- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi University, Nanning, China
- 2Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding, School of Life Science and Engineering, Foshan University, Foshan, China
- 3Department of Biosciences, COMSATS University Islamabad, Islamabad, Pakistan
Rabbits have been domesticated for meat, wool, and fur production, and have also been cherished as a companion, artistic inspiration, and an experimental model to study many human diseases. In the present study, the muscle mass negative regulator gene myostatin (MSTN) was knocked out in rabbits at two novel sites in exon3, and the function of these mutations was determined in subsequent generations. The prominent double muscle phenotype with hyperplasia or hypertrophy of muscle fiber was observed in the MSTN-KO rabbits, and a similar phenotype was confirmed in the F1 generation. Moreover, the average weight of 80-day-old MSTN-KO rabbits (2,452 ± 63 g) was higher than that of wild-type rabbits (2,393.2 ± 106.88 g), and also the bodyweight of MSTN-KO rabbits (3,708 ± 43.06g) was significantly higher (P < 0.001) at the age of 180 days than wild-type (WT) rabbits (3,224 ± 48.64g). In MSTN-KO rabbits, fourteen rabbit pups from the F1 generation and thirteen from the F2 generation stably inherited the induced MSTN gene mutations. Totally, 194 pups were produced in the F1 generation of which 49 were MSTN-KO rabbits, while 47 pups were produced in the F2 generation of which 20 were edited rabbits, and the ratio of edited to wild-type rabbits in the F2 generation was approximately 1:1. Thus, we successfully generated a heritable double muscle buttocks rabbits via myostatin mutation with CRISPR/Cas9 system, which could be valuable in rabbit's meat production and also a useful animal model to study the development of muscles among livestock species and improve their important economic traits as well as the human muscle development-related diseases.
Introduction
Rabbits are long-eared ground-dwelling mammals belonging to the family Leporidae order Lagomorpha and domesticated about 1,400 years ago (1) due to their delicious high-quality meat. They are geographically distributed across the desert, wetland, and tropical forests (2). Rabbits are being reared at the domestic level as a livestock species for meat, wool, or fur production (3), and nowadays rabbit production has become a minor agricultural enterprise in Western European countries such as Italy, Spain, and France (1, 3, 4). The worldwide per capita rabbit meat consumption is 0.242 kg, while in some European countries like Italy the per capita consumption is about 4.39 kg (5, 6). Furthermore, a stable rate of rabbit meat consumption has also been reported from 2000 to 2013 in many European countries (5).
The body growth is primarily regulated through complex interactive pathways that cause cell proliferation and cell enlargement (7). However, several factors including hormonal, genetic, environmental, and nutritional can remarkably predict the growth patterns (8). Myostatin (MSTN) is a member of the transforming growth factor (TGF-β) superfamily which is considered to be a negative regulator through inhibiting muscle development and regeneration (9). MSTN is an extracellular hormone that occurs in the skeletal muscle in an inactive state and restores its activity by binding to the precursor peptide, follistatin 3, and TGF-β binding protein, and transmitting the signals through the receptors (9, 10). The binding affinity of MSTN and ActRIIB could activate a chain of signal transmission that inhibits the myocyte differentiation and proliferation (10–12).
The MSTN gene inactivation might have an effect on muscle development and regeneration since MSTN gene deletion in mice resulted in a double muscle phenomenon (hyperplasia or hypertrophy) and a significant increase in muscle mass (10–12). Natural mutations of MSTN, which have an obvious double muscle phenotypic effect were found in cattle (13, 14), dogs (15), sheep (16, 17), pig (18), and humans (19, 20). CRISPR/Cas9 system is an efficient genome editing tool and has widely been used for functional gene study. Knock out of MSTN with CRISPR/Cas9 system has been processed in sheep (21, 22), goats (23, 24), and pigs (23, 25). Qian et al. have reported the MSTN-KO gene in Meishan pig fetal fibroblasts by engineered zinc-finger proteins and prepared the MSTN-KO pig by somatic cell nuclear transfer technology. In comparison to wild-type Meishan pigs, the lean meat rate of Meishan pigs with MSTN knockout increased by 11.62% (26). In addition, MSTN-KO goats' meat production was 32% higher than the wild-type (27).
Rabbits offer quick breeding sources and meat with excellent nutritive and dietetic properties which contain high-quality protein and low fat and cholesterol contents, that is fine-grained white meat which can substitute chicken, and is also ideal for obese and cardiovascular patients (28). In the present study, we have designed a highly efficient novel regulatory site and additionally verified that the exon 3 region can regulate the MSTN gene. Furthermore, in our study, the rabbits obtained after the MSTN gene editing are able to pass the gene-edited traits to the next-generation normally, and homozygous gene-edited rabbits obtained in the F2 generation have normal production performance and stable double muscle buttock characteristics.
Materials and Methods
Animals
In the present study, rabbits were used to perform all the experiments according to the Principle Guideline for the Use and Care of Laboratory Animals, Guangxi University. Rabbits were kept under controlled conditions at the Animal Center of Guangxi University. A total of 33 rabbits used for ovulation were approximately 6–8 months old and weighed between 3.5 and 4.5 kg.
SgRNA Design and Plasmid Construction
A total of 8 sgRNAs MSTN sites were designed using the website (http://www.genome-engineering.org/) and named as g1, g2, g3, g4, g5, g6, g7, and g8 (Figure 1A). The complementary DNA strand was annealed to develop into a double strand and cloned into the pMD18-hU6-gRNA vector. Primers T76 and T78 were used as marked gene fragments and amplified by targeting two sgRNA vectors. Linearized DNA plasmids and PCR products were extracted and purified using the MEGA shortscrip TM T7 kit (Ambion, USA). The sgRNAs were generated according to the manufacturer's recommendations.
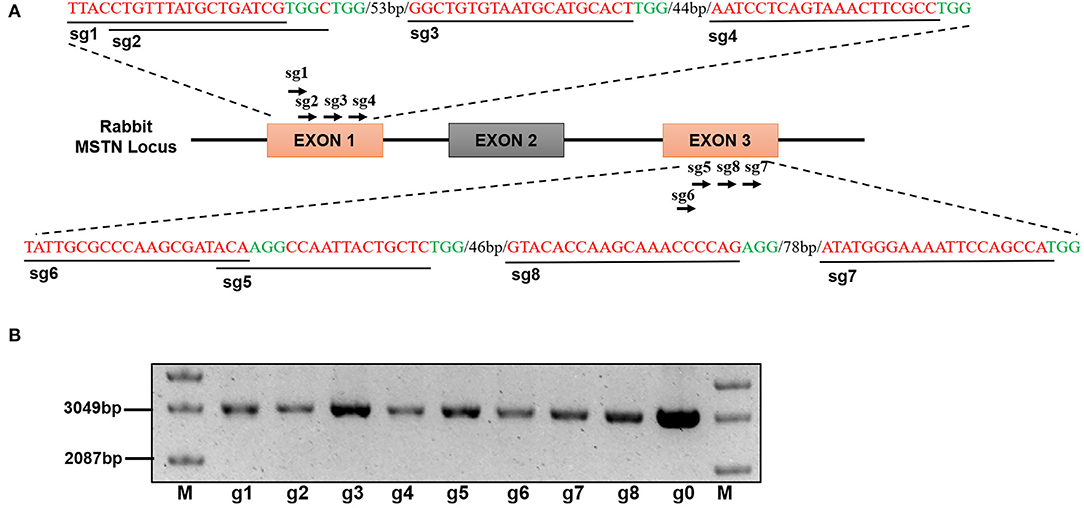
Figure 1. (A) Design and construction of sgRNA Schematic diagram of sgRNA targeting site in the rabbit MSTN gene locus. (B) Electropherogram of eight sgRNA vectors, M: Supercoiled DNA Ladder Marker, g1-g8 refer to phU6-gRNA-Sg1to phU6-gRNA-Sg8 (The picture has been cropped).
Fibroblasts Culturing and Electroporation
Primary rabbit fibroblasts were cultured in 10% (v/v) FBS, 35 mm glutamine, and 1 × DMEM (Gibco). The plasmid was transfected into fibroblasts through electroporation (Gene Pulser Xcell & BIO-RAD) at a pulse of 225 volts for 10 ms. Briefly, 4 μg of pCMV-T7-NLS-hSpCas9-NLS (hSpCas9) DNA plasmid and 2 μg of gRNA plasmid were used to transfect 5 × 105-10 × 105 fibroblasts. After 24 h the culturing media was changed to DMEM containing 10% FBS. Subsequently cultured for 4 days, the DNA was extracted and the MSTN was amplified by PCR and sent to the Beijing Genomics Institute (BGI) for sequencing to analyze the editing effect of the gene fragment. Optimal sgRNA was selected for the following experiment.
Zygote Collection and Blastocyst Genotype Testing
Rabbits (n = 3) in estrus were mated, the female rabbits were anesthetized 16–20 h after mating, and the fertilized eggs were collected from the oviduct with a cell culture medium consisting of M199 (Gibco, USA), 3% bovine serum (Gibco, USA), 5 mM HEPES (Sigma, USA), and 5 mM NaHCO3 (Sigma, USA). Then, the zygotes were microinjected with Cas9 protein and sgRNA and the blastocysts were cultured in vitro. The mutation of the target region was analyzed by PCR and T7EI digestion assay.
Generation of Transgenic Rabbit
The zygote cytoplasm was injected with a 10-pl CRISPR/Cas9 mixture containing 200 ng/μl Cas9 protein (A36497, Hot Fisher, USA), 20 ng/μl sgRNA6, and 20 ng/μl sgRNA8, and the zygotes were transferred back to the oviduct of the recipient rabbit (n = 30). The pregnancy phenomenon was checked 10 days after the transplantation and the pups were born after 30 days.
T7 Endonuclease I (T7EI) Assay
Rabbit ear tissue genomic DNA was extracted using an extraction kit (Quick-DNA Plus, ZYMO RESEARCH) and CRISPR/Cas9-induced mutations were studied by PCR and T7E1 assay. PCR was performed using primers flanking the target site and the product was sequenced (Shanghai Sangon Company, Shanghai, China) and digested with T7EI (NEB, USA) to characterize the mutation.
Off-Target Analysis
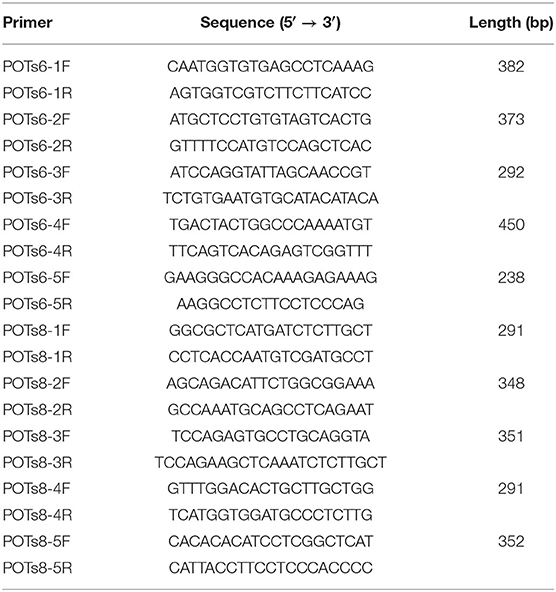
Potential off-target sites of the sgRNAs were assessed using the CRISPR design tool (http://crispor.tefor.net/) and the top 4 potential off-target sites of each sgRNA were selected and amplified by PCR with specific primers (Table 1). PCR products were evaluated by sequencing and T7E1 digestion assay.
Western Blot Analysis
Gluteus maximus tissue samples were collected from 5 MSTN-KO and 5 WT rabbits (anesthesia sampling at 6 months of age), the tissues were ground in liquid nitrogen and 2.5 μl/ml protease inhibitor was added and kept in ice for 30 min. Protein concentration was determined using the Bradford method (Bio-Rad), and 35 μg of protein sample was subjected to a 5–12% separation by SDS-polyacrylamide gel. Anti-MSTN polyclonal antibody (Abcam) and goat anti-rabbit IgG were coupled to horseradish peroxidase (HRP; Santa Cruz, USA) antibodies for protein detection, while β-actin antibody (Santa Cruz) was used as an internal control.
Sample Collection, Physical and Histological Analysis
A total of 10 rabbits, including 5 F0 MSTN-KO and 5 WT, were fed at the same conditions and weaned on the 30th day. Rabbits were kept in separate cages and their body weight was recorded after every 10 days for 200 days. At the age of 6 months, the rabbits were anesthetized with pentobarbital sodium (1 mL/kg) to take gluteal muscle mass tissue. The gluteus maximus tissues of the F0 generation MSTN-KO and WT rabbits were fixed with 4% paraformaldehyde at 4°C. Different concentrations of dehydrated sucrose (30% for 7 h, 40% for 7 h, 45% for 5 h) were also used and frozen at −25°C for histological examination. The tissue slices of 5 μm thickness were first stained with hematoxylin and eosin (H&E) and then examined by a fluorescence inverted microscope (Nikon, Japan). The integral optical density (IOD) analysis of histological sections was performed by using Image-Pro Plus 6.0 software.
Growth Data Recording of Edited Rabbit F1 and F2 Generations
The F0 generation edited rabbits were bred with wild type to obtain the F1 generation rabbits with the edited genotype. There were 3 male parents of edited rabbits and 22 wild-type female rabbits. All the F1 generation-produced edited rabbits were well developed and had significant differences in body weight from the wild type and were reserved for seed use. After sexual maturity, two breeding methods were adopted for breeding with the same genotype, and the edited male rabbits were bred with wild-type female rabbits to produce the F2 generation. Among them, there were 4 male-edited rabbits, 5 female-edited rabbits, and 9 wild-type female rabbits of the F1 generation. Genotype identification was carried out and two genotypes insertion type and deletion type were selected. The F2 generation edited rabbits were crossbred with the same genotype and also with the wild type. The weight recording method is consistent with the F0 generation edited rabbit recording method. Among them, the breeding and farrowing information of the F1 generation male rabbits were recorded.
Statistical Analysis
All the data obtained in this study were analyzed using Graph pad prism software (T-test) and the p < 0.05 was considered statistically significant (29).
Results
The SgRNA Designing and Construction
The exon 1 and 3 of the MSTN gene were targeted to design eight sgRNAs (g1, g2, g3, g4, g5, g6, g7, and g8) which were cloned into the pMD18T vector as shown in Figure 1A. The recombinant vector was named pMD18-hU6-gRNA and the sequence analysis results, as presented in Figure 1B, confirmed that the sgRNA has been successfully inserted into the vector.
Mutational Effect of the SgRNAs in Rabbit Fibroblasts
The rabbit primary fibroblasts were obtained from newborns (Figure 2A) and the mutation efficiency of the designed sgRNAs was confirmed. In g1, g2, g3, and g4 no mutations were generated while the rate of mutation in g5, g6, g7, and g8 was observed to be about 20–45% (Figure 2B). Further, the sgRNAs targeted to exon 3 were significantly better than that of exon 1. The gene alteration proportion of sgRNA6 (45%, 9/20) and sgRNA8 (40%, 8/20) were higher than that of the other gRNAs. Thus, sgRNA6 and sgRNA8 were selected for further experiments.
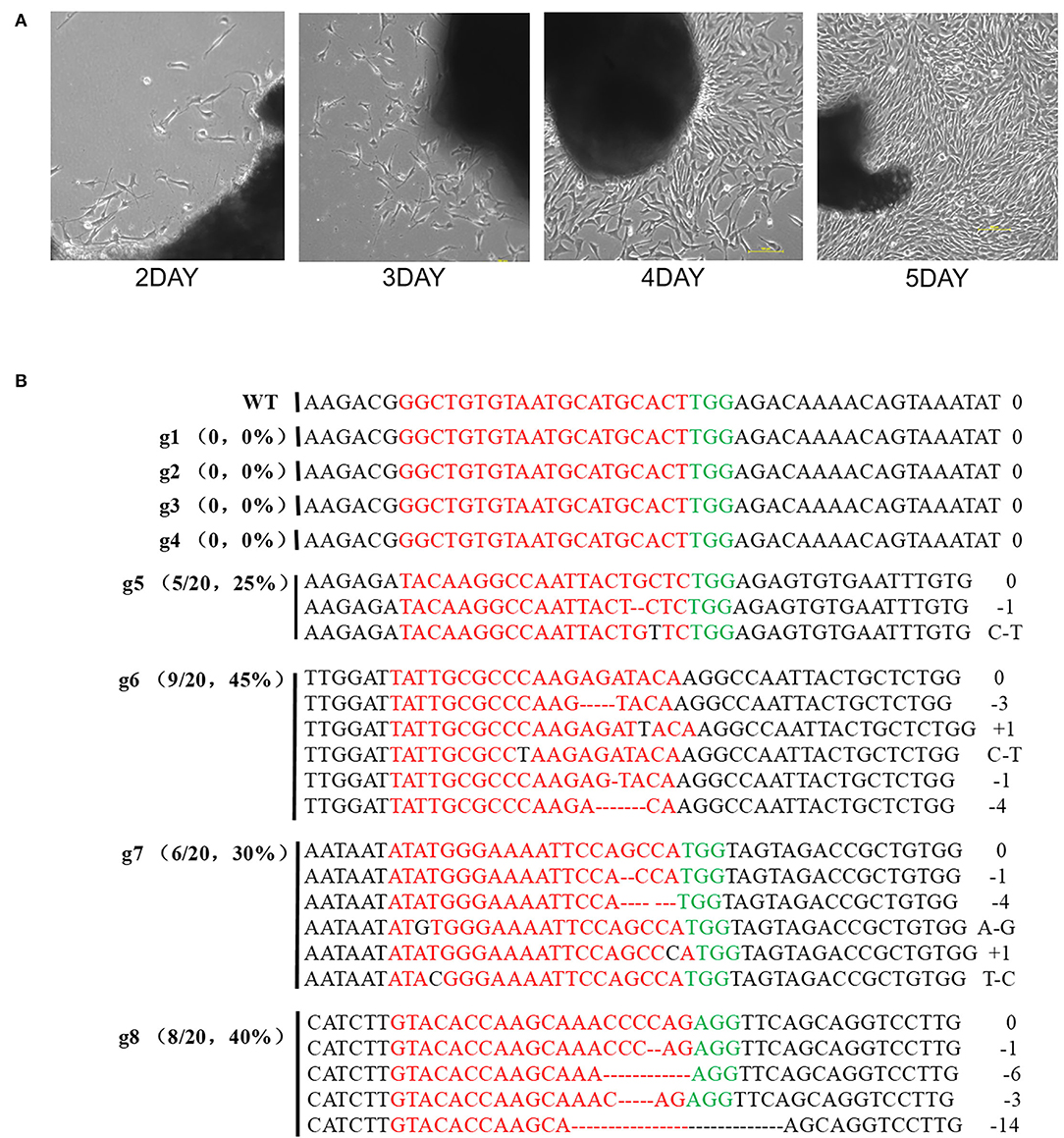
Figure 2. The results of sequencing for mutations at the specific sites in rabbit REFs. (A) Rabbit Primary fibroblast cell in culture from 2days to 5days. (B) Sanger sequence analysis of REFs. g1-g4 target to exon1, g5-g8 target to exon 3. gRNA edited were named on the left. g1-g8 represent different gRNA, PAM sequence is marked in green. The sgRNAs sequence is marked in red. The numbers on the right indicated the type of mutation, with “-” represents deletion of the given number of nucleotides, “+” represents insertion of the given number of nucleotides.
CRISPR/Cas9-Mediated MSTN Site-Specific KNOCK-OUT in Rabbit Zygotes
The CRISPR/Cas9 system containing sgRNA6 and sgRNA8 was injected into the zygotes to verify the site-specific deletion of the MSTN gene. A total of 16 zygotes were used for injection purposes, of which 10 developed to the blastocyst stage through in vitro maturation (Figure 3A). The genome-editing efficiency in the blastocysts was examined by PCR and Sanger sequencing, and the results revealed that 70% (7/10) of the blastocysts had genetic mutations (Figure 3B).
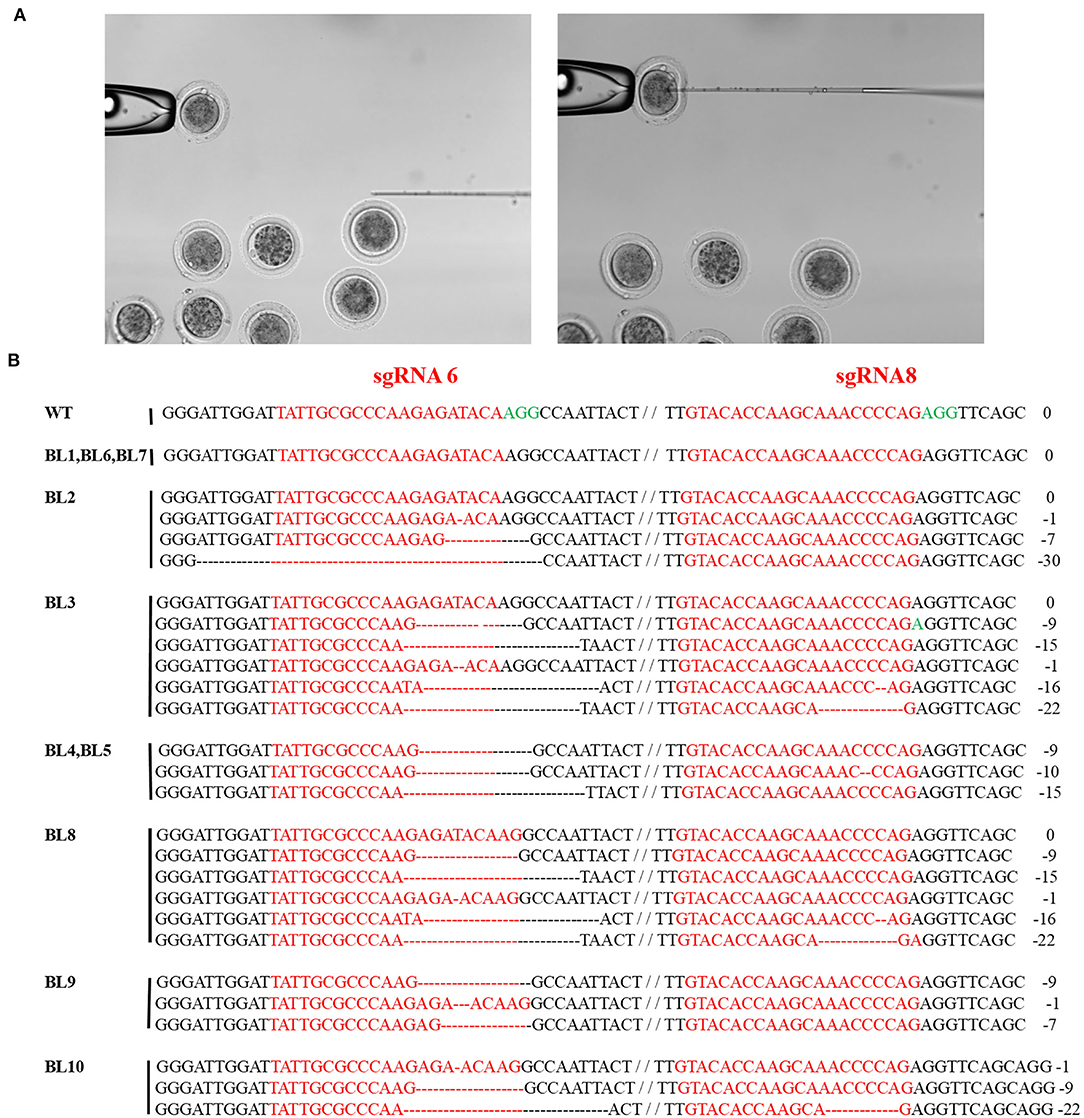
Figure 3. Targeted mutagenesis efficiency of the CRISPR/Cas9 system in rabbit zygotes. (A) Schematic diagram of the microinjection of Cas9 mRNA and sgRNAs into rabbit zygotes. (B) BL1-BL7 represent different blastocysts used in this study. Sanger sequence analysis of zygotes. zygotes edited were named on the left. PAM sequence is marked in green. The sgRNAs sequence is marked in red. The numbers on the right indicated the type of mutation, with “-” represents deletion of the given number of nucleotides.
Generation of MSTN Knock-Out Rabbits
A total of 99 cytoplasmic injected zygotes were transplanted to the oviduct of 10 female receptor rabbits, and 23 rabbit pups were born after full-term gestation (Table 2). Gene-editing was evaluated by T7E1 digestion and Sanger sequencing analysis which presented that 5 of them were MSTN knock-out rabbits (Figure 4) and interestingly, three MSTN+/− rabbits were all males, while two MSTN−/− rabbits were all females. The further T7E1 and sequencing analysis showed that 5 MSTN knock-out rabbits were chimeras.

Figure 4. Generation of F0 MSTN KO rabbits via CRISPR/Cas9. (A) Five mutated rabbits of F0. (B) Sanger sequence analysis of mutated rabbits. Five rabbits edited were named on the left. WT stands for wild type control; PAM sequence is marked in green. The sgRNAs sequence is marked in red. The numbers on the right indicated the type of mutation, with “-” represents deletion of the given number of nucleotides. (C) Upper: The results of sequencing for mutations at the specific sites in Agarose gel electrophoresis of PCR products about a part of pups of F0. Lower: T7E1 cleavage assay for the rabbits (The picture has been cropped).
Off-Target Mutation Analysis of MSTN Knock-Out Rabbits
To detect the presence of off-target mutations in MSTN knockout rabbits, four potential off-target sites were designed. The T7EI digestion and Sanger sequencing results exhibited that no off-target mutation was detected in the MSTN-KO rabbits (Figure 5).
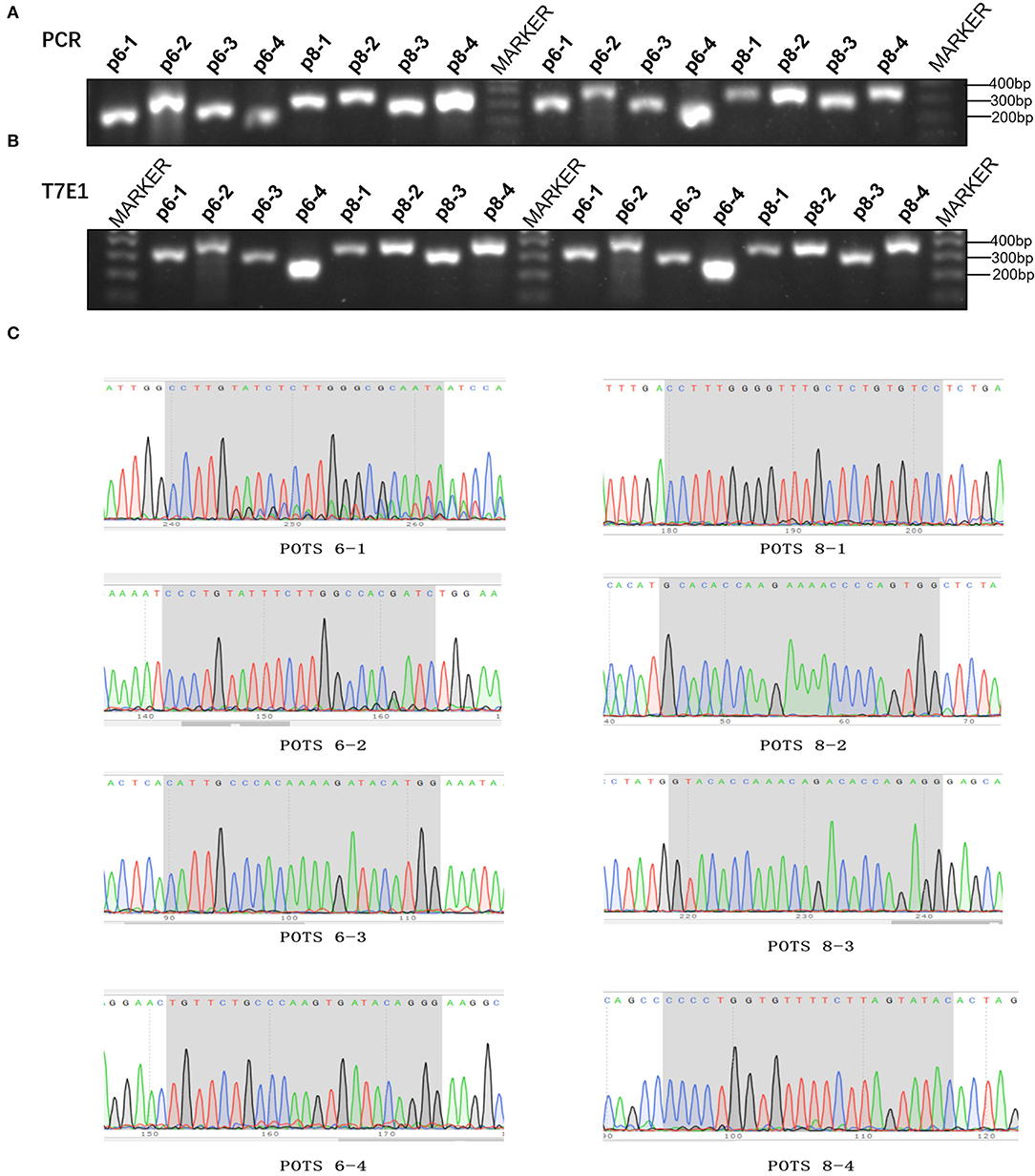
Figure 5. Detection of MSTN modified rabbits. (A) The results of suspicious POTs in Agarose gel electrophoresis of PCR products. M: DL100bp; p6-1–p6-4 represent g6 four suspicious POTs, p8-1–p8-4 represent g8 four suspicious POTs (The picture has been cropped). (B) The results of suspicious POTs in Agarose gel electrophoresis of T7E1. (C) Sanger sequencing for suspicious POTs sites.
Western Blot Analysis of MSTN-KO Rabbits
According to the results of western blot analysis, it was revealed that the expression level of MSTN protein in the muscle of mutant rabbits was lower than that of WT rabbits (Figure 6).
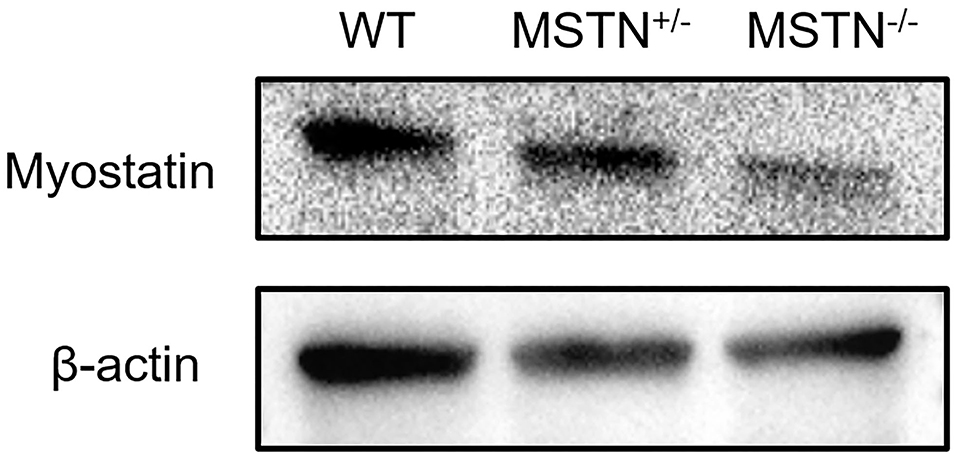
Figure 6. Result of MSTN+/–, MSTN–/– and WT rabbits in Western Blot. Beta-actin served as a loading control (The picture has been cropped).
Morphological Analysis of MSTN-KO Rabbits
The appearance of WT and F0 generation edited rabbits at the age of 3rd and 6th months were observed (Figure 7A). Under the same feeding conditions, the biceps femoris was more developed in the F0-generation edited rabbits than the WT. Moreover, the difference in muscle development of the MSTN-KO rabbit was evaluated using body weight, and data related to weight gain is shown in Supplementary Table S1. No significant difference was observed related to the body weight in the five MSTN-KO rabbits and the control group within the early 90 days (p > 0.05). But after growing to the age of 3rd month, the five MSTN-KO rabbits' weight began to appear slightly higher than that of the control group (Figure 7B) and the “double muscle” phenomenon appeared more obvious in MSTN-KO rabbits. At the age of 140 days, the five MSTN-KO rabbits exhibited a significantly higher body weight than the WT control group rabbits (P < 0.05, n = 5). Furthermore, the bodyweight of MSTN-KO rabbits (3,572 ± 41.40 g) was significantly (P < 0.01) more at 160 days as compared to the control WT rabbit (3,094 ± 69.54 g). Additionally, the weight gain data from two selected genotypes (insertion and deletion type) of the F1 generation rabbit are shown in Supplementary Table S2. A significant weight gain has been observed for both the insertion type and the deletion type in comparison to WT at 140 days, where the bodyweight of insertion type rabbits was 3,398 ± 92.97 g, deletion type rabbits 3,554 ± 110.45 g, and WT rabbits 3,064 ± 80.35 g (P < 0.05, n = 5), respectively. The gluteus of the MSTN-KO rabbits was also bigger than the WT rabbits, and histological analysis showed that in MSTN-KO rabbits the density and diameter of the myofiber were significantly large (Figure 7C). Also, it was found that the phenotypic differences of the edited rabbits were stably inherited in the F2 generation, and Figure 7D shows three young rabbits in the same litter. Similarly, T7E1 enzyme digestion and sequencing identification was used for young rabbit genotyping, and it has been observed that the body size of the edited rabbit was significantly different from that of the WT.
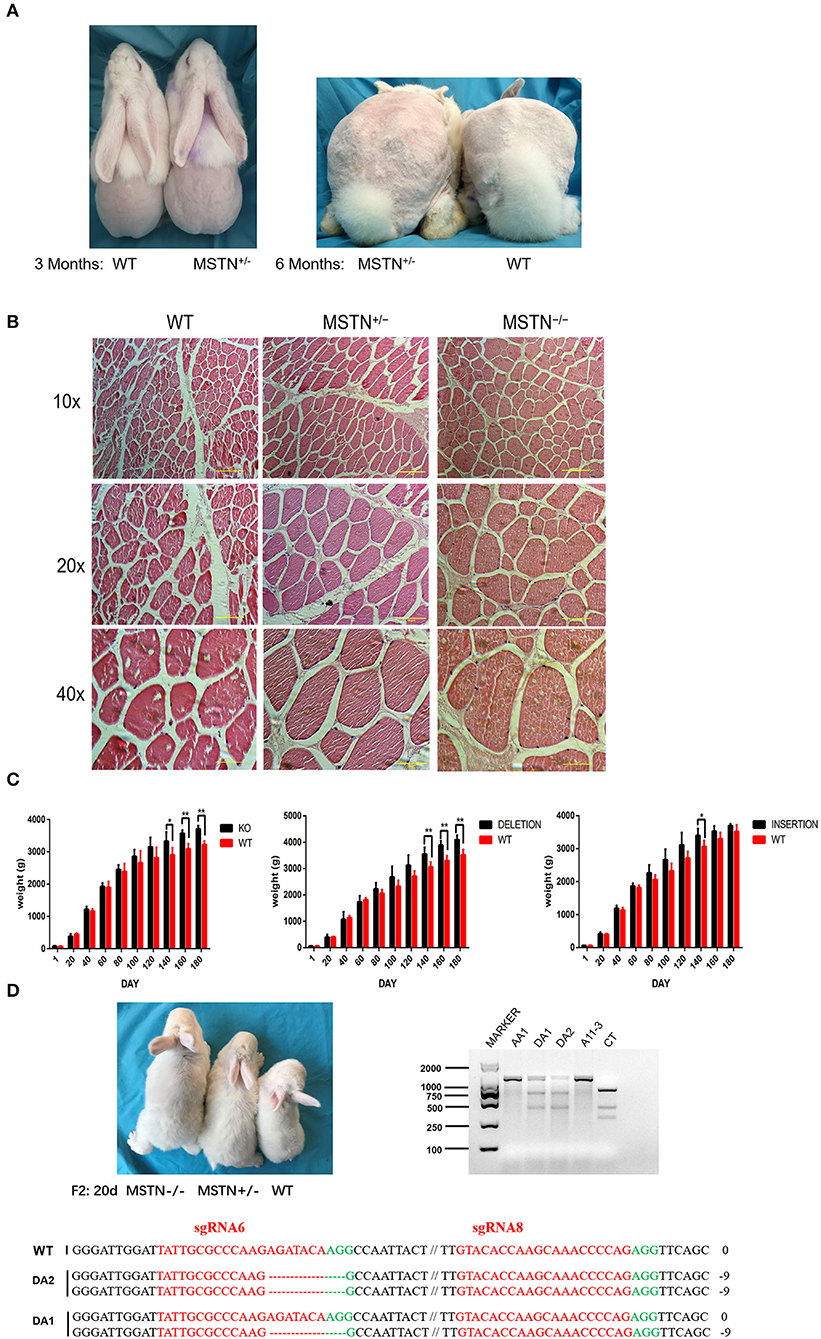
Figure 7. Detection and observation of MSTN-KO rabbits. (A) Comparison of mediated and wild-type rabbits at 3 and 6 months of age. (B) The average body weight of MSTN+/− and WT rabbits from F0 and F1 (n = 5), and the left side shows the weight gain of F0 knockout rabbits and wild rabbits, the middle shows the weight gain of F1 deletion editing rabbits and wild rabbits, and the right side shows the weight gain of F1 insertion editing rabbits and wild rabbits. (C) HandE staining of the muscle fibers from gluteus maximus. (D) Shape difference map and identification results of F2 generation rabbits at the age of 20 days (The picture has been cropped).
Heritability of the MSTN-KO Rabbits
To determine whether the offspring could stably inherit the successfully edited gene fragments from the MSTN-KO, all the F0 generation edited rabbits were crossed with wild-type rabbits. The three MSTN-KO F0 male rabbits were mated with wild rabbits and live F1 rabbit pups were produced. The T-cloning sequencing and T7E1 cleavage assay demonstrated that 28 out of the 53 newborn F1 rabbits carried MSTN mutations, of which 23 were monoallelic, and 5 were biallelic MSTN-KO rabbits (Figure 8). Two genotypes, deletion type and insertion type, were selected from the F1 generation to continue breeding, and the edited rabbits of the F2 generation were obtained. There was no significant difference in the litter size of female rabbits after breeding between F1 generation male rabbits and wild-type female rabbits, and there was no significant difference in the birth weight of offspring (Figure 8). So, there were three homozygous edited rabbits at the age of 1 month, with obvious differences in appearance and body shape. As shown in Figure 8, it is expected that a small number of homozygous edited rabbits obtained from the F2 generation, and in the F3 generation these edited homozygous rabbits continue to produce until a stable expansion thereof be achieved.
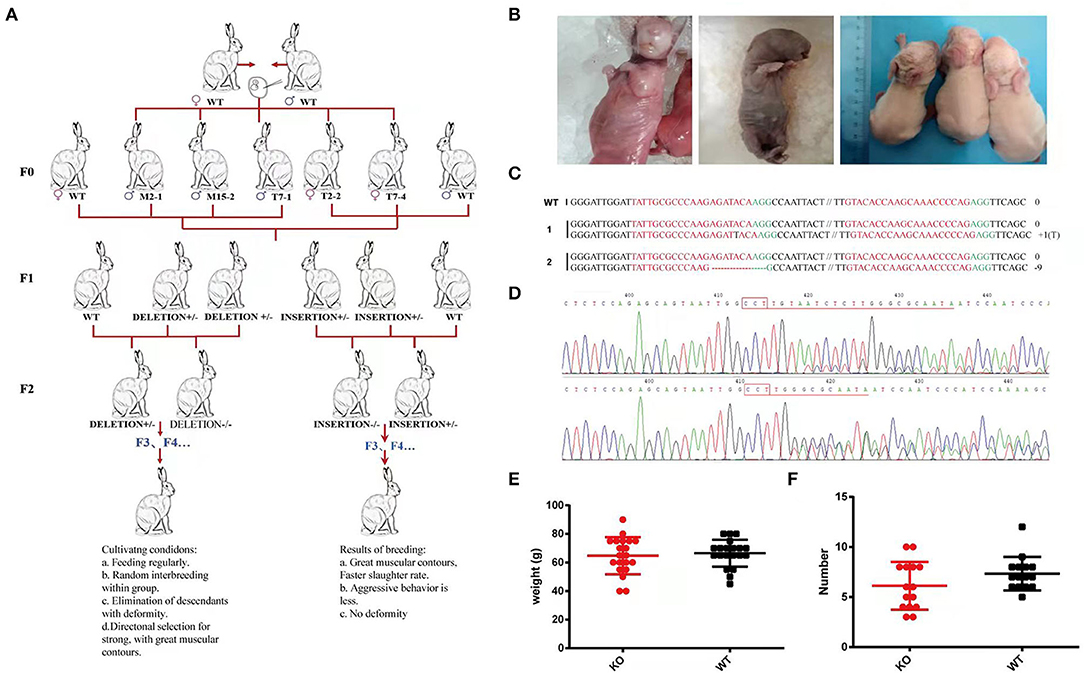
Figure 8. (A) Schematic diagram of MSTN-KO rabbits breeding family. (B) Left: A cub with a large tongue. Right: Difficult to give birth due to the large size of the cubs. (C) Rabbits of F1 were named on the left. 1-2 represent different rabbits, PAM sequence is marked in green. The sgRNAs sequence is marked in red. The numbers on the right indicated the type of mutation, with “-” represents deletion of the given number of nucleotides, “+” represents insertion of the given number of nucleotides. (D) sequence identification. (E) Birth weight of rabbits. (F) Litter size.
Discussion
Humans are entirely reliant on livestock for their daily food supply, which comes in the form of eggs, meat, and milk (30–33). Thus, genetic alteration offers an opportunity for significant production and gains in a short time (34, 35). Since the last decade, many gene-edited organisms have been approved and are being used by the public, such as the gene-edited goats' milk and chicken eggs are being used for drug extraction. Further, the salmon was the first gene-edited species to be approved to be consumed as food, and recently the GalSafe pigs have also been certified to be used for food and medical purpose (36). Rabbit farming is now very well developed all around the world and systematically reared on a large scale, and the global rabbit meat production is reaching 1.8 million metric tons per year (37). As high-quality meat, the demand for rabbit meat is worth looking forward to in the future, but rabbits' muscular development is still a challenge. Therefore, MSTN gene-edited rabbits have the possibility of being approved and used by the public under the advancement of our multi-generation continuous observational research. In the present study, we used CRISPR-Cas9 to delete a long gene fragment of the MSTN gene, which ensured the inactivation of target gene function, and the results of our study presented a successful production of MSTN-KO rabbit with heritable ability, and the loss of the MSTN fragment could lead to muscle growth. In both the F0 and F1 generations, there were significant differences in body weight before it reached the plateau, and the average body weight of the edited rabbits before the slaughter age was also higher than that of the WT rabbits.
Although the increase in muscle mass makes MSTN-KO livestock production more attractive, in these MSTN mutant animals calving difficulties are often caused by various reasons, such as large offspring syndrome (LOS) has been reported in MSTN-deficient animals (15, 38). Compared with other livestock, MSTN gene-edited rabbits rarely report dystocia due to large fetuses, even though in our study the MSTN-KO rabbits exhibited a typical double-muscle phenotype with increased body weight but, at birth, no significant differences have been observed in body weight and size as compared with the control WT. Further, the MSTN-KO rabbits appeared healthy and normal without reproduction difficulties, demonstrating that CRISPR/Cas9 system-generated MSTN-KO rabbits are best suited for studying muscle development and associated diseases, and the results of our study are in line with those of Lv et al. (39).
MSTN gene can code for TGF-beta (transforming growth factor-beta) superfamily ligand that can bind with different TGF-beta receptors and ultimately recruit or activate the SMAD family transcription factors, which regulate the gene expression (40). MSTN is a key regulator of muscle cell proliferation and differentiation throughout muscle development, and MSTN expression signals were detected from the early myogenic stage of embryonic sarcomere formation to the adult skeletal muscle development stage (14, 41, 42). Earlier, the birth weight of MSTN mutant animals has also been reported in sheep, cattle, and goats (43, 44). Wang et al. have also described the MSTN gene-modification and their effects in goats with larger muscle fiber size resulting in enhanced bodyweight (43). Therefore, the MSTN gene knockout might affect muscular development at the embryonic stage (43, 44). Furthermore, it has been stated that MSTN can only express in skeletal muscle during fetal development, thereby controlling the differentiation and proliferation of myoblasts (43, 44). In our study, MSTN-KO rabbits exhibited a typical double-muscle phenotypic trait, but there was no significant difference in body size at birth (P > 0.05) and body weight in the first 60 days compared with the WT group. This might be because of the IGF-1 which is an important positive regulator of muscle cell proliferation and differentiation in the skeletal muscle (45). The IGF1 signaling pathway is critically involved in long-term health regulation, which ultimately plays an essential role to control various homeostatic mechanisms related to growth or development (46). Despite the inhibition of MSTN signal expression, IGF signaling still might upregulate the expression of myostatin in skeletal muscle tissue models, which indicated the presence of an autoregulatory inhibitory loop in a muscular system (47, 48).
Moreover, the muscle fiber hypertrophy, hyperplasia, or a combination thereof triggered by the lack of the MSTN gene subsequently results in increased mammalian muscle production (49–52). The quantity of muscle fibers is mainly determined before birth because the diameter of muscle fibers might be increased after birth (53, 54). The mechanism of muscle hypertrophy and hyperplasia may not occur simultaneously, and it appears that the mechanism by which MSTN mutations enhance the muscle mass varies among species (55, 56). In MSTN mutant mice and goats, the muscle mass gain was caused by muscle fiber hyperplasia and diameter hypertrophy (39, 52, 57), while only muscle fiber hyperplasia was observed in MSTN mutant cattle and pigs (13, 50). The results of the current study presented that the gluteus maximus of the MSTN-KO rabbits was also bigger than the WT rabbits, and histological analysis showed that in MSTN-KO rabbits the density and diameter of the myofiber were significantly large. Therefore, we hypothesized that the enhanced muscular phenotype in the MSTN-KO rabbits is attributed to muscles fiber hyperplasia and/or hypertrophy. Furthermore, previously it has been reported that the muscle mass of MSTN-KO mice was 2–3 times more than that of the WT mice, which was the result of the combined action of muscle fiber hyperplasia and diameter hypertrophy (13, 52). On the other hand, the cattle with double-muscle trait showed that muscle mass increased by 20–25%, and the increased muscle weight appears to be the result of muscle fiber diameter proliferation, rather than an increase in muscle number (13, 50). In this study, compared with WT rabbits, the average body weight of F0 and F1 generation edited rabbits increased by 15 and 15.99% at the age of 180 and 140 days, respectively. The fragment deletion targeting a specific site in MSTN-KO rabbits could suggest that the sgRNA-based CRISPR/Cas9 system might be a useful tool for gene knockout in the mammalian genome (58). Earlier in rabbits, Lv et al. (39) designed two knockout sites at the first exon to edit the MSTN gene (39), while we designed a total of 8 sites in two exons (exon 1 and exon 3) each with four sites. After selection, we determined that the knockout efficiency of the third exon is better than the first exon, thus, we obtained edited rabbits with different knockout sites from already reported MSTN knockout studies. Moreover, at present, most of the experimental research regarding animals' gene-editing is still in the F0 generation (24, 59, 60), including the reported gene editing in rabbits (39). While, no study has yet been performed on the F1 or their subsequent generations, we not only developed MSTN gene-edited rabbits in the F0 generation but also continued the follow-up research in successive F1 and F2 generations with stable heritable mutant populations. Also, the difference between the body type of mutant and the wild type can be observed within 1 month after birth. Gene-editing technology can accelerate the breeding process and improve the meat production of livestock, but the congenital defects of gene edited animals and the uncertainty of trait stability have a great impact on the commercialization of edited animals (51, 61). In our study medium- and long-term breeding can greatly reduce concerns about the phenotypic and genetic stability of MSTN gene-edited animals.
Moreover, various studies reported different types of skeletal-related problems in gene-edited animals such as MSTN gene-edited rabbits had bone deformities, pelvic tilt, and tooth dislocation, while MSTN gene knock-out Meishan pigs had an extrathoracic vertebra (26, 37). Abnormal pelvic structure may cause difficulties in calving in female animals. But, in our study, the MSTN gene-edited rabbits obtained by knocking out the third exon did not show these symptoms in appearance, which are more suitable for the breeding of strains. Additionally, in the previous study, the MSTN-KO animals exhibited the muscular feature, but the MSTN-KO caused severe health issues, such as the fetus having “large tongue syndrome,” which results in milk suckling problems and death (52). In our study, the MSTN-KO rabbits not only exhibited double mushy buttocks but were also able to inherit this trait to subsequent generations without any problem. The majority of MSTN-KO rabbits showed a rapid increase in muscle mass after the age of 60 days, and the MSTN-KO rabbits were healthy and could inherit the knocked-out gene fragments, demonstrating that the MSTN-KO rabbits produced by the CRISPR/Cas9 system are suitable for studying muscle development and related diseases.
Conclusion
The rabbit meat is an enriched source of minerals with low fat and calories and is easy to digest, making it valuable for the consumers, especially for persons suffering from cardiovascular diseases or obesity. Taken together, we developed MSTN-KO rabbits with a typical phenotypic trait of double muscle buttocks that are likely to obtain edited rabbits lines which would help to improve the rabbit's meat production efficiency and promote the development of the rabbit industry. Furthermore, these MSTN-KO rabbits could be a promising tool for studying the development of muscles in other livestock species and improving their important economic trait.
Data Availability Statement
The datasets presented in this study can be found in Genbank repository accession number MZ427879-MZ427887 at: https://www.ncbi.nlm.nih.gov/nuccore/.
Ethics Statement
The animal study was reviewed and approved by Experimental Animal Care and Use Committee of Guangxi University (Permit Code: 2022-GXU-006). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
SR, KC, and QL: conceptualization and resources. YZ, YLZ, and LW: data curation. YLZ, YZ, and ZL: methodology. HR, ZL, and DS: software. QL and KC: supervision. YZ and YLZ: writing—original draft preparation. YLZ, YZ, HR, ZL, DS, SR, QL, and KC: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The present study was granted and supported by the National Natural Science Fund (Grant Nos. 31760648 and 31860638) and Guangxi Natural Science Foundation (Grant No. AB16380042).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.842074/full#supplementary-material
Abbreviations
CRISPR, Clustered regularly interspaced short palindromic repeats; sgRNAs, small guide RNA; FBS, Calf Serum; DMEM, Dulbecco's modified eagle medium.
References
1. Carneiro M, Rubin CJ, Di Palma F, Albert FW, Alföldi J, Barrio AM, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. (2014) 345:1074–9. doi: 10.1126/science.1253714
2. Smith A. T. (2021). “rabbit”. Encyclopedia Britannica. (2021) Available online at: https://www.britannica.com/animal/rabbit. (accessed 24 October 2021).
3. Dalle Zotte A. Rabbit farming for meat purposes. Animal Front. (2014) 4:62–7. doi: 10.2527/af.2014-0035
4. Patton N, Cheeke P. The rabbit as a meat producing animal [Feeding, meat composition]. In: Paper presented at the Proceedings Annual Reciprocal Meat Conference. (1981).
5. Gomer RH. Not being the wrong size. Nat Rev Molec Cell Biol. (2001) 2:48–55. doi: 10.1038/35048058
6. Szendro K, Szabó-Szentgróti E, Szigeti O. Consumers' attitude to consumption of rabbit meat in eight countries depending on the production method and its purchase form. Foods. (2020) 9:654. doi: 10.3390/foods9050654
7. Lui JC, Garrison P, Baron J. Regulation of body growth. Curr Opin Pediatr. (2015) 27:502. doi: 10.1097/MOP.0000000000000235
8. Rodino-Klapac LR, Haidet AM, Kota J, Handy C, Kaspar BK, Mendell JR. Inhibition of myostatin with emphasis on follistatin as a therapy for muscle disease. Muscle Nerve. (2009) 39:283–96. doi: 10.1002/mus.21244
9. McCoy JC, Walker RG, Murray NH, Thompson TB. Crystal structure of the WFIKKN2 follistatin domain reveals insight into how it inhibits growth differentiation factor 8 (GDF8) and GDF11. J Biol Chem. (2019) 294:6333–43. doi: 10.1074/jbc.RA118.005831
10. Wicik Z, Sadkowski T, Jank M, Motyl T. The transcriptomic signature of myostatin inhibitory influence on the differentiation of mouse C2C12 myoblasts. Polish J Veter Sci. (2011) 14. doi: 10.2478/v10181-011-0095-7
11. Nomura T, Ueyama T, Ashihara E, Tateishi K, Asada S, Nakajima N, et al. Skeletal muscle-derived progenitors capable of differentiating into cardiomyocytes proliferate through myostatin-independent TGF-β family signaling. Biochem Biophys Res Commun. (2008) 365:863–9. doi: 10.1016/j.bbrc.2007.11.087
12. Sakamoto K, Kanematsu-Yamaki Y, Kamada Y, Oka M, Ohnishi T, Miwa M, et al. Identification of ligand-selective peptidic ActRIIB-antagonists using phage display technology. Biochem Biophys Rep. (2017) 11:33–9. doi: 10.1016/j.bbrep.2017.06.001
13. Grobet L, Martin LJR, Poncelet D, Pirottin D, Brouwers B, Riquet J, et al. A deletion in the bovine myostatin gene causes the double–muscled phenotype in cattle. Nat Genet. (1997) 17:71–4. doi: 10.1038/ng0997-71
14. Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. (1997) 7:910–5. doi: 10.1101/gr.7.9.910
15. Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. (2007) 3:e79. doi: 10.1371/journal.pgen.0030079
16. Boman IA, Våge DI. An insertion in the coding region of the myostatin (MSTN) gene affects carcass conformation and fatness in the Norwegian Spælsau (Ovis aries). BMC Res Notes. (2009) 2:1–5. doi: 10.1186/1756-0500-2-98
17. Sahu AR, Jeichitra V, Rajendran R, Raja A. Novel report on mutation in exon 3 of myostatin (MSTN) gene in Nilagiri sheep: an endangered breed of South India. Trop Anim Health Prod. (2019) 51:1817–22. doi: 10.1007/s11250-019-01873-7
18. Stinckens A, Luyten T, Bijttebier J, Van den Maagdenberg K, Dieltiens D, Janssens S, et al. Characterization of the complete porcine MSTN gene and expression levels in pig breeds differing in muscularity. Anim Genet. (2008) 39:586–96. doi: 10.1111/j.1365-2052.2008.01774.x
19. Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. New Engl J Med. (2004) 350:2682–8. doi: 10.1056/NEJMoa040933
20. Saunders MA, Good JM, Lawrence EC, Ferrell RE, Li W-H, Nachman MW. Human adaptive evolution at Myostatin (GDF8), a regulator of muscle growth. Am J Human Genet. (2006) 79:1089–97. doi: 10.1086/509707
21. Li H, Wang G, Hao Z, Zhang G, Qing Y, Liu S, et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Sci Rep. (2016) 6:1–12. doi: 10.1038/srep33675
22. Zhang Y, Wang Y, Yulin B, Tang B, Wang M, Zhang C, et al. CRISPR/Cas9-mediated sheep MSTN gene knockout and promote sSMSCs differentiation. J Cell Biochem. (2019) 120:1794–806. doi: 10.1002/jcb.27474
23. Wang X, Yu H, Lei A, Zhou J, Zeng W, Zhu H, et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep. (2015) 5:1–9. doi: 10.1038/srep13878
24. Zhang J, Liu J, Yang W, Cui M, Dai B, Dong Y, et al. Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats. Theriogenology. (2019) 132:1–11. doi: 10.1016/j.theriogenology.2019.03.029
25. Xie S, Li X, Qian L, Cai C, Xiao G, Jiang S, et al. An integrated analysis of mRNA and miRNA in skeletal muscle from myostatin-edited Meishan pigs. Genome. (2019) 62:305–15. doi: 10.1139/gen-2018-0110
26. Qian L, Tang M, Yang J, Wang Q, Cai C, Jiang S, et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci Rep. (2015) 5:1–13. doi: 10.1038/srep14435
27. Zhang J, Cui ML, Nie YW, Dai B, Li FR, Liu DJ, et al. CRISPR/Cas9-mediated specific integration of fat-1 at the goat MSTN locus. FEBS J. (2018) 285:2828–39. doi: 10.1111/febs.14520
28. Dalle Zotte A, Szendro Z. The role of rabbit meat as functional food. Meat Sci. (2011) 88:319–331. doi: 10.1016/j.meatsci.2011.02.017
29. Duan Q, Li Y, Qin F. Research on the protection and mechanism of coptis alkaloids on acetic acid type gastric ulcer. J Phys. (2020) 1629:12028. doi: 10.1088/1742-6596/1629/1/012028
30. Rehman S, Shafique L, Yousuf MR, Liu Q, Ahmed JZ, Riaz H. Spectrophotometric calibration and comparison of different semen evaluation methods in Nili-Ravi buffalo bulls. Pak Vet J. (2019) 39:568–72. doi: 10.29261/pakvetj/2019.073
31. Rehman SU, Nadeem A, Javed M, Hassan FU, Luo X, Khalid RB, et al. Genomic identification, evolution and sequence analysis of the heat-shock protein gene family in buffalo. Genes. (2020) 11:1388. doi: 10.3390/genes11111388
32. Rehman SU, Feng T, Wu S, Luo X, Lei A, Luobu B, et al. Comparative genomics, evolutionary and gene regulatory regions analysis of casein gene family in bubalus bubalis. Front Genet. (2021) 12:662609. doi: 10.3389/fgene.2021.662609
33. Rehman SU, Hassan F-U, Luo X, Li Z, Liu Q. Whole-genome sequencing and characterization of buffalo genetic resources: recent advances and future challenges. Animals Open Access J From MDPI. (2021) 11:904. doi: 10.3390/ani11030904
34. Singh P, Ali SA. Impact of CRISPR-Cas9-based genome engineering in farm animals. Veter Sci. (2021) 8:122. doi: 10.3390/vetsci8070122
35. Wu S, Hassan F-U, Luo Y, Fatima I, Ahmed I, Ihsan A, et al. Comparative genomic characterization of buffalo fibronectin type III domain proteins: exploring the novel role of FNDC5/Irisin as a ligand of gonadal receptors. Biology. (2021) 10:1207. doi: 10.3390/biology10111207
36. Katiyar Kritika S, Burrell Justin C, Laimo Franco A, Browne Kevin D, Bianchi John R, Walters Anneke, et al. Biomanufacturing of axon-based tissue engineered nerve grafts using porcine galsafe neurons. Tissue Eng Part A. (2021) 27:1305–20. doi: 10.1089/ten.TEA.202.0303
37. Zhang T, Lu Y, Song S, Lu R, Zhou M, He Z, et al. ‘Double-muscling’and pelvic tilt phenomena in rabbits with the cystine-knot motif deficiency of myostatin on exon 3. Biosci Rep. (2019) 39:BSR20190207. doi: 10.1042/BSR20190207
38. Lee S-J. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. (2004) 20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836
39. Lv Q, Yuan L, Deng J, Chen M, Wang Y, Zeng J, et al. Efficient generation of myostatin gene mutated rabbit by CRISPR/Cas9. Sci Rep. (2016) 6:1–8. doi: 10.1038/srep25029
40. Alexopoulos T, Vasilieva L, Kontogianni MD, Tenta R, Georgiou A, Stroumpouli E, et al. Myostatin in combination with creatine phosphokinase or albumin may differentiate patients with cirrhosis and sarcopenia. Am J Physiol Gastrointest Liver Physiol. (2021) 321:G543–G551. doi: 10.1152/ajpgi.00184.2021
41. Manceau M, Gros J, Savage K, Thomé V, McPherron A, Paterson B, et al. Myostatin promotes the terminal differentiation of embryonic muscle progenitors. Genes Devel. (2008) 22:668–681. doi: 10.1101/gad.454408
42. Wang X, Niu Y, Zhou J, Zhu H, Ma B, Yu H, et al. CRISPR/Cas9-mediated MSTN disruption and heritable mutagenesis in goats causes increased body mass. Anim Genet. (2018) 49:43–51. doi: 10.1111/age.12626
43. Oldham J, Martyn J, Sharma M, Jeanplong F, Kambadur R, Bass J. Molecular expression of myostatin and MyoD is greater in double-muscled than normal-muscled cattle fetuses. Am J Physiol-Regul Integr Comparat Physiol. (2001) 280:R1488–93. doi: 10.1152/ajpregu.2001.280.5.R1488
44. Valdés JA, Flores S, Fuentes EN, Osorio-Fuentealba C, Jaimovich E, Molina A. IGF-1 induces IP3-dependent calcium signal involved in the regulation of myostatin gene expression mediated by NFAT during myoblast differentiation. J Cell Physiol. (2013) 228:1452–63. doi: 10.1002/jcp.24298
45. Yang W, Zhang Y, Li Y, Wu Z, Zhu D. Myostatin induces cyclin D1 degradation to cause cell cycle arrest through a phosphatidylinositol 3-kinase/AKT/GSK-3β pathway and is antagonized by insulin-like growth factor 1. J Biol Chem. (2007) 282:3799–808. doi: 10.1074/jbc.M610185200
47. Kurokawa M, Sato F, Aramaki S, Soh T, Yamauchi N, Hattori M-A. Monitor of the myostatin autocrine action during differentiation of embryonic chicken myoblasts into myotubes: effect of IGF-I. Mol Cell Biochem. (2009) 331:193–9. doi: 10.1007/s11010-009-0158-6
48. McPherron AC, Lawler AM, Lee S-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature. (1997) 387:83–90. doi: 10.1038/387083a0
49. McPherron AC, Lee S-J. Double muscling in cattle due to mutations in the myostatin gene. Proc Nat Acad Sci. (1997) 94:12457–61. doi: 10.1073/pnas.94.23.12457
50. Wegner J, Albrecht E, Fiedler I, Teuscher F, Papstein H-J, Ender K. Growth-and breed-related changes of muscle fiber characteristics in cattle. J Anim Sci. (2000) 78:1485–96. doi: 10.2527/2000.7861485x
51. Wang K, Ouyang H, Xie Z, Yao C, Guo N, Li M, et al. Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci Rep. (2015) 5:1–11. doi: 10.1038/srep16623
52. Guo R, Wan Y, Xu D, Cui L, Deng M, Zhang G, et al. Generation and evaluation of Myostatin knock-out rabbits and goats using CRISPR/Cas9 system. Sci Rep. (2016) 6:1–10. doi: 10.1038/srep29855
53. Swatland H. Muscle growth in the fetal and neonatal pig. J Anim Sci. (1973) 37:536–45. doi: 10.2527/jas1973.372536x
54. Nishi M, Yasue A, Nishimatu S, Nohno T, Yamaoka T, Itakura M, et al. A missense mutant myostatin causes hyperplasia without hypertrophy in the mouse muscle. Biochem Biophys Res Commun. (2002) 293:247–51. doi: 10.1016/S0006-291X(02)00209-7
55. Zhu X, Hadhazy M, Wehling M, Tidball JG, McNally EM. Dominant negative myostatin produces hypertrophy without hyperplasia in muscle. FEBS Lett. (2000) 474:71–5. doi: 10.1016/S0014-5793(00)01570-2
56. He Z, Zhang T, Jiang L, Zhou M, Wu D, Mei J, et al. Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. (2018) 38. doi: 10.1042/BSR20180742
57. Bi Y, Hua Z, Liu X, Hua W, Ren H, Xiao H, et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci Rep. (2016) 6:1–12. doi: 10.1038/srep31729
58. Zhou J, Wang J, Shen B, Chen L, Su Y, Yang J, et al. Dual sgRNAs facilitate CRISPR/Cas9 mediated mouse genome targeting. FEBS J. (2014) 281:1717–1725. doi: 10.1111/febs.12735
59. Dingwei P, Ruiqiang L, Wu Z, Min W, Xuan S, Jianhua Z, et al. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs. Yi Chuan. (2021) 43:261–70. doi: 10.16288/j.yczz.20-222
60. Gim GM, Kwon DH, Eom KH, Moon J, Park JH, Lee WW, et al. Production of MSTN-mutated cattle without exogenous gene integration using CRISPR-Cas9. Biotechnol J. (2021) 11:e2100198. doi: 10.1002/biot.202100198
Keywords: rabbits, Cas9, MSTN gene, knock-out, double muscle buttocks rabbits
Citation: Zheng Y, Zhang Y, Wu L, Riaz H, Li Z, Shi D, Rehman Su, Liu Q and Cui K (2022) Generation of Heritable Prominent Double Muscle Buttock Rabbits via Novel Site Editing of Myostatin Gene Using CRISPR/Cas9 System. Front. Vet. Sci. 9:842074. doi: 10.3389/fvets.2022.842074
Received: 23 December 2021; Accepted: 18 March 2022;
Published: 20 May 2022.
Edited by:
Marco Milanesi, University of Tuscia, ItalyReviewed by:
Emiliano Lasagna, University of Perugia, ItalyShi-Yi Chen, Sichuan Agricultural University, China
Copyright © 2022 Zheng, Zhang, Wu, Riaz, Li, Shi, Rehman, Liu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saif ur Rehman, c2FpZl9hbGkyOEB5YWhvby5jb20=; Qingyou Liu, cXlsaXUtZ2VuZUBneHUuZWR1LmNu; Kuiqing Cui, a3FjdWlAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Yalin Zheng1†
Yalin Zheng1† Yu Zhang
Yu Zhang Hasan Riaz
Hasan Riaz Zhipeng Li
Zhipeng Li Saif ur Rehman
Saif ur Rehman Qingyou Liu
Qingyou Liu