
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 27 May 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.850466
This article is part of the Research TopicAntimicrobial Use, Antimicrobial Resistance, and the Microbiome in Animals, volume IIView all 13 articles
The aim of this study was to determine the mobile genetic elements involved in the horizontal transfer of erm(T) in Enterococcus faecalis, and its transmission ability in heterologous hosts. A total of 159 erythromycin-resistant enterococci isolates were screened for the presence of macrolide resistance genes by PCR. Whole genome sequencing for erm(T)-carrying E. faecalis E165 was performed. The transmission ability in heterologous hosts was explored by conjugation, transformation, and fitness cost. The erm(T) gene was detected only in an E. faecalis isolate E165 (1/159), which was located on a 4,244-bp small plasmid, designed pE165. Using E. faecalis OG1RF as the recipient strain, pE165 is transferable. Natural transformation experiments using Streptococcus suis P1/7 and Streptococcus mutans UA159 as the recipients indicated it is transmissible, which was also observed by electrotransformation using Staphylococcus aureus RN4220 as a recipient. The erm(T)-carrying pE165 can replicate in the heterologous host including E. faecalis OG1RF, S. suis P1/7, S. mutans UA159, and S. aureus RN4220 and conferred resistance to erythromycin and clindamycin to all hosts. Although there is no disadvantage of pE165 in the recipient strains in growth curve experiments, all the pE165-carrying recipients had a fitness cost compared to the corresponding original recipients in growth competition experiments. In brief, an erm(T)-carrying plasmid was for the first time described in E. faecalis and as transmissible to heterologous hosts.
Macrolides are a class of important natural or semisynthetic antibiotics that bind to the 50S ribosomal subunit and inhibit protein synthesis (1, 2). They have antimicrobial activity against Gram-positive and selected Gram-negative organisms (3, 4). The frequent use of macrolides in medical clinics and animal husbandry is accompanied by increased macrolide resistance, which may result in a failure of the treatment (5).
There are three ways to acquire macrolide resistance: modification of the target site, efflux pump, and drug inactivation (2, 5). Modification of the target site is mediated by 23S rRNA methylation enzymes, encoded by erm genes that confer resistance to macrolides; in the case of constitutive expression, they can also confer resistance to lincosamides and streptogramin B (6–11). Among the different erm genes currently known to occur in the different species of bacteria, erm(A) and erm(B) are most frequent (12, 13), mainly carried by a plasmid, transposons, translocatable units (TUs), and integrative and conjugative elements (ICEs) (13). Ribosomal protection gene msr codes for ABC-F protein confers macrolide and streptogramin B resistance. The mef gene codes for an efflux pump confers macrolide resistance only. Drug inactivation enzymes including phosphotransferases and esterases, which are encoded by mph genes and ere genes, respectively, confer macrolide resistance (13, 14).
Since erm(T) had been detected in Lactobacillus, it has also been described in isolates of the bacterial species Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus gallolyticus, Staphylococcus aureus, Erysipelothrix rhusiopathiae, Haemophilus parasuis, and Enterococcus faecium and Streptococcus suis (15–22), which revealed its widespread presence. However, whether it is transmissible and whether the erm(T)-carrying mobile genetic elements can replicate in a heterologous host had not been fully explored. So, in this study, the presence of erm(T) in enterococci was investigated and the associated mobile genetic elements involved in the horizontal transfer of erm(T) were explored. In addition, the transmission ability and maintenance of erm(T)-carrying plasmid in the heterologous host was elucidated using conjugation, transformation, and fitness cost experiments.
During the routine survey for the presence of erm(T) in enterocci of swine origin, a total of 159 non-duplicate enterococci isolates with erythromycin MICs of no <8 mg/L were collected in July and September 2018 from anal swabs of healthy pigs at two farms in Henan Province, China.
Antimicrobial susceptibility testing (AST) was carried out by broth microdilution according to recommendations given in document M100 (Twenty-Eighth Edition) issued by CLSI (21). S. aureus ATCC 29213 served as the quality control strain.
E. faecalis OG1RF, S. aureus RN4220, Streptococcus suis P1/7, and Streptococcus mutans UA159 served as recipients in conjugation and transformation experiments.
The aforementioned erythromycin-resistant enterococci strains were detected for the presence of macrolide resistance gene erm(T) and other resistance genes by PCR using the primers listed in Table 1 (15, 23–26). PCR products for erm(T) in E. faecalis E165 and its transconjugants and transformants were subjected to Sanger sequencing.
Whole genome DNA of E. faecalis E165 was sequenced by the PacBio RS and Illumina MiSeq platforms (Shanghai Personal Biotechnology Co., Ltd, China). The PacBio sequence reads were assembled with HGAP4 and CANU (Version 1.6), and corrected by Illumina MiSeq with pilon (Version 1.22). The prediction of ORFs and their annotation was performed using Glimmer 3.0.
To investigate whether the erm(T)-carrying plasmid could be transferred into bacteria of the same and other species, plasmid DNA extracted from E. faecalis E165 was obtained by using Qiagen Mini-prep kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
To determine the intraspecies transmissibility of the erm(T)-carrying plasmid pE165, conjugation experiments were performed using E. faecalis E165 as the donor and E. faecalis OG1RF as the recipient as previously described (27). Transconjugants were screened on brain heart infusion (BHI, Oxoid, British) agar supplemented with 32 mg/L rifampicin, 32 mg/L fucidin acid, and 8 mg/L erythromycin. The corresponding transconjugant was designated E. faecalis OG1RF-Tc.
Natural transformation experiment using the recipient S. suis P1/7 was performed as described to investigate the interspecies transmissibility of erm(T)-carrying plasmid pE165 (28). The peptide (GNWGTWVEE) was dissolved in Milli-Q water at a final concentration of 5 mM and was used as a pheromone for the transformation. The erythromycin-susceptible recipient strain S. suis P1/7 was grown overnight in THY broth (3 g Todd-Hewitt Broth and 2 g yeast for 100 ml, Oxoid, British) at 37°C under 5% CO2. The overnight culture was diluted 1:40 into pre-warmed THY broth, and grown at 37°C under 5% CO2 without shaking. Plasmid DNA (1.2 μg) and stock peptide (5 μl) were added when the recipient culture reached an OD600 between 0.035 and 0.058, and then incubated for 2h at 37°C under 5% CO2. The samples were diluted and plated on THA (Todd-Hewitt Broth with 1.3% agar powder, Oxoid, British) containing 8 mg/L erythromycin. The corresponding transformant was designated S. suis P1/7-Tm.
Natural transformation experiment with the recipient S. mutans UA159 was performed following procedures as previously described (29). For the screening of the transformants, THA was supplemented with 8 mg/L erythromycin. The corresponding transformant was designated S. mutans UA159-Tm.
The electrotransformation experiment with recipient strain S. aureus RN4220 was performed as described in a previous study (30). The corresponding transformant was designated S. aureus RN4220-Tm.
All colonies from the selective plates after incubation for 24–48 h at 37°C were further confirmed by erm(A), erm(B), and erm(T) gene detection (15, 23), 16S rRNA sequencing, AST, and multilocus sequence typing (MLST) following the harmonized protocols (http://www.mlst.net/). The plasmids obtained from E. faecalis E165, E. faecalis OG1RF-Tc, S. suis P1/7-Tm, S. mutans UA159-Tm, and S. aureus RN4220-Tm were digested by Sac I and Pac I (New England Biolabs, Inc., USA), and then southern bolt using erm(T) gene as the probe was performed.
Growth kinetics were determined for E. faecalis OG1RF and E. faecalis OG1RF-Tc, S. aureus RN4220 and S. aureus RN4220-Tm, S. suis P1/7 and S. suis P1/7-Tm, and S. mutans UA159 and S. mutans UA159-Tm (19, 31). Volumes of 30 ml BHI broth were inoculated independently with 107 CFU of OG1RF, OG1RF-Tc, RN4220, and RN4220-Tm; cultures were grown for 12 h at 200 rpm and 37°C. Volumes of 30 ml Todd-Hewitt Broth (THB, Oxoid, British) supplemented with 5% fetal calf serum were inoculated independently with 107 CFU of P1/7, P1/7-Tm, UA159, and UA159-Tm; cultures were grown for 12 h at 200 rpm and 37°C. The absorbance at 600 nm was measured every hour.
The fitness cost of pE165 was determined between E. faecalis OG1RF and E. faecalis OG1RF-Tc, S. aureus RN4220 and S. aureus RN4220-Tm, S. suis P1/7 and S. suis P1/7-Tm, and S. mutans UA159 and S. mutans UA159-Tm, as previously described (31, 32), with the following modifications. OG1RF and OG1RF-Tc, RN4220, and RN4220-Tm, were cultured in BHI broth for 24h at 37°C and 200 rpm. P1/7 and P1/7-Tm, UA159 and UA159-Tm, were cultured in THB (supplemented with 5% fetal calf serum) for 24 h at 37°C and 200 rpm. Then 1 × 108 CFU of recipient strain was mixed with 1 × 108 CFU of corresponding transconjugant/ transformant in 30 ml antibiotic-free BHI broth/THB (supplemented with 5% fetal calf serum). The mixtures were grown at 37°C and 200 rpm and diluted at 1:100 to fresh BHI broth/THB (supplemented with 5% fetal calf serum) every 24 h. For each sample, aliquots were plated onto non-selective and erythromycin-containing BHI agar/THA (supplemented with 5% fetal calf serum) plates. The proportion of pE165-carrying strains was calculated by the number of colonies on the selective plate divided by the number of colonies on the non-selective plate.
All erythromycin-resistant strains were investigated for the presence of the macrolide resistance genes by PCR. Of the 159 erythromycin-resistant enterococci isolates, 24(15.1%) contained solely erm(A), 33 (20.8%) contained solely erm(B), and 102 (64.2%) were positive for both erm(A) and erm(B). No strain was erm(C)/erm(F)/erm(G)/erm(X)-positive. A single E. faecalis strain E165 was positive for erm(A), erm(B), and erm(T). This is the first description of the erm(T) gene in E. faecalis.
Whole genome sequencing, assembly, and analysis for E. faecalis E165 showed a 4,244-bp small plasmid (designated pE165) harbored the erm(T) gene, with an average GC content of 33.0%. A total of three open reading frames (ORFs) encoding proteins of >100 amino acids were identified. The erm(T) gene coded for a 244-amino-acid (aa) protein identical to erm(T) of pKKS25 in S. aureus 25 (CAY48681.1) (18), and was highly similar (99.2% amino acid identity, 99.7% DNA sequence identity) to that of pGT633 from Lactobacillus reuteri 100-63 (NG_047838) (Figure 1) (32). The plasmid mobilization protein encoded by the mob gene from pE165 showed a high level of homology (identity≥90.6%) to that encoded by mob genes from Lactococcus garvieae (WP_207144600), E. faecium (HAR1670775.1), S. suis (WP_105139626), S. aureus (CCQ43999), and Escherichia coli (EFG1049274). The replication protein encoded by the rep gene from pE165 exhibited ≥99.5% identities to that encoded by rep genes from E. faecalis (HBI2052878), Listeria monocytogenes (HAB0665403), S. suis (NQK16007), S. agalactiae (WP_228308086), E. faecium (HAZ0989061), Bacillus paranthracis (AHN52261), E. coli (EFG1049261), Lactimicrobium massiliense (WP_108775117), and Amylolactobacillus amylophilus (WP_054746480), which revealed how widespread the erm(T)-carrying pE165-like plasmid is.
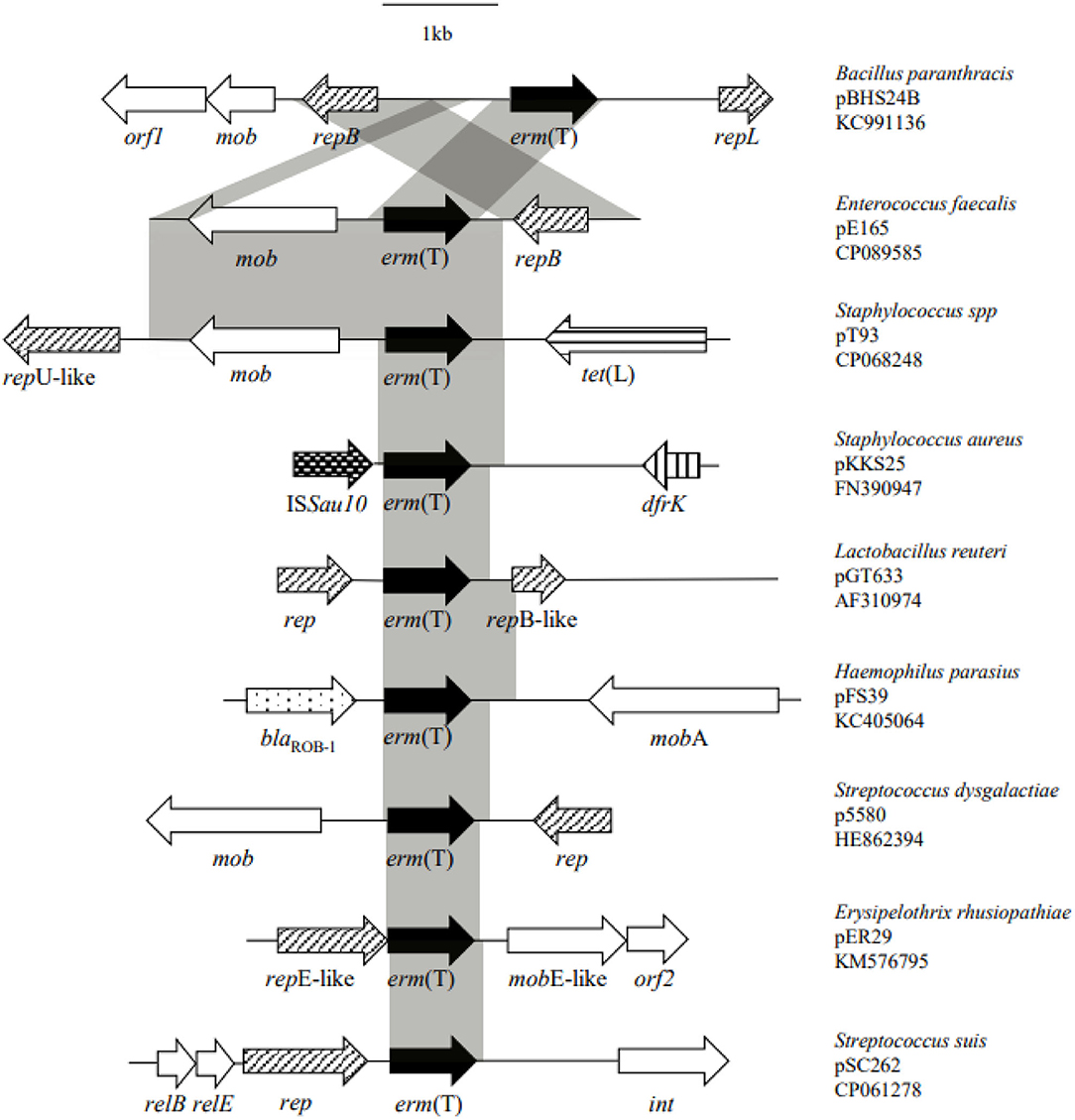
Figure 1. Structural comparison of the genetic environment of erm(T) gene located on pE165 with the genetic environment of erm(T) published previously.
In addition, erm(A) gene carried by Tn6674 was located on the chromosome. Three copies of erm(B) gene were located on a 65,052 bp conjugative plasmid (designated pE165-2) and the conjugative region from pE165-2 exhibits 99% DNA identity to pL15 described in an E. faecalis isolated from swine in Brazil (CP042214). pE165-2 also includes tet(M) and tet(L) conferring resistance to tetracyclines, dfrG conferring resistance to trimethoprim, aacA-aphD conferring resistance to aminoglycosides, and cat encoding chloramphenicol acetyltransferase. A 2,836 bp small plasmid that did not carry any resistance gene was also detected in E. faecalis E165 (designated pE165-3).
Previous studies identified a complete translational attenuator immediately upstream of the erm(T) gene which consisted of two pairs of inverted-repeat sequences of 12 bp each and a reading frame for a regulatory peptide of 19 aa (15, 17, 33). Inducible erm gene expression often required an intact translational attenuator, while deletions or duplications that appeared in the regulatory region would cause constitutive erm gene expression (34). A comparison of the erm(T) regulatory region of pE165 with that of plasmids pRW35 (EU192194) revealed that the erm(T) regulatory region of pE165 had four bp point mutations and one bp deletion in the regulatory peptide ORF. This single nucleotide deletion resulted in a frame shift mutation, which extended the reading frame for the regulatory peptide from 19 aa to 22 aa (Figure 2). In addition, erm(T) regulatory region of pE165 was compared to those of pSC262 and pUR2940 with mutations in previous reports (22, 35), and the results are shown in Supplementary Figures S1, S2.
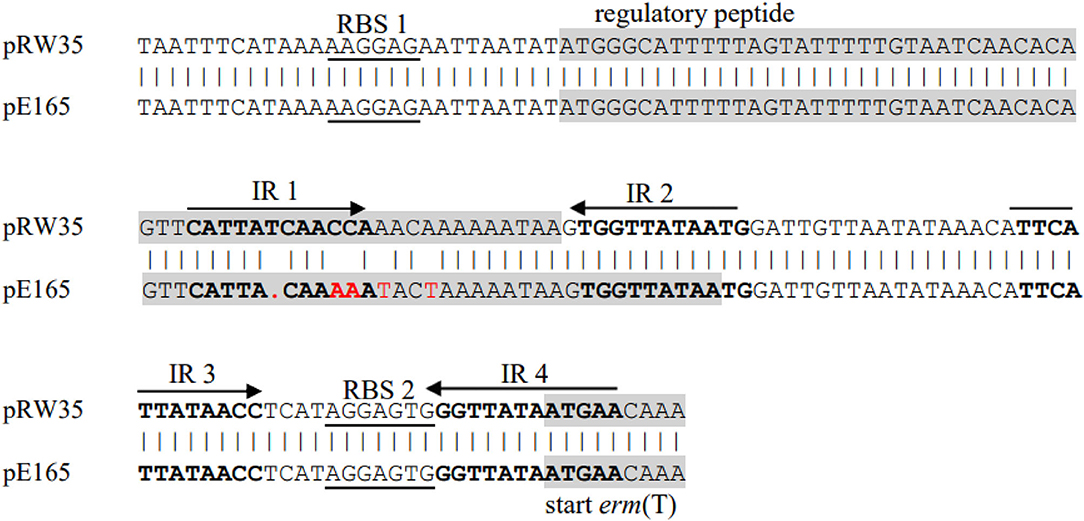
Figure 2. Alignment of erm(T) regulatory region of pRW35.seq (upper line) and erm(T) regulatory region of pE165.seq (lower line). Identity = 97.40%(150/154), Gap = 0.65%(1/155).
Intraspecies transmissibility of pE165 was investigated by conjugation experiments using E. faecalis E165 as the donor and E. faecalis OG1RF as the recipient. The transconjugant OG1RF-Tc was successfully obtained on the selective plates with the transfer frequencies of 1.5 × 10−5. PCR assay was used for the detection of erm(T), erm(A), and erm(B) in transconjugant, and the results revealed that only erm(T) gene was transferred into the recipient. Minimum inhibition concentrations (MICs) of E. faecalis E165, OG1RF, and OG1RF-Tc were determined and are shown in Table 2. Compared to E. faecalis OG1RF, OG1RF-Tc displayed a higher erythromycin MIC (>512 mg/L) and higher clindamycin MIC (256 mg/L).
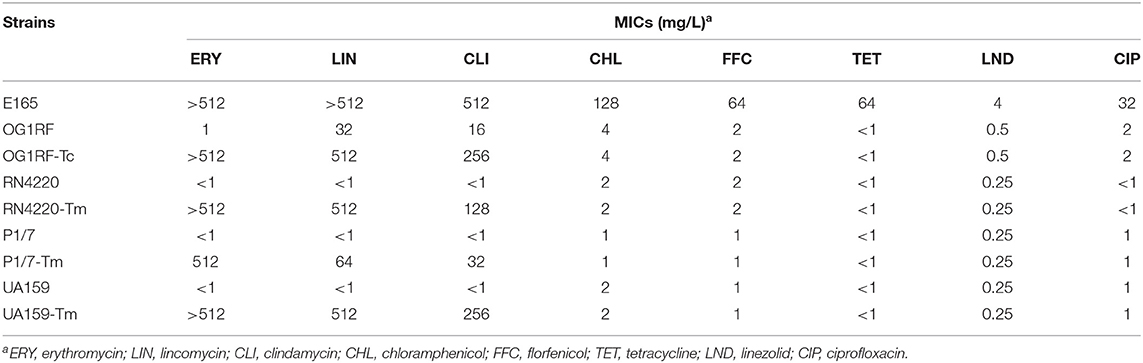
Table 2. MICs of erm(T)-carrying E. faecalis E165, recipient strains E. faecalis OG1RF, S. aureus RN4220, S. suis P1/7, S. mutans UA159, and corresponding transconjugants/transformants.
Interspecies transmissibility of pE165 was investigated by natural transformation using S. suis P1/7 and S. mutans UA159 as the recipients and electrotransformation using S. aureus RN4220 as the recipient. Transformants P1/7-Tm, UA159-Tm, and RN4220-Tm were successfully obtained, and the transfer frequencies were 0.63 × 102 μg−1,2.1 × 102 μg−1 and 4.9 × 104 μg−1. The transformants were confirmed by AST and sequencing of 16S rRNA. PCR assay also revealed that only erm(T) could be detected in these transformants. MICs of E. faecalis E165, S. suis P1/7, S. mutans UA159, S. aureus RN4220, and their transformants are shown in Table 2. S. suis transformant P1/7-Tm displayed a higher erythromycin MIC (>512 mg/L) and a higher clindamycin MIC (32 mg/L) compared with S. suis P1/7. S. mutans transformant UA159-Tm displayed higher erythromycin MIC (>512 mg/L) and clindamycin MIC (256 mg/L) compared with S. mutans UA159. S. aureus transformant RN4220-Tm displayed higher erythromycin MIC (>512 mg/L) and higher clindamycin MIC (128 mg/L) compared with S. aureus RN4220.
The physical map of pE165 and restriction enzyme-digested plasmid profiles of the plasmids from E. faecalis E165, transconjugant E. faecalis OG1RF-Tc, transformants S. suis P1/7-Tm, S. mutans UA159-Tm, and S. aureus RN4220-Tm are shown in Figure 3. The results indicated that only pE165 can transfer into the recipient strains and pE165 can replicate in heterogenous hosts. The result of the southern bolt revealed that erm(T) gene was located on pE165 in heterogenous hosts. The results of AST indicated that pE165 can constitutively express erythromycin- and clindamycin- resistance phenotype in heterogenous hosts.
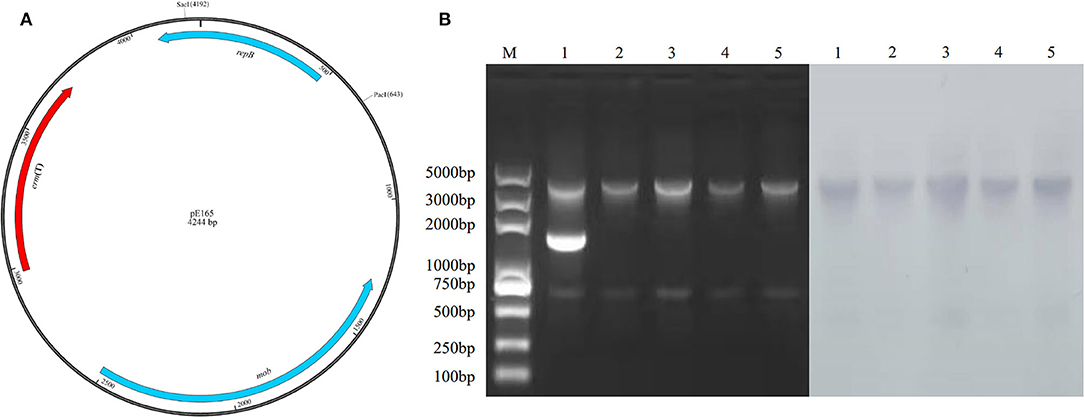
Figure 3. (A) Restriction enzyme cutting sites of Sac I and Pac I on pE165. (B) Restriction enzyme-digested plasmid profiles treated by Sac I, Pac I, and southern blot using erm(T) gene as the probe. M: marker 1; E. faecalis E165, 2; E. faecalis OG1RF-Tc, 3; S. mutans UA159-Tm, 4; S. suis P1/7-Tm, 5; S. aureus RN4220-Tm. The upper and lower bands were the fragments of pE165 digested by Sac I and Pac I. The middle band in line 1 was supercoiled pE165-3.
The growth curve of E. faecalis E165, S. suis P1/7, S. mutans UA159, S. aureus RN4220, and their transconjugants and transformants in the absence of erythromycin are shown in Figure 4. The results showed that no significant fitness burden for pE165-carrying transconjugants and transformants was observed compared with the recipient strains in the absence of selective pressure.
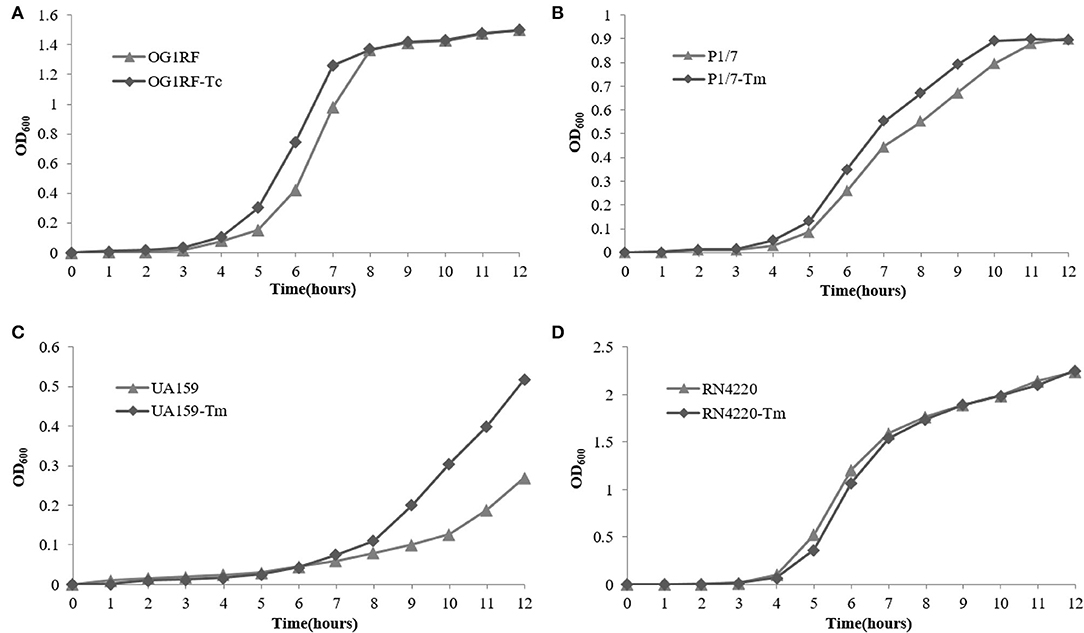
Figure 4. Fitness cost of pE165. (A–D) Comparison of growth kinetics of E. faecalis OG1RF and E. faecalis OG1RF-Tc, S. aureus RN4220 and S. aureus RN4220-Tm, S. suis P1/7 and S. suis P1/7-Tm, S. mutans UA159 and S. mutans UA159-Tm in the absence of erythromycin.
Competition experiments can offer a more discriminative and precise measurement of fitness, and the competitive disadvantage of the fitness burden caused by pE165 can be reflected during all the phases of the growth cycle and in successive cycles. During the competition experiment between E. faecalis OG1RF and OG1RF-Tc, from day 1 on, a successive decrease in the proportion of E. faecalis OG1RF-Tc was observed, and all the colonies tested were pE165 free on day 14 (Figure 5A). From day 1 on, a fast and constant decrease in the proportion of S. suis P1/7-Tm was observed and all the strains were tested pE165 free on day 6 (Figure 5B). In the process of competition experiment between S. mutans UA159 and UA159-Tm, an obvious decrease in the proportion of UA159-Tm was observed. On day 9, all the colonies tested were pE165 free (Figure 5B). For the result of the competition experiment between S. aureus RN4220 and RN4220-Tm, it had an obvious decrease from day 3 on, and RN4220-Tm could not be detected on day 15 (Figure 5A). The above results suggested that all the pE165-carrying transconjugants and transformants had a fitness cost compared to the recipient strains without pE165, but the fitness cost among the different transconjugants and transformants differed.
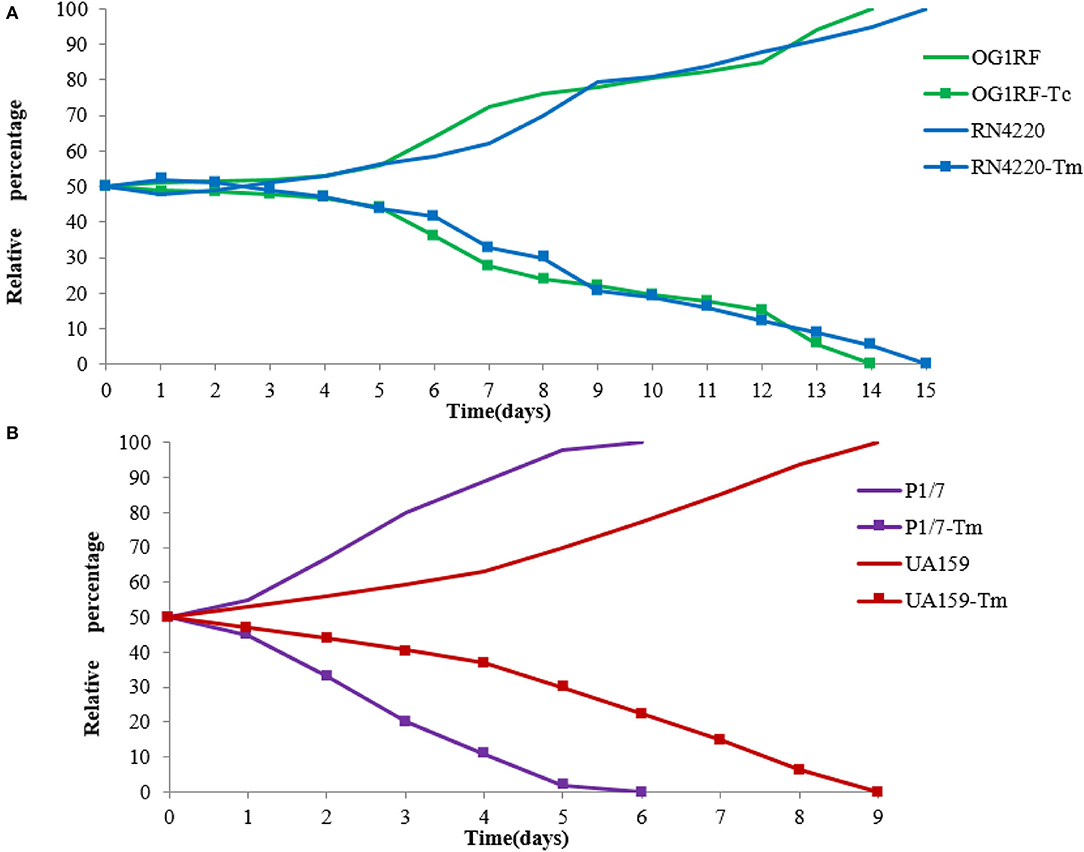
Figure 5. (A) Growth competition experiments between E. faecalis OG1RF and E. faecalis OG1RF-Tc, S. aureus RN4220 and S. aureus RN4220-Tm. (B) Growth competition experiments between S. suis P1/7 and S. suis P1/7-Tm, S. mutans UA159 and S. mutans UA159-Tm. The initial ratio of the recipient strain carrying pE165 to the original recipient strain was 1:1. The results above were averagely calculated from four independent experiments.
Since erm(T) has been reported from Lactobacillus reuteri (33), it had been described in various genera: Lactobacillus, Streptococcus (S. pyogenes, S. suis), S. aureus, E. faecium, and other gram-positive bacteria including E. rhusiopathiae (KM576795.1), even in gram-negative bacteria such as H. parasuis (KC405064.1) and Klebsiella pneumonia (CP040837.1), revealing its widespread presence. Mobile genetic elements play a crucial role in the horizontal gene transfer of erm(T), including plasmids (15, 16, 18, 20, 35–37),transposons (38, 39), and insert sequence (17). erm(T)-positive plasmids of S. agalactiae could efficiently be transferred into group B Streptococcus and in the E. faecalis recipient strain (16).
In this study, erm(T)-positive plasmid pE165 was identified in a E. faecalis strain E165. Whole-genomic sequencing for E165 was performed. The genetic environment of erm(T) in this study was then analyzed by comparing it with similar erm(T) genetic environments published previously (19, 20, 33, 35, 39, 40). The homology analysis of the rep and mob genes located on pE165 suggested that this plasmid had the potential ability to transfer into enterococci and other species. Transformation experiments confirmed that pE165 was successfully transferred into Enterococcus, Streptococcus, and Staphylococcus. Elevated MICs of erythromycin and clindamycin were conferred by erm(T) in the recipient strains. According to recommendations given in CLSI, inducible expression of erm(T) cannot produce clindamycin resistance unless it is induced by erythromycin (41). In this study, the inducible clindamycin resistance tests indicated that the transconjugant E. faecalis OG1RF-Tc, transformants P1/7-Tm, UA159-Tm, and RN4220-Tm were resistant to both erythromycin and clindamycin (Table 2), which revealed the expression of erm(T) in these strains was constitutive. This is also in agreement with the observation that deletions or duplications appeared in the regulatory region of erm(T) in these strains.
It can be found that the proportion of pE165-carrying strains constantly decreased until undetectable in the competition experiments between transformants and original recipients performed by successive culturing in the absence of antibiotic pressure. Although plasmids can mediate the horizontal transmission of resistance genes between bacteria and facilitate their adaptation to the pressure of antibiotics, they also entail a metabolic burden that reduces the competitiveness of the plasmid-carrying clone in the absence of selection (42). Acquisition and maintenance of a plasmid are directly associated with fitness effects on the recipient strain. The constitutive expression of erm(T) will produce a burden (fitness cost) in the recipient strain, and the low prevalence of erm(T) gene in many genera of bacteria may be explained in this way.
The erm(T) gene was first reported in an E. faecalis strain. A 4244 bp erm(T)-positive plasmid pE165 was characterized. The transmissibility of pE165 was investigated between intra- and inter-species. The presence of pE165 greatly elevated the MICs of erythromycin and clindamycin which indicated the expression of erm(T) in the recipient strains was constitutive. Although the fitness cost showed us this plasmid reduced the competitiveness of the host strain, the potential possibility of dissemination of erm(T) among species of bacteria should not be ignored.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below https://www.ncbi.nlm.nih.gov/, CP089585.
X-DD and DL designed the research and supervised the study. X-YL, RY, CX, and YS performed the experiments and analyzed the data. X-YL, RY, and X-DD wrote the manuscript. All authors revised the manuscript and approved the final version for submission.
This work was supported by grants from the Program for Innovative Research Team (in Science and Technology) at the University of Henan Province (No. 18IRTSTHN020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Wanjiang Zhang (Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, P. R. China) for providing recipient strain S. suis P1/7.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.850466/full#supplementary-material
1. Vazquez-Laslop N, Mankin AS. How macrolide antibiotics work. Trends Biochem Sci. (2018) 43:668–84. doi: 10.1016/j.tibs.2018.06.011
2. Dinos GP. The macrolide antibiotic renaissance. Br J Pharmacol. (2017) 174:2967–83. doi: 10.1111/bph.13936
3. Kelly C, Chalmers JD, Crossingham I, Relphl N, Felix LM, Evans DJ, et al. Macrolide antibiotics for bronchiectasis. Cochrane Database Syst Rev. (2018) 3:75. doi: 10.1002/14651858.CD012406.pub2
4. Arsic B, Barber J, Cikos A, Mladenovic M, Stankovic N, Novak P. 16-membered macrolide antibiotics: a review. Int J Antimicrob Agents. (2018) 51:283–98. doi: 10.1016/j.ijantimicag.2017.05.020
5. Torres C, Alonso CA, Ruiz-Ripa L, Leon-Sampedro R, Del Campo R, Coque TM. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol Spectr. (2018) 6:ARBA-0032-2018. doi: 10.1128/microbiolspec.ARBA-0032-2018
6. Schroeder MR, Stephens DS. Macrolide resistance in Streptococcus pneumoniae. Front Cell Infect Microbiol. (2016) 6:9. doi: 10.3389/fcimb.2016.00098
7. Nash KA. Intrinsic macrolide resistance in Mycobacterium smegmatis is conferred by a novel erm Gene, erm(38). Antimicrob Agents Chemother. (2003) 47:3053–60. doi: 10.1128/aac.47.10.3053-3060.2003
8. Strauss C, Hu YM, Coates A, Perreten V. A novel erm(44) gene variant from a human Staphylococcus saprophyticus isolate confers resistance to macrolides and lincosamides but not streptogramins. Antimicrob Agents Chemother. (2017) 61:5. doi: 10.1128/aac.01655-16
9. Huber L, Giguere S, Slovis NM, Alvarez-Narvaez S, Hart KA, Greiter M, et al. The novel and transferable erm(51) gene confers macrolides, lincosamides and streptogramins B (MLSB) resistance to clonal Rhodococcus equi in the environment. Environ Microbiol. (2020) 22:2858–69. doi: 10.1111/1462-2920.15020
10. Qin SS, Wang Y, Zhang QJ, Zhang MJ, Deng FR, Shen ZQ, et al. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother. (2014) 69:964–8. doi: 10.1093/jac/dkt492
11. Zhu Y, Zhang WJ, Liu SG, Schwarz S. Identification of an IS431-derived translocatable unit containing the erm(C) gene in Staphylococcus aureus. J Antimicrob Chemother. (2021) 76:1102–4. doi: 10.1093/jac/dkaa555
12. Portillo A, Ruiz-Larrea F, Zarazaga M, Alonso A, Martinez JL, Torres C. Macrolide resistance genes in Enterococcus spp. Antimicrob Agents Chemother. (2000) 44:967–71. doi: 10.1128/aac.44.4.967-971.2000
13. Fessler AT, Wang Y, Wu CM, Schwarz S. Mobile macrolide resistance genes in staphylococci. Plasmid. (2018) 99:2–10. doi: 10.1016/j.plasmid.2018.05.001
14. Sharkey LKR, Edwards TA, O'Neill AJ. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio. (2016) 7:10. doi: 10.1128/mBio.01975-15
15. Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, BartkuS JM, et al. Plasmid-borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother. (2008) 52:1140–3. doi: 10.1128/aac.01352-07
16. Compain F, Hays C, Touak G, Dmytruk N, Trieu-Cuot P, Joubrel C, et al. Molecular characterization of Streptococcus agalactiae isolates harboring small erm(T)-carrying plasmids. Antimicrob Agents Chemother. (2014) 58:6928–30. doi: 10.1128/aac.03855-14
17. Tsai JC, Hsueh PR, Chen HJ, Tseng SP, Chen PY, Teng LJ. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. Pasteurianus Antimicrob Agents Chemother. (2005) 49:4347–50. doi: 10.1128/aac.49.10.4347-4350.2005
18. Kadlec K, Schwarz S. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother. (2010) 54:915–8. doi: 10.1128/aac.01091-09
19. Xu CW, Zhang AY, Yang CM, Pan Y, Guan ZB, Lei CW, et al. First report of macrolide resistance Gene erm(T) harbored by a novel small plasmid from Erysipelothrix rhusiopathiae. Antimicrob Agents Chemother. (2015) 59:2462–5. doi: 10.1128/aac.00228-15
20. Yang SS, Sun J, Liao XP, Liu BT Li LL, Li L, et al. Co-location of the erm(T) gene and bla(ROB-1) gene on a small plasmid in Haemophilus parasuis of pig origin. J Antimicrob Chemother. (2013) 68:1930–2. doi: 10.1093/jac/dkt112
21. DiPersio LP, DiPersio JR, Frey KC, Beach JA. Prevalence of the erm(T) gene in clinical isolates of erythromycin-resistant group D Streptococcus and Enterococcus. Antimicrob Agents Chemother. (2008) 52:1567–9. doi: 10.1128/aac.01325-07
22. Yu R, Xu Y, Schwarz S, Shang Y, Yuan X, Zhang Y, et al. erm(T)-mediated macrolide-lincosamide resistance in Streptococcus suis. Microbiol Spectr. (2022) 10:e01657–21. doi: 10.1128/spectrum.01657-21
23. Giovanetti E, Brenciani A, Lupidi R, Roberts MC, Varaldo PE. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef (A) or the erm(A) gene. Antimicrob Agents Chemother. (2003) 47:2844–9. doi: 10.1128/aac.47.9.2844-2849.2003
24. Abbas A, Srivastava P, Nirwan PS. Prevalence of MLSB resistance and observation of erm A & erm C genes at a tertiary care hospital. J Clin Diagn Res: JCDR. (2015) 9:DC08–10. doi: 10.7860/jcdr/2015/13584.6112
25. Eitel Z, Soki J, Urban E, Nagy E, Anaerobic ESG. The prevalence of antibiotic resistance genes in Bacteroides fragilis group strains isolated in different European countries. Anaerobe. (2013) 21:43–9. doi: 10.1016/j.anaerobe.2013.03.001
26. Hays C, Lienhard R, Auzou M, Barraud O, Guerin F, Ploy MC, et al. Erm(X)-mediated resistance to macrolides, lincosamides and streptogramins in Actinobaculum schaalii. J Antimicrob Chemother. (2014) 69:2056–60. doi: 10.1093/jac/dku099
27. Hao WB, Shan XX Li DX, Schwarz S, Zhang SM Li XS, et al. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother. (2019) 74:1771–5. doi: 10.1093/jac/dkz109
28. Yu R, Zhang Y, Xu YD, Schwarz S, Li XS, Shang YH, et al. Emergence of a tet(M) variant conferring resistance to tigecycline in Streptococcus suis. Front Vet Sci. (2021) 8:8. doi: 10.3389/fvets.2021.709327
29. Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. (2001) 183:897–908. doi: 10.1128/jb.183.3.897-908.2001
30. Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. (1992) 73:133–8. doi: 10.1016/0378-1097(92)90596-g
31. Millan AS, Garcia-Cobos S, Escudero JA, Hidalgo L, Gutierrez B, Carrilero L, et al. Haemophilus influenzae clinical isolates with plasmid pB1000 bearing bla(ROB-1): fitness cost and interspecies dissemination. Antimicrob Agents Chemother. (2010) 54:1506–11. doi: 10.1128/aac.01489-09
32. Starikova I, Al-Haroni M, Werner G, Roberts AP, Sorum V, Nielsen KM, et al. Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J Antimicrob Chemother. (2013) 68:2755–65. doi: 10.1093/jac/dkt270
33. Tannock GW, Luchansky JB, Miller L, Connell H, Thode-Andersen S, Mercer AA, et al. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. (1994) 31:60–71. doi: 10.1006/plas.1994.1007
34. Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. (1995) 39:797–805. doi: 10.1128/aac.39.4.797
35. Gomez-Sanz E, Kadlec K, Fessler AT, Zarazaga M, Torres C, Schwarz S. Novel erm(T)-carrying Multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother. (2013) 57:3275–82. doi: 10.1128/aac.00171-13
36. Liu BH, Lei CW, Zhang AY, Pan Y, Kong LH, Xiang R, et al. Colocation of the multiresistance gene cfr and the fosfomycin resistance gene fosD on a novel plasmid in Staphylococcus arlettae from a chicken farm. Antimicrob Agents Chemother. (2017) 61:6. doi: 10.1128/aac.01388-17
37. DiPersio LP, DiPersio JR, Beach JA, Loudon AM, Fuchs AM. Identification and characterization of plasmid-borne erm(T) macrolide resistance in group B and group A Streptococcus. Diagn Microbiol Infect Dis. (2011) 71:217–23. doi: 10.1016/j.diagmicrobio.2011.07.010
38. de Vries LE, Valles Y, Agerso Y, Vaishampayan PA, Garcia-Montaner A, Kuehl JV, et al. The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS ONE. (2011) 6:11. doi: 10.1371/journal.pone.0021644
39. Palmieri C, Magi G, Creti R, Baldassarri L, Imperi M, Gherardi G, et al. Interspecies mobilization of an erm(T)-carrying plasmid of Streptococcus dysgalactiae subsp equisimilis by a coresident ICE of the ICESa2603 family. J Antimicrob Chemother. (2013) 68:23–6. doi: 10.1093/jac/dks352
40. Barbosa TM, Phelan RW, Leong D, Morrissey JP, Adams C, Dobson ADW, et al. A novel erythromycin resistance plasmid from Bacillus Sp strain HS24, isolated from the marine sponge haliclona simulans. PLoS ONE. (2014) 9:11. doi: 10.1371/journal.pone.0115583
41. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing-Twenty-Eighth Edition: M100. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
Keywords: Enterococcus faecalis, erythromycin, clindamycin, resistance, fitness cost, transmission
Citation: Li X-Y, Yu R, Xu C, Shang Y, Li D and Du X-D (2022) A Small Multihost Plasmid Carrying erm(T) Identified in Enterococcus faecalis. Front. Vet. Sci. 9:850466. doi: 10.3389/fvets.2022.850466
Received: 07 January 2022; Accepted: 20 April 2022;
Published: 27 May 2022.
Edited by:
Patrick Rik Butaye, Ross University School of Veterinary Medicine, Saint Kitts and NevisReviewed by:
Linda Antionette Bester, University of KwaZulu-Natal, South AfricaCopyright © 2022 Li, Yu, Xu, Shang, Li and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Dang Du, eGRkdUBoZW5hdS5lZHUuY24=; Dexi Li, bGlkZXhpMjAwNkAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.