
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 11 July 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.872205
Porcine reproductive and respiratory syndrome (PRRS) is a contagious disease caused by the porcine reproductive and respiratory syndrome virus (PRRSV). PRRS is also called “blue ear disease” because of the characteristic blue ear in infected sows and piglets. Its main clinical features are reproductive disorders of sows, breathing difficulties in piglets, and fattening in pigs, which cause considerable losses to the swine industry. NSP9, a non-structural protein of PRRSV, plays a vital role in PRRSV replication and virulence because of its RNA-dependent RNA polymerase (RdRp) structure. The NSP9 sequence is highly conserved and contains T cell epitopes, which are beneficial for the development of future vaccines. NSP9 acts as the protein interaction hub between virus and host during PRRSV infection, especially in RNA replication and transcription. Herein, we comprehensively review the application of NSP9 in terms of genetic evolution analysis, interaction with host proteins that affect virus replication, interaction with other viral proteins, pathogenicity, regulation of cellular immune response, antiviral drugs, vaccines, and detection methods. This review can therefore provide innovative ideas and strategies for PRRSV prevention and control.
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious and virulent infectious disease caused by the porcine reproductive and respiratory syndrome virus (PRRSV). PRRS is mainly characterized by reproductive disorders in sows and breathing difficulties in pigs of all ages. It leads to substantial losses in the global swine industry. PRRSV belongs to the order Nidovirale, family Arteriaviridae, and is a single-stranded positive-sense RNA virus with an envelope and a non-segmented genome (1). PRRSV escapes the host immunosuppression reaction after infection and inhibits co-infection with other pathogens. PRRSV is known to frequently mutate and recombine, thereby making it difficult to eliminate and control (2).
The non-structural protein 9 (NSP9), encoded by ORF1b of PRRSV, is an RNA-dependent RNA polymerase (RdRp). RdRp is essential for genome replication and subgenomic (sg mRNA) synthesis and plays a vital role in viral replication (3). PRRSV RNA synthesis includes genomic RNAs (gRNAs) and sgRNAs and requires a negative-strand RNA strand as a template (4). NSP9 alone or with NSP10 cannot activate the synthesis of virus-negative strand RNA (5). The combination of ORF1a and ORF1b is necessary for synthesizing viral negative-strand RNA (6). In PRRSV-infected MARC-145 cells, NSP9 can be identified as a 72–95 kDa band using NSP9 monoclonal antibodies and NSP8 polyclonal antibodies, respectively, which is higher than the actual molecular weight of NSP9. The migration rate of NSP8-9 expressed in vitro is similar to natural NSP9, and NSP9 can co-immunoprecipitate with the NSP8 antibody. NSP4 and NSP2 PLP2 cannot cleave NSP8-9 in vitro, which indicates that NSP8 is the N-terminal extension of NSP9 (7). The RdRp domain is located at the C-terminal of the replicase subunit of NSP9, which contains an upstream domain with an unknown function (8). PRRSV NSP9 is thought to contain NSP8 at the N-terminal and RdRp at the C-terminal (9). The N-terminal region also contains a newly discovered nucleotide transferase domain (NiRAN) which is related to the RdRp of the pancreatic virus (Figure 1).

Figure 1. Structure diagram of NSP9 of PRRSV. N-terminal is linked to NSP8 and contains a newly discovered nucleotide transferase domain (NiRAN) related to pancreatic virus RdRp, and C-terminal contains the RdRP domain.
The most common feature of PRRSV NSP9 is its RdRp structure, which is also present in other Arteriviruses (10). All positive-strand RNA viruses encode an RdRp, which acts as a catalytic subunit of virus replication and transcription complexes, and guides the synthesis of viral RNA along with other viral proteins and sometimes host proteins (11). Severe acute respiratory syndrome coronavirus (SARS-CoV) and equine arteritis virus (EAV) also possess an RdRp, which is a ribozyme for the replication and transcription of their multi-protein complexes (12). RdRp is located in an unusually large replicase subunit for positive-strand ribonucleic acid viruses belonging to nested viruses (13). Bioinformatic analysis of this non-structural protein (NSP) revealed a non-viral characteristic domain (genetic marker) located near the N-terminal of RdRp, and no apparent homolog was identified at other sites. This domain exhibits nucleotidase activity based on its conservative spectrum (13). The homologous polymerase subunit (NSP12) of the distantly related coronaviruses has potent primer-dependent RdRp activity (14). Cytoplasmic replication of a positive-strand RNA virus is related to the characteristic virus membrane structure from host organelles. Double-membrane vesicles are related to EAV RNA synthesis, which can be used to immunolabel EAV NSPs (15).
The NSP9 sequence is highly conserved. One main factor in PRRSV virulence is the interaction with the host or viral proteins to regulate viral replication. This interaction is involved in regulating the cellular immune response, and small interfering RNA (siRNA) targeting NSP9 can lead to the inhibition of virus replication. This review focuses on the evolutionary genetic analysis of NSP9, the influence of the interaction between NSP9 and host proteins or other viral proteins on virus replication, the relationship between NSP9 and pathogenicity, regulation of the cellular immune response by NSP9, the influence of compound drugs and modifications of NSP9 on virus replication, and NSP9 vaccine and detection methods to enhance our understanding of PRRSV replication, and promote the research and development of anti-PRRSV drugs and future vaccines.
Although the NSP9 nucleotide sequence is highly conserved, gene recombination and amino acid changes are observed. Through phylogenetic analysis, Darwich et al. demonstrated that the isolates could be divided into different groups according to the detected ORFs, and found that NSP9 had the highest similarity with the whole genome cluster (16). The NSP9 amino acid homology between the European isolate prototype Lelystad virus (LV) and the North American isolate SDPRRS (01–08) showed a 98.9% similarity, which was the highest among all NSPs (17). Zhao et al. collected 25 PRRSV strains from Guangdong province. Sequence analysis revealed that the homology of the NSP9 gene was as high as 98.1–100.0% in amino acids 8, 64, 112, 205, and 356, which was higher than that of traditional strains (Ch-1a, BJ-4, VR2332, MLV) (18). Li et al. reported genomic variation distributed in the 5′ untranslated region, NSP1b, NSP2, NSP3, NSP5, NSP7, NSP9, NSP10, GP5, and N regions of PRRSV GDQJ strains isolated in Guangdong Province in China, compared to that in the highly pathogenic PRRSV(HP-PRRSV) strains, which was also isolated in China (19). Many mutations were identified in both virulent and non-virulent strains. Two identical amino acid mutations located at positions 519 and 544 of NSP9 were observed between HP-PRRSV and classical PRRSV strains, respectively. The virulent strain has a serine (S) at position 519 and threonine (T) at position 544. In contrast, the attenuated strain had threonine (T) at position 519 and alanine (A) at 544 (20). Furthermore, the 586th amino acid of NSP9 was a conserved T residue in all HP-PRRSV strains. However, A was identified at this position in low-pathogenicity PRRSV (LP-PRRSV). T is present at position 592 in HB-1/3.9 and its wild-type HB-1(sh)/2002, whereas S is present at this position in all other strains (21). NSP9-containing virus replicase has a conserved characteristic sequence motif, a Ser-Asp-Asp (SDD) sequence, among the RdRps of positive-strand RNA viruses. In an attempt to detect whether the conserved SDD motif can tolerate other conserved motifs of RNA viruses and assess the influence of each residue on the catalytic activity of RdRp, Zhou et al. observed that RdRp could maintain its transcription activity only when the serine in the SDD motif was replaced by glycine (G). The other substitutions were lethal mutations leading to a loss in transcription activity (22). The 20 PRRSV NSP9 gene reference strains were retrieved from the NCBI nucleotide database as reference sequences for phylogenetic analysis. Detailed information and the GenBank number of PRRSV reference strains are listed in Table 1. To construct the phylogenetic tree, nucleotide sequences of the target gene were aligned using the ClustalX alignment tool using the MEGA software (ver. 7.0; Center for Evolutionary Medicine and Informatics, Tempe, AZ). Phylogenetic trees were constructed using the Neighbor-joining method with 1,000 bootstrap replicates in MEGA7.0 (Figure 2). The 20 PRRSV NSP9 gene reference strains exhibited relatively short genetic distance. The results showed that the NSP9 sequence was highly conserved, consistent with the previous research results.
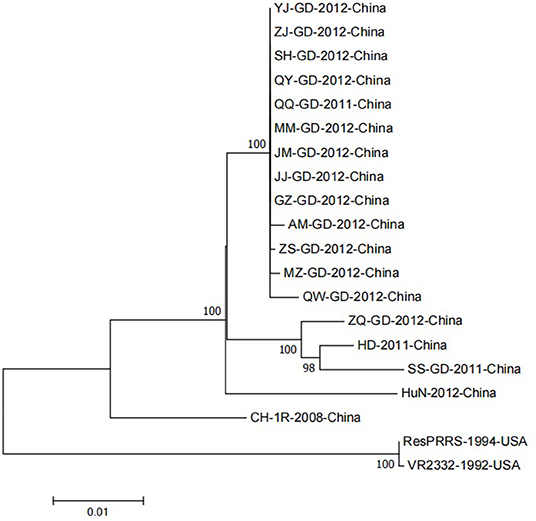
Figure 2. A neighbor-joining phylogenetic tree was constructed based on the NSP9 sequences of 20 PRRSV strains, using MEGA software (ver. 7.0; Center for Evolutionary Medicine and Informatics, Tempe, AZ) with 1000 bootstrap replicates.
Several gene recombination events have been discovered since the NADC30-like strain became the most frequent strain in China; NSP9 has a recombination hotspot sequence (23). Sun et al. reported regular surveillance and field epidemiological studies for PRRSV across China between 2007 and 2019 (24). Genome recombination of Chinese lineage 1 with two dominant clusters (Chinese HP-PRRSVs: lineage 8.7 and VR2332-like: lineage 5.1) was frequently detected (25). Furthermore, recombination hotspots were discovered near NSP9 and ORF2-4 regions of the PRRSV genome. Comparative analyses of all available genome sequences of North American (NA)-type PRRSV in China and the United States between 2014 and 2018 revealed a high frequency of interlineage recombination hot spots in NSP9 and the GP2 to GP3 regions (26). Wang et al. indicated that the HNhx strain, isolated from Henan in 2016, was classified into the NADC30-like PRRSV subgroup (27). Recombination analysis revealed that HNhx resulted from the recombination in NSP4 (nt 5261) to NSP9 (nt 7911) between the NADC30 and highly pathogenic PRRSV vaccine strains circulating in China (28). Chen et al. experimentally determined that HBap4-2018 is a new recombinant PRRSV strain, with HP-PRRSV and NADC30-like PRRSV as the main and secondary parent strains, respectively. Five recombination breakpoints were identified in NSP2, NSP3, NSP5, NSP9, and ORF6, respectively, showing a new recombination pattern in the genome (29). Wang et al. compared and analyzed the complete genome sequences of two PRRSV strains, GXYL1403 and GXNN1839 (30). GXYL1403 is a natural recombinant between sublineages 8.1 (CH-1a-like) and 8.3 (JXA1-like). The recombination site of GXYL1403 is located in NSP9–NSP12 (8961–11181 nt). GXNN1839 is a natural recombinant between the lineage 5 (VR-2332-like) and lineage 1 (NADC30-like) strains. The recombination events occurred in NSP9 (7872-8162 nt) and ORF2 (12587–13282 nt) in the genome of GXNN1839.
Some cellular proteins can interact with PRRSV NSP9, and their interactions can regulate viral replication. In MARC-145 cells and porcine alveolar macrophages (PAMs) infected with PRRSV, some cellular proteins, including interleukin-2 enhancer-binding factor 2 (ILF2), can be translocated from the nucleus to the cytoplasm and co-localize with NSP9 and NSP2 of PRRSV (31). In contrast, ILF2 is only located in the nucleus in non-infected cells. ILF2 knockdown is beneficial for the replication of PRRSV, whereas overexpression of ILF2 reduces the replication of PRRSV, indicating that ILF2 plays a negative regulatory role in the replication of PRRSV (32). Only the full-length annexin A2 (ANXA2) can interact with NSP9 after the MARC-145 cells are infected with PRRSV. RNA may contribute to the interaction between NSP9 and ANXA2. The interaction between NSP9 and ANXA2 was reduced by RNase A treatment. In addition, when ANXA2 was knocked out in MARC-145 cells, the growth of PRRSV was significantly blocked, indicating that the interaction between NSP9 and ANXA2 was beneficial to PRRSV replication (33). The zinc-finger antiviral protein (ZAP) is an effective antiviral protein in cells as ZAP inhibits PRRSV infection during the early stages of replication. ZAP interacts with RdRp, and the active sites are in the zinc finger structural domain of ZAP and the N-terminal amino acid 150–160 region of NSP9 (34). Silencing of DEAD-box RNA helicase 5 (DDX5) in MARC-145 cells significantly inhibited PRRSV replication. In contrast, overexpression of DDX5 in MARC-145 cells enhanced PRRSV replication, indicating that DDX5 may be a cell cofactor, actively and positively regulating PRRSV replication. DDX5 interacts with NSP9, and the active sites are the DEXDc and HELICc domains in DDX5 and the RdRp domain in NSP9 (35). PRRSV infection promoted the expression of nucleotide-binding oligomerization domain-like receptor (NLRX1), whereas downregulation of NLRX1 expression enhanced PRRSV replication in PAMs. NLRX1 inhibits PRRSV replication by inhibiting RNA synthesis, which is realized by the interaction between the leucine-rich repeat (LRR) domain of NLRX1 and the RdRp domain of PRRSV NSP9 (36). PRRSV NSP9 protein interacts with cell retinoblastoma protein (pRb). NSP9 and pRb are co-localized in the cytoplasm of MARC-145 cells and PAMs infected with PRRSV. Overexpression of the pRb gene inhibited PRRSV replication in MARC-145 cells, whereas silencing the pRb gene promoted PRRSV replication in MARC-145 cells. After PRRSV infects MARC-145 cells, NSP9 promotes pRb degradation through the proteasome pathway, demonstrating that the interaction between NSP9 and pRb is beneficial for the replication of PRRSV (37). The NSP1β, NSP4, NSP9, NSP10, and nucleocapsid (N) proteins of PRRSV can interact with the SUMOE2 coupling enzyme Ubc9. Overexpression of Ubc9 inhibited viral genome replication in the early stages of PRRSV infection, whereas siRNA-mediated knockdown of Ubc9 promoted viral replication (38). Understanding the molecular mechanism of the interaction between PRRSV NSP9 and host proteins (Figure 3) can aid in identifying novel antiviral targets and exploring the viral replication mechanism.
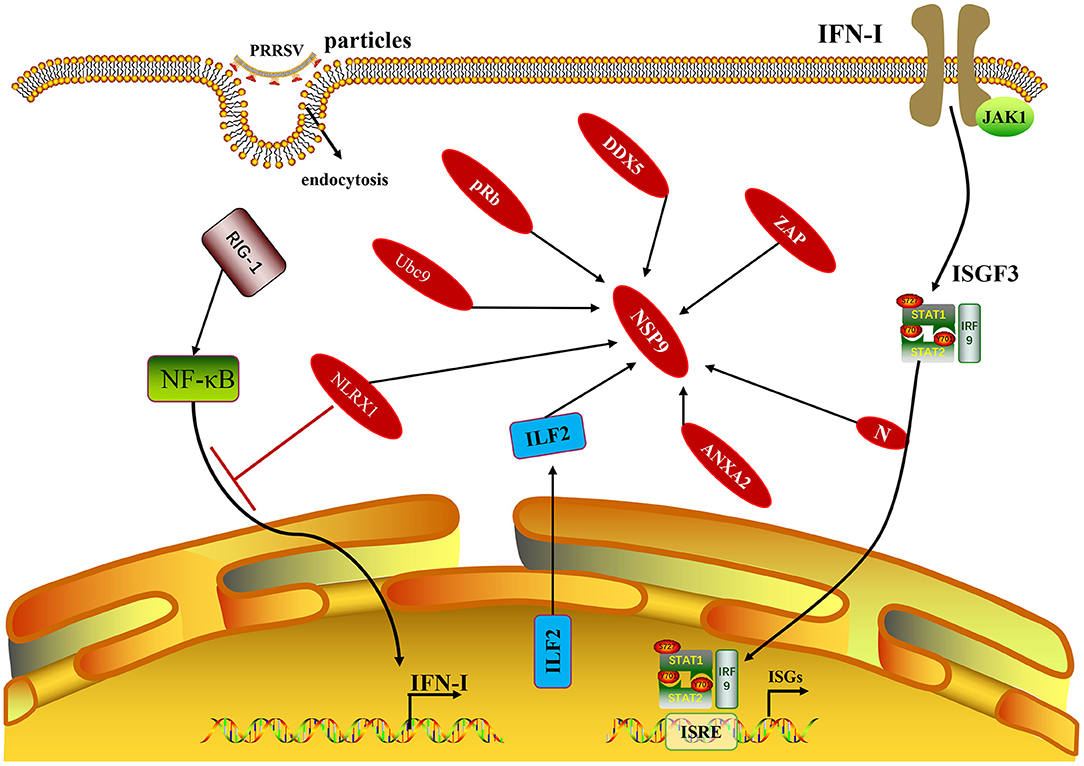
Figure 3. The NSP9 of PRRSV and non-structural proteins with host proteins mentioned in this paragraph. In addition to the host protein, it is also shown that the N protein related to the production of IFN-I also interacts with NSP9.
Nan et al. established a comprehensive schematic diagram of non-structural protein interaction by drawing the interaction network between PRRSV NSPs (39). These interactions are mainly concentrated on NSPs encoded by ORF1b, and the interactions are centered on NSP9, NSP10, and NSP12. These three proteins are mainly interconnected by the transmembrane proteins NSP2, NSP3, and NSP5. NSP9 and NSP12 were identified as two new interaction hubs. Most PRRSV NSPs are related to the viral replication and transcription complex (RTC) in the infection process (40). The membrane-associated NSPs collected by nucleosomes NSP9 and NSP10 in the assembly of the PRRSV RTC may involve the regulatory mechanisms of other viral proteins in the infection process (9). Most of the PRRSV NPSs (NSP1α, NSP1β, NSP3, NSP7α, NSP7β, NSP8, NSP11, and NSP12) can directly bind to NSP9, and the interaction between NSP9 and cytokines may play a role in RNA synthesis in infected cells (41). PRRSV NSP7α interacts with RdRp, and its active site is in the alpha-helix 3 of NSP7α. The mutation of conserved amino acids L69 and F72 causes NSP7α to lose its ability to bind to NSP9, which indicates that these two conserved amino acids are necessary for the interaction between NSP7α and NSP9 (42). The PRRSV N protein binds to NSP9 through protein–protein interactions, and the interaction sites are in two predicted helices formed by 48 residues at the C-terminus of the N protein. Amino acid residues E646, E608, and E611 on NSP9 and amino acid residue E611 on the N protein are predicted to be the critical residues participating in the interaction between N and NSP9. The N protein of PRRSV interacts with the RdRp of NSP9, recruiting the cellular RNA helicase to unravel the double-stranded RNA structure of gRNA, promoting the generation of longer viral sgmRNAs and gRNAs, and participating in the replication and transcription of viral RNA (3).
The HP-PRRSV strain has stronger replication ability and higher pathogenicity than the classical PRRSV strain. However, the molecular mechanism underlying these characteristics is unclear (43). Different HP-PRRSV isolates with high and low pathogenicity were cloned using reverse-genetics technology, and the gene fragments were exchanged between the two strains. The results revealed that NSP9 and NSP10 were the main virulence factors of HP-PRRSV in vivo. NSP9 and NSP10, encoded by HP-PRRSV ORF1b, not only affected the replication efficiency of the virus in vitro and in vivo but were also closely related to the increase in morbidity and the lethal virulence of PRRSV in piglets. This finding helped explain the pathogenesis of HP-PRRSV (5). Two identical amino acid mutations, located at positions 519 and 544 of NSP9, were identified between HP-PRRSV and classical PRRSV strains, respectively. The amino acids at positions 519 and 544 in NSP9 are involved in the replication efficiency of HP-PRRSV and contribute to the enhancement of pathogenicity (20). T586A and T592S of HP-PRRSV NSP9 reduce the replication efficiency of HP-PRRSV and its virulence to piglets, indicating that residues 586 and 592 of NSP9 are the critical sites for determining the lethality of Chinese HP-PRRSV virus (21). These findings significantly contribute to understanding the molecular mechanism of PRRSV replication and pathogenicity and provide a reference to further study PRRSV pathogenicity.
NSP9, along with NSP1, NSP2, NSP5, NSP7, NSP10, and NSP11, are involved in the induction of interferon (IFN)-γ in cells and may be involved in cell-mediated immune responses (44). Cao et al. (45) identified many immunostimulatory cytotoxic T lymphocyte (CTL) epitopes by flow cytometry analysis, including two peptides from GP3 and two peptides from NSP9, which significantly improved the production of the degranulation markers CD107a and IFN-γ in cytotoxic CD4+ T cells, CD8+ T cells, and γδ T cells. NSP9-TMP9 was identified as an immunogenic epitope, which can stimulate the proliferation of specific CD8+ T cells and CTLs, and the expression of IFN-γ in SPF Landrace pigs carrying the SLA-1*1502 allele (46). Autophagy is an important mechanism to maintain cell homeostasis, associated with stress responses, such as immune response and inflammation. It is also an innate defense mechanism against infectious pathogens (47). PRRSV infection can induce autophagy, which can promote PRRSV replication. The PRRSV NSP3, NSP5, and NSP9 can activate intracellular autophagy, and NSP3 and NSP5 induce the formation of autophagic bodies, primarily the cytoplasmic domain of NSP3 (42). However, the specific role of NSP9 in autophagy needs further experimental verification.
The A283 and H421 residues of PRRSV NSP9 play a key role in PRRSV replication by affecting the fidelity of its RdRp and are closely related to the sensitivity of PRRSV to ribavirin (48). The actual usage frequency of each codon pair is greater than the statistical prediction, and such codon pair bias (CPB) affects gene translation. Gao et al. “recombined” the existing codons of HP-PRRSV genes GP5, M, NSP2, and NSP9, which made the CPB of these genes more negative (49) the results showed that the optimization of RdRp significantly reduced PRRSV replication in PAMs. PRRSV infection depends on the existence of lipid rafts in the cell membrane and the virus envelope. Cell lipid rafts play a vital role in the entry, replication, and release of PRRSV. The structure of virus lipid rafts is essential to maintain the integrity of the virus particle core to ensure the infectivity of PRRSV. NSP9 is distributed in the raft, indicating that the raft may be a platform for PRRSV replication; thus, small molecules disturbing the membrane lipid raft can also provide a means for developing antiviral drugs (50). Xie et al. constructed an siRNA expression vector targeting the NSP9 region of PRRSV. This study showed that NSP9, an essential polymerase protein in PRRSV, can be used as an effective antiviral targeting gene (51). Furthermore, siRNA expression vectors, pSi-294 and pSi-1488, can effectively inhibit the expression of the NSP9 protein of PRRSV and consequently inhibit PRRSV replication. Therefore, siRNAs can be valuable in developing new antiviral strategies and offer a candidate drug for basic research on PRRSV. DNA-based short antisense oligonucleotides (AS-ON) can effectively target PRRSV NSP9, thus inhibiting PRRSV replication in MARC-145 cells and PAMs (52). Two types of AS-ONs, modified by locked nucleic acids (LNAs), are more effective in reducing the cytopathic effect (CPE) induced by PRRSV and maintaining cell viability. In a cell culture model, LNA modification prolonged the lifespan of AS-ON. Incorporating LNA into AS-ON technology has a higher therapeutic prospect for PRRS control (53). The water extract from the fruiting body of Cryptococcus obliquus significantly inhibited PRRSV infection by inhibiting PRRSV entry, RNA expression, protein synthesis, cell-to-cell transmission, and PRRSV particle release. However, it did not block the combination of PRRSV and cells. The water extract from the fruiting body of Cryptosporus could directly inhibit the activity of PRRSV RdRp, thus interfering with the synthesis of PRRSV RNA and proteins. The extract effectively inhibited HP-PRRSV infection in vivo, reduced the viral load in serum, and improved the survival rate of pigs infected with HP-PRRSV, indicating that Cryptosporus water extract has potential as an anti-PRRSV agent (54). Camel single-domain antibodies (nanoparticles) represent a promising antiviral method because of their small size, high specificity, and good solubility. The PRRSV NSP9 is crucial for virus replication, and its sequence is relatively conserved, making it a logical antiviral target of PRRSV. NSP9-specific nanoparticles (Nb6) were isolated from a phage display library (VHH) containing only the variable region of the chain antibody of Camellidae heavy. Nb6 maintained its antigen-binding ability when expressed in the cytoplasm. Intracellular Nb6 effectively inhibited PRRSV replication by inhibiting PRRSV genome replication and transcription. Nb6 could also protect MARC-145 cells from a virus-induced CPE and completely block PRRSV replication at a multiplicity of infection of 0.01 or lower (55). Enzyme-linked immunosorbent assay (ELISA) showed that residues Ile588, Asp590, Leu643, Tyr62, Trp105, and Pro107 of Nb6 were the critical sites in the interaction between NSP9 and NB6. Structural analysis of the NSP9–Nb6 complex revealed new amino acid interactions involved in viral RNA replication, which will help guide the development of structure-based anti-PRRSV drugs (4). The cells expressed in prokaryotic expression systems could penetrate MARC-145 cells and PAMs in a time-and dose-dependent manner and inhibited the replication of PRRSV genotype 1 and 2 strains. In addition, Nb6 binds to the C-terminal region of NSP9, which contains two discontinuous regions, NSP9454-551 and NSP9599-646, and overlaps with the predicted catalytic domain of RdRp. These data indicate that TAT-Nb6 has high development potential as an anti-PRRSV drug (41).
NSP9 encodes RdRp in PRRSV, and RdRp only appears in the virus replication process and does not exist in host cells. Therefore, antigens targeting the RdRp fusion protein can distinguish wild strains from inactivated vaccines (18). Immunostimulatory type I CTL epitopes of PRRSV are essential for vaccine development. Many immunostimulatory CTL epitopes have been identified, including two peptides from GP3 and NSP9 and two that inhibit IFN-γ production. These CTL epitopes are expected to contribute to future vaccine development against PRRSV (45, 56). The epitope peptide NSP9-TMP9, based on NSP9, has been identified as a CTL epitope of PRRSV with strong immunogenicity. NSP9-TMP9 can enhance the immune response and produce a specific CTL response. This indicates that the peptide vaccine can confer effective immune protection, demonstrating its immense potential in the development of an effective epitope vaccine (46). Paride et al. identified two highly conserved heptapeptide T cell epitopes in NSP9, which provided an important scientific reference for developing immune agents that can provide extensive cross-protection against various PRRSV strains (57).
Spear et al. established a copy-specific reverse transcription-quantitative polymerase chain reaction (RT-qPCR) detection method based on PRRSV-2 isolates. This technology is aimed at the RNA-dependent RNA polymerase gene (NSP9), which is unique to gRNA and does not exist in subgenomic and heteronuclear RNA (58). This method has proven to be linear in nine orders of magnitude (1010-102 copies) and can be easily applied to detecting multiple PRRSV strains. This method will significantly improve the ability to evaluate and compare PRRSV replication in various tissues and among different strains, including highly pathogenic strains that are of great concern to the global pork industry. Kang et al. established a closed ELISA (b-ELISA) to detect PRRSV NSP9 with the monoclonal antibody 2D6. Since b-ELISA only detects antibodies produced by active PRRSV replication, such as natural infection or live vaccine inoculation, this method can distinguish between antibodies against live and inactivated vaccines in pigs (59).
NSP9 of PRRSV is the core component of the RTC. As the critical enzyme in viral RNA synthesis, its RdRp function is crucial for PRRSV replication. It is the key factor regulating nucleotide selectivity and fidelity in the viral replication process. NSP9 participates in various immunosuppressive reactions. As a newly discovered protein interaction hub, NSP9 interacts with the host cell and self-proteins to regulate virus replication, which is closely related to the improvement of replication efficiency and the formation of pathogenicity of HP-PRRSV. NSP9 can be used as an effective antiviral targeting gene to develop new antiviral strategies and lay a foundation for developing new vaccines and antiviral drugs. The NSP9 gene sequence is highly conserved, and amino acid similarity among different PRRSV strains is high. Based on these characteristics, new and efficient detection reagents, as well as methods for PRRSV, can be developed in the future.
However, PRRSV is constantly undergoing mutation and recombination, which poses significant challenges to vaccine research and development. Currently, the effect of attenuated and inactivated vaccines against PRRSV is not optimal. Therefore, developing new therapeutic strategies with effective antiviral activity is urgently required. PRRSV infections are also characterized by persistent infection, immunosuppression, and complex immune mechanisms. Co-infection with other pathogens increases the mortality rate of pigs. The infection also slows down the growth of pigs, increases feeding costs, reduces production performance, and brings heavy economic losses to the pig industry. Research on the molecular biology of PRRSV is currently progressing rapidly. This research focuses on the screening, transformation, and utilization of immunogenic and pathogenic genes. Understanding the structure of each PRRSV genome and the process of replication in cells is the basis and key to its prevention and control. PRRSV NSP9 plays a vital role in viral replication and virulence. Studying the molecular pathogenic mechanism of PRRSV NSP9 will provide a scientific basis for preventing and controlling this disease. The comprehensive overview of PRRSV NSP9 provided in this review will help further explore the relationship between the virus and its host and offers a reference for the prevention and control of PRRS and the development of new vaccines and antiviral drugs.
HS and HZ collect data and write the original draft. YC and LH complement and improve the manuscript. MZ and NW made the final revision of the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (31902279), Guang Dong Basic and Applied Basic Research Foundation (Nos. 2019a15110056 and 2021A1515110322), and Guangdong Science and Technology Program Project (2017A020208079 and 2021A1515012388).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.cn) for English language editing. We apologize to all colleagues whose contributions were not discussed and cited owing to space constraints.
1. Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, et al. Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. (2016) 4:129–54. doi: 10.1146/annurev-animal-022114-111025
2. Yin B, Qi S, Sha W, Qin H, Liu L, Yun J, et al. Molecular characterization of the Nsp2 and ORF5 (ORF5a) genes of PRRSV strains in nine Provinces of China during 2016–2018. Front Vet Sci. (2021) 8:605832. doi: 10.3389/fvets.2021.605832
3. Liu L, Tian J, Nan H, Tian M, Li Y, Xu X, et al. Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 To regulate viral RNA synthesis. J Virol. (2016) 90:5384–98. doi: 10.1128/JVI.03216-15
4. Wang Y, Li R, Qiao S, Wang J, Liu H, Li Z, et al. Structural characterization of non-structural protein 9 complexed with specific nanobody pinpoints two important residues involved in porcine reproductive and respiratory syndrome virus replication. Front Microbiol. (2020) 11:581856. doi: 10.3389/fmicb.2020.581856
5. Li Y, Zhou L, Zhang J, Ge X, Zhou R, Zheng H, et al. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. (2014) 10:e1004216. doi: 10.1371/journal.ppat.1004216
6. Tang YD, Fang QQ, Liu JT, Wang TY, Wang Y, Tao Y, et al. Open reading frames 1a and 1b of the porcine reproductive and respiratory syndrome virus (PRRSV) collaboratively initiate viral minus-strand RNA synthesis. Biochem Biophys Res Commun. (2016) 477:927–31. doi: 10.1016/j.bbrc.2016.06.161
7. Liu Y, Hu Y, Chai Y, Liu L, Song J, Zhou S, et al. Identification of nonstructural protein 8 as the N-terminus of the RNA-dependent RNA polymerase of porcine reproductive and respiratory syndrome virus. Virol Sin. (2018) 33:429–39. doi: 10.1007/s12250-018-0054-x
8. Fang Y, Snijder EJ. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. (2010) 154:61–76. doi: 10.1016/j.virusres.2010.07.030
9. Song J, Liu Y, Gao P, Hu Y, Chai Y, Zhou S, et al. Mapping the nonstructural protein interaction network of porcine reproductive and respiratory syndrome virus. J Virol. (2018) 92:24. doi: 10.1128/JVI.01112-18
10. Posthuma CC, Te Velthuis AJW, Snijder EJ. Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes. Virus Res. (2017) 234:58–73. doi: 10.1016/j.virusres.2017.01.023
11. Beerens N, Selisko B, Ricagno S, Imbert I, van der Zanden L, Snijder EJ, et al. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J Virol. (2007) 81:8384–95. doi: 10.1128/JVI.00564-07
12. te Velthuis AJ, van den Worm SH, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. (2010) 6:e1001176. doi: 10.1371/journal.ppat.1001176
13. Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GM, Ruben M, Overkleeft HS, et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. (2015) 43:8416–34. doi: 10.1093/nar/gkv838
14. Lehmann KC, Gorbalenya AE, Snijder EJ, Posthuma CC. Arterivirus RNA-dependent RNA polymerase: Vital enzymatic activity remains elusive. Virology. (2016) 487:68–74. doi: 10.1016/j.virol.2015.10.002
15. van Hemert MJ, de Wilde AH, Gorbalenya AE, Snijder EJ. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J Biol Chem. (2008) 283:16525–36. doi: 10.1074/jbc.M708136200
16. Darwich L, Gimeno M, Sibila M, Diaz I, de la Torre E, Dotti S, et al. Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet Microbiol. (2011) 150:49–62. doi: 10.1016/j.vetmic.2011.01.008
17. Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, et al. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. (2004) 100:229–35. doi: 10.1016/j.virusres.2003.12.026
18. Zhao M, Ning Z, Wang H, Huang Z, Zhang M, Zhang G. Sequence analysis of NSP9 gene of 25 PRRSV strains from Guangdong province, subtropical southern China. Virus Genes. (2013) 46:88–96. doi: 10.1007/s11262-012-0842-6
19. Li Y, Xue C, Wang L, Chen X, Chen F, Cao Y. Genomic analysis of two Chinese strains of porcine reproductive and respiratory syndrome viruses with different virulence. Virus Genes. (2010) 40:374–81. doi: 10.1007/s11262-010-0453-z
20. Zhao K, Gao JC, Xiong JY, Guo JC, Yang YB, Jiang CG, et al. Two residues in NSP9 contribute to the enhanced replication and pathogenicity of highly pathogenic porcine reproductive and respiratory syndrome virus. J Virol. (2018) 92:17. doi: 10.1128/JVI.02209-17
21. Xu L, Zhou L, Sun W, Zhang P, Ge X, Guo X, et al. Nonstructural protein 9 residues 586 and 592 are critical sites in determining the replication efficiency and fatal virulence of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Virology. (2018) 517:135–47. doi: 10.1016/j.virol.2018.01.018
22. Zhou Y, Zheng H, Gao F, Tian D, Yuan S. Mutational analysis of the SDD sequence motif of a PRRSV RNA-dependent RNA polymerase. Sci China Life Sci. (2011) 54:870–9. doi: 10.1007/s11427-011-4216-4
23. Liu J, Wei C, Lin Z, Fan J, Xia W, Dai A, et al. Recombination in lineage 1, 3, 5 and 8 of porcine reproductive and respiratory syndrome viruses in China. Infect Genet Evol. (2019) 68:119–26. doi: 10.1016/j.meegid.2018.12.006
24. Sun YK, Chen YJ, Cai Y, Li Q, Xie JX, Liang G, et al. Insights into the evolutionary history and epidemiological characteristics of the emerging lineage 1 porcine reproductive and respiratory syndrome viruses in China. Transbound Emerg Dis. (2020) 67:2630–41. doi: 10.1111/tbed.13613
25. Liu J, Xu Y, Lin Z, Fan J, Dai A, Deng X, et al. Epidemiology investigation of PRRSV discharged by faecal and genetic variation of ORF5. Transbound Emerg Dis. (2021) 68:2334–44. doi: 10.1111/tbed.13894
26. Yu F, Yan Y, Shi M, Liu HZ, Zhang HL, Yang YB, et al. Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014-2018. J Virol. (2020) 94:19. doi: 10.1128/JVI.01813-19
27. Wang LJ, Guo Z, Qiao S, Chen XX, Zhang G. Complete genome sequence of a mosaic NADC30-like porcine reproductive and respiratory syndrome virus in China. Genome Announc. (2016) 4:e01428–16. doi: 10.1128/genomeA.01428-16
28. Wang LJ, Wan B, Guo Z, Qiao S, Li R, Xie S, et al. Genomic analysis of a recombinant NADC30-like porcine reproductive and respiratory syndrome virus in china. Virus Genes. (2018) 54:86–97. doi: 10.1007/s11262-017-1516-1
29. Chen P, Tan X, Lao M, Wu X, Zhao X, Zhou S, et al. The novel PRRSV strain HBap4-2018 with a unique recombinant pattern is highly pathogenic to piglets. Virol Sin. (2021) 36:1611–25. doi: 10.1007/s12250-021-00453-0
30. Wang J, Lin S, Quan D, Wang H, Huang J, Wang Y, et al. Full genomic analysis of new variants of porcine reproductive and respiratory syndrome virus revealed multiple recombination events between different lineages and sublineages. Front Vet Sci. (2020) 7:603. doi: 10.3389/fvets.2020.00603
31. Zhang H, Sha H, Qin L, Wang N, Kong W, Huang L, et al. Research progress in porcine reproductive and respiratory syndrome virus–host protein interactions. Animals. (2022) 12:1381. doi: 10.3390/ani12111381
32. Wen X, Bian T, Zhang Z, Zhou L, Ge X, Han J, et al. Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication. Virol J. (2017) 14:125. doi: 10.1186/s12985-017-0794-5
33. Li J, Guo D, Huang L, Yin M, Liu Q, Wang Y, et al. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res. (2014) 189:106–13. doi: 10.1016/j.virusres.2014.05.015
34. Zhao Y, Song Z, Bai J, Liu X, Nauwynck H, Jiang P. ZAP, a CCCH-type zinc finger protein, inhibits porcine reproductive and respiratory syndrome virus replication and interacts with viral Nsp9. J Virol. (2019) 93:e00001–19. doi: 10.1128/JVI.00001-19
35. Zhao S, Ge X, Wang X, Liu A, Guo X, Zhou L, et al. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. (2015) 195:217–24. doi: 10.1016/j.virusres.2014.10.021
36. Jing H, Song T, Cao S, Sun Y, Wang J, Dong W, et al. Nucleotide-binding oligomerization domain-like receptor X1 restricts porcine reproductive and respiratory syndrome virus-2 replication by interacting with viral Nsp9. Virus Res. (2019) 268:18–26. doi: 10.1016/j.virusres.2019.05.011
37. Dong J, Zhang N, Ge X, Zhou L, Guo X, Yang H. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology. (2014) 464-465:432–40. doi: 10.1016/j.virol.2014.07.036
38. Wang C, Zeng N, Liu S, Miao Q, Zhou L, Ge X, et al. Interaction of porcine reproductive and respiratory syndrome virus proteins with SUMO-conjugating enzyme reveals the SUMOylation of nucleocapsid protein. PLoS ONE. (2017) 12:e0189191. doi: 10.1371/journal.pone.0189191
39. Nan H, Lan J, Tian M, Dong S, Tian J, Liu L, et al. The network of interactions among porcine reproductive and respiratory syndrome virus non-structural proteins. Front Microbiol. (2018) 9:970. doi: 10.3389/fmicb.2018.00970
40. Hu Y, Ke P, Gao P, Zhang Y, Zhou L, Ge X, et al. Identification of an intramolecular switch that controls the interaction of helicase nsp10 with membrane-associated nsp12 of porcine reproductive and respiratory syndrome virus. J Virol. (2021) 95:e0051821. doi: 10.1128/JVI.00518-21
41. Wang L, Zhang L, Huang B, Li K, Hou G, Zhao Q, et al. A nanobody targeting viral nonstructural protein 9 inhibits porcine reproductive and respiratory syndrome virus replication. J Virol. (2019) 93:e01888–18. doi: 10.1128/JVI.01888-18
42. Chen J, Xu X, Tao H, Li Y, Nan H, Wang Y, et al. Structural analysis of porcine reproductive and respiratory syndrome virus non-structural protein 7alpha (NSP7alpha) and identification of its interaction with NSP9. Front Microbiol. (2017) 8:853. doi: 10.3389/fmicb.2017.00853
43. Dong J, Wang G, Liu Y, Shi W, Wu J, Wen H, et al. Quantitative estimation of the replication kinetics of genotype 2 PRRSV strains with different levels of virulence in vitro. J Virol Methods. (2016) 234:87–9. doi: 10.1016/j.jviromet.2016.04.006
44. Rascon-Castelo E, Burgara-Estrella A, Mateu E, Hernandez J. Immunological features of the non-structural proteins of porcine reproductive and respiratory syndrome virus. Viruses. (2015) 7:873–86. doi: 10.3390/v7030873
45. Cao QM, Tian D, Heffron CL, Subramaniam S, Opriessnig T, Foss DL, et al. Cytotoxic T lymphocyte epitopes identified from a contemporary strain of porcine reproductive and respiratory syndrome virus enhance CD4+CD8+ T, CD8+ T, and gammadelta T cell responses. Virology. (2019) 538:35–44. doi: 10.1016/j.virol.2019.09.006
46. Pan X, Zhang N, Wei X, Jiang Y, Chen R, Li Q, et al. Illumination of PRRSV cytotoxic t lymphocyte epitopes by the three-dimensional structure and peptidome of swine lymphocyte antigen class I (SLA-I). Front Immunol. (2019) 10:2995. doi: 10.3389/fimmu.2019.02995
47. Songane M, Kleinnijenhuis J, Netea MG, van Crevel R. The role of autophagy in host defence against Mycobacterium tuberculosis infection. Tuberculosis (Edinb). (2012) 92:388–96. doi: 10.1016/j.tube.2012.05.004
48. Tian D, Meng XJ. Amino acid residues Ala283 and His421 in the RNA-dependent RNA polymerase of porcine reproductive and respiratory syndrome virus play important roles in viral ribavirin sensitivity and quasispecies diversity. J Gen Virol. (2016) 97:53–9. doi: 10.1099/jgv.0.000316
49. Gao L, Wang L, Huang C, Yang L, Guo XK, Yu Z, et al. HP-PRRSV is attenuated by de-optimization of codon pair bias in its RNA-dependent RNA polymerase nsp9 gene. Virology. (2015) 485:135–44. doi: 10.1016/j.virol.2015.07.012
50. Yang Q, Zhang Q, Tang J, Feng WH. Lipid rafts both in cellular membrane and viral envelope are critical for PRRSV efficient infection. Virology. (2015) 484:170–80. doi: 10.1016/j.virol.2015.06.005
51. Xie J, Zhou H, Cui J, Chen Y, Zhang M, Deng S, et al. Inhibition of porcine reproductive and respiratory syndrome virus by specific siRNA targeting Nsp9 gene. Infect Genet Evol. (2014) 28:64–70. doi: 10.1016/j.meegid.2014.08.008
52. Zheng L, Li X, Zhu L, Li W, Bi J, Yang G, et al. Inhibition of porcine reproductive and respiratory syndrome virus replication in vitro using DNA-based short antisense oligonucleotides. BMC Vet Res. (2015) 11:199. doi: 10.1186/s12917-015-0518-2
53. Zhu L, Bi J, Zheng L, Zhao Q, Shu X, Guo G, et al. In vitro inhibition of porcine reproductive and respiratory syndrome virus replication by short antisense oligonucleotides with locked nucleic acid modification. BMC Vet Res. (2018) 14:109. doi: 10.1186/s12917-018-1432-1
54. Gao L, Zhang W, Sun Y, Yang Q, Ren J, Liu J, et al. Cryptoporus volvatus extract inhibits porcine reproductive and respiratory syndrome virus (PRRSV) in vitro and in vivo. PLoS ONE. (2013) 8:e63767. doi: 10.1371/journal.pone.0063767
55. Liu H, Wang Y, Duan H, Zhang A, Liang C, Gao J, et al. An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication. Vet Microbiol. (2015) 181:252–60. doi: 10.1016/j.vetmic.2015.10.021
56. Pan X, Qi J, Zhang N, Li Q, Yin C, Chen R, et al. Complex assembly, crystallization and preliminary X-ray crystallographic studies of the swine major histocompatibility complex molecule SLA-1*1502. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2011) 67:568–71. doi: 10.1107/S174430911100741X
57. Parida R, Choi IS, Peterson DA, Pattnaik AK, Laegreid W, Zuckermann FA, et al. Location of T-cell epitopes in nonstructural proteins 9 and 10 of type-II porcine reproductive and respiratory syndrome virus. Virus Res. (2012) 169:13–21. doi: 10.1016/j.virusres.2012.06.024
58. Spear A, Faaberg KS. Development of a genome copy specific RT-qPCR assay for divergent strains of type 2 porcine reproductive and respiratory syndrome virus. J Virol Methods. (2015) 218:1–6. doi: 10.1016/j.jviromet.2015.02.007
59. Cong Y, Huang Z, Sun Y, Ran W, Zhu L, Yang G, et al. Development and application of a blocking enzyme-linked immunosorbent assay (ELISA) to differentiate antibodies against live and inactivated porcine reproductive and respiratory syndrome virus. Virology. (2013) 444:310–6. doi: 10.1016/j.virol.2013.06.027
Keywords: PRRSV, NSP9, RdRp, protein interaction, pathogenicity
Citation: Sha H, Zhang H, Chen Y, Huang L, Zhao M and Wang N (2022) Research Progress on the NSP9 Protein of Porcine Reproductive and Respiratory Syndrome Virus. Front. Vet. Sci. 9:872205. doi: 10.3389/fvets.2022.872205
Received: 09 February 2022; Accepted: 20 June 2022;
Published: 11 July 2022.
Edited by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Xiaodu Wang, Zhejiang A&F University, ChinaCopyright © 2022 Sha, Zhang, Chen, Huang, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangzong Huang, bGlhbmd6b25naHVhbmdAZm9zdS5lZHUuY24=; Mengmeng Zhao, bWVuZ21lbmd6aGFvMjAyMUBmb3N1LmVkdS5jbg==; Nina Wang, d2FuZ19uaW5hQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.