020-1361
46. Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. (2010) 77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x
47. Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. (2015) 33:125–31. doi: 10.1016/j.ceb.2015.02.003
48. Prüss BM, Wolfe AJ. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol (1994) 12:973–84. doi: 10.1111/j.1365-2958.1994.tb01085.x
49. Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. (2007) 189:5574–81. doi: 10.1128/JB.00564-07
50. Ouidir T, Cosette P, Jouenne T, Hardouin J. Proteomic profiling of lysine acetylation in Pseudomonas aeruginosa reveals the diversity of acetylated proteins. Proteomics. (2015) 15:2152–7. doi: 10.1002/pmic.201500056
51. Yang H, Sha W, Liu Z, Tang T, Liu H, Qin L, et al. Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg Microbes Infect. (2018) 7:34. doi: 10.1038/s41426-018-0032-2
52. Yoshida A, Yoshida M, Kuzuyama T, Nishiyama M, Kosono S. Protein acetylation on 2-isopropylmalate synthase from Thermus thermophilus HB27. Extremophiles. (2019) 23:377–88. doi: 10.1007/s00792-019-01090-y
53. Bhardwaj A, Das S. SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc Natl Acad Sci USA. (2016) 113:E538–47. doi: 10.1073/pnas.1520045113
54. Park SH, Ozden O, Liu G, Song HY, Zhu Y, Yan Y, et al. SIRT2-mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. (2016) 76:3802–12. doi: 10.1158/0008-5472.CAN-15-2498
55. Wang T, Geng SL, Guan YM, Xu WH. Deacetylation of metabolic enzymes by Sirt2 modulates pyruvate homeostasis to extend insect lifespan. Aging. (2018) 10:1053–72. doi: 10.18632/aging.101447
56. Zhang ZH, Zhang H, Wang YR, Liu XL, Huang H, Xu XH. SIRT 1 binding with PKM and NSE and modulate their acetylation and activities. Biochim Biophys Acta Proteins Proteom. (2019) 1867:794–801. doi: 10.1016/j.bbapap.2019.06.003
57. Zhang R, Shen M, Wu C, Chen Y, Lu J, Zhao L, et al. HDAC8-dependent deacetylation of PKM2 directs nuclear localization and glycolysis to promote proliferation in hepatocellular carcinoma. Cell Death Dis. (2020) 11:1036. doi: 10.1038/s41419-020-03212-3
58. Hao L, Park J, Jang HY, Bae EJ, Park BH. Inhibiting protein kinase activity of pyruvate kinase M2 by SIRT2 deacetylase attenuates psoriasis. J Invest Dermatol. (2021) 141:355–63. e356. doi: 10.1016/j.jid.2020.06.024
59. Post DMB, Schilling B, Reinders LM, D'Souza AK, Ketterer MR, Kiel SJ, et al. Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS ONE. (2017) 12:e0179621. doi: 10.1371/journal.pone.0179621
60. Wang MM, You D, Ye BC. Site-specific and kinetic characterization of enzymatic and nonenzymatic protein acetylation in bacteria. Sci Rep. (2017) 7:14790. doi: 10.1038/s41598-017-13897-w
61. Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol Lett. (2004) 235:25–33. doi: 10.1111/j.1574-6968.2004.tb09563.x
62. Gao J, Tian M, Bao Y, Li P, Liu J, Ding J, et al. Pyruvate kinase is necessary for Brucella abortus full virulence in BALB/c mouse. Vet Res. (2016) 47:87. doi: 10.1186/s13567-016-0372-7
63. Peng F, Widmann S, Wünsche A, Duan K, Donovan KA, Dobson RCJ, et al. Effects of beneficial mutations in pykF gene vary over time and across replicate populations in a long-term experiment with bacteria. Mol Biol Evol. (2018) 35:202–10. doi: 10.1093/molbev/msx279
64. Zuo Y, Zhao L, Xu X, Zhang J, Yan Q, Huang L. Mechanisms underlying the virulence regulation of new Vibrio alginolyticus ncRNA Vvrr1 with a comparative proteomic analysis. Emerg Microbes Infect. (2019) 8:1604–18. doi: 10.1080/22221751.2019.1687261
65. Mattevi A, Valentini G, Rizzi M, Speranza ML, Bolognesi M, Coda A. Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition. Structure. (1995) 3:729–41. doi: 10.1016/S0969-2126(01)00207-6
66. Donovan KA, Zhu S, Liuni P, Peng F, Kessans SA, Wilson DJ, et al. Conformational dynamics and allostery in pyruvate kinase. J Biol Chem. (2016) 291:9244–56. doi: 10.1074/jbc.M115.676270
67. Donovan KA, Atkinson SC, Kessans SA, Peng F, Cooper TF, Griffin MD, et al. Grappling with anisotropic data, pseudo-merohedral twinning and pseudo-translational noncrystallographic symmetry: a case study involving pyruvate kinase. Acta Crystallogr D Struct Biol. (2016) 72(Pt 4):512–19. doi: 10.1107/S205979831600142X
68. Schormann N, Hayden KL, Lee P, Banerjee S, Chattopadhyay D. An overview of structure, function, and regulation of pyruvate kinases. Protein Sci. (2019) 28:1771–84. doi: 10.1002/pro.3691
Keywords: Vibrio alginolyticus, lysine deacetylation, PykF, post-translational modification, glycolysis
Citation: Xu Z, Wang L, Wang X, Wan M, Tang M and Ding Y (2022) Characterizing the Effect of the Lysine Deacetylation Modification on Enzyme Activity of Pyruvate Kinase I and Pathogenicity of Vibrio alginolyticus. Front. Vet. Sci. 9:877067. doi: 10.3389/fvets.2022.877067
Received: 16 February 2022; Accepted: 28 April 2022;
Published: 06 June 2022.
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Yu Sang, Oregon Health and Science University, United StatesLiqun Lu, Shanghai Ocean University, China
Copyright © 2022 Xu, Wang, Wang, Wan, Tang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Ding, ZGluZ3l1ZGRkQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Zhou Xu
Zhou Xu Linjing Wang1,2,3†
Linjing Wang1,2,3†